Abstract
Utilizing results of polymerase chain reaction (PCR) testing and subsequent antibody titers, we report on the test characteristics of a PCR screening test for severe acute respiratory syndrome coronavirus 2 among hospital workers. The PCR test was found to be 87% sensitive and 97% specific, with a positive predictive value of 0.98 and a negative predictive value of 0.80.
Keywords: SARS-CoV-2, nasopharyngeal PCR swab, COVID-19
Since the first case of pneumonia was reported in, Wuhan, China, in December 2019 [1], hospital workers have been at risk for exposure to the novel coronavirus currently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2]. During the early days of this pandemic, testing was administered selectively to identify and isolate those infected. Little objective evidence was available to determine the pretest probability and predictive value of these tests. We report a retrospective, cross-sectional, observational study analyzing SARS-CoV-2 health care worker testing at a tertiary care hospital in Long Island, New York.
METHOD
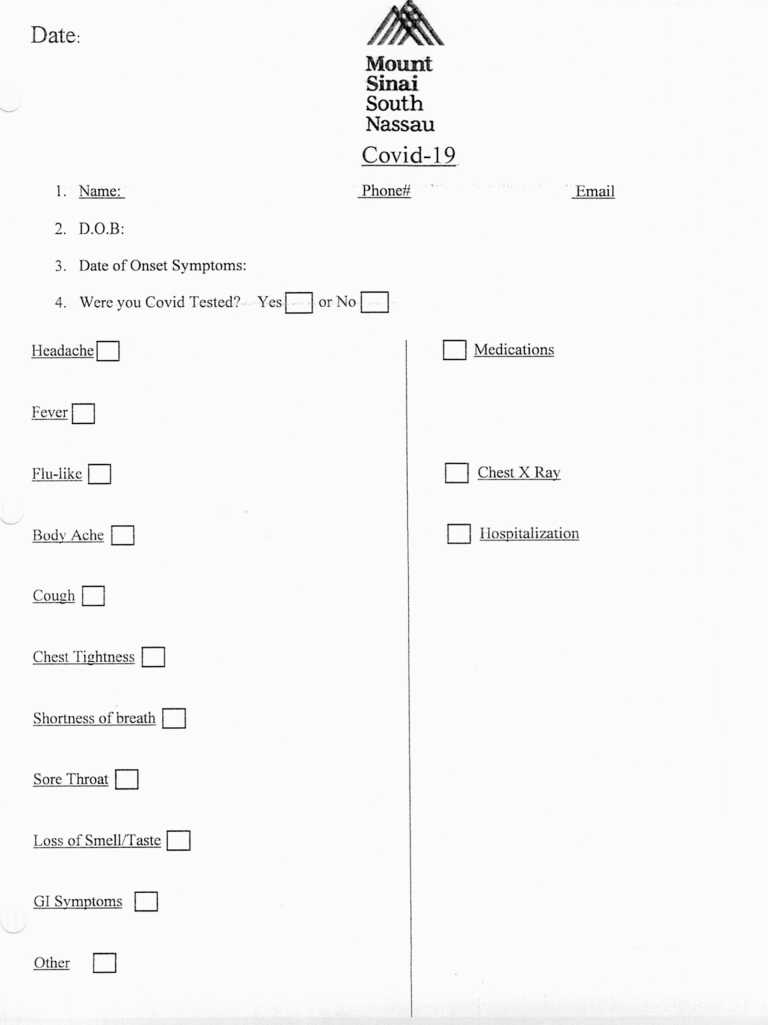
From March 1 to April 30, 2020, Mount Sinai South Nassau tested health care workers with “COVID-19-like” symptoms or significant exposure (defined as at least 15 minutes in close proximity) to a SARS-CoV-2 patient without wearing personal protective equipment using an established reverse transcription polymerase chain reaction (RT-PCR) test on a nasopharyngeal specimen. Due to the limited availability of testing supplies, only symptomatic employees were offered on-site testing. No patient consent statement specific to this testing was obtained, as this was part of the routine care that all patients were receiving. This RT-PCR was performed using the cobas SARS-CoV-2 test on the Roche 6800 utilizing Light Cycler 480 II (Roche) and 2019-novel coronavirus (2019-nCoV) primers and probes (IDT). The limit of detection of the assay is 1 genome copy/μL [3]. Subsequently, they underwent testing for the presence of SARS-CoV-2 IgG antibodies using the Mount Sinai test looking at M spike protein at least 3 weeks after the onset of their symptoms. As shown by Zhou et al. (2020), most patients infected by SARS-CoV-2 develop a peak IgM response by day 9 and an IgG response by 2 weeks [4]. Ni et al. (2020) further characterized the cellular and humoral immune response to SARS-CoV-2 and showed that the neutralizing antibody titers significantly correlated with the numbers of specific T-cell responses to the virus [5]. In turn, the SARS-CoV-2 antibody titers were correlated with in vitro viral neutralization with a specificity of 100%. All staff also completed a questionnaire self-reporting symptoms experienced before the initial RT-PCR screening test using a questionnaire (Figure 1). We analyzed data including date and result of SARS-CoV-2 PCR, symptoms on presentation, and date and result of antibody testing.
Figure 1.
Pre-testing questionnaire for COVID-19 RT-PCR. Abbreviations: Covid-19, coronavirus 2019; D.O.B, date of birth; GI, gastrointestinal.
Symptoms reported were aggregated and compared with SARS-CoV-2 PCR and antibody tests. Symptoms were compared via the 2-tailed z-test, with the threshold for significance defined as a P value of .05.
RESULTS
One hundred five health care workers underwent both SARS-CoV-2 PCR and antibody testing. Sixty-one (58.1%) tested positive using the SARS-CoV-2 PCR test; 44 (41.9%) tested negative. Of the 61 who tested positive, 60 (98.4%) subsequently had antibodies while only 1 (1.6%) had no antibodies. Of the 44 health care workers with a negative SARS-CoV-2 PCR test, 9 (20.5%) were subsequently found to have antibodies. Quantitative titers showed that 94% of subjects developed a robust immune response with a titer >1:320 (chosen as the level accepted by Mount Sinai for plasma donation). At the time of SARS-CoV-2 PCR testing, 76 health care workers reported symptoms, including fevers, chills, cough, dyspnea, wheezing, myalgia, pain, nausea, vomiting, diarrhea, fatigue, anosmia, throat pain, otalgia, tachycardia, and hypersomnia. Statistical analysis of a larger group of employees (n = 461 total tested for antibodies, including these 105 employees) revealed cough and myalgia to be the 2 symptoms significantly different between those with and without antibody (Table 1A). The most common symptom reported was cough. The SARS-CoV-2 PCR test via nasopharyngeal swab was found to be 87% sensitive and 97% specific. The PCR test was also found to have a positive predictive value (PPV) of 0.98 and a negative predictive value (NPV) of 0.80 (Table 1B) among symptomatic patients.
Table 1A.
Distribution of Reported Symptoms and Their Statistical Significance
| Symptom | Total | Pos | Neg | P Value |
|---|---|---|---|---|
| Chills | 6 (0.08) | 4 (0.08) | 2 (0.09) | .864560133 |
| Fever | 27 (0.36) | 17 (0.32) | 10 (0.43) | .33998196 |
| Cough | 60 (0.79) | 46 (0.87) | 14 (0.61) | .010878595 |
| SOB | 23 (0.30) | 15 (0.28) | 8 (0.35) | .572089927 |
| Wheezing | 2 (0.03) | 1 (0.02) | 1 (0.04) | .53806803 |
| Headache | 21 (0.28) | 18 (0.34) | 3 (0.13) | .060999114 |
| Diarrhea | 7 (0.09) | 5 (0.09) | 2 (0.09) | .918556266 |
| Nausea/vomiting | 3 (0.04) | 3 (0.06) | 0 (0.00) | .244338211 |
| Myalgias | 35 (0.46) | 31 (0.58) | 4 (0.17) | .000958949 |
| Fatigue/malaise/weakness | 6 (0.08) | 4 (0.08) | 2 (0.09) | .864560133 |
| Anosmia | 6 (0.08) | 6 (0.11) | 0 (0.00) | .092694007 |
| Sinus | 8 (0.11) | 6 (0.11) | 2 (0.09) | .731918328 |
| Sore throat | 23 (0.30) | 14 (0.26) | 9 (0.39) | .26764694 |
| Lightheaded/dizzy | 3 (0.04) | 1 (0.02) | 2 (0.09) | .161385004 |
| Otalgia | 1 (0.01) | 0 (0.00) | 1 (0.04) | .126489104 |
| Tachycardia | 1 (0.01) | 0 (0.00) | 1 (0.04) | .126489104 |
| Hypersomnia | 1 (0.01) | 0 (0.00) | 1 (0.04) | .126489104 |
Bold indicates symptoms found to be of statistical significance.
Table 1B.
Distribution of Subjects With Both SARS Cov2 PCR and Antibody Result (n = 105)
| Patients | No. | Antibody Positive Titer | Positive Titer <1:320 | Positive Titer ≥1:320 | Antibody Negative Titer |
|---|---|---|---|---|---|
| SARS-CoV-2 | 61 | 60 | 2 | 58 | 1 |
| PCR (+) | |||||
| SARS-CoV-2 | 44 | 9 | 2 | 7 | 35 |
| PCR (-) |
Abbreviations: PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SOB, shortness of breath.
DISCUSSION
Effective use of the RT-PCR test to determine the likelihood of COVID illness is dependent on various factors. The lack of availability of adequate SARS-CoV-2 testing, reports of false-negative tests [6], and variability of symptoms on presentation [7] hinder accurate diagnosis. The high PPV seen is this study reinforces the rationale for PCR testing to identify new active cases. However, one-fifth (20.5%) of symptomatic or significantly exposed staff were incorrectly labeled as not having SARS-CoV-2 after a single negative nasopharyngeal swab. This could be due to subdetectable viral load at the time of screening, poor collection technique, and poor quality of specimen collected (depth of swab and quality of sweep of nasopharyngeal region).
Our study has some notable limitations. Symptoms were self-reported, which does not preclude recall bias, and lacked objective verification. Antibody titers were thus not correlated by symptoms due to the subjective nature of symptom reporting. The calculated specificity was dependent on the assumption that employees did not become infected between the screening test and antibody testing and applies only to symptomatic employees. However, our study shows the value and limitations of relying on a single PCR nasopharyngeal swab to diagnose SARS-CoV2 illness.
Acknowledgments
Financial support. None reported.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. Patient consent specific to this testing was not obtained, as all care was provided as part of routine patient care.
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roche Diagnostics. cobas® SARS-CoV-2 test (for the COVID-19 coronavirus) Available at: https://diagnostics.roche.com/us/en/products/params/cobas-sars-cov-2-test.html. Accessed 13 July 2020.
- 4. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ni L, Ye F, Cheng ML, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 2020; 52:971–7.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 20021493 [Preprint]. Available at: 10.1101/2020.02.11.20021493. Accessed 13 July 2020. [DOI] [Google Scholar]
- 7. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]