Abstract
Purpose of Review
Light microscopy plays an essential role in clinical diagnosis and understanding the pathogenesis of cancer. Conventional bright-field microscope is used to visualize abnormality in tissue architecture and nuclear morphology, but often suffers from many limitations. This review focuses on the potential of new imaging techniques to improve basic and clinical research in pathobiology.
Recent findings
Light microscopy has significantly expanded its ability in resolution, imaging volume, speed and contrast. It now allows 3D high-resolution volumetric imaging of tissue architecture from large tissue and molecular structures at nanometer resolution.
Summary
Pathologists and researchers now have access to various imaging tools to study cancer pathobiology in both breadth and depth. Although clinical adoption of a new technique is slow, the new imaging tools will provide significant new insights and open new avenues for improving early cancer detection, personalized risk assessment and identifying the best treatment strategies.
Keywords: light microscopy, 3D volumetric imaging, super-resolution microscopy, label-free imaging
Introduction
Light microscopy is one of the most important tools for pathological diagnosis and biomedical research. Bright-field light microscopy has been the main technique for pathological evaluation for more than two centuries. The microscopic morphology-based pathology remains the “gold-standard” to identify cancer cells and to specify cancer type. It relies on identifying structural abnormalities in tissue architecture and nuclear morphology on stained cell and tissue visualized under conventional bright-field microscope. Such morphology-based diagnostic features are highly robust, as they are common structural features for a wide variety of tumor types without being confounded by the significant molecular heterogeneity in various stages of carcinogenesis.
Despite the tremendous clinical value of conventional morphology, they have limitations in many clinical scenarios. On one hand, the visualization of conventional morphology requires time-consuming tissue processing (e.g., paraffin-embedding or freezing and tissue section), which is not ideal for surgical applications where real-time “histology” is preferred. On the other hand, conventional morphology also has limited performance in personalized risk stratification for at-risk patients. For example, identification of the earliest precursor lesions in various tumor types play an important role in cancer prevention. Precursor lesions serve as prognostic markers to predict the risk for future development of malignancy. Patients with high-grade dysplasia are likely to progress into aggressive cancer than those with low-grade dysplasia. But in some cases, different types of precursor lesions present similar morphological difference, resulting in significant inter-observer variation [1]. Importantly, most patients with precursor lesions will not develop cancer, so frequent and invasive surveillance of at-risk patients carries financial, physical, and emotional burdens. Similarly, some malignancies are indolent, some are aggressive. But conventional morphology often has limited accuracy in detecting genuinely high-risk patients, or distinguishing aggressive from indolent cases.
In the last decade, advancement in the field of light microscopy has been revolutionary, from the widespread use of confocal/two-photon/non-linear microscopy for three-dimensional (3D) high-resolution tissue imaging, to light-sheet and optical coherence microscopy for 3D volumetric imaging of large tissue and the whole organ, to label-free light scattering and quantitative phase imaging for interrogation of nanoscale cellular architecture, to the emergence of super-resolution microscopy that overcomes the physical diffraction limit to visualize previously invisible molecular structures. This review will discuss the potential of these light microscopy techniques in understanding the pathogenesis of cancer and improving cancer diagnosis.
Clinical samples used for pathological evaluation
As optical signals come from the interaction between light and the imaging object, the knowledge of the sample characteristics is an essential, but often overlooked component for high-resolution microscopic imaging of pathological tissue. The main sources of human samples that are used to study cancer pathogenesis or provide diagnostic, prognostic or therapeutic information are tissue obtained through small biopsies or surgical resection and cells obtained through brushing or body fluid. Although fresh cells and tissue are ideal source to study the underlying pathogenesis of human diseases, their biochemical compositions change quickly after they are removed from human body. For clinical use, these samples have to be preserved in fixatives for short-term use; or prepared as frozen or formalin-fixed and paraffin-embedded (FFPE) tissue blocks for long-term storage. Clinical FFPE tissue preservation is the gold standard for pathological diagnosis. It also provides a large human tissue repository with documented clinical outcome and allows respective evaluation of many diseases, especially for those that would be difficult to study such as precursor lesions that often take decades to progress into malignancy. Therefore, the ability to analyze FFPE is of immense importance to study human diseases [2]. Any imaging techniques that can be tailored for FFPE tissue will be of great advantage that can be easily integrated with the existing clinical workflow.
However, the processing of tissue or cells can change the chemical compositions of tissue and cells and thus alter the optical properties from both intrinsic and exogeneous optical signals in different high-resolution imaging systems. Proper sample preparation is essential to obtain high-quality and reproducible images. For example, fixatives such as alcohol-based solution dehydrate the tissue and cells, thus increasing their refractive index - the intrinsic properties detected by light scattering techniques. The increased refractive index may also induce spherical aberration in high-NA objectives in fluorescence microscopy. Other fixatives (e.g., paraformaldehyde (PFA) and formalin) cross-link proteins, which has been shown to best preserve tissue architecture and morphology. But over-fixation can introduce significant autofluorescence background and mask the epitopes that are essential for immunofluorescence staining. Frozen tissue, on the other hand, has better-preserved molecular constituents such as epitopes, with less autofluorescence background, but suffers from poorly maintained tissue morphology. These factors must be considered for high-resolution imaging of human tissue samples.
Intrinsic vs. exogeneous contrast
In light microscopy, optical contrast is as important as optical resolution that determine the ultimate performance of the imaging system. There are two types of contrast that forms the image - intrinsic contrast or label-free imaging that detect the intrinsic optical signals from light-tissue interaction, and exogeneous contrast that relies on contrast-providing tags (e.g., fluorescent or chromogenic dye) to highlight the molecules of interest in the image. The intrinsic optical signals come from light-tissue interactions, such as scattered light that detects the refractive index variations due to structural or density changes within the samples; autofluorescence, infrared and Raman signals that detect a wide variety of biochemical constituents that have important functional implications of the underlying biological activities; and second-harmonic generation signals that often come from birefringence of extracellular matrix. The biggest advantage of intrinsic optical signal is the minimal sample preparation, as it does not use any chemicals, antibodies or stains, thus immune to the variation introduced by the external reagents. The intrinsic optical signals have been widely used to distinguish normal from cancerous tissue in clinical settings. However, the underlying biology of the detected optical signals can be ambiguous. On the other hand, the exogenous optical contrast is advantageous when there is a well-defined molecule in cancer pathobiology. For example, certain proteins are overexpressed in cancer tissue, so tagging a chromogenic or fluorescent dye to this cancer-specific protein will provide significant diagnostic value. But the staining process is subject to variation, as the accumulation of the exogeneous contrast agent depends on various factors, such as the preservation and accessibility of epitopes, efficiency of antibodies, and penetration of the contrast agent. Further, as most functional molecules can be tagged with a contrast agent, molecular imaging with high-resolution microscopy can serve as important diagnostic tools and provide valuable mechanistic insights into the underlying biology of cancer pathogenesis. Both intrinsic and exogeneous contrast are applied in the following discussion of optical microscopy systems will be divided into three main categories - conventional diffraction-limited 3D microscopy techniques, super-resolution microscopy and other label-free imaging techniques, and their advantages and limitations are summarized in Table 1.
Table 1.
Summary of high-resolution optical microscopy techniques for assessing pathological samples
| Techniques | Representative microscopy system | Advantages | Limitations |
|---|---|---|---|
| 3D microscopy | Confocal microscopy | One of the most versatile instruments; Detects both intrinsic scattered light (refractive index difference) and exogeneous signals (e.g., fluorescence); applicable for both dense and transparent tissue for up to ~200μm for uncleared tissue; Theoretically unlimited depth on cleared tissue |
Resolution is diffraction-limited; Intrinsic contrast often does not clearly distinguish subcellular organelles. |
| Stimulated Raman scattering microscopy | Detects both intrinsic and exogenous Raman scattering; Provides unique molecular information of vibrational mode. |
Resolution is diffraction- limited; Often requires a high laser power to generate strong Raman signals |
|
| Optical coherence microscopy (OCM) | Detects the intrinsic scattering signals due to refractive index mismatch; High speed, large field of view; Low cost. |
Resolution is limited to a few microns; Lack of molecular specificity; Insufficient contrast to distinguish sub-cellular organelles. |
|
| Light-sheet microscopy | Low photobleaching effect and suitable for long-term (days to weeks) imaging of live cells and organelles; High speed and large field of view. |
Requires exogeneous label of fluorophore; Limited penetration depth in dense, uncleared tissue. |
|
| Two-photon/nonlinear microscopy | Provides a high molecular contrast with either autofluorescence or second-harmonic signals (label-free), or labeled fluorophores Provides a deeper penetration depth compared to confocal microscopy. |
Resolution is diffraction-limited; High cost. | |
| Microscopy with UV- surface excitation (MUSE) | Provides a high-contrast visualization of fresh tissue; Quick, simple and low cost; |
Lateral resolution is diffraction-limited; Surface imaging with limited depth resolution of ~10μm. |
|
| Structured illumination microscopy | Provides a 3D image using three structured illumination patterns for intrinsically scattered or fluorophore-labeled tissue; Quick, large field of view and low cost | Resolution is diffraction- limited; The depth resolution is not as good as confocal microscope; It has low signal-to-noise ratio for weak fluorophores. |
|
| Super-resolution microscopy | Super-resolution 3D structured illumination microscopy | Provides a 3D super-resolved image at a resolution of ~120 nm (lateral resolution); Applicable to a wide range of fluorophores; Relatively fast compared to other super-resolution imaging techniques. |
Subject to image reconstruction artifacts for mismatched refractive index of the sample. Limited probing depth to ~4–5μm. |
| STED | Provides 3D super-resolution image at a lateral resolution of ~50 nm; Applicable to a wide range of photostable fluorophores; Achieves deeper probing depth compared to other superresolution imaging techniques. |
Requires high laser power, leading to strong photodamage; High cost. |
|
| STORM | Provides one of the best resolutions of ~10–20 nm; Best potential for a low-cost system. |
Requires long acquisition time of a few minutes; Limited to a small number of photo-switchable fluorophores. | |
| Expansion microscopy | Physically expands the sample, and can be imaged with conventional confocal microscope; Applicable to a wide range of fluorophores. |
Potential for heterogeneous expansion and physical damage to the sample; Reduced brightness. |
|
| Other label-free spectroscopy and imaging techniques | Quantitative phase imaging | Measures optical pathlength difference along the probing depth at nanoscale sensitivity; Label-free imaging, thus simple sample preparation. No photodamage, suitable for long-term imaging. |
Lateral resolution is diffraction-limited; Limited to thin samples (e.g., 4–5 μm). No molecular specificity. |
| Light scattering spectroscopy or microscopy | Detects particle size or optical density ranging from tens of nanometers to several microns; Label-free imaging and no photodamage. |
Lateral resolution is diffraction-limited; No molecular specificity; Data interpretation is based on simplified scattering models that may not fully capture the complexity of tissue properties. |
|
| Partial-wave | Detects refractive index fluctuation within the sample at nanoscale sensitivity; Label-free imaging and no photodamage. |
Lateral resolution is diffraction-limited; No molecular specificity. |
|
| nanoNAM | Detects changes in spatial frequency at nanoscale sensitivity in a depth-resolved manner; Label-free imaging with simple sample preparation. |
Lateral resolution is diffraction-limited; No molecular specificity. |
Conventional microscopy for in-vivo tissue imaging or 3D volumetric imaging of ex-vivo tissue
Conventional high-resolution imaging systems such as confocal microscopy [3], light-sheet microscopy [4], two-photon/nonlinear microscopy [5], optical coherence microscopy [6], structured illumination microscopy [7], stimulated Raman scattering (SRS) microscopy [8], infrared microscopy [9], microscopy with UV-surface excitation (MUSE) [10] have been used for in-vivo imaging to visualize oval tissue and cell morphology in various organs. The resolution of these systems is still limited by diffraction, ranging from ~300 nm to a few microns. They either detect intrinsic tissue properties (e.g., refractive index variation, NADH/FAD, birefringence, intrinsic Raman scattering or absorption signals of macromolecules), or use exogeneous labels to enhance the detection of overall tissue architecture.
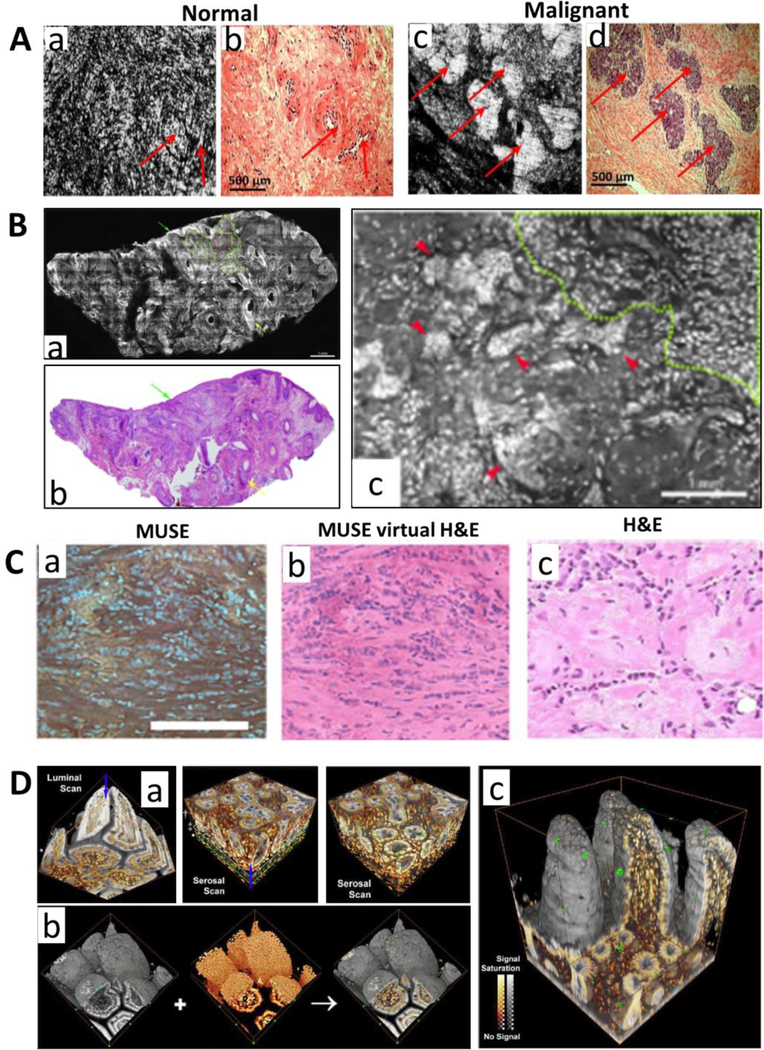
One general goal is for fast visualization of overall tissue morphology with histology-like microscopic images on either freshly resected ex-vivo tissue or without physically removing the tissue. Such capability is very useful to determine which abnormal lesions should be removed, or whether tumors are completely removed at the margin in real time. As tissue is optically thick and scattering, visualization of microscopic structures requires the imaging volume to be confined within several microns. Physical tissue sectioning is currently used in clinical setting, which requires the time-consuming frozen tissue that takes 20–30 minutes or paraffin-embedding process that takes ~10 hours. As many optical microscopy techniques can perform non-invasive optical sectioning, they are ideally suited for 3D microscopic imaging of in-vivo and ex-vivo tissue in real time. For example, confocal reflectance microscopy and optical coherence microscopy have been successfully used for in-vivo imaging of skin [11], ovary [6] and other organs, which detect the intrinsic scattering signals due to the changes in the refractive index of subcellular organelles, and provide label-free high-resolution images that resemble the H&E-stained histology images. An example using optical coherence microscopy is shown in Fig. 1A. In-vivo confocal reflectance microscopy has been cleared by Food and Drug Administration (FDA) for dermatologists to noninvasively visualize cellular structures within the skin. But certain sub-cellular details, especially nuclear morphology, can be difficult to assess from the images without any stains. To overcome this limitation, fluorescence dyes (fluorescein or acridine orange) have been used to enhance the contrast of cell nuclei or overall tissue architecture in the ex-vivo tissue [12, 13] (an example shown in Fig. 1B), but in-vivo tissue imaging using fluorescent dyes is still limited by the very few available fluorescent dyes approved by FDA for safe use in human. Raman scattering based label-free imaging technique also showed the promise for tumor margin assessment with close resemblance to H&E-stained histology images with clear delineation of nuclear details and extracellular matrix in brain tumor [8]. As conventional wide-field microscopy generally lacks 3D sectioning, most techniques discussed above use confocal point-scanning system to reject the out-of-focus background in tissue imaging, which leads to the increased cost and complexity of the imaging system. A nice deviation from these approaches is the recently reported MUSE that uses ultraviolet radiation to naturally “section” the tissue surface due to the short penetration depth of UV has achieved excellent histology-like microscopic images on ex-vivo tissue, using a simple and low-cost wide-field fluorescence microscope [10], as shown in Fig. 1C. Another exception is light-sheet microscopy that uses two orthogonally positioned beams to achieve optical sectioning, which significantly improved the throughput and imaging speed [4].
Figure 1.
(A) Full-field OCT images of (a) normal and (c) malignant ovary; corresponding histology images are shown in (b,d). The matching areas are indicated by red arrows. (B) The acridine orange stained confocal fluorescence microscopy images of basal cell carcinoma. The infiltrative BCC can be readily detected on a wide-field fluorescent confocal mosaic image (b) that correlates with H&E-stained image from a frozen tissue section (a). (c) A zoomed image of (b) that shows nuclear morphology. (C) The wide-field fluorescence image obtained by MUSE (a), converted virtual H&E image (b) and paired FFPE conventional histology (c). (D) (a) Stereo projection of confocal microscopy images of optically cleared mouse ileum with DiD-stained membranes (gray) and PI-stained nuclei (orange). (b) Stereo projection of the confocal “luminal scan” and “serosal scan” for the ileum. Arrows indicate the scan directions. (c) A full-depth, 3D projection of the ileum. Figures A-D are modified with permission from the following sources: [6, 63, 10, 19], respectively.
Besides structural information, some intrinsic optical signals can provide functional information in pathogenesis. For example, NADH and FAD are important indicators for metabolic activities and have been used to monitor the drug resistance in cancer models such as xenograft model [14] and tissue organoid [15], and to image mitochondrial dynamics to improve the in-vivo diagnosis of skin cancer in human patients [16]. The birefringence properties in the extracellular matrix have also shown promising prognostic value in patients with ovarian cancer [17]. Another label-free nonlinear imaging technique provides in-vivo visualization of tumor microenvironment and outline overall tissue morphology that also demonstrate potential diagnostic values in tumor margin detection [5]. Confocal infrared microscopy that uses infrared spectroscopic imaging was also reported to biochemically characterize breast cancer tissue in both epithelial cells and tumor microenvironment [9].
As tissue structure is three-dimensional in nature, a full 3D histopathology mapping of the entire organ or tumor will be valuable for both basic research and pathological diagnosis. But the thickness of these tissues ranges from a few millimeters to several centimeters, which is well beyond the penetration depth (about a few hundred microns) of light microscopy. To achieve the full volumetric 3D high-resolution imaging for a large tissue sample, two approaches are generally used. The first approach uses conventional microscopy technique with physical sectioning, such as knife-edge scanning microscopy [18]; the second approach uses tissue clearing to increase penetration depth, followed by 3D imaging (e.g., confocal or light-sheet microscopy) [4, 19, 20] and an example is shown in Fig. 1D. Both approaches have been widely used in brain imaging, and recently adapted for 3D imaging of pathological tissue. Both chemical processing of tissue and 3D imaging take a long time (e.g., hours to days). This approach shifts the traditional paradigm of 2D pathology of a small region of the resected tissue and opens new avenues of 3D full volumetric histology maps with sub-cellular resolution. Whether sampling the large tissue and 3D pathology provide additional diagnostic value than conventional 2D pathology remain to be proven in the clinical settings.
Super-resolution fluorescence microscopy for imaging molecular structure at nanometer resolution
Super-resolution (SR) fluorescence microscopy has revolutionized biological imaging by overcoming the physical diffraction limit to achieve a spatial resolution down to ~20–100 nm, thereby allowing the observation of subcellular structures that are invisible in conventional light microscopy. Such breakthrough has been recognized by Nobel Prize in 2014. Various SR fluorescence microscopy techniques such as stochastic optical reconstruction microscopy [STORM] [21, 22], (fluorescence) photo-activated localization microscopy [(f)PALM] [23, 24], stimulated emission depletion microscopy (STED) [25], super-resolution structured illumination microscopy (SR-SIM) [26] and recently developed expansion microscopy [27, 28], can now be used to detect molecular structures below the diffraction- limited resolution.
The rationale of using super-resolution imaging system in understanding cancer pathobiology and pathological diagnosis is motivated by our increased understanding of molecular changes in cancer development. It is now established that disrupted genome and epigenome that can alter nuclear architecture and other sub-cellular organelles in all stages of cancer development including in normal precursor cells [29–31]. Correspondingly, the structural transformation of cellular and sub-cellular organelles experiences progressive alterations, ranging from subtle to significant. But conventional biochemistry approaches are largely bulk measurement on the averaged cell population from tissue extract, which lose the spatial context – an important component for pathological diagnosis and pathogenesis of cancer.
The limitations of conventional morphology discussed above are in a large part due to the diffraction-limited resolution of conventional light microscopy. Smaller structural characteristics below 250–500 nm cannot be visualized. Indeed, the currently known abnormal morphology characteristic of neoplastic cells is mostly microscale structural features, such as abnormal tissue architecture, chromatin texture, nuclear size, shape, nuclei-to-cytoplasm ratio, orientation of cells, the prominence of nucleoli, and ploidy. Super-resolution microscopy combines nanometer resolution with molecular specificity, which has a potential as powerful tools to address challenging problems in cancer pathobiology.
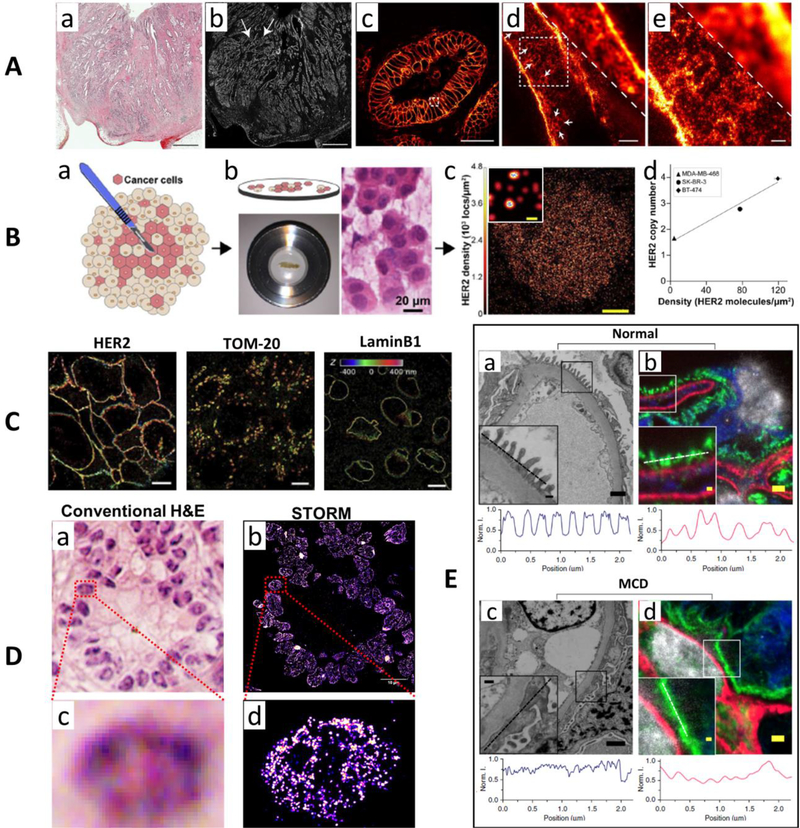
Attempts have been made for imaging pathological tissue using super-resolution techniques, even though it is still at its infancy, with only a handful of studies. For example, 3D-SIM has a resolution of ~120 nm and was used to resolves organizational differences in DNA organization of myeloma cells among normal, multiple myeloma (MM) cells and its precursor cells of monoclonal gammopathy of undetermined significance (MGUS) [32]. The STED-based super-resolution microscopy achieves a resolution of ~50–70 nm and has also been used to visualize nanoscale protein distributions in sections of well-annotated FFPE human rectal cancer tissue [33] (an example shown in Fig. 2A). Single-molecule localization-based approach – PALM, requires the incorporation of photoactivable fluorescent proteins using genetic engineering approaches, it has limited clinical utility in fixed pathological samples. While STORM uses conventional organic dyes and well-established immunofluorescence staining that is highly compatible with pathological tissue and routinely achieves a spatial resolution of ~20–30 nm, one of the best among various super-resolution imaging techniques. It has also been used to image various molecular structures of HER2, TOM20 and lamin B1 in clinical FFPE tissue sample from breast cancer [34] (an example shown in Fig. 2C). STORM was also used with quantitative single-molecule analysis and clinical touch repreparation to precisely quantify HER2 density from patients’ samples that showed a positive correlation with standard FISH-based assay of HER2 copy number [35] (an example shown in Fig. 2B). Further, as tissue section often induces higher background, to ensure robust performance in imaging nuclear architecture, various methods have been developed to remove the heterogeneous background and improve the throughput [36, 37]. Such method has allowed one-to-one mapping between conventional H&E-stained image and STORM-based super-resolution image of chromatin structure – one of the most important sub-cellular components in carcinogenesis, as shown in Fig. 2D, allowing pathological evaluation at molecular scale [38]. In addition, expansion microscopy, distinct from optics- based super-resolution imaging approaches, expands the samples to resolve ultra-structure that is not visible under conventional light microscopy. It has been optimized for clinical samples and demonstrated the potential to detect the kidney tertiary podocyte foot processes in the kidney disease, a feature that was traditionally detected by electron microscopy, as well as the feasibility to improve computation-based diagnosis of breast precursor lesions [39] (an example shown in Fig. 2E).
Figure 2.
(A) Comparison of H&E-stained (a) and the diffraction-limited immunofluorescence-stained confocal microscopy (b) images over a large area of a HER2-positive human rectal cancer tissue. (c) The magnified image of (b) from the area marked with white arrows. (d-e) The comparison of diffraction-limited confocal microscopy with STED super-resolution microscopy images. (B) (a) The workflow for Touch-prep samples for STORM-based super-resolution imaging. (b) Quantitative single-molecule localization microscopy is performed on the excised tumor tissue placed inside an imaging chamber, and H&E stained image of a touch prep sample. (c) STORM image of a HER2-positive cell in patient tissue. (d) The correlation between HER2 copy number from FISH and the average density of HER2 molecules from quantitative analysis of STORM image from three cell lines. (C) 3D-STORM images of HER2, TOM-20 and lamin-B1 on HER2+ tumor FFPE sections. Axial positions are color coded. (D) Comparison of H&E-stained image of human pancreatic tissue (a, c) and corresponding STORM-based super-resolution images (b, d) of facultative heterochromatin structure marked by H3K27me3. (E) Electron microscopy (a, c) and corresponding fluorescence microscopy images after expansion (b, d) on a clinical biopsy sample from a normal human kidney and a patient with a patient with minimal change disease (MCD). The intensity profile along the line cut of the inset is shown below each microscopic image. The expansion microscopy image clearly distinguished the features that are normally detected by electron microscopy. Figures A, B, C and E are modified with permission from the following sources: [33, 35, 34, 39], respectively. Figure D is the previously unpublished data from the authors of this paper.
Super-resolution fluorescence microscopy is far less widely used in the field of pathology than cell biology. The concept of visualizing ultra-structures to improve the pathological diagnosis was well explored in 1970s when electron microscopy first became widely available and became the standard of care in some aspects of renal pathology. But it fell out of favor after a decade due to the emergence of molecular-based approaches such as immunohistochemistry (IHC) resolved ability of the new generation of super-resolution fluorescence microscopy marries the molecular specificity with super-resolved imaging capability, but with similar sample preparation as standard IHC, but at a much higher imaging throughput and cheaper instrument [40] that are not available in electron microscopy. Further, super-resolution fluorescence microscopy is far more powerful, with single-molecule detection sensitivity and the ability to precisely quantify the multiple molecular compositions [35]. Therefore, super-resolution fluorescence microscopy is poised to make a significant impact in the largely unexplored nanoscale regime of molecular structure in pathological evaluation of various diseases and may potentially be used to address many challenges in clinical diagnosis as previously discussed.
Other label-free imaging techniques for interrogation of nanoscale structure
Many label-free imaging techniques, such as autofluorescence/Raman spectroscopy [41, 42], quantitative phase imaging [43], partial-wave spectroscopy [44], light scattering spectroscopy [45], angle-resolved low coherence interferometry (aLCI) [46], spectral encoding of spatial frequency [47], nanoscale nuclear architectural mapping (nanoNAM) [48] also emerge as promising technologies to detect pre-cancerous and cancerous changes in pathological tissue or cells, especially their ability to interrogate the sub-resolution structural abnormalities in cells undergoing early-stage carcinogenesis that are invisible to conventional pathology. Unlike the fluorescence-based imaging approaches that characterize the spatial distribution of the fluorophore, these techniques do not require exogenous labels and quantify the intrinsic structural information, based on scattered light, at various length scales ranging from nanometer to micron.
Light scattering is an intrinsic optical signal from refractive index variation in the cellular and sub-cellular organelles and forms the basis of many label-free imaging techniques. They can quantify the intrinsic architecture at the single-cell level with nanoscale sensitivity. They achieved nanoscale sensitivity through three general approaches: interferometry that detects nanometer optical path length difference, model-based interpretation of scattering signals, and statistical properties of refractive index fluctuation. It is important to note that the nanoscale sensitivity in the label-free imaging is not equivalent to the nanoscale resolution in super-resolution microscopy. In the former case, the image is still diffraction-limited, but the value distinguishes the nanoscale structural difference in a structural parameter (e.g., size, optical path length, refractive index); in the latter case, the image has super-resolved resolution and directly visualizes nanoscale structural features.
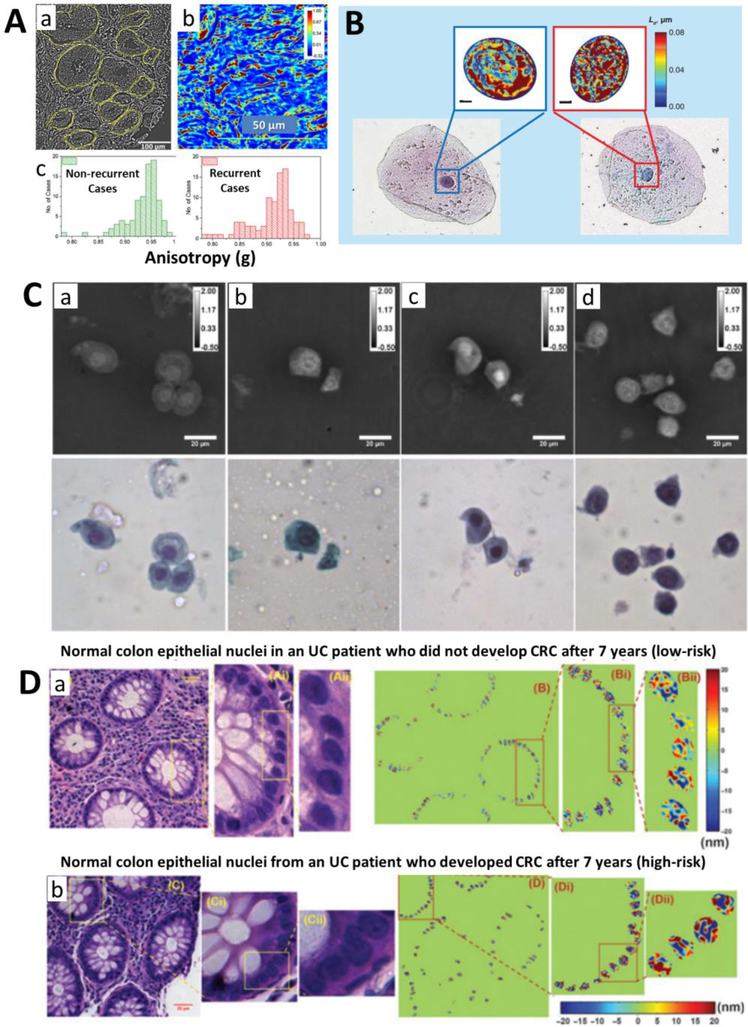
Although label-free imaging techniques lack the specificity to directly visualize the molecular structure at nanoscale resolution (as is the case for super-resolution fluorescence microscopy), subtle nanoscale structural changes that appear invisible to conventional light microscopy can still be detected using simple and low-cost optical set-up with only minimal sample preparation. For example, in early-stage cancer development when the cells still appear normal to pathologists, chromatin structure and associated refractive index distribution in the cell nuclei is already altered due to epigenetic dysregulation [44, 49]. It results in changes in the phase and spatial distribution of light scattering signals that can be picked up by these scattering-based label-free techniques. For example, quantitative phase imaging based on various approaches (e.g., diffraction phase microscopy, digital holographic microscopy) maps the integrated optical pathlength distribution along the tissue section and provides a high-contrast high-resolution images of tissue architecture for unstained tissue. Such information has shown the promise to predict the prostate cancer recurrence [50] (as shown in Fig. 3A) and quantify inflammation [51]. Quantitative phase imaging and its derived dry mass of the cell nuclei showed the potential to improve the cytological diagnosis of urine cytology [52], as shown in Fig. 3C. Partial-wave spectroscopy detects the refractive index fluctuation due to structural heterogeneity and has shown promising clinical data to detecting high-risk lung cancer patients from normal buccal cells based on the field cancerization (an example shown in Fig. 3B) [53, 54]. The aLCI combines low-coherence interferometry with angle-resolved light scattering spectroscopy to quantify the structural information at a depth-resolved manner, and can detect esophageal dysplasia at a high level of sensitivity [55]. In addition, nanoNAM detects the depth-resolved Fourier phase in a configure of spectral-domain optical coherence microscopy [56, 57] has shown the promise to predict the risk of developing colorectal cancer in normal-appearing tissue from patients with inflammatory bowel disease [48, 38], as shown in Fig. 3D. Although much remains to be learned about the underlying molecular mechanisms that drive the minute structural changes in the cells, the ability to accurately quantify the intrinsic structural changes in the cells could be used for many clinical applications.
Figure 3.
(A) The optical anisotropy image (a) calculated based on quantitative phase imaging of a prostate tissue, and the quantitative phase image (b) of a stromal tissue region in the prostate. (c) The histograms distribution of anisotropy values for 89 non-recurrent and 92 recurrent cases. (B) Bright-field microscope images and the corresponding disorder strength maps of the nuclei obtained from partial-wave spectroscopy in histologically normal buccal cells from a healthy patient (left) and a patient with lung cancer (right). (C) The quantitative phase images of unstained urothelial cells and corresponding pap-stained cytology images from patients with a cytologic diagnosis of (a) negative, (b) atypical, (c) suspicious, and (d) positive for urothelial carcinoma. The color bars are in radians. (D) The histology image and corresponding nanoNAM on normal colonic epithelial cells from a patient with ulcerative colitis (UC) who did not develop HGD or colorectal cancer (CRC) after 7 years of follow-up (a, low-risk) and from a UC patient who developed colon adenocarcinoma after 7 years (b, high-risk). Figures A-D are modified with permission from the following sources: [50, 54, 52, 48], respectively.
Discussion and Conclusion
As many microscopy techniques can now visualize or detect molecular-scale structure below 100 nm, the tissue processing methods optimized for conventional microscopy may not meet the demand in the nanoscale regime. For example, fixation may introduce artifacts that are invisible under conventional light microscopy but become obvious with super-resolution microscopy [58–60]. The labeling density that is routinely used for confocal fluorescence microscopy is often insufficient for super-resolution microscopy [61]. As some imaging techniques move towards the clinics, the impact of tissue processing and various pre-analytical variables (e.g., duration of fixation, cold ischemic time) must be carefully assessed based on the established clinical workflow to establish a standardized operating protocol, before large-scale clinical studies. Clinical adoption of such standardized tissue preparation is often driven by clinical benefit of the new technology. For example, due to the strong clinical need to assess the expression level of estrogen/progesterone receptors and HER2, a standardized duration of tissue fixation optimized for IHC has been established in the guidelines of American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) [62].
The revolutionary advancement in the field of light microscopy in the past decade has provided pathologists and biomedical researchers a wide range of powerful imaging tools from 3D volumetric high-resolution of tissue architecture to molecular-level nanometer resolution, to study cancer pathobiology in both breadth and depth. Such fast advances in imaging techniques also generate a huge amount of data that is difficult to process for important diagnostic features with conventional assessment by pathologists. Machine learning has become an essential component in processing the large amount of imaging data and quantitative diagnostic features can be quickly extracted. The molecular-level nanoscale structural features, functional information and 3D high-resolution tissue architecture features characteristic of various stages of tumorigenesis may soon be incorporated to improve our assessment of malignant potential for pre-cancerous lesions or determine the best therapy in cancer diagnosis and guiding personalized treatment. Although clinical adoption of any new technique always takes a long time, the new insights gained from these imaging tools will open new avenues for improving cancer diagnosis, risk stratification and facilitating the development and evaluation of new preventive and therapeutic strategies.
Acknowledgements
We acknowledge the funding support from National Institute of Health Grant Number R01CA185363 and R33CA225494. Due to the large body of literatures, we cannot cover all of the related topics and publications. We acknowledge Dr. Hongbin Ma for preparing Figure 2D. We apologize to researchers whose work is missed in this review.
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest
Jianquan Xu declares no conflict of interest. Yang Liu is the co-inventor for several US patents on light microscopy techniques to analyze nanoscale nuclear architecture for cancer diagnosis and other applications, owned by University of Pittsburgh.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Vennalaganti P, Kanakadandi V, Goldblum JR, Mathur SC, Patil DT, Offerhaus GJ et al. Discordance Among Pathologists in the United States and Europe in Diagnosis of Low-Grade Dysplasia for Patients With Barrett’s Esophagus. Gastroenterology. 2017;152(3):564–70 e4. doi: 10.1053/j.gastro.2016.10.041. [DOI] [PubMed] [Google Scholar]
- 2.Baker M Building better biobanks. Nature. 2012;486:141–6. doi: 10.1038/486141a. [DOI] [PubMed] [Google Scholar]
- 3.Kose K, Gou M, Yelamos O, Cordova M, Rossi AM, Nehal KS et al. Automated video-mosaicking approach for confocal microscopic imaging in vivo: an approach to address challenges in imaging living tissue and extend field of view. Sci Rep. 2017;7(1):10759. doi: 10.1038/s41598-017-11072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glaser AK, Reder NP, Chen Y, McCarty EF, Yin C, Wei L et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nature Biomedical Engineering. 2017;1:0084. doi: 10.1038/s41551-017-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, You S, Tu H, Spillman DR Jr., Chaney EJ, Marjanovic M et al. Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci Adv. 2018;4(12):eaau5603. doi: 10.1126/sciadv.aau5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nandy S, Sanders M, Zhu Q. Classification and analysis of human ovarian tissue using full field optical coherence tomography. Biomed Opt Express. 2016;7(12):5182–7. doi: 10.1364/BOE.7.005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlichenmeyer TC, Wang M, Elfer KN, Brown JQ. Video-rate structured illumination microscopy for high-throughput imaging of large tissue areas. Biomed Opt Express. 2014;5(2):366–77. doi: 10.1364/BOE.5.000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med. 2013;5(201):201ra119. doi: 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal S, Yeh K, Leslie LS, Kenkel S, Kajdacsy-Balla A, Bhargava R. Simultaneous cancer and tumor microenvironment subtyping using confocal infrared microscopy for all-digital molecular histopathology. Proc Natl Acad Sci U S A. 2018;115(25):E5651–E60. doi: 10.1073/pnas.1719551115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fereidouni F, Harmany ZT, Tian M, Todd A, Kintner JA, McPherson JD et al. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat Biomed Eng. 2017;1:957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajadhyaksha M, Marghoob A, Rossi A, Halpern AC, Nehal KS. Reflectance confocal microscopy of skin in vivo: From bench to bedside. Lasers Surg Med. 2017;49(1):7–19. doi: 10.1002/lsm.22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karen JK, Gareau DS, Dusza SW, Tudisco M, Rajadhyaksha M, Nehal KS. Detection of basal cell carcinomas in Mohs excisions with fluorescence confocal mosaicing microscopy. Br J Dermatol. 2009; 160(6): 1242–50. doi: 10.1111/j.1365-2133.2009.09141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragazzi M, Piana S, Longo C, Castagnetti F, Foroni M, Ferrari G et al. Fluorescence confocal microscopy for pathologists. Mod Pathol. 2014;27(3):460–71. doi: 10.1038/modpathol.2013.158. [DOI] [PubMed] [Google Scholar]
- 14.Walsh AJ, Cook RS, Manning HC, Hicks DJ, Lafontant A, Arteaga CL et al. Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res. 2013;73(20):6164–74. doi: 10.1158/0008-5472.CAN-13-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh AJ, Cook RS, Sanders ME, Aurisicchio L, Ciliberto G, Arteaga CL et al. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 2014;74(18):5184–94. doi: 10.1158/0008-5472.CAN-14-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pouli D, Balu M, Alonzo CA, Liu Z, Quinn KP, Rius-Diaz F et al. Imaging mitochondrial dynamics in human skin reveals depth-dependent hypoxia and malignant potential for diagnosis. Sci Transl Med. 2016;8(367):367ra169. doi: 10.1126/scitranslmed.aag2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tilbury KB, Campbell KR, Eliceiri KW, Salih SM, Patankar M, Campagnola PJ. Stromal alterations in ovarian cancers via wavelength dependent Second Harmonic Generation microscopy and optical scattering. BMC Cancer. 2017;17(1):102. doi: 10.1186/s12885-017-3090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayerich D, Abbott L, McCormick B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J Microsc. 2008;231(Pt 1):134–43. doi: 10.1111/j.1365-2818.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- 19.Fu YY, Lin CW, Enikolopov G, Sibley E, Chiang AS, Tang SC. Microtome-free 3-dimensional confocal imaging method for visualization of mouse intestine with subcellular-level resolution. Gastroenterology. 2009;137(2):453–65. doi: 10.1053/j.gastro.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nojima S, Susaki EA, Yoshida K, Takemoto H, Tsujimura N, Iijima S et al. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci Rep. 2017;7(1):9269. doi: 10.1038/s41598-017-09117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods. 2006;3:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172–6. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 23.Hess ST, Girirajan TPK, Mason MD. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophysical Journal. 2006;91:4258–72. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS et al. Imaging Proteins Intracellular at Nanometer Fluorescent Resolution. Science. 2006;313:1642–5. [DOI] [PubMed] [Google Scholar]
- 25.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci U S A. 2000;97:8206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–70. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chozinski TJ, Halpern AR, Okawa H, Kim HJ, Tremel GJ, Wong ROL et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nature Methods. 2016;13:485–8. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nature reviews Genetics. 2016;17:284–99. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feinberg AP. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N Engl J Med. 2018;378(14):1323–34. doi: 10.1056/NEJMra1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nature reviews Genetics. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 32.Sathitruangsak C, Righolt CH, Klewes L, Tammur P, Ilus T, Tamm A et al. Quantitative superresolution microscopy reveals differences in nuclear DNA organization of multiple myeloma and monoclonal gammopathy of undetermined significance. J Cell Biochem. 2015;116(5):704–10. doi: 10.1002/jcb.25030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ilgen P, Stoldt S, Conradi LC, Wurm CA, Ruschoff J, Ghadimi BM et al. STED Super-Resolution Microscopy of Clinical Paraffin-Embedded Human Rectal Cancer Tissue. PLoS One. 2014;9:e101563. doi: 10.1371/journal.pone.0101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creech MK, Wang J, Nan X, Gibbs SL. Superresolution Imaging of Clinical Formalin Fixed Paraffin Embedded Breast Cancer with Single Molecule Localization Microscopy. Scientific reports. 2016;7:40766. doi: 10.1038/srep40766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobin SJ, Wakefield DL, Jones V, Liu X, Schmolze D, Jovanovic-Talisman T. Single molecule localization microscopy coupled with touch preparation for the quantification of trastuzumab-bound HER2. Sci Rep. 2018;8(1):15154. doi: 10.1038/s41598-018-33225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma H, Jiang W, Xu J, Liu Y. Enhanced super-resolution microscopy by extreme value based emitter recovery. bioRxiv. 2018:295261. doi: 10.1101/295261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H, Xu J, Liu Y. WindSTORM: Robust online image processing for high-throughput nanoscopy. Sci Adv. 2019;5:eaaw0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Ma H, Ma H, Jiang W, Duan M, Zhao S et al. Super-resolution imaging reveals the evolution of higher-order chromatin folding in early carcinogenesis. bioRxiv. 2019:672105. doi: 10.1101/672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Bucur O, Irshad H, Chen F, Weins A, Stancu AL et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nature Biotechnology. 2017;35(8):757–64. doi: 10.1038/nbt.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma H, Fu R, Xu J, Liu Y. A simple and cost-effective setup for super-resolution localization microscopy. Scientific Reports. 2017;7:1542. doi: 10.1038/s41598-017-01606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paidi SK, Diaz PM, Dadgar S, Jenkins SV, Quick CM, Griffin RJ et al. Label-Free Raman Spectroscopy Reveals Signatures of Radiation Resistance in the Tumor Microenvironment. Cancer Res. 2019;79(8):2054–64. doi: 10.1158/0008-5472.CAN-18-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgakoudi I, Jacobson BC, Van Dam J, Backman V, Wallace MB, Muller MG et al. Fluorescence, reflectance, and light-scattering spectroscopy for evaluating dysplasia in patients with Barrett’s esophagus. Gastroenterology. 2001;120(7):1620–9. [DOI] [PubMed] [Google Scholar]
- 43.Park Y, Depeursinge C, Popescu G. Quantitative phase imaging in biomedicine. Nature Photonics. 2018;12(10):578–89. doi: 10.1038/s41566-018-0253-x. [DOI] [Google Scholar]
- 44.Subramanian H, Pradhan P, Liu Y, Capoglu IR, Li X, Rogers JD et al. Optical methodology for detecting histologically unapparent nanoscale consequences of genetic alterations in biological cells. Proc Natl Acad Sci U S A. 2008;105:20118–23. doi: 10.1073/pnas.0804723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itzkan I, Qiu L, Fang H, Zaman MM, Vitkin E, Ghiran IC et al. Confocal light absorption and scattering spectroscopic microscopy monitors organelles in live cells with no exogenous labels. Proc Natl Acad Sci U S A. 2007;104:17255–60. doi: 10.1073/pnas.0708669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho D, Drake TK, Smith-McCune KK, Darragh TM, Hwang LY, Wax A. Feasibility of clinical detection of cervical dysplasia using angle-resolved low coherence interferometry measurements of depth-resolved nuclear morphology. Int J Cancer. 2017;140(6):1447–56. doi: 10.1002/ijc.30539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexandrov SA, Uttam S, Bista RK, Staton KD, Liu Y. Spectral encoding of spatial frequency approach for characterization of nanoscale structures. Applied Physics Letters. 2012;101:33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uttam S, Pham HV, LaFace J, Leibowitz B, Yu J, Brand RE et al. Early Prediction of Cancer Progression by Depth-Resolved Nanoscale Mapping of Nuclear Architecture from Unstained Tissue Specimens. Cancer Research. 2015;75:4718–27. doi: 10.1158/0008-5472.CAN-15-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherkezyan L, Stypula-Cyrus Y, Subramanian H, White C, Dela Cruz M, Wali RK et al. Nanoscale changes in chromatin organization represent the initial steps of tumorigenesis: a transmission electron microscopy study. BMC Cancer. 2014;14:189. doi: 10.1186/1471-2407-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sridharan S, Macias V, Tangella K, Kajdacsy-Balla A, Popescu G. Prediction of Prostate Cancer Recurrence Using Quantitative Phase Imaging. Scientific Reports. 2015;5:9976. doi: 10.1038/srep09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenz P, Bettenworth D, Krausewitz P, Bruckner M, Ketelhut S, von Bally G et al. Digital holographic microscopy quantifies the degree of inflammation in experimental colitis. Integr Biol (Camb). 2013;5(3):624–30. doi: 10.1039/c2ib20227a. [DOI] [PubMed] [Google Scholar]
- 52.Pham HV, Pantanowitz L, Liu Y. Quantitative phase imaging to improve the diagnostic accuracy of urine cytology. Cancer Cytopathol. 2016;124(9):641–50. doi: 10.1002/cncy.21734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian H, Roy HK, Pradhan P, Goldberg MJ, Muldoon J, Brand RE et al. Nanoscale Cellular Changes in Field Carcinogenesis Detected by Partial Wave Spectroscopy. Cancer Res. 2009;69:5357–63. doi:Doi 10.1158/0008-5472.Can-08-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauer GM, Stypula-Cyrus Y, Subramanian H, Cherkezyan L, Viswanathan P, Zhang D et al. The transformation of the nuclear nanoarchitecture in human field carcinogenesis. Future Sci OA. 2017;3(3):FSO206. doi: 10.4155/fsoa-2017-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wax A, Terry NG, Dellon ES, Shaheen NJ. Angle-resolved low coherence interferometry for detection of dysplasia in Barrett’s esophagus. Gastroenterology. 2011;141(2):443–7, 7 e1–2. doi: 10.1053/j.gastro.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uttam S, Liu Y. Fourier phase based depth-resolved nanoscale nuclear architecture mapping for cancer detection. Methods. 2018;136:134–51. doi: 10.1016/j.ymeth.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uttam S, Liu Y. Fourier phase in Fourier-domain optical coherence tomography. J Opt Soc Am A Opt Image Sci Vis. 2015;32:2286–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stanly TA, Fritzsche M, Banerji S, Garcia E, Bernardino de la Serna J, Jackson DG et al. Critical importance of appropriate fixation conditions for faithful imaging of receptor microclusters. Biol Open. 2016;5(9):1343–50. doi: 10.1242/bio.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter KN, Revelo NH, Seitz KJ, Helm MS, Sarkar D, Saleeb RS et al. Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy. The EMBO Journal. 2017;37:e201695709. doi: 10.15252/embj.201695709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerr E, Kiyuna T, Boyle S, Saito A, Thomas JS, Bickmore WA. Changes in chromatin structure during processing of wax-embedded tissue sections. Chromosome Res. 2010;18:677–88. doi: 10.1007/s10577-010-9147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu J, Ma H, Liu Y. Stochastic optical reconstruction microscopy (STORM). Current Protocols in Cytometry. 2017;2017:12.46.1–12.46.27. doi: 10.1002/cpcy.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010;134(7):e48–72. doi: 10.1043/1543-2165-134.7.e48. [DOI] [PubMed] [Google Scholar]
- 63.Jain M, Rajadhyaksha M, Nehal K. Implementation of fluorescence confocal mosaicking microscopy by “early adopter” Mohs surgeons and dermatologists: recent progress. J Biomed Opt. 2017;22(2):24002. doi: 10.1117/1.JBO.22.2.024002. [DOI] [PMC free article] [PubMed] [Google Scholar]