Abstract
Unsafe drinking water leads to millions of human deaths each year, while contaminated wastewater discharges are a significant threat to aquatic life. To relieve the burden of unsafe water, we are in search of an inexpensive material that can adsorb pathogenic viruses from drinking water and adsorb toxic residual chlorine from wastewater. To impart virus and chlorine removal abilities to cellulosic materials, we modified the primary hydroxyl group with a positively charged guanidine group, to yield guanidine modified cellulose derivatives. Microcrystalline cellulose (MC) bearing covalently bonded guanidine hydrochloride (MC-GC) and hydrogen-bonded guanidine hydrochloride (MC-GH) were synthesized, and electrospun into nanofibers after blending with the non-ionogenic polyvinyl alcohol (PVA), to produce large pore sized, high surface area membranes. The MC-GC/PVA and MC-GH/PVA nanofibers were stabilized against water dissolution by crosslinking with glutaraldehyde vapor. The water-stable MC-GC/PVA mats were able to remove more than 4 logs of non-enveloped porcine parvovirus (PPV) and enveloped Sindbis virus and reached 58% of chlorine removal. The MC-GC/PVA nanofibers demonstrated better performance for pathogen removal and dechlorination than MC-GH/PVA nanofibers. This first study of MC-GC/PVA electrospun mats for virus removal shows they are highly effective and merit additional research for virus removal.
Keywords: Nanofibrous microfiltration, Water purification, Electrostatic interaction, Electrospinning, Guanidine cation, Polysaccharide
Graphical abstract
(Virus and chemical structure images were created in BioRender.).
Highlights
-
•
Synthetic routes to covalent and H-bonded guanidine on cellulose are given.
-
•
Guanidine functionalized cellulose derivatives were electrospun into nanofibers.
-
•
Glutaraldehyde crosslinking imparted water stability to the electrospun fibers.
-
•
Over 4 log removal value was achieved for an enveloped and non-enveloped virus.
-
•
Approximately 60% of chlorine removal was achieved.
1. Introduction
The World Health Organization (WHO) estimates at least 30% (2 billion people in 2019) of the global population rely on drinking water contaminated with feces [1]. Each year this contaminated water causes millions of human deaths [1]. The major contaminants include pathogenic parasites, bacteria, and viruses. The most common disease-causing waterborne viruses include norovirus, adenovirus, enterovirus, and Hepatitis A and E viruses [2], all of which are non-enveloped viruses. Enveloped viruses possess an additional lipid envelope, and are less stable in water, so they rarely cause waterborne diseases. However, the presence of enveloped severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for the current coronavirus pandemic (i.e., COVID-19), has been detected in municipal wastewater across the globe [[3], [4], [5]]. The occurrence of SARS-CoV-2 in water and in the feces of COVID-19 patients [6,7] have caused concerns for its potential for waterborne transmission, since enveloped coronaviruses can remain infectious for days or even longer in drinking water and wastewater [8].
Currently, municipal systems purify water using a multistep process of coagulation and flocculation, sedimentation, chemical disinfection, and physical membrane filtration [9]. Membrane filtration is an accepted technology for the removal of pathogens, like bacteria and parasites, from drinking water. Pathogens like these are easily removed from drinking water using large pore-sized microfiltration and ultrafiltration membranes at low operating pressure [10]. However, viruses are too small to be removed by a size-exclusion mechanism without a large energy penalty. Advanced systems of membrane filtration, like nanofiltration and reverse osmosis, are able to remove viruses, but require high operating pressures, and also the membranes are easily fouled [10].
Viruses can also be reduced by chemical disinfection. Disinfection is widely used for both drinking water and municipal wastewater treatment [11]. The most common disinfection process is chlorination, which uses compressed chlorine gas or compounds such as sodium hypochlorite and calcium hypochlorite to inactivate the pathogens. When dissolved in water, they can form hypochlorous acid (HClO) and hypochlorite ion (ClO−). The HClO and ClO− can react with natural organic matter to form trihalomethanes and other halogenated disinfection by-products [12,13], which are suspected of being human carcinogens [12] and are toxic to aquatic life [13]. The residual chlorine (1 mg/L) in the treated wastewater is acutely toxic to aquatic organisms [14], while a chlorine level of 5–15 mg/L is typically used for the disinfection of wastewater [11]. Therefore, the Environmental Protection Agency (EPA) requires a dechlorination step to remove free and combined chlorine remaining in the purified wastewater before it is discharged [11]. This is typically accomplished using reducing agents such as sulfur dioxide, sulfite salts, or thiosulfate, which also reduce dissolved oxygen [15]. Activated carbon is also effective in reducing the level of residual chlorine, but it is costly [15].
To help relieve the burden of unsafe water, we are in search of an inexpensive adsorption material that can remove viruses from drinking water, and chlorine from wastewater. The material would ideally form a microfiltration membrane that can clean water with low back pressure and retain high water flux. Cellulosic membranes are of great interest because cellulose is the most abundant natural organic polymer on Earth [16,17], and is non-toxic, biocompatible, biodegradable, biofunctional, and hydrophilic [18]. Cellulose is a polysaccharide containing a linear chain of repeating d-anhydroglucopyranose ring units that are linked by a covalent β-1,4-glycosidic bond [17,18]. For each monomer unit, the reactivity of the primary hydroxyl (-OH) group (-OH on carbon 6) is ten times faster than the secondary –OH groups (-OH on carbon 2 and carbon 3) [18]. Thus, the primary –OH group can be reacted to form a variety of cellulose derivatives to enhance desired properties. Cellulose derivatives have been widely used as materials in the field of water purification, wound healing, drug delivery, food, plastic, and textiles [[16], [17], [18], [19], [20]]. Among the many possible cellulose derivatives, cellulose modified with positively charged groups is the most interesting, due to the excellent antimicrobial and antiviral properties of cationic polymers [19,21].
Among the cationic polymer modified cellulose, cellulose grafted with guanidine groups has shown high activity against bacteria [19]. Cellulose films having 1 wt% of polyhexamethylene guanidine dodecyl benzene, grafted using hydrogen bonding, showed a 99.94% reduction in S. aureus bacteria, and a 96.95% reduction in E. coli bacteria [22]. Similarly, films of bacterial cellulose grafted with polyhexamethylene guanidine hydrochloride, also using hydrogen bonding, showed enhanced activity against the bacteria P. syringae, K. pneumonia, and S. aureus when compared to pure bacterial cellulose films [23]. This result is attributed to the positively charged guanidine group, which can bind to the negatively charged microbial membrane, deforming the cytoplasmic membranes and causing leakage of bacterial cells [24]. Guanidine modified polymers also display antiviral activity. For example, a synthetic polysaccharide that was grafted with the guanidine group by a covalent bond showed a 96.2% antiviral efficiency against a non-enveloped adenovirus [25].
To prepare a microfiltration membrane that can remove viruses and chlorine, a polycationic guanidine modified cellulose must be synthesized that can be electrospun into continuous fibers with a diameter in the nanometer scale [26]. Electrospun nanofiber mats are desired for the adsorption of contaminants in water because the mats have a high surface-to-volume ratio, large pore size in the range of several microns, and ease of fabrication [26]. The high surface-to-volume ratio nanofibers can increase the accessible surface area of the membrane and adsorption capacity. The large pore size can maintain high water flux and low membrane fouling.
In this work, electrospun nanofibers made from guanidine modified microcrystalline cellulose (MC), blended with polyvinyl alcohol (PVA), added to improve fiber spinnability, were created to explore their efficiency in water purification and to determine if covalently bonded guanidine might outperform the more commonly studied hydrogen-bonded guanidine group. It was hypothesized that microfiltration fibers with covalently bonded guanidine groups would have superior virus adsorption than hydrogen-bonded guanidine groups because the covalent bond is more durable and can give the guanidine moiety more stability against changes in pH, ionic strength, and other solution changes. Therefore, this work prepared MC grafted with guanidine hydrochloride via covalent bonds (MC-GC), and hydrogen bonds (MC-GH), and combined these polymers with PVA to form a blend that was electrospun into MC-GC/PVA and MC-GH/PVA nanofibers. These nanofibers were crosslinked with glutaraldehyde vapor to improve water stability, and then their water stability and capacity for virus removal were assessed and compared. Our model viruses were a non-enveloped virus, porcine parvovirus (PPV), and an enveloped virus, Sindbis virus. The ability of MC-GC/PVA and MC-GH/PVA nanofibers to remove chlorine was also evaluated.
2. Results and discussion
2.1. Characterization of cellulose and cellulose derivatives
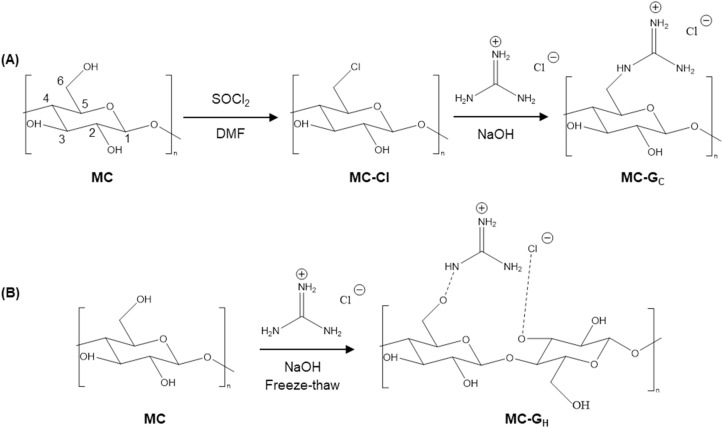
The synthetic route to covalently bonded MC-GC is shown in Scheme 1 A. First, MC is chemically modified with thionyl chloride (SOCl2) to produce chlorodeoxycellulose (MC-Cl). Then, the MC-Cl is reacted with guanidine hydrochloride to obtain the final MC-GC. Fourier transform infrared (FTIR) spectra confirm the chemical modifications in each step. In Fig. 1 , the spectrum of pure MC shows absorptions between 3300 and 3400 cm−1 that are the stretching vibrations of (CH2–OH) and (CH–OH) [27,28]. The spectrum of MC also displays a peak at 1350 cm−1, which is the deformation of the primary and secondary OH groups [27,28]. A comparison of the spectrum of MC-Cl with that of MC shows two new peaks, at 724 and 752 cm−1 [29], which are (C–Cl) stretching vibrations. These new absorptions are accompanied by the expected decreases in the intensity of the bands at 3300-3400 cm−1 and 1350 cm−1 in MC-Cl and MC-GC due to the degree of substitution of the hydroxyl group on carbon 6 by chlorine and subsequently by guanidine. By analyzing the spectrum of the MC-GC and that of pure MC, a characteristic new peak from the guanidine group at 1658 cm−1 [30,31] is found. Also, a single weak band at 3480 cm−1, representing the secondary N–H bond, and the 1255 cm−1 band, corresponding to C–N stretching, support the formation of the covalent bond between MC and guanidine [32]. It is also noted that the peaks in the spectrum of MC-Cl at 724 and 752 cm−1 do not disappear, indicating that the substitution of the chlorine by guanidine was not quantitative.
Scheme 1.
Synthesis route to prepare cellulose derivatives. (A) MC-Cl and MC-GC, and (B) MC-GH.
Fig. 1.
FTIR spectra show the conversion of MC to MC-Cl (C–Cl two new peaks at 724 and 752 cm−1), MC-GC (appearance of C N at 1658 cm−1), and MC-GH (appearance of dominant peak of O–H--- groups at 3332 cm−1 implies strong intramolecular hydrogen bonds, and C N at 1656 cm−1).
The preparation of hydrogen-bonded MC-GH is shown in Scheme 1B. FTIR spectroscopy confirms the successful introduction of the guanidine group into the cellulose and the formation of hydrogen bonding between MC and guanidine hydrochloride (Fig. 1). By comparing the spectrum of MC-GH with that of the unmodified MC, absorption bands at 1656 cm−1 confirm the presence of the guanidine group [30,31]. In addition, the band between 3000 and 3400 cm−1 belongs to the –OH stretching vibration, and the appearance of a dominant peak at 3332 cm−1 indicates the vibrational mode of this broad band is due to strong intramolecular hydrogen bonds [23,33] between MC and guanidine hydrochloride.
Elemental analysis was further used to verify the chemical modifications of cellulose structure. The compositions of the cellulose derivatives are given in Table S1 in the Supplemental Information (SI). In addition to the percentages of elements, Table 1 shows the relative number of moles of carbon, hydrogen, nitrogen, and chlorine for the derivatives, which is calculated based on the weight percentage and can be used to determine the degree of anchored groups to MC. The experimentally determined values of carbon: nitrogen: chlorine for the modified MCs were close to the theoretical values, which indicates a high degree of guanidine substitution, and that many of the cellulose units are modified as the chemical structures shown in Scheme 1. However, the experiment hydrogen content does not match well with the theoretical value, which is likely because the synthesized cellulose derivatives were not able to be 100% purified, and possibly some water and other contaminants may also contribute to the extra hydrogen.
Table 1.
Mole ratio of carbon, hydrogen, nitrogen, and chlorine in the formula of cellulose derivatives based on elemental analysis.
| Sample | Formula | C | H | N | Cl | |
|---|---|---|---|---|---|---|
| Experimental MC-Cl | (C6H9O4Cl)n |
6 | 14 | – | 1 | |
| Theoretical MC-Cl |
6 |
9 |
– |
1 |
||
| Experimental MC-GC | (C7H14O4N3Cl)n |
7 | 29 | 3 | 1 | |
| Theoretical MC-GC |
7 |
14 |
3 |
1 |
||
| Experimental MC-GH |
(C7H16O5N3Cl)n | 8 | 29 | 3 | 1 | |
| Theoretical MC-GH | 7 | 16 | 3 | 1 | ||
2.2. Characterization of MC-GC/PVA and MC-GH/PVA nanofibers
Attempts to electrospin pure MC-GC and MC-GH were unsuccessful, and only nanoparticles were collected on the drum collector. This was likely due to the high repulsion force between ionogenic guanidine molecules impeding the continuous electrospinning fibers. A non-ionogenic partner, PVA [34,35], was added to aid the formation of guanidine modified MC nanofibers. Both MC-GC/PVA and MC-GH/PVA electrospun nanofibers were successfully formed, as shown in Fig. 2 . A blend ratio of 2:1 MC-GC: PVA and MC-GH: PVA was chosen [36], to give the highest amount of guanidine modified cellulose that could be electrospun into fibers for water purification applications.
Fig. 2.
SEM images of electrospun MC-GC/PVA and MC-GH/PVA nanofibers. Glutaraldehyde vapor crosslinked nanofibers, and uncrosslinked nanofibers were tested for water stability by immersing in water for 10 min.
Glutaraldehyde vapor was used to stabilize the MC-GC/PVA and MC-GH/PVA nanofibers against water [37,38]. The stability of crosslinked and uncrosslinked MC-GC/PVA and MC-GH/PVA nanofibers was compared by immersing them in water for 10 min. The morphology of MC-GC/PVA and MC-GH/PVA nanofibers is shown in Fig. 2, and the average fiber diameter of freshly electrospun uncrosslinked and crosslinked fibers is shown in Fig. 3 . Fig. 2 shows that these uncrosslinked MC-GC/PVA and MC-GH/PVA nanofibers swell significantly and tend to dissolve in water. As the swelling continues, the distance between any two polymer chains increases [39]. The blends of both MC-GC and MC-GH, with PVA, are unstable. In contrast, the glutaraldehyde crosslinked MC-GC/PVA, and MC-GH/PVA nanofibers are stable in water. The aldehyde groups of glutaraldehyde reacted with the free hydroxyl groups of MC-GC or MC-GH, and PVA to form crystallization zones [40], which limits the polymer chain mobility [39]. Thus, the glutaraldehyde gave the nanofibers the needed water stability, as confirmed in Fig. 3. The diameter of uncrosslinked MC-GC/PVA (or MC-GH/PVA) nanofibers approximately doubled on immersion in water compared with the diameters of uncrosslinked dry fibers. In contrast, the diameters of crosslinked MC-GC/PVA (or MC-GH/PVA) nanofibers (dry and immersed in water for 10 min) were similar to the diameter of fresh electrospun MC-GC/PVA (or MC-GH/PVA) fibers (uncrosslinked dry). It should be noted that water and even the glutaraldehyde vapor, slightly swelled the crosslinked fibers. The diameter of the crosslinked dry MC-GC/PVA nanofibers (167 ± 21 nm) was also similar to that of the crosslinked dry MC-GH/PVA nanofibers (165 ± 32 nm).
Fig. 3.
Fiber diameter of crosslinked fibers is stable in water, while uncrosslinked fibers approximately double their diameter on immersion in water. The data, taken on fibers from the same batches shown in Fig. 2, and standard deviation shown, are calculated from 50 fibers in SEM-micrographs taken from four different regions on the analyzed specimens.
The pore size of the crosslinked membranes was calculated, based on four SEM images, and found to be 1.1 ± 0.2 μm for MC-GC/PVA, and 1.3 ± 0.3 μm for MC-GH/PVA. The pore sizes of these membranes are significantly reduced, when compared to the uncrosslinked membranes, and coincide with the increase in fiber diameters, because the thicker fibers occupy more space in the network, at the expense of the space for pores. Nevertheless, the membrane pore sizes of 1.1–1.3 μm are well within the range of typical microfiltration membranes (0.1–10 μm) [10] and are much larger than the sizes of viruses, which typically range from 20 to 400 nm [41].
To further characterize the stability of the crosslinked MC-GC/PVA, and MC-GH/PVA nanofibers against water, the fibers were immersed in water for up to 360 min. The morphology of the fibers is shown in Fig. 4 , while the corresponding average fiber diameter, distribution, and swelling degree, calculated from the fiber diameter, are given in Fig. 5 . The images confirm that the crosslinked MC-GC/PVA and MC-GH/PVA nanofibers retain their morphology and do not dissolve within 6 h (360 min) after contact with water. The MC-GC/PVA fibers swelled in water but appeared to approach an equilibrium swelling degree after an approximately 31% increase in diameter, but the MC-GH fibers did not reach swelling equilibrium. Interestingly, it seems like the MC-GC fibers swell more than MC-GH fibers on immersion in water, at least after 360 min immersion time. The reason is not clear, but this may indicate that the MC-GH nanofibers are highly crystalline, or it may be due to mass loss from uncrosslinked fibers.
Fig. 4.
Effect of water immersion on fiber morphology. Crosslinked nanofibers were immersed in water for various time, as shown on the top. Two types of electrospun nanofibers were tested, as shown on the left.
Fig. 5.
Diameter and swelling degree of crosslinked fibers with different water immersion time. Error bars are the standard deviation of 50 fibers that were found on four different SEM-micrographs.
To quantify differences in the dissolution of the nanofibers in water, the weights of MC-GC/PVA, and MC-GH/PVA nanofibers were measured. The density of uncrosslinked and crosslinked MC-GC/PVA and MC-GH/PVA nanofibers before contact with water are almost the same, as shown in SI Table S2. The weight loss percentage of these nanofibers after immersion in water for different times was calculated by Eqn. (1), and the results are shown in Fig. 6 . The weight loss of the crosslinked MC-GC/PVA (and MC-GH/PVA) nanofibers is greatly reduced compared with that of the uncrosslinked nanofibers, at each water contact time. This result confirms the benefit of crosslinking the fibers with glutaraldehyde to improve both the MC-GC/PVA, and MC-GH/PVA nanofibers water stability and is consistent with the prior results shown in Figs. 4 and 5. While the uncrosslinked MC-GH/PVA nanofibers lost more than 16% of their mass after contact with water for 10 min, and then 24% after 360 min, the crosslinked MC-GH/PVA nanofibers lost less than 17% of their total mass after 360 min. The uncrosslinked MC-GC/PVA nanofibers lost less than 13% after 10 min, and this increased to 21% of total mass after 360 min. The crosslinked MC-GC/PVA nanofibers still less than 15% of their total mass even after 360 min. These data confirm that MC-GC/PVA nanofibers have greater structural stability than MC-GH/PVA nanofibers even without crosslinking, but the benefits from the crosslinking are significant.
Fig. 6.
Effect of water immersion on fiber weight. Error bars are the standard deviation of electrospun nanofibers that were measured from three independent experiments.
2.3. Virus removal by MC-GC/PVA and MC-GH/PVA nanofibers
To test the virus removal ability of MC-GC/PVA and MC-GH/PVA nanofibers, two different types of mammalian viruses, porcine parvovirus (PPV), a non-enveloped virus, and Sindbis virus, an enveloped virus, were used. They represent two distinct classes of viruses that can contaminate water [42]. General virus removal techniques are typically assessed for both enveloped and non-enveloped viruses [43]. PPV is one of the smallest known mammalian viruses, having a size of 18–25 nm [44]. It belongs to the Parvoviridae family and has an isoelectric point (pI) of 4.8–5.1 [45]. PPV is difficult to inactivate by solvent-detergent or heat treatment [43] and is most effectively removed by filtration using 20-nm pore-size membranes [46,47]. As already stated, these filtration methods have high backpressures and are prone to fouling. This is why alternative removal techniques are desired and why PPV is a useful model to test alternative methods for the removal of non-enveloped viruses from drinking water, i.e., the difficulty of both inactivating and of removing the virus. Sindbis virus belongs to the Togaviridae family and is slightly larger than PPV at about 70 nm [48]. It has a reported pI of 4.2 [49]. Sindbis virus is also not easily inactivated by mild solvent-detergent [50], due to it having the highest cholesterol and saturated lipid content of the enveloped virus family. Its structure and composition are similar to flaviviruses (for example, hepatitis C, Yellow fever, and dengue virus), all of which cause global public health emergencies [51]. Therefore, Sindbis virus is also a useful model for enveloped virus removal. It should be noted that both PPV and Sindbis viruses carry a net negative charge in drinking water since they both have a pI below 7.
The adsorption capacity of crosslinked MC-GC/PVA and MC-GH/PVA nanofibers was evaluated for both PPV and Sindbis. The results are shown in Fig. 7 . The log removal value (LRV) was used to assess virus removal from drinking water and was calculated by Eqn. (2). The water-stable MC-GC/PVA nanofibers achieved a 4.5 ± 0.1 LRV for PPV, and a 4.3 ± 0.1 LRV was measured for Sindbis virus. However, the water-stable MC-GH/PVA nanofibers only reached a 2.8 ± 0.7 LRV for PPV but did achieve a 4.2 ± 0.0 LRV for Sindbis virus. No virus was removed by the control, which was blank filter paper without nanofibers. The theoretical maximum virus removal is 4.4 LRV in our test, and the EPA requires a 4 LRV for virus removal processes for drinking water [52,53]. Therefore, the MC-GC/PVA nanofibers completely removed both viruses from water and exceeded the EPA virus removal requirement. In comparison, the MC-GH/PVA nanofibers removed less PPV but did meet EPA criteria for removal of Sindbis virus.
Fig. 7.
Virus removal from drinking water by crosslinked nanofibers. PPV and Sindbis virus were incubated with crosslinked MC-GC/PVA and MC-GH/PVA nanofibers for 10 min. Control is blank filter paper without nanofibers. Error bars are the standard deviation of three separate experiments.
The ability to remove viruses using MC-GC/PVA and MC-GH/PVA membranes that have pores that are orders of magnitude greater in size than the viruses themselves shows that the viruses are not being trapped by a size-exclusion mechanism, but by adsorption to the membrane fibers. The adsorption is attributed to the protonated guanidine groups on the MC-GC or MC-GH that can form ionic and hydrogen bonds to the proteins and lipids on the virus surface [54]. In our previous work, a quaternized chitosan derivative (HTCC) was electrospun to give microfiltration membranes, and these nanofibers possessed a positively charged quaternary amine. The HTCC membranes achieved a 3.3 LRV for PPV and a 4.2 LRV for Sindbis virus [37]. The advantage of MC-GC and MC-GH nanofiber membrane over the HTCC nanofiber membrane is attributed to both electrostatic interaction between guanidine cations and negatively charged viruses existed, but also hydrogen bonding could occur between the guanidine group and virus surface protein or envelope.
The data show that the virus removal capacity of the MC-GC/PVA membrane is also higher than that of the MC-GH/PVA membrane. As the ability to adsorb the virus must depend on the immobilized guanidine group, this suggests that either the MC-GC/PVA membrane possesses more guanidine groups, or that the guanidine groups are more effective than those of the MC-GH/PVA membrane, or possibly some combination of these effects. It is highly likely that the positively charged guanidine group is a major factor determining the virus adsorption capacity of the membranes, and the higher stability of the guanidine group immobilized by covalent bonds, compared to hydrogen bonds, likely accounts for most of its greater virus removal. Additionally, the diameter of the MC-GC/PVA nanofibers was slightly larger than that of the MC-GH/PVA nanofibers. An increase in virus adsorption with increased fiber diameter was also observed in our previous work [55]. Overall, the performance of the water-stable MC-GC/PVA nanofibers supports their potential to become an effective and low-cost material for virus removal from drinking water.
2.4. Chlorine removal by MC-GC/PVA and MC-GH/PVA nanofibers
To test the dechlorination ability of MC-GC/PVA and MC-GH/PVA nanofibers, sodium hypochlorite was used to prepare the chlorinated wastewater since chlorine forms hypochlorite ion (ClO−) when dissolved in water. A standard iodometric electrode method [56] was used to measure the concentration of residual chlorine. The chlorine removal is reported as the percentage of chlorine removed from wastewater, as shown in Eqn. (3). The control blank filter paper only removed 18.9% ± 0.4% of the chlorine, while the water-stable MC-GC/PVA nanofibers achieved a 58.1 ± 8.0% of chlorine removal, and the water-stable MC-GH/PVA nanofibers obtained a 57.6 ± 8.0% of chlorine removal.
The EPA requires that the chlorine concentration not exceed 5 mg/L for disinfected wastewater [11]. If the MC-GC/PVA nanofibers were used alone, the residual chlorine after nanofiber adsorption would still be ~2 mg/L, which remains acutely toxic to aquatic organisms (threshold 1 mg/L) [14]. However, the chlorine removal efficiency of electrospun guanidine modified cellulose membrane after only 10 min contact with the water being treated still showed a better performance than that of a Hoechst Celanese hollow fiber membrane with 36% removal for 1-h contact [57]. These results further confirm the potential of these water-stable MC-GC/PVA nanofibers to be used as new material for chlorine removal in wastewater treatment.
3. Conclusions
Guanidine hydrochloride was successfully bonded to MC by two different routes. One route covalently bonded the guanidine to cellulose, giving MC-GC, while the second route bound the guanidine to cellulose using hydrogen bonds, giving MC-GH. The charge density made electrospinning continuous fibers from the pure MC-GC and MC-GH solutions challenging, so PVA, a non-ionogenic polymer, was blended with these solutions, and continuous nanofibers were easily electrospun. The nanofiber mats from the MC-GC/PVA and MC-GH/PVA were stabilized by crosslinking with glutaraldehyde vapor, allowing the crosslinked MC-GC/PVA and MC-GH/PVA nanofibers to retain their structure and morphology in water for 6 h (maximum time tested). The water-stable MC-GC/PVA nanofiber mats achieved a 4.5 ± 0.1 LRV for PPV and a 4.3 ± 0.1 LRV for Sindbis virus. In contrast, the water-stable MC-GH/PVA nanofiber mats only reached a 2.8 ± 0.7 LRV for PPV but reached a 4.2 ± 0.0 LRV for Sindbis virus. The MC-GC/PVA nanofibers completely removed both viruses from water and met the EPA virus removal regulation of 4 LRV. In addition, the water-stable MC-GC/PVA nanofibers achieved a 58.1 ± 8.0% of chlorine removal, while the water-stable MC-GH/PVA nanofibers obtained a 57.6 ± 8.0% of chlorine removal. Therefore, the cellulose nanofiber membranes with covalently bonded guanidine hydrochloride outperformed the mats with hydrogen-bonded guanidine hydrochloride, but the performance was significantly better only against the non-enveloped PPV virus. The improvement is likely because the covalent bond yields a more stable chemical structure that is less vulnerable to changes in solution conditions. Because the mechanism of virus removal is adsorption, this difference may be due to differences in pI and the nature of the electrostatic interactions between the virus surfaces and the guanidine groups.
Providing simple and low-cost methods to purify water, especially in locations without established water-treatment facilities, is critical. This work shows these modified cellulosic materials have the potential to be developed into effective water purification membranes for the removal of pathogenic organisms from drinking water and removal of chlorine from wastewater. It also shows that while hydrogen-bonding the active species to the fiber mat may be simpler, that different results, and in this work, better results were achieved by covalently binding the active species. In the future, the kinetic adsorption and the adsorption isotherms of the guanidine modified cellulose nanofibers should be explored to understand the adsorption mechanism better, and specifically, explain the difference between PPV adsorption and covalently bonded and hydrogen-bonded guanidine groups, and to further evaluate the performance of nanofibers in water purification. A better understanding of these differences can allow even better membranes to be made to achieve the goal of purification of drinking water with the large pore-sized modified cellulose membranes and control waterborne viral disease outbreaks. These membranes can also assist in the second goal of a simple filtration method to remove chlorine from wastewater and reduce the threat to aquatic organisms.
4. Experiment section
4.1. Materials
Microcrystalline cellulose (MC) powder, N,N-dimethylformamide (DMF) (99.8%, anhydrous), thionyl chloride (SOCl2) (reagent grade, 97%), ammonium hydroxide (reagent grade, 28.0–30.0% NH3 basis), sodium hydroxide (NaOH) (ACS reagent, ≥97% pellets), guanidine hydrochloride (≥99.0%), ethanol (ACS reagent, ≤ 0.003% water), polyvinyl alcohol (PVA) (98–99% hydrolyzed, Mw = 146,000–186,000), glutaraldehyde (Grade I, 70% in H2O), thiazolyl blue tetrazolium bromide (MTT salt) (≥97.5%), and sodium dodecyl sulfate (SDS) (for molecular biology, ≥ 98.5%) were all purchased from Sigma-Aldrich (St. Louis, MO). Phosphate buffered saline (PBS) (pH = 7.2) was purchased from Life Technologies (Grand Island, NY). Hydrochloric acid (HCl) (12.1 M) was purchased from VWR (Radnor, PA). A chlorine standard (100 ppm, Cl (C-9A)), iodide reagent (I-55), and acid reagent (A-13) were all purchased from North Central Laboratories (Birnamwood, WI). Sodium hypochlorite (Azone 15) was from Hawkins, Inc (St. Louis, MO). All aqueous solutions were prepared using purified water with a resistivity of ≥18 MΩ cm from a Nanopure filtration system (Fisher Scientific, Pittsburgh, PA).
4.2. Preparation of MC-GC
4.2.1. Synthesis of chlorodeoxycellulose (MC-Cl)
The MC-Cl was synthesized using a previously reported halogenation method [28,58] (Scheme 1A), with minor modifications. First, MC (2.5 g, 15 mmol) was suspended in 50 mL of DMF and then heated at 80 °C for 12 h to activate the cellulose before the reaction. SOCl2 (8.75 mL, 14.4 g, 120 mmol) was slowly added to the MC solution while maintaining the temperature at 80 °C for 4 h under mechanical stirring. Then, the reaction solution was cooled to room temperature, and filtered to obtain the solid MC-Cl. The solid was washed five times with ammonium hydroxide (pH 7, 5 mL, 5 M) and then Nanopure water (100 mL). The solid chlorinated product MC-Cl was dried under vacuum at room temperature for 4 h.
4.2.2. Synthesis of MC-GC
For the preparation of MC with covalently bonded guanidine [59] (Scheme 1A), MC-Cl (0.5 g) was first suspended in 5 mL of Nanopure water in an ice bath for 10 min. The guanidine hydrochloride (0.25 g, 2.6 mmol) was dispersed in 7.5 mL of NaOH (0.8 M) solution that had been pre-cooled in an ice bath for 10 min. Then, the guanidine hydrochloride solution was added dropwise to the cooled MC-Cl suspension. The mixture was stirred in the ice bath for 3 h and stirred at room temperature for an additional 12 h. The reaction solution was filtered, washed five times with Nanopure water (10 mL) and ethanol (10 mL), and dried under vacuum at 70 °C for 4 h to obtain the MC-GC solid.
4.2.3. Dissolution of MC-GC in NaOH
The synthesized MC-GC solid (0.5 g) was further dissolved in 13.45 mL of NaOH (2.3 M) solution [60,61], frozen at −80 °C for 12 h, and thawed at room temperature. A 6.55 mL aliquot of Nanopure water was added to the thawed MC-GC and gently stirred for 15 min to obtain a homogenous light brown gel-liquid (SI Fig. S1A).
4.3. Preparation of MC-GH
To form a hydrogen bond between guanidine and MC [23] (Scheme 1B), the MC (2.0 g, 12 mmol) was first dissolved in 53.8 mL NaOH (2.3 M), then frozen at −80 °C for 12 h, and then thawed at room temperature [60,62]. Guanidine hydrochloride (1.0 g, 10 mmol) was dissolved in 26.2 mL Nanopure water, and then this solution was added to the MC solution with gentle stirring, which was continued for 15–30 min to obtain the MC-GH solution, which was a homogeneous pale-yellow gel-liquid (SI Fig. S1B).
4.4. Electrospinning of MC-GC/PVA and MC-GH/PVA blends
A 10 w/v% PVA solution was prepared by dissolving PVA (1.0 g) in 10 mL of Nanopure water at 85 °C with stirring for 1 h. The homogeneous MC-GC/PVA spinning solution was prepared at a ratio of 2:1 MC-GC: PVA. The total solid content of the MC-GC/PVA spinning solution was 9.2 w/v%, and the final pH was 12. Similarly, the homogeneous MC-GH/PVA spinning solution was also prepared at a ratio 2:1. The total solid content of the MC-GH/PVA spinning solutions was 10 w/v%, and the final pH was 10.
The description of the home-made electrospinning apparatus has been illustrated in previous work [63]. The electrospinning solutions were put into a 3 mL disposal plastic syringe with detachable needles (0.6 mm × 40 mm) (Fisher Scientific, Pittsburgh, PA). The needle was connected to a Glassman positive DC high voltage power supply (High Bridge, NJ) to generate voltages in the range of 0–30 kV, while the ground was connected to a rotation drum collector covered with aluminum foil and operated by an ElectroCraft TorquePower motor (Gallipolis, OH). The electrospun nanofibers were collected on Whatman filter paper circles, which were taped on the aluminum foil and used to support the nanofibers. A multi-speed syringe pump (Braintree Scientific Inc., Braintree, MA) was used to feed the solution at a constant rate.
The MC-GC/PVA and MC-GH/PVA solutions were spun using a flow rate of 4.5 mL/h, and an applied voltage of 20 kV. The tip-to-collector distance was 5 cm, and the rotation speed of the drum collector was 1500 rpm.
4.5. Crosslinking of MC-GC/PVA and MC-GH/PVA nanofibers
The MC-GC/PVA and MC-GH/PVA nanofibers were crosslinked using 30% glutaraldehyde vapor at 37 °C for 4 h [37]. The crosslinked nanofibers were washed with water and dried in a Gold Series DP-32 vacuum drying oven (Ontario, Canada) at 120 °C for 1 h to remove any unreacted glutaraldehyde [37].
4.6. Characterization
Fourier transform infrared (FTIR) spectra were obtained using a PerkinElmer Spectrum One FTIR spectrometer (Shelton, CT). The spectra of MC, MC-Cl, MC-GC, and MC-GH were measured over an absorbance range of 4000–400 cm−1 range with a resolution of 4 cm−1. Elemental analysis was performed by Micro-Analysis Inc. using the following methods: %CHN analysis-combustion/thermal conductivity detector and %Cl–O2 flask combustion/ion chromatography.
To determine the stability of crosslinked electrospun nanofibers against water, the MC-GC/PVA and MC-GH/PVA nanofibers were immersed in room temperature water for the designated time and then dried in the drying oven at 120 °C for 1 h.
The average weight of nanofibers was obtained using the mass of the supporting filter paper before and after collecting the fibers. The density of nanofibers was calculated by the ratio of the weight to the area of the Whatman filter paper. The weight loss percentage was calculated using Eqn. (1):
| (1) |
where w immersed in water is the average weight of nanofibers immersed in water for a corresponding time, and w dry is the initial weight of nanofibers before contact with water.
The nanofiber morphology was observed using a Hitachi S-4700 cold field emission scanning electron microscope (FE-SEM) (Tustin, CA), using an acceleration voltage of 5 kV and magnification ranging from 1000 × to 10,000 × . Due to the non-conductive nature of the nanofibers, they were coated with 3 nm of platinum/palladium with a sputter coater (Hummer Sputtering System, Union City, CA) at a rate of 0.1 nm/min, prior to imaging. Fiber diameter and pore size were determined with the aid of ImageJ software with measuring 50 random fibers, and 25 random pores from four micrographs.
4.7. Virus removal from drinking water
Porcine kidney (PK-13) cells and porcine parvovirus (PPV) strain NADL-2 were a gift from Dr. Ruben Carbonell, North Carolina State University. Baby hamster kidney (BHK-21) cells and Sindbis virus (heat-resistant strain) were a gift from Dr. Raquel Hernandez, North Carolina State University. PPV was propagated in PK-13 cells, and Sindbis virus was propagated in BHK-21 cells [64]. Both types of viruses were titrated with a cell viable MTT assay, as described previously [37,64]. Briefly, either 8 × 104 cells/mL PK-13 cells (to titrate PPV) or 1.1 × 105 cells/mL BHK-21 cells (to titrate Sindbis virus) were seeded into a 96-well plate. The volume added was 100 μL/well. After 1-day incubation, 25 μL/well of virus sample was added to the corresponding host cells in quadruplicate and serially diluted across the plate. After 5 days (for PPV) or 2 days (for Sindbis virus), 10 μL/well of 5 mg/mL MTT salt in PBS (pH 7.2) was added to the plate. After 4 h, 100 μL/well of solubilizing agent, consisting of 10 w/v% SDS in 0.01 M HCl (pH 2.5) was added to the plate. After 4–20 h, the plates were read for absorbance at 550 nm on a Synergy Mx monochromator-based multi-mode microplate reader (Winooski, VT). The virus dilution that killed 50% of the host cells is defined as the virus titer MTT50 [64].
To determine the amount of virus adsorbed to the nanofibers, the log removal value (LRV) was calculated as,
| (2) |
where c f is the virus concentration after water purification, and c i is the initial virus concentration.
For virus removal studies [37], one layer of 0.5 cm2 punched filter paper containing either MC-GC/PVA nanofibers, or MC-GH/PVA nanofibers was placed into a 1.5 mL non-stick surface micro-centrifuge tube, which contained 500 μL of 6 log10 (MTT50/mL) of the virus in water. One layer of the same size of punched blank filter paper without nanofibers was also put into a separate tube as a control. Tubes containing fibers and blank were rotated for 10 min on a Roto-shake Genie rocker (Scientific Industries Inc., Bohemia, NY). Then, the nanofibers were removed from the tubes, and the virus solutions were centrifuged for 30 min at 14,000 rpm in a Sorvall ST16R centrifuge (Thermo Scientific, Pittsburgh, PA) to remove any remaining fibers in the tubes. The supernatant was removed and tested with the MTT assay to determine the concentration of the infectious virus and calculate the LRV.
4.8. Chlorine removal from wastewater
For the chlorine removal study, a standard iodometric electrode method [56] was used to measure the concentration of residual chlorine in the wastewater. This method is based on iodometric measurements of chlorine and requires the addition of an iodide reagent and an acid reagent to a sample before measurement. The iodide will react completely with the chlorine (hypochlorite ion) to form iodine in an acid medium [57]. To measures the iodine concentration, which equals the total residual chlorine concentration before reaction, a Thermo Scientific Orion ion selective electrode (ISE) meter (pH/ISE meter) with a residual chlorine probe (Orion Cat# 977013NWP, with a limit of detection 0.2–20 ppm) was used. The chlorine electrode was calibrated using the manufacturer's instructions with chlorine standards (1, 10, and 20 ppm). The electrode slope of the 3-point standard curve was determined to be 29.1 mV, which was within the reference value range from 25 to 30 mV to indicate the electrode operating normally. Chlorinated wastewater was prepared by the addition of sodium hypochlorite to DI water. To measure the initial concentration of chlorine in the wastewater, a mixture of 1 mL of acid reagent and 1 mL of iodide reagent was added to 100 mL of wastewater, stirred for 10 min, and measured with the chlorine probe. For the dechlorination of wastewater, either the MC-GC/PVA nanofibers or MC-GH/PVA nanofibers were added to the 100 mL wastewater, incubated for 10 min, and taken out. The blank filter paper without nanofibers was the control. The acid and iodide reagents were then added to the treated wastewater, and the final concentration of chlorine was measured. The percentage of chlorine removal was determined by
| (3) |
where c f is the chlorine concentration after nanofibers treatment, and c i is the initial chlorine concentration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Mr. Owen Mills for SEM assistance, Mrs. Melissa Richards at the Ishpeming Area Joint Wastewater Treatment Facility for the chlorine removal test, Dr. Tarun Dam, Dr. Marina Tanasova and Dr. Martin Thompson for the use of their freezer, and Dr. Shiyue Fang for fruitful synthesis discussions. Funding is gratefully received from the Department of Chemistry at Michigan Tech (PAH) and NSF CAREER (CBET-1451959) (CLH).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.carres.2020.108153.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.WHO . World Health Organization; 2019. In: Fact Sheet on Water. [Google Scholar]
- 2.WHO . World Health Organization; 2017. In: Fourth Edition Incorporating the First Addendum. [PubMed] [Google Scholar]
- 3.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. Sci. Total Environ. 2020:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. medRxiv. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 5.Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N. medRxiv. 2020 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 7.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. 2020. Jama. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. 2020. Water Research; p. 115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J.S., Yuan D.X., Weng T.P. Desalination. 2010;263:271–278. [Google Scholar]
- 10.Antony A., Blackbeard J., Leslie G. Crit. Rev. Environ. Sci. Technol. 2012;42:891–933. [Google Scholar]
- 11.EPA . United States Environmental Protection Agency. 1999. Washington, D.C. [Google Scholar]
- 12.Bull R.J. American Water Works Association; 2006. Use of Toxicological and Chemical Models to Prioritize DBP Research. [Google Scholar]
- 13.Hanigan D., Truong L., Simonich M., Tanguay R., Westerhoff P. J. Environ. Sci. 2017;58:302–310. doi: 10.1016/j.jes.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Emmanuel E., Keck G., Blanchard J.M., Vermande P., Perrodin Y. Environ. Int. 2004;30:891–900. doi: 10.1016/j.envint.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Sathasivan A., Herath B.S., Senevirathna S., Kastl G. In: Current Developments in Biotechnology and Bioengineering. Elsevier; 2017. pp. 359–380. [Google Scholar]
- 16.Thakur V.K., Voicu S.I. Carbohydr. Polym. 2016;146:148–165. doi: 10.1016/j.carbpol.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Gopi S., Balakrishnan P., Chandradhara D., Poovathankandy D., Thomas S. Materials Today Chemistry. 2019;13:59–78. [Google Scholar]
- 18.Teixeira M.A., Paiva M.C., Amorim M.T.P. Nanomaterials. 2020;10:557. doi: 10.3390/nano10030557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Y.F., Xia Q.Y., Xiao H.N. Polymers. 2019;11 doi: 10.3390/polym11081283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillay V., Dott C., Choonara Y.E., Tyagi C., Tomar L., Kumar P., du Toit L.C., Ndesendo V.M.K. J. Nanomater. 2013:789289. [Google Scholar]
- 21.Xue Y., Xiao H.N., Zhang Y. Int. J. Mol. Sci. 2015;16:3626–3655. doi: 10.3390/ijms16023626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao C.L., Wu K.J., Yuan W., Zhang Y.M., Wang H.P. Fibers Polym. 2017;18:1040–1047. [Google Scholar]
- 23.Kukharenko O., Bardeau J.F., Zaets I., Ovcharenko L., Tarasyuk O., Porhyn S., Mischenko I., Vovk A., Rogalsky S., Kozyrovska N. Eur. Polym. J. 2014;60:247–254. [Google Scholar]
- 24.Zhou Z.X., Zheng A., Zhong J.J. Acta Biochim. Biophys. Sin. 2011;43:729–737. doi: 10.1093/abbs/gmr067. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y.F., Xue Y., Snow J., Xiao H.N. Macromol. Chem. Phys. 2015;216:511–518. [Google Scholar]
- 26.Suja P.S., Reshmi C.R., Sagitha P., Sujith A. Polym. Rev. 2017;57:467–504. [Google Scholar]
- 27.Silva E.C., Lima L.C.B., Silva F.C., Sousa K.S., Fonseca M.G., Santana S.A.A. Carbohydr. Polym. 2013;92:1203–1210. doi: 10.1016/j.carbpol.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Silva E.C., Santos L.S., Silva M.M.F., Fonseca M.G., Santana S.A.A., Airoldi C. Mater. Res. Ibero-American J. Mater. 2013;16:79–87. [Google Scholar]
- 29.Chen Q., Peng C., Xie H.B., Zhao Z.K., Bao M. RSC Adv. 2015;5:44598–44603. [Google Scholar]
- 30.Goto T., Nakanishi K., Ohashi M. Bull. Chem. Soc. Jpn. 1957;30:723–725. [Google Scholar]
- 31.Nikolaevna G.M., Aleksandrovich S.S., Aleksandrovna A.S., Mihaylovna T.I., Ulzitovna B.L., Babudorjievich B.V., Markovich M.D. J. Appl. Polym. Sci. 2014;131 [Google Scholar]
- 32.Larkin P.J. second ed. ed. Elsevier; 2018. Infrared and Raman Spectroscopy: Principles and Spectral Interpretation. [Google Scholar]
- 33.Marechal Y., Chanzy H. J. Mol. Struct. 2000;523:183–196. [Google Scholar]
- 34.Qi H.S., Sui X.F., Yuan J.Y., Wei Y., Zhang L.N. Macromol. Mater. Eng. 2010;295:695–700. [Google Scholar]
- 35.Zulkifli F.H., Jahir Hussain F.S., Abdull Rasad M.S.B., Mohd Yusoff M. J. Biomater. Appl. 2015;29:1014–1027. doi: 10.1177/0885328214549818. [DOI] [PubMed] [Google Scholar]
- 36.Chahal S., Hussain F.S.J., Yusoff M.B.M. Procedia Eng. 2013;53:683–688. [Google Scholar]
- 37.Mi X., Vijayaragavan K.S., Heldt C.L. Carbohydr. Res. 2014;387:24–29. doi: 10.1016/j.carres.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zulkifli F.H., Shahitha F., Yusuff M.M., Hamidon N.N., Chahal S. Procedia Eng. 2013;53:689–695. [Google Scholar]
- 39.Han B., Li J., Chen C., Xu C., Wickramasinghe S.R. Chem. Eng. Res. Des. 2003;81:1385–1392. [Google Scholar]
- 40.Young R.J., Lovell P.A. CRC press; 2011. Introduction to Polymers. [Google Scholar]
- 41.Payne S. Academic Press; 2017. Viruses: from Understanding to Investigation. [Google Scholar]
- 42.Bosch A. Elsevier; 2007. Human Viruses in Water: Perspectives in Medical Virology. [Google Scholar]
- 43.Kempf C., Stucki M., Boschetti N. Biologicals. 2007;35:35–42. doi: 10.1016/j.biologicals.2006.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrica A., Nardini C., Falbo A., Bailey A.C., Bucci E. Biologicals. 2003;31:165–173. doi: 10.1016/s1045-1056(03)00035-6. [DOI] [PubMed] [Google Scholar]
- 45.Mi X., Bromley E.K., Joshi P.U., Long F., Heldt C.L. Langmuir. 2020;36:370–378. doi: 10.1021/acs.langmuir.9b03070. [DOI] [PubMed] [Google Scholar]
- 46.Furuya K., Murai K., Yokoyama T., Maeno H., Takeda Y., Murozuka T., Wakisaka A., Tanifuji M., Tomono T. Vox Sang. 2006;91:119–125. doi: 10.1111/j.1423-0410.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- 47.Nowak T., Popp B., Roth N.J. Biologicals. 2019;60:85–92. doi: 10.1016/j.biologicals.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Dunbar C.A., Rayaprolu V., Wang J.C.Y., Brown C.J., Leishman E., Jones-Burrage S., Trinidad J.C., Bradshaw H.B., Clemmer D.E., Mukhopadhyay S., Jarrold M.F. ACS Infect. Dis. 2019;5:892–902. doi: 10.1021/acsinfecdis.8b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalrymple J.M., Schlesinger S., Russell P.K. Virology. 1976;69:93–103. doi: 10.1016/0042-6822(76)90197-5. [DOI] [PubMed] [Google Scholar]
- 50.Espindola O.M., Belluci M.S.P., Oliveira B., Liberto M.I.M., Cabral M.C. J. Virol Methods. 2006;134:171–175. doi: 10.1016/j.jviromet.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Gubler D.J. Arch. Med. Res. 2002;33:330–342. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 52.EPA . 2007. in. [Google Scholar]
- 53.EPA . 1944. in. [Google Scholar]
- 54.Meingast C., Heldt C.L. Biotechnol. Prog. 2020;36 doi: 10.1002/btpr.2931. [DOI] [PubMed] [Google Scholar]
- 55.Mi X., Heldt C.L. Colloids Surf. B Biointerfaces. 2014;121:319–324. doi: 10.1016/j.colsurfb.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Baird R.B., Eaton A.D., Rice E.W. 23rd ed. American Public Health Association, American Water Works Association, Water Environment Federation; Washington, DC, USA: 2017. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- 57.Shin C.H., Johnson R. J. Ind. Eng. Chem. 2009;15:613–617. [Google Scholar]
- 58.da Silva E.C., Santana S.A.A., Melo J.C.P., Oliveira F., Airoldi C. J. Therm. Anal. Calorim. 2010;100:315–321. [Google Scholar]
- 59.Porcheddu A., Giacomelli G., Chighine A., Masala S. Org. Lett. 2004;6:4925–4927. doi: 10.1021/ol047926m. [DOI] [PubMed] [Google Scholar]
- 60.Isogai A., Atalla R.H. Cellulose. 1998;5:309–319. [Google Scholar]
- 61.Kostag M., Jedvert K., Achtel C., Heinze T., El Seoud O.A. Molecules. 2018;23 doi: 10.3390/molecules23030511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cai J., Zhang L. Biomacromolecules. 2006;7:183–189. doi: 10.1021/bm0505585. [DOI] [PubMed] [Google Scholar]
- 63.Bai B., Mi X., Xiang X., Heiden P.A., Heldt C.L. Carbohydr. Res. 2013;380:137–142. doi: 10.1016/j.carres.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 64.Heldt C.L., Hernandez R., Mudiganti U., Gurgel P.V., Brown D.T., Carbonell R.G. J. Virol Methods. 2006;135:56–65. doi: 10.1016/j.jviromet.2006.01.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.