Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 21 million people worldwide since August 16, 2020. Compared to PCR and serology tests, SARS-CoV-2 antigen assays are underdeveloped, despite their potential to identify active infection and monitor disease progression.
Methods
We used Single Molecule Array (Simoa) assays to quantitatively detect SARS-CoV-2 spike, S1 subunit, and nucleocapsid antigens in the plasma of patients with coronavirus disease (COVID-19). We studied plasma from 64 patients who were COVID-19 positive, 17 who were COVID-19 negative, and 34 prepandemic patients. Combined with Simoa anti-SARS-CoV-2 serological assays, we quantified changes in 31 SARS-CoV-2 biomarkers in 272 longitudinal plasma samples obtained for 39 patients with COVID-19. Data were analyzed by hierarchical clustering and were compared to longitudinal RT-PCR test results and clinical outcomes.
Results
SARS-CoV-2 S1 and N antigens were detectable in 41 out of 64 COVID-19 positive patients. In these patients, full antigen clearance in plasma was observed a mean ± 95% CI of 5 ± 1 days after seroconversion and nasopharyngeal RT-PCR tests reported positive results for 15 ± 5 days after viral-antigen clearance. Correlation between patients with high concentrations of S1 antigen and ICU admission (77%) and time to intubation (within 1 day) was statistically significant.
Conclusions
The reported SARS-CoV-2 Simoa antigen assay is the first to detect viral antigens in the plasma of patients who were COVID-19 positive to date. These data show that SARS-CoV-2 viral antigens in the blood are associated with disease progression, such as respiratory failure, in COVID-19 cases with severe disease.
Keywords: SARS-CoV-2, viral antigen, serological, longitudinal plasma samples, single molecule arrays
Introduction
The current pandemic of coronavirus disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in over 21 000 000 confirmed cases globally and over 167 000 deaths in the United States alone, as of August 16, 2020 (1). While reverse-transcription polymerase chain reaction (RT-PCR) tests remain the gold standard for diagnosing COVID-19, RT-PCR tests do not provide adequate information on progression of the disease. Furthermore, RT-PCR tests can give positive results for several weeks after a patient has seroconverted or recovered (2). Serological tests can identify individuals who have mounted an immune response but cannot necessarily be used for monitoring disease in the early stages of infection (3–5). SARS-CoV-2 antigen assays can complement PCR and serological tests by identifying active infection or monitoring disease progression by measuring viral antigens in biofluids. Currently, there are 2 FDA approved SARS-CoV-2 antigen tests that detect nucleocapsid in nasopharyngeal (NP) swabs (6, 7). However, both tests provide qualitative results and may only detect viral antigen within the first 5 days of symptom onset. Quantitative and ultra-sensitive SARS-CoV-2 antigen assays could enable detection of viral antigens in blood, saliva, or NP swabs and, in combination with serological assays, could enable analysis of COVID-19 progression from early infection to seroconversion.
To address the need for a quantitative antigen assay, we developed ultra-sensitive Single Molecule Array (Simoa) SARS-CoV-2 antigen assays for S1, S1-S2 extracellular domain (spike), and nucleocapsid (N). The ultra-sensitivity of Simoa enables detection of SARS-CoV-2 antigens in the plasma of COVID-19 positive patients. Additionally, Simoa provides a dynamic range that allows quantification of antigens over a concentration range of 4 orders of magnitude. This precise quantification is advantageous for capturing the wide range of antigen concentrations in COVID-19 patient plasma throughout the course of hospitalization. We combined our Simoa SARS-CoV-2 antigen assays with previously developed Simoa serological assays to monitor SARS-CoV-2 antigens and anti-SARS-CoV-2 immunoglobulins in longitudinal plasma samples of COVID-19 patients. These measurements provide direct evidence of the inverse correlation between anti-SARS-CoV-2 antibody production and viral-antigen clearance from plasma, which provides a unique view of viral infection and immune response from the beginning of hospitalization through recovery or death.
Methods
Plasma Samples
All plasma was collected in purple-top K2-EDTA tubes and centrifuged at 2000 × g at 4 °C for 10 min prior to analysis. COVID-19 positive and negative samples were obtained from adult patients presenting to Brigham and Women’s Hospital or Massachusetts General Hospital. We received 17 samples from patients who tested negative for SARS-CoV-2 using NP RT-PCR. We received 64 samples from patients who tested positive for SARS-CoV-2 using NP RT-PCR. We received 34 prepandemic samples, defined by a collection date before October 1, 2019, from the Mass General Brigham Biobank. Of the prepandemic samples, 20 came from healthy patients and 14 prepandemic samples from sick patients with upper respiratory infections, bacterial pneumonia, viral pneumonia, or unspecified virus positive.
Serial timepoint clinical samples were obtained from patients admitted to Massachusetts General Hospital (n = 67 samples from 19 individual patients) and Brigham and Women’s Hospital (n = 205 samples from 24 patients) diagnosed as SARS-CoV-2 positive by a NP RT-PCR. On average, 7 serial samples per patient were taken 0–30 days after the first SARS-CoV-2 positive NP RT-PCR. All plasma samples were collected under approval of the Mass General Brigham Institutional Review Board for Human Subjects Research.
Simoa Assays
Antigen Assays
Preparation of viral-antigen assay Simoa reagents is in the online Data Supplement. Simoa assays were performed on an HD-X Analyzer (Quanterix) in an automated three-step assay format according to the manufacturer’s instructions and as previously described (8). Plasma samples were diluted 8-fold in Homebrew Detector/Sample Diluent (Quanterix) with Halt Protease Inhibitor Cocktail (ThermoFischer Scientific) and EDTA. Detector antibodies were diluted in Homebrew Detector/Sample Diluent to 0.3 µg/mL, and streptavidin-β-galactosidase (SβG) concentrate (Quanterix) was diluted to 150 pmol/L in SβG Diluent (Quanterix). Antibody-conjugated capture beads were diluted in Bead Diluent, with a total of 500 000 beads per reaction (125 000 S1 beads, 125 000 S2 beads, and 250 000 647-nm dye-encoded helper beads for the S1/S2 multiplex assay, and 125 000 nucleocapsid beads and 375 000 647-nm dye-encoded helper beads for the nucleocapsid assay). All reagents were diluted in plastic bottles that were loaded into the HD-X Analyzer, and all assay steps were performed in an automated manner on the instrument. In each assay, capture beads were incubated with the sample for 15 minutes, detector antibody for 5 minutes, and SβG for 5 minutes, with washing steps in between. The beads were then resuspended in 25 µL of resorufin-β-galactopyranoside and loaded into the microwell array for imaging. Average enzyme per bead and sample concentration values were calculated using the HD-X Analyzer software. All samples were measured in duplicates. Immunoglobulin assays were performed using a similar procedure (online Data Supplement).
Data Analysis
Seroconversion thresholding
Seroconversion classification was determined based upon the early-stage classification model (9) trained using an independent panel of 142 samples positive by RT-PCR SARS-Cov-2 and 200 negative prepandemic controls. The markers for this model were chosen using a fivefold crossvalidation step as previously reported (9). Two cross validations were run: (a) Early-stage cases and prepandemic controls; (b) late-stage cases and all controls. Each training set initially considered all 12 markers (IgG, IgM, and IgA each against S1, Spike, N, and receptor binding domain (RBD)). This crossvalidation yielded 4 markers (IgA S1, IgA Nucleocapsid, IgG Nucleocapsid, and IgG Spike) and exhibited the best performance in the training set. The threshold for a positive test result for the unknown samples was determined based on the cutoff that yielded 100% specificity in the training set.
Cluster Map Analysis
The cluster map analysis was performed on standardized biomarker data. An example (Supplemental Fig. 1) and detailed description of the standardization is provided in the online Data Supplement. Samples were removed if they had any missing values (reduced the number of samples from 272 to 252). Specifically, the cluster map is a hierarchical clustering based on the Ward variance minimization algorithm (10).
Results and Discussion
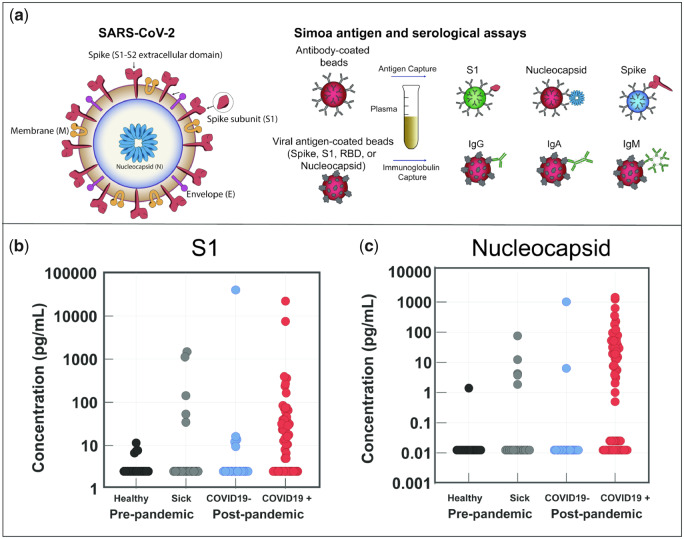
For SARS-CoV-2 antigen Simoa assays, 3 types of dye-encoded paramagnetic bead were functionalized with antibodies against each viral antigen and incubated with plasma samples for multiplexed Simoa measurements (Fig. 1, A, Methods) as described previously (8). The SARS-CoV-2 Simoa antigen assays detect S1, spike, and N antigens with limits of detection (LOD) of 5 pg/mL (0.07 pmol/L), 70 pg/mL (0.39 pmol/L), and 0.02 pg/mL (0.4 fmol/L), respectively (Supplemental Table 2). The range in LODs for each viral-antigen assay is primarily due to differences in affinities of the capture and detection antibodies, as antibody dissociation rate constants are an important factor for Simoa assay sensitivity (11). All SARS-CoV-2 antigen assays were validated in commercial human plasma by spike-and-recovery and dilution-linearity experiments prior to analysis of clinical plasma samples (Supplemental Figs. 2 and 3, Table 3). We measured anti-SARS-CoV-2 immunoglobulins (total IgA, IgM, and IgG) using our recently established SARS-CoV-2 serological assays (8) to correlate viral antigens with immunoglobulin levels (Fig. 1, A). In addition, we developed SARS-CoV-2 serological Simoa assays for IgG1, IgG2, IgG3, and IgG4 detection (online Data Supplement). The combined measurements of 3 SARS-CoV-2 antigens and 7 anti-SARS-CoV-2 immunoglobulin isotypes against 4 SARS-CoV-2 antigens enables quantification of 31 biomarkers from 70 µL of a plasma sample.
Fig. 1.
SARS-CoV-2 antigen and anti-SARS-CoV-2 immunoglobulin detection in plasma. a) Schematic of Simoa detection of SARS-CoV-2 S1, spike, and N antigens and anti-SARS-CoV-2 immunoglobulins IgG, IgA, and IgM. Measurements for all antigens and immunoglobulins can be obtained from a single plasma sample (70 µL). b, c) Simoa SARS-CoV-2 antigen assay results for plasma samples collected from prepandemic healthy patients, prepandemic sick patients, and patients who tested COVID-19 negative and COVID-19 positive. Of the 64 patients, 41 who tested COVID-19 positive show detectable S1 (b) and N concentrations (c) in plasma. Each data point represents the average of 2 replicate measurements.
To probe for the presence of viral antigens in plasma, we tested samples from COVID-19 positive patients using our SARS-CoV-2 Simoa antigen assays for S1, spike, and N. These patients were determined to be COVID-19 positive by NP RT-PCR and all plasma samples were obtained within the first 10 days of the initial NP RT-PCR test. Corresponding immunoglobulin levels are presented in Supplemental Figs. 4 and 5. S1 and N were detected in 41 of 64 COVID-19 positive patients (Fig. 1, B and C), who we identify as “viral-antigen positive.” Despite the presence of S1 and N in some samples, spike was only detectable in 5 of 64 COVID-19 positive patients (Supplemental Fig. 6). Spike may be undetectable in some samples since the LOD is 1 order of magnitude higher than the LOD of the S1 assay. Additionally, in the Simoa assay for spike, the formation of a full immunocomplex depends on spike binding to the S2 subunit capture beads and the S1 subunit detection antibody. Therefore, we hypothesize that free spike antigen in plasma is likely proteolytically cleaved, releasing the S1 subunit, and the remaining spike protein fragment is undetectable by our assay.
Cross-reactivity of the SARS-CoV-2 antigen assays was assessed using plasma samples from 3 control patient cohorts (a): samples from individuals who tested negative for COVID-19 by NP RT-PCR (b), prepandemic samples from healthy individuals, and (c) prepandemic samples collected from sick individuals (Methods). Viral-antigen concentrations are plotted for each individual patient in Supplemental Fig. 7. We observed cross-reactivity that limited the SARS-CoV-2 antigen assays for diagnostic applications and highlighted their use as a complementary technique to RT-PCR diagnosis (Fig. 1, B and C). However, based on our preliminary results, a multiplexed approach for the detection of S1 and N can improve analytical specificity of these assays. A more detailed discussion is provided in the online Data Supplement. We attribute detection of viral antigens in COVID-19 negative patients to either (a) assay cross-reactivity with other coronaviruses or (b) the patients with a negative NP RT-PCR test were COVID-19 positive but received a false negative result (12). We focused on the utility of the SARS-CoV-2 Simoa antigen assays for monitoring disease progression in COVID-19 patients diagnosed by NP RT-PCR. When combined with the diagnostic accuracy of RT-PCR, the ultra-sensitivity of Simoa enabled high-resolution quantification of changes in viral-antigen concentrations over the course of disease in COVID-19 patients. Future studies aim to further optimize capture and detection antibodies, assay conditions, and add additional SARS-CoV-2 antigens to our marker panel to improve analytical specificity for diagnostic applications.
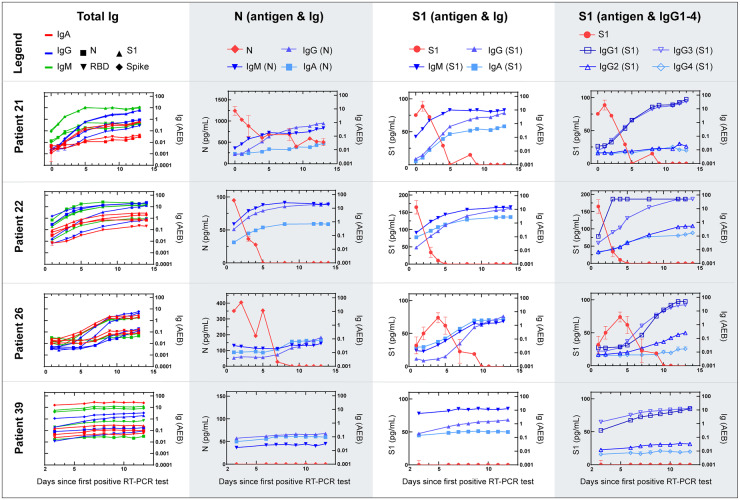
To understand how antigen and immunoglobulin levels change with time after infection, we performed longitudinal studies in COVID-19 positive patients to monitor viral antigen and immunoglobulin levels during the course of hospitalization. We measured viral-antigen concentrations (S1, spike, and N) and immunoglobulin levels (total IgA, IgM, and IgG and IgG1–4) in serial plasma samples from 39 admitted patients at Brigham and Women’s Hospital and Massachusetts General Hospital (patient IDs 1–39, Supplemental Fig. 8). All patients were diagnosed as COVID-19 positive by NP RT-PCR. Longitudinal antigen and immunoglobulin levels for 4 representative patients are plotted in Fig. 2; plots for the remaining 35 patients are shown in the online Data Supplement. In this patient cohort, 25 of 39 patients had detectable concentrations of both S1 and N antigen in their plasma, whereas spike was detectable in only 6 of 39 patients during their hospitalization.
Fig. 2.
Serial data for 4 individual COVID-19 positive patients from admission to clinical recovery or death. Simoa antigen and serological results for serial plasma samples. The total IgA, IgM, and IgG levels against 4 viral antigens: nucleocapsid (N), receptor binding domain (RBD), S1, and spike, for a total of 12 interactions , N antigen concentrations with IgA, IgM, and IgG against N, S1 antigen concentrations with IgA, IgM, and IgG against S1, and S1 antigen concentrations with IgG1-4 against S1. Data points represent the mean concentration or AEB (average enzymes per bead) from 2 replicate measurements and error bars represent the standard error of the mean from 2 replicate measurements.
The ultra-sensitivity of Simoa assays provides quantitative resolution of viral-antigen concentrations and enables us to measure even the earliest stages of antibody production for a detailed view of patient responses over time. For example, in Fig. 2, Patient 21 originally had high concentrations of N (1240 pg/mL) and S1 (75.3 pg/mL) and low concentrations of anti-SARS-CoV-2 immunoglobulins in their plasma on day 0. As patient 21 mounted an antibody response against the virus, indicated by the increase in immunoglobulin levels over time, we observe a decrease in viral-antigen concentrations in plasma. This trend is observed for a majority of viral-antigen positive patients (patients 21, 22, and 26 in Fig. 2 and patients in Supplemental Figs. 10, 12–14, 19–20, S23, 26, 31, 34, 36). As shown in Fig. 2, among patients 21, 22, and 26, once a patient has seroconverted and their plasma immunoglobulin levels reach a steady state, there are typically no detectable concentrations of viral antigen. All patients seroconverted a mean ± 95% CI of 7 ± 1 days after the first NP RT-PCR positive test, in agreement with previous serological studies (5, 13–15). We defined viral-antigen clearance as the first day that both S1 and N were undetectable in a patient’s plasma. For viral-antigen positive patients, full antigen clearance in plasma was observed 5 ± 1 days after seroconversion.
Furthermore, 13 of 15 patients with undetectable viral-antigen concentrations in plasma were already seroconverted at their first NP RT-PCR test (patient 39 in Fig. 2 and Supplemental Figs. 11, 15–17, 18, 24, 27–28, 30–33, 35, 7–44). We propose 3 possibilities for the lack of detectable antigen in some patient plasma: (a) patients are presenting to the hospital after seroconversion, and therefore most viral antigens have been cleared from the plasma, (b) we hypothesize that COVID-19 cases with very severe disease will have viral-antigen leakage into the blood, and therefore patients that do not progress to more severe forms of the disease will not have viral-antigen leakage into the blood, or (c) viral antigens are present but are bound to an immunoglobulin, which blocks a binding epitope of the capture or detection antibody, resulting in an antigen–immunoglobulin complex that is undetectable by our Simoa assays. However, even with no detectable concentrations of viral antigen, 12 of 16 patients were admitted or transferred to the ICU during hospitalization and intubated, indicating that these patients were severe cases. Therefore, we propose a fourth possibility where patients have seroconverted and, despite reaching viral clearance, suffer from severe respiratory damage that inhibits recovery.
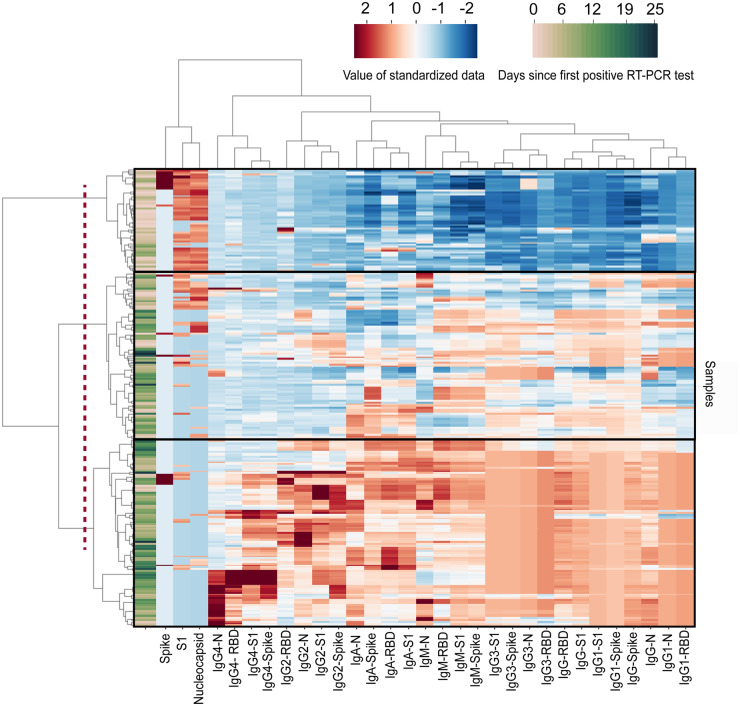
Next, we performed a cluster map analysis to determine if antigen and immunoglobulin levels in COVID-19 positive patients could differentiate different stages of infection across all patients. The cluster map shows 3 dominant branches (Fig. 3). The first branch included a majority of early time points where we observed high concentrations of viral antigens and low levels of antibodies. The second branch showed a transition in patient samples where antigen concentrations began to decrease as total IgM, IgA, and IgG (as well as IgG subclasses IgG1 and IgG3) began to increase. In the third branch, we observed a further reduction in antigen concentrations with small increases in IgG subclasses IgG2 and IgG4. The third branch was observed for later time points where we propose that patients had cleared viral antigens from the plasma and antibodies had reached their steady-state levels. For all 39 patients in this cohort, we observed that IgG1 and IgG3 showed large increases in plasma concentrations over time. In contrast, IgG2 and IgG4 showed little to no changes above background levels over time. Both IgG1 and IgG3 are subclasses of IgG that predominantly respond to viral antigens and mediate neutralization (16–20). Based on these measurements, the IgG response that was mounted by the immune system was primarily mediated by IgG1 and IgG3 for SARS-CoV-2, in agreement with previous serological studies (4, 5). The neutralization effects of IgG1 and IgG3 on viral antigens should be explored in future studies that include correlative neutralization titer assays.
Fig. 3.
Clustergram of Simoa antigen and immunoglobulin results from 252 samples from 39 patients. Cluster map produced by Ward variance minimization algorithm. 252 samples from 39 patients were analyzed as described in the Methods section. Data were standardized after a nonlinear transformation and each row represents a single sample. Days since first RT-PCR test for each sample are presented at the far left of the clustergram. Longitudinal samples are clustered by dendrograms to the left of the clustergram. SARS-CoV-2 antigen and immunoglobulin markers are clustered by dendrograms at the top of the clustergram. The dotted line represents the cutoff that results in 3 branches: branch 1 (top), branch 2 (middle), and branch 3 (bottom). RBD: Receptor binding domain. N: Nucleocapsid.
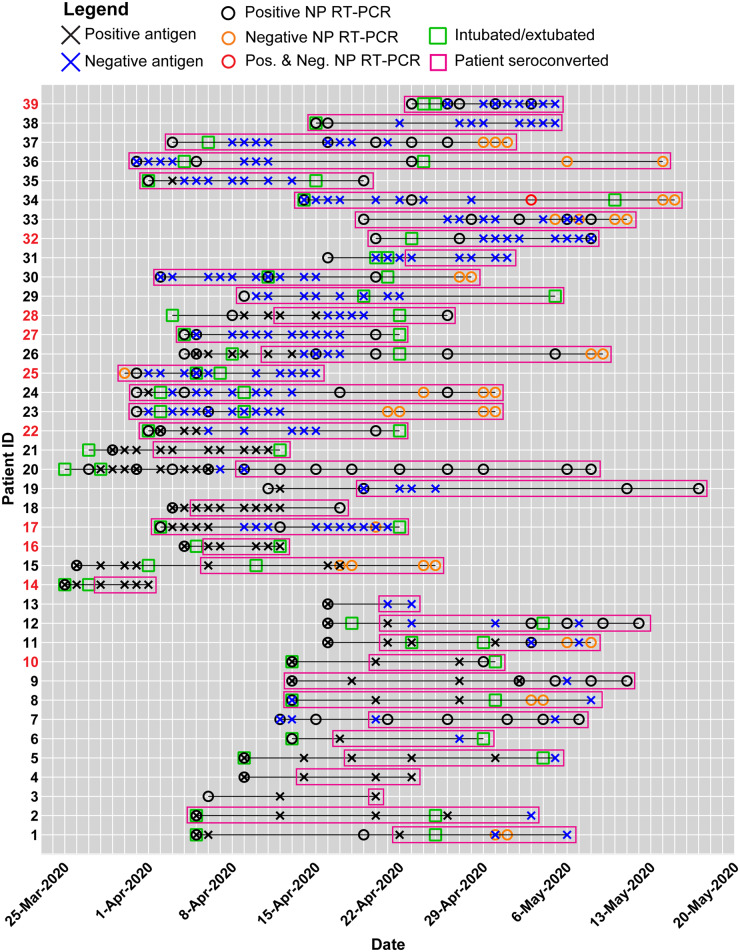
We next explored how antigen clearance compared with longitudinal RT-PCR tests and clinical outcomes. The measured SARS-CoV-2 antigen and immunoglobulin levels in plasma, NP RT-PCR tests, and select clinical outcomes for each of these patients are displayed in Fig. 4, and patient features such as age and gender are summarized in Supplemental Table 4. In this patient cohort, NP RT-PCR tests reported positive results for a mean of 15 ± 5 days after viral-antigen clearance and 18 ± 4 days after seroconversion. Furthermore, several patients never received a negative RT-PCR result before being discharged from the hospital. These observations are in agreement with recent virological studies that confirmed persistence of viral RNA and showed how seroconversion was not immediately followed by a decrease in NP viral RNA in hospitalized patients (21). Because RNA shedding can occur for several weeks after infection and recovery, measurements of viral antigens and immunoglobulins may provide more timely indicators of infectivity compared to viral RNA, but additional studies such as viral load measurements are necessary to corroborate these findings.
Fig. 4.
Summary of clinical data and Simoa SARS-CoV-2 antigen and immunoglobulin assays for individual patients in the longitudinal analysis. A black X indicates a viral-antigen positive test, whereas a blue X indicates a viral-antigen negative test. A black circle indicates a positive NP RT-PCR test, whereas an orange circle indicates a negative NP RT-PCR test. For example, on May 3, 2020, Patient 34 received both a positive and negative NP RT-PCR result, indicated by the red circle. The intubation and extubation dates of each patient are indicated by green squares. The survival outcome for each patient is marked by the patient ID number color. Patient IDs are coded black for recovered patients who were discharged from the hospital, whereas patient IDs are coded red for deceased patients. Pink boxes represent the date of seroconversion for each patient.
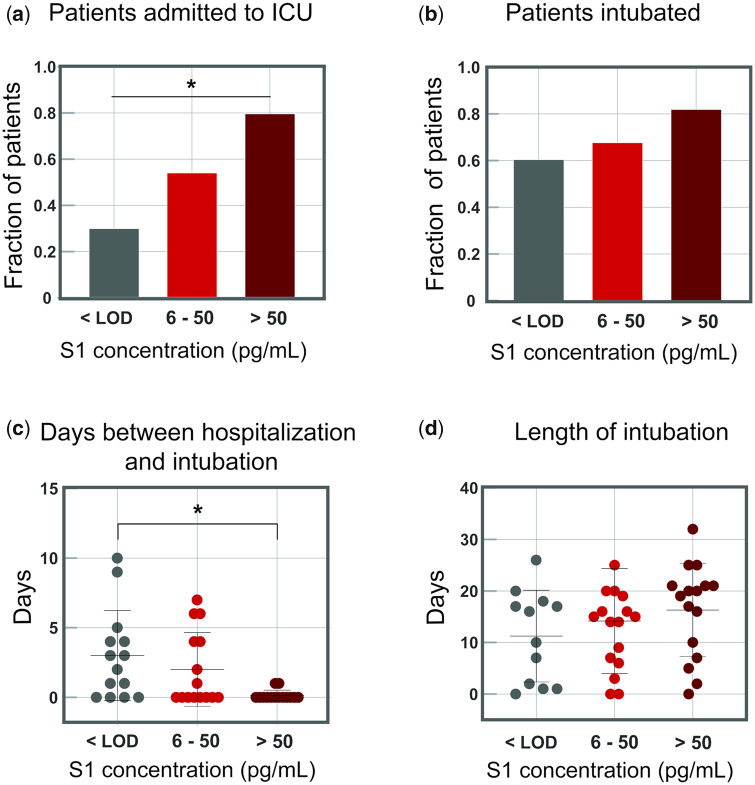
To understand the correlation between SARS-CoV-2 antigen concentrations and disease severity, clinical features for the 64 COVID-19 positive patients (Fig. 5, patient IDs 1–64) were compared with viral-antigen concentrations in plasma. It is important to note that all samples were obtained from patients admitted to the hospital, resulting in a cohort of cases primarily with severe disease. S1 shows higher correlation with clinical severity than N in plasma (Supplemental Tables 5–8, Supplemental Fig. 45). Confounding factors such as age and sex were also assessed and showed no correlation with clinical outcomes (Supplemental Tables 9, 10, Supplemental Fig. 46). Therefore, patients were grouped into 3 categories of S1 concentration: (a) 23 patients with undetectable S1 concentrations (below the limit of detection), (b) 23 patients with low concentrations of S1 (6–50 pg/mL, 0.08–0.65 pmol/L), and (c) 18 patients with high concentrations of S1 (>50 pg/mL, > 0.65 pmol/L). There is a significant difference in rates of ICU admission upon presentation for the 3 patient groups based on S1 concentrations in plasma (P = 0.0107). Patients with zero, low, and high concentrations of S1 were admitted to the ICU upon presentation to the hospital at rates of 30% (7 of 23 patients), 52% (12 of 23 patients), and 77% (14 of 18 patients), respectively (Fig. 5a). Among all COVID-19 positive patients, over 60% were intubated during hospitalization with no statistically significant difference in intubation rates among groups (Fig. 5b). The difference in mean times to intubation between patients with high concentrations of S1 and patients with no detectable S1 is significant (P = 0.0050), where all patients with high concentrations of S1 were intubated within 1 day of hospitalization (Fig. 5c). These results suggest that high S1 concentrations in plasma upon presentation to the hospital correlate with severe cases of COVID-19 that can result in respiratory failure and require immediate intubation. Among all patients, a broad range of intubation periods, up to 32 days, was observed (Fig. 5d). Patients grouped by S1 concentrations, age, and sex showed no statistically significant difference in death rate (Supplemental Tables S9, S10, Supplemental Fig. 46). Although 6 patients showed detectable concentrations of spike protein, there was no correlation between spike concentrations and ICU admission, intubation rate, or death rate. There are potentially more features in these data that will lead to further correlations, but a large cohort of patients that include asymptomatic and mild cases will need to be tested to elucidate other patterns.
Fig. 5.
Indicators of disease severity based on S1 concentrations in plasma for 64 COVID-19 positive patients. COVID-19 positive patients were separated into 3 groups based on S1 concentrations. The cutoff between groups 2 and 3 (50 pg/mL, 0.65 pmol/L) was chosen as 5 standard deviations above the LOD. The fraction of patients admitted to the ICU or who were intubated was calculated for each group independently. a) Fraction of COVID-19 positive patients who were immediately admitted to the ICU upon presentation to the hospital. b) Fraction of COVID-19 positive patients who were intubated during hospitalization. c) Days between date of presentation to the hospital and intubation date for intubated COVID-19 positive patients. d) The length of intubation for intubated COVID-19 positive patients. For all plots, significance indicated by the asterisks (P < 0.05).
Plasma is a readily available biofluid from hospitalized patients but is more difficult to obtain in nonclinical settings. In comparison, saliva is a noninvasive biofluid that is easier to use for wide-scale testing and may be more interesting for monitoring viral-antigen concentrations to understand active viral infection from COVID-19, which is a respiratory disease. We tested 17 saliva samples from patients presenting to the Emergency Department at Brigham and Women’s Hospital who were tested by NP RT-PCR for COVID-19 (Supplemental Figs. 47–49). We found that S1 and N was detectable in 7 of 11 COVID-19 patients when compared to healthy saliva controls. Similar to our observation of antigen and immunoglobulins in plasma, there were 7 of 11 patients with low S1 concentrations and high levels of IgA-S1. One patient showed a background concentration of IgA-S1 and notably high concentration of S1 (135 pg/mL). However, a deep understanding of the correlation between viral antigens and immunoglobulins in saliva will require a larger sample cohort. Nonetheless, these initial results indicate the presence of SARS-CoV-2 antigens and anti-SARS-CoV-2 immunoglobulins in saliva and highlight the potential for adapting our assays to a diagnostic test for COVID-19. Future studies on saliva will include longitudinal sample analysis for mild and severe cases of COVID-19 patients and will explore the potential for developing a saliva-based COVID-19 antigen screening tool.
Conclusion
Using SARS-CoV-2 Simoa assays, we have demonstrated quantitative detection of SARS-CoV-2 antigens and anti-SARS-CoV-2 immunoglobulins in plasma of COVID-19 patients. While detection of N in NP swabs has been cited (6), we present the first report of SARS-CoV-2 spike, S1, and N detection in plasma. The presence of S1 and N in plasma suggests that fragments of virus are entering the bloodstream, potentially due to tissue damage. Although spike is undetectable in most COVID-19 patients, possibly due to proteolytic cleavage, 6 patients showed high concentrations of spike in plasma. No evidence has been reported yet for full viral particles in blood, though we cannot rule out this possibility (22). Nonetheless, severe COVID-19 cases with acute respiratory distress syndrome can result in damage to endothelial cells and vascular leakage (23–26) and we propose that this damage can lead to discharge of viral antigens into the blood. Patients with lung damage can suffer from respiratory failure and require intubation or mechanical ventilation. We also found significant correlation between high S1 concentrations in plasma and time between hospital admission and intubation. Although we hypothesize that asymptomatic or mild cases will likely not show viral-antigen concentrations in plasma, future studies that include mild cases will be used to probe SARS-CoV-2 antigen and antibody concentrations over time in comparison to the severe cases presented here.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Sanjat Kanjilal for collecting saliva samples.
Nonstandard Abbreviations:
- COVID-19
coronavirus disease
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- RT-PCR
reverse-transcription polymerase chain reaction
- NP
nasopharyngeal
- Simoa
single molecule array
- spike
S1–S2 extracellular domain
- N
nucleocapsid
- RBD
receptor binding domain
Author Declaration
A version of this paper was previously posted as a preprint on medRxiv as https://www.medrxiv.org/content/10.1101/2020.07.20.20156372v1.
The anti-SARS-CoV-2 Simoa assays in this publication have been licensed by Brigham and Women’s Hospital to Quanterix Corporation.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
A.F. Ogata, A.M. Maley, C. Wu, T. Gilboa, M. Norman, and D.R. Walt conceived the approach. A.F. Ogata, A.M. Maley, C. Wu, T. Gilboa, M. Norman, and G. Newton performed the experiments, A.F. Ogata, A.M. Maley, C. Wu, T. Gilboa, M. Norman, R. Lazarovits, C.-P. Mao, and T.E. Gibson analyzed the data, M. Chang, K. Nguyen, M. Kamkaew, Q. Zhu, and W.A. Marasco produced and purified the anti-S1 antibody, R.C. Charles and E.T. Ryan collected the samples and performed chart review for patients from Massachusetts General Hospital, C.-P. Mao performed chart review for patients from Brigham and Women’s Hospital. A.F. Ogata, A.M. Maley, and D.R. Walt co-wrote the paper. All authors were involved in designing experiments, reviewing and discussing data, and commented on the manuscript.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
D.R. Walt is an inventor of the Simoa technology, a founder of the company and also serves on its Board of Directors. D.R. Walt’s interests were reviewed and are managed by BWH and Partners HealthCare in accordance with their conflict of interest policies.
Consultant or Advisory Role
None declared.
Stock Ownership
D.R. Walt has a financial interest in Quanterix Corporation, a company that develops an ultra-sensitive digital immunoassay platform.
Honoraria
None declared.
Research Funding
Funding for this work came from a generous donation from Barbara and Amos Hostetter and the Chleck Foundation. This work was also funded through grants from the Massachusetts Consortium on Pathogen Readiness and the Centers for Disease Control and Prevention (U01CK000490). C. Wu, National Institutes of Health (1F32EB029777-01).
Expert Testimony
None declared.
Patents
A.F. Ogata, patent application in progress; A.M. Maley, patent application in progress; T. Gilboa, patent application in progress; M. Norman, patent application in progress; D.R. Walt, many patents.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
References
- 1.World Health Organization. Coronavirus disease (COVID-19) Situation Report 178. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200816-covid-19-sitrep-209.pdf? sfvrsn=5dde1ca2_2 (Accessed August 2020).
- 2. Sethuraman N, Jeremiah SS, Ryo A.. Interpreting diagnostic tests for SARS-CoV-2. JAMA 2020;323:2249–51. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez L, Pekkarinen PT, Lakshmikanth T, et al. Systems-Level Immunomonitoring from Acute to Recovery Phase of Severe COVID-19. Cell Rep Med. 2020;1(5):100078. doi:10.1016/j.xcrm.2020.100078 [DOI] [PMC free article] [PubMed]
- 4. Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 2020;52:971–7.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020;26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mahapatra S, Chandra P.. Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens Bioelectron 2020;165:112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veritor™ System Veritor™ System. 2020. https://www.fda.gov/media/139755/download (Accessed August 2020).
- 8. Rivnak AJ, Rissin DM, Kan CW, Song L, Fishburn MW, Piech T, et al. A fully-automated, six-plex single molecule immunoassay for measuring cytokines in blood. J Immunol Methods 2015;424:20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norman M, Gilboa T, Ogata AF, et al. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat Biomed Eng (2020). 10.1038/s41551-020-00611-x [DOI] [PMC free article] [PubMed]
- 10.The SciPy Community. Scipy-cluster-hierarchy-linkage. https://docs.scipy.org/doc/scipy/reference/generated/scipy.cluster.hierarchy.linkage.html (Accessed July 2020).
- 11. Dinh TL, Ngan KC, Shoemaker CB, Walt DR.. Using antigen-antibody binding kinetic parameters to understand single-molecule array immunoassay performance. Anal Chem 2016;88:11335–9. [DOI] [PubMed] [Google Scholar]
- 12. Steven W, Neeraj P, Aaron SK.. False negative tests for SARS-CoV-2 infection—challenges and implications. N Engl J Med 2020;383:e38. [DOI] [PubMed] [Google Scholar]
- 13. Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26:845–8. [DOI] [PubMed] [Google Scholar]
- 14. Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick L, Rattigan SM, et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. Preprint at https://www.medrxiv.org/content/10.1101/2020.04.14.20065771v1 (2020). [DOI] [PMC free article] [PubMed]
- 15.Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020;584:437--42. [DOI] [PMC free article] [PubMed]
- 16. Vidarsson G, Dekkers G, Rispens T.. IgG subclasses and allotypes: from structure to effector functions. Front Immunol 2014;5:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daley LP, Kutzler MA, Bennett BW, Smith MC, Glaser AL, Appleton JA.. Effector functions of camelid heavy-chain antibodies in immunity to West Nile virus. Clin Vaccine Immunol 2010;17:239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koraka P, Suharti C, Setiati TE, Mairuhu ATA, Van Gorp E, Hach CE, et al. Kinetics of dengue virus-specific serum immunoglobulin classes and subclasses correlate with clinical outcome of infection. J Clin Microbiol 2001;39:4332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scharf O, Golding H, King LR, Eller N, Frazier D, Golding B, et al. Immunoglobulin G3 from polyclonal Human Immunodeficiency Virus (HIV) immune globulin is more potent than other subclasses in neutralizing HIV type 1. J Virol 2001;75:6558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spinsanti LI, Farías AA, Aguilar JJ, del Díaz MP, Contigiani MS.. Immunoglobulin G subclasses in antibody responses to St. Louis encephalitis virus infections. Arch Virol 2011;156:1861–4. [DOI] [PubMed] [Google Scholar]
- 21. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- 22. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Magro C, Mulvey JJ, Berlin D, Nuovo G, Salvatore S, Harp J, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res 2020;220:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amirfakhryan H, Safari F.. Outbreak of SARS-CoV2: pathogenesis of infection and cardiovascular involvement. Hellenic J Cardiol 2020;S1109-9666:30096–8. 10.1016/j.hjc.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016;19:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teuwen LA, Geldhof V, Pasut A, Carmeliet P.. COVID-19: the vasculature unleashed. Nat Rev Immunol 2020;20:448–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.