Abstract
An increasing amount of evidence has demonstrated the importance of microRNAs (miRNAs/miRs) in the tumorigenesis of malignant types of cancer, and data retrieved from The Cancer Genome Atlas database revealed that miR-3690 was upregulated in thyroid cancer (TC). The present study focused on the biological function and mechanism of miR-3690 in TC, demonstrating that miR-3690 expression was significantly elevated in TC cells and clinical tissues. Functional studies indicated that miR-3690 acted as an oncogene in TC by promoting cell proliferation, colony formation and cell cycle progression in association with the increased expression of cyclin E and c-myc. Mechanistically, prediction software indicated that Dickkopf-related protein 3 (DKK3) was a target of miR-3690, which was confirmed by the results of luciferase reporter assays and western blotting. DKK3 silencing abrogated the functions of miR-3690-in on TC cell proliferation. Collectively, the findings of the present study demonstrated that miR-3690 promoted TC cell proliferation and indicated miR-3690 as a potential biomarker and therapeutic target for TC.
Keywords: microRNA-3690, human thyroid cancer, Dickkopf-related protein 3, cell proliferation, the cell cycle
Introduction
Thyroid cancer (TC) is the most common endocrine malignancy and its incidence has increased rapidly in recent years worldwide (1). Due to the absence of obvious symptoms in the early stages of TC, ~30% of patients are diagnosed at a late stage with lymph node metastases or invasion of the surrounding tissues and organs (2). Furthermore, 10–20% of patients exhibit distant metastases and recurrence, decreasing the 10-year survival rate to 40% (3–5). In addition, selected TC cases are not sensitive to radio-iodine treatment, and the subsequent curative effect is poor (6). Therefore, it is of great importance to understand the molecular mechanisms underlying TC in order to develop and improve current therapeutic approaches.
MicroRNAs (miRNAs) are a class of short endogenous non-coding molecules (~22 nucleotides in length) that play key roles in oncogenesis and cancer progression by regulating various biological functions (7–9). Accumulating evidence has suggested that aberrant miRNA expression contributes to multiple physiological and pathological processes during TC (10–12). For example, Boufraqech et al (13) indicated that miR-30a was downregulated in TC, decreasing cellular invasion and migration by regulating lysyl oxidase. In addition, miR-338-3p was revealed to suppress cell proliferation, clonogenicity and metastasis in TC by targeting RAC-γ serine/threonine-protein kinase (14). miR-215 was reported to inhibit TC cell proliferation, migration and invasion via the AKT/glycogen synthase kinase-3β/Snail signaling pathway by targeting brefeldin A-inhibited guanine nucleotide-exchange protein 1 (15). Furthermore, Guan et al (16) indicated that the downregulation of miR-218-2 promoted the invasion and progression of TC by regulating platelet-derived growth factor receptor α and 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase γ-1. The aim of the present study was to systematically investigate the precise role of miR-3690 in TC and elucidate the underlying mechanism, in order to determine the value of miR-3690 as a biomarker for TC.
Materials and methods
Clinical specimens and expression datasets
Clinical tissue samples were collected from 4 male and 4 female patients (mean age, 35±8 years) who underwent total thyroidectomy (cancerous samples were collected from unilateral glandular lobe, and non-cancerous samples were collected from unilateral glandular lobe on the other side) and who were histopathologically diagnosed with TC between January 2016 and January 2017 at Guangzhou First People's Hospital (Guangzhou, China). The samples were immediately cryopreserved in liquid nitrogen and stored at −80°C until further use. In order to investigate the expression of miR-3690 in association with TC, the miR-3690 expression data of 59 patients were downloaded from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) database and subsequently analyzed.
Cell culture
The human thyroid follicular cell line Nthy-ori 3–1, and various human TC cell lines, including TPC-1, SW1736, FRO, K1 and 8505C, were purchased from the National Rodent Laboratory Animal Resource. The TC cell lines were authenticated by STR profiling, and were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 0.1 mg/ml streptomycin. Nthy-ori 3-1 cells were cultured in F-12K medium (Genom Biotech Pvt., Ltd.) containing 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin, and all cells were cultured in a humidified atmosphere at 37°C (5% Co2).
Plasmids, short interfering RNA (siRNA) and transfection
Synthetic miR-3690 (cat. no. miR10018119-1-5), miR-3690-inhibitor (cat. no. miR20018119-1-5), miR-3690-mutant (mut, 5′-GCUUGGACCCAGCGUAGACAAAG-3′) and relative negative control miRNAs (vector: Cat. no. miR1N0000001-1-5, NC: Cat. no. miR2N0000001-1-5) were synthesized by Guangzhou RiboBio Co., Ltd. Cells transfections with 50 nM miRNAs were performed using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions.
Silencing DKK3 siRNAs (cat. no. HSH007526) were synthesized and purified by GeneCopoeia, Inc., and transfections were performed using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. All cells were harvested for subsequent experimentation 48 h post-transfection.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA from the human clinical tissues and cultured cells was extracted using TRIzol® reagent according to the manufacturer's recommendations (Thermo Fisher Scientific, Inc.). Reverse transcription and detection of mRNA (Cyclin E and c-myc) was performed using BlazeTaq™ SYBR® Green qPCR mix 2.0 (GeneCopoeia, Inc.). RT-qPCR was performed to detect miR-3690 expression using the All-in-One™ miRNA qRT-PCR Detection kit 2.0 (GeneCopoeia, Inc, Guangzhou). Thermocycling conditions were as follows: At 95°C for 30 sec, followed by 40 cycles of amplification at 95°C for 5 sec, at 59°C for 30 sec and at 72°C for 30 sec. The following PCR primers were synthesized by GeneCopoeia, Inc.: miR-3690 (cat. no. HmiRQP1976), cyclin E (cat. no. HQP021819) and MYC (cat. no. HQP011597). U6 small nuclear RNA (cat. no. HmiRQP9001) and GAPDH (cat. no. HQP006940) were used as endogenous controls, and mRNA quantification was performed using the 2−ΔΔCq method (17).
MTT and anchorage-independent growth assays
In order to determine the cell proliferation rate, an MTT assay (Sigma-Aldrich; Merck KGaA) was used according to the manufacturer's protocol. The reaction was stopped by 200 µl DMSO; the relative optical density was determined at a wavelength of 490 nm, and the results are expressed as the mean ± standard deviation.
For the anchorage-independent growth assays, transfected K1 cells (1×103 cell per well) were seeded into 2 ml complete medium with 0.3% soft agar (Invitrogen; Thermo Fisher Scientific, Inc.) and overlaid onto 2 ml preset 1.5% agar in the same medium. The cells were cultured at 37°C with 5% CO2 for 2 weeks, the colonies were then stained with 0.05% crystal violet for 10 min at room temperature, and colonies sized >0.1 mm were counted under a microscope (Motic AE30 inverted fluorescence microscope; Microscope Systems Limited; magnification, ×40).
Bioinformatics
The potential target genes of miR-3690 were predicted using Target Scan Human (version 7.1; http://www.targetscan.org/vert_71). Luciferase reporter assays. Based on the binding sites of DDK-3 and miR-3690, which were predicted using TargetScan, wild-type DDK-3 sequences were designed and cloned into the pGL3 luciferase reporter vector (Promega Corporation). Cells were then co-transfected with the vectors and miR-3690, miR-3690-in or miR-3690-mut, using Lipofectamine® 2000 transfection reagent. Following a 48-h incubation, luciferase signals were measured using the Dual luciferase reporter gene assay kit (BioVision, Inc.) according to the manufacturer's protocol. Firefly luciferase activity was normalized to that of Renilla luciferase.
Western blotting
Transcfeted TC cells were lysed in protein RIPA buffer (cat. no. P0013; Beyotime Institute of Biotechnology) according to the manufacturer's protocol. The protein concentration was determined using a bicinchoninic acid protein assay. Proteins (40 µg) were separated via 10% SDS-PAGE and subsequently transferred onto PVDF membranes (EMD Millipore). Membranes were blocked with 5% bovine serum albumin (BSA) for 1 h at room temperature, and incubated with primary antibodies against: DKK3 (1:1,000; cat. no. SAB2701251; Sigma-Aldrich; Merck KGaA), cyclin E (1:1,000; cat. no. 20808; Cell Signaling Technology, Inc.) and c-myc (1:1,000; cat. no. 18583; Cell Signaling Technology, Inc.) overnight at 4°C, as previously described (17). To control sample loading, the membranes were stripped and re-probed with an anti-α-tubulin antibody (1:500; cat. no. SAB5600206; Sigma-Aldrich; Merck KGaA). The membranes were then probed with a peroxidase-conjugated secondary antibody goat-anti-rabbit IgG (1:5,000; ZDR5306 or ZDR5307; Zhongshan Golden Bridge Biotechnology, Inc.), and the protein bands were quantified using Quantity One software version 4.6 (Bio-Rad Laboratories, Inc.).
Bromodeoxyuridine (BrdU) incorporation analysis
To assess cell proliferation, cells were cultured for 24 h following transfection. The medium was then supplemented with 10 h BrdU and the cells were incubated for a further 6 h. The labeled cells were thoroughly rinsed with PBS and incubated with an anti-BrdU antibody (1:500; cat no. 61273; Upstate Biotechnology, Inc.) for 1 h at 37°C, according to the manufacturer's protocol. Images were captured using a laser scanning microscope (magnification, ×100; Axioskop 2 plus; Carl Zeiss AG).
Cell cycle distribution analysis
Following miRNA-transfection for 48 h, the cells were harvested, washed in ice-cold PBS, and fixed with pre-chilled 75% ethanol at 4°C overnight. The cells were stained using a propidium iodide/RNase staining buffer (BD Biosciences) at room temperature for 30 min in the dark, and the cell cycle distribution was analyzed using a FACSCalibur™ flow cytometer (BD Biosciences) according to the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 (SPSS, Inc.). The data were analyzed using a paired Student's t-test for pair-wise comparisons or one-way analysis of variance followed by a post hoc Tukey test for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of miR-3690 in TC clinical tissues and cell lines
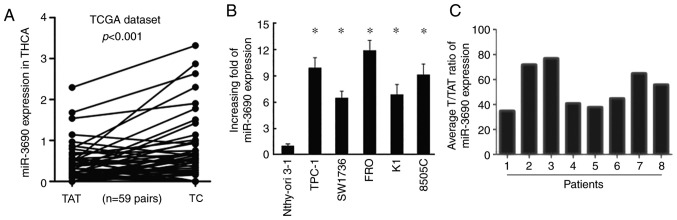
Based on the expression data downloaded from TCGA, the expression level of miR-3690 was found to be higher in TC tissues compared with that in adjacent non-tumor tissues (Fig. 1A). Consistently, miR-3690 expression was also significantly increased in TC cell lines (TPC-1, SW1736, FRO, K1 and 8505C) compared with that in human thyroid follicular cells (Nthy-ori 3-1; Fig. 1B). Furthermore, RT-qPCR analysis of samples from 8 patients with TC revealed significantly increased levels of miR-3690 in TC tissues (T) compared with tumor-adjacent normal tissues (TAT; Fig. 1C).
Figure 1.
Expression of miR-3690 in human TC cell lines and clinical tissues. (A) The expression levels of miR-3690 in TC from TCGA dataset (n=59). (B) Reverse transcription PCR analysis of miR-3690 expression in primary Nthy-ori 3-1 (non-tumor follicular cell line) and human TC cell lines, including TPC-1, SW1736, FRO, K1 and 8505C. (C) Relative miR-3690 expression levels in 8 paired primary TC tissues (TC) and the tumor-adjacent normal tissues (TAT) from the same patient were detected by PCR analysis. Each bar represents the mean of three independent experiments. *P<0.05 vs Nthy-ori 3-1. miR, microRNA; TC, thyroid cancer; TCGA, The Cancer Genome Atlas; TAT, tumor-adjacent normal tissues.
miR-3690 promotes TC cell proliferation, which is counteracted by miR-3690-in
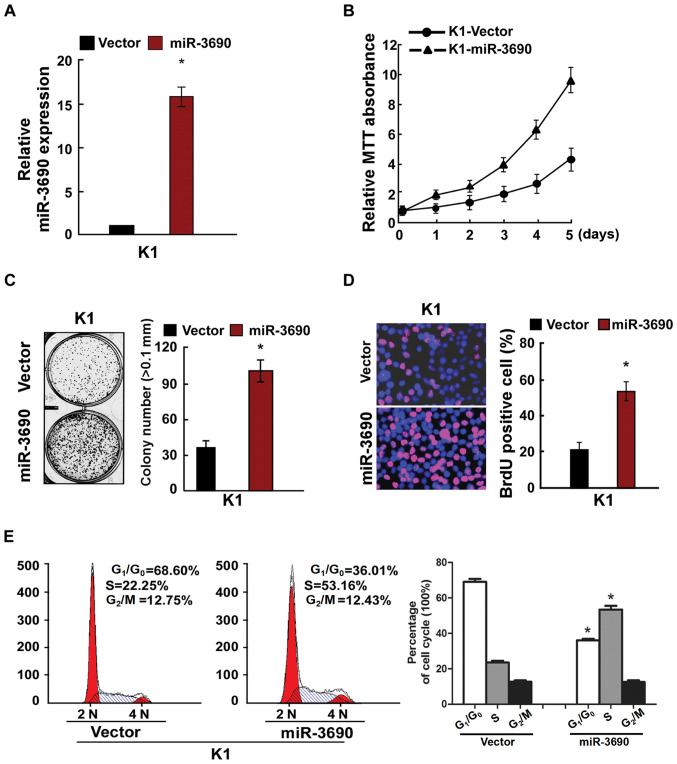
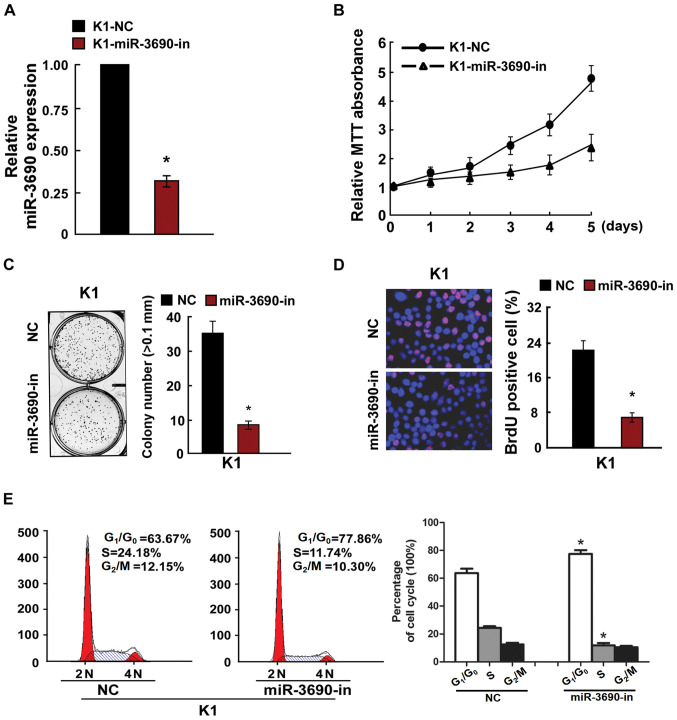
To determine the effects of miR-3690 on cellular proliferation, TC cells were transfected with miR-3690 and assessed using RT-qPCR, MTT and anchorage-independent growth assays, in addition to BrdU labeling, immunofluorescence and cell cycle analysis. Following transfection of K1 cells, the expression level of miR-3690 was found to be significantly upregulated (Fig. 2A), and miR-3690 overexpression facilitated cellular proliferation (Fig. 2B-D). Furthermore, miR-3690 overexpression significantly decreased the number of cells in the G1/G0 phase, but increased the proportion of S phase cells, compared with the control at 48 h post-transfection (Fig. 2E). In turn, miR-3690 expression was successfully inhibited by miR-3690-in transfection in K1 cells (Fig. 3A), which significantly suppressed proliferation and cell cycle progression compared with the control-transfected cells (Fig. 3B-E). Further to the effects of miR-3690 on TC cell proliferation, the results of the MTT and anchorage-independent growth assays revealed that the SW1736 cell line exhibited a lower miR-3690 expression level compared with the K1 line, and thus was selected for further experimentation. As presented in Fig. S1, the results reflected those obtained from K1 cells.
Figure 2.
Upregulation of miR-3690 promoted TC cell proliferation. (A) Validation of miR-3690 expression levels after transfection by PCR analysis. (B) MTT assays revealed that overexpression of miR-3690 promoted proliferation of K1 cells. (C) Upregulation of miR-3690 promoted the anchorage-independent growth of K1 cells. Representative micrographs (left) and quantification of colonies that were >0.1 mm (right). (D) Representative micrographs (left) and quantification (right) of the Brd incorporation assay in K1 cells. (E) Flow cytometric analysis of the indicated TC cell line K1 cells transfected with NC or miR-3690. Each bar represents the mean of three independent experiments. *P<0.05 vs. vector group. TC, thyroid cancer; miR, microRNA; NC, negative control.
Figure 3.
Inhibition of miR-3690 inhibited TC cell proliferation. (A) Validation of miR-3690 expression levels after transfection by PCR analysis. (B) MTT assays revealed that miR-3690-in suppressed growth of K1 cells. (C) Inhibition of miR-3690 inhibited the anchorage-independent growth of K1 cells. Representative micrographs (left) and quantification of colonies that were >0.1 mm (right). (D) Representative micrographs (left) and quantification (right) of the BrdU incorporation assay in K1 cells. (E) Flow cytometric analysis of the indicated TC cell line K1 cells transfected with NC or miR-3690-in. Each bar represents the mean of three independent experiments. *P<0.05 vs. NC group. miR, microRNA; TC, thyroid cancer; NC, negative control; miR-3690-in, microRNA-3690-inhibitor.
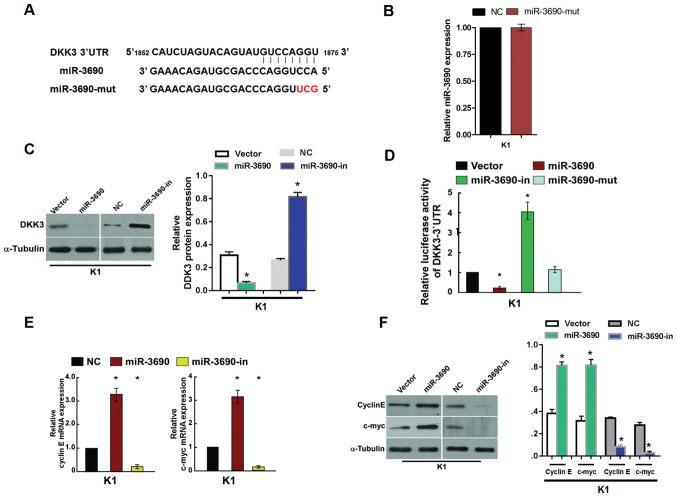
miR-3690 regulates DKK3 via the 3′-UTR and alters cell proliferation and the expression of cell cycle-associated genes
In order to identify the potential targets of miR-3690, a bioinformatics analysis was conducted using TargetScan, and revealed that DKK3 contains a putative binding site for miR-3690 (Fig. 4A). The effect of miR-3690-mut on TC cells was detected via RT-PCR analysis, the result showed that no effects were observed following transfection with miR-3690-mut compared with that in NC (Fig. 4B). To investigate the potential regulatory effect of miR-3690 on DKK3, protein expression levels were examined by western blot analysis in the present study. miR-3690 overexpression significantly decreased the expression levels of the DKK3 protein in K1 cells, while the levels were found to be upregulated following miR-3690-in transfection (Fig. 4C). Furthermore, luciferase reported assays indicated that miR-3690 significantly decreased the luciferase activity of the wild-type 3′-UTR of DKK3, whereas miR-3690-in transfection resulted in increased activity; no suppressive effects were observed following transfection with miR-3690-mut (Fig. 4D). These findings indicate that DKK3 is a direct target of miR-3690.
Figure 4.
miR-3690 suppressed DKK3 expression by directly targeting its 3′-UTR and altered levels of proteins related to cell proliferation and cell cycle in K1 cells. (A) Predicted miR-3690 target sequence in the 3′-UTR of DKK3 (DKK3-3′-UTR) and positions of three mutated nucleotides (red) in the 3′-UTR of miR-3690 (miR-3690-mut). (B) Validation of miR-3690 expression levels after transfection with miR-3690-mut by PCR analysis. (C) Western blotting analysis of DKK3 expression in K1 cells transfected with miR-3690 or the miR-3690-in, α-tubulin served as the loading control. (D) Luciferase reporter assay of the indicated cells transfected with the pGL3-DKK3-3′-UTR reporter and miR-3690 or miR-3690-in or miR-3690-mut. (E) Reverse transcription PCR analysis of expression of Cyclin E and c-myc in indicated K1 cells. (F) Western blotting analysis of expression of Cyclin E and c-myc protein in K1 cells. α-tubulin served as the loading control. *P<0.05 vs. NC group or vector group. miR, microRNA; 3′-UTR, 3′-untranslated region; DKK3, Dickkopf-related protein 3; miR-3690-mut, microRNA-3690-mutant; miR-3690-in, microRNA-3690-inhibitor; NC, negative control.
As miR-3690 promoted cell proliferation and cell cycle progression, regulatory genes associated with these processes were detected. RT-qPCR and western blot analysis revealed elevated mRNA and protein expression levels, respectively, of cyclin E and c-myc in miR-3690-transfected cells, which was consistent with the increased proportion of S phase and decreased proportion of G1/G0 phase cells (Fig. 4E and F). By contrast, decreased expression of cyclin E and c-myc was detected following transfection with miR-3690-in, which was reflected by cell cycle arrest in the G1/G0 phase (Fig. 4D and E).
DKK3 knockdown counteracts miR-3690-in-associated proliferative arrest
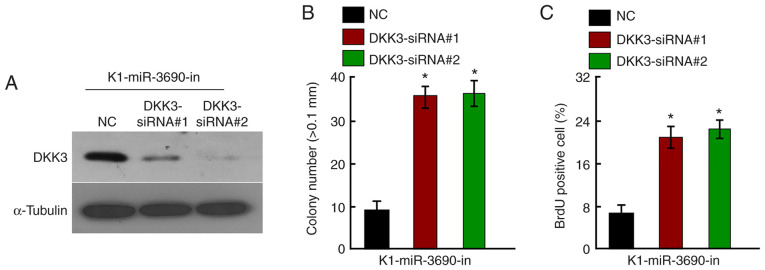
As DKK3 was confirmed to be a direct target of miR-3690, the effects of DKK3 downregulation on miR-3690-in-induced proliferative arrest were investigated. Following treatment with miR-3690-in, DKK3 was knocked down in K1 cells using specific siRNAs; knockdown efficiency was confirmed by western blotting (Fig. 5A). The growth-suppressive effect of miR-3690-in was partially abrogated by DKK3-knockdown, as demonstrated by anchorage-independent growth (Fig. 5B), BrdU labeling and immunofluorescence assays (Fig. 5C). These findings indicate that DKK3 was involved in the miR-3690-in-induced suppression of cellular proliferation.
Figure 5.
Downregulation of DKK3 counteracted the proliferation arrest by miR-3690-in. (A) Western blot analysis verified that silencing DKK3 effectively decreased the expression of DKK3 in miR-3690-in-transfected K1 cells. (B) miR-3690-in-transfected K1 cells after transfection with DKK3-siRNAs promoted the anchorage-independent growth. Representative quantification of colonies that were >0.1 mm. (C) Representative quantification of the BrdU incorporation assay. Each bar represents the mean of three independent experiments. *P<0.05. miR-3690-in, microRNA-3690-inhibitor; DKK3, Dickkopf-related protein 3; siRNA, small interfering-RNA; NC, negative control.
Discussion
Profiling studies have indicated that miRNAs play an essential role in TC development by regulating their target genes. miR-195 was found to suppress tumor growth and metastasis in TC by targeting G1/S-specific cyclin D1 and fibroblast growth factor 2 (18); additionally, miR-577 was reported to inhibit the proliferation, migration and invasive capacity of TC cells by targeting sphingosine kinase 2 (19). The findings of Ye et al (20) indicated that fibronectin 1 was a direct target of miR-139, which inhibits TC progression. Furthermore, Sun et al (21) revealed that miR-144 suppressed TC cell proliferation through WW domain-containing transcription regulator protein 1. Our previous study revealed that miR-639 promoted cell proliferation and cell cycle progression in TC by inhibiting cyclin-dependent kinase inhibitor 1 (22). Data from the present study provide evidence that miR-3690 expression is upregulated in TC tissues from patients and TC cell lines, and also suggest that miR-3690 promotes cell proliferation and cell cycle progression by directly suppressing DKK3.
Human DKK3 is a member of the Dickkopf family, which perturbs the negative regulator Kremen and enhances Wnt signaling via low-density lipoprotein receptor-related protein 6 (23). Previous studies have demonstrated that DKK can act as a tumor promoter or suppressor in certain types of cancer, thereby affecting tumor development and progression. DDK3 has been reported to stimulate pancreatic ductal adenocarcinoma growth and metastasis, as well as resistance to chemotherapy, by activating NF-κB (24). In addition, overexpression of DKK3 reportedly contributes to tumor cell proliferation, invasion, migration and survival in head and neck squamous cell carcinoma by inducing PI3K-Akt signaling (25). Furthermore, Xi et al (26) observed that DKK3 was downregulated in prostatic cancer, and that miR-95-3p contributed to the development of prostatic cancer by repressing DKK3, thus activating the Wnt/β-catenin pathway. In gastric cancer, DKK3 was shown to attenuate CD133-induced NK-cell activation by inhibiting the Erk pathway and immunological synapse formation (27). DKK3 was also found to act as a biomarker and a therapeutic target by modulating Wnt signaling in several types of human cancer (28). However, the role of DKK3 in TC remains understudied. To the best of our knowledge, the present study was the first to demonstrate that DKK3 was a downstream target gene of miR-3690, and is considered to act as a tumor suppressor in TC. Furthermore, DKK3 downregulation counteracted miR-3690-in-associated proliferative arrest.
The present study revealed that miR-3690 promotes cellular proliferation by regulating proliferation- and cell cycle-associated genes, including cyclin E and c-myc. Cyclin E is a key checkpoint protein for G1-to-S phase transition in the cell cycle (29). C-myc is a known master regulator of the cell cycle and plays a critical role in carcinogenesis (30). In gastric carcinoma, downregulated DKK3 (REIC) expression was closely associated with aggressive phenotypes and poor prognosis (31). Ectopic DKK3 expression downregulated β-catenin, cyclin D2 and E expression levels, as well as regulated the cell cycle in gastric carcinoma (31). The results of the present study revealed that downregulation of miR-3690 resulted in cell cycle arrest at the G1/G0 phase in TC.
In conclusion, the present study demonstrated that miR-3690 was upregulated and promoted cancer cell proliferation by directly repressing DKK3. Downregulation of miR-3690 resulted in cell cycle arrest at the G1/G0 phase by inhibiting cyclin E and c-myc, which may affect the development of TC. These findings indicate that miR-3690 may be of value as a therapeutic target in TC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by Guangzhou Science and Technology Plan Project (grant no. 201805010003).
Availability of data and materials
The datasets generated or analyzed during the present study are included in this published article.
Authors' contributions
FS and BX designed the present study. FS, XG, XD and BX drafted the initial manuscript. FS, XG, XD and JF performed the experiments. FS, XG, XD, JF, WC, LS and HX analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Guangzhou First People's Hospital (Guangzhou, China; approval no. 185-01), and written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Zhao H, Huang T, Li H. Risk factors for skip metastasis and lateral lymph node metastasis of papillary thyroid cancer. Surgery. 2019;166:55–60. doi: 10.1016/j.surg.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 3.Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388:2783–2795. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 4.Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA. 2017;317:1338–1348. doi: 10.1001/jama.2017.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12:646–653. doi: 10.1038/nrendo.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner M, Khoury H, Bennetts L, Willet J, Lister J, Berto P, Ehreth J, Badia X, Grimaldi-Bensouda L, Goetghebeur M. Appraising the value of lenvatinib for radio-iodine refractory differentiated thyroid cancer (Rr-Dtc): A multi-country study applying holistic multicriteria decision analysis (Mcda) Value Health. 2015;18:PA477–PA478. doi: 10.1016/j.jval.2015.09.1287. [DOI] [Google Scholar]
- 7.Pei ZJ, Zhang ZG, Hu AX, Yang F, Gai Y. miR-122-5p inhibits tumor cell proliferation and induces apoptosis by targeting MYC in gastric cancer cells. Pharmazie. 2017;72:344–347. doi: 10.1691/ph.2017.6404. [DOI] [PubMed] [Google Scholar]
- 8.Xu FF, Xie WF, Zha GQ, Chen HW, Deng L. MiR-520f promotes cell aggressiveness by regulating fibroblast growth factor 16 in hepatocellular carcinoma. Oncotarget. 2017;8:109546–109558. doi: 10.18632/oncotarget.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang WL, Cao J, Xu B, Yang P, Shen F, Sun Z, Li WL, Wang Q, Liu F. miR-892a regulated PPP2R2A expression and promoted cell proliferation of human colorectal cancer cells. Biomed Pharmacother. 2015;72:119–124. doi: 10.1016/j.biopha.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW, Lin YH, Chi HC, Kuo LM, Lu PH, Lin KH. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer. 2016;64:137–148. doi: 10.1016/j.ejca.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y, Wu W, Zou X, Liu F, Wei T, Zhu J. MiR-26a inhibits thyroid cancer cell proliferation by targeting ARPP19. Am J Cancer Res. 2018;8:1030–1039. [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Shi R, Zhao S, Li X, Lu S, Bu H, Ma X, Su C. E2F8, a direct target of miR-144, promotes papillary thyroid cancer progression via regulating cell cycle. J Exp Clin Cancer Res. 2017;36:40. doi: 10.1186/s13046-017-0504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boufraqech M, Nilubol N, Zhang L, Gara SK, Sadowski SM, Mehta A, He M, Davis S, Dreiling J, Copland JA, et al. miR30a inhibits LOX expression and anaplastic thyroid cancer progression. Cancer Res. 2015;75:367–377. doi: 10.1158/0008-5472.CAN-14-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sui GQ, Fei D, Guo F, Zhen X, Luo Q, Yin S, Wang H. MicroRNA-338-3p inhibits thyroid cancer progression through targeting AKT3. Am J Cancer Res. 2017;7:1177–1187. [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Zhang M, Nie C, Jia J, Wang F, Yu J, Bi W, Liu B, Sheng R, He G, et al. miR-215 suppresses papillary thyroid cancer proliferation, migration, and invasion through the AKT/GSK-3β/Snail signaling by targeting ARFGEF1. Cell Death Dis. 2019;10:195. doi: 10.1038/s41419-019-1444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan H, Wei G, Wu J, Fang D, Liao Z, Xiao H, Li M, Li Y. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1334–E1344. doi: 10.1210/jc.2013-1053. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Yin Y, Hong S, Yu S, Huang Y, Chen S, Liu Y, Zhang Q, Li Y, Xiao H. MiR-195 inhibits tumor growth and metastasis in papillary thyroid carcinoma cell lines by targeting CCND1 and FGF2. Int J Endocrinol. 2017;2017:6180425. doi: 10.1155/2017/6180425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue KC, Hu DD, Zhao L, Li N, Shen HY. MiR-577 inhibits papillary thyroid carcinoma cell proliferation, migration and invasion by targeting SphK2. Eur Rev Med Pharmacol Sci. 2017;21:3794–3800. [PubMed] [Google Scholar]
- 20.Ye Y, Zhuang J, Wang G, He S, Ni J, Xia W. MicroRNA-139 targets fibronectin 1 to inhibit papillary thyroid carcinoma progression. Oncol Lett. 2017;14:7799–7806. doi: 10.3892/ol.2017.7201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Sun W, Lan X, Wang Z, Dong W, He L, Zhang T, Zhang P, Zhang H. MicroRNA-144 inhibits proliferation by targeting WW domain-containing transcription regulator protein 1 in papillary thyroid cancer. Oncol Lett. 2018;15:1007–1013. doi: 10.3892/ol.2017.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei ST, Shen F, Chen JW, Feng JH, Cai WS, Shen L, Hu ZW, Xu B. MiR-639 promoted cell proliferation and cell cycle in human thyroid cancer by suppressing CDKN1A expression. Biomed Pharmacother. 2016;84:1834–1840. doi: 10.1016/j.biopha.2016.10.087. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari N, Ranftl R, Chicherova I, Slaven ND, Moeendarbary E, Farrugia AJ, Lam M, Semiannikova M, Westergaard MCW, Tchou J, et al. Dickkopf-3 links HSF1 and YAP/TAZ signalling to control aggressive behaviours in cancer-associated fibroblasts. Nat Commun. 2019;10:130. doi: 10.1038/s41467-018-07987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou L, Husted H, Moore T, Lu M, Deng D, Liu Y, Ramachandran V, Arumugam T, Niehrs C, Wang H, et al. Suppression of stromal-derived Dickkopf-3 (DKK3) inhibits tumor progression and prolongs survival in pancreatic ductal adenocarcinoma. Sci Transl Med. 2018;10:eaat3487. doi: 10.1126/scitranslmed.aat3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katase N, Nishimatsu SI, Yamauchi A, Yamamura M, Terada K, Itadani M, Okada N, Hassan NMM, Nagatsuka H, Ikeda T, et al. DKK3 overexpression increases the malignant properties of head and neck squamous cell carcinoma cells. Oncol Res. 2018;26:45–58. doi: 10.3727/096504017X14926874596386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xi M, Cheng L, Hua W, Zhou YL, Gao QL, Yang JX, Qi SY. MicroRNA-95-3p promoted the development of prostatic cancer via regulating DKK3 and activating Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23:1002–1011. doi: 10.26355/eurrev_201902_16987. [DOI] [PubMed] [Google Scholar]
- 27.Xia P, Xu XY. DKK3 attenuates the cytotoxic effect of natural killer cells on CD133 + gastric cancer cells. Mol Carcinog. 2017;56:1712–1721. doi: 10.1002/mc.22628. [DOI] [PubMed] [Google Scholar]
- 28.Hamzehzadeh L, Caraglia M, Atkin SL, Sahebkar A. Dickkopf homolog 3 (DKK3): A candidate for detection and treatment of cancers? J Cell Physiol. 2018;233:4595–4605. doi: 10.1002/jcp.26313. [DOI] [PubMed] [Google Scholar]
- 29.Jing L, Gong M, Lu X, Jiang Y, Li H, Cheng W. LINC01127 promotes the development of ovarian tumors by regulating the cell cycle. Am J Transl Res. 2019;11:406–417. [PMC free article] [PubMed] [Google Scholar]
- 30.Elliott B, Millena AC, Matyunina L, Zhang M, Zou J, Wang G, Zhang Q, Bowen N, Eaton V, Webb G, et al. Essential role of JunD in cell proliferation is mediated via MYC signaling in prostate cancer cells. Cancer Lett. 2019;448:155–167. doi: 10.1016/j.canlet.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu XY, Xia P, Yu M, Nie XC, Yang X, Xing YN, Liu YP, Takano Y, Zheng HC. The roles of REIC gene and its encoding product in gastric carcinoma. Cell Cycle. 2012;11:1414–1431. doi: 10.4161/cc.19823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed during the present study are included in this published article.