Abstract
Many recent studies have revealed exquisite subcellular localization of proteins, DNA, and other molecules within bacterial cells, giving credence to the concept of prokaryotic anatomy. Common sites for localized components are the poles of rod-shaped cells, which are dynamically modified in composition and function in order to control cellular physiology. An impressively diverse array of mechanisms underlies bacterial polarity, including oscillatory systems, phospho-signaling pathways, the sensing of membrane curvature, and the integration of cell cycle regulators with polar maturation.
Introduction
Over the past decade, the development of highly sensitive light microscopes, together with variably colored and brightly fluorescent marker proteins for specific labeling, has allowed biologists to surmount many of the challenges in visualizing components within tiny bacterial specimens. From this, we have learned that even the smallest and simplest forms of life could be dissected, given an appropriately sized scalpel, into discrete anatomical parts that carry out unique functions. A recent genome scale screen of Caulobacter crescentus [1] revealed that at least 10% of the encoded proteins are localized to a specific region of the cell, indicating that spatial organization can be a prominent feature of prokaryotic physiology.
Among all of the bacteria that have been studied by fluorescent protein labeling, one of the most common localization patterns is protein accumulation at the cell poles, which are defined as the end points on the long axis of a rod shaped cell. These are locations for site-specific assembly of surface organelles such as flagella, pili, and virulence factor secretion systems, allowing the cell to orient itself for directional motility and interaction with surfaces [2]. The poles can also direct the organization of elements in the cytoplasm, by forming anchor points for chromosome orientation [3], controlling the placement of the division septum [4], and serving as platforms for phospho-signaling complexes that control the timing of cell cycle progression [5]. In some species, different protein complexes are targeted to each pole, thereby establishing polar asymmetry. When this includes proteins that control cellular functions, cell division results in the formation of genetically identical but functionally distinct daughter cells [6], thus enabling a form of multicellularity. In this work we examine the mechanisms that underlie the establishment of bacterial cell polarity, highlighting examples across the bacterial kingdom. That polarity is so widespread across species, and accomplished by such widely varying mechanisms, suggests that the development of polarity has been a recurring theme in bacterial evolution, and a fundamental step in the advancement of life beyond primordial stages.
Oscillating polarity
A well-characterized example of bacterial polarity is the Min system, which has been extensively studied in E. coli (Figure 1a). Two components, MinC and MinD, form a complex that oscillates between poles with a periodicity of about 20 s [7,8]. MinC performs the functional output of this system, by preventing the assembly of division plane polymers near the cell poles [9], and spatial regulation is achieved by the concerted activities of MinD and a third component, MinE. MinD is an ATPase that is membrane associated when bound to ATP [10], and forms helical structures that follow the contour of the plasma membrane [11]. Membrane-associated MinD–ATP interacts with a ring of MinE, which induces ATP hydrolysis and subsequent release of the entire complex into the cytosol [12,13]. Surprisingly, MinD and MinE generate self-sustaining waves of membrane association and dissociation over a flat membrane in vitro [14•,15••], suggesting that oscillatory behavior in vivo can be fully explained by these components. Mathematical models propose that MinD molecules associate to form localized concentrations that are dynamically unstable in the presence of MinE [16]. Next generation models should account for the influence of helical or polymeric structures of Min components [17], and the recent discovery that MinE activity involves direct interaction with the cell membrane [18•].
Figure 1.
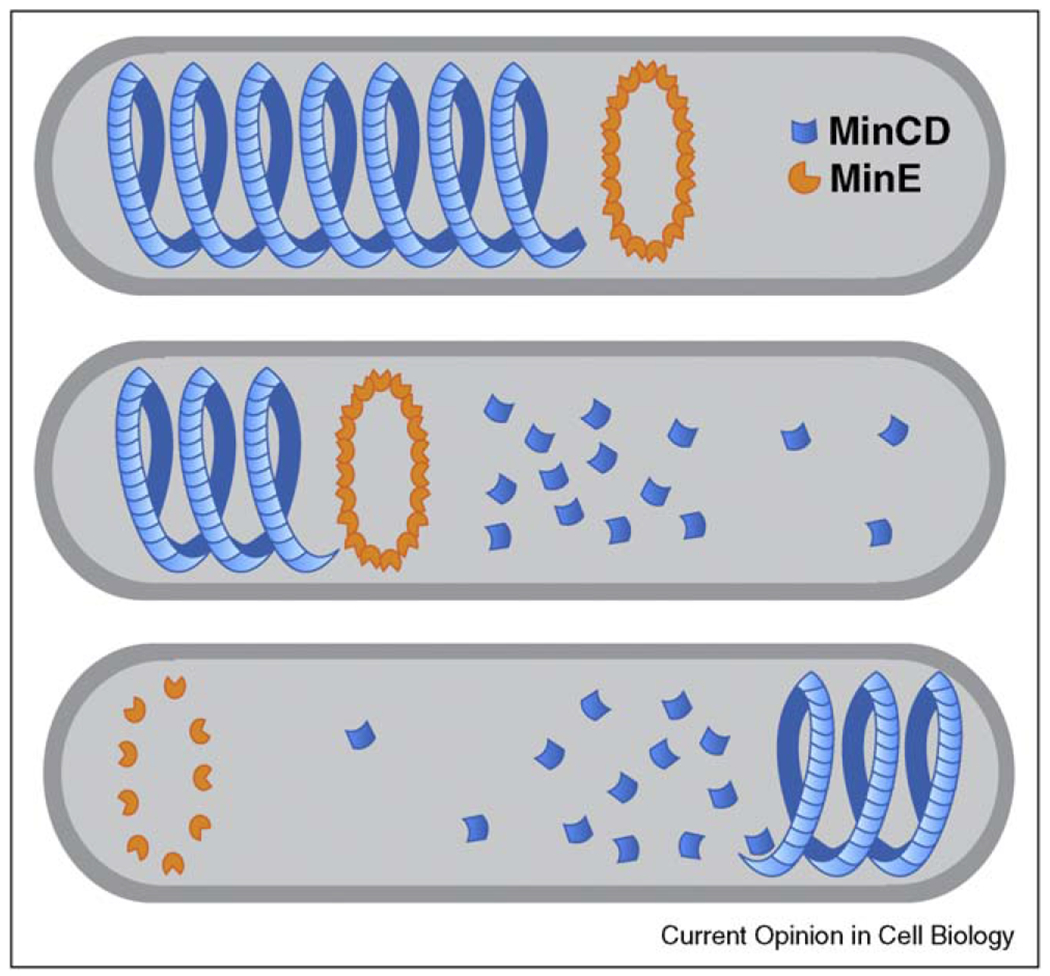
The Min system and oscillating polar asymmetry in E. coli. The dynamically unstable MinCD helix interacts with MinE, resulting in disassembly of the structure (upper panels). Subsequently, MinCD re-assembles in an area of low MinE concentration at the opposite pole of the cell (lower panel).
Oscillating polarity is also observed in Myxococcus xanthus, which reverses direction of movement over approximately 10-min intervals [19]. Several polar oscillating proteins have been discovered, including some that are directly involved in motility [20], and regulatory proteins that control motor activity. Oscillating regulators include the Ras-like G-protein MglA and its cognate GTPase activating protein MglB [21••]. MglB stimulates the conversion of pole-localized MglA-GTP to non-localized MglA-GDP, in a manner that is similar to the Min system. Dynamic localization of MglA also requires the input of a two-component phospho-signaling module, called Frz, that is oscillatory in itself [22,23] (D Kaiser, unpublished data), but the mechanistic linkage between Frz oscillations and MglA/MglB dynamics has not been described.
Establishing polarity de novo
Whereas oscillatory systems perpetuate indefinitely, other mechanisms are used to establish and maintain fixed asymmetry in cells that are initially symmetric. This occurs during sporulation in Bacillus subtills, when the position of the division plane switches from the center of the cell (as occurs during normal growth) to a site near the pole (Figure 2a). The smaller of the two daughter cells is programmed to develop into a spore, and the larger serves as a mother cell, providing nutrients to the developing prespore. At the beginning of sporulation, division plane components are found near both poles, though only one site is chosen for completion [24,25]. Here, asymmetry is generated by the mechanics of division plane assembly, which is strictly limited to only one site at a time, perhaps due to the titration of a limiting factor [26]. In order to preserve asymmetry, the cell must prevent the formation of a second division site at the opposite pole. This is accomplished through a complex signaling pathway, originating in the prespore compartment and sent to the mother cell, that controls the activity of the transcription factors σF and σE, which regulate many compartment-specific genes [27]. Mutations that block or even delay the relay of this signal into the mother cell form a second division site near the opposite pole, and the resulting symmetry yields spores with reduced viability [28••].
Figure 2.
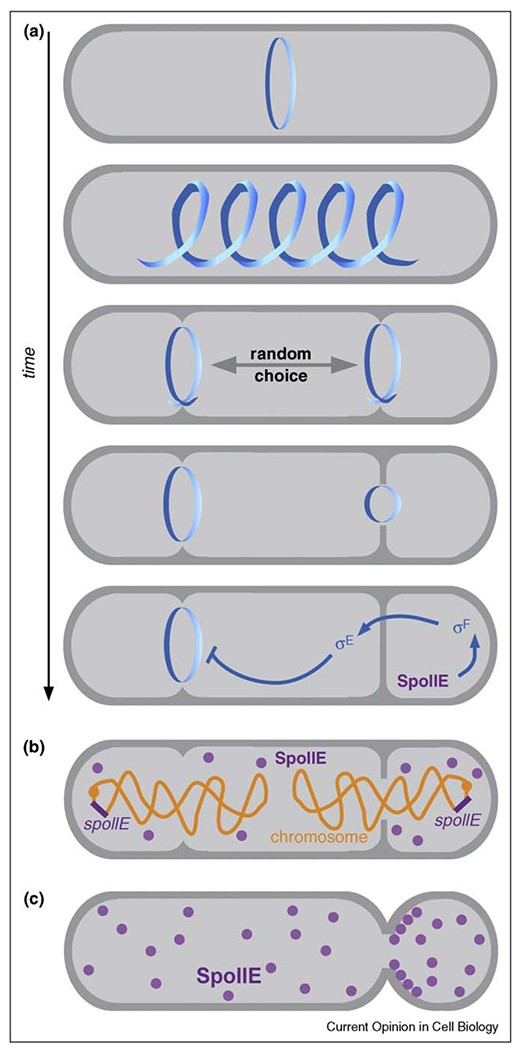
Establishing polarity in Bacillus subtilis. (a) FtsZ is re-organized into two symmetrically positioned rings upon entry into sporulation (upper panels), and after one ring is chosen for completion (arrow), asymmetry is reinforced by a feedback signaling pathway that controls compartment-specific gene expression (lower panel). (b) The genetic asymmetry model for spore fate determination. At the early stages of sporulation, only the origin-proximal third of the chromosome is present in the forespore. Origin-distal gene expression is limited to the mother cell while origin-proximal gene expression is concentrated in the forespore. (c) SpoIIE concentration could determine spore fate. SpoIIE is initially targeted to the division plane, and if it is equally shared between mother cell and forespore compartments, the relative concentration of SpoIIE will be higher in the smaller forespore compartment following septation. SpoIIE may also have specific affinity for the forespore side of the division plane.
Why is σF turned on in the prespore compartment and not in the mother cell? In one model, the compartments experience a temporary asymmetry in gene expression (Figure 2b), because the origin-proximal region of the chromosome is anchored to the cell pole in the prespore and the remainder of the chromosome is pulled through a translocation channel in the septal membrane over the course of approximately 20 min. During this time, the expression of a σF inducing factor from a locus near the origin could be restricted to the forespore [29,30]. A different model (Figure 2c) holds that the localization propensities of the σF inducer SpoIIE cause it to become concentrated in the prespore. SpoIIE is targeted to the division plane, and because this structure is shared between prespore and mother cell, the relative differences in cytoplasmic volume will yield a higher concentration of SpoIIE in the smaller compartment [31]. These models are not mutually exclusive, and may work in concert to promote cell fate determination.
Targeting proteins to the poles
The systems described above are notable in that their mechanisms are not directly anchored at the cell poles. However, many aspects of bacterial cell polarity depend on physical connection to the pole [5,32], usually in the form of localized multiprotein complexes. In some cases, the polar localization of one protein can be perturbed by eliminating another that performs an earlier step in complex assembly. However, the question of localization mechanism then resorts to the upstream protein. It is difficult to imagine that this chain could continue very far before encountering some other type of interaction that underlies the affinity between a protein and the cell pole. Several recent reports have shed light on these potential ‘ultimate causes’ for polar localization.
Ramamurthi et al. [33••] provided compelling evidence that membrane geometry can be a determinant for polar localization (Figure 3). They showed that convex membrane curvature in vivo is necessary and sufficient for recruiting SpoVM, a 26 amino acid spore coat protein from B. subtilis, and that purified SpoVM preferentially interacts with phospholipid vesicles that have the same diameter as that of spores. Given the short length of the peptide in relation to the radius ofmembrane curvature, it is difficult to imagine that one molecule could directly sense vesicle shape. However, SpoVM was recruited to membranes in a cooperative manner, and the authors suggest that clusters of proteins could produce a larger structure that is sensitive to membrane geometry. Subsequent reports revealed that another B. subtilis protein, DivIVA, exhibits similar affinity for concave membrane curvature [34••,35]. This is particularly significant for bacterial polarity, as DivIVA recruits other proteins to the cell poles and the division plane, and also anchors the chromosome origin to the poles during sporulation. The N-terminal 60 amino acids of DivIVA are necessary and sufficient for detecting concave membranes, and form a coiled coil domain capped by a hydrophobic tip for membrane insertion [36•]. However, unlike full length DivIVA [37], the N-terminal domain does not form oligomers, and the question of how individual molecules sense membrane curvature remains.
Figure 3.
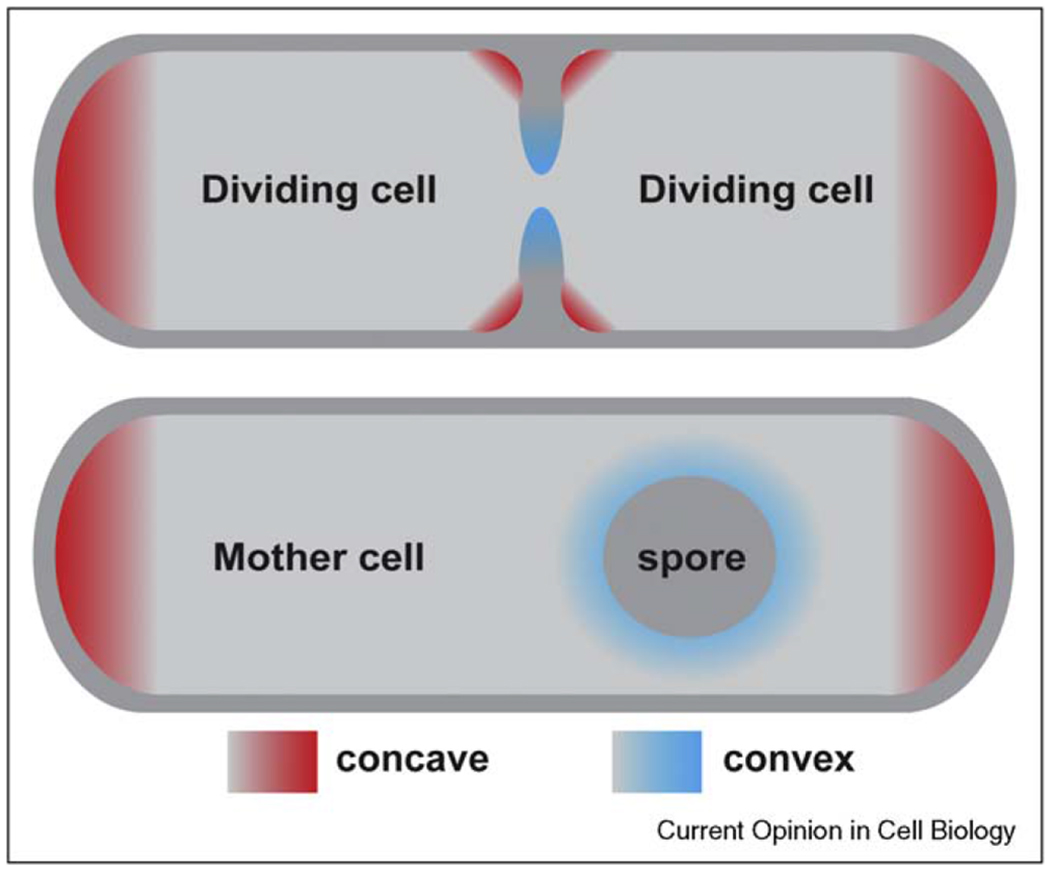
Convex and concave membrane curvatures in a bacterial cell. Some membrane-associated proteins have specific affinity for convex or concave membrane surfaces.
Instead of sensing curvature directly, some proteins could interact with specialized lipids that accumulate in curved membranes. Anionic phospholipids, including cardiolipin, are concentrated at the poles of E. coli cells [38•], where they could form distinct lipid domains [39]. Although the subcellular localization of DivIVA and other polar proteins is unperturbed in cardiolipin synthase mutants [34••,40], the osmosensory transmembrane protein ProP exhibits reduced polar localization in cardiolipin-deficient E. coli [41]. In bacteria, ProP is the only clear example of lipid-dependent polar localization, and because the lipid synthase mutant used to provide evidence for this interaction could have indirect effects on ProP or the cell pole, additional verification is needed to substantiate this potential localization mechanism.
A third possible mechanism for connecting proteins to the pole is through interaction with peptidoglycan (PG), which encases the cell membrane in a covalently linked network of polymeric strands. Because PG linkages are progressively modified, chemical labeling methods have been used to distinguish between old and newly synthesized material, revealing that the division planes are active sites of new PG synthesis [42,43]. Since the division planes become new cell poles upon completion of cytokinesis (Figure 4a), polar localization could be achieved through affinity for certain PG species, or for enzymes that catalyze PG modifications. Many PG modifying enzymes are directly targeted to the division plane [44], yet there is little evidence for PG interaction motifs in polar proteins. Two potential exceptions are the Caulobacter proteins PleA and SpmX, which have domains that are similar to PG hydrolyzing enzymes and are upstream components in polar multiprotein complex assembly pathways [45••,46].
Figure 4.
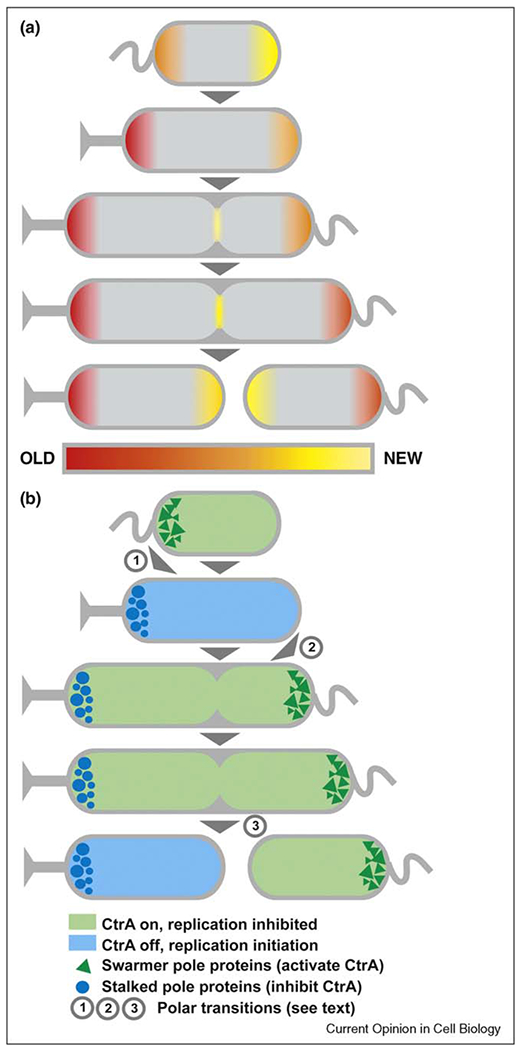
Polar maturation in Caulobacter crescentus. (a) Cell poles ‘age’ over the course of cell division. (b) In C. crescentus, polar aging is coupled with the timing of chromosome replication, transitions in polar protein composition, and cell cycle progression. Important polar transition periods are numbered (1: regulatory proteins are switched at swarmer to stalked pole transition; 2: assembly of cell cycle regulators at new pole; 3: compartmentalization and formation of new poles at cell division). Different sets of polar regulatory proteins promote (blue shading) or inhibit (green shading) chromosome replication by controlling the activity of a master regulator, CtrA.
Polar maturation
The patterning of PG synthesis suggests that the poles of rod-shaped bacteria can be distinguished on the basis of age (Figure 4a). In C. crescentus, the poles experience age-dependent transitions in protein composition, activity, and morphology, and because one of the cell poles is always younger than the other, this constitutes a robust mechanism for generating polar asymmetry [47]. In order for this system to work, the aging of the cell poles must be coordinated with the mechanism that drives polar transformation. This is accomplished by placing cell cycle regulators at the poles, which effectively couples polar maturation with cell cycle progression.
A group of polar cell cycle regulators regulate CtrA, a transcription factor for over 90 cell cycle regulated genes, and a direct repressor of DNA replication initiation [48]. Its activity is inhibited by de-phosphorylation and timely degradation just before the onset of chromosome replication, mediated by proteins that are localized to the old pole during a period called the swarmer to stalked cell transition (Figure 4b step 1; [49]). DNA replication is limited to once per cell cycle because CtrA activating factors are targeted to the new pole shortly thereafter. DNA replication itself triggers the localization of proteins to the new pole (Figure 4b step 2; [50,51•]). Thus, polar signaling proteins are integrated with cell cycle progression in a feedback loop that inhibits DNA replication until cell division separates positive and negative regulators into separate compartments (Figure 4b step 3). Many of the connections in this circuit await further analysis: the mechanism that couples DNA replication initiation to the accumulation of proteins at the new pole is unknown, as are the events that govern the turnover of polar proteins at the swarmer to stalked cell transition.
The study of polar maturation in C. crescentus led to a surprising connection between the division plane and polar localization. Two studies identified TipN as a factor that is required for the correct placement of polar features, and made the surprising discovery that this transmembrane protein is initially targeted to the division plane, and achieves polar localization simply by remaining in place after cell division transforms the division plane into a new pole [52,53]. TipN is an important multifunctional protein that recruits flagellar components to the new pole and works with the machinery that drives chromosome segregation [54,55]. Given the initial localization of TipN, it appears that the ‘ultimate causes’ of polar maturation in C. crescentus lie in the formation of the division plane. Since the localization of every known division plane component is dependent on FtsZ, a cytoskeletal protein that polymerizes at the site of cell division [56,57], and FtsZ is among the most evolutionarily conserved of all bacterial proteins, it appears that this aspect of cell polarity is rooted in ancient cellular mechanisms.
Conclusions
This review describes four general mechanisms underlying bacterial cell polarity. Oscillatory mechanisms undergo continuous reversal, implying that no permanent marks are used to identify one pole versus the other (‘Oscillating polarity’ section). In stochastic mechanisms, both poles initially appear equivalent, and the choice of one pole over the other is made permanent by feedback pathways that reinforce polarity (‘Establishing polarity de novo’ section). Targeting proteins to the pole can involve the recognition of membrane geometry or special locations marked by lipid or PG subdomains (‘Targeting proteins to the poles’ section), and poles can be differentially marked on the basis of age (‘Polar maturation’ section). Several different species were offered as examples, but this does not imply that each prokaryotic lineage has a unique mechanism for achieving polarity. The use of polar age as a marker appears widespread, as it affects the localization of chemoreceptors in a wide variety of species [58]. Thus, some organisms can employ multiple mechanisms for generating cell polarity, and the type of mechanism may be related to the function of the polar proteins.
Recently, in vitro analyses of polar proteins in the context of synthetic membranes have made significant strides in understanding polarity at a biochemical level. These experimental systems have revealed direct connections between polar proteins and curved lipid membranes [33••,34••], and demonstrated that oscillatory behavior can be fully reconstituted by two proteins and a membrane surface [15••]. Given the relative simplicity of prokaryotic systems, with a limited number of components operating in a small space, there is hope for achieving a deep understanding of cell polarity in the near future. This will require the synthesis of structural information with molecular dynamics, perhaps through the application of single molecule microscopy, and sufficient quantitative data to enable physical modeling through computational algorithms.
Acknowledgements
We thank Dale Kaiser and Rich Losick for helpful discussions and advice. The National Institute of Health supported the work of AL (F32HL090263), GB (F32GM080008), and LS (R01GM32506).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z: Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A 2009, 106:7858–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiley, Sherwood, Wolverton (Eds): Prescott’s Microbiology. McGraw-Hill; 2010. [Google Scholar]
- 3.Toro E, Shapiro L: Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol 2010, 2:a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothfield L, Taghbalout A, Shih YL: Spatial control of bacterial division-site placement. Nat Rev Microbiol 2005, 3:959–968. [DOI] [PubMed] [Google Scholar]
- 5.Goley ED, Toro E, McAdams HH, Shapiro L: Dynamic chromosome organization and protein localization coordinate the regulatory circuitry that drives the bacterial cell cycle. Cold Spring Harb Symp Quant Biol 2009, 74:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanbichler M: Spatial regulation in Caulobacter crescentus. Curr Opin Microbiol 2009, 12:715–721. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z, Lutkenhaus J: Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 1999, 34:82–90. [DOI] [PubMed] [Google Scholar]
- 8.Raskin DM, de Boer PA: Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci U S A 1999, 96: 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen B, Lutkenhaus J: The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol 2009, 72:410–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szeto TH, Rowland SL, Habrukowich CL, King GF: The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem 2003, 278:40050–40056. [DOI] [PubMed] [Google Scholar]
- 11.Shih YL, Le T, Rothfield L: Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci U S A 2003, 100:7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, Gogol EP, Lutkenhaus J: Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci U S A 2002, 99:6761–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lackner LL, Raskin DM, de Boer PA: ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol 2003, 185:735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.•.Ivanov V, Mizuuchi K: Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci U S A 2010, 107:8071–8078. [DOI] [PMC free article] [PubMed] [Google Scholar]; Building on the in vitro system developed by Loose et al., this study considers physical parameters that more closely approximate the behavior on the Min system in vivo.
- 15.••.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P: Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 2008, 320:789–792. [DOI] [PubMed] [Google Scholar]; This is the first study to demonstrate that MinD, MinE, and a phospholipid membrane are sufficient for the establishment of a self-propagating system.
- 16.Kruse K, Howard M, Margolin W: An experimentalist’s guide to computational modelling of the Min system. Mol Microbiol 2007, 63:1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suefuji K, Valluzzi R, RayChaudhuri D: Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc Natl Acad Sci U S A 2002, 99:16776–16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.•.Hsieh CW, Lin TY, Lai HM, Lin CC, Hsieh TS, Shih YL: Direct MinE–membrane interaction contributes to the proper localization of MinDE in E. coli. Mol Microbiol 2010, 75:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work provides novel insight for an emerging model that describes the interactions of MinD, MinE, and membrane at a molecular level.
- 19.Kaiser D: Myxococcus-from single-cell polarity to complex multicellular patterns. Annu Rev Genet 2008, 42:109–130. [DOI] [PubMed] [Google Scholar]
- 20.Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Hone A, Maier B, Hoppert M, Sogaard-Andersen L: Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol 2009, 74:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.••.Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L: Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]; The GAP/GTPase properties of MglA and MglB are directly demostrated through biochemistry, and fluorescence microscopy reveals that their oscillating localization patterns are coordinated with reversals in cell motility.
- 22.Igoshin OA, Goldbeter A, Kaiser D, Oster G: A biochemical oscillator explains several aspects of Myxococcus xanthus behavior during development. Proc Natl Acad Sci U S A 2004, 101:15760–15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mignot T, Merlie JP Jr, Zusman DR: Two localization motifs mediate polar residence of FrzS during cell movement and reversals of Myxococcus xanthus. Mol Microbiol 2007, 65:363–372. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Yehuda S, Losick R: Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 2002, 109:257–266. [DOI] [PubMed] [Google Scholar]
- 25.Levin PA, Losick R: Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev 1996,10:478–488. [DOI] [PubMed] [Google Scholar]
- 26.Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K: A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol 1999, 31:1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barak I, Wilkinson AJ: Where asymmetry in gene expression originates. Mol Microbiol 2005, 57:611–620. [DOI] [PubMed] [Google Scholar]
- 28.••.Eldar A, Chary VK, Xenopoulos P, Fontes ME, Loson OC, Dworkin J, Piggot PJ, Elowitz MB: Partial penetrance facilitates developmental evolution in bacteria. Nature 2009,460:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]; The feedback signaling mechanism was slowed by altering the chromosomal location of a key regulator or changing the promoter to reduce its expression. The results show that the feedback relay must be rapid in order to prevent a second division septum.
- 29.Dworkin J, Losick R: Differential gene expression governed by chromosomal spatial asymmetry. Cell 2001, 107:339–346. [DOI] [PubMed] [Google Scholar]
- 30.McBride SM, Rubio A, Wang L, Haldenwang WG: Contributions of protein structure and gene position to the compartmentalization of the regulatory proteins sigma(E) and SpoIIE in sporulating Bacillus subtilis. Mol Microbiol 2005, 57:434–451. [DOI] [PubMed] [Google Scholar]
- 31.Iber D: A computational analysis of the impact of the transient genetic imbalance on compartmentalized gene expression during sporulation in Bacillus subtilis. J Mol Biol 2006, 360:15–20. [DOI] [PubMed] [Google Scholar]
- 32.Dworkin J: Cellular polarity in prokaryotic organisms. Cold Spring Harb Perspect Biol 2009, 1 :a003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.••.Ramamurthi KS, Lecuyer S, Stone HA, Losick R: Geometric cue for protein localization in a bacterium. Science 2009, 323:1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]; This was the first study to provide evidence that a bacterial protein could sense membrane curvature directly.
- 34.••.Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW: Localisation of DivIVA by targeting to negatively curved membranes. EMBO J 2009, 28:2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]; The membrane interaction properties of purified DivIVA and the 60 amino acid N-terminal domain are demonstrated in vitro, mutliple experiments demonstrate specific affinity for concave membrane curvature in vivo.
- 35.Ramamurthi KS, Losick R: Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A 2009, 106:13541–13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.•.Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, Veprintsev DB, Hamoen LW, Lowe J: Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J 2010, 29:1988–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of DivIVA structure provides atomic level detail and critical insight into its interaction with the membrane.
- 37.Stahlberg H, Kutejova E, Muchova K, Gregorini M, Lustig A, Muller SA, Olivieri V, Engel A, Wilkinson AJ, Barak I: Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol Microbiol 2004, 52:1281–1290. [DOI] [PubMed] [Google Scholar]
- 38.•.Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA: Phosphatidic acid and N-acylphosphatidylethanolamine form membrane domains in Escherichia coli mutant lacking cardiolipin and phosphatidylglycerol. J Biol Chem 2009, 284:2990–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work shows that E. coli segregates anionic phospholipids into polar regions, and suggests that the weak phenotype of cardiolipin synthase mutants is compensated by the presence of other anionic phospholipids at the poles.
- 39.Mukhopadhyay R, Huang KC, Wingreen NS: Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J 2008, 95:1034–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romantsov T, Battle AR, Hendel JL, Martinac B, Wood JM: Protein localization in Escherichia coli cells: comparison of the cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J Bacteriol 2010, 192:912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM: Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol 2007, 64:1455–1465. [DOI] [PubMed] [Google Scholar]
- 42.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C: The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 2007, 64:938–952. [DOI] [PubMed] [Google Scholar]
- 43.de Pedro MA, Quintela JC, Holtje JV, Schwarz H: Murein segregation in Escherichia coli. J Bacteriol 1997, 179:2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T: Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 2005, 55:1631–1645. [DOI] [PubMed] [Google Scholar]
- 45.••.Radhakrishnan SK, Thanbichler M, Viollier PH: The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev 2008, 22:212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]; A murein hydrolase domain in SpmX is necessary and sufficient for localization to the old pole. A transmembrane protein, SpmX is an upstream component in an assembly pathway for cell cycle regulatory proteins on the cytoplasmic face of the old pole.
- 46.Viollier PH, Shapiro L: A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol Microbiol 2003, 49:331–345. [DOI] [PubMed] [Google Scholar]
- 47.Goley ED, Iniesta AA, Shapiro L: Cell cycle regulation in Caulobacter: location, location, location. J Cell Sol 2007, 120:3501–3507. [DOI] [PubMed] [Google Scholar]
- 48.Marczynski GT, Shapiro L: Control of chromosome replication in Caulobacter crescentus. Annu Rev Microbiol 2002, 56:625–656. [DOI] [PubMed] [Google Scholar]
- 49.Iniesta AA, Shapiro L: A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci U S A 2008, 105:16602–16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong SH, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 2010, 76:173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.•.Iniesta AA, Hillson NJ, Shapiro L: Polar remodelling and histidine • kinase activation essential for Caulobacter cell cycle progression is dependent on DNA replication initiation. J Bacteriol 2010, 192:3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that a key cell cycle regulator, involved in activating CtrA and preventing a second round of DNA replication, is targeted to the new pole in response to the chromosome replication initiation.
- 52.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH: Bacterial birth scar proteins mark future flagellum assembly site. Cell 2006, 124:1025–1037. [DOI] [PubMed] [Google Scholar]
- 53.Lam H, Schofield WB, Jacobs-Wagner C: A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 2006, 124:1011–1023. [DOI] [PubMed] [Google Scholar]
- 54.Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L: Aspindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 2010, 12:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schofield WB, Lim HC, Jacobs-Wagner C: Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J 2010, 29:3068–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goehring NW, Beckwith J: Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol 2005, 15:R514–526. [DOI] [PubMed] [Google Scholar]
- 57.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA: Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol 2009, 191:4186–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ping L: The asymmetric flagellar distribution and motility of Escherichia coli. J Mol Biol 2010, 397:906–916. [DOI] [PubMed] [Google Scholar]


