Abstract
The rise in antibiotic-resistant bacteria and contamination of water bodies is a serious issue that demands immense attention of scientific acumen. Here, we examined the pervasiveness of ESBL producing bacteria in Dal Lake and Wular Lake of Kashmir valley, India. Isolates were screened for antibiotic, heavy metal resistant elements, and their coexistence with mobile genetic elements. Out of two hundred one isolates screened, thirty-eight were found positive for ESBL production. Antibiotic profiling of ESBL positive isolates with 16 different drugs representing β-lactam or -non-β-lactam, exhibited multidrug resistance phenotype among 55% isolates. Molecular characterization revealed the occurrence of drug resistance determinants blaTEM, AmpC, qnrS, and heavy metal resistance genes (MRGs) merB, merP, merT, silE, silP, silS, and arsC. Furthermore, mobile genetic elements IntI, SulI, ISecp1, TN3, TN21 were also detected. Conjugation assay confirmed the transfer of different ARGs, HMRGs, and mobile elements in recipient Escherichia coli J53 AZR strain. Plasmid incompatibility studies showed blaTEM to be associated with Inc groups B/O, HI1, HI2, I1, N, FIA, and FIB. Co-occurrence of blaTEM, HMRGs, and mobile elements from the aquatic milieu of Kashmir, India has not been reported so far. From this study, the detection of the blaTEM gene in the bacteria Bacillus simplex and Brevibacterium frigoritolerans are found for the first time. Considering all the facts it becomes crucial to conduct studies in natural aquatic environments that could help depict the epidemiological situations in which the resistance mechanism might have clinical relevance.
Keywords: Antibiotic resistance, blaTEM, Mobile elements, HMRGs, Kashmir valley, Aquatic environment
1. Introduction
Infections due to drug-resistant bacteria are of serious concern worldwide. Every year more than 700,000 deaths occur globally, because of such deadly infections. Moreover, it was estimated that by the year 2050 the worldwide resistance would lead to 10 million deaths per year which is higher than the mortality rate of cancer (O’Neill, 2016). In India mortality rate caused by infectious diseases is about 416.75 per 100,000 persons (Laxminarayan and Chaudhury, 2016). Due to constant anthropogenic interference, the aquatic environment is considered as a pool of antibiotic resistance genes (ARGs) as these water bodies receive effluents from domestic, hospital sewage and from agricultural runoffs which act as the reservoirs of antibiotic resistance. Therefore, the presence of bacterial population, antibiotics, and nutrients from sewage furnish a selective pressure for the evolution of resistance in the aquatic environment (Parvez and Khan, 2018). Due to un-wise and frequent usage of antibiotics, community-acquired and nosocomial infections by Extended-Spectrum β Lactamases (ESBLs) producing bacteria are at the rise, known to grant resistance towards all available β-lactam antibiotics (Singh et al., 2018).
With the high consumption of antibiotics in India, the proportion of ESBL producing bacteria in the clinical and aquatic environment is uprising (Siddiqui et al., 2018). According to a scoping report on antimicrobial resistance (2017), published by the Department of Biotechnology, India, only 4% of publications were from environmental samples. There is paucity in the information available regarding epidemiology and the advent of resistance in response to β-lactams exposure of clinically relevant dosage regimens of cephalosporins and carbapenems (Gandra et al., 2017). Combating infections by antibiotics is an endless race due to bacterial acquisition of resistance genes from diverse environments. Unabated use of antibiotics causes environment contamination that promotes the prevalence and dissemination of antimicrobial resistance (Sultan et al., 2018).
Natural water bodies of Kashmir valley are, known for their aesthetic, cultural, and socio-economic values which are constantly deteriorating due to anthropogenic disturbance. Unabated discharge of waste from domestic, hospital, and agricultural sources into lakes has adversely affected the water quality. With no baseline data available on the resistance profile of bacterial inhabitants from pristine lakes, active surveillance and epidemiological studies are imperative. Identification of predominant factors involved in the emergence and mobilization of resistance determinants are of critical concern. Dal Lake and Wular Lake plays a dynamic role in the conservation of biodiversity. Wular Lake is counted as Asia’s largest freshwater Lake. Acknowledging the significance of the wetland for its biodiversity and socio-economic values; it was identified as a Wetland of international importance under the Ramsar Convention in 1990. Dal Lake is famous globally for tourism and its houseboats. Nevertheless, these lakes have deteriorated due to ecological stress from human activities (Saleem et al., 2011, Sheikh et al., 2014, Romshoo et al., 2011, Romshoo and Rashid, 2014), that has turned these water bodies into the reservoirs of antibiotic and heavy metal resistant bacteria (Kumar et al., 2013, Tacao et al., 2012, Sanderson et al., 2016, Lenart-Boron, 2017). Moreover, the dissemination of antibiotic resistance genes from antibiotic-resistant to susceptible bacterial strain may lead to the emergence of a new population of antibiotic-resistant bacteria (Parvez and Khan, 2018). The current study was undertaken to investigate the dominance of ESBL producing bacteria in Dal Lake &Wular Lake of Kashmir valley, India. ESBL producing bacteria were further screened for the presence of different antibiotic resistance genes blaTEM, AmpC, qnrS, and heavy metal resistance determinants merB, merP, merT, silE, silP, silS and arsC along with mobile genetic elements IntI, SulI, ISecp1, TN3, and TN21. Studies on the transfer of ARGs and HMGs through conjugation have also been carried out.
2. Material and methodology
2.1. Sampling and isolation of bacteria
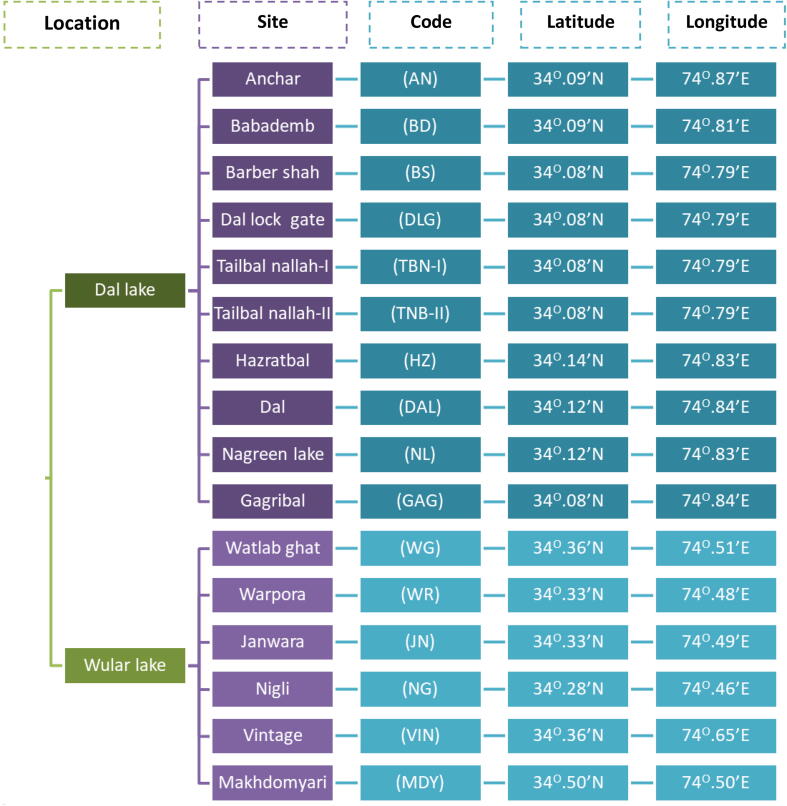
During 2015 and 2016, 16 different sites spread across Dal Lake and Wular Lake of Kashmir valley, India, was selected for the sample collection (Fig. 4). The collection of water samples was done aseptically 100 m away from major sewage discharge points, pH and temperature was checked on the spot. Further, collected samples were subjected to serial dilution, followed by bacteria count using the plate count method on Luria Agar. The visible colonies were carefully taken for re-culture and plated for ESBL activity.
Fig. 4.
Different sampling sites across Dal lake and Wular lake in Kashmir valley, India.
2.2. Screening of bacterial isolates for ESBL production
All the isolated bacteria were tested primarily for ESBL production using a preliminary test (disc diffusion) and as per clinical and laboratory standards institute (CLSI, 2016), for third-generation cephalosporins such as cefotaxime, ceftazidime, and ceftriaxone. Moreover, all positive isolates were further tested for confirmation by using a phenotypic disk confirmatory test (CLSI, 2016). For this test antibiotic disc of cefotaxime, and ceftazidime were used alone and along with clavulanic acid. The results for these tests were predicted following CLSI guidelines, 2016 (Siddiqui et al., 2018). For ESBL confirmation, K. pneumoniae was chosen as positive control and E. coli as a negative control.
2.3. Antimicrobial susceptibility profile
For antimicrobial susceptibility, we selected sixteen antibiotics from ten different classes (Siddiqui et al., 2018). Multidrug-resistant (MDR) isolates were chosen based on their resistance against three or more than three classes of antibiotics (Magiorakos et al., 2011). Similarly, the multiple antibiotic resistance (MAR) index of selected sample sites was determined as previously described (Krumperman 1983). Besides, minimum inhibitory concentration (MIC) of seven antibiotics was determined against ESBL positive isolates using 96-well microtitre plates by the microdilution process approved by CLSI. The optical density was noted down at 600 nm using a microplate reader (Azam et al., 2016).
2.4. Isolation and amplification of DNA
The genomic DNA isolation was carried out using Phenol: Chloroform: Isoamyl alcohol method. The plasmid DNA extraction was done using the alkaline lysis method, followed by PCR amplification by the method as described by Siddiqui et al. (2018). Using gene-specific primers (Table 4), the amplified products were verified by running on agarose gel (1%). Each PCR product of a particular gene was compared with a DNA marker.
Table 4.
Primers used for amplification of different genes.
| Gene | Primer name | Primer sequence | Product size (bp) | References/Accession No. |
|---|---|---|---|---|
| 16S rRNA | RR-F RR-R |
5′- GGCGGACGGGTGAGTAATG −3′ 5′- CGATTACTAGCGATTCCGACTTC −3′ |
1250 | This study |
| blaTEM | TEM-F TEM-R |
5′-ATGAGTATTMAACATTTYCGTGTCGCC-3′ 5′-TTACCAATGCTTAATCAGTGAGGCACCTATC-3′ |
861 | This study |
| AmpC | AMP-F AMP-R |
5′-GATCGTTCTGCCGCTGTG-3′ 5′-GGGCAGCAAATGTGGAGCAA-3 |
271 | Corvec et al. (2007) |
| qnrA | QA-F QA-R |
5′-AGAGGATTTCTCACGCCAGG-3′ 5′-TGCCAGGCACAGATCTTGAC-3′ |
580 | Cattoir et al. (2007) |
| qnrB | QB-F QB-R |
5′-GATCGTGAAAGCCAGAAAGG-3′ 5′-ATGAGCAACGATGCCTGGTA-3′ |
476 | Kim et al. (2009) |
| qnrS | QS-F QS-R |
5′-GCAAGTTCATTGAACAGGGT-3′ 5′-TCTAAACCGTCGAGTTCGGCG-3′ |
428 | Cattoir et al. (2007) |
| merB | MB-F MB-R |
5′-ATGAAGCTCGCCCCATATATT-3′ 5′-TCACGGTGTCCTAGATGACAT |
640 | This study |
| merP | MP-F MP-R |
5′-ATGAAGAAACTGTTTGCCTCC-3′ 5′-TCACTGCTTGACGCTGGACG-3′ |
272 | This study |
| merT | MT-F MT-R |
5′-TTAATAGAAAAATGGAACGAC-3′ 5′-ATGTCTGAACCACAAAACGGG-3′ |
355 | This study |
| silE | SE-F SE-R |
5′-GTACTCCCCCGGACATCACTAATT −3′ 5′-GGCCAGACTGACCGTTATT-3′ |
410 | Percival et al. (2008) |
| silP | SP-F SP-R |
5′-GGCGATAAGCTCCGCATCAGA-3′ 5′-TCCACT TTT TCAAGACGCTCA-3′ |
524 | Kremera and Hoffmann (2012) |
| silS | SS-F SS-R |
5′-GGAGATCCCGGATGCATAGCAA-3′ 5′-GTTTGCTGCATGACAGGCTAA AGACATC-3′ |
1500 | Percival et al. (2008) |
| arsC | AR-F AR-R |
5′-GTAATACGCTGGAGATGATCCG-3′ 5′-TTTTCCTGCTTCATCAACGAC-3′ |
409 | Sunita et al. (2012) |
| CS | 5′CS 3′CS |
5′-GGCATCCAAGCAGCAAG-3′ 5′-AAGCAGACTTGACCTGA-3′ |
530 | M73819 |
| IntI |
IntI- F IntI -R |
5′-CTACCTCTCACTAGTGAGGGGCGG-3′ 5′-GGGCAGCAGCGAAGTCGAGGC-3′ |
845 | U12338 |
| SulI | SulI-F SulI-R |
5′-ATGGTGACGGTGTTCGGCAT-3′ 5′CTAGGCATGATCTAACCCTC-3′ |
800 | Galimand et al. (2003) |
| ISecp1 | IS-F IS-R |
5′-TAAAAAACACAGGTGGAATTTAG-3′ 5′-CCAGGAACCACGGAGC-3′ |
1000 | This study |
| TN3 | Tn3 P-1 Tn3 P-2 Tn3 P-3 |
5′-AACTGATCTTCCTGACCGTC- 3′ 5′-TATGACCGATACGGCAGGTG- 3′ 5′-TCAGCAATGAACGGACCAGC- 3′ |
752 | Heffron et al. (1979) |
| TN21 | Tn21 P-1 Tn21 P-2 Tn21 P-3 |
5′-AGAAAGTTCGTCCTGGGCTG- 3′ 5′-TACTGCCGCGCATCAAGATC-3′ 5′-GGCCAAGGACAAGAACCTGT-3′ |
1000 | Kim et al. (2009), Brown et al. (1985) |
2.5. Characterization of ESBL genes and Identification of bacterial isolates
ESBL phenotype isolates were screened for blaTEM gene with the help of specific primer sets at SciGenome Lab, Cochin. Similarly, the sequences were analyzed and compared to the sequences in GenBank for the characterization of a particular variant present.
Isolates with blaTEM genotype were sent for 16S rRNA gene sequencing (SciGenome Lab, Cochin). The primers used are listed in Table 4. The sequences received were analyzed using the BLAST algorithm.
2.6. Detection of plasmid-mediated quinolone resistance and AmpC genes
The positive isolates of ESBL (blaTEM) production were analyzed for qnrA, qnrB, and qnrS followed by AmpC promoter using a specific set of primers.
2.7. Detection of metal resistance determinants
The co-existence of blaTEM positive isolates were checked for metal resistance determinants using specific primers of variant genes and plasmid DNA template. Further, silE, silP, and silS genes for silver resistance were subjected to amplification by the following method of Percival et al. (2008). The amplified products were tested for the possible amalgamations of ESBLs (blaTEM) and sil genes besides the determinants merB, merP, and merT genes, that confer the resistance against mercury. Arsenic resistant (arsC) gene was also amplified using the primer sequence from previously available literature (Sunita et al., 2012).
2.8. Determination of heavy metal tolerance
All the isolates harboring silver, mercury, and arsenic resistance determinants were subjected to the determination of corresponding heavy metal tolerance. MIC was observed by performing broth micro-dilution assay for silver nitrate (AgNO3), mercuric chloride (HgCl2), and sodium arsenate (Na3AsO4). For determination of minimum inhibitory concentrations, serial dilutions (256–2 mg/L) of heavy metals were prepared in 96 well microtitre plates containing 200 µl of Luria broth, (LB, HIMedia) and O.D. was accustomed to 0.5 McFarland.
2.9. Characterization and genetic environment analysis of blaTEM
Isolates having blaTEM were checked for integron analysis using PCR amplification of a conserved segment (CS 3′/5′) along with IntI, Sul1 using published primers (Galimand et al., 2003, M73819, U12338) and simultaneously (insertion sequence, transposons) ISecp1, TN3, and TN21 (Mood et al., 2015, Sheikh et al., 2014, Brown et al., 1985) were also analyzed using a specific set of primers.
2.10. Plasmid conjugation and replicon typing
Selected isolates and azide resistant Escherichia coli J53 strain were subjected to conjugation, to identify the resistant markers of a plasmid (Walsh et al., 2011). Transconjugants of the strains were further screened on Luria-Bertani agar added with cefotaxime (10 μg/ml) and sodium azide (100 μg/ml). The transconjugant carrying resistant markers, mobile elements, gene cassettes, heavy metals were inveterated by PCR amplification.
The presence of different plasmid incompatibility (Inc) groups was established by PCR based replicon typing (PBRT) for all the blaTEM carrying isolates. Transconjugants were also checked for the same (Carattoli et al., 2006). In brief, plasmid DNA was obtained by an alkaline lysis method followed by PCR amplification using different primers.
2.11. Nucleotide sequences accession numbers
A total of eleven partial 16S rRNA gene sequences and eleven blaTEM gene sequences were deposited in GenBank with following accession numbers, MH271258, MH271259, MH271273, MH271289, MH271261, MH271280, MH271260, MH271282, MH271283, MH271279, MH271287, TEM-116: MH742418.1, MH742419.1, MH742420.1, MH742421.1, MH742422.1, MH742423.1, MH742424.1, MH742425.1, TEM-1: MH742415.1, MH742416.1, MH742417.1.
3. Results
3.1. Isolation and screening of ESBL producing bacteria
A total of two hundred and one bacteria were isolated from different samples collected from sixteen diverse sampling sites of Wular Lake & Dal Lakes of Kashmir valley. Thirty-eight isolates turned positive for ESBL production was confirmed by phenotypic disc confirmatory tests. Notably, from thirty-eight ESBL producers, 11 isolates turned to carry blaTEM.
3.2. Antibiotic susceptibility profile of ESBL(blaTEM)producing isolates
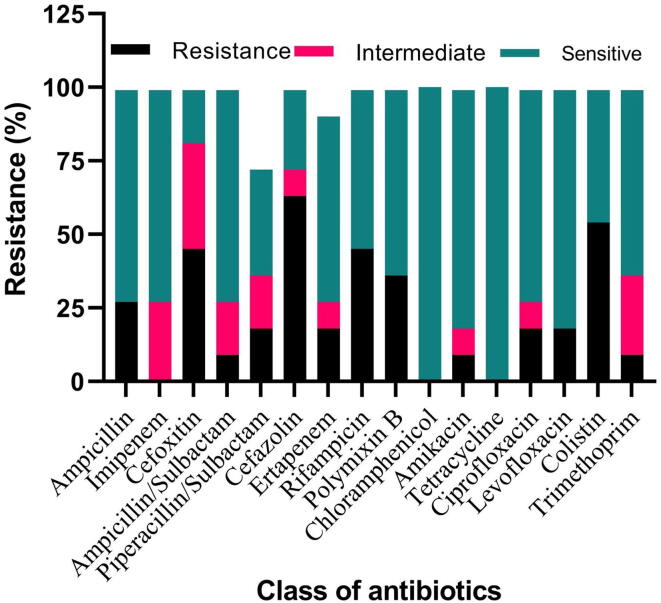
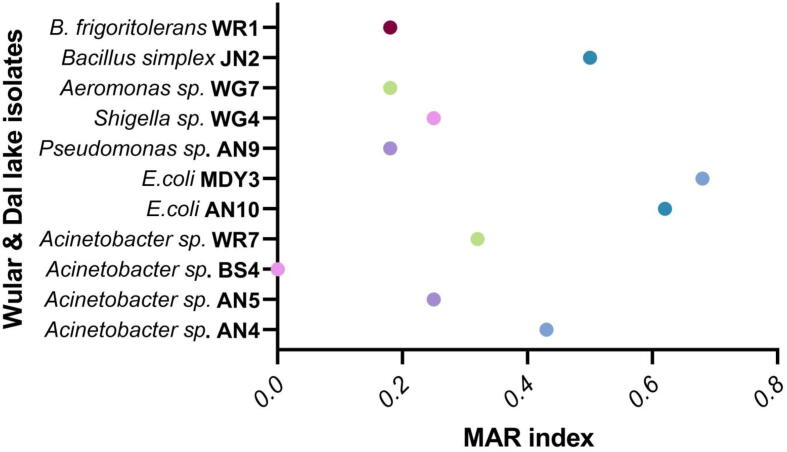
Fig. 1 depicts the antimicrobial susceptibility pattern of 11 blaTEM positive isolates against sixteen antibiotics. A vast number of bacteria strains were found resistant to β-lactams except for tetracycline, imipenem, and chloramphenicol. The maximum resistance was perceived for cefazolin (63%) followed by colistin (54%). Resistance to cefoxitin, rifampicin, ampicillin, ertapenem, piperacillin/tazobactam, ciprofloxacin, levofloxacin, polymixin B, amikacin and trimethoprim was observed as 45, 45, 27, 18, 18, 18, 18, 18, 9 and 9% respectively. Besides, 55% of ESBL positive strains showed MAR index greater than 0.2. Out of eleven ESBL (blaTEM) carrying isolates, seven strains depicted 0.2 to 0.62 MAR index. The MAR index of different isolates was found to be AN4 (0.43), AN5 (0.25), BS4 (0), WR7 (0.32), AN10 (0.62), MDY3 (0.68), AN9 (0.18), WG4 (0.25) WG7 (0.18), JN2 (0.5) and WR1 (0.18). We have obtained a value >0.2 for all the isolates except 3 isolates BS4, AN9, and WR1 (Fig. 2). Table 1 shows the MIC of ESBL positive isolates depicting the TEM genotype.
Fig. 1.
Antimicrobial susceptibility profile of 11 blaTEM positive isolates against sixteen different antibiotics.
Fig. 2.
Multilpe antibiotic resistance (MAR) index values of bacterial isolates tested against sixteen different antibiotics reaging 0–0.68.
Table 1.
MIC values for blaTEM producing bacteria isolated from Kashmir Valley, India.
| Isolates | MIC value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ (μg/ml) | CTX (µg/ml) | CIP (µg/ml) | C (µg/ml) | TR (µg/ml) | AMP (µg/ml) | CL (µg/ml) | AgNO3 (mg/l) | Na3ASO4 (mg/l) | HgCl2 (mg/l) | |
| AN9 | <2 | <2 | <2 | <2 | <2 | 8 | <2 | 8 | 64 | 16 |
| AN5 | <2 | <2 | <2 | <2 | 16 | <2 | <2 | 8 | 32 | 2 |
| BS4 | <2 | <2 | <2 | <2 | <2 | 64 | <2 | 8 | 128 | 16 |
| WR7 | 128 | 256 | <2 | 8 | 256 | 8 | 8 | 16 | 16 | 8 |
| AN10 | 256 | 256 | <2 | 4 | >256 | 256 | 8 | 4 | 64 | 64 |
| MDY3 | >256 | 256 | 256 | 16 | 32 | >256 | 16 | 8 | 256 | 16 |
| AN9 | 32 | <2 | <2 | 4 | <2 | 32 | >256 | 8 | 256 | 8 |
| WG4 | 256 | 32 | <2 | <2 | <2 | 16 | 256 | 2 | 128 | 2 |
| WG7 | 256 | 64 | <2 | 8 | <2 | <2 | >256 | 2 | 256 | 16 |
| JN2 | >256 | 256 | 32 | 16 | 256 | 8 | 4 | 16 | 128 | 16 |
| WR1 | 8 | 128 | 4 | 8 | 16 | 4 | >256 | 8 | 64 | 8 |
The antibiotics used for Minimum Inhibitory Concentration (MIC) determination were ceftazidime (CAZ), cefotaxime (CTX), ciprofloxacin (CIP), chloramphenicol (C), trimethoprim (TR), ampicillin (AMP) and colistin (CL) for heavy metals silver nitrate (AgNO3), mercuric chloride (HgCl2) and sodium arsenate (Na3AsO4) was used. Reference values as per Clinical laboratory Standard Institute (CLSI) guidelines for MIC determination of resistance, intermediate and sensitive phenotype are mentioned CAZ: ≥16, 8, ≤4, CTX: ≥4, 2, ≤1, CIP: ≥4, 2, ≤1, C: ≥32, 18, ≤8, TR: ≥16, ≥8, ≤8, AMP: ≥32, 16, ≤8, CL: ≥2, 2, ≤1. Reference values for the permissible limits of heavy metals in water were adapted from the available literature for Arsenic (As): 0.02 mg/L, Mercury (Hg): 0.01 mg/L Adopted from Singh, et al. 2011 (Indian Journal of Pharmacology) and for Silver (Ag): 0.1 mg/L Adopted from Alan B. G. Lansdown 2010 (Advances in Pharmacological Sciences).
3.3. Molecular characterization of ESBL isolates
The isolated strains rendering the blaTEM gene were characterized by 16S rRNA gene sequencing. The identified bacteria are Acinetobacter sp. (4), E. coli (2), Pseudomonas sp. (1), Shigella sp. (1), Aeromonas sp. (1), Brevibacterium frigoritolerans (1) and Bacillus simplex (1).
3.4. Identification of antibiotic resistance genes
PCR amplification, and the sequencing inveterate that out of thirty-eight isolates, eleven harbored blaTEM (three blaTEM-1 and eight blaTEM-116). These sequences were submitted to GenBank. Further, AmpC was detected in ten isolates including Acinetobacter sp. (Four), E. coli (two), Pseudomonas sp. (one), Shigella sp. (one), Aeromonas sp. (one) and Brevibacterium frigoritolerans (one). Further, qnrS was detected in seven isolates including, Acinetobacter sp. (four), Pseudomonas sp., (one) Aeromonas sp. (one), and Brevibacterium frigoritolerans (one) as shown in Table 2. Nevertheless, blaSHV, qnrA, and qnrB did not amplify in any isolate. However, the conjugation experiment reinforced the presence of resistant determinants on the plasmid.
Table 2.
Genetic profile of bacterial isolates obtained from Dal and Wular Lake, India.
| Strain name | I. ARGs |
II. Silver (HMRG) |
III. Arsenic (HMRG) |
IV. Mercury (HMRG) |
V. Integron (MGE) |
VI. Insertion seq (MGE) |
VII. Transposon (MGE) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | P | S | B | P | T | CS | IntI | SuLI | (ISecp1) | TN3 | TN21 | |||
| Acinetobacter sp. AN4 | blaTEM,qnrS, AmpC | – | – | – | – | – | – | – | – | – | – | – | – | + |
| Acinetobacter sp. AN5 | blaTEM,qnrS, AmpC | – | + | – | – | + | + | + | + | + | + | + | – | – |
| Acinetobacter sp. BS4 | blaTEM,qnrS, AmpC | – | – | – | – | – | – | – | + | + | + | – | – | + |
| Acinetobacter sp. WR7 | blaTEM,qnrS, AmpC | – | + | – | + | – | + | – | + | + | + | – | + | + |
| E.coli AN10 | blaTEM,AmpC | + | + | + | + | + | + | + | + | + | – | + | + | + |
| E.coliMDY3 | blaTEM,AmpC | + | + | – | – | + | + | + | – | – | – | + | – | – |
| Pseudomonas sp. AN9 | blaTEM,qnrS, AmpC | + | – | – | + | + | + | + | + | + | + | + | + | + |
| Shigella sp. WG4 | blaTEM,AmpC | + | + | – | – | + | + | + | + | – | – | – | + | – |
| Aeromonas sp. WG7 | blaTEM,qnrS, AmpC | + | – | – | + | + | + | + | + | + | + | + | + | + |
| Bacillus simplex JN2 | blaTEM | – | + | – | – | – | – | – | + | + | + | + | – | – |
| BrevibacteriumfrigoritoleransWR1 | blaTEM,qnrS, AmpC | + | + | + | – | + | + | + | + | + | + | – | – | + |
(+) sign indicates presence of a target gene in a particular isolate, (–) indicates absence of a targeted gene.Antibiotic resistance genes (ARGs), Heavy metal resistance genes (HMRG), Mobile genetic element (MGE).
3.5. Metal resistance determinants
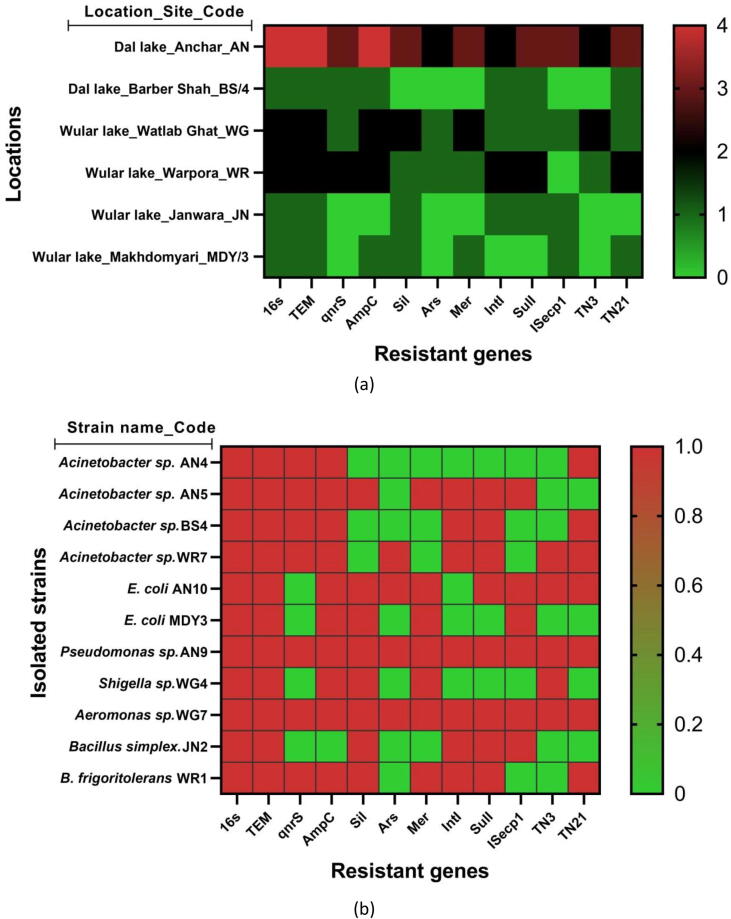
The eleven isolates harboring blaTEM genes were also screened for silver (sil), mercury (mer), and arsenic (ars) resistant determinants. The PCR amplification was accomplished for mer, sil, and ars, the genes for these metals were found in eight, nine, and four isolates, respectively (Fig. 3). Of the 11 isolates tested, it was found that two isolates of Acinetobacter sp. (AN4, BS4) were negative for all the silver genes tested viz. silE, silP, and silS, however, all the three genes were found in E.coli (AN10). Further, arsC was found in four isolates such as Acinetobacter sp. (WR7), E.coli (AN10), Pseudomonas sp. (AN9), Aeromonas sp. (WG7) and seven isolates each one of Acinetobacter sp. (AN5), two of E. coli (AN10), Pseudomonas sp. (MDY3), Shigella sp. (AN9), Aeromonas sp. (WH4), Brevibacterium frigoritolerans (WR1) were found positive for all mercury resistant genes (merB, merP, and merT). But two Acinetobacter sp. and one Bacillus simplex (AN4, BS4, and JN2) isolates were found negative for mercury resistant genes (Table 2).
Fig. 3.
Heatmap of antibiotic resistant phenotype and genotypes of the seuqnces blaTEM isolates. (a). Location based heatmap. (b). Strain based heatmap.
3.6. Susceptibility toward metal resistance
The isolates having metal resistant gene(s) mer, sil, and arsC were further checked for tolerance against different concentrations of HgCl2, AgNO3, and Na3AsO4. The MIC value of HgCl2 was recorded between the ranges of 2–16 mg/l. We observed that the isolates tolerate exposure of AgNO3 between ranges of 2 to 16 mg/l, and for Na3AsO4, the MIC was obtained between 16 and >256 mg/l range (Table 1).
3.7. Genetic environment analysis of blaTEM
Bacterial isolates with blaTEM genotype were screened for the genetic environment. PCR based genetic environment analysis showed that genetic elements IntI, SulI, ISecp1, TN3, and TN21 were present in 9, 7, 6, 5, and 7 isolates respectively (Table 2). However, all the isolates except AN4 (Acinetobacter sp.) and MDY3 (E.coli) were PCR positive for 5′/3′ CS, intI1, and Sul1 gene, demonstrating the presence of Class 1 integron. ISecp1 was found positive for 6 isolates two E.coli and one each of Acinetobacter sp., Pseudomonas sp., Aeromonas sp., Bacillus simplex. TN3 was detected in 5 isolates viz. Pseudomonas sp., E.coli, Acinetobacter sp., Shigella sp., and Aeromonas sp. Similarly, 7 isolates three Acinetobacter sp., one each of E. coli, Pseudomonas sp., Aeromonas sp., Brevibacterium frigoritolerans were positive for TN21 as shown in Fig. 3.
3.8. Replicon typing
Replicon typing shows that more than three plasmid incompatibility types were found to co-exist among all the isolates carrying blaTEM. Moreover, four isolates of Acinetobacter sp. carry plasmid incompatibility groups (B/O, HI1, HI2, I1, N, FIA, FIB), two E.coli (B/O, HI1, HI2, I1, N, F, FIA) and one each of Pseudomonas sp. (F, HI1, HI2, I1, N), Shigella sp. (B/O, HI1, HI2, I1, N), Aeromonas sp. (F, HI1, HI2, I1, N, FIA), Bacillus simplex (B/O, HI1, HI2, I1, N) and Brevibacterium frigoritolerans (F, HI1, HI2, I1, N).
3.9. Conjugation experiment
To determine whether blaTEM, AmpC, qnrS, IntI, SulI, ISecp1, TN3, TN21, and heavy metal resistance determinants mer, sil, and arsC genes transfer by conjugation, AN10 and WR7 isolates were selected. Using different primers sets, we amplified the transconjugant plasmids. Our results depicted that both the isolates AN10 and WR7 have blaTEM gene present. While qnrS was found in WR7 only, and AmpC gene was detected in both isolates. Besides, AN10 was found positive for all metal resistant genes, however, none of the metal resistant genes was found positive for transconjugant WR7. Furthermore, IntI was amplified successfully in both the transconjugants, nevertheless, none was found positive for SulI. ISecp1 could not be detected in any of the transconjugants. Transposon TN3 and TN21 resistance genes were present in both the transconjugants. After successful conjugation experiment, the plasmid of transconjugants was isolated and after getting positive amplification of PCR, it was confirmed that donor bacterial isolates (E.coli, Acinetobacter sp.) could transfer their resistant determinants like blaTEM, AmpC, qnrS, mobile elements (IntI, TN3, TN21) and metal resistant genes (silE, silP and silS, merB, merP and merT, and arsC) via conjugation. The MIC value of different antibiotics and metal for transconjugants are given in Table 3.
Table 3.
MIC values for blaTEM producing transconjugants isolated from Kashmir valley, India.
| Isolates | MIC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ (μg/ml) | CTX (µg/ml) | CIP (µg/ml) | C (µg/ml) | TR (µg/ml) | AMP (µg/ml) | CL (µg/ml) | AgNO3 (mg/l) | Na3ASO4 (mg/l) | HgCl2 (mg/l) | |
| E. coli AN10 | 256 | 256 | <2 | 4 | >256 | 256 | 8 | 4 | 64 | 64 |
| AN10/Transconjugant (J53AZR) | 256 | 32 | <2 | 4 | 128 | 16 | 2 | 2 | 64 | 64 |
| Acinetobacter sp.WR7 | 128 | 256 | <2 | 8 | 256 | 8 | 8 | 16 | 16 | 8 |
| WR7/Transconjugant (J53AZR) | 128 | 256 | <2 | 2 | 64 | 8 | 4 | 8 | 8 | 2 |
For the result analysis Reference values of CLSI guidelines were used for MIC determination of resistance, intermediate and sensitive phenotype the values are mentioned as CAZ: ≥16, 8, ≤4, CTX: ≥4, 2, ≤1, CIP: ≥4, 2, ≤1, C: ≥32, 18, ≤8, TR: ≥16, ≥8, ≤8, AMP: ≥32, 16, ≤8, CL: ≥2, 2, ≤1. Reference values for the permissible limits of heavy metals in water were adapted from the available literature for Arsenic (As): 0.02 mg/L,Mercury (Hg): 0.01 mg/L Adopted from Singh, et al. 2011 (Indian Journal of Pharmacology) and for Silver (Ag): 0.1 mg/L Adopted from Alan B. G. Lansdown 2010 (Advances in Pharmacological Sciences)
4. Discussion
The existence of ESBL positive bacteria in an open aquatic environment influenced by anthropogenic activities have been reported globally (Maloo et al., 2017). The genes responsible for ESBL production are linked with genetic elements like transposons, integrons, and heavy metal determinants, thereby, complicating the situation further as these elements expedite the transmission of resistance determinants among different species of bacteria (Singh et al., 2018, Azam et al., 2016, Bajaj et al., 2015). In this study, antibiotic-resistant genes were identified, including TEM, AmpC, qnrS, various genetic elements like ISecp1, TN3, TN21, IntI, and SulI among bacterial isolates from pristine lakes of Kashmir valley, India. The co-existence of antibiotics and different heavy metal resistant genes (mer, sil, and ars) were also determined. Of the total 201 bacterial isolates obtained from 16 different sampling sites, 38 were found to be ESBLs producers. The presence of ESBL producers along with mobile elements and metal resistance determinants are in line with earlier reports (Singh et al., 2018, Siddiqui et al., 2018, Bajaj et al., 2015, Azam et al., 2016). Resistance to cefazolin, colistin, cefoxitin, rifampicin, ampicillin, polymixin B, ciprofloxacin, levofloxacin, ertapenem, piperacillin/tazobactam, amikacin, trimethoprim, and ampicillin/sulbactam, was observed among 63, 54, 45, 45, 27, 18, 18, 18, 18, 9, and 9% isolates, respectively. These results corroborate with previous studies (Siddiqui et al., 2018, Azam et al., 2016). A report carried on antimicrobial resistance in water bodies of Kashmir valley revealed that about 7% of E. coli are resistant to cephalosporins, third-generation antibiotics (Rather et al., 2013). Another report published by Dr. Nisarul Hassan (2016) stated that 80% of bacteria isolated from SKIMS hospital ICUs were resistant to imipenem. Among the resistant bacteria, the most common isolates found were E coli, Klebsiella, Pseudomonas, and Acinetobacter. These isolates showed 100% resistance to ceftriaxone. The colistin resistance was observed higher than reported previously (Siddiqui et al., 2018, Vivant et al., 2016), which is worrisome as this antibiotic is one of the last resorts against multidrug-resistant bacteria. In this study, a complete sensitivity pattern was observed for antibiotics like imipenem, chloramphenicol, and tetracycline. All the isolates carrying blaTEM gene were identified by molecular characterization. The identified bacteria were Acinetobacter sp. (4), E. coli (2), Pseudomonas sp. (1), Shigella sp. (1), Aeromonas sp. (1), Brevibacterium frigoritolerans (1) and Bacillus simplex (1). We observed the domination of Acinetobacter sp. in contract with the previous findings (Kittinger et al., 2017), reporting Acinetobacter sp., as in higher prevalence with antibiotic resistance in freshwater bodies of Austria. The 16S rRNA nucleotide sequences of bacterial isolates showed 99–100% homology with previously submitted clinical and environmental isolates indicating that bacteria in this study might have originated from domestic sewage and or hospital effluent. Our findings are also in line with the study conducted on water samples from Dal Lake (Saleem et al., 2011).
Sequence analysis of blaTEM gene depicted that blaTEM-116 (8/11) is the most prevalent type including blaTEM-1 (3/11), these results are in line with one of the reports published earlier (Maravic et al., 2016). Based on the literature survey, the occurrence of blaTEM-116 in Brevibacterium frigoritolerans and Bacillus simplex from this study is the first report. The presence of AmpC positive isolates corroborates with the previous finding carried out from the urban river environment (Bajaj et al., 2015). Oteo et al., et al. (2010) also reported a very high proportion of the AmpC positive ESBL producers (95.2%) in the environmental setting. The results of this finding correlated with the results of clinical specimens from SKIMS Kashmir, where 27.1% of AmpC producing isolates were obtained (Ahmad et al., 2010). qnrS gene was obtained in 63% isolates in this study, similarly, the occurrence of PMQR and ESBL genes have been reported in studies from China and Iran (Li et al., 2014, Mood et al., 2015). We found that the co-existence of blaTEM and heavy metal resistance elements viz. mercury resistant genes, silver resistance genes, and arsenic resistance gene imparting resistance to a wide range of antibiotics, however, not to ignore mobile genetic elements. Therefore, the co-existence of antibiotic and metal resistance determinants on the same conjugative plasmids results in their transfer and as such maintenance among indigenous community members (Seiler et al., 2012). Mercury resistance is considered to be regulated by mer operon genes and has frequently been observed linked genetically to antibiotic resistance genes (McIntosh et al., 2008). The co-occurrence of ESBLs and mer operon genes has been previously reported among human and avian samples from Sweden and the aquatic environment from India (Sutterlin et al., 2014, Azam et al., 2016). E. coli isolated from Dal Lake showed a high incidence of mercury resistances (Murtaza et al., 2002). Various genes present in ars operon are responsible for the emergence of arsenic resistance. Among these genes, arsC is a very important determinant that provides arsenate resistance by reducing it to arsenite. Our results corroborate with the previous study carried out among isolates from municipal wastewater in Italy, where co-occurrence of antibiotic resistance and arsC gene was observed (Di Cesare et al., 2016). Reports from Kaur et al., 2009, Sunita et al., 2012 also showed the presence of arsC gene among the bacterial isolates from freshwater bodies in India. The association of mobile elements with blaTEM gene was also studied where positive amplification for IntI 81, SulI 63, ISecp1 54, TN3 45, and TN21 63% isolates was achieved. In the present study, TN3 and TN21 were found associated with blaTEM gene, similar results were also reported in the past (Nicolas et al., 2015). Perez et al. (2018) showed the presence of ISEcp1, intI1 with blaTEM gene (ISecp1- 88.5, and intI -94%). Plasmid incompatibility studies showed where Inc groups like B/O, HI1, HI2, I1, N, FIA, FIB to be associated with blaTEM. A study by Marcade et al. (2009) showed an association of blaTEM with Inc type N, B/O, FIA, FIB, HI1, HI2, I1 which supports our data.
5. Conclusion
In this study, we reported multidrug-resistant bacteria with ESBL, metal resistance, and genetic elements contemporaneous in the aquatic environment of Kashmir valley. The overall resistance pattern of ESBL positive isolates towards the presence of antibiotics, β-lactamase gene, metal resistant genes, and mobile elements was found to be similar to several reports from across the globe. Two variants of blaTEM viz. blaTEM-116 and blaTEM-1 have been detected in this report. Moreover, the existence of blaTEM gene in Bacillus simplex and Brevibacterium frigoritolerans are new reports. Detection of mobilome, metal, and antibiotic-resistant determinant from the aquatic environment highlights the co-existence and co-transfer of these elements possibly between different bacteria through horizontal gene transfer. Further research regarding mobile genetic elements fortifies these elements in the expansion of multidrug-resistant bacteria. The study necessitates the need to explore and investigate aquatic ecosystems that provides a hotspot of antibiotic resistance genes.
Author contributions
IS, AA Data curation, conceptualized, designed, and conducted the experimental work. IS, AA, FAG, QMRH, and IAR contributed to data analysis and validation. IS, AA, FAG, JSMS, and IAR contributed to formal analysis and visualization. IAR and JSMS carried out project administration and funding acquisition. IS wrote the first draft, IAR and QMRH edited and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant no. (DF-704-130-1441). The authors, therefore, gratefully acknowledge DSR technical and financial support.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ahmad P.M., Thokar M.A., Fomda B.A., Ahmed K. Extended Spectrum-β-Lactamase producing Klebsiella pneumoniae at a tertiary care setup in Kashmir, India: Comparative phenotypic detection and antimicrobial susceptibility pattern. Rev. Infect. 2010;1:124–133. [Google Scholar]
- Azam M., Jan A.T., Haq Q.M.R. blaCTX-M-152, a novel variant of CTX-M-group-25, identified in a study performed on the prevalence of multidrug resistance among natural inhabitants of river Yamuna, India. Front. Microbiol. 2016;7:1–13. doi: 10.3389/fmicb.2016.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj P., Singh N.S., Kanaujia P.K., Virdi J.S. Distribution and molecular characterization of genes encoding CTX-M and AmpC β-lactamases in Escherichiacoli isolated from an Indian urban aquatic environment. Sci. Total Environ. 2015;505:350–356. doi: 10.1016/j.scitotenv.2014.09.084. [DOI] [PubMed] [Google Scholar]
- Brown N.L., Winnie J.N., Fritzinger D., Pridmore R.D. The nucleotide sequence of the tnpA gene completes the sequence of the Pseudomonas transposon Tn501. Nucleic Acids Res. 1985;13:5657–5669. doi: 10.1093/nar/13.15.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Miriagou V., Bertini A. Replicon typing of plasmids encoding resistance to newer β-lactams. Emerg. Infect. Dis. 2006;12:1145–1148. doi: 10.3201/eid1207.051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V., Poirel L., Rotimi V., Soussy C.J., Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007;60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- Corvec S., Prodhomme A., Giraudeau C., Dauvergne S., Reynaud A., Caroff N. Most Escherichia coli strains overproducing chromosomal AmpC beta-lactamase belong to phylogenetic group A. J. Antimicrob. Chemother. 2007;60:872–876. doi: 10.1093/jac/dkm284. [DOI] [PubMed] [Google Scholar]
- Di Cesare A., Eckert E.M., D'Urso S., Bertoni R., Gillan D.C., Wattiez R., Corno G. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal waste water treatment plants. Water Res. 2016;1:208–214. doi: 10.1016/j.watres.2016.02.049. [DOI] [PubMed] [Google Scholar]
- Galimand M., Courvalin P., Lambert T. Plasmid mediated high-level resistance to aminoglycosides in Enterobacteriaceaedue to 16S rRNA methylation. Antimicrob. Agents Chemother. 2003;47:2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandra, S., Joshi, J., Trett, A., Lamkang, A.S., Laxminarayan, R., 2017. Scoping report on antimicrobial resistance in India.
- Heffron F., McCarthy B.J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: Three genes and three sites involved in transposition of Tn3. Cell. 1979;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kaur S., Kamli M.R., Ali A. Diversity of arsenate reductase genes (arsC Genes) from arsenic-resistant environmental isolates of E. coli. Curr. Microbiol. 2009;59:288–294. doi: 10.1007/s00284-009-9432-9. [DOI] [PubMed] [Google Scholar]
- Kim H.B., Park C.H., Kim C.J., Kim E.C., Jacoby G.A., Hooper D.C. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob. Agents Chemother. 2009;53:639–645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittinger C., Kirschner A., Lipp M., Baumert R., Mascher F., Farnleitner A.H., Zarfel G.E. Antibiotic resistance of Acinetobacter spp. isolates from the river Danube: susceptibility stays high. Int. J. Environ. Res. Public Health. 2017;15:52. doi: 10.3390/ijerph15010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremera A.N., Hoffmann H. Subtractive hybridization yields a silver resistance determinant unique to nosocomial pathogens in the Enterobacter cloacae complex. J. Clin. Microbiol. 2012;50:3249–3257. doi: 10.1128/JCM.00885-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tripathi V.R., Garg S.K. Antibiotic resistance and genetic diversity in water-borne Enterobacteriaceae isolates from recreational and drinking water sources. Int. J. Environ. Sci. Technol. 2013;10:789–798. [Google Scholar]
- Laxminarayan R., Chaudhury R.R. Antibiotic resistance in India: drivers and opportunities for action. PLoS Med. 2016;13:e1001974. doi: 10.1371/journal.pmed.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart-Boron A. Antimicrobial resistance and prevalence of extended-spectrum beta-lactamase genes in Escherichia coli from major rivers in Podhale, southern Poland. Int. J. Environ. Sci. Technol. 2017;14:241. [Google Scholar]
- Li L., Wang B., Feng S., Li J., Wu C., Wang Y., Ruan X., Zeng M. Prevalence and characteristics of extended-spectrum β-lactamase and plasmid-mediated fluoroquinolone resistance genes in Escherichia coli isolated from chickens in Anhui province, China. PLoS ONE. 2014;9:e104356. doi: 10.1371/journal.pone.0104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.P., Srinivasan A., Carey R.B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2011;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Maloo A., Fulke A.B., Mulani N., Sukumaran S., Ram A. Pathogenic multiple antimicrobial resistant Escherichia coli serotypes in recreational waters of Mumbai, India: a potential public health risk. Environ. Sci. Pollut. Res. 2017;24:11504–11517. doi: 10.1007/s11356-017-8760-8. [DOI] [PubMed] [Google Scholar]
- Maravic A., Skocibusic M., Fredotovic T., Samanic I., Cvjetan S., Knezovic M., Puizina J. Urban riverine environment is a source of multidrug-resistant and ESBL-producing clinically important Acinetobacter spp. Environ. Sci. Pollut. Res. Int. 2016;23:3525–3535. doi: 10.1007/s11356-015-5586-0. [DOI] [PubMed] [Google Scholar]
- Marcade G., Deschamps C., Boyd A., Gautier V., Picard B., Branger C., Denamur E., Arlet G. Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2009;63:67–71. doi: 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- McIntosh D., Cunningham M., Ji B., Fekete F.A., Parry E.M., Clark S.E., Zalinger Z.B., Gilg I.C., Danner G.R. Johnson, K.A., Beattie, M., Ritchie, R. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonassalmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 2008;61:1221–1228. doi: 10.1093/jac/dkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mood E.H., Meshkat Z., Izadi N., Rezaei M., Jamehdar S.A., Nasab M.N. Prevalence of quinolone resistance genes among extended-spectrum β-lactamase-producing Escherichia coli in Mashhad, Iran. Jundishapur J. Microbiol. 2015;8:e16217. doi: 10.5812/jjm.16217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza I., Dutt A., Ali A. Relationship between the persistence of mer operon sequences in Escherichia coli and their resistance to mercury. Curr. Microbiol. 2002;44:178–183. doi: 10.1007/s00284-001-0085-6. [DOI] [PubMed] [Google Scholar]
- Nicolas E., Lambin M., Dandoy D., Galloy C., Nguyen N., Oger C.A., Hallet B. The Tn3-family of replicative transposons. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0060-2014. MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- O’Neill, J., 2016. The review on antimicrobial resistance. In: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. [Available at: http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf].
- Oteo J., Perez-Vazquez M., Campos J. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 2010;23:320–326. doi: 10.1097/qco.0b013e3283398dc1. [DOI] [PubMed] [Google Scholar]
- Parvez S., Khan A.U. Hospital sewage water: a reservoir for variants of New Delhi metallo-β-lactamase (NDM)- and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Int. J. Antimicrob. Agents. 2018;51:82–88. doi: 10.1016/j.ijantimicag.2017.08.032. [DOI] [PubMed] [Google Scholar]
- Percival S.L., Woods E., Nutekpor M., Bowler P., Radford A., Cochrane C. Prevalence of silver resistance in bacteria isolated from diabetic foot ulcers and efficacy of silver-containing wound dressings. Ostomy Wound Manag. 2008;54:30–40. [PubMed] [Google Scholar]
- Rather T.A., Hussain S.A., Bhat S.A., Shah S.N., Arshid S., Shahnawaz M. Antibiotic sensitivity of E. coli and Salmonella isolated from different water sources in Kashmir, India. Comp. Clin. Pathol. 2013;22:729–731. [Google Scholar]
- Romshoo S.A., Ali N., Rashid I. Geoinformatics for characterizing and understanding the spatio-temporal dynamics (1969 to 2008) of Hokersar Wetland in Kashmir Himalayas. Int. J. Phys. Sci. 2011;6:1026–1038. [Google Scholar]
- Romshoo S.A., Rashid I. Assessing the impacts of changing land cover and climate on Hokersar wetland in Indian Himalayas. Arabian J. Geosci. 2014;7:143–160. [Google Scholar]
- Saleem S., Kamili A.N., Kakru D.K., Bandh S.A., Ganai B.A. Isolation, identification and seasonal distribution of bacteria in Dal Lake, Kashmir. Int. J. Environ. Sci. 2011;2:1. [Google Scholar]
- Sanderson H., Fricker C., Brown R.S., Majury A., Liss S.N. Antibiotic resistance genes as an emerging environmental contaminant. Environ. Rev. 2016;24:205–218. [Google Scholar]
- Seiler C., Berendonk T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:399. doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J.A., Jeelani G., Gavali R.S., Shah R.A. Weathering and anthropogenic influences on the water and sediment chemistry of Wular Lake, Kashmir Himalaya. Environ. Earth Sci. 2014;71:2837–2846. [Google Scholar]
- Siddiqui M.T., Mondal A.H., Sultan I., Ali A., Haq Q.M.R. Co-occurrence of ESBLs and silver resistance determinants among bacterial isolates inhabiting polluted stretch of river Yamuna, India. Int. J. Environ. Sci. Technol. 2018;16:5611–5622. [Google Scholar]
- Singh N.S., Singhal N., Virdi J.S. Genetic environment of blaTEM-1, blaCTX-M-15, blaCMY-42 and characterization of integrons of Escherichia coli isolated from an Indian Urban Aquatic Environment. Front. Microbiol. 2018;9:382. doi: 10.3389/fmicb.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan I., Rahman S., Jan A.T., Siddiqui M.T., Mondal A.H., Haq Q.M.R. Antibiotics, resistome and resistance mechanisms: a bacterial perspective. Front. Microbiol. 2018;9:2066. doi: 10.3389/fmicb.2018.02066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunita M.S.L., Prashant S., Chari P.V.B., Rao S.N., Balaravi P., Kishor P.B.K. Molecular identification of arsenic-resistant estuarine bacteria and characterization of their ars genotype. Ecotoxicology. 2012:202–212. doi: 10.1007/s10646-011-0779-x. [DOI] [PubMed] [Google Scholar]
- Sutterlin S., Edquist P., Sandegren L., Adler M., Tangden T. Silver resistance genes are overrepresented among Escherichia coli isolates with CTX-M production. Appl. Environ. Microbiol. 2014;80:6863–6869. doi: 10.1128/AEM.01803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacao M., Correia A., Hentriques I. Resistance to broad-spectrum antibiotics in aquatic systems: anthropogenic activities modulate the dissemination of blaCTX-M like genes. Appl. Environ. Microbiol. 2012;78:4134–4140. doi: 10.1128/AEM.00359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivant A.L., Boutin C., Prost-Boucle S., Papias S., Hartmann A., Depret G., Ziebal C., Le Roux S., Pourcher A.M. Free water surface constructed wetlands limit the dissemination of extended spectrum β-lactamase producing Escherichia coli in the natural environment. Water Res. 2016;104:178–188. doi: 10.1016/j.watres.2016.08.015. [DOI] [PubMed] [Google Scholar]
- Walsh T.R., Weeks J., Livermore D.M., Toleman M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]