Abstract
Serum microRNAs (miRNAs) have been implicated as noninvasive biomarkers for lung cancer diagnosis. However, there are no sensitive and specific biomarkers for the detection of radiotherapy-related non-small cell lung cancer (NSCLC) metastasis. The present study aimed to investigate the role of three serum miRNAs, namely miRNA (miR)-130a, miR-25 and miR-191*, in diagnosing NSCLC, and their biological functions in radiation-mediated development of metastatic properties in A549 cells. To determine this, serum samples were collected from 84 patients with NSCLC and 42 age- and sex-matched healthy controls. Differential expression of serum miRNAs was analyzed by quantitative PCR. Significant associations between miRNA expression and overall survival of patients with NSCLC were identified using the Cox proportional regression model. A receiver operating characteristic curve was generated to evaluate diagnostic accuracy. The functions of miR-130a, miR-25 and miR-191* in lung cancer cells were studied by transfecting A549 cells with miRNA mimics and inhibitors. The results of the present study demonstrated that the expression levels of miR-130a, miR-25 and miR-191* in the serum of patients with NSCLC were increased compared with those in healthy controls, and these increases were associated with advanced age (≥60 years), radiotherapy, histological type (squamous carcinoma), low survival rate and low median survival time. Additionally, irradiation induced the upregulation of miR-130a, miR-25 and miR-191* expression in A549 cells in vitro and in a xenograft mouse model. Irradiation also promoted the invasiveness of A549 cells in vitro and metastasis in vivo. In conclusion, miR-130a, miR-25 and miR-191* may be potential biomarkers for the diagnosis of patients with NSCLC and may serve oncogenic roles in radiation-mediated metastasis of NSCLC.
Keywords: microRNA, non-small cell lung cancer, irradiation, metastasis, biomarker
Introduction
Lung cancer is considered to be the leading cause of death, as well as the leading cause of cancer-associated deaths worldwide (1). It is estimated that 12.7 million new cases of lung cancer are diagnosed each year, and that it causes 7.6 million deaths worldwide annually (2). Data from the Surveillance, Epidemiology and End Results database have revealed that >57% of patients with lung cancer exhibit distant metastasis upon diagnosis (3). According to the latest data released by the National Cancer Registration Center, lung cancer is also the leading cause of cancer-associated deaths in China, with incidence and mortality rates of 53.86 and 43.41/100,000 people per year, respectively (4). Among all lung cancer subtypes, non-small cell lung cancer (NSCLC) accounts for >80% of lung cancer cases (5–7). The 5-year overall survival rate of patients with NSCLC has remained as low (~16.1%) over the past two decades due to high rates of recurrence and distant metastasis (8,9). Therefore, there is an urgent requirement to identify diagnostic and prognostic biomarkers for NSCLC to predict the occurrence of metastasis and to guide treatment strategy.
MicroRNAs (miRNAs) are crucial post-transcriptional regulators involved in numerous disease processes, including tumorigenesis and metastasis (10–13). Altered expression of various miRNAs has been reported in NSCLC and demonstrated to be associated with carcinogenesis, tumor progression and treatment outcome (14–17). In addition, miRNAs can be detected in various human biofluids, such as blood, serum and plasma, and thus have the potential to be used as biomarkers for the diagnosis of NSCLC (18–21). Previous studies have suggested that miRNA expression patterns may be used to identify different cancer subtypes, for example, distinguishing lung squamous cell carcinomas (SCCs) from adenocarcinomas (ADCs) (22–24). However, existing literature regarding the association of miRNAs with tumorigenesis, outcome of clinical management and metastasis is considerably varied (25–27); for example, miR-25 can serve different roles in the pathogenesis of different types of cancer, exerting both tumor suppressor (26) and cancer promoting effects (25,27). Furthermore, the underlying mechanisms of miRNA involvement in the tumorigenesis, metastasis and recurrence of lung cancer are not well understood, particularly regarding radio-resistance following radiotherapy of patients with lung cancer. Our previous study demonstrated that 2- and 4-Gy irradiation induced upregulated expression of miR-130a, miR-25 and miR-191*, and promoted the metastatic properties of A549 cells in vitro and in vivo by upregulating the expression of vascular endothelial growth factor (VEGF) and C-C chemokine receptor type 7 (CCR-7). The aim of the present study was to investigate the diagnostic and functional roles of miR-130a, miR-25 and miR-191* in NSCLC and to determine their carcinogenic effects that enhance tumor metastasis in vitro and in vivo.
Materials and methods
Patients
A total of 84 patients diagnosed with NSCLC at the PLA Rocket Force Characteristic Medical Center between January 2014 and April 2016 were enrolled in the present study. The diagnosis of all patients was confirmed by cytology or histopathology assessment. The inclusion criteria were as follows: Complete medical records, physical examination, fibrobronchoscopy, chest, head and abdomen CT or abdominal ultrasound scan, whole-body bone scan, final pathological diagnosis and clear surgical histopathological diagnosis. The exclusion criteria were as follows: Neurological abnormality, severe dysfunction of the heart, liver, kidney or other organs, diabetes and hypertension. The patient clinicopathological characteristics are presented in Table I. Parallel information was collected from 42 age- and sex-matched patients who did not exhibit abnormalities upon physical examination at the PLA Rocket Force Characteristic Medical Center in May 2014. The routine laboratory and imaging tests performed included complete blood count, baseline electrolyte measurement, blood chemistry profiling and chest X-rays.
Table I.
General information and clinical characteristics of patients with NSCLC, and statistical differences of miR expression levels in patients with different clinical characteristics.
| P-value | ||||
|---|---|---|---|---|
| Category | n (%) | miR-130a | miR-25 | miR-191-3p |
| Sex | ||||
| Male | 58 (69) | 0.477 | 0.180 | 0.102 |
| Female | 26 (31) | |||
| Age, years | ||||
| <60 | 36 (43) | 0.762 | 0.035a | 0.058 |
| ≥60 | 48 (57) | |||
| Smoking status | ||||
| Yes | 42 (50) | 0.254 | 0.188 | 0.115 |
| No | 42 (50) | |||
| Radiotherapy | ||||
| Yes | 33 (39) | 0.004b | 0.003b | 0.047a |
| No | 51 (61) | |||
| Histological type | ||||
| Adenocarcinoma | 46 (55) | 0.584 | 0.013a | 0.006b |
| Squamous cell carcinoma | 38 (45) | |||
| Lymph node metastasis | ||||
| Yes | 66 (79) | 0.887 | 0.200 | 0.639 |
| No | 18 (21) | |||
| Extracranial metastasis | ||||
| Yes | 67 (80) | 0.777 | 0.534 | 0.773 |
| No | 17 (20) | |||
| BM | ||||
| Yes | 22 (26) | 0.559 | 0.542 | 0.535 |
| No | 62 (74) | |||
| No. of BM | ||||
| 0 | 62 (74) | 0.529 | 0.871 | 0.670 |
| 1 | 5 (6) | |||
| ≥2 | 17 (20) | |||
| Maximum diameter of BM, cm | ||||
| 0 | 62 (74) | 0.448 | 0.305 | 0.121 |
| ≤2 | 11 (13) | |||
| >2 | 11 (13) | |||
| TNM T stage | ||||
| T1 | 3 (4) | 0.378 | 0.733 | 0.157 |
| T2 | 0 (0) | |||
| T3 | 5 (6) | |||
| T4 | 76 (90) | |||
| N stage | ||||
| N0 | 18 (21) | 0.235 | 0.222 | 0.234 |
| N1 | 17 (20) | |||
| N2 | 24 (29) | |||
| N3 | 25 (30) | |||
| Survival status | ||||
| Death | 26 (31) | 0.054 | 0.016a | 0.033a |
| Survival | 58 (69) | |||
| ECOG performance status | ||||
| 1 | 47 (56) | 0.836 | 0.989 | 0.935 |
| 2 | 37 (44) | |||
P<0.05
P<0.01 determined using the Mann-Whitney U test. BM, brain metastasis; TNM, Tumor-Node-Metastasis; N, Node; ECOG, Eastern Cooperative Oncology Group.
Follow-up
All patients were followed up by outpatient visits or telephone between June 2016 and June 2018, and no patients were lost to the follow-up. The date of diagnosis was set as the start of follow-up, and survival time was assessed monthly. The period between the date of diagnosis and death due to any cause was considered as the overall survival time. Patients who passed away due to non-neoplastic causes and those who were alive at the end of follow-up were included as censored data that met statistical requirements.
Cell culture
The human bronchioloalveolar lung carcinoma cell line A549 was obtained from the cell bank of Peking Union Medical College Hospital. A549 cells were cultured at 37°C in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin; Gibco; Thermo Fisher Scientific, Inc.) in a humidified chamber at 5% CO2. Cells were seeded at 5×105 cells per 60-mm culture dish.
Irradiation and experimental groups
Following 24-h culture, A549 cells in the exponential growth phase were irradiated with 2 or 4 Gy at a rate of 442.89 cGy/min in a linear accelerator (source distance, 100 cm; 6 MeV; Elekta Instrument AB). The A549 cells treated with each dose of radiation were divided into 3 subgroups and cultured for 2, 12 or 24 h. Non-irradiated A549 cells were collected at the indicated time points and used as negative controls.
Animal model
A total of 30 female athymic BALB/c nu/nu mice (6–8 weeks; weight, ~18 g) were obtained from Vital River Laboratories and housed in specific pathogen-free conditions with free access to food and water, temperature of 20–25°C, relative humidity of 55–65% and a 12-h light/dark cycle in the Beijing Laboratory Animal Research Center (Beijing Institute of Science and Technology, Beijing, China). Mice were injected with 5×105 A549 cells irradiated with 0-, 2- or 4-Gy X-rays via the tail vein, and were separated into the 0-, 2- and 4-Gy groups. The mice in the 0-Gy group were divided into 3 groups according to the time point of observation, with 6 mice in each group, and mice that did not undergo inoculation of A549 were used as controls. Mice were sacrificed 3, 6 or 10 weeks after inoculation, and lung and sera samples were harvested and stored at −80°C.
RNA isolation
RNA isolation from serum
Total RNA were extracted from the serum of patients and nude mice using a mirVana™PARIS™ Kit (cat. no. 1556; Ambion; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, ≤625 µl serum was mixed with an equal volume of 2X denaturing solution at room temperature and incubated on ice for 5 min. Subsequently, an equal volume of acid-phenol: Chloroform was added for extraction of the aqueous phase.
RNA extraction from A549 cells and lung tissue of nude mice
The cells were washed twice with PBS prior to the addition of 600 µl lysis/binding buffer (mirVana miRNA Isolation kit; cat. no. 1561; Ambion; Thermo Fisher Scientific, Inc.) directly to the culture plate or flask. The lysates were manually harvested using a sterile cell scraper and transferred to a 2-ml tube. The samples were stored at −80°C unless RNA was immediately extracted according to the manufacturer's instructions (Ambion; Thermo Fisher Scientific, Inc.).
For murine lung tissue, 0.2 g tissue was ground into a powder in liquid nitrogen with a pre-cooled mortar and pestle. The tissue was mixed with 1 ml lysis/binding buffer, followed by acid-phenol: Chloroform extraction. Ethanol was added to the samples prior to passing them through a filter cartridge containing a glass-fiber filter for RNA immobilization. The filter was washed with miRNA wash solution 1/2/3 three times, and the RNA was eluted with 30 µl elution solution (low ionic-strength solution), with both solutions included in the aforementioned kit.
Reverse transcription-quantitative (RT-q)PCR quantification of miRNA expression
To quantify miRNA expression in A549 cells, the TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was performed according to the manufacturer's instructions. The template cDNA was synthesized from total miRNA using a TaqMan™ MicroRNA Reverse Transcription kit (cat. no. 4366596; Applied Biosystems; Thermo Fisher Scientific, Inc.) and quantified by qPCR using the following parameters: Hold at 50°C for 2 min, denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 60 sec. The TaqMan® Universal PCR Master mix II (cat. no. 4440038; Applied Biosystems; Thermo Fisher Scientific, Inc.) was used with a LightCycler 480 real-time PCR system (Roche Molecular Systems, Inc.). The primers and probes of the three miRNAs were purchased from Thermo Fisher Scientific, Inc. (TaqMan® microRNA assay; cat. no. 4427975; Applied Biosystems; Thermo Fisher Scientific, Inc.) U6 RNA was used for internal normalization. Relative expression was calculated using the following equations: Relative gene expression=2−∆∆Cq; -∆∆Cq=(Cq gene of interest-Cq internal control gene)treated -(Cq gene of interest-Cq internal control gene)untreated (6,28).
miRNA transfection
A total of 5×105 A549 cells were seeded per 60-mm plate 18–24 h prior to the transfection. The medium was replaced with antibiotic-free medium 6–12 h before transfection. Synthetic miRNA mimics and inhibitors (miR-130a, miR-25 and miR-191*) and the negative control (cat. nos. 4464084, 4464066, 4464058 and 4464076; Thermo Fisher Scientific, Inc.) were transfected into 70–80% confluent A549 cells using the Lipofectamine® RNAiMAX (cat. no. 13778030; Invitrogen; Thermo Fisher Scientific, Inc.) reagent according to the manufacturer's instructions. The prepared mixtures of Lipofectamine® RNAiMAX and miRNA mimics/inhibitor diluted in Opti-MEM (cat. no. 31985-062; Invitrogen; Thermo Fisher Scientific, Inc.) were incubated separately for 5 min at room temperature, then added to adherent A549 cells and incubated at 37°C for 24 h. After 24 h transfection, cells were harvested for the extraction of total RNA and protein.
Transwell invasion assay
Cell invasion was measured using 24-well micro chemotaxis chambers with a membrane pore size of 8.0 µm (EMD Millipore) pre-coated with 10 µg/ml Matrigel at 37°C for 30 min (BD Biosciences). A549 cells transfected with miRNA mimics or inhibitors were resuspended in 100 µl serum-free DMEM (cat. no. 10566016; Gibco; Thermo Fisher Scientific, Inc.) and seeded into the upper well of each chamber. A total of 100 µl DMEM containing 10% FBS and 10% fibronectin (BD Biosciences) was loaded into the lower well.
Cells were incubated at 37°C for 22 h. Cells in the upper well that had not migrated were scraped off using a cotton swab, and the cells that had migrated through the membrane were fixed with 4% paraformaldehyde for 20 min and stained with Giemsa for 10 min, both at room temperature. Fixed cells were imaged under a phase-contrast microscope (magnification ×400) for six fields and counted for statistical analysis.
Protein extraction and western blotting
Transfected A549 cells were washed twice in ice-cold PBS and lysed in RIPA lysis buffer with protease inhibitors (50 nmol/l HEPES, pH 7.5, 150 nmol/l NaCl, 1% glycerol, 1% Triton, protease inhibitor cocktail). The protein concentration was determined using a BCA Protein Assay kit (Beyotime Institute of Biotechnology). A total of 25 µg protein was loaded per sample for SDS-PAGE (12% acrylamide) and transferred to polyvinylidene difluoride membranes (EMD Millipore). The membranes were blocked using 5% skimmed milk in TBS-Tween (TBST; 0.05% Tween-20) at room temperature for 1 h, and then incubated with antibodies against VEGF (1:800; cat. no. 34-4300; Sigma-Aldrich; Merck KGaA), CCR-7 (1:2,000; cat. no. ab32527; Abcam) or tubulin (1:2,000; cat. no. SC-12462; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature, the membrane was washed 3 times with TBST (10 min each), followed by incubation with horseradish peroxidase-conjugated mouse anti-rabbit secondary antibody for 1 h at room temperature (1:1,000; cat. no. sc-2357; Santa Cruz Biotechnology, Inc.). The signal was captured using a Super Signal West Pico chemiluminescent substrate (cat. no. 34080; Pierce; Thermo Fisher Scientific, Inc.) to visualize the bands, and Image-Pro Plus 6.0 software (Media Cybernetics, Inc.) was used for grayscale analysis.
Statistical analysis
Data are presented as the mean ± SD. Data were processed using SPSS version 19.0 (IBM Corp.). The Mann-Whitney U test was used to determine the statistical difference between the expression levels of the three miRNAs and different clinical characteristics. Associations between clinical characteristics and miRNA expression were assessed by logistic regression analysis, and survival was analyzed using the Kaplan-Meier method with Cox regression. One-way ANOVA followed by Dunnett's post hoc test was used to evaluated the statistical differences of the expression levels of the three miRNAs or VEGF or CCR-7 mRNA/protein expression at different time points of A549 cells with 0-, 2- and 4-Gy X-ray irradiation or with miRNA transfection. Kruskal-Wallis test followed by Tukey's or Dunnett's post hoc tests was used to evaluate the statistical differences of three miRNAs expression in lung tissues and serum of nude mice injected with A549 cells with different doses of X-ray irradiation. Receiver operating characteristic curves were used to evaluate the diagnostic value of miR-130a, miR-25 and miR-191* in patients with NSCLC. P<0.05 was considered to indicate a statistically significant difference.
Results
Associations between clinical features of patients with NSCLC and the expression of miR-130a, miR-25 and miR-191*
A total of 84 patients (58 male and 26 female) with lung cancer with a mean age of 60.78 years (range, 37–87) were enrolled in the present study. According to the International Association for the Study of Lung Cancer Staging Project (29), various clinical stages were present among the enrolled patients, including three cases at stage I, five cases at stage III and 76 cases at stage IV. Detailed clinical characteristics of the patients are presented in Table I. Notably, the expression of miR-25 and miR-191* was significantly higher in patients treated with radiotherapy compared with those who did not receive radiotherapy treatment, in patients with SCC compared with those with ADC, and in patients who succumbed to NSCLC or the associated complications during the follow-up period compared with those who survived (P<0.05 and P<0.01; Table I; Fig. S1B and C). miR-130a expression levels were significantly different between the radiotherapy and non-radiotherapy groups (P=0.004; Fig. S1A), and there was also a significant difference in miR-25 expression between patients aged ≥60 and <60 years (P=0.035; Table I; Fig. S1B). There was no association between miR-130a, miR-25 or miR-191* expression and sex, smoking status, lymph node metastasis, extracranial metastasis, brain metastasis (BM), number of BMs, maximum diameter of BM, Tumor-Node-Metastasis (TNM) T stage (30), node (N) stage (30) or Eastern Cooperative Oncology Group (ECOG) (31,32) performance status (P>0.05; Table I).
Logistic regression analysis of clinical characteristics and the expression of miR-130a, miR-25 and miR-191* in the serum of patients with NSCLC
Based on the optimal cut-off values for miR-130a, miR-25 and miR-191* (Data S1 and Fig. S2), logistic regression analysis was performed to determine the association between the expression of each miRNA and clinical characteristics. The results indicated that miR-130a expression was associated with radiotherapy (P=0.021), but not sex, age, smoking status, histological type, survival status, lymph-node metastasis, BM, extracranial metastasis or ECOG value. By contrast, miR-25 and miR-191* expression levels were associated with radiotherapy (P=0.026 and P=0.014, respectively), histological type (P=0.010 and P=0.005, respectively) and survival status (P=0.020 and P=0.043, respectively) (Table II). The expression of miR-25 was also significantly associated with age (P=0.019).
Table II.
Logistic regression analysis of factors that affect the expression of miR-130a, miR-25 and miR-191* in the serum of patients with non-small cell lung cancer.
| 95% CI of Exp(B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| microRNA | Clinical characteristic | B | SE | Wald | P-value | Exp(B) | Lower | Upper |
| miR-130a | Radiotherapy | −1.228 | 0.533 | 5.315 | 0.021a | 0.293 | 0.103 | 0.832 |
| Age | 1.252 | 0.534 | 5.486 | 0.019a | 3.496 | 1.227 | 9.963 | |
| Radiotherapy | −1.268 | 0.570 | 4.948 | 0.026a | 0.282 | 0.092 | 0.860 | |
| miR-25 | Histological type | 1.551 | 0.605 | 6.570 | 0.010a | 4.715 | 1.440 | 15.431 |
| Survival status | −1.118 | 0.480 | 5.428 | 0.020a | 0.327 | 0.128 | 0.837 | |
| miR-191* | Histological type | 1.344 | 0.483 | 7.750 | 0.005b | 3.836 | 1.489 | 9.884 |
| Radiotherapy | −1.326 | 0.537 | 6.101 | 0.014a | 0.266 | 0.093 | 0.761 | |
| Survival status | −1.034 | 0.511 | 4.097 | 0.043a | 0.356 | 0.131 | 0.968 | |
P<0.05
P<0.01. miR, microRNA; B, regression coefficient; SE, standard error; Exp(B) odds ratio.
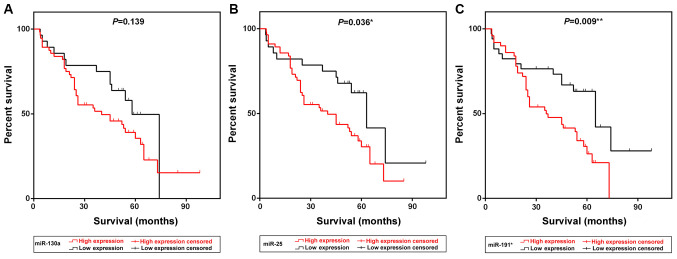
Kaplan-Meier survival curve analysis demonstrated that patients exhibiting high expression of miR-25 and miR-191* presented with low survival rates compared with patients with low expression of the two miRNAs (P=0.036 and P=0.009, respectively; Fig. 1B and C). The median survival time of patients in the high miR-25 and miR-191* expression groups was 40 and 36 months, respectively, which was notably lower compared with the survival times of patients in the low expression groups [63 (P=0.036) and 65 (P=0.009) months, respectively; Table III]. Multivariate Cox survival analysis indicated that radiotherapy (P=0.024), ECOG status (P=0.008) and miR-191* expression (P=0.034) were independent prognostic factors for NSCLC in this cohort (Table IV).
Figure 1.
Kaplan-Meier survival curves of patients with NSCLC grouped by miR-130a, miR-25 and miR-191* expression levels. (A-C) Kaplan-Meier curves indicating the survival of patients with NSCLC exhibiting low or high expression of (A) miR-130a, (B) miR-25 and (C) miR-191*. NSCLC, non-small cell lung cancer; miR, microRNA.
Table III.
Analysis of miR-130a, miR-25 and miR-191* expression and survival time of patients with non-small cell lung cancer.
| 95% CI of HR | ||||||
|---|---|---|---|---|---|---|
| Expression levels | Median survival time, months | HR | Lower | Upper | Chi-square | P-value |
| miR-130a | ||||||
| High | 40 | 1.584 | 0.878 | 2.77 | 2.188 | 0.139 |
| Low | 58 | 0.631 | 0.361 | 1.14 | ||
| miR-25 | ||||||
| High | 40 | 1.942 | 1.057 | 3.287 | 4.409 | 0.036a |
| Low | 63 | 0.515 | 0.304 | 0.945 | ||
| miR-191* | ||||||
| High | 36 | 2.12 | 1.235 | 3.71 | 6.789 | 0.009b |
| Low | 65 | 0.472 | 0.27 | 0.81 | ||
P<0.05
P<0.01. miR, microRNA; HR, hazard ratio.
Table IV.
Multivariate analysis of clinicopathological features, miR-130a, miR-25 and miR-191* expression and prognosis of patients with non-small cell lung cancer.
| 95% CI of Exp(B) | |||||||
|---|---|---|---|---|---|---|---|
| Factor | B | SE | Wald | P-value | Exp(B) | Lower | Upper |
| Age (<60 years vs. ≥60 years) | −0.093 | 0.338 | 0.075 | 0.784 | 0.912 | 0.470 | 1.767 |
| Sex (male vs. female) | −0.506 | 0.455 | 1.238 | 0.266 | 0.603 | 0.247 | 1.470 |
| Smoking status (yes vs. no) | −0.360 | 0.404 | 0.707 | 0.372 | 0.697 | 0.316 | 1.538 |
| Radiotherapy (yes vs. no) | 0.808 | 0.357 | 5.110 | 0.024a | 2.242 | 1.113 | 4.516 |
| Histological type (LAD vs. LSCC) | −0.215 | 0.394 | 0.298 | 0.585 | 0.806 | 0.372 | 1.746 |
| Lymph node metastasis (yes vs. no) | −0.173 | 0.385 | 0.202 | 0.653 | 0.841 | 0.395 | 1.789 |
| Extracranial metastasis (yes vs. no) | −0.273 | 0.410 | 0.443 | 0.506 | 0.761 | 0.340 | 1.701 |
| Brain metastasis (yes vs. no) | −0.261 | 0.396 | 0.436 | 0.509 | 0.770 | 0.355 | 1.672 |
| ECOG (score 1 vs. 2) | −0.876 | 0.328 | 7.133 | 0.008b | 0.416 | 0.219 | 0.792 |
| miR-130a expression (up- vs. downregulation) | −0.406 | 0.450 | 0.812 | 0.368 | 0.666 | 0.276 | 1.611 |
| miR-25 expression (up- vs. downregulation) | −0.723 | 0.487 | 2.200 | 0.138 | 0.485 | 0.187 | 1.261 |
| miR-191* expression (up- vs. downregulation) | −0.825 | 0.389 | 4.509 | 0.034a | 0.438 | 0.205 | 0.938 |
P<0.05
P<0.01. B, regression coefficient; SE, standard error; Exp(B) odds ratio; miR, microRNA; ECOG, Eastern Cooperative Oncology Group; LAD, lung adenocarcinoma; LSCC, lung squamous cell cancer.
Two- and 4-Gy X-ray irradiation upregulates the expression of miR-130a, miR-25 and miR-191* in A549 cells in vitro and in vivo
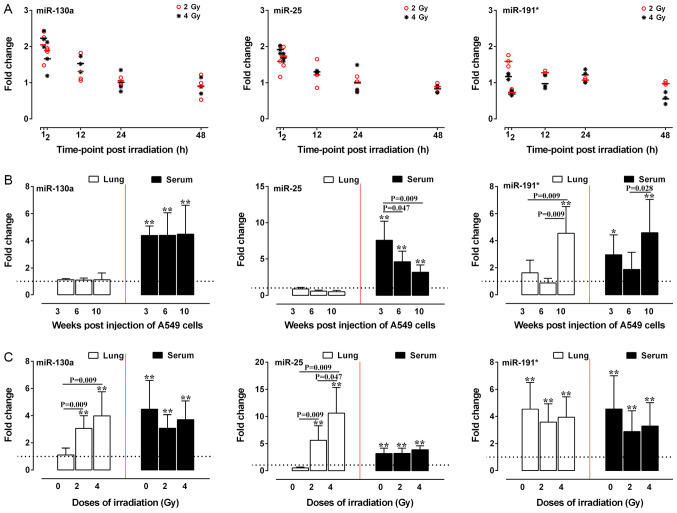
The aforementioned results indicated that radiotherapy, histological type and prognosis were associated with the expression of miR-130a, miR-25 and miR-191* in patients with NSCLC. Therefore, the expression levels of miR-130a, miR-25 and miR-191* in A549 cells treated with different doses of irradiation were examined in vitro and in vivo. At 2 h post-2-Gy irradiation, the expression levels of miR-130a and miR-25 were significantly increased in A549 cells (P<0.05 and P<0.01), whereas the expression levels of miR-191* were significantly increased at 12 h post-irradiation compared with those in the respective control groups (Fig. 2A). At 2 h post-4-Gy irradiation, the expression levels of miR-191* and miR-25 were also significantly increased (P<0.05 and P<0.01), whereas the expression levels of miR-130a were significantly increased at 12 h post-irradiation compared with those in the respective control groups (P<0.05) (Fig. 2A).
Figure 2.
Relative expression of miR-130a, miR-25 and miR-191* in irradiated A549 in vitro and in vivo. (A) Scatter plots of miR-130a, miR-25 and miR-191* expression at various time-points after 2- or 4-Gy irradiation of A549 cells. (B) Relative expression of miR-130a, miR-25 and miR-191* in the lung and serum of nude mice 3, 6 and 10 weeks after injection with A549 cells. (C) The relative expression levels of miR-130a, miR-25 and miR-191* in the lung and serum of nude mice 10 weeks after injection with A549 cells irradiated with 0-, 2- and 4-Gy X-ray. Reverse transcription-quantitative PCR was performed in triplicate, and each experiment was repeated ≥3 times. The dotted line represents the level of the control group. *P<0.05 and **P<0.01 vs. control. miR, microRNA.
For the in vivo study, 6 mice each in the 0-Gy group were sacrificed at 3, 6 and 10 weeks, and 5 mice each in the 2- and 4-Gy groups were sacrificed at 10 weeks after injection. At 3, 6 and 10 weeks after A549 cell injection, miR-130a and miR-25 expression was significantly increased in mice serum samples, but there was no significant increase in lung tissue expression of these miRNAs compared with in the control mice. The expression levels of miR-191* were increased in the serum of nude mice at 3 and 10 weeks after injection when the mice exhibited visible tumors scattered in the lungs (data not shown), while the miR-191* expression in the lung tissues of nude mice was upregulated at 10 weeks after injection compared with that of the control group (P<0.05; Fig. 2B). Both 2- or 4-Gy irradiation resulted in significant upregulation of the expression levels of all three miRNAs at 10 weeks post injection in the serum and lung tissue compared with those in the control group (P<0.01; Fig. 2C). Notably, the expression levels of miR-130a and miR-25 in the lung tissues of mice treated with 2- and 4-Gy irradiation exhibited a dose-dependent tendency to increase compared with those of mice injected with non-irradiated A549 cells (P<0.05 and P<0.01; Fig. 2C).
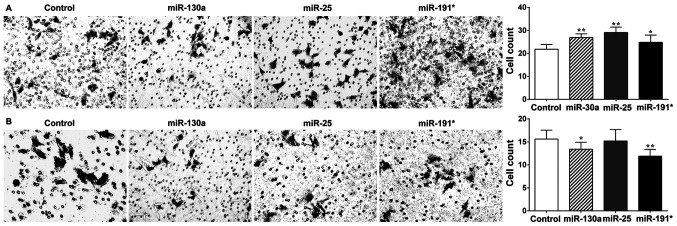
Roles of miR-130a, miR-25 and miR-191* in A549 cell invasion in vitro
To delineate the roles of the three miRNAs in response to X-ray irradiation, the effects of miR-130a, miR-25 and miR-191* on the invasive capability of A549 cells were assessed in vitro following transfection with miRNA mimics and inhibitors. Transfection efficiency was verified by qPCR (data not shown). The results of the cell invasion assay demonstrated that the invasiveness of A549 cells transfected with miR-130a, miR-25 and miR-191* mimics was significantly increased compared with that of the control group (P<0.05 and P<0.01; Fig. 3A). Additionally, transfection of A549 cells with miR-130a and miR-191* inhibitors significantly reduced invasiveness (P<0.05; Fig. 3B), while transfection of A549 cells with miR-25 inhibitor had no significant effect on cell invasiveness.
Figure 3.
Effects of miR-130a, miR-25 and miR-191* on the invasion of A549 cells. The invasiveness of A549 cells transfected with miR-130a, miR-25 and miR-191* mimics and inhibitors was assessed by Transwell assay. (A) The invasiveness of A549 cells transfected with miR-130a, miR-25 and miR-191* mimics. (B) The invasiveness of A549 cells transfected with miR-130a, miR-25 and miR-191* inhibitors. *P<0.05 and **P<0.01 vs. control. miR, microRNA.
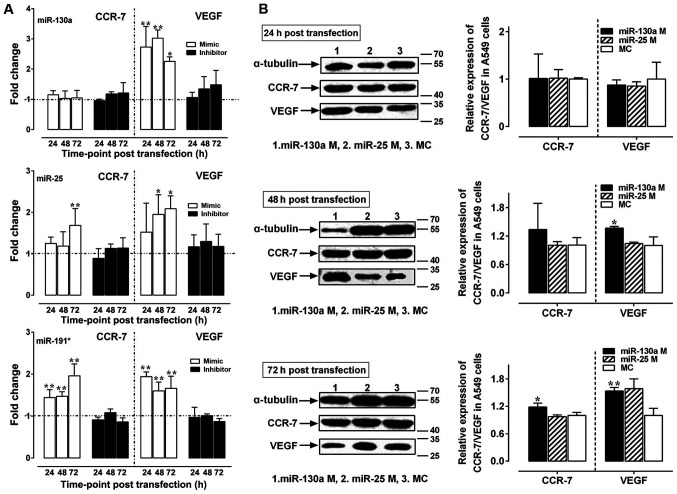
Our previous study has demonstrated that 2- and 4-Gy X-rays promote the invasion of A549 cells in vitro and in vivo mainly by upregulating the expression of VEGF and CCR-7 mRNA (33). Following A549 cell exposure to 2- and 4-Gy X-ray irradiation, the expression of miR-130a was positively correlated with the expression of VEGF and CCR-7 mRNA (P<0.05), while the expression of miR-25 and miR-191* was not significantly correlated with the expression of VEGF/CCR-7 mRNA (P>0.05) (34). Therefore, the effects of miR-130a, miR-25 and miR-191* on the expression of VEGF and CCR-7 in X-ray-irradiated A549 cells were subsequently investigated. Upregulation of VEGF expression levels was observed following transfection of all three miRNA mimics compared with those in the control groups (P<0.05 and P<0.01; Fig. 4A), whereas upregulation of CCR-7 expression was observed in the miR-25 mimics group at 72 h and in the miR-191* mimics group at 24, 48 and 72 h post-transfection (P<0.05 and P<0.01; Fig. 4A). Compared with those in the negative control group, protein expression levels of VEGF were upregulated at 48 and 72 h post-transfection with miR-130a mimics (P<0.05 and P<0.01, respectively; Fig. 4B), whereas the expression of CCR-7 was upregulated only at 72 h post-transfection with miR-130a mimics (P<0.05; Fig. 4B). By contrast, no significant changes in the mRNA (Fig. 4A) or protein expression levels (data not shown) of VEGF or CCR-7 were observed in the three miRNA inhibitor transfection groups.
Figure 4.
Relative expression of VEGF/CCR-7 mRNA and protein following transfection with miR-130a, miR-25 and miR-191* mimics and inhibitors. (A) The relative expression of VEGF and CCR-7 mRNA in A549 cells transfected with miR-130a, miR-25 and miR-191* mimics and inhibitors. The dotted line represents the level of the control group. (B) The relative expression of VEGF and CCR-7 protein in A549 cells transfected with miR-130a and miR-25 mimics. Western blotting and reverse transcription-quantitative PCR were performed in triplicate, and each experiment was repeated ≥3 times. *P<0.05 and **P<0.01 vs. control. VEGF, vascular endothelial growth factor; miR, microRNA; CCR-7, C-C chemokine receptor type 7; MC, mimics control.
Discussion
Although a large number of circulating miRNAs have been studied as potential biomarkers for NSCLC (7,20,35,36), the value of detecting miRNAs in the blood for predicting the prognosis of patients following radiotherapy has not yet been defined. The results of the present study demonstrated that upregulation of miR-130a, miR-25 and miR-191* expression in the serum of patients with NSCLC was associated with radiotherapy, histological type, low survival rate and low median survival time. These results suggested that these miRNAs may serve roles in predicting the prognosis of NSCLC. Existing miRNA biomarkers are used as indicators for the early diagnosis in NSCLC (36,37), carcinogenesis in gastric cancer and NSCLC (38,39), metastasis (40,41) in lung adenocarcinoma and NSCLC, disease recurrence in adrenocortical cancer (42), drug/radiotherapy-resistance (43) and prognosis in NSCLC (39). Geng et al (37) have identified five miRNAs (miR-20a, miR-223, miR-21, miR-221 and miR-45) in the plasma for early-stage NSCLC screening. Xu et al (44) have reported that miR-130a is upregulated in the plasma of patients with NSCLC compared with healthy controls, but not associated with poor prognosis. The expression of miR-25 is increased in lung tissues of patients with ADC with an epidermal growth factor receptor mutation, which is frequent in non-smoking patients (45). In addition, previous studies have indicated that the expression of miR-25 is significantly upregulated in NSCLC tissues compared with in paired adjacent tissues and negatively correlated with the protein expression of regulator of G protein signaling 3 (46,47). In consistence with previous reports, the results of the present study suggested that patients with NSCLC exhibiting high expression of miR-25 and miR-191* in the serum exhibited lower survival rates compared with patients with low expression of these miRNAs. The shorter median survival times of patients exhibiting high expression of miR-25 and miR-191* indicates a role for these miRNAs in predicting poor prognosis. In addition, miR-191* expression was demonstrated to be an independent prognostic factor in patients with NSCLC in the present study.
Further analysis demonstrated that the expression of the three miRNAs in patients with SCCs or those who had undergone radiotherapy was significantly higher compared with that in patients with ADCs or those who had not undergone radiotherapy, respectively. This suggested that miR-130a, miR-25 and miR-191* may have a potential role in predicting the response to radiotherapy and in differentiating SCCs from ADCs among patients with NSCLC. Radiotherapy remains essential as a tool for the treatment of lung cancer, particularly in patients with local or advanced metastases for whom surgical treatment is not an option (48). However, radiotherapy of primary tumors has been demonstrated to accelerate metastasis in mice (49), thus limiting the efficacy of radiotherapy. In addition, ionizing radiation promotes the migration and invasion of a variety of tumor cells, including glioma, neck squamous cell carcinoma, pancreatic cancer and lung cancer cells (50). The underlying mechanisms of this effect may involve the tumor microenvironment, cell-cell junctions, extracellular matrix junction, protease secretion and induction of epithelial-mesenchymal transition (51–56). To investigate the potential role of miRNAs in regulating the response to radiotherapy, the present study analyzed the effects of 2- and 4-Gy X-ray irradiation on the expression of miR-130a, miR-25 and miR-191* in A549 cells in vitro and in vivo. The results demonstrated that 2- and 4-Gy irradiation promoted the expression of these three miRNAs, as well as the invasiveness of A549 cells in vitro. Further in vivo experiments revealed that irradiation promoted metastasis in A549-cell xenograft mice, accompanied by high expression of miR-130a, miR-25 and miR-191* in the serum. These results indicated functional roles of the three miRNAs in radiation-mediated metastasis and suggested that they may be potential biomarkers of lung cancer metastasis induced by radiotherapy.
miRNAs have been demonstrated to regulate invasion and metastasis via a variety of mechanisms, including the regulation of oncogene and/or tumor suppressor gene expression (57,58), tumor metastasis-related gene expression (59,60) and tumor angiogenesis-related gene expression (61). VEGF and CCR-7 are involved in metastasis, and have been clinically demonstrated to be associated with metastasis and prognosis (62–65). The present study aimed to further study the involvement of miR-130a, miR-25 and miR-191* in radiation-mediated metastasis using A549 cells in vitro. miR-130a, miR-25 and miR-191* mimics promoted the invasion of A549 cells, whereas miR-130a and miR-191* inhibitors impeded their invasiveness. The miR-130a mimics also increased the expression of VEGF mRNA and protein in A549 cells at 24, 48 and 72 h post-transfection, and the expression of CCR-7 protein was upregulated at 72 h post-transfection. miR-25 and miR-191* mimic transfection increased the expression of CCR-7 and VEGF only at the mRNA level. Overall, these results suggested that miR-130a, miR-25 and miR-191* may promote the invasion of A549 cells by increasing VEGF and CCR-7 expression.
The results of the present study demonstrated the significance of miR-130a, miR-25 and miR-191* in NSCLC, particularly regarding diagnosis and tumor metastasis. However, certain limitations remain in the present study. The number of patients included in this study was relatively small, and the stage distribution of the patients was uneven, which may have caused selection bias. Furthermore, due to the large time-span of sample collection, the follow-up time was extended accordingly, which may also introduce bias. To address these issues, a larger cohort of patients should be included in future studies, with a shorter follow-up duration. In addition, the present study only used one cell line, and including multiple cell lines in future studies will help eliminate concerns that the observations made in the in vitro and xenograft experiments were cell line-specific.
In conclusion, the results of the present study demonstrated that the expression levels of miR-130a, miR-25 and miR-191* were elevated in the serum of patients with NSCLC compared with that of healthy control subjects, and that this was associated with patient prognosis and response to radiotherapy. Therefore, these miRNAs exhibit potential for use as biomarkers to predict the prognosis and radiotherapy outcome for patients with NSCLC. In vitro and in vivo study confirmed the tumorigenic roles of miR-130a, miR-25 and miR-191* in radiation-mediated xenograft metastasis in mice, supporting their possible clinical and functional roles in radiation-mediated metastasis of lung cancer.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Na Jia (Beijing Institute of Microbiology and Epidemiology, Beijing, China) for critical review of this manuscript, Ms. Ying Yang (Biolancet Technology, Ltd., Beijing, China) for performing the microRNA profile assay and Mr. Li-gang An (Beijing Laboratory Animal Research Center, Beijing, China) for technical assistance.
Glossary
Abbreviations
- ADC
adenocarcinoma
- BM
brain metastasis
- CCR-7
C-C chemokine receptor type 7
- ECOG
Eastern Cooperative Oncology Group
- miRNA
microRNA
- NSCLC
non-small-cell lung cancer
- SCC
squamous cell carcinoma
- TNM
Tumor-Node-Metastasis
- VEGF
vascular endothelial growth factor
Funding
The present study was supported by the China Postdoctoral Science Foundation (grant no. 2014M552667), the National Natural Science Foundation of China (grant no. 31770914) and the Major Program of Military Logistics (grant no. AEP17J001).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JL, JA and QSJ conceived and designed the study. JL, JA, AML and JG performed the experiments. ZXL, GLZ, WWH and HMC participated in the acquisition and interpretation of data, drafted the work and revised it critically for important intellectual content. JL, YDZ, WWH and QSJ analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All studies involving animals were performed according to a protocol approved by the Animal Welfare Ethics Committee of Beijing Experimental Animal Research Center [Beijing, China; approval no. ShiYanXueKe (SYXK) (Beijing) 2011-0006]. Ethics approval for the study involving human samples was provided by the PLA Rocket Force Characteristic Medical Center Ethics Committee (approval no. KeYan 2015038). Written informed consent was obtained from all participants who donated lung tissue and serum samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the surveillance, epidemiology, and end results (SEER) program. Oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: Adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84:13–22. doi: 10.1016/j.lungcan.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 7.Xu FX, Su YL, Zhang H, Kong JY, Yu H, Qian BY. Prognostic implications for high expression of miR-25 in lung adenocarcinomas of female non-smokers. Asian Pac J Cancer Prev. 2014;15:1197–1203. doi: 10.7314/APJCP.2014.15.3.1197. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Yang G, Zhou M, Smith M, Ge H, Boreham J, Hu Y, Peto R, Wang J, Chen Z. Body mass index and mortality from lung cancer in smokers and nonsmokers: A nationally representative prospective study of 220,000 men in China. Int J Cancer. 2009;125:2136–2143. doi: 10.1002/ijc.24527. [DOI] [PubMed] [Google Scholar]
- 9.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 10.Mitra R, Sun J, Zhao Z. MicroRNA regulation in cancer: One arm or two arms? Int J Cancer. 2015;137:1516–1518. doi: 10.1002/ijc.29512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18:184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 12.Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, Bari M, Singha RS, Malakar AK, Chakraborty S. Interplay between miRNAs and human diseases. J Cell Physiol. 2018;233:2007–2018. doi: 10.1002/jcp.25854. [DOI] [PubMed] [Google Scholar]
- 13.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 14.Jiang LP, Zhu ZT, Zhang Y, He CY. Downregulation of microRNA-330 correlates with the radiation sensitivity and prognosis of patients with brain metastasis from lung cancer. Cell Physiol Biochem. 2017;42:2220–2229. doi: 10.1159/000479996. [DOI] [PubMed] [Google Scholar]
- 15.Dong J, Zhang Z, Gu T, Xu SF, Dong LX, Li X, Fu BH, Fu ZZ. The role of microRNA-21 in predicting brain metastases from non-small cell lung cancer. Onco Targets Ther. 2017;10:185–194. doi: 10.2147/OTT.S116619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG, Luo JC. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat Oncol. 2013;8:146. doi: 10.1186/1748-717X-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang LP, He CY, Zhu ZT. Role of microRNA-21 in radiosensitivity in non-small cell lung cancer cells by targeting PDCD4 gene. Oncotarget. 2017;8:23675–23689. doi: 10.18632/oncotarget.15644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powrozek T, Krawczyk P, Kowalski DM, Kuźnar-Kamińska B, Winiarczyk K, Olszyna-Serementa M, Batura-Gabryel H, Milanowski J. Application of plasma circulating microRNA-448, 506, 4316, and 4478 analysis for non-invasive diagnosis of lung cancer. Tumour Biol. 2016;37:2049–2055. doi: 10.1007/s13277-015-3971-4. [DOI] [PubMed] [Google Scholar]
- 19.Matikas A, Syrigos KN, Agelaki S. Circulating biomarkers in non-small-cell lung cancer: Current status and future challenges. Clin Lung Cancer. 2016;17:507–516. doi: 10.1016/j.cllc.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Wang Y, Zhang Q, Tang L, Liu X, Dai Y, Xiao L, Huang S, Chen L, Guo Z, et al. MicroRNA-486 as a biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS One. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greystoke A, Ayub M, Rothwell DG, Morris D, Burt D, Hodgkinson CL, Morrow CJ, Smith N, Aung K, Valle J, et al. Development of a circulating miRNA assay to monitor tumor burden: From mouse to man. Mol Oncol. 2016;10:282–291. doi: 10.1016/j.molonc.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 25.Wu T, Hu H, Zhang T, Jiang L, Li X, Liu S, Zheng C, Yan G, Chen W, Ning Y, et al. MiR-25 promotes cell proliferation, migration, ad invasion of non-small cell lung cancer by targeting the LATS2/YAP signaling pathway. Oxid Med cell Longev. 2019;18:9719723. doi: 10.1155/2019/9719723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, Wang W, Zhang L, Zhang X, Tang Q, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting smad7. Cancer Lett. 2013;335:168–174. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Pan H, Qian Y, Zhou W, Liu X. miR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2018;17:4. doi: 10.1186/s12943-017-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Lee EJ, Jiang J. High-throughput real-time PCR. Methods Mol Biol. 2008;429:89–98. doi: 10.1007/978-1-60327-040-3_7. [DOI] [PubMed] [Google Scholar]
- 29.Funai K, Kawase A, Mizuno K, Koyama S, Shiiya N. 8th edition tumor, node, and metastasis T-stage prognosis discrepancies: Solid component diameter predicts prognosis better than invasive component diameter. Cancer (Basel) 2020;12:1577. doi: 10.3390/cancers12061577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rami-Porta R, Bolejack V, Crowley J, Ball D, Kim J, Lyons G, Rice T, Suzuki K, Thomas CF, Jr, Travis WD, et al. The IASLC lung cancer staging project: Proposals for the revisions of the T descriptors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2015;10:990–1003. doi: 10.1097/JTO.0000000000000559. [DOI] [PubMed] [Google Scholar]
- 31.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH, Eastern Cooperative Oncology Group Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 32.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-Carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 33.Lv J, Jiang QS, Song XJ, Wang CL, Guo LJ, Wang SN, Li FS, Hu WW. Role of VEGF-A/C and CCR-7 in the enhanced metastasis of A549 cells induced by 2 and 4 Gy X-rays in vitro and in vivo. Sci China Tech Sci. 2014;57:990–997. doi: 10.1007/s11431-014-5517-1. [DOI] [Google Scholar]
- 34.Lv J, Wang SN, Song XJ, Li X, He R, Yu HJ, Chen S, Wang L, Jiang QS. The impact of X-rays on the expressions of miR-130a/miR-25 and its potential role in the enhanced metastasis of A549 cell lines in vitro induced by X-rays. Sci China Technol Sci. 2013;56:2243–2249. doi: 10.1007/s11431-013-5290-6. [DOI] [Google Scholar]
- 35.Doval DC, Deshpande R, Dhabhar B, Babu KG, Prabhash K, Chopra R, Sripada PV, Deshmukh C, Suryavanshi M. Liquid biopsy: A potential and promising diagnostic tool for advanced stage non-small cell lung cancer patients. Indian J Cancer. 2017;54(Suppl):S25–S30. doi: 10.4103/ijc.IJC_514_17. [DOI] [PubMed] [Google Scholar]
- 36.Han Y, Li H. miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC) J Thorac Dis. 2018;10:3119–3131. doi: 10.21037/jtd.2018.05.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Li R, Yu H, Wang R, Shen Z. MicroRNA-130a is an oncomir suppressing the expression of CRMP4 in gastric cancer. Onco Targets Ther. 2017;10:3893–3905. doi: 10.2147/OTT.S139443. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol. 2017;15:175. doi: 10.1186/s12957-017-1244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daugaard I, Veno MT, Yan Y, Kjeldsen TE, Lamy P, Hager H, Kjems J, Hansen LL. Small RNA sequencing reveals metastasis-related microRNAs in lung adenocarcinoma. Oncotarget. 2017;8:27047–27061. doi: 10.18632/oncotarget.15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Li Y, Liu J, Fan Y, Li X, Dong M, Liu H, Chen J. Expression levels of microRNA-145 and microRNA-10b are associated with metastasis in non-small cell lung cancer. Cancer Biol Ther. 2016;17:272–279. doi: 10.1080/15384047.2016.1139242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chabre O, Libe R, Assie G, Barreau O, Bertherat J, Bertagna X, Feige JJ, Cherradi N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr Relat Cancer. 2013;20:579–594. doi: 10.1530/ERC-13-0051. [DOI] [PubMed] [Google Scholar]
- 43.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X, Zhu S, Tao Z, Ye S. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Med. 2018;7:21–31. doi: 10.1002/cam4.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: Correlation with mutational status. Mod Pathol. 2010;23:1577–1582. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- 46.Wu T, Chen W, Kong D, Li X, Lu H, Liu S, Wang J, Du L, Kong Q, Huang X, Lu Z. miR-25 targets the modulator of apoptosis 1 gene in lung cancer. Carcinogenesis. 2015;36:925–935. doi: 10.1093/carcin/bgv068. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z, Wu Y, Meng Q, Xia Z. Elevated microRNA-25 inhibits cell apoptosis in lung cancer by targeting RGS3. In vitro cell. Dev Biol Anim. 2016;52:62–67. doi: 10.1007/s11626-015-9947-2. [DOI] [PubMed] [Google Scholar]
- 48.Deek MP, Kim S, Yue N, Baby R, Ahmed I, Zou W, Langenfeld J, Aisner J, Jabbour SK. Modern radiotherapy using image guidance for unresectable non-small cell lung cancer can improve outcomes in patients treated with chemoradiation therapy. J Thorac Dis. 2016;8:2602–2609. doi: 10.21037/jtd.2016.08.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartford AC, Gohongi T, Fukumura D, Jain RK. Irradiation of a primary tumor, unlike surgical removal, enhances angiogenesis suppression at a distal site: Potential role of host-tumor interaction. Cancer Res. 2000;60:2128–2131. [PubMed] [Google Scholar]
- 50.Moncharmont C, Levy A, Guy JB, Falk AT, Guilbert M, Trone JC, Alphonse G, Gilormini M, Ardail D, Toillon RA, et al. Radiation-Enhanced cell migration/invasion process: A review. Crit Rev Oncol Hematol. 2014;92:133–142. doi: 10.1016/j.critrevonc.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, Luo H. Circulating miR-125b as a marker predicting chemoresistance in breast cancer. PLoS One. 2012;7:e34210. doi: 10.1371/journal.pone.0034210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manne U, Shanmugam C, Bovell L, Katkoori VR, Bumpers HL. miRNAs as biomarkers for management of patients with colorectal cancer. Biomark Med. 2010;4:761–770. doi: 10.2217/bmm.10.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cha HJ, Shin S, Yoo H, Lee EM, Bae S, Yang KH, Lee SJ, Park IC, Jin YW, An S. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int J Oncol. 2009;34:1661–1668. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- 55.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, Rosenberg A. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 56.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 58.Sayed D, Rane S, Lypowy J, He M, Chen LY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.e08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23:2592–2597. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X, Zhang Y, Hosaka K, Andersson P, Wang J, Tholander F, Cao Z, Morikawa H, Tegnér J, Yang Y, et al. VEGF-B promotes cancer metastasis through a VEGF-A-independent mechanism and serves as a marker of poor prognosis for cancer patients. Proc Natl Acad Sci USA. 2015;112:E2900–E2909. doi: 10.1073/pnas.1503500112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jean C, Chen XL, Nam JO, Tancioni I, Uryu S, Lawson C, Ward KK, Walsh CT, Miller NL, Ghassemian M, et al. Inhibition of endothelial FAK activity prevents tumor metastasis by enhancing barrier function. J Cell Biol. 2014;204:247–263. doi: 10.1083/jcb.201307067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji RC. Lymph nodes and cancer metastasis: New perspectives on the role of intranodal lymphatic sinuses. Int J Mol Sci. 2016;18:51. doi: 10.3390/ijms18010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Wu BQ, Geng H, Xu ML, Zhong HH. Association of chemokine and chemokine receptor expression with the invasion and metastasis of lung carcinoma. Oncol Lett. 2015;10:1315–1322. doi: 10.3892/ol.2015.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.