Abstract
Upper respiratory tract infections with Equid Herpesvirus 1 (EHV-1) typically result in a peripheral blood mononuclear cell-associated viremia, which can lead to vasculopathy in the central nervous system. Primary EHV-1 infection also likely establishes latency in trigeminal ganglia (TG) via retrograde axonal transport and in respiratory tract-associated lymphatic tissue. However, latency establishment and reactivation are poorly understood. To characterize the pathogenesis of EHV-1 latency establishment and maintenance, two separate groups of yearling horses were experimentally infected intranasally with EHV-1, strain Ab4, and euthanized 30 days post infection (dpi), (n = 9) and 70 dpi (n = 6). During necropsy, TG, sympathetic trunk (ST), retropharyngeal and mesenteric lymph nodes (RLn, MesLn) and kidney samples were collected. Viral DNA was detected by quantitative PCR (qPCR) in TG, ST, RLn, and MesLn samples in horses 30 and 70 dpi. The number of positive TG, RLn and MesLn samples was reduced when comparing horses 30 and 70 dpi and the viral copy number in TG and RLn significantly declined from 30 to 70 dpi. EHV-1 late gene glycoprotein B reverse transcriptase PCR and IHC results for viral protein were consistently negative, thus lytic replication was excluded in the present study. Mild inflammation could be detected in all neural tissue samples and inflammatory infiltrates mainly consisted of CD3+ T-lymphocytes (T-cells), frequently localized in close proximity to neuronal cell bodies. To identify latently infected cell types, in situ hybridization (ISH, RNAScope®) detecting viral DNA was used on selected qPCR- positive neural tissue sections. In ganglia 30 dpi, EHV-1 ISH signal was located in the neurons of TG and ST, but also in non-neuronal support or interstitial cells surrounding the neuron. In contrast, distinct EHV-1 signal could only be observed in neurons of TG 70 dpi. Overall, detection of latent EHV-1 in abdominal tissue samples and non-neuronal cell localization suggests, that EHV-1 uses T-cells during viremia as alternative route toward latency locations in addition to retrograde neuronal transport. We therefore hypothesize that EHV-1 follows the same latency pathways as its close relative human pathogen Varicella Zoster Virus.
Keywords: EHV-1, horses, latency, Alphaherpesviruses, pathogenesis, trigeminal ganglia, lymphocytes
Introduction
Equid Herpesvirus 1 (EHV-1) belongs to the family of Alphaherpesvirinae and is a clinically important herpesvirus in horses that causes respiratory disease, myeloencephalopathy and abortions worldwide. EHV-1 is typically spread by direct horse-to-horse contact through respiratory tract secretions and replicates in the upper respiratory tract epithelium following initial infection. Subsequently, EHV-1 infects mononuclear cells, enters the blood circulation and is thereby disseminated widely to secondary infection sites, such as the central nervous system (CNS) or the uterus (1–3). The main cell type infected during cell-associated viremia are monocytes and T-lymphocytes (T-cells), followed by B-lymphocytes (B-cells) (4–7).
During or shortly after acute lytic infection and replication, the virus starts to establish a life-long chronic persistent infection in the host. This latent infection is an evolutionary advantage found in all alphaherpesviruses, as it allows the virus to remain in the host undetected from the immune system while maintaining its capability for reactivation (8–10). A successful latent infection is described to be strongly dependent on a well-balanced host-virus interaction (11, 12), and neurons are thought to be excellent locations for latency establishment, as they represent a stable, long-living cell population. Furthermore, neurons are not known to express viral antigen, which is an important advantage for the virus allowing it to evade the immune system (3). During latency, viral DNA is present in the nucleus of infected cells in a non-integrated form, without lytic transcription and translation processes (13–15). In addition, there is growing evidence, that T-cells and satellite cells surrounding neurons are playing key roles in maintaining Alphaherpesvirus latency supposedly by interacting directly with viral transcription (12, 16).
EHV-1 has been shown to establish latency in the trigeminal ganglia (TG), like its relatives Herpes Simplex Virus 1 (HSV-1), Bovine Herpesvirus 1 (BoHV-1) and Varicella Zoster Virus (VZV) (8, 10, 12, 17) but also in respiratory associated lymphoid tissue (RALT) and circulating CD8+ T-cells (18–22). Furthermore, our research group has recently shown that 70 days post infection (dpi), chronic persistent EHV-1 infection is not limited to TG and RALT, but EHV-1 DNA can also be detected (via qPCR) in various other sensory- (spinal cord dorsal root ganglia), sympathetic and parasympathetic- ganglia as well as in abdominal lymphoid tissue (mesenteric lymph node, spleen) (23). We reported wide distribution of chronic persistent EHV-1 infection beyond viremia. Important remaining questions are how the virus gets to these sites of presumed latency, and which cells predominantly harbor the virus at these locations; e.g., which cells are preferred for latency establishment.
Most Alphaherpesviruses gain access to the sensory nerve endings in the vicinity of the primary infection sites and use retrograde axonal transport to reach the neuronal cell bodies. For VZV, which, like EHV-1, belongs to the subfamily of Varicellovirinae, it is thought that VZV is transported to the sensory ganglia during viremia and in VZV-infected T-cells, as well as by axonal transportation (24, 25). EHV-1 tropism for T-cells is also well described and it is proposed to be important for viral dissemination throughout the host and for bypassing viral clearance by the immune system (7, 22, 26). Interestingly, while EHV-1 gene transcription and protein expression is active during viral replication in epithelial cells, during viremia, the virus seems to be carried in a non-replicative state, until secondary locations, such as CNS or uterine endothelium, are reached and virus transfer occurs (7). However, it is unclear if EHV-1 is also transported to ganglia in the same manner and uses this route for successful latency establishment.
In the present study, we compared EHV-1 viral load and state of infection in neural and lymphatic tissue (based on previously identified locations at 70 dpi) (23) with a timepoint closer to acute infection and viremia (30 dpi). We hypothesized that EHV-1 chronic persistent infection is influenced by time passed after inoculation. Furthermore, we wanted to investigate latency establishment and maintenance on a cellular level by using in situ hybridization (ISH) to localize viral DNA within neural tissue.
Materials and Methods
Animals
Tissue samples used in this study were obtained from a total of fifteen yearling horses of both sexes (six females and nine males). All horses were clinically healthy and screened for EHV-1 and EHV-4 serum neutralizing antibodies using serum neutralization (SN) tests prior to infection. Only horses with a titer <4 for EHV-1 and <40 for EHV-4 were selected for the study. Animals were fed twice a day and had ad libitum access to water. Horses were housed together in a naturally ventilated barn throughout the experiment. The maintenance and experimental protocols were reviewed and approved by Michigan State University Institutional Animal Care and Use Committee.
Experimental Design
Two separate experimental groups of horses were established (group 1: n = 9, group 2: n = 6) and infected by intranasal instillation of 5 × 107 plaque forming units (PFU) of EHV-1 in 10 ml of saline of the neuropathogenic EHV-1 strain Ab4 (D752 variant). This virus was previously isolated from a quadriplegic mare (27). Horses in group 1 were euthanized 30 dpi, while horses in group 2 were euthanized 70 dpi.
Clinical Data, Nasal Virus Shedding, and Viremia
Collection of clinical data, nasal viral shedding and viremia are described in detail elsewhere (28). Briefly, clinical examinations were performed and nasal swabs for viral DNA isolation were collected prior to infection (at day −5 and −3) and daily from day 1 to 14, and every other day from day 14 to 21 dpi. Blood samples were taken prior to infection (at day −5) and daily from 1 to 10 dpi for detection of cell-associated viremia. Nasal viral shedding and viremia were analyzed by real-time PCR (qPCR) using specific probes and primers for EHV-1 glycoprotein B (1).
Necropsy and Tissue Collection
All horses were euthanized by sedation with detomidine (0.012 mg/kg, i.v.) followed by administration of pentobarbital (380 mg/kg, i.v.) and immediately (within 10–15 min) transported to the Veterinary Diagnostic Laboratory at MSU for necropsy and sample collection. Abdominal sympathetic trunk ganglia (ST), TG, retropharyngeal lymph nodes (RLn) and mesenteric lymph nodes (MesLn) were collected from all animals. For two horses (one in each infection group), RLn samples were not available for the study. Furthermore, kidneys were collected from all animals as negative tissue controls, hypothesizing, that this sample type represents no target for EHV-1. Time from euthanasia to completed tissue collection was <70 min. All tissues were fixed in 5% paraformaldehyde for 24 h, followed by routine tissue processing and paraffin embedding.
DNA/RNA Extraction
Viral DNA and total RNA from paraformaldehyde-fixed-paraffin-embedded (PFPE) tissue was extracted using AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany). Briefly, five serial sections of 4 μm (20 μm in total) were used and processed according to the manufacturer's protocol. Extraction from DNA-free and RNA-free water was included as quality control during extraction. For neural tissue, Hematoxylin and Eosin (H&E) slides were screened for the presence of ganglia cell bodies before DNA/RNA isolation. All RNA extraction samples were tested with qPCR for absence of genomic DNA using primers and probe for equine glyceraldehyde-3-phosphate dehydrogenase (eqGAPDH) as previously published (29). Genomic DNA positive RNA samples were treated with the RQ1 Rnase-free Dnase Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions and re-tested afterwards, as described above. Only samples negative for genomic DNA were converted to complementary DNA (cDNA) using the Quantinova Reverse Transcription Kit (Qiagen, Hilden, Germany) with random hexamer primers according to the manufacturer's protocol.
Real-Time PCR
For detection of EHV-1 genomic DNA and late gene mRNA, a qPCR assay targeting a 106 bp long region of the glycoprotein B gene (open reading frame 33) was performed as previously published (1). Forward and reverse primers were used in a final concentration of 450 nM and the probe in a final concentration of 100 nM. Sequences for primers and probes used in this study are listed in Table 1.
Table 1.
Primers and probes for qPCR used in this study.
| eGAPDH | |
| egapdh (F) | 5′- GCCATCACTGCCACCCAG-3′ |
| egapdh (R) | 5′- TGGCAGCACCAGTAGAAGCA-3′ |
| egapdh (probe) | 5′[FAM]- AGGGGCTGCCCAGAACATCATCC - [TAMRA]3′ |
| B2M | |
| B2M (F) | 5′-ATGGAAAGCCAAATTTCCTG-3′ |
| B2M (R) | 5′-ACCGGTCGACTTTCATCTTC-3′ |
| B2M (probe) | 5′[HEX]-TGGGTTCCATCCGCCTGAGA –[BHQ1]3′ |
| gB | |
| gB (F) | 5′-CATACGTCCCTGTCCGACAGAT-3′ |
| gB (R) | 5′-GGTACTCGGCCTTTGACGAA-3′ |
| gB (probe) | 5′[FAM]- GGTACTCGGCCTTTGACGAA -[BHQ1]3′ |
All qPCR reactions were performed in a total reaction volume of 20 μl using 10 μl 1 × SensiFAST™ Probe Lo-ROX Kit (Bioline, Luckenwalde, Germany) and 5 μl of the template. All samples were analyzed in duplicates and amplified with the following thermal profile: initial 95°C step for 2 min, followed by 40 cycles of 95°C for 10 s and 60°C for 60 s (and hold 60°C for 60 s).
For absolute quantification, DNA and mRNA results were compared to a standard curve generated with cloned EHV-1 oligonucleotides that were kindly provided by W. Azab and N. Osterrieder. Viral copy numbers were then normalized to a standard curve generated with oligonucleotides specific to the housekeeping gene equine B2M, as previously described (30). Viral DNA and mRNA concentrations were expressed as copies per 106 cells, considering that each diploid eukaryotic cell contains two copies of B2M gene (30). Positive EHV-1 DNA (extracted from lung of an EHV-1 aborted fetus) and horse DNA (extracted from equine liver) were included as positive controls for EHV-1 gB and B2M qPCR assay. DNA-free and RNA-free water was used as negative control.
Hematoxylin and Eosin Staining & Histology Grading
For Histological evaluation, 4 μm thick sections of the PFPE blocks were routinely stained with Hematoxylin and Eosin (H&E). Ganglion cells, satellite cells and nerve fibers were evaluated for the presence of histopathological changes and inflammatory infiltrations were assessed and scored as mild, moderate or severe, if present.
Immunohistochemistry
Immunohistochemistry (IHC) was conducted on samples positive for EHV-1 genomic DNA using EHV-1/EHV-4 polyclonal caprine antiserum (VMRD, Pullman, USA) cross-reactive with EHV-1 and EHV-4 antigen (Ag). Serial section of PFPE blocks were deparaffinized and antigen retrieval was performed by incubation in citrate buffer (0.1 M, pH6.0) and heating in a microwave oven (700 W) for 20 min. Endogenous peroxidase was quenched through incubation with H2O2 (1%, 15 min) and subsequently nonspecific binding of proteins was blocked with blocking buffer containing rabbit serum (1:10, 30 min), followed by incubation with the primary antibody (Ab) against EHV-1/EHV-4 Ag (VMRD, Pullman, USA, polyclonal, goat, 1:1,600) for 1 h at room temperature. Subsequently, samples were incubated with rabbit anti-goat biotinylated Ab (Vector laboratories LTD, Burlingame, USA), then incubated with avidin-biotin-complex (1:100, 30 min) and visualized by 3,3'- diaminobenzidine (DAB) (Vector laboratories LTD, Burlingame, USA). Counterstaining was performed with Mayers Hemalum. Lung tissue from an EHV-1 aborted fetus was included as positive control. As a negative control, the primary Ab was omitted and replaced by the corresponding antibody dilutent (blocking buffer containing rabbit serum). Non-neuronal cells were characterized on selected neural samples using antibodies directed against CD3 (DAKO, Hamburg, Germany, polyclonal, rabbit, 1:500), CD20 (Thermo Scientific Labvision, Fremont CA, USA, polyclonal, rabbit, 1:1,000) and S-100 (DAKO, Hamburg, Germany, polyclonal, rabbit, 1:6,000) using routine methods. Briefly, pre-treatment was performed as described for EHV-1/EHV-4 with blocking buffer containing goat serum, followed by incubation with the correspondent primary Ab overnight at 4°C. Incubation with goat anti-rabbit biotinylated Ab, detection and counterstaining was performed as described for EHV-1/EHV-4 Ab.
In situ Hybridization
In situ hybridization detecting viral DNA was performed by RNAScope® technology (Advanced Cell Diagnostics, Inc., USA) as previously described (31–33) using the RNAScope® 2.5 Detection Kit (Red) and the EHV-1 probe (V-EHV-1-ORF33, Cat.no.552651) targeting the region between nucleotides 61485-62416 of AY665713.1. Briefly, 4 μm serial sections were cut from PFPE blocks of selected EHV-1 qPCR positive TG (group 1, n = 3, group 2, n = 4) and ST (group 1, n = 4; group 2, n = 3). Subsequently, deparaffinization, pretreatment and hybridization were performed according to the manufacturer's protocol using the provided pretreatment solutions and wash buffer. All incubation steps were performed in a humidity control tray and a HybEZTM oven (Advanced Cell Diagnostics, Inc., USA). Following hybridization the signal was detected using Fast Red as chromogen provided by the manufacturer (RedB:RedA, 1:60 ratio). Counterstaining was performed using 50% Gill's Hematoxylin 1 (American MasterTech, Lodi, CA), followed by bluing with tap water and 0.02% ammonium hydroxide water. Slides were air dried and mounted with Xylene and EcoMount (EcoMount, Biocare Medical, Concord, CA).
EHV-1 qPCR positive (lung of an EHV-1 aborted fetus) and qPCR negative (TG) tissues were used as controls and an EHV-1-scrambled probe was designed targeting the same region as the target probe but with minor sequence alteration. For technical assay control, the housekeeping gene Peptidylprolyl isomerase B (PPIB) was used as positive control and the bacterial gene dihydrodipicolinate reductase (dapB) was included as negative control according to the manufacturer's protocol.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 6.01 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). Differences in the number of EHV-1 qPCR positive samples between the two infection groups were evaluated using Fisher's Exact Test. Mann-Whitney U test was used to compare significant differences in viral load between the two infection groups. P-values < 0.05 were considered statistically significant.
Results
Clinical Data, Viral Nasal Shedding, and Viremia
Animals in both infection groups showed respiratory symptoms and a classical bi-phasic fever post infection. Viral DNA amounts in nasal shedding and cell-associated viremia were detected in both groups for multiple days post infection. Overall, no significant differences were observed when comparing the two groups (data not shown) (28).
Detection of EHV-1 gB DNA and Characterization of Viral Activity
EHV-1 DNA was detected at both timepoints post infection in TG, ST, RLn, and MesLn samples. Viral distribution and -load for both groups are shown in Table 2. Kidney samples were consistently negative for EHV-1 DNA in both groups (data not shown). Lytic replication of EHV-1 was excluded for each EHV-1 DNA positive sample using reverse transcription PCR (RT-PCR) for gB mRNA and IHC for viral antigen. Specific gB mRNA amplification signal or viral antigen could not be detected in EHV-1 DNA positive tissue samples. Therefore, we concluded that there was no lytic EHV-1 infection in any of the examined tissue samples. Two out of 15 retropharyngeal lymph nodes were not available for this study and for horses 70 dpi, 3/6 sympathetic trunk ganglia samples could not be included in further analysis due to the absence of neural cell bodies in the collected tissue sample.
Table 2.
Distribution of EHV-1 genomic DNA 30 and 70 days post infection (dpi) (EHV-1 gDNA copies/106 cells).
| dpi | Horse ID | TG | ST | RLn | MesLn |
|---|---|---|---|---|---|
| 30 | 1,598 | – | – | 1.43 x 104 | 3.81 x 102 |
| 1,619 | 1.42 × 104 | – | 2.19 × 103 | 6.35 × 101 | |
| 1,620 | 3.13 × 103 | – | 7.72 × 101 | 7.89 × 102 | |
| 1,651 | 5.41 × 103 | 1.98 × 105 | n.d. | 6.64 × 103 | |
| 1,621 | 2.58 × 104 | – | 1.40 × 103 | 6.85 × 102 | |
| 1,636 | 1.10 × 104 | – | 7.30 × 102 | 1.43 × 102 | |
| 1,638 | 2.03 × 103 | 1.80 × 104 | 7.53 × 102 | 2.81 × 102 | |
| 1,628 | 3.05 × 103 | 3.27 × 104 | 3.63 × 103 | 6.45 × 101 | |
| 1,629 | 4.03 × 103 | 5.07 × 103 | 1.97 × 102 | – | |
| 70 | 905 | 4.19 × 101 | 3.82 × 104 | – | – |
| 909 | – | n.d. | n.d. | – | |
| 913 | – | – | – | 1.72 × 103 | |
| 900 | 9.93 × 101 | n.d. | – | – | |
| 914 | 2.11 × 101 | 1.57 × 104 | 2.62 × 102 | 1.47 × 103 | |
| 908 | 3.51 × 101 | 2.55 × 103 | – | 7.48 × 102 |
–, negative; n.d., not determined; TG, trigeminal ganglia; ST, sympathetic trunk ganglia; RLn, retrophrayngeal lymph node; MesLn, mesenteric lymph node.
Comparison of Viral Distribution and Copy Number at 30 and 70 dpi
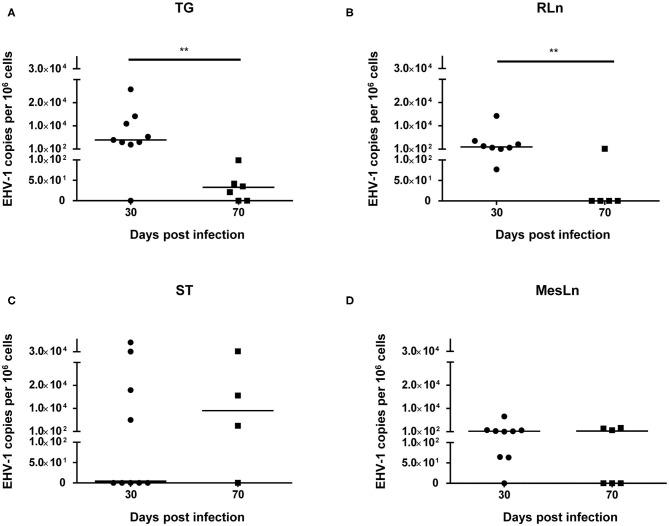
When comparing samples from horses 30 dpi with samples from horses 70 dpi (Table 2), a reduction of the percentage of positive tissue samples could be detected for TG, RLn, and MesLn samples, but this difference was only statistically significant for RLn samples. All of the available RLn samples at day 30 pi were positive (8/8), while only 1/5 was positive at day 70 pi. There were 8/9 positive TG and MesLn samples 30 dpi compared to 4/6 and 3/6 positive samples by 70 dpi, but the difference in percentage of positive samples for this group was not statistically significant. For ST, 4/9 in the 30 dpi group were positive for EHV-1 DNA and 3/4 samples were positive at day 70 pi, but this difference was not statistically significant. The viral copy number in both TG and RLn samples was significantly higher at 30 dpi when compared to samples collected at 70 dpi (Figures 1A,B). No statistically significant difference in viral copy number could be observed in ST or MesLn samples (Figures 1C,D).
Figure 1.
Significant differences (**P < 0.01) in viral copy numbers in (A) trigeminal ganglia and (B) retropharyngeal lymph nodes at 30 dpi compared to 70 dpi. Viral copy numbers in (C) sympathetic trunk and (D) mesenteric lymph nodes at 30 dpi compared to 70 dpi. Line indicates the median.
Histopathological Evaluation and Cellular Characterization
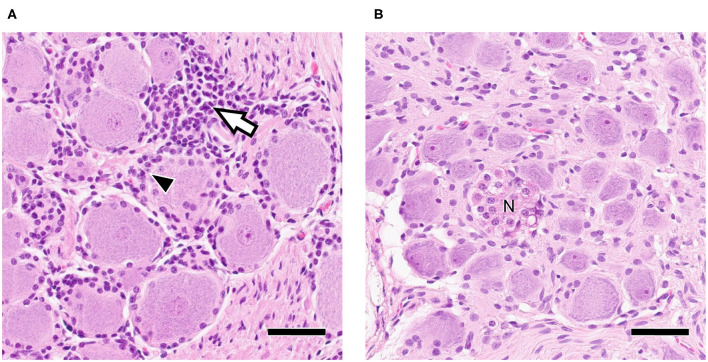
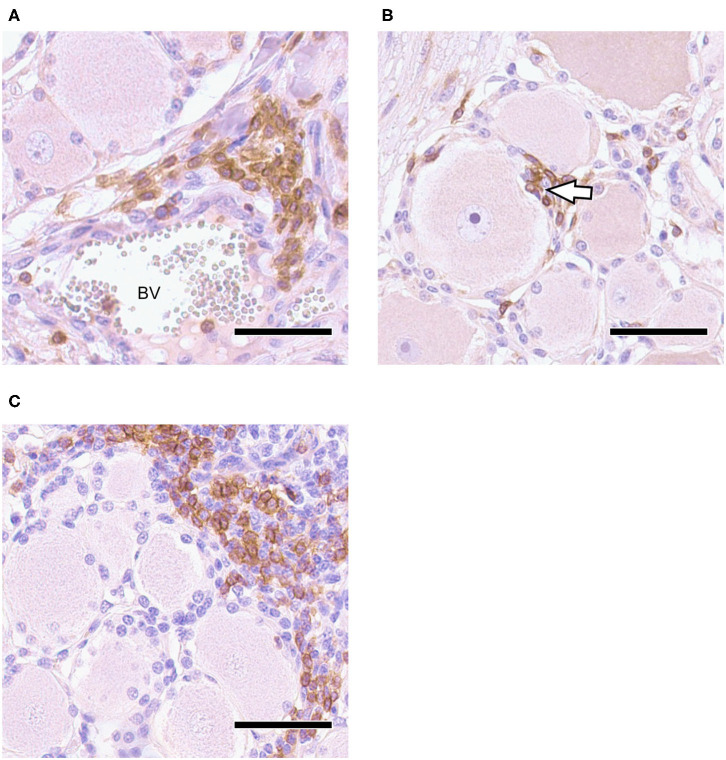
Neural tissue sections were analyzed for histopathological changes at 30 and 70 dpi. All TG and 7/12 ST had mild inflammatory infiltrations and mild satellitosis (Figure 2A) at both 30 and 70 dpi. In addition, Nageotte's bodies, which are a sign for neuronal degeneration (34) were detected in TG and in ST occasionally in both groups but no differences in numbers were observed when comparing time points (Figure 2B). No differences in severity of inflammation could be observed when comparing the two groups. Further characterization of ganglia at 30 dpi by IHC revealed that inflammatory infiltrates consisted of mainly CD3+ T-cells (Figures 3A,B). Few CD20+ B-cells were detected (Figure 3C).
Figure 2.
(A) Trigeminal Ganglion 30 dpi: Moderate infiltration of inflammatory cells (white arrow) and satellitosis (arrowhead); (B) Sympathetic trunk ganglion 30 dpi: Nageotte's body (N) as sign of neuronal degeneration; Bars = 50 μm.
Figure 3.
Trigeminal Ganglion 30 dpi, IHC for CD3 and CD20, DAB with Mayer's hemalum counterstaining: (A) CD3+ T-cell infiltration in vicinity of blood vessel (BV); (B) CD3+ T-cells penetrate neuron-satellite sheet (arrowhead); (C) localized infiltrates of CD20+ B-cells associated with non-labeling T-cells; bars = 50 μm.
Detection of EHV-1 gB DNA by in situ Hybridization
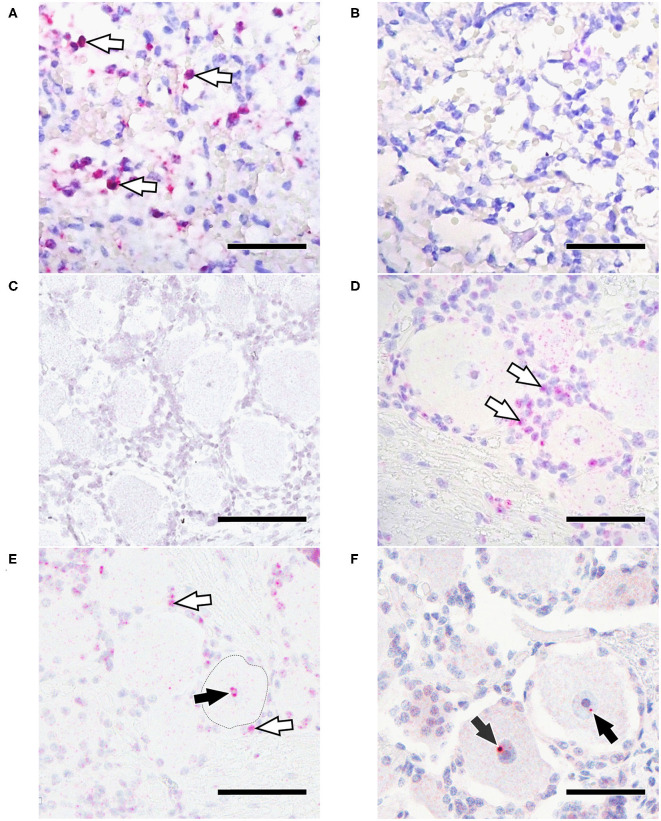
The EHV-1 gB probe detected large number of positive cells in the known EHV-1 positive control (Figure 4A), while the negative control probe (a.k.a. EHV-1 scrambled probe), showed no signal (Figure 4B). In addition, no signal was detected with the EHV-1 gB probe in a trigeminal ganglion from an EHV-1 negative horse (as determined by qPCR), which was euthanized for unrelated reasons (Figure 4C).
Figure 4.
In situ hybridization for EHV-1 gB. (A) Positive control with positive labeling (arrowheads); (B) Positive control tissue with no signal using an EHV-1 scrambled probe; (C) TG from a EHV-1 qPCR negative horse lacks positive ISH signal; (D) EHV-1 qPCR positive TG 30 dpi with strong signal in non-neuronal cells (arrowheads); (E) EHV-1 qPCR positive TG 30 dpi with strong in non-neuronal cells (white arrowheads) and nucleus of the neuron (black arrowhead); (F) EHV-1 positive TG 70 dpi with positive signal in the nucleus of the neuron (arrowhead). Bars = 50 μm.
A positive signal could be detected in 6/7 TG samples and 4/7 ST samples that had tested positive for EHV-1 by qPCR. Results are shown in Table 3 and Figures 4D–F. At 30 dpi, all tested TG (3/3) had a positive ISH signal in ganglion cells as well as strong concurrent labeling in non-neuronal cells surrounding ganglia (Figures 4D,E). Within neurons, the ISH signal was located in the nucleus and the cytoplasm. For ST at 30 dpi, 3/4 were positive by ISH with signal being detected in the cytoplasm of neuronal cell bodies and in 2/4 samples also in non-neuronal cells. At 70 dpi, an EHV-1 hybridization signal was detected in the nuclei of ganglia cell bodies from 3/4 TG, but not in non-neuronal cells (Figure 4F). For ST, only 1/3 samples from horses euthanized at 70 dpi had a positive signal in the cytoplasm of the neurons but was also positive in non-neuronal cells.
Table 3.
Cellular visualization of EHV-1 DNA in trigeminal ganglia and sympathetic trunk ganglia using in situ hybridization.
| 30 dpi | 70 dpi | |||||||
|---|---|---|---|---|---|---|---|---|
| Viral localization (ISH) | Viral localization (ISH) | |||||||
| Tissue | Horse ID | Viral load gDNA gB | Neuron | Non-neuronal cells | Horse ID | Viral load gDNA gB | Neuron | Non-neuronal cells |
| TG | 1,651 | 5.41 × 103 | + | + | 905 | 4.19 × 101 | + | – |
| 1,621 | 2.58 × 104 | + | + | 900 | 9.93 × 101 | + | – | |
| 1,628 | 3.05 × 103 | + | + | 914 | 2.11 × 101 | + | – | |
| 908 | 3.51 × 101 | – | – | |||||
| ST | 1,651 | 1.98 × 105 | + | + | 905 | 3.82 × 104 | + | + |
| 1,638 | 1.80 × 104 | – | – | 914 | 1.57 × 104 | – | – | |
| 1,628 | 3.27 × 104 | + | – | 908 | 2.55 × 103 | – | – | |
| 1,629 | 5.07 × 103 | + | + | |||||
Viral load expressed in EHV-1 gDNA copies/106cells based on qPCR.
dpi, days post infection; ISH, in situ hybridization; TG, trigeminal ganglia; ST, sympathetic trunk ganglia; +, positive ISH signal; –, no ISH signal.
Discussion
This study compares neuropathogenic EHV-1 strain Ab4 distribution and persistence in selected lymphoid and neural tissues collected at different time points (30 and 70 days) following experimental infection. We detected EHV-1 in non-neuronal support or interstitial cells of trigeminal and sympathetic trunk ganglia. These findings provide new insight in EHV-1 latency establishment as we propose that in addition to retrograde neuronal transport, EHV-1 uses mononuclear cells during viremia as alternative route toward latency locations.
The current understanding is that EHV-1 latency is established in the trigeminal ganglia and respiratory associated lymphoid tissue during primary respiratory tract infection and/or during cell-associated viremia (8, 9, 21). Both of our sample collection timepoints were chosen after active viral replication and viremia had ceased in all horses. Typically, this occurs around 14 days after experimental infection (1, 35–38), which was also the case in all horses sampled for this study. Furthermore, at both collection time points, neither gB (late gene) mRNA, nor viral protein could be detected, favoring the idea that EHV-1 was present in a quiescent state in both EHV-1 DNA positive tissue sets, as defined previously (15). Thirty days post infection, EHV-1 (gB) DNA was detected at high copy number in TG, ST, RLn and in MesLn samples. While TG and RLn are considered the most common latency locations described in previous studies, we also show latent EHV-1 to be present in abdominal neuronal and lymphoid tissues. This also confirms our previous results, where we showed viral distribution in various neuronal and lymphoid tissues, and not just in the vicinity of primary infection at day 70 post infection (23). These findings raise the question of how the virus is transported to abdominal locations, as a thorough understanding of EHV-1 pathogenesis is crucial to further define the balance between latency and reactivation.
The pathogenesis of EHV-1 latency establishment is currently best studied for the trigeminal ganglia but overall poorly understood. EHV-1 is thought to follow the strategy of other Alphaherpesviruses and to reach neuronal cell bodies via retrograde axonal transport (3, 8, 9, 39), following primary infection of the upper respiratory tract. Alphaherpesviruses are known to enter sensory nerve endings and are transported actively by using cellular molecular motor proteins to the ganglion cell body (40). Here, the virus translocates to the nucleus of the neuron and becomes latent after a short initial lytic replication cycle (12). While this pathway seems to be likely for EHV-1 latency establishment in neuronal tissue in the vicinity of the primary infection sites, retrograde axonal transport to abdominal sympathetic trunk ganglia would be challenging as the virus would have to travel very long distances and potentially overcome multiple synapses. VZV, a close relative to EHV-1, establishes latency in TG and dorsal root ganglia (DRG) but also infects various sensory and autonomic ganglia (41–46). Recently, there is growing evidence that VZV most likely uses infected lymphocytes as alternative route to reach ganglion neurons during cell-associated viremia (24, 25, 47).
In order to get a better understanding of the pathways and cellular interplays of EHV-1 latency establishment, we used in situ hybridization to determine the cellular localization of latent EHV-1 DNA in TG and ST samples. While in samples collected 30 dpi a positive EHV-1 ISH signal was noticed in the neurons of TG and ST, a strong signal was also detected in the (non-neuronal) support and interstitial cells. These findings suggest that there exists an alternative pathway for EHV-1 latency establishment, which might be similar to VZV. Numerous studies described neurons as main infected cell types in ganglia during latency for HSV-1, BoHV-1 and VZV (48–51). However, only few studies tried to determine latent EHV-1 localization in ganglia. Baxi et al. claimed latent EHV-1 in trigeminal neurons by using RNA in situ hybridization to detect potential latency associated transcripts (9). Neurons represent a stable cell population where the virus is able to hide lifelong in the host (52, 53) without being detected by the immune system. The viral DNA translocates to the nucleus of the infected neuron and persists in a circular episome, not integrated into the host DNA. In addition to the nuclear localization of viral DNA, we could also detect ISH signal frequently in the cytoplasm of TG and ST neurons. This cytoplasmatic signal could not be detected in samples with the EHV-1 scrambled (negative) probe or with the technical assay control probe provided by the manufacturer. Therefore, unspecific probe binding seems unlikely. Similarly, cytoplasmatic signal of VZV DNA could be found in neurons, where the virus reactivates, and lytic infection occurs (54). IHC for lytic protein and for late gene (gB) mRNA could not be detected in the present study, and thus we exclude an overall active infection. However, latent infection is not exclusively silent and occasional transcription of lytic genes is described for HSV-1 and VZV latent infection at low levels (55). Therefore, we might have detected low levels of viral DNA or gB mRNA in the cytoplasm in the present study. This phenomenon needs further investigation, as we did not detect viral gB mRNA via RTqPCR and assume an overall higher sensitivity for the RTqPCR assay. However, RNAScope® assay has been described to be highly sensitive due to the unique selective amplification scheme in combination with a serial target probe design (31, 56).
To the best of our knowledge, EHV-1 has not been detected in non-neuronal (support or interstitial) cells within ganglia previously. Non-neuronal cells are mainly composed of glial cells of the peripheral nervous system (satellite glial cells) and mononuclear cells. Each neuronal cell body is enveloped by several satellite cells forming a functional unit. This is important for the regulation of extracellular chemical conditions of the neuron and providing physical support (34, 57). Furthermore, satellite cells act as antigen presenting cells during infections, attract T-cells and can interact with the neuron by signaling processes (57, 58). In acute human herpesvirus infections, the barrier formation of satellite cells around the neurons seems to protect the neuron from virus spread (57). However, it has been shown that this barrier is a flexible wall, where macrophages still can penetrate (59) and infiltrating T-cells tend to come in close contact to the neuron-satellite sheath (16). Furthermore, satellite cells are infected during productive VZV infection and seem to transfer virus to neurons by cell-to-cell fusions in vivo mouse models (60). To further differentiate lymphocytes from satellite cells in the present study, we used CD3+, CD20+ and S-100 IHC staining. We could show that besides local CD3+ and CD20+ lymphocytic inflammatory infiltrates, CD3+ T-cells also associate with satellite cells and tend to squeeze into the satellite sheath around neurons. This phenomenon was not detected for CD20+ B-cells. These results suggest that the EHV-1 ISH signal from non-neuronal cells surrounding neurons in the current study, might mainly attributed to T-cells and satellite cells.
T-cell tropism is well described for EHV-1 and suggested to be a key strategy for immune evasion and dissemination in the host (4, 7, 19, 21, 61, 62). During primary viral infection, EHV-1 seems to initially infect monocytes (4, 5, 63), while T-cells are the preferred cell type during viremia (6, 7, 61) to transport the virus to secondary target organs like the CNS or uterus. Among the circulating EHV-1 strains, the neurovirulent strain Ab4 has been shown to infect immune cells efficiently and rapidly following replication in the respiratory epithelium and subsequently result in a longer viremia with higher viral loads compared to non-neuropathogenic strain counterparts (5, 36, 64). It has been suggested that viral replication is not productive or restricted in carrier cells (65) or viral capsids are accumulated in the nucleus of the carrier cell, but viral egress is hampered until the endothelium of a secondary target organ is reached (7). This has also been described for VZV (11) and it has been shown that migration markers on T-cells are still intact following VZV infection which enables the virus to be transported to target sites (66). Interestingly, while there was still distinct EHV-1 ISH signal in the nucleus of the trigeminal ganglia at 70 dpi, no signal could be detected in non-neuronal cells at this time point. Moreover, there was a significant decrease in viral load when comparing TG 30 and 70 dpi. For HSV-1 infections, it has been shown that the number of T-cells in ganglia declines continuously between days 21 and 92 p.i. (16). Apoptosis of infected lymphocytes and degeneration of some neurons could be reason for the decrease of viral load in ganglia in the present study. However, it might also be evidence for the existence of a persistently infected EHV-1 memory T-cell pool, where T-cells are retrieved from ganglia, (re-)circulate in the blood and home to lymphoid tissue, where the virus remains stationary latent and/or starts new re-circulation. However, in our previous study no or only very low amounts of EHV-1 DNA was present in peripheral blood mononuclear cells (PBMC) at day 70 pi (unpublished data). This may imply that the majority of latently infected mononuclear cells are in tissue sites and it is challenging to detect very low amounts of silent virus in the circulating PBMCs using our methods as the collection time point represents a moment in time in the circulating latent infected cell pool. Further studies involving methods with higher sensitivity, e.g., Next Generation Sequencing, are needed to finally define the role and type of mononuclear cells in EHV-1 latency establishment.
Furthermore, latency in lymphatic tissue has been described as an advantage for the virus, as infected T-cells can enter rapidly the blood circulation to be transported to primary infection sites for replication, once reactivation occurs (19). In the present study, we report EHV-1 DNA in RLn and MesLn at 30 and 70 dpi. Respiratory associated lymphoid tissue has been previously described as latency location for EHV-1 (19–21, 67) and EHV-1 DNA could also be detected in MesLn and in the spleen (19, 23, 67). When comparing results 30 and 70 dpi, it was somewhat surprising, that the number of positive retropharyngeal lymph node samples decreased by 80% over time. While apoptosis of some EHV-1 infected T-cells is likely contributing to this decrease, positive T-cells could also migrate from lymph nodes toward neurons and/or other lymphatic tissue and therefore deplete positive signal in lymph nodes over time. A neurotropic re-circulation could also explain the findings in sympathetic trunk ganglia 70 dpi, where positive EHV-1 non-neuronal cells could still be detected.
In addition, while EHV-1 strain Ab4 is known to strongly dysregulate the host immune response by increasing inflammation and resulting in a high quantity of circulating infected cells, other EHV-1 strains may act differently in their immune evasive strategies (7, 68). Therefore, latency establishment and reactivation also may be strain dependent and more research is necessary to reveal the latently infected host pool in the populations worldwide.
Taken together, the present study confirms previous findings, identifying EHV-1 as both neurotropic and lymphotropic. EHV-1 most likely uses retrograde axonal transport as a direct pathway from the upper respiratory epithelium toward the TG, where latency is established in the nucleus of the neuron after a short replication cycle. Furthermore, we hypothesize, that T-cell tropism assists EHV-1 in its neurotropism and additionally provides latency establishment in lymphatic tissues other than respiratory associated lymphoid tissue. The navigation to neuronal structures throughout the body may enable the virus to establish latency in numerous ganglia, but further studies in random horse populations are required to elucidate if other neuronal structures than the TG can be repeatedly confirmed as alternative latency locations.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by Michigan State University Institutional Animal Care and Use Committee.
Author Contributions
KG and SS carried out the experiments and data analysis. KG wrote the manuscript with support from GSH and LG. GSH developed the experimental design of the horse infection experiment and conducted the clinical study. MK and DS conducted the necropsy and sample collection and GSH, KG, and LG contributed to sample collection. KM contributed to interpretation of histopathological and in situ hybridization results and performed figure formatting. FL and JS helped with the design and performance of the in situ hybridization experiment. FL, JS, and RF contributed to the interpretation of the results. LG supervised the project. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank S. Baur, C. Haupt, and E. Fink for their excellent technical support as well as J. Hengesbach and S. Griffin for their very helpful assistance during necropsy and sample collection.
Footnotes
Funding. This study was made possible through a grant (no. 2016#306) by the Grayson Jockey Club Research Foundation, Inc., Lexington, KY.
References
- 1.Hussey SB, Clark R, Lunn KF, Breathnach C, Soboll G, Whalley JM, et al. Detection and quantification of equine herpesvirus-1 viremia and nasal shedding by real-time polymerase chain reaction. J Vet Diagn Invest. (2006) 18:335–42. 10.1177/104063870601800403 [DOI] [PubMed] [Google Scholar]
- 2.Goehring LS, van Maanen C, Berendsen M, Cullinane A, de Groot RJ, Rottier PJM, et al. Experimental infection with neuropathogenic equid herpesvirus type 1. (EHV-1) in adult horses. Vet J. (2010) 186:180–7. 10.1016/j.tvjl.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 3.Slater J. Equine herpesviruses. In: Sellon DC, and Long MT, editor. Equine Infectious Diseases. St. Louis, MO: Elsevier; (2014). p. 151-168.e158. [Google Scholar]
- 4.van der Meulen KM, Nauwynck HJ, Buddaert W, Pensaert MB. Replication of equine herpesvirus type 1 in freshly isolated equine peripheral blood mononuclear cells and changes in susceptibility following mitogen stimulation. J Gen Virol. (2000) 81:21–5. 10.1099/0022-1317-81-1-21 [DOI] [PubMed] [Google Scholar]
- 5.Vandekerckhove AP, Glorieux S, Gryspeerdt AC, Steukers L, Duchateau L, Osterrieder N, et al. Replication kinetics of neurovirulent versus non-neurovirulent equine herpesvirus type 1 strains in equine nasal mucosal explants. J Gen Virol. (2010) 91:2019–28. 10.1099/vir.0.019257-0 [DOI] [PubMed] [Google Scholar]
- 6.Wilsterman S, Soboll-Hussey G, Lunn D, Ashton L, Callan R, Hussey S, et al. Equine herpesvirus-1 infected peripheral blood mononuclear cell subpopulations during viremia. Vet Microbiol. (2011) 149:40–7. 10.1016/j.vetmic.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 7.Poelaert KC, Van Cleemput J, Laval K, Favoreel HW, Couck L, Van den Broeck W, et al. Equine herpesvirus 1 Bridles T lymphocytes to reach its target organs. J Virol. (2019) 93:e02098–18. 10.1128/JVI.02098-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slater JD, Borchers K, Thackray AM, Field HJ. The trigeminal ganglion is a location for equine herpesvirus 1 latency and reactivation in the horse. J Gen Virol. (1994) 75:2007–16. 10.1099/0022-1317-75-8-2007 [DOI] [PubMed] [Google Scholar]
- 9.Baxi MK, Efstathiou S, Lawrence G, Whalley JM, Slater JD, Field HJ. The detection of latency-associated transcripts of equine herpesvirus 1 in ganglionic neurons. J Gen Virol. (1995) 76:3113–8. 10.1099/0022-1317-76-12-3113 [DOI] [PubMed] [Google Scholar]
- 10.Efstathiou S, Preston CM. Towards an understanding of the molecular basis of herpes simplex virus latency. Virus Res. (2005) 111:108–19. 10.1016/j.virusres.2005.04.017 [DOI] [PubMed] [Google Scholar]
- 11.Reichelt M, Wang L, Sommer M, Perrino J, Nour AM, Sen N, et al. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against Varicella-Zoster Virus. PLOS Pathog. (2011) 7:e1001266. 10.1371/journal.ppat.1001266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones C. Bovine Herpes Virus 1 (BHV-1) and Herpes Simplex Virus Type 1 (HSV-1) promote survival of latently infected sensory neurons, in part by inhibiting apoptosis. J Cell Death. (2013) 6:1–16. 10.4137/JCD.S10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramakrishnan R, Poliani PL, Levine M, Glorioso JC, Fink DJ. Detection of herpes simplex virus type 1 latency-associated transcript expression in trigeminal ganglia by in situ reverse transcriptase PCR. J Virol. (1996) 70:6519–23. 10.1128/JVI.70.9.6519-6523.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pusterla N, Hussey SB, Mapes S, Johnson C, Collier JR, Hill J, et al. Molecular investigation of the viral kinetics of equine herpesvirus-1 in blood and nasal secretions of horses after corticosteroid-induced recrudescence of latent infection. J Vet Int Med. (2010) 24:1153–7. 10.1111/j.1939-1676.2010.0554.x [DOI] [PubMed] [Google Scholar]
- 15.Pusterla N, Mapes S, Wilson WD. Prevalence of equine herpesvirus type 1 in trigeminal ganglia and submandibular lymph nodes of equids examined postmortem. Vet Rec. (2010) 167:376–9. 10.1136/vr.c3748 [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Tang Q, Hendricks RL. Inflammatory infiltration of the trigeminal ganglion after herpes simplex virus type 1 corneal infection. J Virol. (1996) 70:264–71. 10.1128/JVI.70.1.264-271.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilden DH, Vafai A, Shtram Y, Becker Y, Devlin M, Wellish M. Varicella-zoster virus DNA in human sensory ganglia. Nature. (1983) 306:478–80. 10.1038/306478a0 [DOI] [PubMed] [Google Scholar]
- 18.Edington N, Bridges CG, Patel JR. Endothelial cell infection and thrombosis in paralysis caused by equid herpesvirus-1: equine stroke. Arch Virol. (1986) 90:111–24. 10.1007/BF01314149 [DOI] [PubMed] [Google Scholar]
- 19.Welch HM, Bridges CG, Lyon AM, Griffiths L, Edington N. Latent equid herpesviruses 1 and 4: detection and distinction using the polymerase chain reaction and co-cultivation from lymphoid tissues. J Gen Virol. (1992) 73:261–8. 10.1099/0022-1317-73-2-261 [DOI] [PubMed] [Google Scholar]
- 20.Kydd JH, Smith KC, Hannant D, Livesay GJ, Mumford JA. Distribution of equid herpesvirus-1 (EHV-1) in respiratory tract associated lymphoid tissue: implications for cellular immunity. Equine Vet J. (1994) 26:470–3. 10.1111/j.2042-3306.1994.tb04052.x [DOI] [PubMed] [Google Scholar]
- 21.Chesters PM, Allsop R, Purewal A, Edington N. Detection of latency-associated transcripts of equid herpesvirus 1 in equine leukocytes but not in trigeminal ganglia. J Virol. (1997) 71:3437–43. 10.1128/JVI.71.5.3437-3443.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith DJ, Iqbal J, Purewal A, Hamblin AS, Edington N. In vitro reactivation of latent equid herpesvirus-1 from CD5+/CD8+ leukocytes indirectly by IL-2 or chorionic gonadotrophin. J Gen Virol. (1998) 79:2997–3004. 10.1099/0022-1317-79-12-2997 [DOI] [PubMed] [Google Scholar]
- 23.Goehring L. Latency in Equid Herpesvirus-1 purpose-infected Horses, in 2017 ACVIM Forum Research Report Program. J Vet Int Med. (2017) 31:1598–9. 10.1111/jvim.14784 [DOI] [Google Scholar]
- 24.Grigoryan S, Yee MB, Glick Y, Gerber D, Kepten E, Garini Y, et al. Direct transfer of viral and cellular proteins from varicella-zoster virus-infected non-neuronal cells to human axons. PLoS ONE. (2015) 10:e0126081. 10.1371/journal.pone.0126081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depledge DP, Sadaoka T, Ouwendijk WJD. Molecular aspects of Varicella-Zoster virus latency. Viruses. (2018) 10:349. 10.3390/v10070349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kydd JH, Townsend HGG, Hannant D. The equine immune response to equine herpesvirus-1: the virus and its vaccines. Vet Immunol Immunopathol. (2006) 111:15–30. 10.1016/j.vetimm.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 27.Crowhurst FA, Dickinson G, Burrows R. An outbreak of paresis in mares and geldings associated with equid herpesvirus 1. Vet Rec. (1981) 109:527–8. [PubMed] [Google Scholar]
- 28.Holz CL, Nelli RK, Wilson ME, Zarski LM, Azab W, Baumgardner R, et al. Viral genes and cellular markers associated with neurological complications during herpesvirus infections. J Gen Virol. (2017) 98:1439–54. 10.1099/jgv.0.000773 [DOI] [PubMed] [Google Scholar]
- 29.Pusterla N, Wilson WD, Conrad PA, Barr BC, Ferraro GL, Daft BM, et al. Cytokine gene signatures in neural tissue of horses with equine protozoal myeloencephalitis or equine herpes type 1 myeloencephalopathy. Vet Rec. (2006) 159:341–6. 10.1136/vr.159.11.341 [DOI] [PubMed] [Google Scholar]
- 30.Abdelgawad A, Damiani A, Ho SYW, Strauss G, Szentiks CA, East ML, et al. Zebra alphaherpesviruses (EHV-1 and EHV-9): genetic diversity, latency and co-infections. Viruses. (2016) 8:262. 10.3390/v8090262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. (2012) 14:22–9. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabinski TM, Kneynsberg A, Manfredsson FP, Kanaan NM. A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS ONE. (2015) 10:e0120120. 10.1371/journal.pone.0120120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennington MR, Cossic BG, Perkins GA, Duffy C, Duhamel GE, Van de Walle GR. First demonstration of equid gammaherpesviruses within the gastric mucosal epithelium of horses. Virus Res. (2017) 242:30–6. 10.1016/j.virusres.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 34.Pannese E. The satellite cells of the sensory ganglia. Adv Anat Embryol Cell Biol. (1981) 65:1–111. 10.1007/978-3-642-67750-2_1 [DOI] [PubMed] [Google Scholar]
- 35.Gibson J, Slater J, Awan A, Field H. Pathogenesis of equine herpesvirus-1 in specific pathogen-free foals: primary and secondary infections and reactivation. Arch Virol. (1992) 123:351–66. 10.1007/BF01317269 [DOI] [PubMed] [Google Scholar]
- 36.Allen GP. Risk factors for development of neurologic disease after experimental exposure to equine herpesvirus-1 in horses. Am J Vet Res. (2008) 69:1595–600. 10.2460/ajvr.69.12.1595 [DOI] [PubMed] [Google Scholar]
- 37.Lunn DP, Davis-Poynter N, Flaminio MJ, Horohov DW, Osterrieder K, Pusterla N, et al. Equine herpesvirus-1 consensus statement. J Vet Intern Med. (2009) 23:450–61. 10.1111/j.1939-1676.2009.0304.x [DOI] [PubMed] [Google Scholar]
- 38.Pusterla N, Wilson WD, Mapes S, Finno C, Isbell D, Arthur RM, et al. Characterization of viral loads, strain and state of equine herpesvirus-1 using real-time PCR in horses following natural exposure at a racetrack in California. Vet J. (2009) 179:230–9. 10.1016/j.tvjl.2007.09.018 [DOI] [PubMed] [Google Scholar]
- 39.Pusterla N, Mapes S, David Wilson W. Prevalence of latent alpha-herpesviruses in Thoroughbred racing horses. Vet J. (2012) 193:579–82. 10.1016/j.tvjl.2012.01.030 [DOI] [PubMed] [Google Scholar]
- 40.Diefenbach RJ, Miranda-Saksena M, Douglas MW, Cunningham AL. Transport and egress of herpes simplex virus in neurons. Rev Med Virol. (2008) 18:35–51. 10.1002/rmv.560 [DOI] [PubMed] [Google Scholar]
- 41.Gilden DH, Rozenman Y, Murray R, Devlin M, Vafai A. Detection of varicella-zoster virus nucleic acid in neurons of normal human thoracic ganglia. Ann Neurol. (1987) 22:377–80. 10.1002/ana.410220315 [DOI] [PubMed] [Google Scholar]
- 42.Furuta Y, Takasu T, Fukuda S, Sato-Matsumura KC, Inuyama Y, Hondo R, et al. Detection of varicella-zoster virus DNA in human geniculate ganglia by polymerase chain reaction. J Infect Dis. (1992) 166:1157–9. 10.1093/infdis/166.5.1157 [DOI] [PubMed] [Google Scholar]
- 43.Mahalingam R, Wellish MC, Dueland AN, Cohrs RJ, Gilden DH. Localization of herpes simplex virus and varicella zoster virus DNA in human ganglia. Ann Neurol. (1992) 31:444–8. 10.1002/ana.410310417 [DOI] [PubMed] [Google Scholar]
- 44.Kennedy PGE, Grinfeld E, Gow JW. Latent varicella–zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc Natl Acad Sci. (1998) 95:4658–62. 10.1073/pnas.95.8.4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy PGE, Grinfeld E, Gow JW. Latent Varicella-Zoster Virus in human dorsal root ganglia. Virology. (1999) 258:451–4. 10.1006/viro.1999.9745 [DOI] [PubMed] [Google Scholar]
- 46.Nagel MA, Rempel A, Huntington J, Kim F, Choe A, Gilden D. Frequency and abundance of alphaherpesvirus DNA in human thoracic sympathetic ganglia. J Virol. (2014) 88:8189–92. 10.1128/JVI.01070-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zerboni L, Arvin AM. The pathogenesis of Varicella-Zoster virus neurotropism and infection. In: Reiss CS, editor. Neurotropic Viral Infections: Volume 2: Neurotropic Retroviruses, DNA Viruses, Immunity and Transmission. Cham: Springer International Publishing; (2016). [Google Scholar]
- 48.Croen KD, Ostrove JM, Dragovic LJ, Straus SE. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc Natl Acad Sci USA. (1988) 85:9773–7. 10.1073/pnas.85.24.9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai GY, Pizer LI, Levin MJ. Fractionation of neurons and satellite cells from human sensory ganglia in order to study herpesvirus latency. J Virol Methods. (2002) 104:21–32. 10.1016/S0166-0934(02)00032-0 [DOI] [PubMed] [Google Scholar]
- 50.Levin MJ, Cai GY, Manchak MD, Pizer LI. Varicella-zoster virus DNA in cells isolated from human trigeminal ganglia. J Virol. (2003) 77:6979–87. 10.1128/JVI.77.12.6979-6987.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci USA. (2005) 102:16055–9. 10.1073/pnas.0505850102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T, Khanna KM, Carriere BN, Hendricks RL. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. J Virol. (2001) 75:11178–84. 10.1128/JVI.75.22.11178-11184.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bloom DC. Chapter two - alphaherpesvirus latency: a dynamic state of transcription and reactivation. In: Kielian M, Maramorosch K, Mettenleiter TC, editors. Advances in Virus Research. Cambridge, MA: Academic Press; (2016) p. 53–80. [DOI] [PubMed] [Google Scholar]
- 54.Lungu O, Annunziato PW, Gershon A, Staugaitis SM, Josefson D, LaRussa P, et al. Reactivated and latent varicella-zoster virus in human dorsal root ganglia. Proc Natl Acad Sci USA. (1995) 92:10980–4. 10.1073/pnas.92.24.10980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy PGE, Rovnak J, Badani H, Cohrs RJ. A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation. J General Virol. (2015) 96:1581–602. 10.1099/vir.0.000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carossino M, Loynachan AT, MacLachlan NJ, Drew C, Shuck KM, Timoney PJ, et al. Detection of equine arteritis virus by two chromogenic RNA in situ hybridization assays (conventional and RNAscope®) and assessment of their performance in tissues from aborted equine fetuses. Arch Virol. (2016) 161:3125–36. 10.1007/s00705-016-3014-5 [DOI] [PubMed] [Google Scholar]
- 57.Hanani M. Satellite glial cells in sensory ganglia: from form to function. Brain Res Rev. (2005) 48:457–76. 10.1016/j.brainresrev.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 58.van Velzen M, Laman JD, Kleinjan A, Poot A, Osterhaus AD, Verjans GM. Neuron-interacting satellite glial cells in human trigeminal ganglia have an APC phenotype. J Immunol. (2009) 183:2456–61. 10.4049/jimmunol.0900890 [DOI] [PubMed] [Google Scholar]
- 59.Hu P, McLachlan E. Macrophage and lymphocyte invasion of dorsal root ganglia after peripheral nerve lesions in the rat. Neuroscience. (2002) 112:23–38. 10.1016/S0306-4522(02)00065-9 [DOI] [PubMed] [Google Scholar]
- 60.Reichelt M, Zerboni L, Arvin AM. Mechanisms of Varicella-Zoster virus neuropathogenesis in human dorsal root ganglia. J Virol. (2008) 82:3971–83. 10.1128/JVI.02592-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCulloch J, Williamson SA, Powis SJ, Edington N. The effect of EHV-1 infection upon circulating leucocyte populations in the natural equine host. Vet Microbiol. (1993) 37:147–61. 10.1016/0378-1135(93)90189-E [DOI] [PubMed] [Google Scholar]
- 62.Kydd JH, Smith K, Hannant D, Livesay GJ, Mumford JA. Distribution of Equid herpesvirus-1 (EHV-1) in the respiratory tract of ponies: implications for vaccination strategies. Equine Vet J. (1994) 26:466–9. 10.1111/j.2042-3306.1994.tb04051.x [DOI] [PubMed] [Google Scholar]
- 63.Gryspeerdt AC, Vandekerckhove AP, Baghi HB, Van de Walle GR, Nauwynck HJ. Expression of late viral proteins is restricted in nasal mucosal leucocytes but not in epithelial cells during early-stage equine herpes virus-1 infection. Vet J. (2012) 193:576–8. 10.1016/j.tvjl.2012.01.022 [DOI] [PubMed] [Google Scholar]
- 64.Goodman LB, Loregian A, Perkins GA, Nugent J, Buckles EL, Mercorelli B, et al. A point mutation in a herpesvirus polymerase determines neuropathogenicity. PLoS Pathog. (2007) 3:e160. 10.1371/journal.ppat.0030160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laval K, Favoreel HW, Nauwynck HJ. Equine herpesvirus type 1 replication is delayed in CD172a+ monocytic cells and controlled by histone deacetylases. J General Virol. (2015) 96:118–30. 10.1099/vir.0.067363-0 [DOI] [PubMed] [Google Scholar]
- 66.Ku CC, Padilla JA, Grose C, Butcher EC, Arvin AM. Tropism of varicella-zoster virus for human tonsillar CD4(+) T lymphocytes that express activation, memory, and skin homing markers. J Virol. (2002) 76:11425–33. 10.1128/JVI.76.22.11425-11433.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edington N, Welch HM, Griffiths L. The prevalence of latent Equid herpesviruses in the tissues of 40 abattoir horses. Equine Vet J. (1994) 26:140–2. 10.1111/j.2042-3306.1994.tb04353.x [DOI] [PubMed] [Google Scholar]
- 68.Wagner B, Wimer C, Freer H, Osterrieder N, Erb HN. Infection of peripheral blood mononuclear cells with neuropathogenic equine herpesvirus type-1 strain Ab4 reveals intact interferon-alpha induction and induces suppression of anti-inflammatory interleukin-10 responses in comparison to other viral strains. Vet Immunol Immunopathol. (2011) 143:116–24. 10.1016/j.vetimm.2011.06.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.