ABSTRACT
Influenza virus and coronaviruses continue to cause pandemics across the globe. We now have a greater understanding of their functions. Unfortunately, the number of drugs in our armory to defend us against them is inadequate. This may require us to think about what mechanisms to address. Here, we review the biological properties of these viruses, their genetic evolution and antiviral therapies that can be used or have been attempted. We will describe several classes of drugs such as serine protease inhibitors, heparin, heparan sulfate receptor inhibitors, chelating agents, immunomodulators and many others. We also briefly describe some of the drug repurposing efforts that have taken place in an effort to rapidly identify molecules to treat patients with COVID-19. While we put a heavy emphasis on the past and present efforts, we also provide some thoughts about what we need to do to prepare for respiratory viral threats in the future.
Keywords: RNA viruses, coronaviruses, SARS-Cov-2, influenza, chloroquine, serine protease inhibitors, heparan sulfate glycoconjugate
Influenza virus and coronaviruses continue to cause pandemics across the globe and here we discuss their common and different properties.
INTRODUCTION
With regularity we face previously unknown strains of virulent respiratory viruses that are life-threatening for large numbers of people. Perhaps the most well known and recent are the pandemics associated with influenza viruses and coronaviruses that have been in contact with humans for millennia.
It is generally thought that the first large outbreak of a respiratory infection with clinical symptoms similar to those of influenza was described in detail by Hippocrates in the year 412 BC as contagious cough of Perinthus (Kuszewski and Brydak 2000; Pappas, Kiriaze and Falagas 2008). Next, a detailed written report of an epidemic respiratory disease similar to influenza was noted in England and named peasant fever and lasted from 1173 to 1174 (Potter 2001). The first pandemic of influenza was clearly documented in 1580 (Potter 2001; Daly, Gustafson and Kendall 2007). In the 16th century, this infection was named influenza (from the Latin influentia, influence), as this disease was considered a bad influence from the heavens (Broxmeyer 2006). Since that time, no less than 31 pandemics of influenza have been documented, including 3 in the 20th century and 1 in the 21st century (Kilbourne 2006; Daly, Gustafson and Kendall 2007; Al-Muharrmi 2010) (Table 1).
Table 1.
Influenza pandemics in the last 100 years.
| Name of the pandemic | Years | Strain | Number of deaths (millions) |
|---|---|---|---|
| Spanish flu | 1918–1920 | H1N1 | 40–50 |
| Asian flu | 1957–1958 | H2N2 | 1–2 |
| Hong Kong flu | 1968–1970 | H3N2 | 0.5–2 |
| Swine flu | 2009–2010 | H1N1 | 0.5 |
Although a targeted search for pathogens able to produce an epidemic/pandemic of acute respiratory infections started in the late 19th century (Pfeiffer 1893; Olitsky and Gates 1921a,b), it was not until 1933 that the influenza A virus was selected (myxovirus influenza) (Smith, Andrewes and Laidlaw 1933). Influenza B and C viruses were identified in 1940 and 1947, respectively (Francis 1940; Taylor 1949), and the influenza D virus was isolated and characterized recently in 2011 (Hause et al. 2013; Ducatez, Pelletier and Meyer 2015).
Coronaviruses are also very common (Suzuki et al. 2005; Koetz et al. 2006; Sloots et al. 2006; Zhao et al. 2008) and seem to have been in contact with humans from the earliest of times (Wertheim et al. 2013). Until recently, it was thought that coronavirus infections with symptoms of the common cold cause between 15% and 35% seasonal acute respiratory diseases. Children become infected at a rate of five to seven times more often than adults (McIntosh et al. 1970; Callow et al. 1990; Holmes 2001). In humans, respiratory infections can be caused by two species of α-coronaviruses (229E and NL63) and two species of β-coronaviruses (OC43 and HKU1) (Gaunt et al. 2010). In addition, veterinary specialists have known for a long time that coronaviruses cause fatal respiratory and gastrointestinal infections in animals (Pensaert 1999). Coronaviruses were only recently acknowledged as a potential biological hazard as they are a challenge for medicine. In recent decades, new pandemic strains of coronaviruses have often appeared, which are frequently fatal for humans. These include severe acute respiratory syndrome-related coronavirus (SARS-CoV, which occurred from 2002 to 2004), Middle East respiratory syndrome-related coronavirus (MERS-CoV, which was identified in 2012) and most recently the new pneumonia coronavirus (SARS-CoV-2, which is the ongoing outbreak that was identified in 2019) (Table 2). In all cases, these three viruses cause severe bronchiolitis and pneumonia, often with fatal outcomes (Cherry 2004; Ramadan and Shaib 2019; Hui et al. 2020).
Table 2.
Coronavirus epidemics and pandemics in recent years.
| Name of the epidemic/pandemic | Years | Strain | Number of deaths (hundreds) |
|---|---|---|---|
| 2002–2004 SARS outbreak | 2002–2004 | SARS-CoV-1 | 774 |
| 2012 Middle East respiratory syndrome coronavirus outbreak | 2012–present | MERS-CoV | 862 |
| (as of 13 January 2020, WHO) | |||
| COVID-19 pandemic | 2019–present | SARS-CoV-2 | 280 431 |
| (as of 9 May 2020, WHO) |
Human coronaviruses were for the first time isolated from a patient with acute respiratory diseases in 1965 (Hamre and Procknow 1966; Tyrrell and Bynoe 1966). Their characteristic corona seen under the electronic microscope was reflected in the name coronaviruses (Tyrrell et al. 1975). During the next three decades (until the pandemic strains appeared), the coronaviruses were not of any special interest for most scientists.
It is apparent that pandemic outbreaks of respiratory viral infections represented a danger for humanity in the past, and there are no reasons to believe that they would not repeat in the future. It is as yet impossible to predict the time and place of the start of a new pandemic as well as the virulence of pandemic viral strains. However, there are certain factors that increase the potential for these viruses to spill over from other species (Bobrowski et al. 2020; Gomes and Ruiz 2020; Johnson et al. 2020).
BIOLOGICAL PROPERTIES OF INFLUENZA VIRUSES AND CORONAVIRUSES
Influenza viruses belong to the orthomyxoviruses family (Orhtomyxoviridae, RNA viruses with segmented genome) and are represented by four monotypic genera: influenza A viruses (Alpha influenzavirus), influenza B viruses (Beta influenzavirus), influenza C viruses (Gamma influenzavirus) and influenza D viruses (Delta influenzavirus); each genus contains only one type of eponymous virus. It is understood that only type A viruses have pandemic potential (Bouvier and Palese 2008; Spickler 2016; King et al. 2018). Influenza A viruses are further classified into subtypes, depending on the antigenic properties of hemagglutinin (HA; a glycoprotein of the viral envelope that ensures the recognition of target cells and binding of viral particles to the terminal residues of sialic acids of the glycoproteins of plasma membranes of epithelial cells) and neuraminidase (NA; exo-α-sialidase catalyzing the splitting of glycoside bonds of the terminal residues of sialic acids of oligosaccharides, glycoproteins and glycolipids, thus providing release of newly formed influenza virions from the infected cells).
There are 18 known types of hemagglutinin (H1–H18) and 11 identified serotypes of neuraminidase (N1–N11). Therefore, in theory, 198 diverse combinations of these proteins (and thus subtypes of the influenza A virus) are possible (Skehel 2009; Tong et al. 2013; Quan et al. 2016; Kosik and Yewdell 2019; Zhao et al. 2019); of them, >120 combinations have been identified in nature (Tsai and Chen, 2011; Rejmanek et al. 2015).
There are eight negative polar segments of RNA genome of the influenza virus that code at least 10 structural and 9 regulatory proteins (Varga et al. 2011; Muramoto et al. 2013; Hutchinson et al. 2014; Vasin et al. 2014). Some uncertainty regarding the proteome of the influenza A viruses is related to the fact that, unlike most RNA viruses, the transcription and translation of the genome of these viruses take place in the nucleus and not in the cytoplasm of infected cells. This permits influenza A viruses (Fig. 1A) to use the cellular splicing machinery to form splice variants of viral mRNA(messenger RiboNucleuc Acid). In addition, to widen their proteome, the influenza A viruses are probably using alternative open reading frames.
Figure 1.
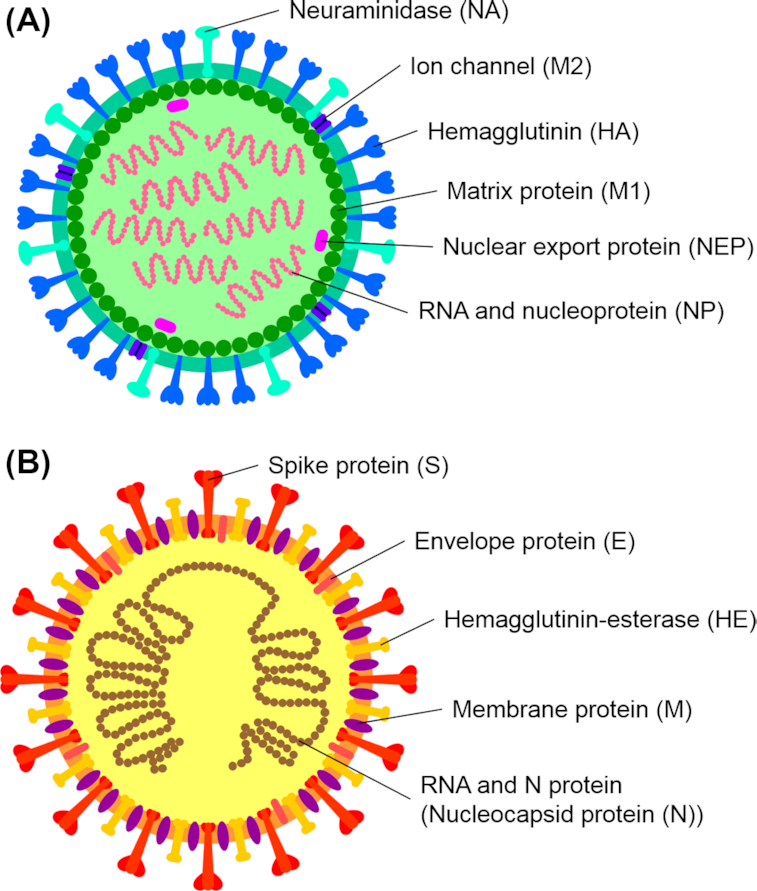
(A) Structural elements of the influenza A virus. (B) Structural elements of the coronavirus (based on betacoronavirus subgroup A).
Most viral proteins are located inside the lipid envelope, while only HA, NA (in the molar relation about 10:1; Mitnaul et al. 2000) and M2 proteins, which are built into the virion envelope and present antigenic determinants, are available for immune antibodies (Kosik and Yewdell 2019). HA and NA molecules are highly glycosylated proteins, which give them functional activity and provide for immune evasion by shielding antigenic determinants (Kim et al. 2018; York, Stevens and Alymova 2019).
Unlike influenza viruses, coronaviruses are enveloped RNA viruses (with non-segmented positive polar RNA) of the Nidovirales order, Coronaviridae family and Orthocoronavirinae subfamily (Fehr and Perlman 2015). Coronaviral virions have a spherical shape with the typical bulbous projections (Neuman et al. 2006; Bárcena et al. 2009). The viral envelope is made of a bilipid layer where S, M and E proteins are fixed (Lai and Cavanagh 1997; de Haan and Rottier 2005) (Fig. 1B).
The S protein functions in the form of highly glycosylated 3D complexes (Zheng et al. 2018; Parsons et al. 2019) provide the interaction of the virion with the receptors of epithelial cells followed by the internalization of the viral genome (Li 2016). The S protein also is known as the spike protein of SARS-CoV-2 which crystal structure described (Wang et al. 2020b).
The M protein functions in the form of a dimer with a glycolyzed N-terminal ectodomain (Nal et al. 2005) and can be present in two different conformations. The conformers of this glycoprotein ensure the correct assembly and formation of a viral particle (Neuman et al. 2011).
The E protein is a transmembrane protein that is present in low quantities and has several functions, namely, in virion assembly, envelope forming and release of a viral particle from the cell. There is indirect evidence that it has the structure of a glycoprotein (Schoeman and Fielding 2019).
The N protein is the only protein present inside the virion; it is responsible for the viral genome packaging (McBride, van Zyl and Fielding 2014).
The fact that deserves particular attention is that the proteins of the envelopes of both influenza A viruses and coronaviruses are made up of glycoproteins.
An influenza virus enters a cell during a process that involves several steps. A critically important moment in the lifecycle of an influenza virus is the recognition of the specific cellular receptors that are glycoproteins or glycolipids containing a terminal α2,6- or α2,3-sialic acid in the glycan (Leung et al. 2012; Byrd-Leotis, Cummings and Steinhauer 2017). When viral HA binds sialic glycoproteins or glycolipids on the plasma membrane of an epithelial cell, this results in the initiation of several mechanisms of endocytosis that quickly lead to the formation of endosomes, each of which contains a viral particle (Chardonnet and Dales 1970; Matlin et al. 1981; Kartenbeck, Stukenbrok and Helenius 1989; Rojek, Perez and Kunz 2008; Nanbo et al. 2010; Watanabe, Watanabe and Kawaoka 2010; Boulant, Stanifer and Lozach 2015).
The next step of the internalization is the release of the viral genome (RNA segments) into the cellular cytoplasm; this phase depends on the activity of Na+/K+-ATPase located in the endosomal membrane, which functions as a proton pump. Na+/K+-ATPase is responsible for the acidification of the internal environment of endosomes/lysosomes (to pH 5.0) (Cain, Sipe and Murphy 1989). The acidification of the internal endosomal medium, i.e. the accumulation of protons (Н+) inside the endosomes, helps the tetramers of the M2 protein of the viral envelop to realize its potential as a protonophore (Sugrue and Hay 1991; Pinto, Holsinger and Lamb 1992; Manzoor, Igarashi and Takada 2017). When hydrogen ions enter a viral particle, it mediates conformational changes and decomposition of the structural components of the viral envelope, which finally leads to an increase in the lability of its genome (Shibata et al. 1983; Yoshimura and Ohnishi 1984). However, the fusion of the viral envelope membrane and the endosomal membrane, which releases the RNA genome of the virus into the cellular cytoplasm, is possible only with the participation of the viral HA after the previous proteolytic processing with serine (secretory trypsin-like) proteases (Klink et al. 1975; Lazarowitz and Choppin 1975; Tashiro et al. 1987; Steinhauer 1999; Kido et al. 2008).
The translocation of RNA segments of the influenza viral genome from the cytoplasm to the nucleus is necessary for their replication, during which viral mRNA exits the nucleus to synthetize viral proteins in the cytoplasm. The viral self-assembly takes place at the apical surface of the plasma membrane of epithelial cells, where HA and NA molecules are concentrated (Samji 2009; Dou et al. 2018).
The process of internalization of coronaviruses is determined by the functional activity of the S protein (widely known as the spike protein) of the viral envelope. The S protein of a coronavirus is a highly glycosylated supramolecular structure that enables the fixation of viral particles on the plasma membrane of epithelial cells, followed by the release of their RNA into the cellular cytoplasm (Li 2016; Watanabe et al. 2020). Each S protein has two receptor-binding domains located on its S1-subunit; these domains interact with either specific proteins or sialoglycans of the epithelial cells (Li 2012; Shahwan et al. 2013; Hulswit et al. 2019). For example, MERS-CoV preferentially binds the α2,3-bonded sialic acid (and to a lesser degree the α2,6-bonded sialic acid) (Li et al. 2017). It seems that SARS-CoV-2 has the same affinity for the α2,3-sialic acid conjugates (Ou et al. 2020).
After that, the internalization of the viral genome may proceed by endocytosis of the virion (which is in many respects a similar process to the internalization of the influenza viruses) or by the fusion of the membrane of a coronaviral envelope with the plasma membrane of an epithelial cell, without the formation of endosomes (directly on the plasma membrane). In any case, the release of the viral RNA into the cellular cytoplasm is preceded by the proteolytic (provided by serine proteases) cleavage of S1-subunit and modulation of the S2-subunit of the S protein (Bosch et al. 2003; Belouzard, Chu and Whittaker 2009; Simmons et al. 2013; Heurich et al. 2014; Zumla et al. 2016).
In the cytoplasm of an epithelial cell, the viral RNA genome functions as mRNA, where the complex of replication and transcription is responsible for both RNA genome replication and synthesis of mRNA of structural viral proteins (Sola et al. 2015; Nakagawa, Lokugamage and Makino 2016). After the posttranslational glycosylation in the Golgi apparatus cisternae (Nal et al. 2005; Tseng et al. 2010), newly synthesized coronaviral proteins enter the cytoplasm and ensure the self-assembly of viral particles. The latter particles migrate to the cellular membrane inside the cisternae and are released from the cell by exocytosis (Fehr and Perlman 2015; Lim et al. 2016).
Taking into account the importance of serine proteases, glycoproteins and glycolipids in the lifecycle of influenza viruses and coronaviruses, it seems logical to suggest that the factors that modulate the profile of glycosylation of proteins and lipids of epithelial cells and viruses, as well as control the activity of serine proteases on the epithelial lining of respiratory ways, may significantly limit the virulence of influenza viruses and coronaviruses and represent therapeutic drug targets.
GENETIC EVOLUTION OF INFLUENZA A VIRUSES AND CORONAVIRUSES
When influenza viruses circulate in their natural reservoirs, they are characterized by high genetic variability that is reflected in the formation of quasi-subtypes (immunologically different antigenic variants) of type A viruses (Barbezange et al. 2018). This biological characteristic is called antigenic drift (Taubenberger and Kash 2010) and it is explained by the fact that RNA-dependent RNA-polymerase of influenza viruses does not have an active corrective site (Steinhauer, de la Torre and Holland 1989; Cheung et al. 2014), which results in a high frequency of point mutations in the process of RNA genome replication (300 times higher than during the replication of bacterial DNA genome) (Drake 1993). Another distinctive characteristic is the high mutational tolerance of glycoproteins of viral envelopes, i.e. the ability of HA and NA to maintain their functional activity in case of significant changes in the primary structure of the polypeptide chain (Thyagarajan and Bloom 2014; Visher et al. 2016).
An important and prevalent phenomenon in the evolution of influenza A viruses is so-called antigenic shift (Holmes et al. 2005; Dugan et al. 2008). The antigenic shift is the interchange of RNA segments of viral genome that code the HA and/or NA structure in case of simultaneous infection of a cell by several strains of the influenza A virus (Taubenberger and Kash 2010). It is the antigenic shift that permits new subtypes of influenza A virus to overcome cross-species barriers (Scholtissek et al. 1978; Garten et al. 2009).
Unlike other RNA viruses, the coronavirus genome replication involves RNA-dependent RNA-polymerase that has 3′-exonuclease corrective activity (Smith, Sexton and Denison 2014). With the objective of immune evasion in humans and maintenance of the genotype in the Homosapiens population, as has been demonstrated for the coronaviral strain HCoV-OC43, coronaviruses also maintain the antigenic drift (Ren et al. 2015). In addition, the genome of coronaviruses uses RNA–RNA recombination for its evolution (Keck et al. 1988; Huang et al. 2016; Forni et al. 2017). Homologous RNA recombination represents a redistribution of the genetic material by interchange of RNA segments in the conditions of co-infection (Makino et al. 1986; Lai 1990; Lai and Cavanagh 1997). In addition to evasion from the host immune reactions, RNA recombination lets coronaviruses change the profile of virulence and tissue affinity as well as overcome cross-species barriers (Haijema, Volders and Rottier 2003; Stavrinides and Guttman 2004).
High genetic and phenotypic variability of influenza A viruses and coronaviruses can lead to a situation where these pathogenic agents obtain resistance to specific therapeutics as well as to the sudden appearance of new virulent pandemic strains.
PANDEMIC RESPIRATORY VIRAL INFECTIONS AND THE PROBLEM OF PNEUMONIA
The influenza pandemic in 1918–1920 became the most fatal disease-related event in human history (to date), which resulted in the death of >50 million people (Johnson and Mueller 2002). The mortality during pandemics of influenza and coronaviral infections is largely associated with pneumonia (Morens, Taubenberger and Fauci 2008; Metersky et al. 2012; Yin and Wunderink 2018; Al-Baadani et al. 2019). Primary viral pneumonias are often complicated by bacterial co-infection as they transform to viral-bacterial and bacterial pneumonias (Oswald, Shooter and Curwen 1958; Bisno et al. 1971; Palacios et al. 2009; Gill et al. 2010; Martín-Loeches et al. 2011; Cillóniz et al. 2012). The statement by Louis Cruveilhier expressed in 1919 is still common in expert circles: ‘The influenza awards a sentence, and it is bacterial flora that carries it out’ (Cruveilhier 1919).
The clinical picture of severe viral respiratory infections often presents with symptoms of primary viral pneumonia. The development of primary viral pneumonia in case of a viral respiratory infection is probably related to co-expression of glycoproteins and glycolipids that contain glycans with terminal α2,3-linked sialic acid (which plays the role of respiratory virus receptor), and to the transmembrane serine protease TMPRSS2 (which itself plays a role in proteolytically activating viral HA and S protein) of the epithelial cells of alveoli and bronchioles (Ibricevic et al. 2006; Shinya et al. 2006; Kumlin et al. 2008; Bertram et al. 2010; Limburg et al. 2019; Tortorici et al. 2019).
The vulnerability to bacterial co-infection during respiratory viral pandemics is associated with multiple factors: virus-induced dysbiosis and disruption of barrier function of the epithelial lining of respiratory airways (Pittet et al. 2010; Ellis et al. 2015; Nita-Lazar et al. 2015; Hanada et al. 2018; Sencio et al. 2020); virus-induced dysfunction of effector immune cells (McNamee and Harmsen 2006; Small et al. 2010; Ghoneim, Thomas and McCullers 2013; Sun and Metzger 2014) and immunosuppressive activity of cytokines in relation to antibacterial immunity (Cao et al. 2014; van der Sluijs et al. 2004; Shepardson et al. 2019); and virus-associated dysfunction of alveolar-capillary barrier (McAuley et al. 2007; Henkel et al. 2010; Short et al. 2016; Kamal, Alymova and York 2018) and suppression of activity of ion channels that are responsible for the absorption of fluid from the alveolar lumen (Carlson et al. 2010; Peteranderl et al. 2016; Brand et al. 2018).
Pneumonias associated with respiratory viral infections are an independent factor in disease severity and mortality (Maruyama et al. 2016; Ishiguro et al. 2017). This means that the main problem of severe viral infections, in the past as well as in the present, has been the problem of viral, viral-bacterial and secondary bacterial pneumonias.
ANTIVIRAL THERAPY
The biology of influenza viruses and coronaviruses inevitably leads to the appearance of new pandemic strains; it is impossible to predict the moment of their development, genomic variability and antigenic properties. This means that pandemics of new respiratory infections will always start in the absence of immune prophylactics and treatments. This underlines the necessity of prior research and development of treatments for the prevention and treatment of respiratory viral infections and in particular for coronaviruses and influenza A viruses. Several antiviral drugs that will be described herein are presented in Table 3.
Table 3.
Chemical structures of selected drugs described in this review.
| INN | Chemical structure | Brand name | Key reference |
|---|---|---|---|
| Tilorone |
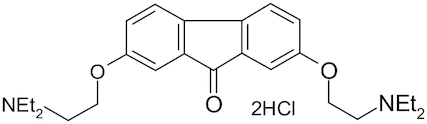
|
Amixin, Lavomax | Ekins et al. 2020; Jeon et al. 2020 |
| Meglumine acridine acetate |
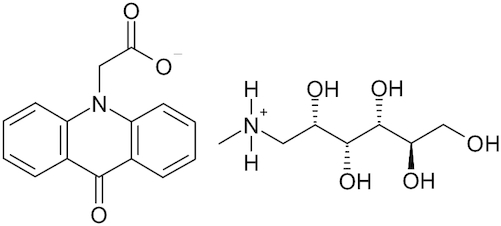
|
Cycloferon | Ekins et al. 2020 |
| Oseltamivir |
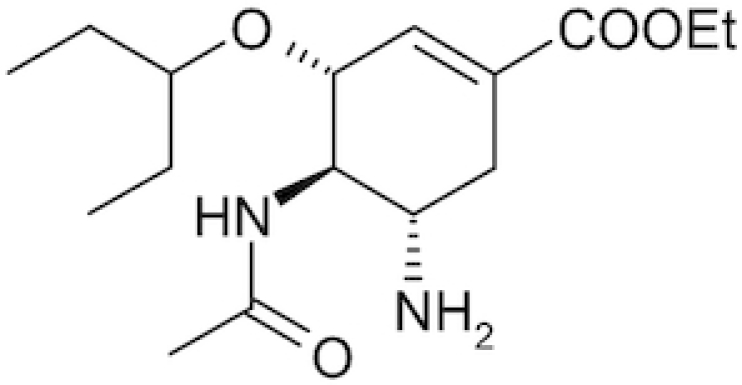
|
Tamiflu | Neupane et al. 2020 |
| Ribavirin |
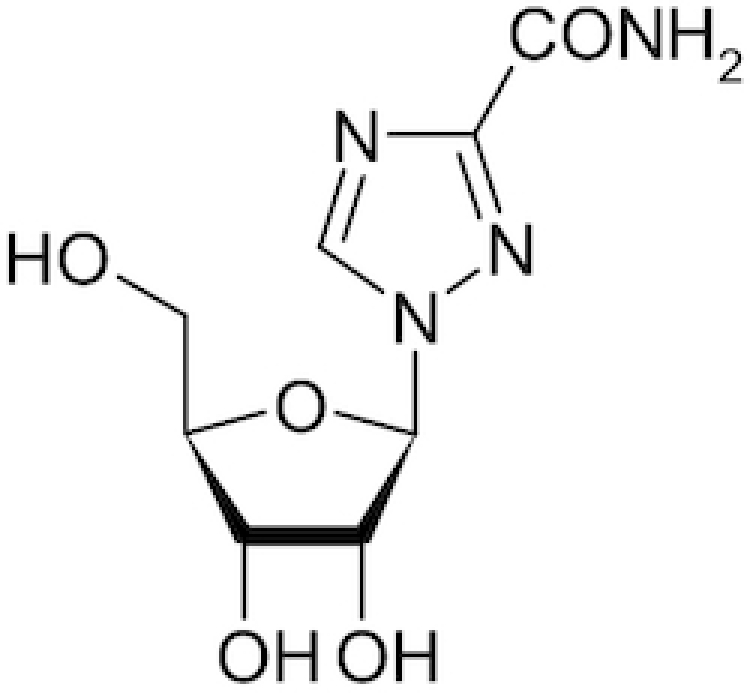
|
Copegus, Rebetol, Ribasphere, Vilona, Virazole | Neupane et al. 2020 |
| Inosine Pranobex |
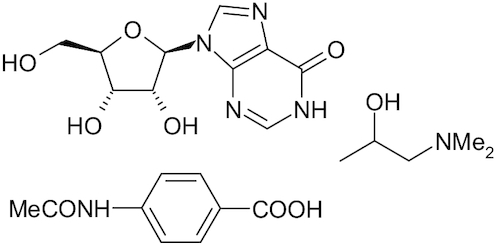
|
Methisoprinol | Sliva, Pantzartzi and Votava 2019 |
| Quercetin |
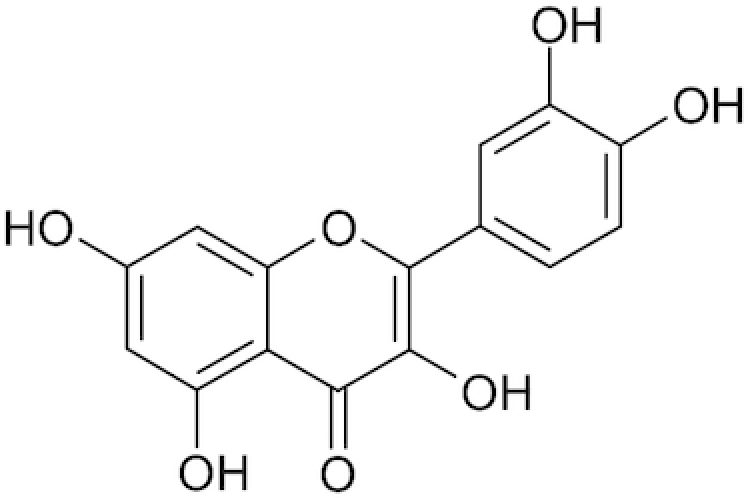
|
Zakaryan et al. 2017 | |
| Ambroxol |
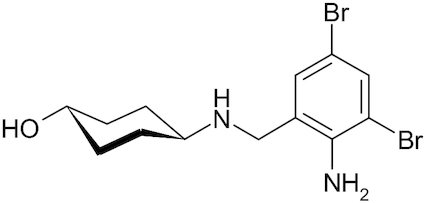
|
Muciclar, Mucosolvan, Mucobrox, Mucol, Lasolvan, Mucoangin, Surbronc, Brontex, Ambolar, Lysopain | Yang et al. 2002; Yamaya et al. 2014 |
| Allopurinol |
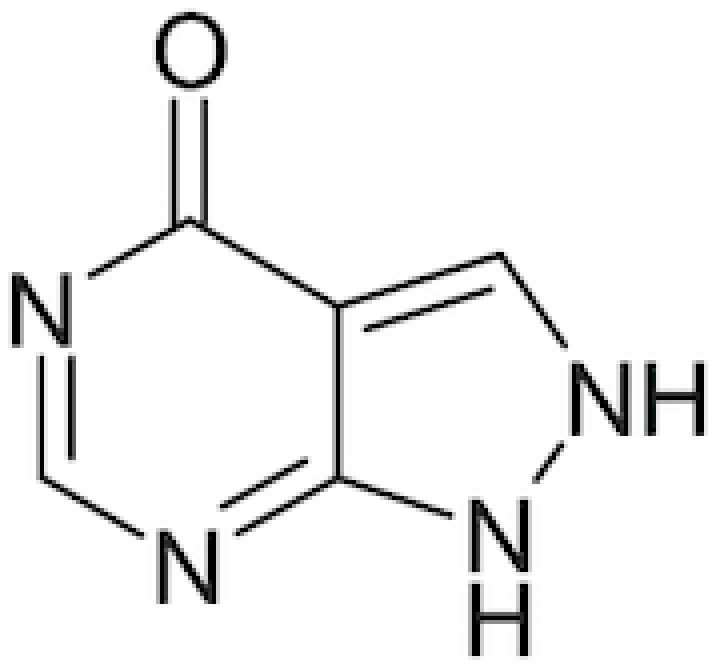
|
Allohexal, Allosig, Milurit, Alloril, Progout, Ürikoliz, Zyloprim, Zyloric, Zyrik and Aluron | Pacher, Novorozhkin and Szabo 2006; George and Struthers 2009 |
| Melatonin |
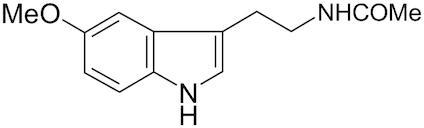
|
Reiter et al. 2017 | |
| Deferoxamine |
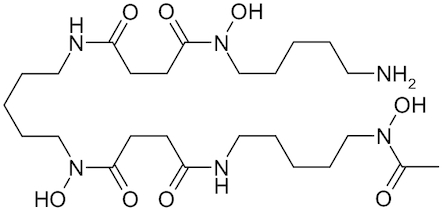
|
Desferal | Borg and Schaich 1986; Klebanoff et al. 1989; Dulchavsky et al. 1996; Niihara et al. 2002; Francisco et al. 2010 |
| Mexidol |
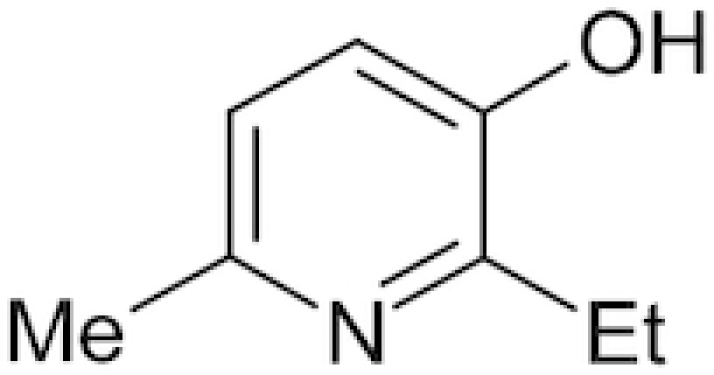
|
Emoxipine, Emoxypin, Epigid | Pavelkina, Yerovichenkov and Pak 2010 |
| Chloroquine |
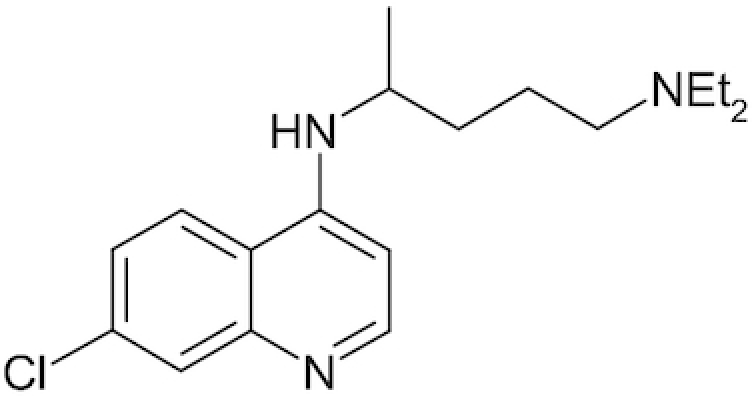
|
Chloroquine FNA, Resochin, Dawaquin, Lariago, Delagil | Jeon et al. 2020; Jin et al. 2020; Liu et al. 2020 |
| Hydroxychloroquine |
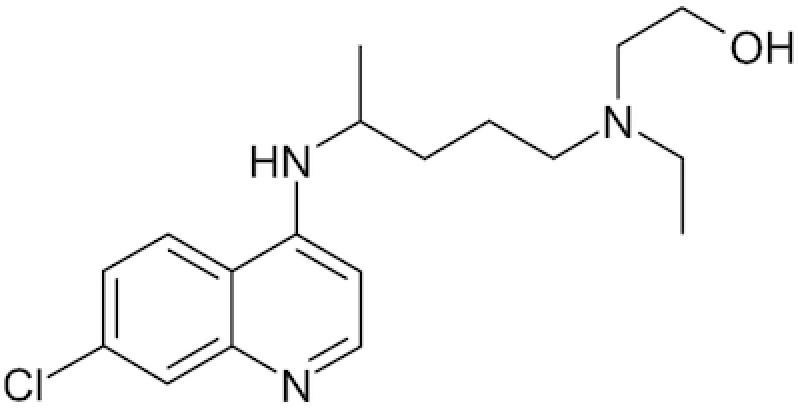
|
Plaquenil, Hydroquin, Axemal, Dolquine, Quensyl, Quinoric, Immard | Liu et al. 2020 |
| Mefloquine |
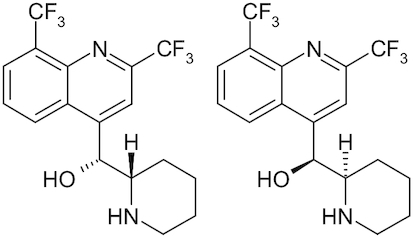
|
Lariam | |
| Remdesivir |
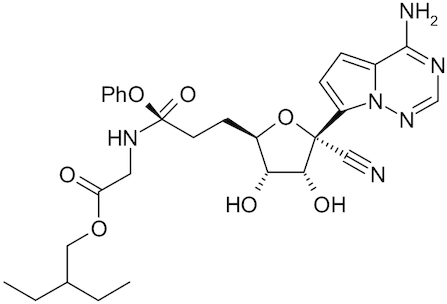
|
GS-5734 | Lu 2020; Wang et al. 2020a; Zhang et al. 2020 |
| Ivermectin |
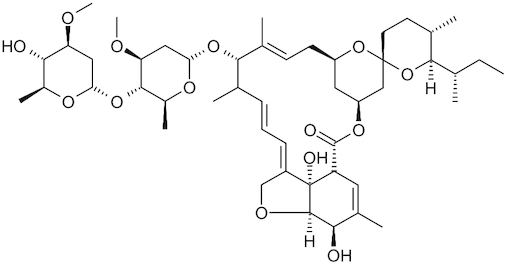
|
Stromectol | Caly et al. 2020 |
The nature of RNA viruses suggests that systemic interferon alfa-2b might be effective as non-specific background therapy, taking into account the weakened state of patients. The efficacy of topical interferon solutions is doubtful, but they may be considered in case of local symptoms (rhinitis, pharyngitis etc.). Usage of systemic interferon inducers such as tilorone and cycloferon (Ekins et al. 2020; Ekins and Madrid 2020) may result in secondary immunosuppression 10–14 days later, which can lead to another infection. Background antiviral therapy also includes targeted agents that affect enzymes of the viral genome replication; this includes oseltamivir, and the most potent (but also most toxic of this group) ribavirin, as well as other novel targeted antiviral medications. Anti-replicative activity has been observed for inosine pranobex (Sliva, Pantzartzi and Votava 2019), a purine derivative that is active against influenza A and B viruses.
The current knowledge of the viral nature and pathogenetic properties of the infectious process allows us to consider the possibility of using adjuvant agents, the efficacy of which has been observed in different studies (Ekins, Lane and Madrid 2020).
It is well known that serine proteases participate in the process of internalization of coronaviruses and influenza A viruses into the epithelial cells (Simmons 2013; Garten et al. 2015). The activity of trypsin-like proteinases in the upper respiratory tract significantly depends on the activity of inhibitors of secretory leucoproteinases and in the lower respiratory tract it depends on the surfactant (Kido et al. 2004). Therefore, therapeutics that induce the expression of inhibitors of secretory leucoproteinases and surfactant may significantly inhibit the multicyclic replication of RNA viruses (including influenza and coronaviruses).
Quercetin has such properties. In the micromolar range, in addition to antioxidant effects, it can chelate metals of mixed valency (Gholampour and Saki 2019), stimulate the expression of antioxidant enzymes (Chen et al. 2017), provide direct reduction of free radicals of fatty acid residues of phospholipids and oxidized forms of vitamin E (Ozgen, Kilinc and Selamoglu 2016; Chepur et al. 2020), inhibit the activity of serine proteases (Xue et al. 2017; Jo et al. 2019) and shield the active center of HA of the influenza A virus (Wu et al. 2015), which gives it a wide range of antiviral effects (Zakaryan et al. 2017). However, in our opinion, this compound is highly promiscuous and not a good drug candidate.
Ambroxol (trans-4-[[(2-amino-3,5-dibromophenyl)methyl] amino]cyclohexanolhydrochlo-ride) also deserves attention as an additional antiviral agent (Yang et al. 2002). The in vitro inhibitory effects of ambroxol on influenza virus were described in 2014 (Yamaya et al. 2014). The spectrum of pharmacological activity of ambroxol, in addition to its mucolytic effects (Rogers 2007), includes antibacterial and anti-biofilm effects (Lu et al. 2010; Li et al. 2011; Cabral-Romero et al. 2013; Cataldi et al. 2014); the ability to serve as chemical chaperones (Bendikov-Bar et al. 2013; Sanchez-Martinez et al. 2016), modulate surfactant secretion (Yang et al. 2002; Seifart et al. 2005), provide anti-inflammatory (Gibbs et al. 1999; Beeh et al. 2008; Gupta 2010) and antioxidant action (Nowak et al. 1994; Stetinová, Herout and Kvetina 2004); and the ability to locally (in the respiratory airways) stimulate the secretion of IgA and IgG (Yang et al. 2002) as well as to provide a local anesthetic effect (Kern and Weiser 2015). Due to these diverse effects and high oral bioavailability (Jauch et al. 1978), ambroxol may be included in a list of medications used for the treatment of viral pneumonias.
An important role in the pathogenesis of respiratory infections is being played by the virus-induced oxidative stress (Schwarz 1996; Lin et al. 2006; Liu et al. 2017; Khomich et al. 2018). Xanthine oxidoreductase has an important role in the appearance of the symptoms and complications of virus-associated pneumonias. Xanthine oxidoreductase is a cytosolic enzyme of purine catabolism (Frederiks and Vreeling-Sindelarova 2002; Agarwal, Banerjee and Banerjee 2011) and its activity strongly increases in hypoxic conditions (Poss et al. 1996; Terada et al. 1997; Linder et al. 2003) as well as under the influence of proinflammatory mediators and cytokines (Page et al. 1998; Brandes et al. 1999). In pathological conditions, xanthine oxidoreductase is released from the cells to the blood (predominantly in oxidase form; Spiekermann et al. 2003) and fixates at the luminal surface of the plasma membrane of endothelial cells in the area of the inflammation by physical/chemical interaction with glycosaminoglycans (Akaike et al. 1990; Adachi et al. 1993; Rouquette et al. 1998). Xanthine oxidoreductase located on the cytoplasmic membrane of endothelial cells produces a superoxide anion radical in the process of purine oxidation, and at the same time may redux nitrite and nitrate anions to the nitrogen oxide (NO•) at another active site (Jansson et al. 2008; Cantu-Medellin and Kelley 2013), i.e. it can recycle this vasodilating agent. Local production of the prooxidative complex (O2–•, H2O2, NO•, ONOO–) is potentially very dangerous, especially in the vascular bed of the lungs. Nevertheless, the attempts of using allopurinol, an inhibitor of xanthine oxidoreductase (Pacher, Novorozhkin and Szabo 2006; George and Struthers 2009), for the treatment of influenza A virus-induced pneumonia in daily doses of 5–50 mg/kg have failed. Allopurinol has not shown any effects on the evolution and outcomes of the viral infection (Dolganova and Sharonov 1997). Lack of therapeutic effect in this case is associated with the fact that after the inhibition of (Mo-Co)-containing center of the enzyme by allopurinol, the NADH-oxidative and nitrite-/nitrate-reductive activities of xanthine oxidoreductase, which are realized at the FAD-dependent site of the enzyme, were not affected (Harris and Massey 1997; Doel et al. 2001; Boueiz, Damarla and Hassoun 2008). As there are still no approved medications able to inhibit the FAD-dependent activity of xanthine oxidoreductase, administration of heparin seems feasible as prophylaxis of pulmonary embolism with the objective of the desorption of xanthine oxidoreductase from the cytoplasmic membrane of endothelial cells (Povalyaev 2014; Obi et al. 2019).
Another significant source of the active forms and metabolites of oxygen during respiratory viral infections is mitochondria (To et al. 2020). Melatonin is a mitochondrial antioxidant (Reiter et al. 2017) with anti-inflammatory and immunomodulatory activity and has noticeable positive effects on the evolution and outcomes of viral infections under experimental conditions (Srinivasan, Mohamed and Kato 2012; Silvestri and Rossi 2013; Tan et al. 2014; Huang et al. 2019; Zhang et al. 2020). Melatonin is also widely used to promote sleep, so this may be undesirable in an antiviral during the daytime.
The superoxide anion radical may act on organic and inorganic compounds, depending on their chemical properties, as an oxidant (E0O2–•/H2O2 ꞊ +0.89 V) or a reductant (E0O2/O2–•꞊ –0.16 V) (Wood 1987, 1988). The reductive properties of the superoxide radical is produced in the area of inflammation during viral pneumonias. This may occur via reduction of ferric ions after their release from complexes with biomacromolecules. For example, iron in a molecule of ferritin is represented by Fe3+ ions, which under the influence of the superoxide anion radical transforms into Fe2+ and leaves the aforementioned protein (Biemond et al. 1984; Bolann and Ulvik 1987). In the presence of free ferric ions and partially reduced forms of oxygen, the conditions are created for a kind of catalytic reactor for redox catabolic production of prooxidants, especially very toxic hydroxyl radicals (Morris, Earl and Trenam 1995). This condition of a biological system is extremely dangerous because in the presence of free ferric ions, biological fluids lose their antibacterial properties (Bullen, Ward and Rogers 1991; Griffiths 1991; Sritharan 2006). The elimination of free ferric ions from the biological media of a body is a life/death issue in case of viral pneumonias. There were earlier attempts to use available complexones (for example, deferoxamine) to bind ferric ions during viral pneumonia; contrary to the expected, not only did they show no positive effects on the pathological process, but they also led to increased mortality (Dolganova and Sharanov 1997). The explanation of this paradox is that deferoxamine (desferal) has approximately the same affinity constant for ferric ions as siderophores of microorganisms (Hallaway et al. 1989; Askwith, de Silva and Kaplan 1996); for this reason, it is unable to limit the availability of Fe3+ for pathogenic organisms (Kim, Park and Shin 2007; Cassat and Skaar 2013). At the same time, it seems that ferric ions chelated by deferoxamine do not completely lose their ability to redox transformation and thus support the reactions of Fenton and Osipov (Borg and Schaich 1986; Klebanoff et al. 1989; Dulchavsky et al. 1996; Niihara et al. 2002; Francisco et al. 2010).
In contrast, 2-ethyl-6-methyl-3-hydroxypyridine succinate (mexidol, emoxipine) has noticeable iron chelating activity (Andrusishina et al. 2014), antioxidative activity (Voronina 2001) and the ability to inhibit serine proteases and matrix metalloproteases (Akhmedov, Budylgin and Dolgikh 2010). Mexidol has many such biological effects and has been proposed for the effective use as a supportive agent in the treatment of pneumonia (Ilyashenko et al. 2001; Luzhnikov et al. 2006) and viral infections (Pavelkina, Yerovichenkov and Pak 2010).
In clinical practice, chloroquine has been widely used as a safe, effective and affordable medication for more than seven decades (since 1947; Solomon and Lee 2009). It is used in the forms of phosphate, hydrochloride and sulfate for the following indications: treatment and prevention of malaria (Mengesha and Makonnen 1999; Bello, Chika and Bello 2010; Waqar, Khushdil and Haque 2016); treatment of leprosy (Meinão et al. 1996; Bezerra et al. 2005; Gordon et al. 2018); as an anti-inflammatory agent in patients with rheumatoid arthritis (Augustijns et al. 1992; Schrezenmeier and Dorner 2020); treatment of antiphospholipid syndrome (Tektonidou et al. 2019); treatment of Sjogren's syndrome (Vivino et al. 2016; Shivakumar et al. 2018; Lee et al. 2019); treatment of amoebic hepatitis and hepatic abscesses (Sodeman et al. 1951; Cohen and Reynolds 1975); cancer treatment as sensitizing agent (Solomon and Lee 2009; Maycotte et al. 2012; Kimura et al. 2013); and treatment of metabolic syndrome (Kastan, Semenkovich and Schneider 2008; McGill et al. 2019) and inflammatory diseases of bacterial nature (in synergy with antibiotics (Crowle and May 1990; Feurle et al. 2012; Jagadeesh, Saivisveswar and Revankar 2014; Son and Chung 2014).
Chloroquine and its many analogs (such as hydroxychloroquine etc.) have properties of weak acidic amines in unprotonated form as they easily permeate cellular membranes (Chinappi et al. 2010) and after the protonation accumulate in closed cellular compartments with acidic pH (i.e. endosomes or lysosomes) (Vincent et al. 2005). The level of chloroquine in such compartments may be >100 times higher than its concentration in the cell (de Duve et al. 1974). Chloroquine may stay in the isolated intracellular compartments for hundreds of hours (Schrezenmeier and Dorner 2020). Accumulating in endosomes/lysosomes, chloroquine shifts the pH to alkali (Homewood et al. 1972; Ohkuma and Poole 1978; Al-Bari 2017) and inhibits diverse ATPases, including Н+-ATPase (V-ATPase), which defines the acidification of the environment of endosomes and cisternae of the Golgi apparatus (Chandra et al. 1992; Bhattacharyya and Sen 1999; Holliday 2017). It is possible that these many phenomena define the blockade of the release of RNA genome of influenza viruses from the lipoproteins of their envelopes (Shibata et al. 1983), which results in the inhibition of viral replication (Ooi et al. 2006; Di Trani et al. 2007). The ability of chloroquine to inhibit the acidification of endosomes that contain respiratory viruses, and thus to block the release of their RNA genomes and following replication, may partially explain its antiviral activity. Chloroquine also has high antiviral activity against not only influenza A viruses (internalized in the endosomes) but also coronaviruses (Keyaerts et al. 2004; Vincent et al. 2005; Ooi et al. 2006; Yan et al. 2013; de Wilde et al. 2014; Kearney 2020), which are almost exclusively internalized by membrane fusion, i.e. without the formation of endosomes (Matsuyama et al. 2005).
Of the three types of biological aperiodic polymers (nucleic acids, polypeptides and carbohydrates), aperiodic polymers of carbohydrates (glycans and oligosaccharides) have the highest information capacity, due to their structural properties. This ensures high specificity of ligand–receptor interactions of oligosaccharide conjugates. But the structure of glycans in the eukaryotic genome is coded indirectly. Oligosaccharides are synthetized in the cisternae of Golgi apparatus with the support of secondary protein matrices that form functional heterogenic associations (conveyor lines) of glycosyltransferases (Chepur et al. 2019). Obviously, the spatial structure of such matrix protein molecules and thus their affinity to the enzymes of glycan synthesis may quickly and significantly change under the influence of the dynamics in the pH and oxidative-reductive potential in the cisternae of Golgi apparatus.
For this reason, it is important that chloroquine is able to change the redox status of a cell (Giovanella et al. 2015) and decrease the concentration of protons (increase the рН) in the cisternae of Golgi apparatus by suppression of ATPase activity, including Н+-ATPase (Reaves and Banting 1994; Hassinen et al. 2011). The function of the Golgi apparatus that is considered most sensitive to pH changes is the synthesis of aperiodic oligosaccharides (Kellokumpu 2019). A pH increase by 0.2 inside the Golgi apparatus is associated with a disruption in terminal α2,3-sialylation of both N-linked and O-conjugated glycans (Rivinoja et al. 2006, 2009). It seems that aberrant glycosylation after the decrease in acidity of intraluminal environment of Golgi complex cisternae is associated with рН-induced changes in the topology/location of glycosyltransferases in multienzyme complexes of aperiodic oligosaccharides synthesis.
As all participants of the interaction between human cells and respiratory RNA viruses (glycoproteins and glycolipids) are richly decorated by glycans with terminal sialic acids, which are recognized by the viral particles as specific receptors, the chloroquine-induced disruption of the processes of sialylation/glycosylation of cellular and viral participants of this interaction is reflected in its antiviral effects.
The participation of glycans in viral adhesion and proliferation are extremely important. A wide array of viruses, including coronaviruses (Milewska et al., 2014, 2018; Szczepanski et al. 2019), use a common heparan sulfate-dependent mechanism of the attachment to a cellular membrane. Inhibitors of this attachment could therefore prevent and treat infections. The N,N′-bisheteryl derivative of dispyrotripiperasine, pyrimidine dispirotripiperasinium, became the first synthetic small molecule (Schmidtke, Wutzler and Makarov 2004; Novoselova et al. 2017) broad spectrum inhibitor of the replication of viruses of different families that use heparan sulfate to attach to and/or enter a host cell. The inhibition is via mimicking the binding of specific structural parts of heparan sulfate. This investigational class of compounds opens new opportunities for the inhibition of the process of viral transmission, for example, by using them to prevent infection by herpes simplex virus type I.
A method of prevention and treatment of aspiration pneumonias and ventilator-associated pneumonias may be adapted for virus-associated pneumonias. The method involves hypoosmotic (to 200–250 mM) conditioning of red blood cells (RBCs) of autogenic blood in a solution of a broad-spectrum antibiotic, with the addition of dimethyl sulfoxide (DMSO) and heparin. This approach avoids hemolysis and uses autogenic RBCs as an intravenous depot for the delivery of antibiotics to the area of inflammation (pneumonia), where the tonicity of blood is normalized due to swelling. DMSO increases the fluidity (decreases the microviscosity) and permeability of cellular RBC membranes, which helps to deliver antibiotic into the cell. A proposed dose of DMSO (0.3–0.4 ml) does not affect morphology or functional properties of blood cells (Gurtovenko and Anwar 2007). In addition, DMSO inhibits the activation of proinflammatory transcription factors NF-κB, AP-1 and expression of adhesion molecules ICAM-1 (Chang, Albarillo and Schumer 2001), blocks transcription of the IL-1, IL-6, IL-8 genes, as well as activation of the inflammasomes NLRP3 (Ahn et al. 2014; Elisia et al. 2016), and has noticeable antioxidant activity in extremely low concentrations (Jia et al. 2010; Sanmartín-Suárez et al. 2011).
From the earliest days of the current outbreak of SARS-CoV-2, there has been considerable focus on drug repurposing. A bibliometric analysis of drug repurposing has described the many FDA-approved drugs that have been tested for other indications. This analysis highlighted chloroquine as one of the most repurposed drugs as it has been tested against hundreds of diseases (Baker et al. 2018). Not surprisingly, chloroquine has also been identified by several groups (in China, South Korea and the United States) (Jeon et al. 2020; Jin et al. 2020; Liu et al. 2020) to have micromolar activity against SARS-CoV-2. Remdesivir, which had previously failed in clinical trials for Ebola (Mulangu et al. 2019) but had also recently shown activity against MERS in rhesus macaques (de Wit et al. 2020), was tested in vitro against SARS-CoV-2 and shown to be active. Both these drugs (and closely related analogs) are already in many clinical trials globally. There are numerous other drugs proposed, including a broad array of nucleoside analogs, neuraminidase inhibitors, peptides, RNA synthesis inhibitors, anti-inflammatory drugs as well as traditional Chinese medicines (Lu 2020; Wang et al. 2020a; Zhang and Liu 2020). In just a few months, many papers and preprints have described one or more molecules with in vitro data against the virus. To date, there are likely >100 drugs that have been tested and described with in vitro IC50 data in cells from these studies (Caly et al. 2020; Choy et al. 2020; Jeon et al. 2020; Jin et al. 2020; Liu et al. 2020; Yamamoto et al. 2020). These cover large natural product molecules like ivermectin (Caly et al. 2020) through to an array of small molecules that are primarily lysosomotropic drugs (Weston et al. 2020). Most of these studies use Vero cells for testing and this animal cell type may not be an ideal. We await seeing how the wider use of human cells may impact the discovery of other inhibitors of this virus. Additionally, some of these molecules identified may be impractical due to off-target effects or not being able to be used at concentrations similar to their original indication.
CONCLUSIONS
Respiratory RNA viruses are anthropozoonotic infectious pathogens that have natural reservoirs and form dynamic genetic pools. Such a genetic pool suggests the interchange or spillover of genetic material between the genomes of familial RNA viruses of humans and animals. This inevitably leads to the appearance of new, highly virulent strains of pathogens and it is impossible to predict the moment of such appearance and antigenic properties of these strains. This means that epidemics of new respiratory RNA viral infections will always begin in the absence of medications for their immune-mediated prevention or treatment. This underlines the necessity of continuing to perform research and development of antivirals and other therapeutic drugs that could be used in the treatment of respiratory RNA viral infections. This review has focused on the past and present efforts at addressing these viruses. Clearly, our future will be very much defined by such viral outbreaks if we are not able to identify broad-spectrum antivirals or vaccines. Looking at the past research may provide some important clues as to how we can identify such therapeutics. The reliance on a single magic bullet for every disease may be unrealistic and we therefore need to consider the combination of diverse antiviral treatments as we currently do for HIV and HBV. Considering molecules that are traditionally not considered ‘antivirals’ may also be critical to open our eyes to accessing additional targets and mechanisms. Host-targeted mechanisms may also be of interest such as those that stimulate the immune system. Clearly, we are seeing many drugs that are lysosomotropic; while long-term use of such molecules may be detrimental, short-term use may prevent viral entry and protect the individual. There is certainly much more research that can be performed to understand how combinations of drugs for these respiratory viruses may work together. While interest in antiviral research and development has apparently languished for decades, the COVID-19 may permanently change that. If we continue to ignore such viruses, the cost will be unimaginable and continue to hold back human progress. We will now see a rebirth in interest and perhaps significant investment in developing antivirals. For years, there have been few major drug companies dominating this field. What we have seen with viruses should also serve to remind us that we also face great pressures such as drug resistance for other classes of drugs like antibiotics. This review should remind us that we need to be ready for the next outbreak and that means having a plentiful supply of drugs that can potentially address any new virus we are faced with. A relatively small investment in this science could pay big dividends for the future in preventing catastrophic pandemics, limiting the global financial depressions that result and providing a degree of security for humanity. We cannot neglect these or other viruses for they provide other insights that could ultimately be useful in healthcare and beyond.
ACKNOWLEDGMENTS
The authors kindly acknowledge Anna Egorova for help in manuscript preparation.
FUNDING
We kindly acknowledge the funding from NIH NINDS 1R01NS102164-01 and NIAID R41AI13456.
Contributor Information
Vadim Makarov, Federal Research Center Fundamentals of Biotechnology of the Russian Academy of Sciences, 33-2 Leninsky Prospect, Moscow 119071, Russia.
Olga Riabova, Federal Research Center Fundamentals of Biotechnology of the Russian Academy of Sciences, 33-2 Leninsky Prospect, Moscow 119071, Russia.
Sean Ekins, Collaborations Pharmaceuticals, Inc., 840 Main Campus Drive, Lab 3510, Raleigh, NC 27606, USA.
Nikolay Pluzhnikov, State Research Institute of Military Medicine of the Ministry of Defence of the Russian Federation, St Petersburg 195043, Russia.
Sergei Chepur, State Research Institute of Military Medicine of the Ministry of Defence of the Russian Federation, St Petersburg 195043, Russia.
Conflicts of interest
None declared.
REFERENCES
- Adachi T, Fukushima T, Usami Y et al. Binding of human xanthine oxidase to sulphated glycosaminoglycans on the endothelial-cell surface. Biochem J. 1993;289:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Banerjee A, Banerjee UC. Xanthine oxidoreductase: a journey from purine metabolism to cardiovascular excitation–contraction coupling. Crit Rev Biotechnol. 2011;31:264–80. [DOI] [PubMed] [Google Scholar]
- Ahn H, Kim J, Jeung EB et al. Dimethyl sulfoxide inhibits NLRP3 activation. Immunobiology. 2014;219:315–22. [DOI] [PubMed] [Google Scholar]
- Akaike T, Ando M, Oda T et al. Dependence on O2-generation by xanthine oxidase of pathogenesis of influenza virus infection in mice. J Clin Invest. 1990;85:739–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmedov VA, Budylgin AV, Dolgikh VT. The matrix metalloproteinase 9 (MMP-9) and TIMP-1 activities in patients with chronic and recurrent pancreatitis. Eksp Klin Gastroenterol. 2010;6:11–3. [PubMed] [Google Scholar]
- Al-Baadani AM, Elzein FE, Alhemyadi SA et al. Characteristics and outcome of viral pneumonia caused by influenza and Middle East respiratory syndrome-coronavirus infections: a 4-year experience from a tertiary care center. Ann Thorac Med. 2019;14:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bari MAA. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspect. 2017;5:e00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Muharrmi Z. Understanding the influenza AH1N1 2009 pandemic. Sultan Qaboos Univ Med J. 2010;10:187–95. [PMC free article] [PubMed] [Google Scholar]
- Andrusishina IN, Vazhnichaya EM, Donchenko EA et al. Agent for the treatment of iron overload and hemochromatosis. Russian Federation patent RU 2557959C1 2014.
- Askwith CC, de Silva D, Kaplan J. Molecular biology of iron acquisition in Saccharomyces cerevisiae. Mol Microbiol. 1996;20:27–34. [DOI] [PubMed] [Google Scholar]
- Augustijns P, Geusens P, Verbeke N. Chloroquine levels in blood during chronic treatment of patients with rheumatoid arthritis. Eur J Clin Pharmacol. 1992;42:429–33. [DOI] [PubMed] [Google Scholar]
- Baker NC, Ekins S, Williams AJ et al. A bibliometric review of drug repurposing. Drug Discov Today. 2018;23:661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbezange C, Jones L, Blanc H et al. Seasonal genetic drift of human influenza A virus quasispecies revealed by deep sequencing. Front Microbiol. 2018;9:2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeh KM, Beier J, Esperester A et al. Antiinflammatory properties of ambroxol. Eur J Med Res. 2008;13:557–62. [PubMed] [Google Scholar]
- Bello SO, Chika A, Bello AY. Is chloroquine better than artemisinin combination therapy as first line treatment in adult Nigerians with uncomplicated malaria? A cost effectiveness analysis. Afr J Infect Dis. 2010;4:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106:5871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendikov-Bar I, Maor G, Filocamo M et al. Ambroxol as a pharmacological chaperone for mutant glucocerebrosidase. Blood Cells Mol Dis. 2013;50:141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S, Glowacka I, Blazejewska P et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol. 2010;84:10016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra EL, Vilar MJ, da Trindado Neto PB et al. Double-blind, randomized, controlled clinical trial of clofazimine compared with chloroquine in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52:3073–8. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D, Sen PC. The effect of binding of chlorpromazine and chloroquine to ion transporting ATPases. Mol Cell Biochem. 1999;198:179–85. [DOI] [PubMed] [Google Scholar]
- Biemond P, van Eijk HG, Swaak AJ et al. Iron mobilization from ferritin by superoxide derived from stimulated polymorphonuclear leukocytes. Possible mechanism in inflammation diseases. J Clin Invest. 1984;73:1576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno AL, Griffin JP, Van Epps KA et al. Pneumonia and Hong Kong influenza: a prospective study of the 1968–1969 epidemic. Am J Med Sci. 1971;261:251–63. [DOI] [PubMed] [Google Scholar]
- Bobrowski T, Melo-Filho CC, Korn D et al. Learning from history: do not flatten the curve of antiviral research!. Drug Discov Today. 2020:S1359–6446:30285–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolann BJ, Ulvik RJ. Release of iron from ferritin by xanthine oxidase. Role of the superoxide radical. Biochem J. 1987;243:55–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg DC, Schaich KM. Prooxidant action of desferrioxamine: Fenton-like production of hydroxyl radicals by reduced ferrioxamine. J Free Radic Biol Med. 1986;2:237–43. [DOI] [PubMed] [Google Scholar]
- Bosch BJ, van der Zee R, de Haan CA et al. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boueiz A, Damarla M, Hassoun PM. Xanthine oxidoreductase in respiratory and cardiovascular disorder. Am J Physiol Lung Cell Mol Physiol. 2008;294:L830–40. [DOI] [PubMed] [Google Scholar]
- Boulant S, Stanifer M, Lozach PY. Dynamics of virus–receptor interactions in virus binding, signaling, and endocytosis. Viruses. 2015;7:2794–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26:D49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes RP, Koddenberg G, Gwinner W et al. Role of increased production of superoxide anions by NAD(P)H oxidase and xanthine oxidase in prolonged endotoxemia. Hypertension. 1999;33:1243–9. [DOI] [PubMed] [Google Scholar]
- Brand JD, Lazrak A, Trombley JE et al. Influenza-mediated reduction of lung epithelial ion channel activity leads to dysregulated pulmonary fluid homeostasis. JCI Insight. 2018;3:123467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer L. Bird flu, influenza and 1918: the case for mutant Avian tuberculosis. Med Hypotheses. 2006;67:1006–15. [DOI] [PubMed] [Google Scholar]
- Bullen JJ, Ward CG, Rogers HJ. The critical role of iron in some clinical infections. Eur J Clin Microbiol Infect Dis. 1991;10:613–7. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L, Cummings RD, Steinhauer DA. The interplay between the host receptor and influenza virus hemagglutinin and neuraminidase. Int J Mol Sci. 2017;18:E1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena M, Oostergetel GT, Bartelink W et al. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc Natl Acad Sci USA. 2009;106:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral-Romero C, Martínez-Sanmiguel JJ, Reséndez-Pérez D et al. Antibacterial and anti-biofilm activities of ambroxol against oral bacteria. Pharma Innovation J. 2013;2:52–8. [Google Scholar]
- Cain CC, Sipe DM, Murphy RF. Regulation of еndocytic pH by the Na+,K+-ATPase in living cells. Proc Natl Acad Sci USA. 1989;86:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow KA, Parry HF, Sergeant M et al. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105:435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L, Druce JD, Catton MG et al. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu-Medellin N, Kelley EE. Xanthine oxidoreductase-catalyzed reduction of nitrite to nitric oxide: insights regarding where, when and how. Nitric Oxide. 2013;34:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wang D, Xu F et al. Activation of IL-27 signaling promotes development of postinfluenza pneumococcal pneumonia. EMBO Mol Med. 2014;6:120–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CM, Turpin EA, Moser LA et al. Transforming growth factor-β: activation by neuraminidase and role in highly pathogenic H5N1 influenza pathogenesis. PLoS Pathog. 2010;6:e1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi M, Sblendorio V, Leo A et al. Biofilm-dependent airway infections: a role for ambroxol? Pulm Pharmacol Ther. 2014;28:98–108. [DOI] [PubMed] [Google Scholar]
- Chandra S, Adhikary G, Sikdar R et al. The in vivo inhibition of transport enzyme activities by chloroquine in different organs of rat is reversible. Mol Cell Biochem. 1992;118:15–21. [DOI] [PubMed] [Google Scholar]
- Chang CK, Albarillo MV, Schumer W. Therapeutic effect of dimethyl sulfoxide on ICAM-1 gene expression and activation of NF-κB and AP-1 in septic rats. J Surg Res. 2001;95:181–7. [DOI] [PubMed] [Google Scholar]
- Chardonnet Y, Dales S. Early events in the interaction of adenoviruses with Hela cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology. 1970;40:462–77. [DOI] [PubMed] [Google Scholar]
- Chen BH, Park JH, Ahn JH et al. Pretreated quercetin protects gerbil hippocampal CA1 pyramidal neurons from transient cerebral ischemic injury by increasing the expression of antioxidant enzymes. Neural Regen Res. 2017;12:220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepur SV, Pluzhnikov NN, Saiganov SA et al. Mechanisms of implementation of alpha-tocopherol antioxidant effects. Adv Current Biol. 2020;140:149–65. [Google Scholar]
- Chepur SV, Pluzhnikov NN, Saiganov SA et al. The hypothesis of the aperiodic polysaccharides matrix synthesis. Adv Current Biol. 2019;139:583–93. [Google Scholar]
- Cherry JD. The chronology of the 2002–2003 SARS mini pandemic. Paediatr Respir Rev. 2004;5:262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung PP, Watson SJ, Choy KT et al. Generation and characterization of influenza A viruses with altered polymerase fidelity. Nat Commun. 2014;5:4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinappi M, Via A, Marcatili P et al. On the mechanism of chloroquine resistance in Plasmodium falciparum. PLoS One. 2010;5:e14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy KT, Wong AY, Kaewpreedee P et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillóniz C, Ewig S, Menéndez R et al. Bacterial co-infection with H1N1 infection in patients admitted with community acquired pneumonia. J Infect. 2012;65:223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HG, Reynolds TB. Comparison of metronidazole and chloroquine for the treatment of amoebic liver abscess. A controlled trial. Gastroenterology. 1975;69:35–41. [PubMed] [Google Scholar]
- Crowle AJ, May MH. Inhibition of tubercle bacilli in cultured human macrophages by chloroquine used alone and in combination with streptomycin, isoniazid, pyrazinamide, and two metabolites of vitamin D3. Antimicrob Agents Chemother. 1990;34:2217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruveilhier L. Action du serum antipneumococcique au cours de la pneumonie et dans les complications de la grippe. Ann Inst Pasteur. 1919;33:448–61. [Google Scholar]
- Daly P, Gustafson R, Kendall P. Introduction to pandemic influenza. BC Med J. 2007;49:240–4. [Google Scholar]
- de Duve C, de Barsy T, Poole B et al. Commentary. Lysosomotropic agents. Biochem Pharmacol. 1974;23:2495–531. [DOI] [PubMed] [Google Scholar]
- de Haan CA, Rottier PJ. Molecular interactions in the assembly of coronaviruses. Adv Virus Res. 2005;64:165–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde AH, Jochmans D, Posthuma CC et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58:4875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Feldmann F, Cronin J et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci USA. 2020;117:6771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Trani L, Savarino A, Campitelli L et al. Different pH requirements are associated with divergent inhibitory effects of chloroquine on human and avian influenza A viruses. Virol J. 2007;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doel JJ, Godberg BL, Eisenthal R et al. Reduction of organic nitrates catalyzed by xanthine oxidoreductase under anaerobic conditions. Biochim Biophys Acta. 2001;1527:81–7. [DOI] [PubMed] [Google Scholar]
- Dolganova A, Sharonov BP. Application of various antioxidants in the treatment of influenza. Braz J Med Biol Res. 1997;30:1333–6. [DOI] [PubMed] [Google Scholar]
- Dou D, Revol R, Östbye H et al. Influenza A virus cell entry, replication, virion assembly and movement. Front Immunol. 2018;9:1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA. 1993;90:4171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Pelletier C, Meyer G. Influenza D virus in cattle, France, 2011–2014. Emerg Infect Dis. 2015;21:368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulchavsky SA, Davidson SB, Cullen WJ et al. Effects of deferoxamine on H2O2-induced oxidative stress in isolated rat heart. Basic Res Cardiol. 1996;91:418–24. [DOI] [PubMed] [Google Scholar]
- Ekins S, Lane TR, Madrid PB. Tilorone: a broad-spectrum antiviral invented in the USA and commercialized in Russia and beyond. Pharm Res. 2020;37:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Madrid PB. Tilorone: a broad-spectrum antiviral for emerging viruses. Antimicrob Agents Chemother. 2020;64:e00440–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Mottin M, Ramos P et al. Déjà vu: stimulating open drug discovery for SARS-CoV-2. Drug Discov Today. 2020;25:928–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisia I, Nakamura H, Lam V et al. DMSO represses inflammatory cytokine production from human blood cells and reduces autoimmune arthritis. PLoS One. 2016;11:e0152538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis GT, Davidson S, Crotta S et al. TRAIL+ monocytes and monocyte-related cells cause lung damage and thereby increase susceptibility to influenza-Streptococcus pneumoniae coinfection. EMBO Rep. 2015;16:1203–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurle GE, Moos V, Schneider T et al. The combination of chloroquine and micocycline, a therapeutic option in cerebrospinal infection of Whipple`s disease refractory to treatment with ceftriaxone, meropenem and co-trimoxazole. J Antimicrob Chemother. 2012;67:1295–6. [DOI] [PubMed] [Google Scholar]
- Forni D, Cagliani R, Clerici M et al. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco AF, de Abreu Vieira PM, Arantes JM et al. Increase of reactive oxygen species by desferrioxamine during experimental Chagas' disease. Redox Rep. 2010;15:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T. A new type of virus from epidemic influenza. Science. 1940;92:405–8. [DOI] [PubMed] [Google Scholar]
- Frederiks WM, Vreeling-Sindelarova H. Ultrastructural localization of xanthine oxidoreductase activity in isolated rat liver cells. Acta Histochem. 2002;104:29–37. [DOI] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA et al. Antigenic and genetic characteristics of swine-origin A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten W, Braden C, Arendt A et al. Influenza virus activating host proteases: identification, localization and inhibitors as potential therapeutics. Eur J Cell Biol. 2015;94:375–83. [DOI] [PubMed] [Google Scholar]
- Gaunt ER, Hardie A, Claas ECJ et al. Epidemiology and clinical presentation of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Struthers AD. Role of urate, xanthine oxidase and the effects of allopurinol in vascular oxidative stress. Vasc Health Risk Manag. 2009;5:265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholampour F, Saki N. Hepatic and renal protective effects of quercetin in ferrous sulfate-induced toxicity. Gen Physiol Biophys. 2019;38:27–38. [DOI] [PubMed] [Google Scholar]
- Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfection. J Immunol. 2013;191:1250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs BF, Schmutzler W, Vollrath IB et al. Ambroxol inhibits the release of histamine, leukotrienes and cytokines from human leukocytes and mast cells. Inflamm Res. 1999;48:86–93. [DOI] [PubMed] [Google Scholar]
- Gill JR, Sheng ZM, Ely SF et al. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134:235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanella F, Ferreira GK, de Prá SD et al. Effects of primaquine and chloroquine on oxidative stress parameters in rats. An Acad Bras Cienc. 2015;87:1487–96. [DOI] [PubMed] [Google Scholar]
- Gomes C, Ruiz J. Research inequities: avoiding the next pandemic. Pathog Glob Health. 2020;1–2.. Advance online publication, DOI: 10.1080/20477724.2020.1802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C, Amissah-Arthur MB, Gayed M et al. The British Society for Rheumatology guideline for the management of systemic lupus erythematosus in adults. Rheumatology (Oxford). 2018;57:e1–45. [DOI] [PubMed] [Google Scholar]
- Griffiths E. Iron and bacterial virulence – a brief overview. Biol Met. 1991;4:7–13. [DOI] [PubMed] [Google Scholar]
- Gupta PR. Ambroxol – resurgence of an old molecule as an anti-inflammatory agent in chronic obstructive airway diseases. Lung India. 2010;27:46–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtovenko AA, Anwar J. Modulating the structure and properties of cell membranes: the molecular mechanism of action of dimethyl sulfoxide. J Phys Chem B. 2007;111:10453–60. [DOI] [PubMed] [Google Scholar]
- Haijema BJ, Volders H, Rottier PJ. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J Virol. 2003;77:4528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallaway PE, Eaton JW, Panter SS et al. Modulation of deferoxamine toxicity and clearance by covalent attachment to biocompatible polymers. Proc Natl Acad Sci USA. 1989;86:10108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–3. [DOI] [PubMed] [Google Scholar]
- Hanada S, Pirzadeh M, Carver KY et al. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CM, Massey V. The reaction of reduced xanthine dehydrogenase with molecular oxygen. Reaction kinetics and measurement of superoxide radical. J Biol Chem. 1997;272:8370–9. [DOI] [PubMed] [Google Scholar]
- Hassinen A, Pujol FM, Kokkonen N et al. Functional organization of Golgi N- and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J Biol Chem. 2011;286:38329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Ducatez M, Collin EA et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C virus. PLoS Pathog. 2013;9:e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel M, Mitzner D, Henklein P et al. The proapoptotic influenza A virus protein PB1-F2 forms a nonselective ion channels. PLoS One. 2010;5:e11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A, Hofmann-Winkler H, Gierer S et al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday LS. Vacuolar H-ATPases (V-ATPases) as therapeutic targets: a brief review and recent developments. Biotarget. 2017;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Ghedin E, Miller N et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KV. Coronaviruses. In: Fields Virology, 4th edn Philadelphia: Lippincott Williams and Wilkins, 2001, 1187–1203. [Google Scholar]
- Homewood CA, Warhurst DC, Peters W et al. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235:50–2. [DOI] [PubMed] [Google Scholar]
- Huang C, Liu WJ, Xu W et al. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. 2016;12:e1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Liao CL, Chen SJ et al. Melatonin possesses an anti-influenza potential through its immune modulatory effect. J. Funct Foods. 2019;58:189–98. [Google Scholar]
- Hui DS, I Azhar E, Madani TA et al. The continuing 2019-nCoV epidemic threat of a novel coronaviruses to global health – the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit RJG, Lang Y, Bakkers MJG et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acid via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci USA. 2019;116:2681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Charles PD, Hester SS et al. Conserved and host-specific features of influenza virion architecture. Nat Commun. 2014;5:4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibricevic A, Pekosz A, Walter MJ et al. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol. 2006;80:7469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyashenko KK, Luzhnikov EA, Abakumov MM et al. Sklifosovsky Research Institute for Emergency Medicine, assignees. Russian Federation patent RU 2205641C2 2001.
- Ishiguro T, Kagiyama N, Uozumi R et al. Clinical characteristics of influenza-associated pneumonia of adults: clinical features and factors contributing to severity and mortality. Yale J Biol Med. 2017;90:165–181. [PMC free article] [PubMed] [Google Scholar]
- Jagadeesh K, Saivisveswar KN, Revankar SP. Efficacy of chloroquine against Escherichia Coli and Proteus vulgaris: an in vitro study. Sch J App Med Sci. 2014;2:3046–50. [Google Scholar]
- Jansson EA, Huang L, Malkey R et al. A mammalian functional reductase that regulates nitrite and nitric oxide homeostasis. Nat Chem Biol. 2008;4:411–7. [DOI] [PubMed] [Google Scholar]
- Jauch R, Bozler G, Hammer R et al. Ambroxol, studies of biotransformation in man and determination in biological samples. Arzneimittelforschung. 1978;28:904–11. [PubMed] [Google Scholar]
- Jeon S, Ko M, Lee J et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 2020:AAC.00819–20., DOI: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Zhu H, Li Y et al. Potent inhibition of peroxynitrite-induced DNA strand breakage and hydroxyl radical formation by dimethyl sulfoxide at very low concentration. Exp Biol Med (Maywood). 2010;235:614–22. [DOI] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y et al. Structure of Mpro from 1 COVID-19 virus and discovery of its inhibitors. Nature. 2020, DOI: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Johnson CK, Hitchens PL, Pandit PS et al. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc Biol Sci. 2020;287:20192736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–15. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim H, Kim S et al. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem Biol Drug Des. 2019;94:2023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal RP, Alymova IV, York IA. Evolution and virulence of influenza A virus protein PB1-F2. Int J Mol Sci. 2018;19:E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Stukenbrok H, Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol. 1989;109:2721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Semenkovich CF, Schneider J. Use of chloroquine to treat metabolic syndrome. United States patent US 20080319010A1 2008.
- Kearney J. Chloroquine as a potential treatment and prevention measure for the 2019 novel coronavirus: a review. Preprints: 2020030275 [Preprint] 2020; [cited 2020 May 07]. Available from: preprints.org/manuscript/202003.0275/v1.
- Keck JG, Matsushima GK, Makino S et al. In vivo RNA–RNA recombination of coronavirus in mouse brain. J Virol. 1988;62:1810–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellokumpu S. Golgi, pH, ion and redox homeostasis: how much do they really matter? Front Cell Dev Biol. 2019;7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern KU, Weiser T. Topical ambroxol for the treatment of neuropathic pain. An initial clinical observation. Schmerz. 2015;29:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyaerts E, Vijgen L, Maes P et al. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun. 2004;323:264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khomich OA, Kochetkov SN, Bartosch B et al. Redox biology of respiratory viral infections. Viruses. 2018;10:E392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido H, Okumura Y, Takahashi E et al. Host envelope glycoprotein processing proteases are indispensable for entry into human cells by seasonal and highly pathogenic avian influenza viruses. J Mol Genet Med. 2008;3:167–75. [PMC free article] [PubMed] [Google Scholar]
- Kido H, Okumura Y, Yamada H et al. Secretory leukoprotease inhibitor and pulmonary surfactant serve as principal defenses against influenza A virus infection in the airway and chemical agents up-regulating their levels may have therapeutic potential. Biol Chem. 2004;385:1029–34. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Park YJ, Shin SH. A widespread deferoxamine-mediated iron-uptake system in Vibrio vulnificus. J Infect Dis. 2007;196:1537–45. [DOI] [PubMed] [Google Scholar]
- Kim P, Jang YH, Kwon SB et al. Glycosylation of hemagglutinin and neuraminidase of influenza A virus as signature for ecological spillover and adaptation among influenza reservoirs. Viruses. 2018;10:E183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A et al. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73:3–7. [DOI] [PubMed] [Google Scholar]
- King AMQ, Lefkowitz EJ, Mushegian AR et al. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch Virol. 2018;163:2601–31. [DOI] [PubMed] [Google Scholar]
- Klebanoff SJ, Waltersdorph AM, Michel BR et al. Oxygen-based free radicals generation by ferrous ions and deferoxamine. J Biol Chem. 1989;264:19765–71. [PubMed] [Google Scholar]
- Klink HD, Rott R, Orlich M et al. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–39. [DOI] [PubMed] [Google Scholar]
- Koetz A, Nilsson P, Lindén M et al. Detection of human coronavirus NL63, human metapneumovirus and respiratory syncytial virus in children with respiratory tract infections in south-west Sweden. Clin Microbiol Infect. 2006;12:1089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik I, Yewdell JW. Influenza hemagglutinin and neuraminidase: Yin-Yang proteins coevolving to thwart immunity. Viruses. 2019;11:E346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumlin U, Olofsson S, Dimock K et al. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir Viruses. 2008;2:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszewski K, Brydak L. The epidemiology and history of influenza. Biomed Pharmacother. 2000;54:188–95. [DOI] [PubMed] [Google Scholar]
- Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–33. [DOI] [PubMed] [Google Scholar]
- Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–54. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Shin S, Yoon SG et al. The effect of chloroquine on the development of dry eye in Sjogren syndrome animal model. Invest Ophthalmol Vis Sci. 2019;60:3708–16. [DOI] [PubMed] [Google Scholar]
- Leung HS, Li OT, Chan RW et al. Entry of influenza A virus with a α2,6-linked sialic acid binding preference requires host fibronectin. J Virol. 2012;86:10704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang W, Hu L et al. Effect of ambroxol on pneumonia caused by Pseudomonas aeruginosawith biofilm formation in an endotracheal intubation rat model. Chemotherapy. 2011;57:173–80. [DOI] [PubMed] [Google Scholar]
- Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virol. 2012;86:2856–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limburg H, Harbig A, Bestle D et al. TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J Virol. 2019;93:e00649–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YX, Ng YL, Tam JP et al. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Lin KH, Hsieh TH et al. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol Med Microbiol. 2006;46:375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder N, Martelin E, Lapatto R et al. Posttranslational inactivation of human xanthine oxidoreductase by oxygen under standard cell culture conditions. Am J Physiol Cell Physiol. 2003;285:C48–55. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao R, Xu M et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Chen F, Liu T et al. The role of oxidative stress in influenza virus infection. Microbes Infect. 2017;19:580–6. [DOI] [PubMed] [Google Scholar]
- Li W, Hulswit RJG, Widjaja I et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci USA. 2017;114:E8508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends. 2020;14:69–71. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yu J, Yang X et al. Ambroxol interferes with Pseudomonas aeruginosa quorum sensing. Int J Antimicrob Agents. 2010;36:211–5. [DOI] [PubMed] [Google Scholar]
- Luzhnikov EA, Ilyashenko KK, Pinchuk TP et al. The use of ‘Mexidol’ in comprehensive treating patients with acute exogenous poisoning. Bull Exp Biol Med. 2006;Supl. 1:190–8. [Google Scholar]
- Makino S, Keck JG, Stohlman SA et al. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986;57:729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor R, Igarashi M, Takada A. Influenza A virus M2 protein: roles from ingress to egress. Int J Mol Sci. 2017;18:E2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Loeches I, Sanchez-Corral A, Diaz E et al. Community-acquired respiratory coinfection in critically ill patients with pandemic 2009 influenza A(H1N1) virus. Chest. 2011;139:555–62. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Fujisawa T, Suga S et al. Outcomes and prognostic features of patients with influenza requiring hospitalization and receiving early antiviral therapy: a prospective multicenter cohort study. Chest. 2016;149:526–34. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A et al. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol. 1981;91:601–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Ujike M, Morikawa S et al. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci USA. 2005;102:12543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycotte P, Aryal S, Cummings CT et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Hornung F, Boyd KL et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill JB, Johnson M, Hurst S et al. Low dose chloroquine decreases insulin resistance in human metabolic syndrome but does not reduce carotid intima-media thickness. Diabetol Metab Syndr. 2019;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K, Kapikian AZ, Turner HC et al. Seroepidemiological studies of coronavirus infection in adults and children. Am J Epidemiol. 1970;91:585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee LA, Harmsen AG. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect Immun. 2006;74:6707–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinão IM, Sato EI, Andrade LE et al. Controlled trial with chloroquine diphosphate in systemic lupus erythematosus. Lupus. 1996;5:237–41. [DOI] [PubMed] [Google Scholar]
- Mengesha T, Makonnen E. Comparative efficacy and safety of chloroquine and alternative antimalarial drugs: a meta-analysis fromsix African countries. East Afr Med J. 1999;76:314–9. [PubMed] [Google Scholar]
- Metersky ML, Masterton RG, Lode H et al. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis. 2012;16:e321–31. [DOI] [PubMed] [Google Scholar]
- Milewska A, Nowak P, Owczarek K et al. Entry of human coronavirus NL63 into the cell. J Virol. 2018;92:e01933–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A, Zarebski M, Nowak P et al. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88:13221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnaul LJ, Matrosovich MN, Castrucci MR et al. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol. 2000;74:6015–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CJ, Earl JR, Trenam C, Blake DR. Reactive oxygen species and iron – a dangerous partnership in inflammation. Int J Biochem Cell Biol. 1995;27:109–22. [DOI] [PubMed] [Google Scholar]
- Mulangu S, Dodd LE, Davey RT Jr. et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto Y, Noda T, Kawakami E et al. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. 2013;87:2455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Lokugamage KG, Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res. 2016;96:165–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B, Chan C, Kien F et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol. 2005;86:1423–34. [DOI] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman BW, Adair BD, Yoshioka C et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80:7918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman BW, Kiss G, Kunding AH et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane K, Ahmed Z, Pervez H et al. Potential Treatment Options for COVID-19: A Comprehensive Review of Global Pharmacological Development Efforts. Cureus. 2020;12:e8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihara Y, Ge J, Shalev O et al. Desferrioxamine decreases NAD redox potential of intact red blood cells: evidence for desferrioxamine as an inducer of oxidant stress in red blood cells. BMC Clin Pharmacol. 2002;2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar M, Banerjee A, Feng C et al. Desialylation of airway epithelial cells during influenza virus infection enhances pneumococcal adhesion via galectins binding. Mol Immunol. 2015;65:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselova EA, Riabova OB, Leneva IA et al. Antiretroviral activity of a novel pyrimidyl-di(diazaspiroalkane) derivative. Acta Naturae. 2017;9:105–7. [PMC free article] [PubMed] [Google Scholar]
- Nowak D, Antczak A, Król M et al. Antioxidant properties of ambroxol. Free Radic Biol Med. 1994;16:517–22. [DOI] [PubMed] [Google Scholar]
- Obi AT, Tignanelli CJ, Jacobs BN et al. Empirical systemic anticoagulation is associated with decreased venous thromboembolism in critically ill influenza A H1N1 acute respiratory distress syndrome patients. J Vasc Surg Venous Lymphat Disord. 2019;7:317–24. [DOI] [PubMed] [Google Scholar]
- Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA. 1978;75:3327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olitsky PK, Gates FL. Experimental studies of the nasopharyngeal secretions from influenza patients. II. Filterability and resistance to glycerol. J Exp Med. 1921b;33:361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olitsky PK, Gates FL. Experimental studies of the nasopharyngeal secretions from influenza patients: I. Transmission experiments with nasopharyngeal washings. J Exp Med. 1921a;33:125–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi EE, Chew JS, Loh JP et al. In vitro inhibition of human influenza A virus replication by chloroquine. Virol J. 2006;3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald NC, Shooter RA, Curwen MP. Pneumonia complicating Asian influenza. Br Med J. 1958;2:1305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgen S, Kilinc OK, Selamoglu Z. Antioxidant activity of quercetin: a mechanistic review. Turkish JAF Sci Tech. 2016;4:1134–8. [Google Scholar]
- Pacher P, Novorozhkin A, Szabo C. Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol. Pharmacol Rev. 2006;58:87–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S, Powell D, Benboubetra M et al. Xanthine oxidoreductase in human mammary epithelial cells: activation in response to inflammatory cytokines. Biochim Biophys Acta. 1998;1381:191–202. [DOI] [PubMed] [Google Scholar]
- Palacios G, Hornig M, Cisterna D et al. Streptococcus pneumoniae coinfection is correlated with the severity of HINI pandemic influenza. PLoS One. 2009;4:e8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas G, Kiriaze IJ., Falagas ME. Insights into infectious disease in the era of Hippocrates. Int J Infect Dis. 2008;12:347–50. [DOI] [PubMed] [Google Scholar]
- Parsons L, Bouwman KM, Azurmendi H et al. Glycosylation of the viral attachment protein of avian coronavirus is essential for host cell and receptor binding. J Biol Chem. 2019;294:7797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelkina V, Yerovichenkov A, Pak S. Optimization of pathogenetic therapy in the diseases of viral and bacterial etiology. Farmateka. 2010;4:64–71. [Google Scholar]
- Pensaert MB. Porcine epidemic diarrhea. : Straw BE, D'Allaire S, Mengeling WL et al. (eds.) Diseases of Swine, 8th edn Ames, Iowa: Iowa State University Press, 1999, 179–85. [Google Scholar]
- Peteranderl C, Morales-Nebreda L, Selvakumar B et al. Macrophage-epithelial paracrine crosstalk inhibits lung edema clearance during influenza infection. J Clin Invest. 2016;126:1566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RFJ. Die Aetiologie der Influenza. Zeitschrift für Hygiene und Infektionskrankheiten. 1893;13:357–86. [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–28. [DOI] [PubMed] [Google Scholar]
- Pittet LA, Hall-Stoodley L, Rutkowski MR et al. Influenza virus infection decreases tracheal mucociliary velocity and clearance of Streptococcus pneumonia. Am J Respir Cell Mol Biol. 2010;42:450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss WB, Huecksteadt TP, Panus PC et al. Regulation of xanthine dehydrogenase and xanthine oxidase activity by hypoxia. Am J Physiol. 1996;270:L941–6. [DOI] [PubMed] [Google Scholar]
- Potter CW. A history of influenza. J Appl Microbiol. 2001;91:572–9. [DOI] [PubMed] [Google Scholar]
- Povalyaev D. The efficacy of adjuvant use low molecular weight heparins in patients with community-acquired pneumonia. Eur Respir J. 2014;44:P2503. [Google Scholar]
- Quan FS, Lee YT, Kim KH et al. Progress in developing virus-like particle influenza vaccines. Expert Rev Vaccines. 2016;15:1281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan N, Shaib H. Middle East respiratory syndrome coronavirus (MERS-CoV): a review. Germs. 2019;9:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves B, Banting G. Vacuolar ATPase inactivation blocks recycling to the trans-Golgi network from the plasma membrane. FEBS Lett. 1994;345:61–6. [DOI] [PubMed] [Google Scholar]
- Reiter RJ, Rosales-Corral S, Tan DX et al. Melatonin as a mitochondria-targeted antioxidant: one of evolution's best ideas. Cell Mol Life Sci. 2017;74:3863–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejmanek D, Hosseini PR, Mazet JA et al. Evolutionary dynamics and global diversity of influenza A virus. J Virol. 2015;89:10993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Zhang Y, Li J et al. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci Rep. 2015;5:11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivinoja A, Hassinen A, Kokkonen N et al. Elevated Golgi pH impairs terminal N-glycosylation by inducing mislocalization of Golgi glycosyltransferases. J Cell Physiol. 2009;220:144–54. [DOI] [PubMed] [Google Scholar]
- Rivinoja A, Kokkonen N, Kellokumpu I et al. Elevated Golgi pH in breast and colorectal cancer cells correlates with the expression of oncofetal carbohydrate T-antigen. J Cell Physiol. 2006;208:167–74. [DOI] [PubMed] [Google Scholar]
- Rogers DF. Mucoactive agents for airway mucus hypersecretory diseases. Respir Care. 2007;52:1176–93. [PubMed] [Google Scholar]
- Rojek JM, Perez M, Kunz S. Cellular entry of lymphocytic choriomeningitis virus. J Virol. 2008;82:1505–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette M, Page S, Bryant R et al. Xanthine oxidoreductase is asymmetrically localized on the outer surface of human endothelial and epithelial cells in culture. FEBS Lett. 1998;426:397–401. [DOI] [PubMed] [Google Scholar]
- Samji T. Influenza A: understanding the viral life cycle. Yale J Biol Med. 2009;82:153–9. [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martinez A, Beavan M, Gegg ME et al. Parkinson disease-linked GBA mutation effects reversed by molecular chaperones in human cell and fly models. Sci Rep. 2016;6:31380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín-Suárez C, Soto-Otero R, Sánchez-Sellero I et al. Antioxidant properties of dimethyl sulfoxide and its viability as a solvent in the evaluation of neuroprotective antioxidants. J Pharmacol Toxicol Methods. 2011;63:209–15. [DOI] [PubMed] [Google Scholar]
- Schmidtke M, Wutzler P, Makarov V. Novel opportunities to study and block interactions between viruses and cell surface heparan sulfates. Lett Drug Design Discov. 2004;1:35–44. [Google Scholar]
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C, Rohde W, Von Hoyningen V et al. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. [DOI] [PubMed] [Google Scholar]
- Schrezenmeier E, Dorner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16:155–66. [DOI] [PubMed] [Google Scholar]
- Schwarz KB. Oxidative stress during viral infection: a review. Free Radic Biol Med. 1996;21:641–9. [DOI] [PubMed] [Google Scholar]
- Seifart C, Clostermann U, Seifart U et al. Cell-specific modulation of surfactant proteins by ambroxol treatment. Toxicol Appl Pharmacol. 2005;203:27–35. [DOI] [PubMed] [Google Scholar]
- Sencio V, Barthelemy A, Tavares LP et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020;30:2934–47.e6. [DOI] [PubMed] [Google Scholar]
- Shahwan K, Hesse M, Mork AK et al. Sialic acid binding properties of soluble coronavirus spike (S1) proteins: differences between infectious bronchitis virus and transmissible gastroenteritis virus. Viruses. 2013;5:1924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepardson K, Larson K, Cho H et al. A novel role for PDZ-binding motif of influenza A virus nonstructural protein 1 in regulation of host susceptibility to postinfluenza bacterial superinfections. Viral Immunol. 2019;32:131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Aoki H, Tsurumi T et al. Mechanisms of uncoating of influenza B virus in MDCK cells: action of chloroquine. J Gen Virol. 1983;64:1149–56. [DOI] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–6. [DOI] [PubMed] [Google Scholar]
- Shivakumar S, Panigrahi T, Shetty R et al. Chloroquine protects human corneal epithelial cells from desiccation stress induced inflammation without altering the autophagy flux. Biomed Res Int. 2018;2018:7627329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KR, Kasper J, van der Aa S et al. Influenza virus damages the alveolar barrier by disrupting epithelial tight junctions. Eur Respir J. 2016;47:954–66. [DOI] [PubMed] [Google Scholar]
- Silvestri M, Rossi GA. Melatonin: its possible role in the management of viral infections – a brief review. Ital J Pediatr. 2013;39:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Zmora P, Gierer S et al. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J. An overview of influenza haemagglutinin and neuraminidase. Biologicals. 2009;37:177–8. [DOI] [PubMed] [Google Scholar]
- Sliva J, Pantzartzi CN, Votava M. Inosine pranobex: a key player in the game against a wide range of viral infections and non-infectious diseases. Adv Ther. 2019;36:1878–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots TP, McErlean P, Speicher DJ et al. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CL, Shaler CR, McCormick S et al. Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J Immunol. 2010;184:2048–56. [DOI] [PubMed] [Google Scholar]
- Smith EC, Sexton NR, Denison MR. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol. 2014;1:111–32. [DOI] [PubMed] [Google Scholar]
- Smith W, Andrewes CH, Laidlaw PP. A virus obtained from influenza patients. Lancet. 1933;222:66–8. [Google Scholar]
- Sodeman WA, Doerner AA, Gordon EM et al. Chloroquine in hepatic amebiasis. Ann Intern Med. 1951;35:331–41. [DOI] [PubMed] [Google Scholar]
- Sola I, Almazán F, Zúñiga S et al. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2:265–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–33. [DOI] [PubMed] [Google Scholar]
- Son D, Chung MH. In vitro synergism between chloroquine and antibiotics against Orientia tsutsugamushi. Infect Chemother. 2014;46:182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler AR. Influenza. Flu, grippe, avian influenza, grippe aviaire, fowl plaque, swine influenza, hog flu, pig flu, equine influenza, canine influenza February 01, 2016. [cited 2020 April 07]. http://www.cfsph.iastate.edu/Factsheets/pdfs/influenza.pdf.
- Spiekermann S, Landmesser U, Dikalov S et al. Electron spin resonance characterization of vascular xanthine and NAD(P)H oxidase activity in patients with coronary artery disease. Circulation. 2003;107:1383–9. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Mohamed M, Kato H. Melatonin in bacterial and viral infections with focus on sepsis: a review. Recent Pat Endocr Metab Immune Drug Discov. 2012;6:30–9. [DOI] [PubMed] [Google Scholar]
- Sritharan M. Iron and bacterial virulence. Indian J Med Microbiol. 2006;24:163–4. [PubMed] [Google Scholar]
- Stavrinides J, Guttman DS. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J Virol. 2004;78:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA, de la Torre JC, Holland JJ. High nucleotide substitution error frequencies in clonal pools of vesicular stomatitis virus. J Virol. 1989;63:2063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. [DOI] [PubMed] [Google Scholar]
- Stetinová V, Herout V, Kvetina J. In vitro and in vivo antioxidant activity of ambroxol. Clin Exp Med. 2004;4:152–8. [DOI] [PubMed] [Google Scholar]
- Sugrue RJ, Hay AJ. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Metzger DW. Influenza infection suppresses NADPH oxidase-dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin-resistant Staphylococcus aureus infection. J Immunol. 2014;192:3301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Okamoto M, Ohmi A et al. Detection of human coronavirus-NL63 in children in Japan. Pediatr Infect Dis J. 2005;24:645–6. [DOI] [PubMed] [Google Scholar]
- Szczepanski A, Owczarek K, Bzowska M et al. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: receptors and attachment factors. Viruses. 2019;11:E328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DX, Kormaz A, Reiter RJ et al. Ebola virus disease: potential use of melatonin as a treatment. J Pineal Res. 2014;57:381–4. [DOI] [PubMed] [Google Scholar]
- Tashiro M, Ciborowski P, Reinacher M et al. Synergistic role of staphylococcal proteases in the induction of influenza virus pathogenicity. Virology. 1987;157:421–30. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation and pandemic formation. Cell Host Microbe. 2010;7:440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM. Studies on survival of influenza virus between epidemics and antigenic variants of the virus. Am J Public Health Nations Health. 1949;39:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tektonidou MG, Andreoli L, Limper M et al. EULAR recommendations for the management of antiphospholipid syndrome in adults. Ann Rheum Dis. 2019;78:1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada LS, Piermattei D, Shibao GN et al. Hypoxia regulates xanthine dehydrogenase activity at pre- and posttranslational levels. Arch Biochem Biophys. 1997;348:163–8. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. Elife. 2014;3:e03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To EE, Erlich JR, Liong F et al. Mitochondrial reactive oxygen species contribute to pathological inflammation during influenza A virus infection in mice. Antioxid Redox Signal. 2020;32:929–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici MA, Walls AC, Lang Y et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26:481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KN, Chen GW. Influenza genome diversity and evolution. Microbes Infect. 2011;13:479–88. [DOI] [PubMed] [Google Scholar]
- Tseng YT, Wang SM, Huang KJ et al. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J Biol Chem. 2010;285:12862–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell DA, Almeida JD, Cunningham CH et al. Coronaviridae. Intervirology. 1975;5:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell DA, Bynoe ML. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966;1:76–7. [DOI] [PubMed] [Google Scholar]
- van der Sluijs KF, van Elden LJ, Nijhuis M et al. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J Immunol. 2004;172:7603–9. [DOI] [PubMed] [Google Scholar]
- Varga ZT, Ramos I, Hai R et al. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7:e1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasin AV, Temkina OA, Egorov VV et al. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 2014;185:53–63. [DOI] [PubMed] [Google Scholar]
- Vincent MJ, Bergeron E, Benjannet S et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visher E, Whitefield SE, McCrone JT et al. The mutational robustness of influenza A virus. PLoS Pathog. 2016;12:e1005856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivino FB, Carsons SE, Foulks G et al. New treatment guidelines for Sjogren's disease. Rheum Dis Clin North Am. 2016;42:531–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina TA. Antioxidant mexidol. The basic neuropsychotropic effects and mechanism of action. Psychopharmakol Biol Narkol. 2001;1:2–12. [Google Scholar]
- Wang M, Cao R, Zhang L et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020a;30:269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Wu L et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020b:S0092–8674(20)30338-X., DOI: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waqar T, Khushdil A, Haque K. Efficacy of chloroquine as a first line agent in the treatment of uncomplicated malaria due to Plasmodium vivax in children and treatment practices in Pakistan: a pilot study. J Pak Med Assoc. 2016;66:30–3. [PubMed] [Google Scholar]
- Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Berndsen ZT, Raghwani J et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11:2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Chu DKW, Peiris JSM et al. A case for the ancient origin of coronaviruses. J Virol. 2013;87:7039–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S, Haupt R, Logue J et al. FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro. bioRxiv 2020.03.25.008482 [Preprint] 2020; [cited 2020 May 06]. Available from: biorxiv.org/content/10.1101/2020.03.25.008482v1.
- Wood PM. The potential diagram for oxygen at pH 7. Biochem J. 1988;253:287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood PM. The two redox potentials for oxygen reduction to superoxide. Trends Biochem Sci. 1987;12:250–1. [Google Scholar]
- Wu W, Li R, Li X et al. Quercetin as an antiviral agent inhibits influenza A virus (IAV) entry. Viruses. 2015;8:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G, Gong L, Yuan C et al. A structural mechanism of flavonoids in inhibiting serine proteases. Food Funct. 2017;8:2437–43. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Matsuyama S, Hoshino T et al. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. bioRxiv 2020.04.06.026476 [Preprint] 2020; [cited 2020 May 06]. Available from: biorxiv.org/content/10.1101/2020.04.06.026476v1.
- Yamaya M, Nishimura H, Nadine LK et al. Ambroxol inhibits rhinovirus infection in primary cultures of human tracheal epithelial cells. Arch Pharm Res. 2014;37:520–9. [DOI] [PubMed] [Google Scholar]
- Yang B, Yao DF, Ohuchi M et al. Ambroxol suppresses influenza-virus proliferation in the mouse airway by increasing antiviral factor levels. Eur Respir J. 2002;19:952–8. [DOI] [PubMed] [Google Scholar]
- Yan Y, Zou Z, Sun Y et al. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York IA, Stevens J, Alymova IV. Influenza virus N-linked glycosylation and innate immunity. Biosci Rep. 2019;39:BSR20171505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Ohnishi S. Uncoating of influenza virus in endosomes. J Virol. 1984;51:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan H, Arabyan E, Oo A et al. Flavonoids: promising natural compounds against viral infection. Arch Virol. 2017;162:2539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu Y. Potential interventions for novel coronavirus in China: a systemic review. J Med Virol. 2020;92:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang X, Ni L et al. COVID-19: melatonin as potential adjuvant treatment. Life Sci. 2020;250:117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Sun L, Xiong J et al. Semiaquatic mammals might be intermediate hosts to spread avian influenza virus from avian to human. Sci Rep. 2019;9:11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Li S, Xue F et al. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J Virol. 2008;82:8647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yamada Y, Fung TS et al. Identification of N-linked glycosylation sites in the spike protein and their functional impact on the replication and infectivity of coronavirus infectious bronchitis virus in cell culture. Virology. 2018;513:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Chan JF, Azhar EI et al. Coronaviruses – drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–47. [DOI] [PMC free article] [PubMed] [Google Scholar]


