Abstract
Background
Plasmid-mediated resistance to the last-resort drugs: carbapenems and colistin is an emerging public health threat. The studies on the prevalence and co-expression of resistant genes among livestock and human pathogens are rare in Nepal. This is the first study in Nepal exploring the prevalence and co-existence of colistin resistance gene, mcr-1 along with carbapenemase resistance gene, OXA-48 in Escherichia coli isolated from poultry and clinical specimens.
Methods
A total of 240 rectal swabs from chickens of five different poultry farms of Kathmandu valley and 705 mid-stream urine samples from human subjects attending Kantipur Hospital, Kathmandu were collected between August, 2018 and March, 2019. Rectal swabs and urine specimens were cultured. E. coli isolated from the specimens were screened for antimicrobial susceptibility testing (AST) using disk diffusion method’. Minimum inhibitory concentration (MIC) of colistin was determined by agar dilution method using 0.5 µg/ml to 32 µg/ml. The E. coli isolates were first screened for mcr-1 followed by screening for OXA-48 genes using conventional Polymerase chain reaction (PCR).
Results
Of the total samples analyzed, E. coli was isolated from 31.7% (76/240) of poultry and 7.9% (56/705) of clinical specimens. In AST, 80% (61/76) of E. coli from poultry and 79% (44/56) from clinical specimens were MDR. The phenotypic prevalence of colistin resistance in poultry specimens were 31.6% (24/76) and clinical specimens were 21.4% (12/56). In PCR assay, 27.6% (21/76) of poultry and 19.6% (11/56) of clinical isolates had colistin resistant mcr-1 gene. MICs value of E. coli isolates ranged from 4 to 32 (µg/ml) in both clinical and poultry isolates. Prevalence of co-existing carbapenem resistance gene, OXA-48, among colistin resistant mcr-1 positive isolates was 38% (8/21) in poultry specimens and 18.2% (2/11) in clinical specimens.
Conclusions
The high prevalence of colistin and carbapenem resistant genes, and their co-existence in plasmid DNA of E. coli isolates in this study suggests the possible spread to other animal, human and environmental pathogens. Molecular methods in addition to the conventional diagnostics in laboratories can help in early diagnosis, effective management and control of their potential transmission.
Keywords: mcr-1, OXA-48, Colistin resistant E. coli, MDR, Polymerase chain reaction
Background
The plasmid-encoded colistin resistant gene, mcr-1 was first reported in E. coli isolates from livestock and human specimens in China [1]. Until the discovery of mcr-1, all reported polymyxin resistance mechanisms were chromosomally mediated, due to mutation and regulatory changes [2], and had never been reported to occur via horizontal gene transfer [3]. Two earlier mechanisms: natural and phenotypic mechanisms were suggested among colistin resistant strains, the former occurring via mutations of bacterial genomes while the latter was the result of adaptive mechanism [3]. A number of mcr-1 strains have been reported worldwide among several species of Enterobacteriaceae in a short span of time since its first report in 2016 [4].
mcr-1 gene is not only limited to E. coli but also has been spreading among other members of Enterobacteriaceae co-existing with other resistance genes [5]. Several retrospective studies performed worldwide showed that mcr-1 had been circulating undetected for at least 20 years [6]. Subsequent findings of 11 new genetic variant of mcr-1 across different countries show increasing divergence of colistin resistance mechanism [7]. There is a strong association of ISApl1 genomic insertion site and mcr-1, which causes demographic expansion and global distribution [2]. Furthermore, insertion of mcr-1 into E. coli chromosomes may enable it to become intrinsically resistant, which is expected to become more prevalent in future [8].
mcr-1 genes carrying colistin resistant E. coli isolates are challenging ‘one health’ concept as these strains have been isolated from humans, animals, and environ-ments from all continents [9]. Finding of mcr-1 gene carrying bacteria in natural environments shows possibility of transference of mcr-1 carrying Enterobacteriaceae to humans via food chain [10].
There is a growing number of findings reporting the rapid surge of carbapenemase including blaVIM-1, blaNDM-1 and OXA-48 among carbapenem resistance Enterobacteriaceae (CRE) in human beings [1, 11, 12], though the CRE is still rare in animals [13]. The emergence and global spread of co-existing carbapenem resistance with plasmid-mediated colistin resistance (mcr-1) in gram-negative bacterial pathogens, particularly among the members of Enterobacteriaceae can be catastrophic [13, 14]. There is an increasing concern on the co-existence of colistin and carbapenem resistances in Enterobacteriaceae from human and animal samples, because this combination can limit the therapeutic options in the treatment of MDR bacteria [4, 15].
In Nepal, there is a pervasive use of antibiotics in food animals farming as growth promoters in addition to indiscriminate over the counter use among humans [16, 17]. The colistin resistance can increase in exponential proportion because of its widespread use in food animals and human beings and has been supported by the detection of mcr-1 gene from environment, food, animals and human beings [18]. Although global reports on colistin resistance in human and environment are increasing [19], there is only one previous report till date from Nepal. Recent studies reported from Nepal showed 26.66% of colistin-resistant E. coli harbored mcr-1 gene isolated from the chicken meats [20] and 33.3% of carbapenem resistant E. coli harbored OXA-48 isolated from urine samples [21]. The main objective of this study was to explore the prevalence and co-existence of colistin resistance gene, mcr-1 along with carbapenemase resistance gene, OXA-48 in Escherichia coli isolated from poultry farms located in Kathmandu, Kavrepalanchok and Bhaktapur and human specimens (urine) from patients attending Kantipur Hospital, Tinkune, Kathmandu between August, 2018 and March, 2019.
Methods
Study design and study sample
This cross-sectional study was carried out between August 2018 and March 2019 in Bhaktapur Kavrepalanchok, and Kathmandu districts. The clinical specimens were collected from Kantipur Hospital, Tinkune Kathmandu. The study subjects included patients of all ages and both gender (male and female) attending the hospital with suspicion of UTI and all of the participants provided written informed consent to participate in the study. A total of 705 mid-stream urine samples from patients (male, N = 315 and female, N = 390) suspected of urinary tract infection (UTI) were collected in a sterile, clean, well-labeled, and leak proof container with no visible signs of contamination and were transported to laboratory [22].
The poultry specimens were collected from selected poultry farms in Kavrepalanchok (Panauti), Bhaktapur (Darjeeling Height, Duwakot), Kathmandu (Sundarijal and Dhakal Gau). A total of 240 rectal swabs of chicken were collected and inoculated into buffered peptone water, kept in insulated ice-cold box and were transported to laboratory within 1 hour.
Culture of specimens and identification of the isolates
The samples were observed for macroscopic, microscopic and culture characteristics. Urine samples were inoculated on Cysteine Lactose Electrolyte Deficient (CLED) (Hi Media, India) agar using a standard calibrated loop (4 mm). The collected rectal swabs with inoculated tubes were incubated for 18–24 h at 37 °C, and were sub-cultured on MacConkey agar (MA) (Hi Media, India). Inoculated CLED media plates were incubated aerobically at 37 °C for 18–24 h. Thus, obtained colony growth of gram-negative rods suspected of E. coli were further sub-cultured aerobically on Nutrient Agar (NA) from both MA and CLED media plates. E. coli from both specimens was identified on the basis of colony morphology, staining, biochemical tests and a greenish-metallic sheen of colonies formed on Eosin Methylene Blue (EMB) agar [23, 24].
Antibiotic susceptibility test (AST)
All isolates of E. coli were tested for antibiotic susceptibility using modified Kirby Bauer disc diffusion method based on the Clinical and Laboratory Standard Institute guidelines [25]. Isolates were tested for resistance against amoxiclave (30 µg), ceftazidime (30 µg), cefixime (5 µg), ciprofloxacin (5 µg), gentamicin (10 µg), imipenem (10 µg), meropenem (10 µg), piperacillin/tazobactam (100/10 µg) and tetracycline (30 µg) (HiMedia, India). Results were interpreted as sensitive, intermediate and resistant [25]. Isolates showing non-susceptibility (either resistance or intermediate) to at least one antimicrobial agent in three or more of the categories were considered as multi-drug resistant (MDR) [26].
Screening of colistin resistant E. coli
In AST, minimum inhibitory concentration (MIC) for colistin was determined by agar and broth micro-dilution methods using 0.5 µg/ml to 32 µg/ml. E. coli isolates showing visible growth on colistin concentration of 2 µg/ml or more than 2 µg/ml were detected as colistin resistant isolates [25].
Extraction of plasmid DNA and PCR amplification of colistin resistance gene (mcr-1)
The plasmid DNA was extracted using alkaline lysis method. The extracted plasmids were then suspended in TE buffer, labeled well and stored at – 20 °C [27]. mcr-1 gene of plasmid carried by E. coli isolates was amplified using primer pairs CLR5-F (5ʹCGGTCAGTCCGTTTGTTC-3ʹ) and CLR5-R (5ʹ-CTTGGTCGGTCTGTA GGG-3ʹ) as forward and reverse primers [1]. Reaction volume was set as 25 µl by adding 21 µl of 1X master mix, 0.5 µl each of forward and reverse primer and 3 µl of DNA template. The optimized PCR amplification of mcr-1 gene is 95 °C for 15 min to activate hot-star; 30 cycle of denaturation at 94 °C for 30 s; annealing at 57 °C for 1:30 min; extension at 72º for 1:30 min, and final extension at 72 °C for 10 min and holding at 4 °C for 10 min.
PCR amplification of carbapenem resistance gene (OXA-48)
OXA-48 gene responsible for the resistance of carbapenem drug was amplified using primer pairs as forward sequence OXA-48_forward primers (FP) (5′GCTTGATCGCCCTCGATT-3′) and reverse sequence OXA-48_reverse primers (RP) (3′GATTTGCTCCGTGGCCGAAA-5′). Reaction volume was set as 25 µl by adding 21 µl of 1× master mix, 0.5 µl each of forward and reverse primer and 3 µl of DNA template. The optimized PCR amplification of OXA-48 gene is 95 °C for 15 min to activate the hotstar, 30 cycle of denaturation at 94 °C for 30 s, annealing at 57 °C for 1:30 min and extension at 72º for 1:30 min and final extension at 72 °C for 10 min and holding at 4 °C for 10 min [28].
Agarose gel electrophoresis
The agarose gel electrophoresis of extracted plasmid DNA and amplified PCR product was performed, followed by confirmation of mcr-1 and OXA-48 gene by visualizing in UV transillumination [28].
Results
Distribution of samples and prevalence of E. coli in poultry and clinical specimens
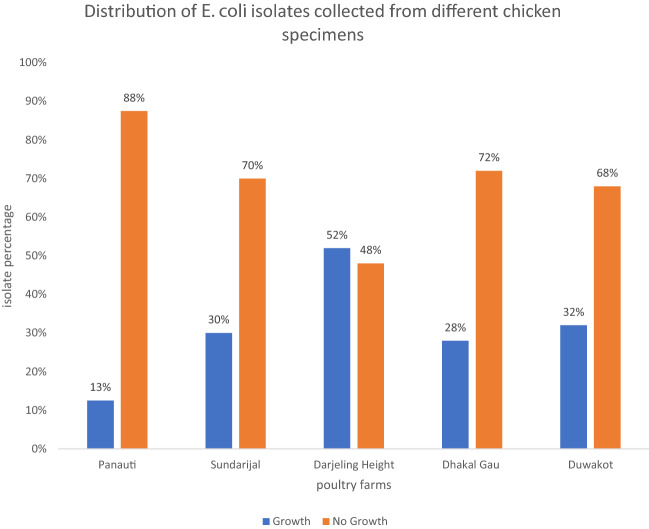
The distribution of poultry samples according to farms with previous history of vaccination, colistin antibiotic use, number of samples collected, number of E. coli isolates and the presence of colistin resistant isolates are illustrated in Table 1. The growth pattern observed in samples from each farm is presented in Fig. 1. Overall, the prevalence of E. coli in poultry specimens was (31.7%;76/240) whereas it was (7.9%; 56/705) in clinical specimens.
Table 1.
Farm wise epidemiological data of colistin resistant E. coli
| District | Poultry sites | Number of samples taken | Vaccination | Use of colistin | E. coli isolated | Colistin resistant E. coli isolated |
|---|---|---|---|---|---|---|
| Kavrepalanchok | Panauti | 40 | + | – | 5 | 2 |
| Bhaktapur | Duwakot | 50 | + | – | 16 | 9 |
| Darjeling Height | 50 | + | + | 26 | 6 | |
| Kathmandu | Sundarijal | 50 | + | + | 15 | 2 |
| Dhakal Gau | 50 | + | + | 14 | 2 | |
| Total | 240 | 76 | 21 |
Fig. 1.
Farm wise growth distribution of E. coli isolates
Among 705 urine specimens, (18.4%; 130/705) showed bacterial growth. Of 130 bacterial growth, (43%;56/130) were E. coli. On age wise distribution of UTI patients, 44.6% (58/130) were from age group (16–45) years, (32.3%;42/130) were from age group > 45 years and (23.1%; 30/130) were from age group (0–15) years respectively.
Antibiotic susceptibility test (AST) of E. coli isolates
Among the total of 76 E. coli isolates from poultry, highest resistance was found in tetracycline (67.1%; 51/76) followed by amoxiclave (55.3%; 42/76), ciprofloxacin (50%;38/76), nalidixic acid (50%;38/76) and imipenem (32.9%;25/76). Similarly, among the total of 56 E. coli isolates from clinical specimen, highest number of isolates were resistant to cefixime (71.6%;40/56) followed by ceftazidime (66.1%;37/56), ciprofloxacin (53.6%;30/56) and piperacillin/ tazobactam (39.3%;22/56) (Table 2).
Table 2.
Antibiotic resistance profile of E. coli isolated from poultry and clinical specimens
| Antibiotics | Resistance (%) | |
|---|---|---|
| Poultry (n = 76) | Clinical (n = 56) | |
| Amoxiclave (30 µg) | 42 (55.3) | 3 (5.4) |
| Ceftazidime (30µ g) | 4 (5.3) | 37 (66.1) |
| Cefixime (5 µg) | 10 (13.2) | 40 (71.4) |
| Ciprofloxacin (5 µg) | 38 (50) | 30 (53.6) |
| Gentamicin (10 µg) | 21 (27.6) | 12 (21.4) |
| Imipenem (10 µg) | 25 (32.9) | 2 (3.6) |
| Meropenem (10 µg) | 2 (2.6) | 0 |
| Nalidixic acid (30 µg) | 38 (50) | 11 (19.6) |
| Piperacillin/Tazobactam (100/10 µg) | 0 | 22 (39.3) |
| Tetracycline (30 µg) | 51 (67.1) | 21 (37.5) |
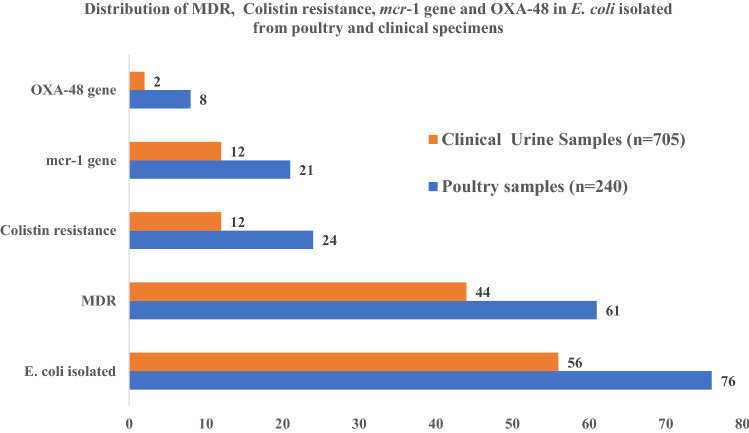
In AST, (80%; 61/76) of poultry specimens and (79%; 44/56) of clinical specimens were MDR (Fig. 2).
Fig. 2.
Distribution of MDR, colistin resistance, mcr-1 gene and OXA-48 in E. coli
Antimicrobial resistance phenotypes of clinical and poultry E. coli isolates
Antibiotic resistance pattern of MDR E. coli isolates were determined by 12 different antibiotic resistance phenotypes (R- phenotypes). Among them the most resistant pattern in poultry E. coli isolates were imipenem-tetracycline-amoxiclave (21%; 16/76) followed by imipenem-ciprofloxacin-amoxiclav (19.7%; 15/76) and ciprofloxacin- tetracycline-nalidixic (18.4%; 14/76). Among clinical E. coli isolates, the most resistant pattern was ceftazidime-cefixime-tetracycline (21.4%; 12/56) followed by cefixime-nalidixic acid-tetracycline (8.9%; 5/56) (Table 3).
Table 3.
Antibiotic resistance phenotypes of E. coli isolated from poultry and clinical specimens
| S.N | Antimicrobial resistance pattern | Poultry E. coli Resistant isolates N (%) |
Clinical E. coli Resistant isolates N (%) |
|---|---|---|---|
| 1 | CIP/TE/NA | 14 (18.4) | 2 (3.6%) |
| 2 | CIP/GEN/AMC | 9 (11.8) | |
| 3 | IMP/TE/AMC | 16 (21) | |
| 4 | IMP/CIP/AMC | 15 (19.7) | |
| 5 | CFM/NA/TE | 3 (4) | 5 (8.9%) |
| 6 | GEN/NA/AMC | 10 (13.2) | |
| 7 | IMP/CAZ/CIP | 1 (1.3) | 1 (1.8%) |
| 8 | CAZ/CFM/TE | 2 (2.6) | 12 (21.4%) |
| 9 | GEN/IMP/CIP | 3 (4) | 1 (1.8%) |
| 10 | CIP/TE/GEN/AMC | 4 (5.2) | |
| 11 | TE/AMC/GEN/NA | 6 (8) | |
| 12 | CIP/TE/AMC/NA | 8 (10.5) |
AMC Amoxiclave, CAZ Ceftazidime, CFM Cefixime, CIP Ciprofloxacin, GEN Gentamicin, IMP Imipenem, NA Nalidixic acid, TE Tetracycline
Determination of MIC and colistin resistant E. coli isolates
MICs value of E. coli isolates ranged from 4 to 32 (µg/ml) in both clinical and poultry E. coli isolates (Table 4). In this assay, 31.6% (24/76) of poultry isolates and 21.43% (12/76) of clinical isolates were confirmed as colistin resistant (Fig. 2) (Table 5).
Table 4.
MIC colistin resistant E. coli isolates of poultry and clinical specimens
| S. No | Colistin concentration (MIC) |
Resistant isolates | P -value | |
|---|---|---|---|---|
| Poultry (n = 24) | Clinical (n = 12) | |||
| 1 | 4 (µg/ml) | 4 (16.7) | 3 (25) | 0.8 |
| 2 | 8 (µg/ml) | 5 (20.8) | 2 (16.7) | |
| 3 | 16 (µg/ml) | 7 (29.2) | 4 (33.3) | |
| 4 | 32 (µg/ml) | 8 (33.3) | 3 (25) | |
Table 5.
Distribution of colistin resistance, mcr-1 gene and OXA-48 gene among E. coli isolates of poultry and clinical specimens
| Colistin resistant E. coli isolates | mcr-1 gene among colistin resistant E. coli isolates | OXA-48 gene positive isolates in mcr-1 gene positive E. coli | |||
|---|---|---|---|---|---|
| Poultry (n = 76) | Clinical (n = 56) | Poultry (n = 24) | Clinical (n = 12) | Poultry (n = 21) | Clinical (n = 11) |
| 24 (31.6%) | 12 (21.4%) | 21 (87.5%) | 11 (91.6%) | 8 (38%) | 2 (18.2%) |
Prevalence of mcr-1 gene among colistin resistant clinical and poultry E. coli isolates
All E. coli isolates were screened for plasmid mediated mcr-1 gene using conventional PCR. In PCR assay, colistin resistance was found to be 27.6% in poultry isolates and 19.6% in clinical isolates. Among phenotypic colistin resistance, 87.5% (21/24) E. coli isolates from poultry specimens were tested positive for mcr-1 gene and 91.6% (11/12) of the clinical isolates (E. coli) were tested positive for mcr-1 gene (Table 5). The mcr-1 with 309 bp size is presented in Fig. 3.
Fig. 3.
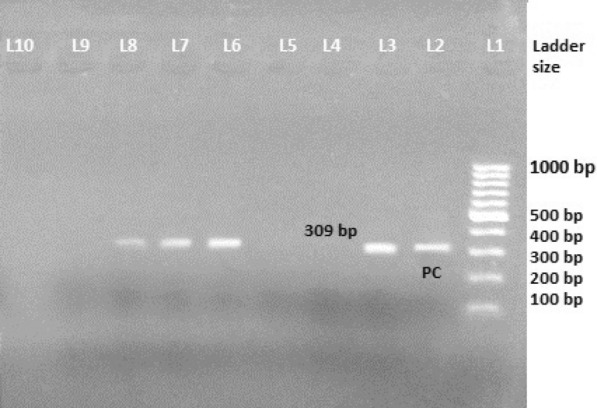
mcr-1 gene in colistin resistant E. coli (Lane1, marker DNA (100–1000 bp), Lane L2 (positive control), L3, L6, L7 and L8 mcr-1 positive (309 bp)
OXA-48 gene among mcr-1 positive colistin resistant E. coli isolates
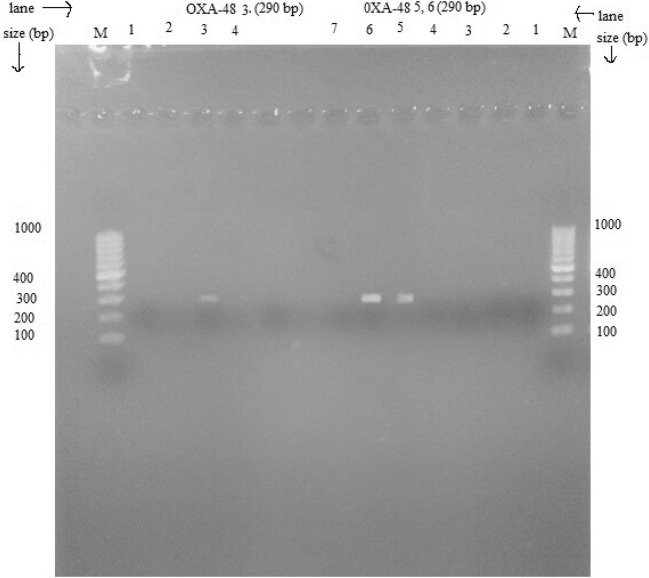
In this assay, 38% (8/21) of poultry isolates and 18.2% (2/11) of clinical isolates had carbapenem resistant OXA-48 gene (Table 5). The amplified OXA-48 gene with 290 bp is illustrated in Fig. 4.
Fig. 4.
OXA-48 gene among mcr-1 positive E. coli isolates (Lane M, marker DNA (100–1000 bp), lane 5, 6 and 3, OXA-48 gene (290 bp)
Discussion
This is the first report of plasmid mediated colistin resistance mcr-1 gene and OXA-48 gene among mcr-1 positive colistin resistant E. coli isolates from clinical and poultry specimens of Nepal. In this study, using PCR assay, colistin resistance was found to be 27.6% in poultry isolates and 19.6% in clinical isolates. The emergence and spread of CRE has obliged clinicians to use colistin—a drug notorious for its toxicity [29]. Colistin is the last resort drugs for these resistant isolates [30]. Increasingly over the recent years, morbidity, mortality and the economic consequences associated with colistin resistance are reported [31]. mcr-1 recently identified as resistance and transferable gene has been recovered from healthy carriers, food, environmental sources and clinical isolates [14]. Most of the mcr-1 positive strains have been proven to be carrying different carbapenemase genes such as blaNDM-9, blaNDM-5, blaVIM-1, blaOXA-48-type, and blaKPC- 2 [32]. This combination further limits the therapeutic options because both last resort drugs: colistin and carbapenems are challenged by this phenomenon [4]. Our study also confirmed the substantial presence of mcr-1 and blaOXA-48 among isolates of human and animal (poultry) origin.
In our study, 32% E. coli were isolated from poultry specimens consistent with a previous study that reported 36.4% growth of E. coli from poultry fed with colistin in Central Nepal [20]. Research conducted in China showed higher (80%) growth of E. coli from rectal swab of food animals [33]. The increased and variable rate of colistin resistance may be due to quality, concentration and extensive use of colistin in the livestock farming [34]. In clinical samples, E. coli remains as the most frequently isolated organisms which is consistent with several previous studies from Nepal [35–38]. The higher load of E. coli in urine samples may be due to their main role in urinary tract infections [35].
AST of poultry isolates showed more than 50% of resistance to most of the antibiotics used in this study. Similar resistance pattern was screened in China [2]. Another study reported from Iran had showed 77.7% resistance to ciprofloxacin, 33.3% resistance to gentamicin but no resistance to Imipenem [39]. A study from China reported higher rate of resistance to tetracycline (90%), cefixime (71.4%), and ceftazidime (66.1%) in E. coli isolates [33]. Another study from Pakistan reported higher resistance to gentamicin (77%) and ciprofloxacin (65%) in clinical isolates of Enterobacteriaceae [40]. Higher (79%) prevalence of multi-drug resistant isolates in our study is consistent with previous studies from Nepal [21, 41] and Pakistan [40]. The higher load of MDR isolates may be due to significant antibiotic pressure in the environment, irrational dose regimens, use in food animals and transmission of resistant isolates between people, animal and the environment [40]. In our study, 80% (61 out of 76) of E. coli from poultry were found to be MDR. Our finding was consistent with a study from Bharatpur, Nepal that showed MDR rate as 79.6% in poultry meat [42]. However, similar study from Bangladesh reported 100% MDR in E. coli isolates [43]. Open access between poultry farms and communities, unhygienic practice, inappropriate use of antibiotics, are some of the reasons attributed for emerging resistance patterns including MDRs in Nepal [33, 43, 44].
In this study, MIC value of colistin was observed up to 32 mg/L and found similar (4–32 µg/ml) dose in both poultry and clinical specimen. Most of the isolates of clinical specimen showed MIC value of 16 μg/ml while those E. coli isolates from poultry showed 32 μg/ml. Studies from Vietnam have reported lower MIC value of clinical isolates as 4 (µg/ml) [45] and MICs range of 4 to 16 μg/ml in E. coli isolates were reported from Chinese University Hospital [46]. The greater MIC value revealed these E. coli isolates as non-wild type [47]. The presence of multiple systems of resistance and multiple copy number of plasmids carrying mcr-1 gene may have played role in increasing the MIC value of colistin [48]. A multi-country study has reported consistent findings with our study [49]. A previous study from Germany has confirmed a presence of four plasmid carrying mcr-1 gene within E. coli isolates from patients, which indicated the role of mcr-1 gene for increased MIC of colistin [15].
In this study, the true occurrence of mcr-1 gene in poultry isolates is 27.6%. Similar result was reported by a study in Iran [50] while a study from Bangladesh reported a higher prevalence (94%) of colistin resistance in ESBLs E. coli isolates in poultry [51]. In this study, among 12 colistin resistant isolates, 11 (91.6%) were mcr-1 positive. The prevalence of mcr-1 gene is 19.6%. The findings of our study were in line with a study from Italy (8.3% resistance to colistin in hospital surfaces) [52]. The findings of our study, however differed from studies reported from China (low level of colistin resistance) [5], Denmark (higher rate of colistin resistance and presence of 87.5% mcr-1 gene among colistin resistant Enterobacteriaceae) [50], and Korea (14.3% colistin resistance) [9]. Nonetheless, these reports have shown the increasing resistance trend globally.
In this study, 38% and 18% of mcr-1 positive isolates from poultry and human specimens respectively harbored OXA-48 genes. This scenario is increasing in Asia and other continent in E. coli isolates [53, 54]. Most of the carbapenem resistance reports are concerned about CRE in clinical settings while this study reports CRE among mcr-1 positive isolates in clinical setting and poultry as well [55]. The presence of both OXA-48 and mcr-1 gene within a clinical E. coli isolates carried by plasmid is serious threat to public health. This study meanwhile could not predict whether both types were carried by the same plasmid.
Strengths and limitations
This is the first study from Nepal that investigated the co-existence of mcr-1 and OXA-48. Since poultry remains one of the major sources of food in Nepal, use of antibiotics and its concurrent contribution in development of Antimicrobial Resistance (AMR) warrants an urgent attention to embrace ‘one health approach’ in Nepal. The findings of this study will be a fundamental reference for policy makers and clinicians to be informed about the characteristics and prevalence of colistin resistance which can subsequently guide the optimal treatment, use of antibiotics and infection control.
While our data clearly showed the presence of plasmid mediated mcr-1 and OXA-48 in both settings: poultry and humans, the findings could have been strengthened by conducting surveillance targeted at regional and national level for a whole picture for Nepal. Lack of whole genome sequencing in this study could not confirm the origin and transferability of the genes from animals to humans or vice-versa. Further studies using multi-locus sequence typing could be useful for epidemiological investigations.
Conclusion
One fourth and one fifth of the plasmid mediated colistin resistance genes in E. coli from poultry specimens and clinical specimens indicate high burden of colistin resistance in Nepal. Furthermore, co-existence of colistin and carbapenem resistant genes; and their co-existence in plasmid DNA of E. coli isolates in this study suggests the possible spread to other animal, human and environmental pathogens. Molecular methods can aid in early diagnosis, effective management and potential control of transmission. One health approach is critical to fight against MDR that may have been cross-contaminated from environment, animal food and human beings.
Acknowledgements
We would also like to express our sincere gratitude to all the farmers of poultry farms for providing study samples; all laboratory staff at Kantipur Hospital; Central Department of Microbiology, TU for their technical support and NHRC and UGC for financial support of this work. Above all, we are grateful to the patients for their involvement in this study.
Abbreviations
- AST
Antimicrobial susceptibility test
- CAMPS
Cationic antimicrobial cyclic polypeptide
- CPE
Cytopathic effect
- CRE
Carbapenem resistant E. coli
- CLSI
Clinical laboratory standard institute
- ESBL
Extended spectrum β-lactamase
- EUCAST
European committee on antimicrobial susceptibility testing
- GNB
Gram negative bacteria
- ICU
Intensive care unit
- kDa
Kilo-Dalton
- LPS
Lipopolysaccharide
- MCR
Mediated colistin resistance
- MDR
Multi-drug resistance
- MIC
Minimum inhibitory concentration
- ORF
Open reading frame
- PCR
Polymerase chain reaction
- UTI
Urinary tract infection
- WHO
World health organization
- XDR
Extensive/xeno-drug resistance
Authors’ contributions
BM and UTS performed sampling and laboratory experiment; NS, BCM, UTS, NA and PG contributed in the supervision of the experiment; UTS, KRR and MRB were involved in data analysis. UTS, NA, KRR and PG contributed to the initial study concept, design, development and funding acquisition; BM, UTS and BD contributed in initial draft of the manuscript. KRR and BA were involved in amendment and drafting of several versions of the manuscript. All authors read and approved the final manuscript.
Funding
This research was partially supported by NHRC and partially by University Grants Commission, Nepal (UGC-Nepal), Bhaktapur through Faculty Research Grant. We declare that this research is free from interest of funding providers.
Ethics approval and consent to participate
This study was approved by Ethical Review Board (ERB) of Nepal Health Research Council (NHRC), Kathmandu, Nepal (Reg. No. 494/2018). Written consent was applicable to literate people while verbal consent was approached for the rest subjects. Parents/Guardians were interviewed in case of children. The research was in compliance with the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bijaya Muktan and Upendra Thapa Shrestha contributed equally to this work
References
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Chen L, Wang J, Butaye P, Huang K, Qiu H, Zhang X, Gong W, Wang C. Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC Res Notes. 2018;11(1):143. doi: 10.1186/s13104-018-3255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mello MMVJ, Simoes FV, Freire SV, Moraes R, Santos RP. Microbial resistance to colistin in consequence of mutations in the mcr-1 gene of Escherichia coli. MOJ Public Health. 2018;7(2):59–63. [Google Scholar]
- 4.Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis. 2016;16(3):281. doi: 10.1016/S1473-3099(16)00006-2. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Liu F, Lin IY, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):146–147. doi: 10.1016/S1473-3099(15)00533-2. [DOI] [PubMed] [Google Scholar]
- 6.Zurfluh K, Nuesch-Inderbinen M, Klumpp J, Poirel L, Nordmann P, Stephan R. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob Resist Infect Control. 2017;6:91. doi: 10.1186/s13756-017-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao R, Hu Y, Li Z, et al. Dissemination and mechanism for the mcr-1 colistin resistance. PLoS Pathog. 2016;12(11):e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu R, Yi LX, Yu LF, Wang J, Liu Y, Chen X, Lv L, Yang J, Liu JH. Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front Microbiol. 2018;9:331. doi: 10.3389/fmicb.2018.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon EJ, Hong JS, Yang JW, Lee KJ, Lee H, Jeong SH. Detection of mcr-1 plasmids in enterobacteriaceae isolates from human specimens: comparison with those in Escherichia coli isolates from livestock in Korea. Ann Lab Med. 2018;38(6):555–562. doi: 10.3343/alm.2018.38.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Yu L, Chen X, Zhi C, Yao X, Liu Y, Wu S, Guo Z, Yi L, Zeng Z, et al. High prevalence of colistin resistance and mcr-1 Gene in Escherichia coli isolated from food animals in China. Front Microbiol. 2017;8:562. doi: 10.3389/fmicb.2017.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al Bayssari C, Olaitan AO, Dabboussi F, Hamze M, Rolain JM. Emergence of OXA-48-producing Escherichia coli clone ST38 in fowl. Antimicrob Agents Chemother. 2015;59(1):745–746. doi: 10.1128/AAC.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun SD, Ahmed MF, El-Adawy H, Hotzel H, Engelmann I, Weiss D, Monecke S, Ehricht R. Surveillance of extended-spectrum beta-lactamase-producing Escherichia coli in dairy cattle farms in the Nile Delta. Egypt. Front Microbiol. 2016;7:1020. doi: 10.3389/fmicb.2016.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Eur Surveill. 2016;21(9):30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 15.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, et al. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis. 2016;16(3):282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 16.Pokharel S, Raut S, Adhikari B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob Health. 2019;4(6):e002104. doi: 10.1136/bmjgh-2019-002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pokharel S, Adhikari B. Antimicrobial resistance and over the counter use of drugs in Nepal. J Glob Health. 2020;10(1):010360. doi: 10.7189/jogh.10.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhouma M, Beaudry F, Letellier A. Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents. 2016;48:119–126. doi: 10.1016/j.ijantimicag.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Elbediwi M, Li Y, Paudyal N, et al. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980–2018) Microorganisms. 2019;7:461. doi: 10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi PR, Thummeepak R, Leungtongkam U, et al. The emergence of colistin-resistant Escherichia coli in chicken meats in Nepal. FEMS Microbiol Lett. 2019;366(20):fnz237. doi: 10.1093/femsle/fnz237. [DOI] [PubMed] [Google Scholar]
- 21.Gurung S, Kafle S, Dhungel B, et al. Detection of OXA-48 gene in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from urine samples. Infect Drug Resist. 2020;13:2311–2321. doi: 10.2147/IDR.S259967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isenberg HD. Clinical microbiology procedures handbook. 2. Washington, DC: ASM Press; 2004. [Google Scholar]
- 23.Cheesbrough M. District laboratory practice in tropical countries. 2. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 24.Forbes B, Sahm DF, Weissfelt SA. Bailey and Scott;s diagnostic microbiology. London: Mosby Publication; 2007. [Google Scholar]
- 25.Clinical and Laboratory Standards Institute (2018). Performance standards for antimicrobial susceptibility testing. 28th edition Informational supplement M100-S28. Wayne PCalsi.
- 26.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 28.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 29.Dhariwal AK, Tullu MS. Colistin: re-emergence of the ‘forgotten’ antimicrobial agent. J Postgrad Med. 2013;59(3):208–215. doi: 10.4103/0022-3859.118040. [DOI] [PubMed] [Google Scholar]
- 30.El-Sayed Ahmed MAE, Zhong LL, Shen C, Yang Y, Doi Y, Tian GB. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019) Emerg Microbes Infect. 2020;9(1):868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capone A, Giannella M, Fortini D, Giordano A, Meledandri M, Ballardini M, Venditti M, Bordi E, Capozzi D, Balice MP, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013;19(1):E23–E30. doi: 10.1111/1469-0691.12070. [DOI] [PubMed] [Google Scholar]
- 32.Sonnevend A, Ghazawi A, Alqahtani M, Shibl A, Jamal W, Hashmey R, Pal T. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis. 2016;50:85–90. doi: 10.1016/j.ijid.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Qiu J, Jiang Z, Ju Z, Zhao X, Yang J, Guo H, Sun S. Molecular and phenotypic characteristics of Escherichia coli isolates from Farmed Minks in Zhucheng, China. Biomed Res Int. 2019;2019:3917841. doi: 10.1155/2019/3917841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen NT, Nguyen HM, Nguyen CV, Nguyen TV, Nguyen MT, Thai HQ, Ho MH, Thwaites G, Ngo HT, Baker S, et al. Use of colistin and other critical antimicrobials on pig and chicken farms in Southern Vietnam and its association with resistance in commensal Escherichia coli bacteria. Appl Environ Microbiol. 2016;82(13):3727–3735. doi: 10.1128/AEM.00337-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayastha K, Dhungel B, Karki S, Adhikari B, Banjara MR, Rijal KR, Ghimire P. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species in pediatric patients visiting International Friendship Children’s Hospital, Kathmandu, Nepal. Infect Dis (Auckl). 2020;13:1178633720909798. doi: 10.1177/1178633720909798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhungana K, Awal BK, Dhungel B, Sharma S, Banjara MR, Rijal KR. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo betalactamae (MBL) producing Gram negative bacteria isolated from different clinical samples in a Transplant Center, Kathmandu, Nepal. ASMI. 2019;2(12):60–69. [Google Scholar]
- 37.Guragin N, Pradhan A, Dhungel B, Banjara MR, Rijal KR, Ghimire P. Extended spectrum B-lactamase producing Gram Negative bacterial isolates from urine of patients visiting Everest Hospital, Kathmandu, Nepal. TUJM. 2019;6(1):26–31. [Google Scholar]
- 38.Raut S, Rijal KR, Khatiwada S, et al. Trend and characteristics of Acinetobacter baumannii infections in patients attending Universal College of Medical Sciences, Bhairahawa, Western Nepal: a longitudinal study of 2018. Infect Drug Resist. 2020;13:1631–1641. doi: 10.2147/IDR.S257851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pishnian Z, Haeili M, Feizi A. Prevalence and molecular determinants of colistin resistance among commensal Enterobacteriaceae isolated from poultry in northwest of Iran. Gut Pathog. 2019;11:2. doi: 10.1186/s13099-019-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jafri SA, Qasim M, Masoud MS, Rahman MU, Izhar M, Kazmi S. Antibiotic resistance of E. coli isolates from urine samples of Urinary Tract Infection (UTI) patients in Pakistan. Bioinformation. 2014;10(7):419–422. doi: 10.6026/97320630010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adhikari RP, Shrestha S, Rai JR, Amatya R. Antimicrobial resistance patterns in clinical isolates of enterobacteriaceae from a tertiary care hospital, Kathmandu, Nepal. Nepalese Med J. 2018;1(2):74–78. doi: 10.3126/nmj.v1i2.21578. [DOI] [Google Scholar]
- 42.Shrestha A, Bajracharya AM, Subedi H, Turha RS, Kafle S, Sharma S, Neupane S, Chaudhary DK. Multi-drug resistance and extended spectrum beta lactamase producing Gram negative bacteria from chicken meat in Bharatpur Metropolitan, Nepal. BMC Res Notes. 2017;10(1):574. doi: 10.1186/s13104-017-2917-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarker MS, Mannan MS, Ali MY, Bayzid M, Ahad A, Bupasha ZB. Antibiotic resistance of Escherichia coli isolated from broilers sold at live bird markets in Chattogram, Bangladesh. J Adv Vet Anim Res. 2019;6(3):272–277. doi: 10.5455/javar.2019.f344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Med Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tada T, Nhung PH, Shimada K, Tsuchiya M, Phuong DM, Anh NQ, Ohmagari N, Kirikae T. Emergence of colistin-resistant Escherichia coli clinical isolates harboring mcr-1 in Vietnam. Int J Infect Dis. 2017;63:72–73. doi: 10.1016/j.ijid.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 46.He QW, Xu XH, Lan FJ, Zhao ZC, Wu ZY, Cao YP, Li B. Molecular characteristic of mcr-1 producing Escherichia coli in a Chinese university hospital. Ann Clin Microbiol Antimicrob. 2017;16(1):32. doi: 10.1186/s12941-017-0207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moennighoff C, Thomas N, Nienhaus F, et al. Phenotypic antimicrobial resistance in Escherichia coli strains isolated from swine husbandries in North Western Germany—temporal patterns in samples from laboratory practice from 2006 to 2017. BMC Vet Res. 2020;16(1):37. doi: 10.1186/s12917-020-2268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Migura-Garcia L, González-López JJ, Martinez-Urtaza J, et al. mcr-colistin resistance genes mobilized by Inc X4, IncHI2, and IncI2 plasmids in Escherichia coli of pigs and white stork in Spain. Front Microbiol. 2020;10:3072. doi: 10.3389/fmicb.2019.03072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadjadj L, Riziki T, Zhu Y, Li J, Diene SM, Rolain JM. Study of mcr-1 gene-mediated colistin resistance in enterobacteriaceae isolated from humans and animals in different countries. Genes (Basel). 2017;8(12):394. doi: 10.3390/genes8120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alba P, Leekitcharoenphon P, Franco A, Feltrin F, Ianzano A, Caprioli A, Stravino F, Hendriksen RS, Bortolaia V, Battisti A. Molecular epidemiology of mcr-encoded colistin resistance in Enterobacteriaceae from food-producing animals in Italy revealed through the EU harmonized antimicrobial resistance monitoring. Front Microbiol. 2018;9:1217. doi: 10.3389/fmicb.2018.01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amin MB, Sraboni AS, Hossain MI, et al. Occurrence and genetic characteristics of mcr-1-positive colistin-resistant E. coli from poultry environments in Bangladesh. J Glob Antimicrob Resist. 2020;22:546–552. doi: 10.1016/j.jgar.2020.03.028. [DOI] [PubMed] [Google Scholar]
- 52.Caselli E, D’Accolti M, Soffritti I, Piffanelli M, Mazzacane S. Spread of mcr-1-driven colistin resistance on hospital surfaces. Italy. Emerg Infect Dis. 2018;24(9):1752–1753. doi: 10.3201/eid2409.171386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen CW, Tang HJ, Chen CC, Lu YC, Chen HJ, Su BA, Weng TC, Chuang YC, Lai CC. The microbiological characteristics of carbapenem-resistant Enterobacteriaceae carrying the mcr-1 gene. J Clin Med. 2019;8(2):261. doi: 10.3390/jcm8020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manohar P, Shanthini T, Ayyanar R, Bozdogan B, Wilson A, Tamhankar AJ, Nachimuthu R, Lopes BS. The distribution of carbapenem- and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol. 2017;66(7):874–883. doi: 10.1099/jmm.0.000508. [DOI] [PubMed] [Google Scholar]
- 55.Huang H, Dong N, Shu L, et al. Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014–2019. Emerg Microbes Infect. 2020;9(1):237–245. doi: 10.1080/22221751.2020.1717380. [DOI] [PMC free article] [PubMed] [Google Scholar]