Abstract
Background
Few studies have compared the yield of reverse transcription polymerase chain reaction (RT-PCR) assays in nasopharyngeal swabs, oropharyngeal swabs, and sputum for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection.
Methods
We conducted an observational study in Beijing Ditan Hospital, China. Specimens including nasopharyngeal swabs, oropharyngeal swabs, and sputum from confirmed coronavirus 2019 patients were collected for RT-PCR testing. Disease duration was calculated from the date of symptom onset to the date of specimen collection and divided into 3 groups: ≤14 days, 14–21 days, and >21 days. We compared positive rates across the 3 specimens collected. The kappa coefficient was used to evaluate the consistency of RT-PCR results between different specimens.
Results
A total of 291 specimens were collected and tested from 43 confirmed patients. Among specimens collected with a disease duration of ≤14 days, the positive rate was highest in sputum (79.2%); this rate was significantly higher than that in nasopharyngeal swabs (37.5%; P = .003) and oropharyngeal swabs (20.8%; P < .001). Similar findings were observed with the disease durations of 14–21 days and >21 days. The consistency of testing results between nasopharyngeal swabs and oropharyngeal swabs was low with the disease durations of ≤14 days and >21 days. The consistency between the sputum and oropharyngeal swabs and between the sputum and nasopharyngeal swabs was very low across all 3 disease durations, with statistical significance.
Conclusions
Compared with nasopharyngeal swabs and oropharyngeal swabs, sputum had the highest yield of SARS-CoV-2 detection. Nasopharyngeal swabs and oropharyngeal swabs had a similar yield. If sputum is not feasible, a nasopharyngeal swab can be recommended for the detection of SARS-CoV-2, and early testing is needed.
Keywords: nasopharyngeal swabs, oropharyngeal swabs, positive rate, SARS-CoV-2, sputum
Since the novel coronavirus disease 2019 (COVID-19) outbreak in Wuhan, China, in December 2019, COVID-19 has caused more than 6 million infections around the world (as of the end of May) [1]. The World Health Organization (WHO) has declared the COVID-19 outbreak a global pandemic [2]. Real-time fluorescence polymerase chain reaction (RT-PCR) testing of respiratory specimens for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA is currently widely used for case diagnosis and to guide the duration of patient isolation from hospitalization [3]. However, it has been reported that there are discrepancies in detection results using different respiratory specimens [4–6]. The differences in viral presentation among COVID-19 patients at different disease stages could also play a role [7].
By far, oropharyngeal and nasopharyngeal swabs, as well as sputum, are the recommended respiratory specimens for the detection of SARS-CoV-2. A recent study in Wuhan showed that in suspected individuals the positive rate of RT-PCR tests in sputum (76.9%) is significantly higher than the positive rate of oropharyngeal swabs (44.2%), but the nasopharyngeal swab was not included in their study [8]. Wang K et al. reported that in 68 patients with COVID-19, SARS-CoV-2 RNA was detected from nasopharyngeal swabs in 48 patients (70.6%) and from sputum specimens in 30 patients (44.1%) [9]. According to the study of Wang X et al., the positive rate was quite different between nasopharyngeal (32.9%) and oropharyngeal swabs (9.3%) among inpatients with COVID-19 [6]. Notably, dry cough was reported to be common in patients with COVID-19, making sputum collection difficult in many clinical settings [10, 11], and coughing up sputum can increase the chance of health care workers’ exposure. To seek a more applicable specimen for the detection of SARS-CoV-2 in order to efficiently use the limited public health resources under the current pandemic situation, we would like to propose a study to compare the positive rates of RT-PCR testing in nasopharyngeal swabs, oropharyngeal swabs, and sputum for the detection of SARS-CoV-2.
METHOD
Study Site and Study Population
An observational study was conducted at Beijing Ditan Hospital, which is 1 of the 3 designated medical centers to treat and manage suspected and confirmed COVID-19 patients in Beijing. We collected nasopharyngeal swabs, oropharyngeal swabs, and sputum from February 2 to February 19 from 43 confirmed COVID-19 patients, who were admitted to our hospital from January 21 to February 19, 2020. All 43 patients were diagnosed according to Chinese management guidelines for COVID-19 [3] (they met the criteria for suspected cases and had at least 2 positive results by RT-PCR assay for SARS-CoV-2 or a genetic sequence that matched SARS-CoV-2). Each patient had an electronic case file to collect basic demographics, medical history, and biochemical and radiological test results during hospitalization.
Sample Collection
We collected a nasopharyngeal swab, oropharyngeal swab, and sputum at the same sampling times, and each patient could be sampled more than once during their disease course. The nasopharyngeal swab and oropharyngeal swab were both collected using the Applied Cell Nasopharyngeal Swab with a flocking head. The nasopharyngeal swab was collected from a single nostril, and the oropharyngeal swab was collected from both sides of the throat according to detailed sampling videos [12] (http://www.cslm.org.cn/cn/news.asp?id=74.html). When collecting sputum specimens, patients were instructed to breathe in deeply and cough hard, then produce sputum from deep inside the chest. To control the quality, all specimens were collected at 8 am by trained nurses. Specimens were stored in a collection tube with 5 mL of virus preservation solution and transferred to the laboratory within 2–3 hours by a biosafety box at a temperature of 0°C~4°C.
Laboratory Testing
RT-PCT testing was conducted in the P2+ laboratory of Beijing Ditan Hospital, which is the officially recognized laboratory for confirmation of SARS-CoV-2 infection. Viral RNA was extracted within 2 hours using the QIAamp Viral RNA Mini Kit with primers and probes targeting the open reading frame 1ab (ORF1ab) and nucleocapsid protein (N): ORF1ab: forward primer CCCTGTGGGTTTTACACTTAA; reverse primer ACGATTGTGCATCAGCTGA; and the probe 5’-VIC-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′. N gene: forward primer GGGGAACTTCTCCTGCTAGAAT; reverse primer CAGACATTTTGCTCTCAAGCTG; and the probe 5’-FAM- TTGCTGCTGCTTGACAGATT-TAMRA-3′. Reaction system and amplification conditions were performed according to the manufacturer’s specifications (Shanghai BioGerm Medical Technology Co. Ltd., Shanghai, China). The result was considered valid when the cycle threshold (Ct) value of the reference gene was ≤38. The result was considered positive when the Ct values of both target genes were ≤38. If 1 of the target genes had a Ct value ≤38 and the other >38, it was interpreted as single-gene positive. A single-gene positive was also considered positive in our study.
Definitions
The positive rate of the RT-PCR testing was calculated as the number of specimens that tested positive divided by the total number of specimens tested. The positive rate was compared among the 3 specimens collected at the 3 disease durations.
Disease duration was calculated from the date of symptom onset to the date of sample collection and was used to divide patients into 3 groups: ≤14 days, 14–21 days, and >21 days.
Statistical Analysis
Continuous variables were described as means (SDs) or medians (interquartile ranges [IQRs]). Categorical variables were described as counts and percentages. The chi-square test and Fisher exact test were used for comparison, and Bonferroni adjustment was used to account for multiple comparisons between different sample types (P < .016 was considered statistically significant). A kappa coefficient (KC) was used to evaluate testing consistency across 3 different specimens (highly consistent if KC ≥ 0.75, consistent if KC ≥ 0.4, and low consistent if KC < 0.4; P < .05 was considered statistically significant). All analyses were conducted with SAS statistical software (version 9.2).
Patient Consent Statement
Each patient included in this study signed an informed consent form when they were admitted to the hospital, and any information of patients was anonymized throughout the study and writing process. The design of our study has been reviewed and approved by the Ethics Review Board of Beijing Ditan Hospital (IRB 2020-011-02).
RESULTS
Basic Characteristics
We conducted sampling 97 times (24 times at ≤14 days, 33 times at 14–21 days, and 40 times at >21 days) and collected a nasopharyngeal swab, oropharyngeal swab, and sputum specimen each time. A total of 291 specimens from 43 confirmed COVID-19 patients, including 97 of each type of specimen, were collected and tested. Among 43 included patients, 21 (48.8%) patients were male and the average age (SD) was 43.8 (17.1) years. Thirty-six patients (83.7%) had mild illness, 6 patients had severe illness, and 1 patient was critically ill (Supplementary Table 1).
Comparison of the Testing Yield
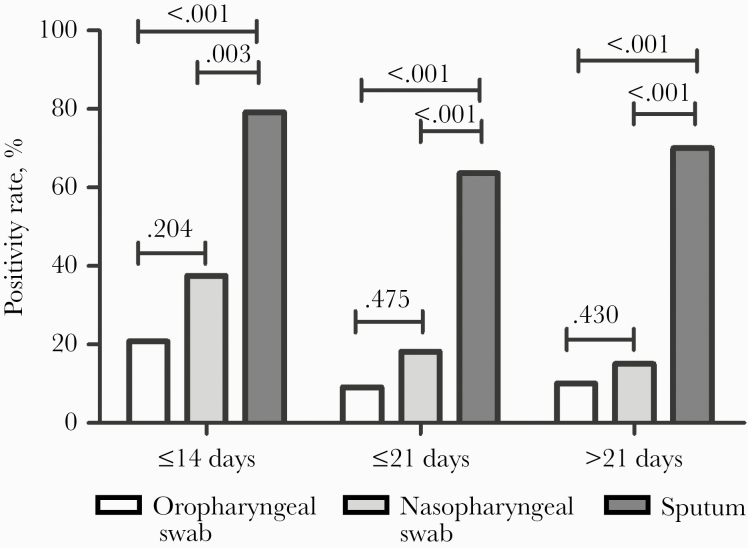
As shown in Figure 1, the positive rate of RT-PCR testing in sputum was the highest, followed by nasopharyngeal swabs and oropharyngeal swabs across the 3 disease durations. In the disease duration of ≤14 days, the positive rate in sputum (79.2%) was significantly higher than that in nasopharyngeal swabs (37.5%; P = .003) and oropharyngeal swabs (20.8%; P < .001). In the disease duration of 14–21 days, the highest positive rate was also found in sputum (63.6%); it was significantly higher than that in nasopharyngeal swabs (18.2%; P < .001) and oropharyngeal swabs (9.1%; P < .001). A similar finding was observed in the disease duration of >21 days, that the positive rate was highest in sputum (70.0%) and significantly higher than that in nasopharyngeal swabs (15.0%; P < .001) and oropharyngeal swabs (10.0%; P < .001).
Figure 1.
The positive rate of various specimens at different disease durations.
We also compared the positive rates between nasopharyngeal swabs and oropharyngeal swabs. There was no statistical significance between the disease durations, even though the positive rates seemed generally higher in nasopharyngeal swabs than in oropharyngeal swabs (Figure 1).
Consistency Test
Tables 1–3 show the KC value of the pairwise comparison of 3 different specimens collected across 3 disease durations. For nasopharyngeal swabs and oropharyngeal swabs, the testing consistency was low in the disease durations of ≤14 days and >21 days, with KC values of 0.22 (P = .157) and 0.09 (P = .480), respectively. However, the differences in positive rates were not statistically significant. In the disease duration 14–21 days, the testing consistency between the 2 specimens was higher but not significantly so (KC, 0.62; P = .083).
Table 1. .
The Consistency Between Nasopharyngeal Swabs and Oropharyngeal Swabs
| Nasopharyngeal Swabs | ||||
|---|---|---|---|---|
| Oropharyngeal Swabs | Positive | Negative | Kappa | P |
| ≤ 14 d | 0.22 (0.16 to 0.6) | .157 | ||
| Positive | 3 (33.3) | 2 (13.3) | ||
| Negativity | 6 (66.7) | 13 (86.7) | ||
| ≤21 d | 0.62 (0.24 to 1.00) | .083 | ||
| Positive | 3 (50.0) | 0 (0.0) | ||
| Negativity | 3 (50.0) | 27 (100.0) | ||
| >21 d | 0.09 (0.27 to 0.45) | .480 | ||
| Positive | 1 (16.7) | 3 (8.8) | ||
| Negativity | 5 (83.3) | 31 (91.2) | ||
Table 3. .
The Consistency Between Sputum Specimens and Nasopharyngeal Swabs
| Sputum Specimens | ||||
|---|---|---|---|---|
| Nasopharyngeal Swabs | Positive | Negative | Kappa | P |
| ≤14 d | 0.13 (0.13 to 0.38) | .004 | ||
| Positive | 8 (42.1) | 1 (20.0) | ||
| Negativity | 11 (57.9) | 4 (80.0) | ||
| ≤21 d | 0.12 (0.08 to 0.32) | .000 | ||
| Positive | 5 (23.8) | 1 (8.3) | ||
| Negativity | 16 (76.2) | 11 (91.7) | ||
| >21 d | –0.09 (–0.28 to 0.09) | <.001 | ||
| Positive | 3 (10.7) | 3 (25.0) | ||
| Negativity | 25 (89.3) | 9 (75.0) | ||
Between sputum and oropharyngeal swabs, the testing consistency was very low across all 3 disease durations with statistical significance. The KC values in the disease durations of ≤14 days, 14–21 days, and >21 days were 0.13 (P = .000), 0.11 (P < .001), and 0.02 (P < .001), respectively (Table 2). Similar findings were observed in the consistency test between sputum and nasopharyngeal swabs, that there was a low consistency across the 3 disease durations, with statistical significance. The KC values were 0.13 (P = .004), 0.12 (P = .000), and –0.09 (P < .001) for the disease durations of ≤14 days, 14–21 days, and >21 days, respectively (Table 3).
Table 2. .
The Consistency Between Sputum Specimens and Oropharyngeal Swabs
| Sputum Specimens | ||||
|---|---|---|---|---|
| Oropharyngeal Swabs | Positive | Negative | Kappa | P |
| ≤14 d | 0.13 (0.02 to 0.27) | .000 | ||
| Positive | 5 (26.3) | 0.0 (0.0) | ||
| Negativity | 14 (73.7) | 5 (100.0) | ||
| ≤21 d | 0.11 (0.02 to 0.23) | <.001 | ||
| Positive | 3 (14.3) | 0 (0.0) | ||
| Negativity | 18 (85.7) | 12 (100.0) | ||
| >21 d | 0.02 (–0.11 to 0.14) | <.001 | ||
| Positive | 3 (10.7) | 1 (8.3) | ||
| Negativity | 25 (89.3) | 11 (91.7) | ||
DISCUSSION
In our study, we found that the positive rate in sputum was significantly higher than that in nasopharyngeal swabs and oropharyngeal swabs in any disease duration we defined. It is similar to that of other respiratory viruses, such as influenza A and respiratory syncytial virus, with a detection rate in sputum that was significantly higher than that in nasopharyngeal swabs [13]. Several articles have revealed that the viral load of SARS-CoV-2 in sputum was significantly higher than that in both throat swabs and nasopharyngeal swabs [14, 15], while a report of 28 patients from the Korean Cohort Study on COVID-19 showed that viral shedding was higher in nasopharyngeal swabs than sputum [16]. We found that the positive rate was slightly higher in nasopharyngeal swabs than in oropharyngeal swabs. This is consistent with the research of Wang X et al., in which a higher positive rate was observed in nasopharyngeal swabs than oropharyngeal swabs (P = .000) [6]. In another study conducted by the Chinese Center for Disease Control and Prevention (CCDC), the detection of SARS-CoV-2 also showed a higher positive rate (5/8; 63%) in nasopharyngeal swabs than oropharyngeal swabs (126/398; 32%) [5], but the number of nasopharyngeal swabs was very small, making the research inconclusive. In our results, the difference in positive rates between nasopharyngeal swabs and oropharyngeal swabs was not statistically significant, which may be related to our small sample size.
The discrepancies of consistency tests between sputum and nasopharyngeal swabs, and between sputum and oropharyngeal swabs, indicate that sputum could be a more reliable specimen for SARS-CoV-2 detection. However, not all patients with COVID-19 produce sputum [11]. In many situations, sputum is hard to collect, especially among patients with dry cough. The majority of patients with COVID-19 are mild, but the virus has high infectivity [17], so induced sputum in patients might cause further transmission. Therefore, oropharyngeal swabs and nasopharyngeal swabs are the most commonly used specimens for SARS-CoV-2 detection in China and the United States [18, 19]. However, the collection of oropharyngeal swabs or nasopharyngeal swabs requires the patient to open their mouth wide enough that it might cause discomfort and epistaxis [12, 19]. The close contact between health care workers and patients creates a high risk of transmission.
As the pandemic is still accelerating, the global public health system is being challenged. Because of the lack of specific antiviral treatment and a vaccine for COVID-19, it is critical to efficiently use limited public health resources to detect SARS-CoV-2. The choice of specimen is not only based on the efficiency of viral detection, but also the cost and available expertise. Researchers have reported that there is a high expression of ACE-2 receptor of SARS-CoV-2 on cell surfaces in the tongue and salivary tissues [20], and the positive rates for saliva reported in different studies were 31%–91% [21, 22]. Considering that sputum is not a common clinical manifestation of COVID-19 and that there is a probability of collecting saliva as sputum based on the method of sputum sampling, researchers have demonstrated the potential for saliva to be an ideal specimen type for the detection of SARS-CoV-2 [23]. In our study, we found that nasopharyngeal swabs had a slightly higher yield of SARS-CoV-2 detection compared with oropharyngeal swabs. The collection of nasopharyngeal swabs is considered to be more convenient and safer than the collection of oropharyngeal swabs for health care workers, which made us change our policy in favor of nasopharyngeal swabs for testing. Currently, in our hospital, we try to use nasopharyngeal swabs for SARS-CoV-2 detection instead of oropharyngeal swabs when sputum is not available. The outcomes of this strategy change will be published shortly.
According to our study, the positive rates using nasopharyngeal swabs and oropharyngeal swabs were not satisfactory among our confirmed patients. In Wang X et al.’s study, false-negative results occurred in the late stage of hospitalization, especially in oropharyngeal swabs [6]. Chen et al. reported that a certain proportion of COVID-19 patients had positive RT-PCR results for SARS-CoV-2 in the sputum or feces after oropharyngeal swabs became negative [24]. These findings suggest that we should be cautious when using negative RT-PCR assays with oropharyngeal swabs as the criterion to rule out infection or to determine COVID-19 cure. We also found that not only sputum but also nasopharyngeal and oropharyngeal swabs had a better yield when collected within 14 days of symptom onset. When the duration after symptom onset was >21 days, the profitability of harvest was lower for all types of samples collected. Studies have suggested that viral shedding from upper respiratory tract specimens may reach its peak during the early days after illness onset [4, 16]. Early testing might be critical to increase the yield for SARS-CoV-2 detection in nasopharyngeal and oropharyngeal swabs, especially when sputum is not available [5]. It is possible that some of these patients are already in the convalescence phase, and that is why the negative tests are actually true negative. Further studies are needed to confirm our hypothesis. Our current study included all 3 commonly used respiratory specimens, with different disease durations; we compared the positive rates and evaluated testing consistency across these 3 specimens simultaneously, which enabled us to add more comprehensive information to the literature.
There are several limitations to our study. First, the generalizability of our study results might be restricted by the single-center source and the small number of patients included in our study. Patients with severe disease were somewhat under-represented (n = 7), and the sample size was too small to stratify analysis based on disease severity. Further studies are needed that include multiple centers and large numbers of patients. Second, we did not detect the viral load in these specimens, which is a more representative indicator of the viral distribution. The Ct value of the samples was also not available in this study to provide semiquantified virus levels. Third, the patients we included and tested mostly had mild to severe disease. The results cannot be generalized to asymptomatic patients. Other types and larger sizes of specimens for SARS-CoV-2 detection need to be further studied.
CONCLUSIONS
In conclusion, compared with nasopharyngeal swabs and oropharyngeal swabs, sputum had the highest yield in RT-PCR testing for SARS-CoV-2 detection. Nasopharyngeal swabs and oropharyngeal swabs had similar yields of SARS-CoV-2 detection. When sputum is hard to get, nasopharyngeal swabs could be a better option, and early testing might be needed.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We appreciate the support of the Beijing Science and Technology Commission. We acknowledge all the health care workers involved in the diagnosis and treatment of patients in China.
Financial support. This work was funded by Beijing Ditan Hospital, Capital Medical University.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. F.Z., H.Z., and G.W. had the idea for and designed the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Y.Z., J.W., D.L., Y.W., and J.B. contributed to sample acquisition and data collection. Y.W., J.G., C.L., F.Y., and L.Y. contributed to sample testing. G.G., S.W., and D.Y. contributed to the diagnosis and treatment of patients. G.W. contributed to the statistical analysis. H.Z. and M.C. contributed to data analysis and data interpretation. M.C. and L.M.W. contributed to the writing of the article. All authors reviewed and approved the final version.
References
- 1. World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19-1 June 2020. Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---01-june-2020. Accessed 1 June 2020. [Google Scholar]
- 2. World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19–23 March 2020. Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---23-march-2020. Accessed 23 March 2020. [Google Scholar]
- 3. National Health Commission of the People’s Republic of China. Chinese COVID-19 diagnosis and treatment plan (sixth edition), 2020-02-19. Available at: http://www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2.shtml. Accessed 19 February 2020. [Google Scholar]
- 4. The COVID-19 Investigation Team. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 2020; 2:861–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X, Tan L, Wang X, et al. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int J Infect Dis 2020; 94:107–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan Y, Zhang D, Yang P, et al. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 2020; 20:411–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin C, Xiang J, Yan M, et al. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19). Clin Chem Lab Med 2020; 58:1089–94. [DOI] [PubMed] [Google Scholar]
- 9. Wang K, Zhang X, Sun J, et al. Differences of SARS-CoV-2 shedding duration in sputum and nasopharyngeal swab specimens among adult inpatients with COVID-19. Chest 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan China. JAMA 2020; 323:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marty FM, Chen K, Verrill KA. How to obtain a nasopharyngeal swab specimen. N Engl J Med 2020; 382:e76. [DOI] [PubMed] [Google Scholar]
- 13. Jeong JH, Kim KH, Jeong SH, et al. Comparison of sputum and nasopharyngeal swabs for detection of respiratory viruses. J Med Virol 2014; 86:2122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 15. Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis 2020; 71:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim ES, Chin BS, Kang CK, et al. Clinical course and outcomes of patients with severe acute respiratory syndrome coronavirus 2 infection: a preliminary report of the first 28 patients from the Korean Cohort Study on COVID-19. J Korean Med Sci 2020; 35:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020; 382:1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323:1239–42. [DOI] [PubMed] [Google Scholar]
- 19. Gupta K, Bellino PM, Charness ME. Adverse effects of nasopharyngeal swabs: three-dimensional printed versus commercial swabs. Infect Control Hosp Epidemiol 2020; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci 2020; 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baghizadeh FM. What dentists need to know about COVID-19. Oral Oncol 2020; 105:104741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pasomsub E, Watcharananan SP, Boonyawat K, et al. Saliva sample as a noninvasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. To KK, Tsang OT, Yip CC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 2020; 71:841–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen C, Gao G, Xu Y, et al. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann Intern Med 2020; 172:832–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.