Abstract
Enterobacterales are widely distributed in the gastro-intestinal system of animals and may cause opportunistic infections. Worse still, multidrug-resistant Enterobacterales also poses a serious threat to public health. Tn7-like transposons have been found in several species of the Enterobacterales order and play an important role in dissemination of antibiotic resistance. This study aimed to investigate the distribution and genetic characterization of Tn7-like transposons in Enterobacterales isolates from food animals and their association with antibiotic resistance. Enterobacterales isolated from the samples were identified and classified according to the 16S rDNA sequence. Tn7-like transposons and associated integrons were detected by polymerase chain reaction (PCR) and sequencing. The antibiotic resistance of each Tn7-like transposon positive isolate was detected according to the Kirby-Bauer disk diffusion method. Then, six representative strains were selected to study the genetic environment by whole-genome sequencing (WGS). In total, we isolated 377 Tn7-like transposons positive strains of Enterobacterales. Class 2 integrons were detected in 99.5% of the isolates, and there were high frequency mutation sites especially in base 535, a stop mutation. Many isolates (54.9%) were multidrug-resistant and observed high resistance rates to trimethoprim/sulfamethoxazole and streptomycin. Among these strains, we found three new types of Tn7-like transposons, named Tn6813, Tn6814, and Tn6765. This is the first comprehensive survey that shows Tn7-like transposons in Enterobacterales from animals used for food production in different regions of China. This study also provides an insight into the horizontal transfer of resistance genes associated with Tn7-like transposons.
Keywords: Tn7-like transposons, Enterobacterales, antibiotic resistance, class 2 integrons, animals used for food production
Introduction
Several genera and species among the Enterobacterales are widely distributed in humans and other animals (Sassone-Corsi et al., 2016). The Enterobacterales order includes Escherichia, Salmonella, Klebsiella, Proteus, Morganella, and Providencia among other genera. Although Enterobacterales often comprise less than 1% of a healthy intestine’s microbiota, some Enterobacterales are also encountered in the inflamed gut and cause urinary tract infections, septicaemia, pneumonia, and other intra-abdominal infections (Lupo et al., 2013; Toombs-Ruane et al., 2017). To prevent and treat these infections caused by Enterobacterales, many antibiotics are usually used in animals. However, nonsystematic use or misuse of antibiotics have resulted in the emergence of antibiotic-resistant variants and a corresponding increase in the failure rate of these agents for treating bacterial diseases (Marshall and Levy, 2011).
Antimicrobial resistance (AMR) in Enterobacterales has emerged as a problem in both human and veterinary medicine (Livermore, 2009; von Tippelskirch et al., 2018), and the interest of the scientific community in the presence and circulation of resistant organisms from animals used for food production has also increased due to the important public health implications (Meunier et al., 2006; Cortes et al., 2010; Goncalves et al., 2010).
A common form of resistance dissemination in Enterobacterales is mediated by transposons. Tn7-like transposons, as the Tn7 transposon derivatives, have been found in several species of the Enterobacterales order, such as Proteus mirabilis and Morganella morganii (Chen et al., 2018, 2019). This kind of transposons carries a great diversity of antimicrobial resistance genes (ARGs).
Highly conserved, the sequences of the two ends of the Tn7 transposon are encoding transposition module and class 2 integron system. The transposition module encodes five proteins required for two transposition pathways, TnsA, TnsB, TnsC, TnsD, and TnsE (TnsABCDE; Peters and Craig, 2001b). The class 2 integron of Tn7 transposon has an organization similar to that of the class 1 integron and carries three resistance gene cassettes—aadA1, sat, and dfrA1 (Sundstrom et al., 1988, 1991; Sundstrom and Skold, 1990)—close to an open reading frame (ORF), intI2. intI2 has premature translation termination due to mutation of base 535 encoding integrase from C to T, thus with no function. Although these gene cassettes are fixed in Tn7 transposons due to mutations in the homologous recombinase, they can be rearranged in hosts expressing the relevant recombinase, resulting in other combinations of antibiotic resistance genes (Hansson et al., 2002).
The Tn7-like transposons, as important mobile platforms to transfer bacterial AMR, transfer various resistance genes among bacteria through their transposase, promoting the horizontal spread of drug resistance in bacteria. However, there is a paucity of data on the comprehensive analysis of Tn7-like transposons in antibiotic resistant Enterobacterales isolates. In this study, we have determined the incidence of Tn7-like transposons and their associated integrons and dealt with detailed genetic characterization of three novel transposons with complex mosaic structures in a collection of random Enterobacterales isolates collected from animals used for food production in China. Additionally, the possible association between the occurrence of Tn7-like transposons and antibiotic resistance phenotypes was also determined by statistical analysis.
Materials and Methods
Isolation and Identification of Enterobacterales
A total of 1,474 consecutive and unduplicated clinical isolates of Enterobacterales were collected from animals used for food production on 10 poultry and 12 swine farms in eight provinces in China between June 2018 and January 2020 (Table 1). All the isolates were presumptively identified through phenotypic methods, including colony morphology on MacConkey Agar (Land Bridge, Beijing, China) or Eosin-Methylene Blue Agar (Land Bridge, Beijing, China). The identification of these isolates was later confirmed using 16S rDNA gene sequencing. All Enterobacterales strains were stored at −20°C in 25% glycerol.
Table 1.
Bacterial strains isolated from 2018 to 2020.
| Source | Species | No. of strains | Note |
|---|---|---|---|
| Pigs | Escherichia coli | 413 | Fecal sample, cloacal swab, drinking water, or small intestine of swines from 12 swine farms in six different provinces of China. |
| Proteus spp. | 208 | ||
| Providencia spp. | 164 | ||
| Morganella morganii | 146 | ||
| Klebsiella pneumoniae | 141 | ||
| Salmonella enterica | 82 | ||
| Total | 1,154 | ||
| Chicken | Escherichia coli | 121 | Fecal sample, cloacal swab, drinking water, or small intestine of chicken from 10 poultry farms in five different provinces of China. |
| Proteus spp. | 53 | ||
| Providencia spp. | 46 | ||
| Klebsiella pneumoniae | 24 | ||
| Salmonella enterica | 76 | ||
| Total | 320 |
The animal study was reviewed and approved by the College of Life Science, Sichuan University affiliation ethics committee, and all efforts were made to minimize animal suffering.
Detection of Tn7-Like Transposons by Polymerase Chain Reaction
All 1,474 Enterobacterales isolates in this study were screened for the presence of Tn7-like transposons using a PCR-based method targeting tnsA, tnsB, and tnsC genes, which encode three conserved Tn7 transposases (Peters and Craig, 2001b; Peters, 2014). The isolates were grown overnight (18–24 h) in Brain Heart Infusion (BHI) broth (Oxoid, Basingstoke, UK) at 37°C with a rotation speed of 200 rpm, and the DNA template was prepared using the boiling method (Lévesque et al., 1995). The PCR mixture was prepared with a final volume of 20 μl, containing 1 μl of template DNA, 8 μl ddH2O, 10 μl Taq PCR MasterMix, and 0.5 μl each primer. The specific primers for detecting tnsA, tnsB, and tnsC genes are shown in Supplementary Table S1. Positive PCR products were sequenced by Chengdu Sangon Biological Engineering Technology & Services Co, Ltd.
Phenotypic Evaluation of Antibiotic Resistance
The antibiotic resistance profile of all Tn7-like transposons positive isolates were determined according to the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2017) guidelines. The following antimicrobials (all discs from Oxoid, Basingstoke, UK) were used: amikacin (AMK, 30 μg), imipenem (IPM, 10 μg), cefoxitin (FOX, 30 μg), ciprofloxacin (CIP, 5 μg), streptomycin (STR, 10 μg), gentamicin (GEN, 10 μg), florfenicol (FLR, 30 μg), aztreonam (ATM, 30 μg), ceftazidime (CAZ, 30 μg), and trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg). Escherichia coli ATCC 25922 was used as a quality control strain.
Incidence of the Integrons and Associated ARGs
The isolated Tn7-like transposons positive Enterobacterales strains were checked for the presence of intl2 integrase genes by PCR, using primers (Supplementary Table S1) and methodology described previously (Cocchi et al., 2007; Rehman et al., 2017). These mutation site sequences of intI2 were visualized with Weblogo1.
WGS and Analysis
We also analyzed genetic environment of Tn7-like transposons among strains exhibiting unique resistance phenotypes. The whole genome of the Tn7-like transposons positive strain was sequenced using Illumina MiSeq with a 200-fold sequencing depth and Nanopore PromethION platform with a 100-fold sequencing depth (Novogene Technology Co., Beijing, China). Genome assembly was carried out by de novo assembly with Unicycler v0.4.7, and the sequence was annotated using the NCBI Prokaryotic Genome Annotation Pipeline v4.2. Mobile elements, resistance genes, and other features were annotated by INTEGRALL (Moura et al., 2009), ISfinder (Siguier et al., 2006), ResFinder (Kleinheinz et al., 2014), PlasmidFinder (Carattoli et al., 2014), and the Tn Number Registry (Roberts et al., 2008) online databases, and the analysis was conducted using the BLAST program2.
Horizontal Transfer and Stability of Tn7-Like Transposons
Conjugation was performed using rifampin-resistant E. coli EC600 as the recipient and the P. mirabilis SCBX1.1 isolate as the donor with selection on Salmonella Shigella agar (Land Bridge, Beijing, China) plates containing 300 μg/ml rifampicin and 10.24 μg/ml flufenicol. Successful horizontal transfer of plasmid p1.1.2 containing Tn7-like transposon was confirmed using antibiotic sensitivity test and PCR, and then, the conjugation frequency was calculated as transconjugants divided by number of donors (Dong et al., 2019). The stability of Tn6765, Tn6813, and Tn6814 was determined by passage in BHI broth lacking antibiotics as it was described previously (Sandegren et al., 2012).
Statistical Analysis
Variables are expressed as percentages (%). All statistical analyses were conducted with the GraphPad Prism 8 software. Chi square test for samples were used. Value of p < 0.05 was considered significant.
GenBank Accession Numbers
The complete sequences of p1.1.1 (CP047113), p1.1.2 (CP047114), and all Tn7-like transposons, Tn6763 (MN641830), Tn6764 (MN628641), Tn6817 (MT469878), Tn6813 (MT469876), Tn6814 (MT469877), and Tn6765 (MT503200), identified in the present study were submitted to NCBI GenBank.
Results
Incidence of Tn7-Like Transposons in Enterobacterales
A total of 1,474 strains of Enterobacterales were isolated through analysis of samples collected from Sichuan, Hainan, Chongqing, Shandong, Hebei, Xizang, Liaoning, and Anhui provinces. Of the 1,474 Enterobacterales strains examined, 377 strains contained Tn7-like transposons. They included 128 (24.0%) E. coli, 150 (57.5%) Proteus spp., 48 (22.9%) Providencia spp., 21 (14.4%) Morganella morganii, 17 (10.3%) Klebsiella pneumoniae, and 13 (8.2%) Salmonella enterica. Statistical analysis showed that the prevalence of Tn7-like transposons was different among different bacteria genera (p < 0.0001; Chi square test). The Tn7-like transposons positive rate of Proteus spp. was significantly higher than other bacteria’s (p < 0.0001; Chi square test; Table 2).
Table 2.
The separation rate of Tn7-like transposons in Enterobacterales of different genera.
| Species | Total isolates | Tn7-like-positive isolates | Separation rate, % |
|---|---|---|---|
| E. coli | 534 | 128 | 24.0b |
| Proteus spp. | 261 | 150 | 57.5a |
| Providencia spp. | 210 | 48 | 22.9bc |
| M. morganii | 146 | 21 | 14.4cd |
| K. pneumoniae | 165 | 17 | 10.3d |
| S. enterica | 158 | 13 | 8.2d |
Chi-square test was used to analyze the data. Data labeled with entirely different letters were significantly different (p < 0.05).
Antimicrobial Resistance Phenotypes
Among the isolates, 207 (54.9%) were multi-drug resistant (MDR, resistant to at least three different classes of antibiotics). Proteus spp. and S. enterica had the higher multidrug resistance rates of 59.3 and 61.5%, respectively. Notably, high resistance rates were observed for streptomycin (87.8%) and trimethoprim/sulfamethoxazole (74.0%), followed by the rates for florfenicol (60.5%), gentamicin (24.4%), and ciprofloxacin (22%). Among them, resistance rates to gentamicin and ciprofloxacin were higher in Proteus spp. (30.7 and 28.0%) and S. enterica (38.5 and 23.1%). Low resistance rates to amikacin (6.1%), ceftazidime (5.8%), cefoxitin (5.8%), and aztreonam (4.2%) were detected, whereas S. enterica was more resistant than other Enterobacterales strains to ceftazidime and aztreonam (38.5 and 46.2%). Resistance to imipenem (1.6%) was only observed in E. coli, and most of the Enterobacterales isolates were highly susceptible to imipenem (Table 3). No significant difference was found between the antibiotic resistant profiles of isolates from pigs and chicken (Supplementary Table S2).
Table 3.
Rates of resistance to antimicrobial agents of Tn7-like transposons positive isolates.
| Antimicrobial agent | Breakpoints (mm) | % resistant isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R(≤) | I | S(≥) | E. coli (n = 128) | Proteus spp. (n = 150) | Providencia spp. (n = 48) | M. morganii (n = 21) | K. pneumoniae (n = 17) | S. enterica (n = 13) | Total (n = 377) | |
| Gentamicin (GEN, 10 μg) | 12 | 13–14 | 15 | 24.2 | 30.7 | 12.5 | 9.5 | 11.8 | 38.5 | 24.4 |
| Streptomycin (STR, 10 μg) | 11 | 12–14 | 15 | 84.4 | 90.0 | 95.8 | 90.5 | 70.6 | 84.6 | 87.8 |
| Florfenicol (FLR, 30 μg) | 14 | 15–18 | 19 | 60.9 | 59.3 | 56.3 | 76.2 | 64.7 | 53.9 | 60.5 |
| Ceftazidime (CAZ, 30 μg) | 17 | 18–20 | 21 | 6.3 | 3.3 | 8.3 | 0.0 | 0.0 | 38.5 | 5.8 |
| Imipenem (IPM, 10 μg) | 19 | 20–22 | 23 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 |
| Cefoxitin (FOX, 30 μg) | 14 | 15–17 | 18 | 7.0 | 6.0 | 2.1 | 0.0 | 11.8 | 7.7 | 5.8 |
| Trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg) | 10 | 11–15 | 16 | 68.0 | 85.3 | 75.0 | 52.4 | 41.2 | 76.9 | 74.0 |
| Aztreonam (ATM,30 μg) | 17 | 18–20 | 21 | 3.9 | 1.3 | 4.2 | 0.0 | 5.9 | 46.2 | 4.2 |
| Ciprofloxacin (CIP, 5 μg) | 15 | 16–20 | 21 | 22.7 | 28.0 | 16.7 | 4.8 | 0.0 | 23.1 | 22.0 |
| Amikacin (AMK, 30 μg) | 14 | 15–16 | 17 | 6.3 | 7.3 | 4.2 | 4.8 | 5.9 | 0.0 | 6.1 |
| Multi-drug resistant (MDR) | 55.5 | 59.3 | 43.8 | 42.9 | 52.9 | 61.5 | 54.9 | |||
R, resistant; I, intermediate; S, susceptible.
Distribution of Tn7-Like Transposons Associated Integrons
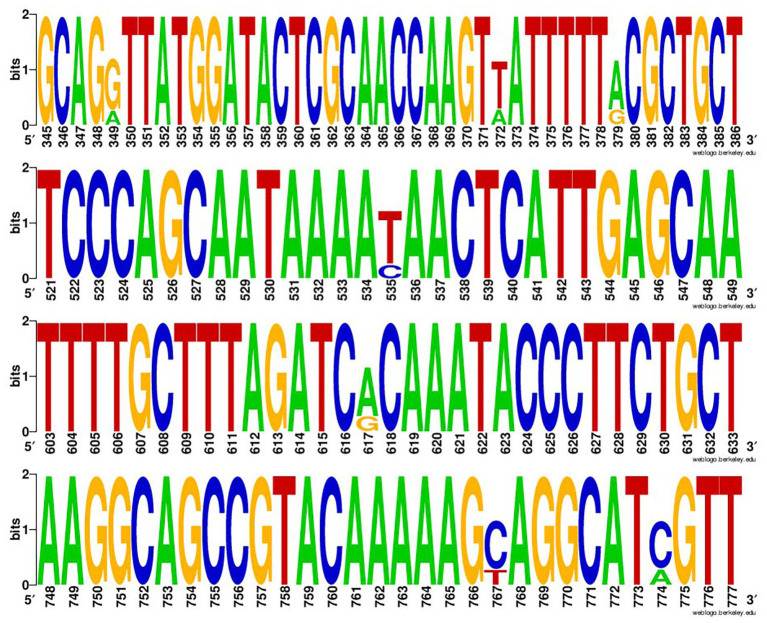
The positive rate of class 2 integrons in 377 Enterobacterales strains was 99.5% (two strains of E. coli lacked intI2). Through sequencing and sequence comparison, the results showed that intI2 usually had mutation sites, and most of them were at position 349, 372, 379, 535, 617, 767, and 774, among which 535 base mutations (C mutated to T) were terminating mutations (Figure 1).
Figure 1.
The Weblogo of repeats of intI2. The sequences were each mutation site of the integrase gene of class 2 integrons.
Genetic Characterization of Tn7-Like Transposons
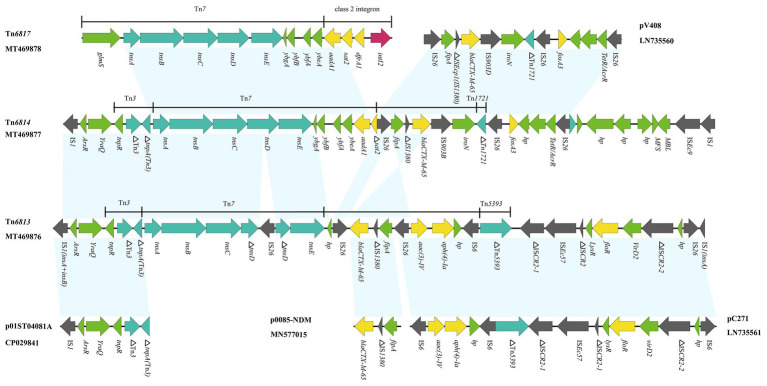
Among the Tn7-like transposons carrying strains, six multi-drug resistant isolates were randomly selected for whole-genome sequencing, and yielded six completed Tn7-like transposons genetic structures. According to the transposon nomenclature3, we designated them Tn6763, Tn6764, Tn6765, Tn6813, Tn6814, and Tn6817. Tn6763 (GenBank accession number MN641830), Tn6764 (GenBank accession number MN628641), and Tn6817 (GenBank accession number MT469878) were located on the chromosomes of P. mirabilis, E. coli, and S. enterica, respectively. They were typical Tn7 transposons comprising the transposition module (tnsA, tnsB, tnsC, tnsD, and tnsE), three gene cassettes (aadA1, sat2, and dfrA1), and an inactive class 2 integrase gene (Figures 2, 3).
Figure 2.
Genetic structure of Tn6813, Tn6814, and Tn6817. The physical maps were generated using Easyfig 2.2.3 and DNAMAN Version 8.0. Linear comparison of Tn6813 region in E. coli strain SFE8 with Tn6814 region in E. coli strain SCZE5, and Tn6817 region in S. enterica strain SCFS4. Genes and open reading frames (ORFs) are shown as arrows, and their orientations of transcription are indicated by the arrowheads. Horizontal lines, different regions corresponding to Tn7, Tn1721, Tn3, Tn5393, and integrons. Antimicrobial resistance genes are in yellow, transposase are in blue, and integrase genes are in red. The IS elements are indicated by gray arrows. Other functions or putative proteins are in green. Shared regions with 99% identity are indicated by shading.
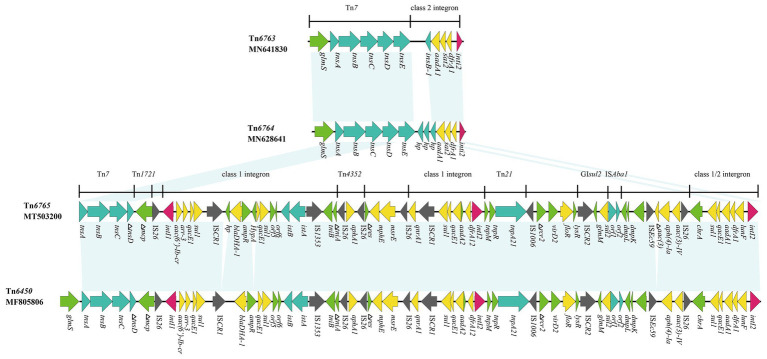
Figure 3.
Genetic structure of Tn6763, Tn6764, and Tn6765. The physical maps were generated using Easyfig 2.2.3 and DNAMAN Version 8.0. Linear comparison of Tn6765 region in p1.1.2 with Tn6763 region in P. mirabilis strain, Tn6764 region in E. coli strain and Tn6450 region in P. mirabilis strain SNYG17 (GenBank accession number MF805806). Genes and ORFs are shown as arrows, and their orientations of transcription are indicated by the arrowheads. Horizontal lines, different regions corresponding to Tn7, Tn1721, Tn21, and integrons. Antimicrobial resistance genes are in yellow, transposase are in blue, and integrase genes are in red. The IS elements are indicated by gray arrows. Other functions or putative proteins are in green. Shared regions with 99% identity are indicated by shading.
Tn6813 (GenBank accession number MT469876) and Tn6814 (GenBank accession number MT469877) were novel Tn7-like transposons in chromosomes, acquired from E. coli SFE8 and SCZE5, respectively. Tn6813 was 32,688 bp in size and Tn6814 was 32,874 bp. The structures of the two Tn7-like transposons were respectively surrounded by two IS1 elements, while one IS1 of Tn6813 was incomplete with missing C-terminus. Transposition module (tnsA, tnsB, tnsC, tnsD, and tnsE) of Tn7 in Tn6814 was intact, but tnsD in Tn6813 was truncated by an IS26 element. Compared to canonical Tn7, the area’s downstream of transposition modules of Tn6813 and Tn6814 differed by deletions involving class 2 integrons and its associated gene cassettes. Actually, Tn6813 had no intI2 and no gene cassettes (dfrA1-sat2-aadA1), and Tn6814 had only an aadA1 and an incomplete sat2 gene left (Figure 2).
Tn6765 (GenBank accession number MT503200) was located on a plasmid of P. mirabilis SCBX1.1, named p1.1.2 (GenBank accession number CP047114). Sequence analysis showed that 19 resistance genes, except cfr and erm(B), were carried by the Tn7-like transposon (Figure 3). The novel MDR transposon harbored different resistance genes, including bla DHA-1 (cephalosporin resistance), qnrA1 (fluoroquinolone), aac(6’)-Ib-cr (fluoroquinolone and aminoglycosides), floR (chloramphenicol/florfenicol), mphE and msrE (macrolide), and lunF (lincosamide) genes. However, the cfr and erm(B) gene was carried by another 12,795 bp plasmid p1.1.1 (Supplementary Figure S1; GenBank accession number CP047113), which existed in the same strain as p1.1.2 (Figure 3).
Tn6765 was 64,752 bp (corresponding to bases 47,309–112,060 in GenBank accession number CP047114) with an average GC content of 52.94% that differed from that of the rest of the P. mirabilis plasmid (GC content, 44.35%). It had partial characteristics of the Tn7 transposon, which contained transposase genes tnsA, tnsB, and tnsC. But its transposition module lost tnsE gene and the tnsD was truncated by insertion of the inverted repeat (IR mcp) of Tn1721 (Figure 3).
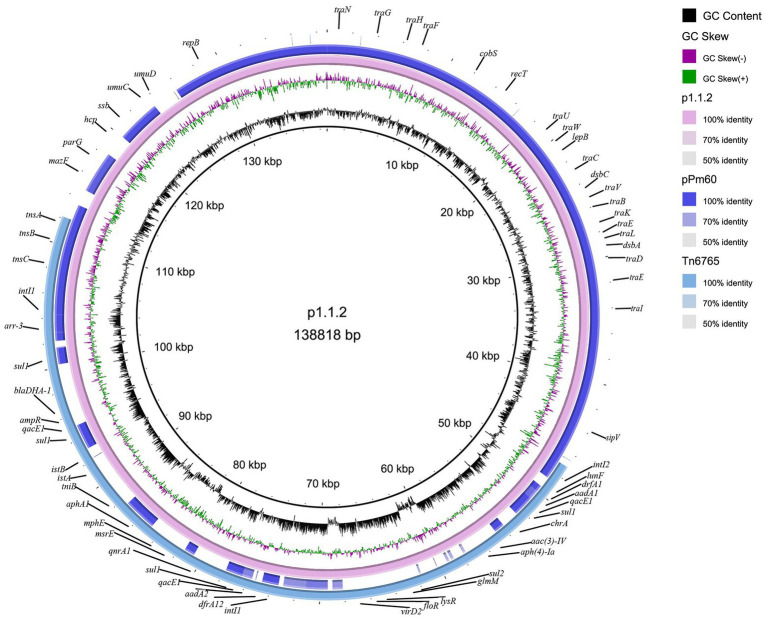
The plasmid carrying Tn6765, named as p1.1.2, was 138,818 bp in size and had a GC content of 44.35%. Blastn results showed that, except the Tn6765 region, other regions of plasmid p1.1.2. showed 99.98% similarity and 91% coverage with nucleic acid sequence of P. mirabilis plasmid pPm60 (GenBank accession number MG516911), which also carried the Tn7-like transposon published in NCBI database (Figure 4).
Figure 4.
Circular representation of plasmid p1.1.2. The physical map of p1.1.2 was generated using BRIG v0.95.
Horizontal Transfer and Stability of Tn7-Like Transposons
Double-antibiotics Salmonella Shigella agar plates (300 μg/ml rifampicin + 10.24 μg/ml flufenicol) were used to screen the transconjugant. The results of drug sensitivity, electrophoresis, and sequencing showed that the plasmid p1.1.2 could be successfully transferred to E. coli EC600 (Supplementary Table S3). A novel Tn7-like transposon (Tn6765) transconjugant was obtained through the conjugative transfer test and at a frequency of 1.5 × 10−4 transconjugants per donor (average of three independent determinations). The stability of Tn6765, Tn6813, and Tn6814 was determined by picking 100 clones in the twenty first passage to detect the presence of Tn6765, Tn6813, and Tn6814, respectively. The results of all PCRs were positive, meaning that Tn6765, Tn6813, and Tn6814 can be stably inherited in the bacteria.
Discussion
The Tn7-like transposons are important mobile elements to transfer bacterial AMR. Compared with information obtained from studies about the transposition mechanism of the Tn7-like transposons, available data on the comprehensive analysis of Tn7-like transposons in Enterobacterales isolates are still inadequate. Herein, we determined the incidence of Tn7-like transposons in Enterobacterales isolates obtained from several farms of chicken and swine raised for meat purpose. Among the 1,474 Enterobacterales isolates, 377 strains that carry Tn7-like transposons were identified. In different genus of bacteria, the separation rate of Tn7-like transposons was different (p < 0.0001; Chi square test; Table 2). The positive rate of Tn7-like transposons in Proteus spp. (57.5%) was significantly higher than other bacteria’s (p < 0.0001; Chi square test). This finding was consistent with previous reports that genomes of Proteus spp. exhibited strong plasticity facilitating high-frequency insertion of mobile genetic elements like Tn7-like transposons (Dong et al., 2019; de Curraize et al., 2020; Gu et al., 2020). In addition, E. coli strains were ubiquitous commensal bacteria and abundant in the intestine of humans and animals (Zhang et al., 2013; Paitan, 2018). Therefore, its proportion in Tn7-like transposons positive Enterobacterales isolates was also large (Table 2). Since E. coli, Proteus spp. and some of other Enterobacterales species were opportunistic pathogens, even healthy food animals could be asymptomatic carriers of the bacteria and may cause an impact on human health. Our results provide evidence of the Enterobacterales strains as a possible reservoir of Tn7-like transposons, a risk that deserves our attention.
Antimicrobial susceptibility testing indicated that Enterobacterales strains carrying Tn7-like transposons exhibited high resistance to a variety of antibiotics, with a 54.9% multi-drug resistance rate. Relatively high rates of MDR in Proteus spp. (59.3%) and S. enterica (61.5%) strains may be due to the prevalence of multiple mobile elements in both bacteria, which was confirmed in previous studies (Beutlich et al., 2011; Murgia et al., 2015; Schultz et al., 2015; Lei et al., 2018). This also implied that Tn7-like transposons were likely to occur simultaneously with other mobile elements. High resistance to streptomycin (87.8%) and trimethoprim/sulfamethoxazole (74.0%) can be attributed to the intI2-associated resistance gene cassette (aadA1, sat2, and dfrA1) carried by Tn7-like transposons (Tietze and Brevet, 1991; Kaushik et al., 2019). The gene cassettes of intI2 contained the aminoglycoside adenyltransferase (aadA1 and aadA2), dihydrofolate reductase (dfrA1), and streptothricin acetyltransferase (sat2) encoding genes, which are responsible for streptomycin-spectinomycin, trimethoprim, and streptothricin resistance, respectively (Zhang et al., 2019). Through sequencing, we found that there were multiple mutated sites in intI2, among which 535 base mutations (C mutated to T) were terminating mutations (Figure 2). They led to premature termination of integrase gene translation, making its gene cassette sequence show a high degree of stability, usually aadA1, sat2, and dfrA1 (Deng et al., 2015). Moreover, we knew from workers on the farms that spectinomycin is widely used against the respiratory or enteric infections in swine and chicken.
Compared with the resistance rates to streptomycin and trimethoprim/sulfamethoxazole, relatively low resistance rates to florfenicol (60.5%), gentamicin (24.4%), and ciprofloxacin (22%) were detected in the Tn7-like transposons positive strains. Most of them showed susceptibility to amikacin, ceftazidime, cefoxitin, aztreonam, and imipenem. The fact also emphasizes the importance of standardization of clinical drug application to prolong the efficacy of new drugs. The emergence of severe drug-resistant status in China is largely related to the abuse of antibiotics as feed additives in veterinary clinical practice and the transfer of movable components carrying drug-resistant genes between isolates, which has become a common problem in both human and veterinary clinical practice (Rahmani et al., 2013). Obviously, for Enterobacterales, carriage of Tn7-like transposons, each containing a set of resistance genes, may increase the chances of horizontal transfer of multiple resistance determinants to susceptible strains and may in turn bring unique advantages to the host and enable them to survive a strong antimicrobial selection pressure especially in poultry and livestock farm settings. Tn7-like transposons have been so successful at spreading into diverse relevant taxa that they could be used as a proxy for antibiotics pollution (Flores et al., 1992; Cleaver and Wickstrom, 2000).
In this study, six completed Tn7-like transposons genetic structures were identified by WGS analysis, which showed that they included three typical Tn7 transposons (Tn6763, Tn6764, and Tn6817) and three novel Tn7-like transposons (Tn6813, Tn6814, and Tn6765). The IS26 segment in Tn6814 showed high level homology to the segment characterized in E. coli plasmid pV408 (GenBank accession number LN735560), and it harbored different mobile genetic elements (complete or truncated) and two resistance genes (bla CTX-M-65 and fosA3). The bla CTX-M-65-containing segment (IS26-fipA-IS1380-bla CTX-M-65-IS903B) in Tn6814 also showed nucleotide identity to the corresponding genetic structure that habored bla CTX-M-65 gene in Tn6813, with the exception of one IS26 sequence replaced by IS903B sequence in Tn6814 (Figure 2). Interestingly, the organisms of Tn6813 and Tn6814 came from the same swine farm, and therefore, the bla CTX-M-65 segment bounded by two ISs in Tn6814 could be derived from Tn6813 or vice versa and likely by ISs-mediated mobilization.
Another resistance region bounded by IS26 in Tn6814 was identical to M1 module of pC271 in E. coli (GenBank accession number LN735561; Riccobono et al., 2015), containing resistance determinants to aminoglycosides [aac(3)-IV and aph(4)-Ia] and florfenicol (floR). Diverse mobile genetic elements in Tn6814 included four intact ISs (three copies of IS26, an IS903B, and an ISEc9) and one incomplete IS1380, while six intact ISs (four copies of IS26, an IS6, and an ISEc57) and five incomplete elements (three ISCR2, an IS1380, and a Tn5393) were inserted in Tn6813 (Figure 2). From the findings of various IS inserted sequences in these novel transposons, it is indicated that an increase in the diversity of Tn7-like transposons was prompted by the ISs-mediated homologous recombination.
Tn6765 was highly homologous with Tn6450 (GenBank accession number MF805806), which was located on the chromosome detected by Chen et al. (2018). The similarity of nucleic acid sequence was more than 99%, and the coverage was 96%. The differences between the two were mainly in the following four aspects: (i) Tn6765 had a hypotheical protein gene before bla DHA-1; (ii) an HypA gene was inserted between ampR and qacE1 in Tn6765; (iii) an incomplete aminoglycoside N(3)-acetyltransferase encoding gene was inserted between ISEc59 and aph(4)-Ia; and (iv) Tn6450 was inserted downstream of the glmS gene, encoding glucosamine 6-phosphate synthase and surrounded by 5-bp direct repeats (CCAAT), whereas Tn6765 was located on the plasmid, there is no glms gene and repeated sequences at both ends (Figures 3, 4). The carrier difference between Tn6765 and Tn6450 indicated that Tn7-like can be transmitted alternately on the chromosome and plasmid by cutting and inserting. To make matters worse, Tn6450 comes from a chicken source (Chen et al., 2018) and Tn6765 from a swine source suggested that Tn7-like transposons can be transmitted from one animal to another with bacterial hosts.
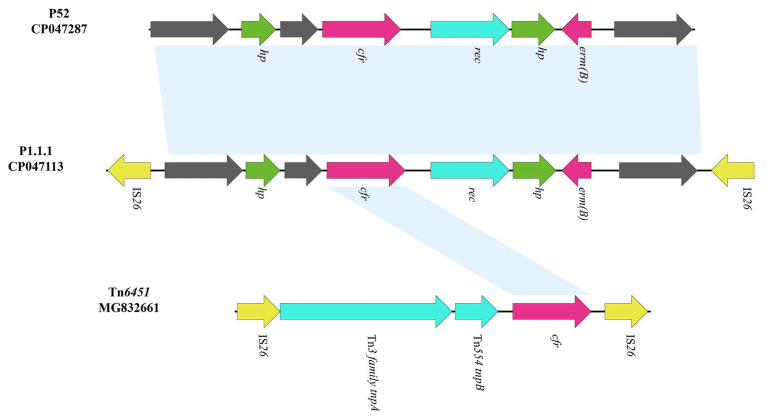
The cfr-containing segment of plasmid p1.1.1 (corresponding to bases 8,925 to 12,726 to 4,624 in GenBank accession number CP047113) harbors a genetic structure, showing homology to the cfr segment characterized in P. cibarius G11 (Genebank accession number CP047287), which is partially differ from the existence of IS26-cfr-▵Tn554 tnpB-▵Tn3 family tnpA-IS26 section in another Tn7-like transposon, Tn6451 (GenBank accession number MG832661; Chen et al., 2019; Figure 5). Although the cfr section of p1.1.1 and Tn6451 is slightly different, this also suggests an evolutionary direction of the Tn6765 carrying strain.
Figure 5.
Genetic structure of cfr-containing region and in p1.1.1. The physical maps were generated using Easyfig 2.2.3 and DNAMAN Version 8.0. Linear comparison of the cfr-containing region in p1.1.1 with the cfr-containing region in P. cibarius strain G11 (Genebank number CP047287) and in M. morganii strain BCMM24 (Genebank number MG832661). Genes and ORFs are shown as arrows, and their orientations of transcription are indicated by the arrowheads. Antimicrobial resistance genes are in red. The IS elements are indicated by yellow arrows. Putative proteins are in green. Shared regions with 99% identity are indicated by shading.
By conducting mating (conjugation) experiments, the Tn6765-carrying plasmid was successfully transferred to EC600. Although Tn6813 and Tn6814 located on the chromosomes could not be transferred by conjugation, the stability experiment showed that they could stably exist in the hosts. The “core transposition machinery” of Tn7-like transposon consists of the transposase proteins TnsA and TnsB along with a regulator protein, TnsC (Choi et al., 2013). This core machinery is directed by one of two target selecting proteins, TnsD or TnsE. Transposition with TnsABC+TnsD has evolved to maximize the efficiency of vertical transmission of the element by directing transposition into the chromosome. Transposition with TnsABC+TnsE occurs preferentially into mobile plasmids through the ability of the TnsE protein to recognize features found enriched during DNA replication on the lagging-strand template (Wolkow et al., 1996; Peters and Craig, 2000, 2001a). Therefore, it cannot rule out the possibility that these Tn7-like transposons located on the chromosome will target transposition into mobile plasmids, facilitating the spread of Tn7-like transposons between bacteria during the propagation of the hosts.
By comparing the genetic structure of different Tn7-like transposons, we can speculate that the multidrug-resistant Tn7-like transposons have a certain evolutionary relationship, which has contributed to Tn7-like transposons playing a vital role in the field of storing resistance genes. Although the assembly was original, each of these Tn7-like transposons, or parts thereof, was identical to those found in other plasmids or chromosomes. This was related to the fact that Tn7-like transposons can transfer between strains and accumulate genetic material via mobile genetic elements.
Conclusion
This study is the first report to comprehensively analyze the incidence and antibiotic resistance characteristics of Tn7-like transposons in Enterobacterales isolates from livestock and poultry in China, and it presented detailed genetic characterization of three novel Tn7-like transposons Tn6765, Tn6813, and Tn6814, which were integrated into plasmids or chromosomes from E. coli and P. mirabilis. Multiple antibiotic resistance genes in Tn7-like transposons, considering that they were found in chicken and swine, are highly worrisome and may become a serious threat by spreading in other nearby animals, humans, and the environment. Therefore, robust measures should be taken to control the spread and emergence of mobile genetic resistance determinants in animals used for food production in China and the world, and our study provides an important reference for this.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by College of Life Science, Sichuan University affiliation ethics committee, and all efforts were made to minimize animal suffering.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication. JH conceived the study and drafted the manuscript. JH and CL collected samples and conducted data statistics. JH, CL, and PC performed experiments. HW supervised the research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Adam P. Roberts (University College London) for helpful suggestionsregarding the nomenclature of Tn6813, Tn6814, Tn6817, Tn6763, Tn6764 andTn6765.
This manuscript has been released as a pre-print at ResearchSquare, (He et al., 2020).
Funding. This work was supported by the National Natural Science Foundation of China (Grant No. 31830098) and the National Key Research and Development Program of China (Grant No. 2016YFD0501608).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02049/full#supplementary-material
References
- Beutlich J., Jahn S., Malorny B., Hauser E., Huhn S., Schroeter A., et al. (2011). Antimicrobial resistance and virulence determinants in European Salmonella genomic island 1-positive Salmonella enterica isolates from different origins. Appl. Environ. Microbiol. 77, 5655–5664. 10.1128/AEM.00425-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. 10.1128/AAC.02412-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. P., Lei C. W., Kong L. H., Zeng J. X., Zhang X. Z., Liu B. H., et al. (2018). Tn6450, a novel multidrug resistance transposon characterized in a Proteus mirabilis isolate from chicken in China. Antimicrob. Agents Chemother. 62, e02192–e02217. 10.1128/AAC.02192-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lei C., Zuo L., Kong L., Kang Z., Zeng J., et al. (2019). A novel cfr-carrying Tn7 transposon derivative characterized in Morganella morganii of swine origin in China. J. Antimicrob. Chemother. 74, 603–606. 10.1093/jac/dky494, PMID: [DOI] [PubMed] [Google Scholar]
- Choi K. Y., Li Y., Sarnovsky R., Craig N. L. (2013). Direct interaction between the TnsA and TnsB subunits controls the heteromeric Tn7 transposase. Proc. Natl. Acad. Sci. U. S. A. 110, E2038–E2045. 10.1073/pnas.1305716110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver S. H., Wickstrom E. (2000). Transposon Tn7 gene insertion into an evolutionarily conserved human homolog of Escherichia coli attTn7. Gene 254, 37–44. 10.1016/s0378-1119(00)00283-3, PMID: [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2017). Performance standards for antimicrobial susceptibility testing M02-A12, M07-A10, and M11-A8. 27th Edn. Philadelphia, PA: Clinical and Laboratory Standards Institute, 282. [Google Scholar]
- Cocchi S., Grasselli E., Gutacker M., Benagli C., Convert M., Piffaretti J. C. (2007). Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol. Med. Microbiol. 50, 126–132. 10.1111/j.1574-695X.2007.00242.x, PMID: [DOI] [PubMed] [Google Scholar]
- Cortes P., Blanc V., Mora A., Dahbi G., Blanco J. E., Blanco M., et al. (2010). Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76, 2799–2805. 10.1128/AEM.02421-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Curraize C., Siebor E., Varin V., Neuwirth C., Hall R. M. (2020). Two new SGI1-LK variants found in Proteus mirabilis and evolution of the SGI1-HKL group of Salmonella genomic Islands. mSphere 5, e00875–e00919. 10.1128/mSphere.00875-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., et al. (2015). Resistance integrons: class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 14:45. 10.1186/s12941-015-0100-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D., Li M., Liu Z., Feng J., Jia N., Zhao H., et al. (2019). Characterization of a NDM-1‐ encoding plasmid pHFK418-NDM from a clinical Proteus mirabilis isolate harboring two novel transposons, Tn6624 and Tn6625. Front. Microbiol. 10:2030. 10.3389/fmicb.2019.02030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C. C., Cotterill S., Lichtenstein C. P. (1992). Overproduction of four functionally active proteins, TnsA, B, C, and D, required for Tn7 transposition to its attachment site, attTn7. Plasmid 28, 80–85. 10.1016/0147-619x(92)90038-c, PMID: [DOI] [PubMed] [Google Scholar]
- Goncalves A., Torres C., Silva N., Carneiro C., Radhouani H., Coelho C., et al. (2010). Genetic characterization of extended-spectrum beta-lactamases in Escherichia coli isolates of pigs from a Portuguese intensive swine farm. Foodborne Pathog. Dis. 7, 1569–1573. 10.1089/fpd.2010.0598, PMID: [DOI] [PubMed] [Google Scholar]
- Gu W., Wang W., Tong P., Liu C., Jia J., Lu C., et al. (2020). Comparative genomic analysis of Proteus spp. isolated from tree shrews indicated unexpectedly high genetic diversity. PLoS One 15:e0229125. 10.1371/journal.pone.0229125, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson K., Sundstrom L., Pelletier A., Roy P. H. (2002). IntI2 integron integrase in Tn7. J. Bacteriol. 184, 1712–1721. 10.1128/jb.184.6.1712-1721.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Li C., Cui P., Wang H. (2020). Characterization of Tn7-like transposons and its related antibiotic resistance among Enterobacteriaceae from livestock and poultry in China. Research Square [Preprint]. 10.21203/rs.3.rs-20496/v1, PMID: 11872723 [DOI] [Google Scholar]
- Kaushik M., Khare N., Kumar S., Gulati P. (2019). High prevalence of antibiotic resistance and integrons in Escherichia coli isolated from Urban river water, India. Microb. Drug Resist. 25, 359–370. 10.1089/mdr.2018.0194, PMID: [DOI] [PubMed] [Google Scholar]
- Kleinheinz K. A., Joensen K. G., Larsen M. V. (2014). Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. 10.4161/bact.27943, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C. W., Chen Y. P., Kang Z. Z., Kong L. H., Wang H. N. (2018). Characterization of a novel SXT/R391 integrative and conjugative element carrying cfr, blaCTX-M-65, fosA3, and aac(6')-Ib-cr in Proteus mirabilis. Antimicrob. Agents Chemother. 62, e00849–e00918. 10.1128/AAC.00849-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque C., Piché L., Larose C., Roy P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191. 10.1128/aac.39.1.185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M. (2009). Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl. 1), i29–i36. 10.1093/jac/dkp255, PMID: [DOI] [PubMed] [Google Scholar]
- Lupo A., Papp-Wallace K. M., Sendi P., Bonomo R. A., Endimiani A. (2013). Non-phenotypic tests to detect and characterize antibiotic resistance mechanisms in Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 77, 179–194. 10.1016/j.diagmicrobio.2013.06.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B. M., Levy S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. 10.1128/CMR.00002-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D., Jouy E., Lazizzera C., Kobisch M., Madec J. Y. (2006). CTX-M-1‐ and CTX-M-15-type beta-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28, 402–407. 10.1016/j.ijantimicag.2006.08.016, PMID: [DOI] [PubMed] [Google Scholar]
- Moura A., Soares M., Pereira C., Leitao N., Henriques I., Correia A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. 10.1093/bioinformatics/btp105, PMID: [DOI] [PubMed] [Google Scholar]
- Murgia M., Bouchrif B., Timinouni M., Al-Qahtani A., Al-Ahdal M. N., Cappuccinelli P., et al. (2015). Antibiotic resistance determinants and genetic analysis of Salmonella enterica isolated from food in Morocco. Int. J. Food Microbiol. 215, 31–39. 10.1016/j.ijfoodmicro.2015.08.003, PMID: [DOI] [PubMed] [Google Scholar]
- Paitan Y. (2018). Current trends in antimicrobial resistance of Escherichia coli. Curr. Top. Microbiol. Immunol. 416, 181–211. 10.1007/82_2018_110, PMID: [DOI] [PubMed] [Google Scholar]
- Peters J. E. (2014). Tn7. Microbiol. Spectr. 2:MDNA3-0010-2014. 10.1128/microbiolspec.MDNA3-0010-2014, PMID: [DOI] [PubMed] [Google Scholar]
- Peters J. E., Craig N. L. (2000). Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell 6, 573–582. 10.1016/s1097-2765(00)00056-3, PMID: [DOI] [PubMed] [Google Scholar]
- Peters J. E., Craig N. L. (2001a). Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 15, 737–747. 10.1101/gad.870201, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. E., Craig N. L. (2001b). Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2, 806–814. 10.1038/35099006, PMID: [DOI] [PubMed] [Google Scholar]
- Rahmani M., Peighambari S. M., Svendsen C. A., Cavaco L. M., Agerso Y., Hendriksen R. S. (2013). Molecular clonality and antimicrobial resistance in Salmonella enterica serovars Enteritidis and Infantis from broilers in three northern regions of Iran. BMC Vet. Res. 9:66. 10.1186/1746-6148-9-66, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman M. U., Zhang H., Huang S., Iqbal M. K., Mehmood K., Luo H., et al. (2017). Characteristics of integrons and associated gene cassettes in antibiotic-resistant Escherichia coli isolated from free-ranging food animals in China. J. Food Sci. 82, 1902–1907. 10.1111/1750-3841.13795, PMID: [DOI] [PubMed] [Google Scholar]
- Riccobono E., Di Pilato V., Di Maggio T., Revollo C., Bartoloni A., Pallecchi L., et al. (2015). Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum beta-lactamase in the Bolivian Chaco region. Antimicrob. Agents Chemother. 59, 5340–5347. 10.1128/AAC.00589-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. P., Chandler M., Courvalin P., Guedon G., Mullany P., Pembroke T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. 10.1016/j.plasmid.2008.08.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L., Linkevicius M., Lytsy B., Melhus A., Andersson D. I. (2012). Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67, 74–83. 10.1093/jac/dkr405, PMID: [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi M., Nuccio S. P., Liu H., Hernandez D., Vu C. T., Takahashi A. A., et al. (2016). Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283. 10.1038/nature20557, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E., Haenni M., Mereghetti L., Siebor E., Neuwirth C., Madec J. Y., et al. (2015). Survey of multidrug resistance integrative mobilizable elements SGI1 and PGI1 in Proteus mirabilis in humans and dogs in France, 2010-13. J. Antimicrob. Chemother. 70, 2543–2546. 10.1093/jac/dkv154, PMID: [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. 10.1093/nar/gkj014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom L., Radstrom P., Swedberg G., Skold O. (1988). Site-specific recombination promotes linkage between trimethoprim‐ and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet. 213, 191–201. 10.1007/BF00339581, PMID: [DOI] [PubMed] [Google Scholar]
- Sundstrom L., Roy P. H., Skold O. (1991). Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173, 3025–3028. 10.1128/jb.173.9.3025-3028.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom L., Skold O. (1990). The dhfrI trimethoprim resistance gene of Tn7 can be found at specific sites in other genetic surroundings. Antimicrob. Agents Chemother. 34, 642–650. 10.1128/aac.34.4.642, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze E., Brevet J. (1991). The trimethoprim resistance transposon Tn7 contains a cryptic streptothricin resistance gene. Plasmid 25, 217–220. 10.1016/0147-619x(91)90015-o, PMID: [DOI] [PubMed] [Google Scholar]
- Toombs-Ruane L. J., Benschop J., Burgess S., Priest P., Murdoch D. R., French N. P. (2017). Multidrug resistant Enterobacteriaceae in New Zealand: a current perspective. N. Z. Vet. J. 65, 62–70. 10.1080/00480169.2016.1269621, PMID: [DOI] [PubMed] [Google Scholar]
- von Tippelskirch P., Golz G., Projahn M., Daehre K., Friese A., Roesler U., et al. (2018). Prevalence and quantitative analysis of ESBL and AmpC beta-lactamase producing Enterobacteriaceae in broiler chicken during slaughter in Germany. Int. J. Food Microbiol. 281, 82–89. 10.1016/j.ijfoodmicro.2018.05.022, PMID: [DOI] [PubMed] [Google Scholar]
- Wolkow C. A., DeBoy R. T., Craig N. L. (1996). Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10, 2145–2157. 10.1101/gad.10.17.2145, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang S., Yang H., Rehman M. U., Yang K., Dong M., Yang J., et al. (2019). Class 1 integrons as predominant carriers in Escherichia coli isolates from waterfowls in Hainan, China. Ecotoxicol. Environ. Saf. 183:109514. 10.1016/j.ecoenv.2019.109514, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang R. Q., Ying G. G., Su H. C., Zhou L. J., Liu Y. S. (2013). Antibiotic resistance and genetic diversity of Escherichia coli isolates from traditional and integrated aquaculture in South China. J. Environ. Sci. Health B 48, 999–1013. 10.1080/03601234.2013.816611, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.