ABSTRACT
Coronavirus disease 2019 (COVID-19) is a viral pneumonia, responsible for the recent pandemic, and originated from Wuhan, China, in December 2019. The causative agent of the outbreak was identified as coronavirus and designated as severe acute respiratory syndrome coronavirus 2 (SARS- CoV-2). Few years back, the severe acute respiratory syndrome coronavirus (SARS- CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) were reported to be highly pathogenic and caused severe infections in humans. In the current situation SARS-CoV-2 has become the third highly pathogenic coronavirus that is responsible for the present outbreak in human population. At the time of this review, there were more than 14 007 791 confirmed COVID-19 patients which associated with over 597 105 deaths in more then 216 countries across the globe (as reported by World Health Organization). In this review we have discussed about SARS-CoV, MERS-CoV and SARC-CoV-2, their reservoirs, role of spike proteins and immunogenicity. We have also covered the diagnosis, therapeutics and vaccine status of SARS-CoV-2.
Keywords: SARS-CoV, MERS-CoV, SARS-CoV-2, COVID 19, spike protein and therapeutics
An updated review of emerging cornoaviruses SARS-CoV, MERS-CoV, SARS-CoV-2, and potential therapeutic and preventive approaches.
INTRODUCTION
On December 31, 2019, several cases of severe pneumonia were reported from Wuhan, China. The causative agent of the outbreak was identified as Betacoronavirus. Genome sequencing revealed that it is closely related to the SARS-CoV (Severe Acute Respiratory Syndrome Coronavirus) which had emerged in 2003, and is designated as SARS-CoV-2 (Gorbalenya et al. 2020; Zhou et al. 2020). In a very short duration, more than 80 000 infectious cases including more than 3000 deaths were reported in China as on March 15, 2020. At the time of this review (18 May, 2020), the disease, termed as COVID-19 (Corona Virus Disease 2019), had already become pandemic and spread to more than 216 countries and territories, including community transmissions in countries like the United States, Germany, France, Spain, Japan, Singapore, South Korea, Iran, Italy and India. As on July 19, more than 14 007 791 cases and 597 105 deaths had been reported globally, with the rapid growth of numbers in many countries. For the up-to-date information about COVID-19, visit the World Health organization (WHO) website (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
The bats are likely to be the origin of SARS-CoV-2, but the role of an intermediate host cannot be ruled out at this stage. Initial studies showed that SARS-CoV-2, can use angiotensin-converting enzyme 2 (ACE2) from bats, cats, civet cats, swine, ferrets, non-human primates (NHPs) and humans as a receptor (Letko, Marzi and Munster 2020; Wan et al. 2020a; Zhou et al. 2020). A pet dog in Hong Kong and a tiger in Bronx Zoo in the United State of America tested positive with SARS-CoV-2 infection, indicating that canine ACE2 can also be recognized by SARS-CoV-2. Pangolins, which are endangered animals and are illegally imported into southern China (Guangdong and Guangxi provinces), have been considered as a potential intermediate host (Lam et al. 2020; Zhang et al. 2020b). The initial reports showed that in most of the COVID-19 cases there was mild to moderate infection. However, approximately 20% of the cases were reported severe (Chen et al. 2020; Wang et al. 2020a). In this review, we will discuss about SARS-CoV, MERS-CoV and SARC-CoV-2. We have also discussed about various reservoirs, associated with them. In the end, we have covered the role of spike proteins and their immunogenicity along with the diagnosis, therapeutics and vaccine status of SARS-CoV-2.
CORONAVIRUSES: SARS-CoV, MERS-CoV and SARC-CoV-2
Zoonotic coronaviruses are becoming a global concern as there was emergence of earlier two coronaviruses, SARS-CoV and MERS-CoV (Middle East Respiratory Syndrome Coronavirus) which created a havoc and recently emergence of the third highly pathogenic SARC-CoV-2. It has been observed that members of the family Coronaviridae are known to infect a wide range of vertebrates and humans. Before the outbreak of SARS (Severe Acute Respiratory Syndrome), only two coronaviruses including hCoV-229E and hCoV-OC43 were known to infect humans. However, post-SARS outbreak, the SARS coronavirus (SARS-CoV), human coronavirus hCoV-NL63, human coronavirus hCoV-HKU1 and MERS-CoV have been isolated from humans. Similar to SARS-CoV and MERS-CoV, the newly isolated SARC-CoV-2 is highly pathogenic in humans and causes severe acute respiratory distress (Shi, Guo and Rottier 2016).
The genomes of coronaviruses consist of a positive and single-stranded RNA genome of about 30 kb. The 5′ terminus encodes the enzyme viral replicase/transcriptase, which is involved in virus replication, whereas the 3′ terminus encodes viral structural proteins and virus group specific accessory proteins. Functional studies of these viral proteins in detail are essential for antiviral drug screening and vaccine development.
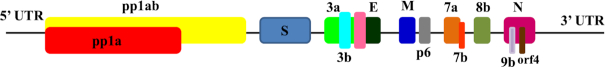
The earliest available genome sequencing data of SARS-CoV-2 made it possible to compare it with the genomes of SARS-CoV and other coronaviruses. Results showed that SARS-CoV-2 belongs to the genus Betacoronavirus and subgenus Sarbecovirus, which also includes SARS-CoV. However, the MERS-CoV belongs to another subgenus, Merbecovirus (Lu et al. 2020; Zhou et al. 2020). The comparison study also showed that there is 79% nucleotide similarity between SARS-CoV-2 and SARS-CoV. The essential surface glycoprotein of SARS-CoV-2 known as spike (S) protein, essential for host cell receptor binding, showed only 72% similarity with SARS-CoV at the nucleotide level. The genomic organization of SARS-CoV-2 resembles those of other betacoronaviruses, including 5’-ORF1ab-S (Surface glycoprotein)-ORF3a-E (Envelope)-M (Membrane glycoprotein)-N-3’ as shown in Fig. 1. Comparative genome analysis of RaTG13, a virus from a Rhinolophusaffinis (i.e. horseshoe) bat sampled from Yunnan province in China in 2013, with SARS-CoV-2, showed that SARS-CoV-2 has 96% similarity at the nucleotide sequence level (Zhou et al. 2020). Although the SARS-CoV-2 and RaTG13 have high similarity, yet they differ in some genomic features, such as SARS-CoV-2 contains a polybasic (furin) cleavage site insertion (residues PRRA) between the S1 and S2 subunits of the surface glycoprotein S protein (Coutard et al. 2020). Polybasic insertion may increase the infectivity of the virus, as it is absent in other related betacoronaviruses. However, similar polybasic insertions are observed in different human coronaviruses, such as HCoV-HKU1 and highly virulent strains of avian influenza viruses. Therefore, whether polybasic insertion between S1 and S2 subunits of S protein occurs due to the natural evolution in SARS-CoV-2 or by other means is going to be the topic of debate in the future. However, an independent insertion(s) of the amino acids PAA at the S1/S2 cleavage site was also observed in aRmYN02 virus (having 72% similarity in spike protein and 97% similarity in replicase nucleotide) isolated from Rhinolophus bat in Yunnan province in mid-2019, indicating that these insertion events may be a natural part of ongoing coronavirus evolution (Zhou et al. 2020).
Figure 1.
Schematic representation of the genomic organization of SARS-CoV-2. The orf1ab and orf1a encodes pp1ab and pp1a nonstructural proteins, respectively. The structural proteins are encoded by the structural genes, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) genes.
The receptor-binding domain (RBD) of S protein is essential to interact with the ACE2 receptor present on the surface of the target cells of the host. Therefore, comparative sequence analysis was performed and results showed that there is 85% similarity in RBD between SARS-CoV-2 and RaTG13, but they share only one amino acid among the six key amino acid residues. Further, due to the proteinaceous nature of spike, structural comparisons were also performed, suggesting that the RBD domain of the SARS-CoV-2 is well suited to interact with the human ACE2 receptor. Interestingly the same receptor was also utilized by SARS-CoV to cause infection (Wrapp et al. 2020).
The SARS-CoV-2 is closely related to SARS-CoV and MERS-CoV having bat as reservoirs, but there are huge biological differences in the former as compared to the other two. The SARS-CoV-2 is markedly more infectious and has very different epidemiological dynamics. Moreover, the MERS-CoV has never been able to fully adapt to human transmission (Sabir et al. 2016), whereas there is the remarkable local and global spread of SARS-CoV-2.
As in the case of SARS and MERS, the intermediate host including civets and camels, respectively, played an important role and may be considered as a true reservoir host (Sabir et al. 2016). Therefore, due to the ecological separation between a bat (reservoir) and humans, ‘intermediate’ or ‘amplifying’ mammalian host is a must to acquire mutations in SARS-CoV-2, and is essential for the efficient human transmission.
To determine the intermediate host, it is essential to perform a wider sampling of animals that live close to human populations or available in wet markets for human consumption. Surprisingly, there was discovery of viruses, closely related to SARS-CoV-2 from the Malayan pangolins (Manisjavanica) that are illegally imported into southern China (Guangdong and Guangxi provinces). It has been observed that RBD domain of Guangdong pangolin viruses are particularly closely related to SARS-CoV-2. There is a 97% amino acid sequence similarity and contain all six critical key mutations that are essential for binding to the ACE2 receptor in these viruses. However, the rest of the genome is highly divergent from SARS-CoV-2.
Hence, the evolution of coronaviruses in animal reservoirs as well as in intermediate hosts is required to explain the emergence of SARS-CoV-2 in humans. It might be possible that due to its asymptomatic infection, the virus could have acquired some of its essential mutations during a period the ‘‘cryptic’’ spread in humans before it was first detected in December 2019.
Recombination is another possibility, which cannot be ruled out as sarbeviruses, and coronaviruses experience widespread recombination. The genome of sarbeviruses experience recombination at multiple locations, including spike protein. There are studies, which showed that recombination does occur among SARS-CoV-2, RaTG13 and the Guangdong pangolin CoVs (Lam et al. 2020). The genome of RmYN02 too has been impacted by recombination (Zhou et al. 2020). Because of the small recombinant region, which may likely change as we increase the sample size of viruses related to SARS-CoV-2, it would be difficult to determine the pattern and genomic ancestry of recombination.
RESERVOIRS
A total of 8422 cases with 916 deaths in 29 countries including China were reported due to the human respiratory disease during 2002–2003, caused by the SARS-CoV. The studies showed that bats acted as a natural reservoir of SARS-CoV that caused the outbreaks (Chan-Yeung and Xu 2003). Later, the SARS-CoV like similar antibody and genomic sequences were also discovered in Rhinolophus bat, such as in R. ferrumequinum, R. pearsoni, R. sinicus, R. pusillus and R. macrotis (Lau et al. 2005). The comparative study of the genomes revealed that bat SARS-like CoVs (SL-CoV) have 78–92% nucleotide sequence identities with SARS-CoV and also among themselves, and hence display great genetic diversity. Further, the phylogenetic analysis pointed out that Rhinolophus bat might be the direct progenitor of human SARS-CoV (Hon et al. 2008).
Various SARS-CoV groups were isolated in different epidemic periods and hosts. Several methods have been adopted to investigate the selective pressure. Results have shown that the most functional proteins of SARS-CoV adopted the stepwise adaptive evolutionary pathway. For example, the spike protein showed strong positive selection in the early as well as middle phases, and not in the late phase. However, the replicase enzyme experienced positive selection only in humans, and assembly proteins experienced the same in the middle and late phases. Interestingly, no such positive selection was observed in any proteins of bat SARS-like-CoV. However, specific amino acid sites that may be the targets of positive selection in each group were identified (Tang et al. 2009).
Later in 2010, a study suggested the presence of two distinct genotypes of Bt-SLCoV in R. sinicus (i.e. Rp3/Rs672 and HKU3/Rs806). The results also showed the evidence for the recombinant origin of Rp3 and Rs672. The phylogenetic study showed that their major parent has a relatively closer relationship with Hu-SCoVs. Therefore, there may be a possibility for the presence of a Bt-SCoV lineage in R. sinicus, that may have Hu-SCoVs as their direct ancestor, as reported earlier (Hon et al. 2008). However, these speculations are based on studies done on limited strains only, therefore an extensive analysis for the prevalence of such genotype is required for its credibility (Yuan et al. 2010).
In 2012, globally, 64 human cases were confirmed resulting in 38 deaths by June 17, 2013, by a disease having symptoms similar to SARS, that emerged in Saudi Arabia (WHO 2013). Later, it was found that the disease was caused by a virus designated as a novel human coronavirus, MERS-CoV, phylogenetic data showed that it belonged to lineage C of the Betacoronavirusgenus and was highly similar to bat coronaviruses HKU4 (Tylonycterispachypus) and HKU5 (Pipistrelluspipistrellus; Lau et al. 2013). Comparative genomic results showed that MERS-CoV has a 50% nucleotide identity in the entire genome with HKU4 and HKU5. Moreover, the RNA dependent RNA polymerase (RdRp) gene has 82% nucleotide identity. Later, while studying the mode of entry into the cell it was confirmed that MERS-CoV uses dipeptidyl peptidase 4 (DPPIV), also known as CD26. As DPPIV is evolutionarily conserved among mammals, therefore MERS-CoV can infect a broad range of mammalian cells (humans, pigs, monkeys and bats) and may be efficient in cross-host transmission (Raj et al. 2013).
Similar to the case for SARS-CoV and MERS-CoV (Li et al. 2005), the bat is still a probable species for origin of SARS-CoV-2, as it shares 96% whole-genome identity with a bat CoV, BatCoV RaTG13, from Rhinolophusaffinis from Yunnan Province (Zhou et al. 2020). However, SARS-CoV and MERS-CoV before entering humans pass through intermediate hosts, such as civets or camels(Cui, Li and Shi 2019). This fact indicates that SARS-CoV-2 was probably transmitted to humans by other animals. By comparing the overall genome identity, it was concluded that Pangolin-CoV genome sequence is 90.55% identical to RaTG13 and 91.02% identical to SARS-CoV-2. However, there was 96.2% identity between SARS-CoV-2 and RaTG13 (Zhou et al. 2020). Other SAR like CoVs (SL-CoVs) are also showed similarity with Pangolin-CoV, as its was 85.65% similar to ZXC21 and 85.01% with ZC45.
In a comparative genome analysis between Pangolin-CoV and SARS-CoV-2 (GenBank: MN908947), result showed 45.8–100% coverage range (average coverage 76.9%). Moreover, Pangolin-CoV genes shared high average nucleotide (93.2%) and amino acid identity (94.1%) with SARS-CoV-2 (GenBank MN908947). Similar results were obtained when Pangolin-CoV genes were compared with RaTG13 where 92.8% nucleotide and 93.5% amino acid identity was observed (Zhang, Wu and Zhang 2020). Interestingly, some of the Pangolin-CoV genes showed higher amino acid sequence identity to SARS-CoV-2 genes than to RaTG13genes. For example orf1b of Pangolin-CoV 73.4%, the spike (S) protein 97.5%, orf7a 96.9% and orf10 is 97.3% identical to SARS-CoV-2. Similarly, orf1b 72.8%, the spike (S) protein 95.4%, orf7a 93.6% and orf10 is 94.6% identical to RaTG13. The high S protein amino acid identity governs the functional similarity between SARS-CoV-2 and Pangolin-CoV.
A comprehensive phylogenetic analysis was performed based on the nucleotide sequences of whole-genome sequence, RNA-dependent RNA polymerase gene (RdRp), non-structural protein genes ORF1a and ORF1b, and main structural proteins encoded by the S and M genes. Results showed that in all phylogenetic trees, Pangolin-CoV, RaTG13 and SARS-CoV-2 were clustered into a well-supported group designated as ‘‘SARS-CoV-2 group’’ which represents a novel Betacoronavirus group. However, within this group, RaTG13 and SARS-CoV-2 were grouped together, and Pangolin-CoV was their closest common ancestor (Zhang, Wu and Zhang 2020). Recently, an extensive study including localized genomic analysis and the pattern of evolutionary recombination was done. The results showed that the strong purifying selection among coronaviruses from distinct host species as well as cross-species infections is responsible for the origin of SARS CoV-2 (Li et al. 2020). Therefore, we may summarize the origin and intermediate hosts of SARS-CoV, MERS-CoV and SARS-CoV-2 as shown in Fig. 2.
Figure 2.
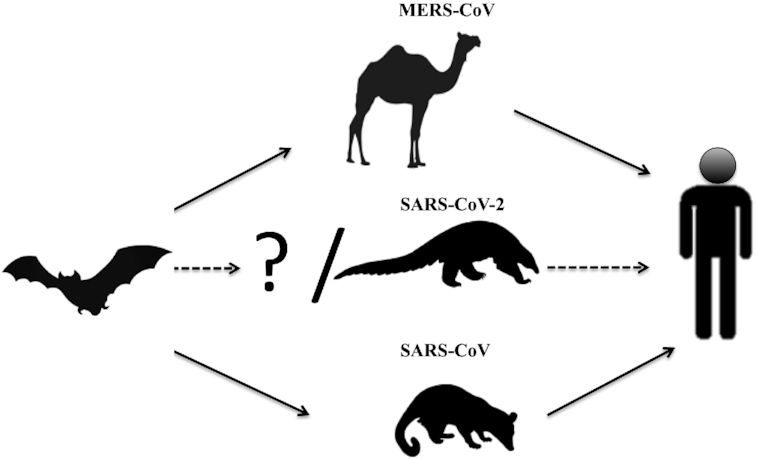
Prospective interspecies transmission routes of MERS-CoV, SARS-CoV-2 and SARS-CoV.
ROLE OF SPIKE PROTEIN AND ITS IMMUNOGENICITY
SARS-CoV is responsible to cause severe acute respiratory syndrome. SARS-CoV utilizes angiotensin-converting enzyme 2 (ACE2) receptor present on the surface of host cells, as shown in Fig. 3. SARC like-CoVs and SARS-CoVs have identical genetic organizations with high sequence identities. The schematic representation of spike protein (S) from SARS-CoV-2 is shown in Fig. 4. However, there is some important exception at the N’ terminus of spike protein (S), essential for receptor binding in CoVs. There is a study to investigate the receptor usage by full-length S of SL-CoV, SARS-CoV and a series of S chimeras. Different ACE2 receptors from human, civet, or horseshoe bat were expressed in cell lines by using human immunodeficiency virus-based pseudovirus system. Several important observations were made in the study. First, the SL-CoV S was unable to use any of the three receptors. Second, the SARS-CoV S was unable to enter the cells expressing bat ACE2. Third, the chimeric S enters the cells with different efficiencies for different constructs via human ACE2. Fourth, a minimal insert region (amino acids 310–518) was sufficient to convert the SL-CoV S from non-ACE2 binding to human ACE2 binding, indicating that the SL-CoV S is largely compatible with SARS-CoV S protein, both in structure and function (Ren et al. 2008).
Figure 3.
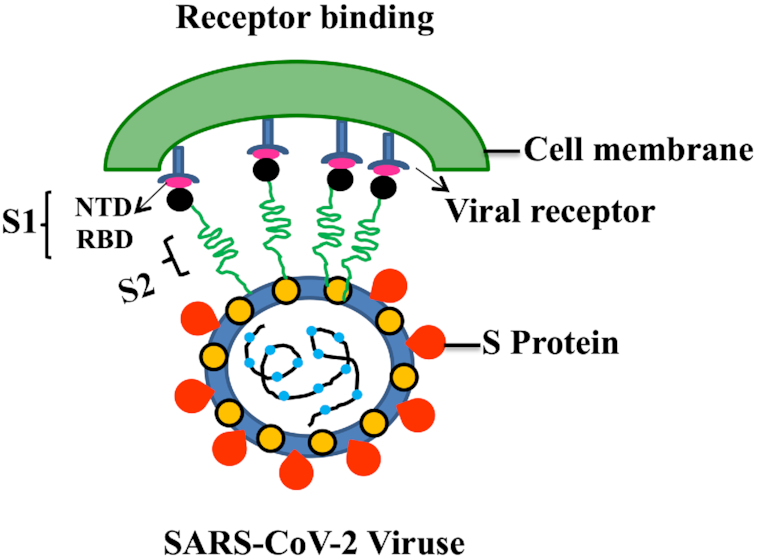
Schematic representation SARS-CoV-2 intercation with human receptor. The SARS-CoV-2 binds to a ACE2 through the receptor-binding domain (RBD) in the S1 domain of S protein, followed by fusion with cell membrane.
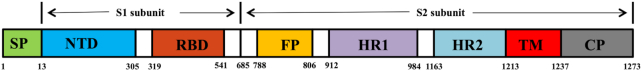
Figure 4.
Diagrammatic representation of functional domains of S protein in SAS-CoV-2. The SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide, HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CP, cytoplasmic domain.
Detailed structural study of human SARS-CoV RBD complexed with human ACE2 receptors was performed. Results revealed that it is the truncations in the receptor-binding motif (RBM) region of SL-CoV spike protein, which abolished its human ACE2-binding ability (Li 2008)). Therefore, we may hypothesize that the SL-CoV found in horseshoe bats is not the direct ancestor of human SARS-CoV. Moreover, it has been observed that the human SARS-CoV, as well as its closely related civet SARS-CoV spike proteins, were not able to use a horseshoe bat (R. pearsoni) ACE2 as a receptor for cell entry (Ren et al. 2008). These findings highlight a critical missing link (an intermediate host) in the bat-to-civet/human transmission chain of SARS- CoV (Hou et al. 2010).
An earlier study showed that ACE2 from horseshoe bat could not function as a receptor for SARS-CoV. However, changing 3 amino acids (40, 41 and 42 amino acids) from SHE to FYQ was found adequate to convert the nonfunctional bat ACE2 into a fully active receptor for SARS-CoV. Further, an ACE2 molecule from a fruit bat, which naturally has the FYQ motif, supports SARS-CoV entry into the cells thus causing infection. This result indicates that there must be a wide host range for SARS-CoV-related viruses among different bat populations (Yuan et al. 2010).
In the case of SARS-CoV-2, the structural bioinformatics approaches accurately predicted that SARS-CoV-2 spikes bind human ACE2 (Wan et al. 2020b). When cell lines over-expressed the transmembrane protein ‘angiotensin-converting enzyme 2’ (ACE2) from humans, bats, pig or civet cats and were infected with SARS- CoV-2, results showed that they became hypersensitized to infection, thus indicating that ACE2 is a SARS-CoV-2 receptor (Zhou et al. 2020). The binding studies also revealed that receptor-binding domains on the SARS-CoV-2 S proteins have a high affinity to human ACE2 (Wrapp et al. 2020) which makes it more virulent. However, apart from ACE2 interaction, the N-terminal domain (NTD) of the SARS-CoV-2 S proteins may show binding to alternative host-cell receptors (Zhou et al. 2020).
SARS-CoV-2 S proteins have also acquired a furin protease cleavage site, by acquiring several basic residues (RRAR/S). The SARS-CoV-2 furin substrate site facilitates the prime cleavage step, which further sensitizes S proteins for subsequent activation of cleavages occurring on susceptible target cells, and finally facilitates virus to enter the cells and cause infection (Qing and Gallagher 2020).
Human SARS-CoV and SARS-like coronavirus (SL-CoV) in bats have a similar genomic organization; therefore their corresponding gene products are highly conserved. As far as S protein is concerned, it has only a 63–64% sequence identity at the N-terminal region. It is the N-terminal region of coronavirus S protein that is responsible for receptor interaction. When the immunogenicity of the SL-CoV S protein was analyzed and compared with that of SARS-CoV, results revealed that they shared only a limited number of immunogenic epitopes in their S proteins. Moreover, major neutralization epitopes were also different (Zhou et al. 2009).
In another study, a pseudovirus expressing full-length SL-CoV S protein was used to raise mouse sera and monoclonal antibody. Series of constructs expressing truncated S protein were prepared and analyzed with ELISA, as well as western blot. Results showed that amino acids 280–455 and amino acids 561–666 are two immunogenic determinants in mice. Further, it was also shown that 280–455 amino acids are more immunogenic, as it was recognized by polyclonal as well as monoclonal antibodies. Earlier studies also showed that amino acids 528–635 from SARS-CoV are immunodominant determinants (He et al. 2004). Due to the high sequence similarity with SL-CoV S protein in the same region, the amino acids 561–666 of S protein also demonstrated immune response in mouse (Zhou et al. 2013).
In a cross-reactivity test with antibodies against RBD domain SARS-CoV, some of the SL-CoV strains (WIVI) have shown positive results whereas some strains (SHC014) failed too. This difference in reactivity is due to the low sequence identity in the RBD domain of SHC014 and high sequence identity in the RBD domain of WIVI with RBD domain of SARS-CoV (Zeng et al. 2017).
DIAGNOSIS AND THERAPEUTICS FOR SARS-CoV-2 INFECTION
Detection of novel coronavirus is done by different molecular biology techniques including real-time reverse transcription PCR (rRT-PCR), reverse transcription PCR (RT-PCR), reverse transcription loop-mediated isothermal amplification (RT-LAMP), multiplex nucleic acid amplification, real-time RT-LAMP and microarray-based assays (Zhang et al. 2020). WHO also recommended a pan-coronavirus assay for characterization and confirmation.).
Viral culture and RT-PCR are among the most efficient and reliable methods for the diagnosis of SARS-CoV-2 infection. These methods are time consuming and generally takes hours to detect the nucleic acid and many days to isolate the virus from the samples. Apart from that, specialized equipments and expertise are also required. To overcome these limitations, rapid diagnosis of SARS-CoV-2 infection can be done with rapid antigen detection (RAD) tests. In RAD tests, the immobilized SARS-CoV-2 antibody on the device can detect viral antigen in the sample. The results of RAD tests are prompt and interpreted without specialized instrument. Hence, RAD tests could be beneficial reduce the workload in diagnostic laboratories and hospitals (Mak et al. 2020). However, as per the WHO, RAD tests for SARS − COV-2 antigen detection, further needs evaluation and is not recommended for clinical diagnosis (Laboratory Testing Strategy Recommendations for COVID-19: Interim Guidance2020).
The immune response to SARS-CoV2 in the early weeks of the infection can be detected using Enzyme-Linked Immunosorbant Assay (ELISA), automated chemiluminescence immunoassay (CLIA), and lateral flow immunoassay (LFIA), plaque reduction neutralization tests (PRNT), or a combination of these methods (Espejo et al. 2020). The most commonly antigens used in these assays were the spike glycoprotein S1 including the receptor binding domain (Jin et al. 2020), the nucleocapsid protein or both (Pang et al. 2020).
Application of inhibitor to halt virus interactions with the host may be one of the prophylactic methods. In this direction, an engineered pan-Cov fusion inhibitor has been designed and designated as EK1 peptide. It has shown promising results in mice by inhibiting the infection in five human coronaviruses, including SARS-CoV, MERS-CoV and three bat-SL-CoVs (Xia et al. 2019). It has also been reported that intranasal application of engineered EK1 peptide before or post viral infection showed protection in human DPP4-transgenic mice against MERS-CoV infection, indicating its potential prophylactic and therapeutic effect.
Another approach is designing of neutralizing antibody, which may block the interaction with the host cell. The S proteins of SARS-CoV and MERS-CoV are immunogenic. The RBD domains of SARS-CoV and MERS-CoV are known to have non-sequential epitopes that induce a more potent neutralizing antibody and give protection against SARS-CoV and MERS-CoV (Du et al. 2009; Zhou et al. 2019). The modification on the structural basis for MERS-CoV S-RBD amino acid has improved the efficacy against MERS-CoV infection (Zhou et al. 2019). Therefore, we may suggest that SARS-CoV-2 S-RBD or modified S-RBD of another related coronavirus could be used as target to develop a vaccine against SARS-CoV-2.
Recently, neutralizing monoclonal antibodies and nanobodies against the RBD domain of S protein showed protection against SARS-CoV and MERS-CoV (Du et al. 2009; Zhou et al. 2019). Although the NTD and S2 unit of S protein from SARS-CoV and/or MERS-CoV was also studied to develop neutralizing antibodies, but the efficacy was found to be very low (Du et al. 2009). Therefore, RBD of S protein SARS-CoV-2 would be a key target for developing neutralizing antibodies as shown in Fig. 5.
Figure 5.
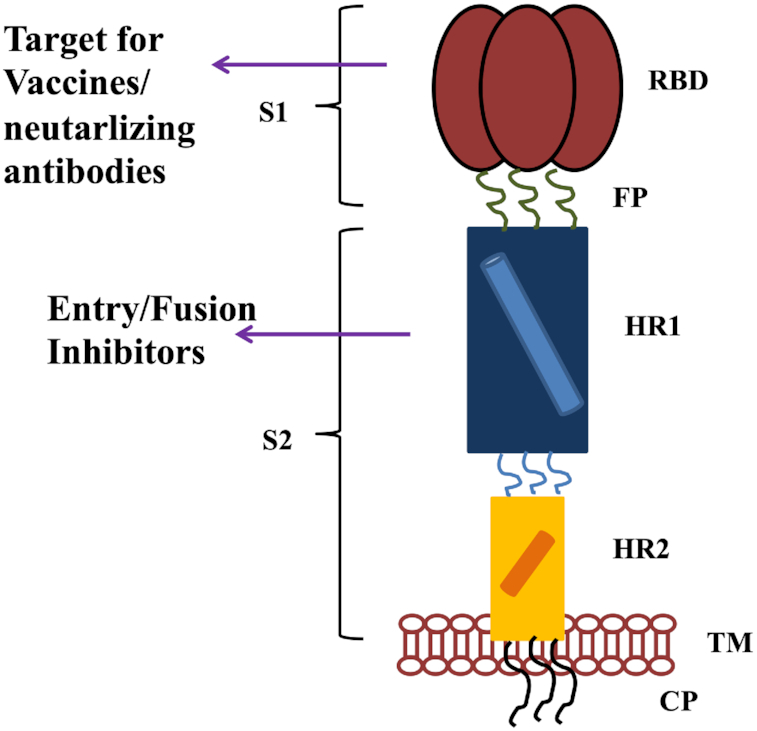
Diagrammatic representation of SARS-CoV-2 targets, for the neutralizing antibodies, vaccines design and various entry/fusion inhibitors.
Cross protection by the antibodies developed against SARS-CoV, has been observed against bat-SL-CoV-W1V1 and bat-SL- CoV-SHC014 (Zeng et al. 2017). Therefore, the development of cross-neutralizing antibodies can be another possible way for urgent prevention and treatment of SARS-CoV-2 infection.
Currently, plasma therapy in which polyclonal antibodies from recovered SARS-CoV-2-infected patients have been used to treat SARS-CoV-2 infection is also being considered.
Researchers are working hard to develop monoclonal Abs (mAbs) and once such antibodies are produced, the next step will involve in vitro testing for neutralizing and/or cross-neutralizing activity as well as in vivo evaluation for protective efficacy in available COVID-19 animal models. Preclinical and clinical trials testing the safety and and efficacy before they are approved for clinical applications are also necessary.
Recently, 1000 memory B cells specific to SARS-CoV-2 S1 or RBD (receptor binding domain) have been purified. Among these, 178 antibodies showed positive results in antigen binding assays with the top 17 binders having EC50 below 1 nM specific for RBD. Further, among 11 neutralizing antibodies, eight of them have shown an IC50 value within 10 nM, whereas 414–1 best among all have IC50 of 1.75 nM. In epitope mapping, three main epitopes recognized by monoclonal antibodies have been identified in RBD domain. Interestingly, 515–5 monoclonal antibody from same study, also showed cross-neutralizing property in the SARS-CoV pseudovirus assay (Wan et al. 2020a). In another study, 61 SARS-CoV-2-neutralizing monoclonal antibodies were isolated from five infected patients. 19 among them have shown positive result in in vitro neutralization assay and nine among them shown 50% virus-inhibition at the concentrations of 1–9 ng/mL. Epitope mapping showed that receptor-binding domain (RBD) and the N-terminal domain (NTD), both are immunogenic in nature. Further, structural studies of these monoloclonal antibodies have proven that one is targeting RBD, second one is targeting NTD and a third bridging RBD and NTD. Therefore, several of these monoclonal antibodies are promising candidates for clinical development as potential therapeutic and/or prophylactic agents against SARS-CoV-2 (L et al. 2020).
Due to the high sequence identity of S protein between SARS-CoV-2 and its closely related SARS-CoV (Zhou et al. 2020), SARS-CoV nAbs have been tested for its cross-reactivity and/or cross-neutralizing activity against SARS-CoV-2 infection. Interestingly, a SARS-CoV RBD-specific human neutralizing mAb, CR3022, have shown the binding of SARS- CoV-2 RBD with high affinity and may recognize an epitope on the RBD that does not overlap with the ACE2-binding site (Tian et al. 2020). Further, SARS-CoV-2 entry and infection may be blocked by cross-reacting the sera isolated by convalescent SARS patients or from animals specific for SARS-CoVS1 (Hoffmann et al. 2020). Moreover, it has been observed that polyclonal antibodies against the RBD domain of SARS-CoV have been cross-reacted with the RBD protein of SARS-CoV-2. They cross-neutralized SARS-CoV-2 infection in HEK293T cells expressing the human ACE2 receptor. Such findings may open new avenues for the potential development of SARS- CoV RBD-based vaccines that might eventually prevent SARS-CoV-2 and SARS- CoV infection (Tai et al. 2020). It may be possible that SARS-CoV RBD-targeting neutralizing antibodies could be applied for treatment/prophylaxis of SARS-CoV-2 infection in the current absence of a specific vaccine against SARS-CoV-2.
Remdesivir has been recently recognized as a promising antiviral drug in cultured cells, mice and nonhuman primate (NHP) models, against RNA viruses including SARS-CoV and MERS-CoV (Sheahan et al. 2017). It is currently under clinical trials for the treatment of Ebola virus infection (Mulangu et al. 2019). Recently studies have shown that EC90 value of remdesivir against 2019-nCoV in Vero E6 cells was 1.76 μM. These data suggest that its working concentration is likely to be achieved in NHPs (Wang et al. 2020)
Remdesivir have shown the efficient in vitro antiviral activity against SARS-CoV-2. However, the controversial evidence of clinical improvement in severe COVID-19 patients has been reported recently in France. The five COVID-19 patients admitted in ICU and treated with remdesivir. Treatment showed significant reduction of SARS-CoV-2 viral load from upper respiratory in most of the cases, however but two patients died with active SARS-CoV-2 replication in their lower respiratory tract. Remdesivir treatment was interrupted for its side effects among four patients due the complexity in such critically ill patients (Dubert et al. 2020).
The first COVID-19 case in the United States was intravenously treated with remdesivir (IV) (Holshue et al. 2020). Within 24 h of remdesivir treatment, the patient showed recovery sign. As the viral loads was decreasing before remdesivir treatment, therefore it cannot be determined if further viral load reduction and clinical improvement were as the direct result of remdesivir treatment. In another study, the compassionate use of remdesivir (N = 53) reported 68% improved oxygenation, 47% discharge and 13% death. This study was not most significantly as it lacks of a paired control group (Grein et al. 2020). Recent study at the National Institutes of Health (NIH), showed preliminary results of the Adaptive COVID-19 Treatment Trial (ACTT, N = 1063). In this randomized controlled trial (RCT), remdesivir treatment showed 31% faster time to recovery as comparative to the placebo group (P < 0.001). The mortality rate was also showed reduction in remdesivir group, however it was not statistically significant (8% vs. 11.6%, P = 0.059). So far, remdesivir has not shown any significant benefit in the reduction of mortality rate. Currently, remdesivir is recommended by the NIH for hospitalized severe COVID-19 cases as defined by oxygenation needs (Clinical management of COVID-19).
Another drug, like chloroquine (C), has recently been reported as a potential broad-spectrum antiviral drug (Savarino et al. 2006). It inhibits the virus infection by increasing endosomal pH, which is essential for virus/cell fusion, as well as by interfering with the glycosylation of cellular receptors of SARS-CoV (Vincent et al. 2005). Recent studies demonstrated that chloroquine is effective at both entry, as well as at post-entry stages of the SARS-CoV-2 infection in Vero E6 cells (Wang et al. 2020). Apart from antiviral activity, chloroquine also has immune-modulating activity. Therefore, it may synergistically enhance antiviral effect in vivo. Chloroquine gets widely distributed in the whole body after oral administration, including lungs. The EC90 value of chloroquinein Vero E6 cells against the SARS-CoV-2 was 6.90μM, therefore it could be clinically achievable (Wang et al. 2020).
The effect of hydroxychloroquine (HCQ) and chloroquine (CQ) in vitro was also tested by Yao et al. In a systematic way, they had divided the experiment into two phases: treatment study and prophylaxis. In the treatment study, they determined the EC50 values for chloroquine. Results showed that it was 23.90 µM and 5.47 µM at 24 and 48 h, respectively. However, in the case ofhydroxychloroquine,the EC50 values were 6.14 µM and 0.72 µM at 24 and 48 h, respectively. On the other hand in the prophylaxis study for chloroquine, the EC50 values were more than100 µM and 18.01 µM at 24 and 48 h, respectively. Similarly, for hydroxychloroquine, the EC50 values were 6.25 µM and 5.85 µM at 24 and 48 h, respectively (Yao et al. 2020). Hence, they found that hydroxychloroquine is more effective in vitro than chloroquine for both prophylaxis and treatment.
A study in United States, where Covid-19 patients hospitalized within 24 h of diagnosis was treated with hydroxychloroquine alone (HCQ) or with hydroxychloroquine and azithromycin (HCQ + AZM) or no HCQ as treatments. Among patients, there was no significant reduction in mortality rate or in the need of ventilation with hydroxychloroquine alone or with hydroxychloroquine and azithromycin (Magagnoli et al. 2020).
A New York hospital stated the QTc prolongation associated with HCQ + AZM (n = 84; Chorin et al. 2020). It was amplified from a baseline of 435 ± 24 ms to a maximal value of 463 ± 32 ms (P < 0.001) on day 3.6 ± 1.6 of the treatment. Till date, researchers present conflicting data's related to the treatment with CQ and HCQ. Therefore, significant randomized control tests (RCTs) with improved study designs are required to examine the efficacy and the clinical benefits of HCQ/CQ treatment over its risks. Currently, the NIH recommendation are against CQ/HCQ and HCQ + AZM treatment for COVID-19, except for clinical trials. Due to the potential toxicity, NIH recommendations are also against the high-dose of CQ (600 mg) twice daily for 10 days in all settings (Coronavirus Disease 2019 (COVID-19) Treatment Guidelines).
Nitazoxanide has demonstrated potent in vitro activity against SARS CoV-2, with an EC50 at 48 h of 2.12 μM in Vero E6 cells (Wang et al. 2020). This potent activity is consistent with EC50 values for nitazoxanide and its active metabolite, tizoxanide, against MERS-CoV in LLC-MK2 cells where EC50 values of 0.92 µM and 0.83 μM respectively, have been demonstrated (Rossignol 2016).
Dexamethasone, a synthetic glucocorticoid, has anti-inflammatory and immunosuppressive properties. There is a hyper inflammatory response involved in the clinical course of patients with pneumonia due to SARS-CoV-2. The elevated level of C-reactive protein (CRP) in SARS-CoV-2 patients has significantly decreased from 129.52 to 40.73 mg/L at time of discharge. 71% of the patients were discharged home with a mean length of stay of 7.8 days. None of the patients had escalation of care, leading to mechanical ventilation (Selvaraj et al. 2020)
Recently, a randomized, controlled clinical trial in the United Kingdom save the lives of people seriously ill with COVID-19 when treated with dexamethasone. Results showed the reduction of number of death by one-third (Ledford 2020). Dexamethasone may be useful for the short-term in severe SARS-CoV-2 patients as it inhibit the protective function of T cells and block B cells from making antibodies (Theoharides and Conti 2020).
VACCINE STATUS
The development of successful vaccine for humans can take several years. As no coronavirus vaccines are available in the market as of now, therefore, the development of a vaccine for the first time can be difficult and time-consuming. However, a mRNA-based vaccine has been co-developed by Moderna and the Vaccine Research Center at the National Institutes of Health. In this vaccine, the target antigen's mRNA, encapsulated in lipid nanoparticles are injected into vaccinee and antigen expresses in vivo. The phase I clinical trial has been recently started (ClinicalTrials.gov: NCT04283461). Curevac is also using the same platform but they are still in the pre-clinical phase.
Apart from this, there are various other approaches including DNA vaccine, recombinant protein-based vaccine, recombinant vector vaccine, inactivated vaccine and attenuated vaccine. Different research companies, Universities and Institutes are targeting S protein of SARS-CoV-2 to develop recombinant protein-based vaccines including NovavaxExpresS2ion, iBio, Sichuan Clover Biopharmaceutical, Baylor College of Medicine and the University of Queensland. Similarly, Cansino Biologics, Geovax Vaxart and the University of Oxford are using viral-vector- based vaccine platform especially focused on the S protein. Applied DNA Sciences and Inovio are using DNA vaccines platform again focused on the S protein (Amanat Fatima 2020). Apart from the above-mentioned platforms, the whole microorganism based vaccine platform like inactivated and attenuated virus vaccine is also in consideration. Codagenix with the Serum Institute of India is using live attenuated vaccine platforms. The recombinant vector-based platform (adenovirus vector) is adopted by Johnson and Johnson, and on the other hand, Sanofi is also using the same platform (recombinant influenza vector) to develop a vaccine against SARS-CoV-2. At this stage, it is difficult to predict the best platform for a vaccine against SARS-CoV-2 as all the above mentioned platforms have some advantages as well as disadvantages (Amanat Fatima 2020).
CONCLUSION
Coronaviruses have shown the capability to jump species boundaries and adapt to new hosts. Therefore, we may face more such kind of outbreaks in the future. Role of the intermediate host is also of major importance, as they provide direct pathway for virus transmission in humans. The enormous diversity of viruses in animals and their ongoing evolution makes it important to limit our exposure to animal pathogens as much as possible. Based on the metagenomic data it is predicted that the Pangolin-CoV is most closely related to SARS-CoV-2. Pangolin-CoV genome showed 91.02% nucleotide identity with the SARS-CoV-2 genome. Due to a very limited knowledge of this novel virus, it is difficult to explain the significant number of amino acid substitutions that occurred between the SARS-CoV-2 and SARS or SL-CoVs. For example, in SARS-S-CoV, six mutations occurred in the regions other than that of the RBD domain, but interestingly no amino acid substitutions were present in the receptor-binding motifs that directly interact with human receptor ACE2 protein. Therefore, such differences that could affect SARS-CoV-2 transmission property as compared to SARS-CoV are of importance for future investigation. SARS-CoV-2 continues to infect people globally; therefore it is imperative to develop new, safe, accurate, fast and simple new technologies for detecting SARS-CoV-2. Apart from diagnosis, effective prophylactic and therapeutic agents are also required to control and prevent infection. Various therapeutic agents including dexamethasone have shown promising results in the in vitro studies to control infection. However, there is an urgency to develop vaccine against coronaviruses. In this direction, studies on neutralizing antibodies from SARS-CoV and MERS-CoV against S protein and its many fragments including S1-NTD, RBD and S2 may provide important guidelines for development of vaccine against SARS-CoV-2. Apart from neutralizing antibody against S protein, other approaches including DNA vaccine, recombinant vector vaccine, inactivated vaccine and attenuated vaccine are also in pipeline to develop vaccines against SARS-CoV-2.
So far, the traditional public health measures including detection of active cases, isolation of such cases, tracing of all contacts and their quarantine, maintaining social distancing, as well as community quarantine were found to be successful. Only after this pandemic ends, we will be in a position to assess the social, health and economic impact of such a massive outbreak. Therefore, we must learn lessons for our future from such outbreaks, as new viruses will keep coming.
Contributor Information
Jitendra Singh Rathore, School of Biotechnology, Gautam Buddha University, Yamuna Expressway, Greater Noida, Uttar Pradesh 210312, India.
Chaitali Ghosh, Department of Zoology, Gargi College, University of Delhi, New Delhi, Delhi 110049 India.
Conflicts of interest
None declared.
REFERENCES
- Amanat Fatima KF. SARS-CoV-2 vaccines: status report. Cell. 2020;52:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Yeung M, Xu RH. SARS: epidemiology. Respirology (Carlton, Vic.). 2003;8:S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X et al. . Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorin E, Dai M, Shulman E et al. . The QT interval in patients with COVID-19 treated with hydroxychloroquine and azithromycin. Nat Med. 2020;26:808–9. [DOI] [PubMed] [Google Scholar]
- Clinical management of COVID-19 https://www.who.int/publications/i/item/clinical-management-of-covid-19. [Google Scholar]
- Coronavirus Disease 2019; (COVID-19) Treatment Guidelines. https://www.covid19treatmentguidelines.nih.gov/. [Google Scholar]
- Coutard B, Valle C, de Lamballerie X et al. . The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubert M, Visseaux B, Isernia V et al. . Case reports study of the first five patients COVID-19 treated with remdesivir in France. Int J Infect Dis. 2020;98:290–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, He Y, Zhou Y et al. . The spike protein of SARS-CoV - A target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo AP, Akgun Y, Al Mana AF et al. . Review of current advances in serologic testing for COVID-19. AJCP Rev Artic Am J Clin Pathol. 2020;XX:0–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS et al. . The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J, Ohmagari N, Shin D et al. . Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhou Y, Wu H et al. . Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol. 2004;173:4050–7. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S et al. . SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue ML, DeBolt C, Lindquist S et al. . First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C-C, Lam T-Y, Shi Z-L et al. . Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J Virol. 2008;82:1819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Peng C, Yu M et al. . Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch Virol. 2010;155:1563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang M, Zuo Z et al. . Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboratory Testing Strategy Recommendations for COVID-19: Interim Guidance, 2020. https://apps.who.int/iris/handle/10665/331509. [Google Scholar]
- Lam TT-Y, Shum MH-H, Zhu H-C et al. . Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. 2020, DOI: 10.1101/2020.02.13.945485. [Google Scholar]
- Lau SKP, Li KSM, Tsang AKL et al. . Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol. 2013;87:8638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Woo PCY, Li KSM et al. . Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469. [DOI] [PubMed] [Google Scholar]
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome Coronavirus infections. J Virol. 2008;82:6984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lihong L, Pengfei W, Manoj S N et al. . Potent neutralizing monoclonal antibodies directed to multiple epitopes on the SARS-CoV-2 spike. bioRxiv Prepr Serv Biol. 2020, DOI: 10.1101/2020.06.17.153486. [Google Scholar]
- Li W, Shi Z, Yu M et al. . Bats are natural reservoirs of SARS-like coronaviruses. Science (80-). 2005;310:676–9. [DOI] [PubMed] [Google Scholar]
- Li X, Giorgi EE, Marichannegowda MH et al. . Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci Adv. 2020;6:eabb9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J et al. . Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagnoli J, Narendran S, Pereira F et al. . Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Med. 2020;0, DOI: 10.1016/j.medj.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak GC, Cheng PK, Lau SS et al. . Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S, Dodd LE, Davey RT et al. . A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Wang MX, Ang IYH et al. . Potential rapid diagnostics, vaccine and therapeutics for 2019 Novel Coronavirus (2019-nCoV): a systematic review. J Clin Med. 2020;9:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E, Gallagher T. SARS coronavirus redux. Trends Immunol. 2020;41:271–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Mou H, Smits SL et al. . Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Qu X, Li W et al. . Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-Like cronavirus of bat origin. J Virol. 2008;82:1899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol JF. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016;9:227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir JSM, Lam TTY, Ahmed MMM et al. . Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science (80-). 2016;351:81–4. [DOI] [PubMed] [Google Scholar]
- Savarino A, Di Trani L, Donatelli I et al. . New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj V, Dapaah-Afriyie K, Finn A et al. . Short-term corticosteroids in SARS-CoV2 patients: hospitalists’ perspective. medRxiv. 2020, DOI: 10.1101/2020.06.19.20109173. [Google Scholar]
- Sheahan TP, Sims AC, Graham RL et al. . Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZL, Guo D, Rottier PJM. Coronavirus: epidemiology, genome replication and the interactions with their hosts. Virol Sin. 2016;31:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W, He L, Zhang X et al. . Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Li G, Vasilakis N et al. . Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol Biol. 2009;9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theoharides TC, Conti P. Dexamethasone for COVID-19? Not so fast. J Biol Regul Homeost Agents. 2020;34, DOI: 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- Tian X, Li C, Huang A et al. . Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent MJ, Bergeron E, Benjannet S et al. . Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cao R, Zhang L et al. . Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Xing S, Ding L et al. . Human IgG neutralizing monoclonal antibodies block SARS-CoV-2 infection. Cell Rep. 2020a;0:107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Shang J, Graham R et al. . Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020b;94, DOI: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO | Novel coronavirus summary and literature update – as of 17 May2013. [Google Scholar]
- Wrapp D, Wang N, Corbett KS et al. . Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-). 2020;367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Yan L, Xu W et al. . A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5:eaav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Ye F, Zhang M et al. . In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Clin Infect Dis, 2020;71:732–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Hon CC, Li Y et al. . Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J Gen Virol. 2010;91:1058–62. [DOI] [PubMed] [Google Scholar]
- Zeng LP, Ge XY, Peng C et al. . Cross-neutralization of SARS coronavirus-specific antibodies against bat SARS-like coronaviruses. Sci China Life Sci. 2017;60:1399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang L, Deng X et al. . Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92:408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wu Q, Zhang Z. Probable Pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1346–51e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Han Z, Wang LF et al. . Identification of immunogenic determinants of the spike protein of SARS-like coronavirus. Virol Sin. 2013;28:92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Han Z, Wang LF et al. . Immunogenicity difference between the SARS coronavirus and the bat SARS-like coronavirus spike (S) proteins. Biochem Biophys Res Commun. 2009;387:326–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Lou YX, Wang XG et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang Y, Huang J et al. . Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. 2019;11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]