Abstract
More than 24 million infections with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were confirmed globally by September 2020. While polymerase chain reaction–based assays are used for diagnosis, there is a need for high-throughput, rapid serologic methods. A Luminex binding assay was developed and used to assess simultaneously the presence of coronavirus disease 2019 (COVID-19)–specific antibodies in human serum and plasma. Clear differentiation was achieved between specimens from infected and uninfected subjects, and a wide range of serum/plasma antibody levels was delineated in infected subjects. All 25 specimens from 18 patients with COVID-19 were positive in the assays with both the trimeric spike and the receptor-binding domain proteins. None of the 13 specimens from uninfected subjects displayed antibodies to either antigen. There was a highly statistically significant difference between the antibody levels of COVID-19–infected and –uninfected specimens (P < .0001). This high-throughput antibody assay is accurate, requires only 2.5 hours, and uses 5 ng of antigen per test.
Keywords: coronavirus, SARS-CoV-2, COVID-19, antibodies, antibody assay
Rapid serologic assays are needed to detect antibodies in individuals infected with or recovering from severe acute respiratory syndrome coronavirus 2. The assay described is advantageous in that it is highly accurate, requires only 2.5 hours, uses little antigen/test, and constitutes a high-throughput platform.
Reverse transcription polymerase chain reaction (RT-PCR) techniques are currently used for the qualitative detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acids in specimens from the upper and lower respiratory tracts [1, 2]. While molecular testing has been well established in clinical laboratories throughout the world for 2 decades, testing of serum and other bodily fluids for antibodies (Abs) to infectious diseases such as syphilis, typhoid, and diphtheria has been used for more than a century [3, 4].
Antibody assays are most useful for identifying individuals who have been previously exposed to a particular pathogen. As such, they can be especially valuable, for example, for identification of persons who have had asymptomatic viral infections and for those who have recovered and would no longer be positive in tests for viral nucleic acids. Antibody tests in the context of the current SARS-CoV-2 pandemic are also critical for serosurveillance, for identification of donors for COVID-19 convalescent plasma therapy, and to identify individuals who are potentially immune to reinfection, though this has not yet been established. Antibody assays thus fill an essential gap both during and after the current SARS-CoV-2 pandemic. In fact, 1 study found that, depending on the time of testing postinfection, the combined use of RT-PCR and Ab positivity provided an advantage over either test alone [5].
We and others [5–7] have described tests for assessing the presence of Abs to SARS-CoV-2 in serum and plasma using the enzyme-linked immunosorbent assay (ELISA) platform with a recombinant form of the S protein of the virus and the central portion of this molecule identified as the receptor binding domain (RBD), consisting of amino acids 319–541 of the S protein [7–9]. We report here a bead-based multiplex immunoassay in which bead sets labeled with 2 different fluorochrome signatures are coated with the soluble recombinant S protein or RBD. The 2 coated bead sets are incubated simultaneously with serum or plasma, then with biotinylated antihuman total immunoglobulin Abs, and finally with phycoerythrin (PE)–labeled streptavidin. The readout is performed with a laser-based instrument. This is a high-throughput assay that offers the advantages of being able to prepare the antigen-coated beads for thousands of tests in a single day and using 20-fold less antigen than is required for ELISA. It also allows assessment of Abs to RBD and S protein simultaneously, thus providing a screen and titrated confirmatory test in a single step. In the setting of hospitals and regional reference laboratories, results on thousands of specimens per day can be generated.
MATERIALS AND METHODS
Recombinant Proteins
The recombinant S and RBD proteins were produced as previously described [7] in Expi293F cells (ThermoFisher) by transfections of purified DNA using an ExpiFectamine Transfection Kit (ThermoFisher). The soluble version of the spike protein included the S protein ectodomain (amino acids 1–1213), a C-terminal thrombin cleavage site, a T4 foldon trimerization domain, and a hexahistidine tag. The protein sequence was modified to remove the polybasic cleavage site (RRAR to A) and 2 stabilizing mutations (K986P and V987P, wild-type numbering). The RBD (amino acids 319–541) also contained a hexahistidine tag. Supernatants from transfected cells were harvested on day 3 posttransfection by centrifugation of the culture at 4000g for 20 minutes. Supernatant was then incubated with 6 mL Ni-NTA agarose (Qiagen) for 1–2 hours at room temperature. Next, gravity flow columns were used to collect the Ni-NTA agarose and the protein was eluted. Each protein was concentrated in Amicon centrifugal units (EMD Millipore) and resuspended in phosphate-buffered saline (PBS).
Human Samples
Banked serum samples were obtained from study participants enrolled in 2 institutional review board (IRB)–approved longitudinal observational protocols (Icahn School of Medicine at Mount Sinai; principal investigator: Dr V. Simon; IRB-16-00772 and IRB-16-00791). All participants provided written consent at study enrollment and agreed to sample banking and future research use of their banked biospecimen. Specimens from these protocols included sera from 4 participants with documented SARS-CoV-2 infection: specimens from P1 (3 time points), P2 (2 time points), and P3 and P4 (1 time point each). In addition, sera were used that had been collected from 3 healthy donors (N1, N2, and N3) in October and November 2019, prior to the spread of SARS-CoV-2 in the United States. Four additional de-identified serum specimens (P5–P8) were provided by the Clinical Pathology Division of the Department of Pathology, Molecular and Cell-Based Medicine at the Icahn School of Medicine at Mount Sinai. Twenty-four plasma specimens were also used from the Division of Transfusion Medicine of the Department of Pathology, Molecular and Cell-Based Medicine: 14 were segments from citrated plasma units collected from convalescent COVID-19 subjects that were destined for transfusion to SARS-CoV-2–infected subjects (TF1–TF14). A pool of 0.1 mL from each of these specimens was created (convalescent plasma pool) to use as a positive control. An additional 10 specimens were derived from the general blood bank plasma inventory (N4–N13), which represented standard nonconvalescent plasma donors. A pool of 0.1 mL from each of these specimens was created (nonconvalescent plasma pool) and used as a negative control. Table 1 provides an overview of the specimens studied.
Table 1.
Characteristics of Patients Whose Specimens Were Tested
| Specimen IDa | SARS-CoV-2 PCR Result | Type of Specimen | Symptoms Suggestive of COVID-19 | Days After Symptom Onset and Serum Collection | COVID-19 Severity |
|---|---|---|---|---|---|
| P1a | Positive | Serum | Yes | 8 | Severe |
| P1b | Positive | Serum | Yes | 11 | Severe |
| P1c | Positive | Serum | Yes | 15 | Severe |
| P2a | Positive | Serum | Yes | 7 | Severe |
| P2b | Positive | Serum | Yes | 10 | Severe |
| P3 | Positive | Serum | Yes | 22 | Mild |
| P4 | Inconclusive | Serum | Yes | 21 | Mild |
| P5b | Positive | Serum | Unknown | Unknown | Unknown |
| P6b | Positive | Serum | Unknown | Unknown | Unknown |
| P7b | Positive | Serum | Unknown | Unknown | Unknown |
| P8b | Positive | Serum | Unknown | Unknown | Unknown |
| TF1–TF14 | Positive | Plasma | Unknown | Unknown | Unknown |
| Convalescent plasma pool | Positive | Plasma | Unknown | Unknown | Unknown |
| N1 | NA; collected Oct 2019 | Plasma | NA | NA | NA |
| N2 | NA; collected Nov 2019 | Plasma | NA | NA | NA |
| N3 | NA; collected Oct 2019 | Plasma | NA | NA | NA |
| N4–N13c | Unknown | Plasma | NA | NA | NA |
| Nonconvalescent plasma poolc | Unknown | Plasma | NA | NA | NA |
Abbreviations: COVID-19, coronavirus disease 2019; NA, not applicable or not available; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
aSpecimens from P: sera of patients infected with SARS-CoV-2; N: normal, uninfected individual; TF: citrated plasma from units to be used for transfusion.
bP5–P8 were de-identified.
cN4–N13 were derived from the general blood bank plasma inventory.
Luminex Binding Ab Assay
The SARS-CoV-2 antigens included a soluble recombinant trimerized form of the S protein and a recombinant RBD protein produced in Expi293F mammalian cells as described previously (mSpike, mRBD) [7]. Each antigen was covalently coupled individually to a uniquely labeled fluorochrome carboxylated xMAP bead set at 4.0 μg protein/million beads using a 2-step carbodiimide reaction with the xMAP Ab Coupling Kit according to the manufacturer’s instructions (Luminex, Austin, Texas). The coupled beads were pelleted, resuspended at 5 × 105 beads/mL in storage buffer (PBS, 0.1% bovine serum albumin [BSA], 0.02% Tween-20, and 0.05% sodium azide, pH 7.4), and stored at –80°C.
The beads needed for a single run (2500 beads/well × number of wells) were diluted in assay buffer (PBS, 0.1% BSA, 0.02% Tween-20) to a volume that delivered 2500 beads to each well in an aliquot of 50 μL/well. Serum/plasma was diluted in PBS, added as 50 μL/well to the wells containing the beads, and incubated at room temperature for 1 hour on a plate shaker at 600 rpm. After 2 washes with assay buffer, 100 μL/well of biotinylated antihuman total immunoglobulin (Abcam, catalog number ab97158) at 2 μg/mL was added and incubated for 30 minutes at room temperature on a plate shaker. After 2 washes, 100 μL/well of streptavidin-PE at 1 μg/mL was added (BioLegend Catalog number 405204) followed by a 30-minute incubation at room temperature on a plate shaker. After 2 additional washes, 100 μL of assay buffer/well was added and put on a shaker to resuspend the beads. The plate was read with a Luminex Flexmap 3D instrument. Samples were tested in duplicate and the results were recorded as mean fluorescence intensity (MFI). Serum and plasma were used for the experiments herein. Sera from uninfected individuals, drawn before the start of the COVID-19 epidemic served as negative controls, as was plasma derived from the blood bank. Figures were generated using GraphPad Prism 7.02 software.
RESULTS
A total of 25 serum and plasma specimens were tested from 18 different patients with confirmed SARS-CoV-2 infection. Sera from 3 uninfected individuals were used that had been banked as part of an ongoing longitudinal study prior to the COVID-19 outbreak (N1–N3), and an additional 10 plasma were obtained from blood bank donors (N4–N13). All assays for COVID-19 Abs were performed in a Biosafety Level 2 laboratory as the specimens were heat-inactivated; normal blood precautions that are employed when handling all human specimens were used for these studies.
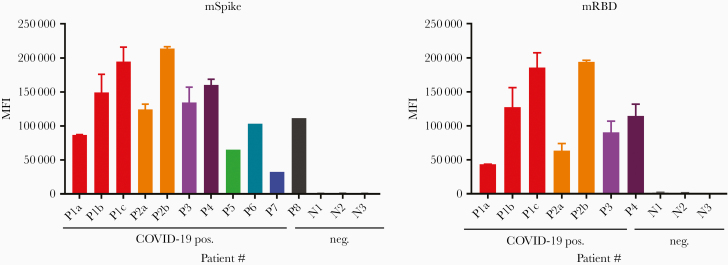
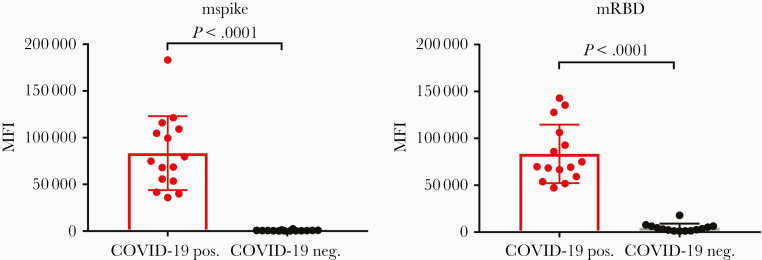
The specimens were screened for the presence of Abs reactive with fluorochrome-labeled magnetic beads coated with soluble recombinant trimeric mammalian cell–expressed S protein (mSpike) or mRBD followed by incubation with a secondary biotinylated Ab that detects total immunoglobulin and streptavidin-PE. All specimens from COVID-19 patients reacted strongly with both mRBD and mSpike. Results are shown in Figure 1 for sera from 4 patients and 3 controls, and in Figure 2 for plasma from 14 infected subjects and 13 controls, as well as from the positive and negative plasma pools. The data shown in these figures reflect a wide range of values for Abs in patients’ specimens: the range of Ab levels in the plasma specimens diluted 1:200 from COVID-19 patients against mSpike ranged from 35 888 to 121 479, and with RBD, from 47 173 to 142 998 MFI (Figure 2). The background activities in the specimens from the control subjects also showed a range of values (1316–2480 for mSpike and 1005–17 947 for mRBD). Two of the specimens from the nonconvalescent controls (N4 and N10) displayed unusually high MFI values against either (but not both) mSpike or mRBD. Since these came from the general blood bank plasma inventory, these 2 specimens may have been derived from donors who had unknowingly been infected with SARS-CoV-2. The likelihood that their Abs represented Abs that were cross-reactive due to common endemic coronaviruses is unlikely based on a previous study showing that prepandemic samples that were positive for spike proteins of endemic human coronaviruses 229E and NL63 (responsible for a large proportion of common colds each year) were negative for Abs reactive with SARS-CoV-2 Spike and RBD [7]. As shown in Figure 2, the MFI values were significantly higher in COVID-19–positive than in COVID-19–negative specimens (P < .0001 by the Mann–Whitney test, for both Spike and RBD). There was essentially no reactivity in the absence of serum or plasma (data not shown).
Figure 1.
Screening for the presence of severe acute respiratory syndrome coronavirus 2 antibodies in specimens from coronavirus disease 2019–infected (pos.) and –uninfected (neg.) humans. Assays were run using the S protein produced in mammalian cells (mSpike, left) and mRBD (right). Results are shown using sera tested at a dilution of 1:200. For specimens from 4 patients (P1–P4) run against both antigens, the data shown are the mean + standard deviation of 2–5 experiments. For 4 patients (P5–P8), the specimens were run only against the mSpike in a single experiment. Abbreviations: COVID-19, coronavirus disease 2019; MFI, mean fluorescence intensity; mRBD, mammalian-derived Receptor Binding Domain polypeptide; mSpike, mammalian-derived Spike protein.
Figure 2.
Screening for the presence of severe acute respiratory syndrome coronavirus 2 antibodies in plasma from coronavirus disease 2019 (COVID-19)–positive subjects (COVID-19 pos.) and from plasma derived from the general blood bank plasma inventory (COVID-19 neg.). Assays were run using the S protein produced in mammalian cells: mSpike (left) and mRBD (right). Results are shown using plasma tested at a dilution of 1:200. Statistical analysis using the Mann–Whitney test is shown. Medians with error bars representing standard deviation are indicated. Abbreviations: COVID-19, coronavirus disease 2019; MFI, mean fluorescence intensity; mRBD, mammalian-derived Receptor Binding Domain polypeptide; mSpike, mammalian-derived Spike protein.
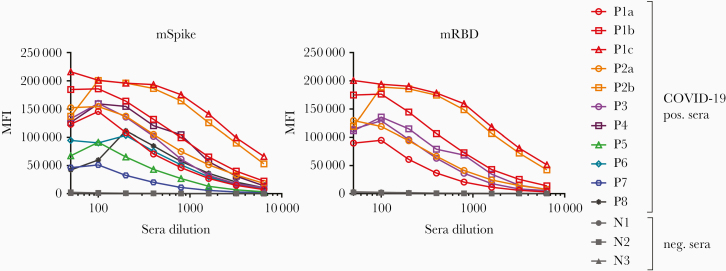
To determine the dynamic range of the assay and the titer of Abs present in COVID-19–positive sera, titrations were performed with sera from patients and from uninfected subjects using mSpike and mRBD. The titration curves are shown in Figure 3. A prozone effect (the phenomenon in which a value below the peak is observed at low dilutions of a specimen) is suggested by the curves generated by 6 of the sera tested against the mSpike. The prozone phenomenon was less apparent when using the mRBD antigen. Generally, the maximum MFI values were achieved with serum dilutions of 1:100 or 1:200. As noted above, the level of Abs varied greatly between patient specimens. Of interest, titration curves show that the specimens with the highest Ab levels (P1c and P2b) were from 2 different patients (P1 and P2), who presented with severe disease at the time of the blood draw (Table 1). Further study is needed to ascertain how and whether disease severity affects the level and type of Abs induced. However, it is notable that the level of Abs increased over a few days in each of the 2 patients from whom longitudinal specimens were available (P1 and P2; Table 1 and Figure 1). The factors that affect the variation in Ab responses (including severity of disease, length of infection, sex, and genetics) are yet to be determined and will require a large panel of specimens from patients accompanied by clinical and demographic data.
Figure 3.
Titration of coronavirus disease 2019 (COVID-19)–positive/negative sera. Specimens were diluted at 2-fold dilutions from 1:50 to 1:6400 and tested using the Luminex assay described herein. Titration curves are shown for sera from 11 specimens from 8 infected patients using the mSpike as antigen (left) and for sera from 3 patients tested vs mRBD (right). Abbreviations: COVID-19, coronavirus disease 2019; MFI, mean fluorescence intensity; mRBD, mammalian-derived Receptor Binding Domain polypeptide; mSpike, mammalian-derived Spike protein.
Discussion
A rapid throughput test for SARS-CoV-2 Abs such as the Luminex bead binding Ab assay described here can serve as an essential tool for identifying individuals infected with SARS-CoV-2 who have seroconverted and those who have been infected and recovered. The Ab assay will be a particularly useful assay for identifying individuals who have had asymptomatic infections, including children [10–12]. Given that there appears to be a relatively high prevalence of asymptomatic individuals who have or have had SARS-CoV-2 infection, Ab assays fill an essential gap both during and after the current SARS-CoV-2 pandemic for identifying the true prevalence of infection in a given population. Since the Luminex Ab assay can simultaneously test qualitatively and quantitatively for RBD and S protein Abs, and can be performed in ≤2.5 hours (see the Materials and Methods and Figure 4) with 5–10 ng of antigen per test, it provides a platform that will result in cost savings and the processing of large numbers of samples per day.
Figure 4.
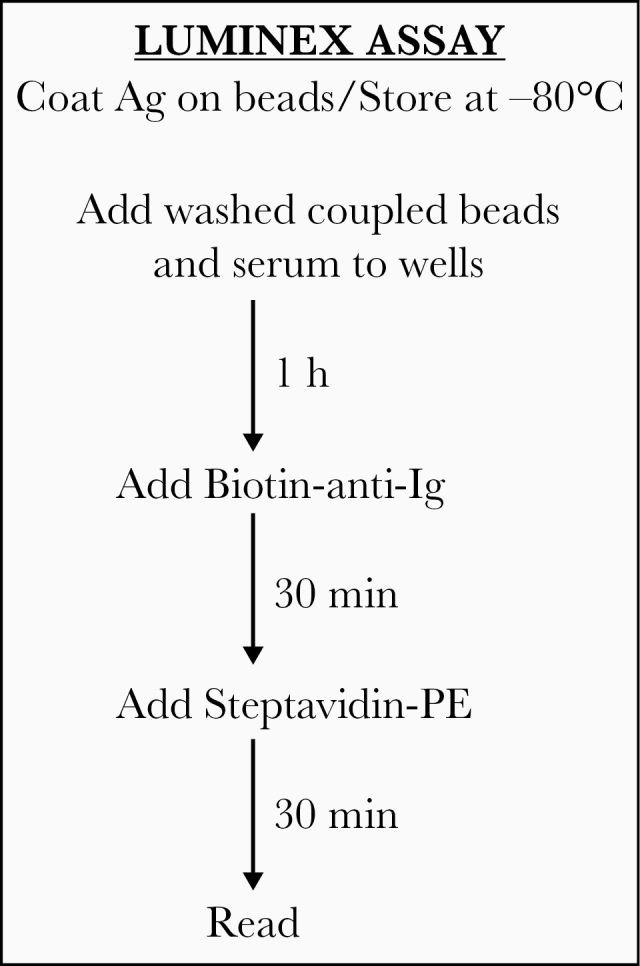
Steps and time required for detection of antibodies to the mSpike or mRBD of severe acute respiratory syndrome coronavirus 2 antigens using the Luminex antibody binding assay described herein. Washing steps, delineated in the Materials and Methods, add approximately 30 minutes, resulting in a test requiring approximately 2.5 hours. Abbreviations: Ag, antigen; Ig, immunoglobulin; PE, phycoerythrin.
Ten weeks after this manuscript had been published online as a preprint (https://doi.org/10.1101/2020.04.14.20059501), Luminex Corporation announced that it had submitted an Emergency Use Authorization for its xMAP SARS-CoV-2 multiantigen immunoglobulin G (IgG) assay. This assay detects human IgG Abs in serum and plasma specific for 3 SARS-CoV-2 antigens (S1 subunit of the spike protein, RBD, and nucleocapsid protein) [13]. The COVID-19 antibody assay described in this publication has the advantage of detecting not only IgG, but also total immunoglobulin, including immunoglobulin M (IgM) and immunoglobulin A (IgA). Several publications suggest that seroconversion to IgA and IgM against SARS-CoV-2 develops earlier compared to IgG [14]. Thus, the multibead assay described here should be able to detect anti–SARS-CoV-2 Abs more reliably earlier after infection than the commercially available SARS-CoV-2 assay from Luminex.
Using the SARS-CoV-2 nucleoprotein is expected to induce higher cross-reactivity to other coronaviruses compared to the spike protein, owing to sequence homology of the viral nucleoprotein, with a sequence homology between SARS-CoV-1 and SARS-CoV-2 for spike, RBD, and nucleocapsid protein being 76%, 50%, and 90%, respectively [15, 16]. With trimeric spike and spike-derived antigens like RBD, a higher level of specificity can be achieved, and the use of both the monomeric RBD and the trimeric spike as antigens enables detection of Abs that can specifically block entry via ACE2, as well as Abs against conformational epitopes on the envelope spike.
The Ab assay described here uses antigen-coated fluorochrome-labeled magnetic beads that can be prepared in bulk and used for at least 4 weeks, simplifying the work load and logistics in the laboratory. It is worth noting that, using various human immunodeficiency virus (HIV) proteins and peptides, we have used antigen-coated xMAP beads stored under sterile conditions as described in the Materials and Methods, for 4 or more weeks [17]. Currently, we have used xMAP beads coated with the mSpike or mRBD proteins for 3 weeks; barring degradation of the proteins at –80°C under sterile conditions, it is anticipated that they would be stable and useful for several weeks to months. This means that the antigen-coated beads could be prepared and stored in batches, providing reagents for several thousands of tests.
A concentration of 4 μg/106 beads was used for bead coating in the experiments described above, as that concentration had been used previously with various proteins; a concentration of 2 μg protein/106 beads gave comparable values, but <2 μg protein/106 beads gave lower MFI values in titration curves with sera from infected individuals (data not shown). Given that the S and RBD recombinant proteins are currently in limited supply, the use of the lower level of protein for coating is advantageous; coating at 4 and 2 μg/million beads results in the use of a 10 or 5 ng antigen/test, respectively—a level considerably less than that required for most ELISA assays. Further modifications are underway to detect separately immunoglobulin isotypes, to decrease the time and concentration of antigen required, and to determine if other bodily fluids, such as urine and/or saliva, can be used in this assay. Both urine and saliva Ab tests are used for diagnostics in other diseases such as HIV [18, 19].
This assay can be scaled up for use in hospital and reference laboratories. A high-throughput assay such as this can be used for a multitude of purposes including as a diagnostic assay in hospitals and reference laboratories, serosurveillance of communities, screening of healthcare workers who may have developed immunity to SARS-CoV-2, identification of donors for COVID-19 convalescent plasma therapy, and selection of individuals who are potentially immune to reinfection. High-throughput serologic tests like the one described here will also be helpful in surveying infection-fatality rates (vs case fatality rates). These have to be model-based in the absence of broad serological data, and a test such as that described herein could be useful in this important public health role.
Notes
Acknowledgments. We are extremely grateful to the COVID-19 patients for their contribution to this research and wish for them a full recovery. This manuscript was previously published as a preprint on medRxiv on 17 April 2020 (https://doi.org/10.1101/2020.04.14.20059501).
Financial support. This work was supported by the Microbiology Laboratory Clinical Services at the Mount Sinai Health System and the Translational Science Hub at the Mount Sinai Health System, National Institutes of Health (grant number U54TR001433); the Personalized Virology Initiative and philanthropic donations (to V. S.); the Department of Medicine (to S. Z. P.) at Icahn School of Medicine at Mount Sina; the National Institutes of Health grant U54TR001433, the National Institute of Allergy and Infectious Diseases Centers of Excellence for Influenza Research and Surveillance contract HHSN272201400008C (to F. K.,V. S.), grant AI139290 (to C. E. H., S. Z. P.) and grant AI136916 (V. S.); the Department of Veterans Affairs Merit Review Grant I01BX003860 (to C. E. H., S. Z. P., S. W.) and Research Career Scientist Award 1IK6BX004607 (to C. E. H.).
Potential conflicts of interest. The Icahn School of Medicine at Mount Sinai has decided it will submit a patent application based on the method described herein. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Coronavirus Virtual Webinar Series, 17 June 2020; broadcast online by labroots.com.
References
- 1. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020; 382:1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Storch GA. Diagnostic virology. Clin Infect Dis 2000; 31:739–51. [DOI] [PubMed] [Google Scholar]
- 4. National Museum of American History–Behring Collection. The antibody initiative: diagnosing disease with antibodies Available at: https://americanhistory.si.edu/collections/object-groups/antibody-initiative/diagnostics. Accessed 10 August 2020.
- 5. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019 [manuscript published online ahead of print 28 March 2020]. Clin Infect Dis 2020. doi:10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 7. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berry JD, Hay K, Rini JM, et al. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs 2010; 2:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020; 584:115–9. [DOI] [PubMed] [Google Scholar]
- 10. Kimball A, Hatfield KM, Arons M, et al. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility—King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep 2020; 69:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishiura H, Kobayashi T, Miyama T, et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int J Infect Dis 2020; 94:154–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis 2020; 20:689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luminex Corporation. xMAP SARS-CoV-2 multi-antigen IgG assay Available at: https://www.luminexcorp.com/xmap-sars-cov-2-antibody-testing/#overview. Accessed 10 August 2020.
- 14. Ma H, Zeng W, He H, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol 2020; 17:773–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 2020; 432:3309–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020; 27:671–80 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hessell AJ, Powell R, Jiang X, et al. Multimeric epitope-scaffold HIV vaccines target V1V2 and differentially tune polyfunctional antibody responses. Cell Rep 2019; 28:877–95.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao YZ, Hosein B, Borkowsky W, et al. Antibodies to human immunodeficiency virus type 1 in the urine specimens of HIV-1-seropositive individuals. AIDS Res Hum Retroviruses 1989; 5:311–9. [DOI] [PubMed] [Google Scholar]
- 19. Archibald DW, Hebert CA. Salivary detection of HIV-1 antibodies using recombinant HIV-1 peptides. Viral Immunol 1991; 4:17–22. [DOI] [PubMed] [Google Scholar]