Abstract
Novel DNA materials promise unpredictable perspectives for applications in cell biology. The realization of DNA-hydrogels built by a controlled association of DNA nanostars, whose binding can be tuned with minor changes in the nucleotide sequences, has been recently described. DNA hydrogels, with specific gelation properties that can be reassambled in desired culture media supplemented with drugs, RNA, DNA molecules and other bioactive compounds offer the opportunity to develop a novel nanomaterial for the delivery of single or multiple drugs in tumor tissues as an innovative and promising strategy. We provide here a comprehensive description of different, recently realized DNA-gels with the perspective of stimulating their biomedical application. Finally, we discuss the possibility to design sophisticated 3D tissue-like DNA-gels incorporating cell spheroids or single cells for the assembly of a novel kind of cellular matrix as a preclinical investigation for the implementation of tools for in vivo delivery of bioactive molecules.
Keywords: nanomaterials, DNA-GEL, gelation, biomedical applications, delivery of bioactive molecules
Introduction
The well-understood reversibility and programmability of the Watson-Crick pairing interactions have promoted DNA as a leading component for the realization of bottom-up materials at the nanoscale (Seeman, 2016). The further support of robust synthesis methods, allowing to encipher a purposeful order of nucleotides in the DNA sequences, has encouraged the construction of several systems that respond to external stimuli such as temperature, pH, and presence of host DNA strands, among others (Murakami and Maeda, 2005; Condon, 2006; Cheng et al., 2009; Gao et al., 2015; Cherkasov et al., 2020). DNA has thus become an appealing material for the realization of 2D and 3D ordered structures (Winfree et al., 1998; Seeman, 2003; Zheng et al., 2009) as well as for the design of purposeful objects with predetermined shapes via the DNA Origami (Rothemund, 2006) or for the construction of hybrid materials that combine DNA with other systems, such as metallic nanoparticles (Macfarlane et al., 2011; Chao et al., 2015; Wang et al., 2015). When dissolving short DNA strands (oligomers) in an aqueous solution, segments of complementary sequences pair to form double-helical complexes, whose thermodynamic stability depends on the number of complementary bases. DNA nanotechnology exploits this general phenomenon by properly designing the oligomer sequences to build the desired structures. When the oligomers form an extended network, the resulting material is named DNA hydrogel. Such water-swollen DNA networks can reach macroscopic dimensions by mixing bulk quantities (≈100 μM) of oligomer sequences of nanometric size. The opportunity to play with a vast number of different oligomers makes it possible to generate DNA hydrogels differing in their local structure and functionality, from disordered networks resulting upon the spontaneous binding of randomly occurring sequences (Bellini et al., 2012) to the controlled formation of ordered networks composed of identical DNA nanoparticles, exploiting what is today called hierarchical multi-step self-assembly (Figures 1A, B). Typically, gelation stems from a two-step aggregation process. In the first step, the DNA oligomers pair to form suitably designed branched DNA structures. A possible example is shown in Figure 1A, where four selected sequences hybridize to form a nanostar with four arms. This aggregation process is followed by an additional step in which the binding of the sticky overhangs – single-stranded DNA sequences located at the end of each arm – occurs (Figure 1B). This mechanism allows for the realization of extended supramolecular structures with interesting behavior. The possibility to design specific bio-functional sequences with stimuli-responsive properties led to the construction of biocompatible DNA-hydrogels for biomedical applications, which are stable under physiological conditions. To date, these systems have been mainly applied to fields such as immunotherapy (Nishikawa et al., 2014; Ishii-Mizuno et al., 2017; Umeki et al., 2017; Shao et al., 2018; Komura et al., 2020), tissue engineering, drug and small molecules delivery (Um et al., 2006; Nishikawa et al., 2011; Nishikawa et al., 2014; Song et al., 2015b; Ishii-Mizuno et al., 2017; Umeki et al., 2017; Liu et al., 2018; Wei et al., 2019; Yue et al., 2019). Indeed, DNA-hydrogels represent a novel 3D scaffold in biomedical research, in addition to other natural and synthetic hydrogels - networks of polymers in an aqueous microenvironment - that have been developed and are very promising for biomedical applications (Tibbitt and Anseth, 2009; Gu and Mooney, 2016). These biomaterials, usually developed as injectable hydrogels, can be used depending on the specific biological purpose, from drug delivery (Ma et al., 2015; Pan et al., 2018) to tissue engineering for regenerative medicine (Keskar et al., 2009; Cheng et al., 2013; Purcell et al., 2014; Parmar et al., 2015). Therefore, structural, chemical, and mechanical similarities with the extracellular matrix (ECM) should be reproduced, such as the natural pore size, mechanical strength, cellular attachment, molecular response, biodegradability, biocompatibility, and solute transport (O’Brien, 2011; Thiele et al., 2014; Caliari and Burdick, 2016; Tsou et al., 2016; Naahidi et al., 2017). In the following, we will discuss, among others: the possibility to realize DNA-based materials that form a gel only around body temperature (Bomboi et al., 2016), which makes them ideal for the encapsulation of therapeutic oligonucleotides or drugs, as well as for favoring the interactions with specific proteins and RNA sequences; gels that do not cause an immune reaction, allowing the cell to live in the gel matrix for days (Um et al., 2006); gels with reconfigurable properties, able to restructure their topology via exchange reactions. Such systems are appealing for their self-healing and stress-realising properties, being thus promising for in vivo engineering applications, for cell transplantation and encapsulation (Um et al., 2006) as well as for the protein production (Park et al., 2009). The fight for cancer is coping with less toxic and more targeted therapy to avoid or lower the dangerous effects induced by the massive cytotoxic therapies. However, after aggressive therapies cancer cells often relapse with a worse and more aggressive malignant phenotype. In this context, Anakoinosis as an innovative therapeutic approach is intended to re-equilibrate tissue homeostasis. Current therapies, either classic antiblastic or more specific targeted therapies have the bias of selecting resistant tumor clones causing failure in the control of advanced disease. It is becoming increasingly evident that cellular damage induced by cancer progression is affected by tumor environment. Anakoinosis, the therapeutic approach based on reprogramming malignant cells in the frame of an intercellular communication and activation of endogenous signaling, has been proposed to achieve a more effective control of cancer cells progression. In this respect, there is an expanding research focused on finding the conditions and the effectors that can reactivate cellular homeostasis. In this field, DNA-gels might be applied successfully for their versatility. DNA-hydrogels will possibly be employed at preliclinical level in order to modify the intercellular communication and obtain a novel equilibrium in cellular homeostasis. DNA hydrogels composed of DNA NS can be dissolved in desired culture supplemented media, may incorporate different bioactive molecules, compounds, drugs and can provide an in situ platform for their targeted slow release inside tissues. In addition, the DNA-gel structure may also provide an ideal substrate for resetting the cellular homeostasis for redifferentiation of transformed cells. In this review, we discuss (i) the impact of DNA-hydrogels in cell environments, (ii) the potential applications of these materials, (iii) the in vivo analysis of DNA-gels and their applications to human cells, and (iv) possible future applications of these novel materials in the therapeutic field.
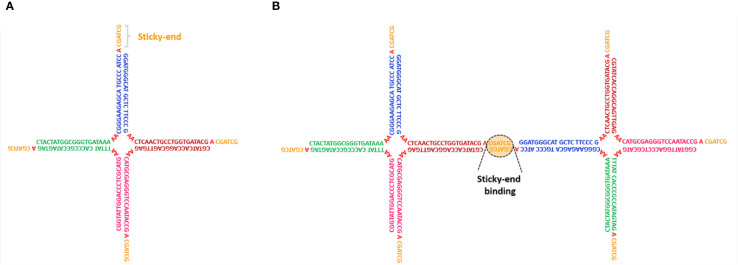
Figure 1.
(A) Schematic representation of a limited-valence NS with functionality f=4, obtained by the self-assembly of four specific DNA sequences. Different colors highlight complementary DNA regions that selectively hybridize, forming the ds-sections that constitute the particle arm. Each arm terminates with a self-complementary sticky-end recognition sequence that allows NS-NS interactions (B).
DNA-Nanostars Hydrogels
In recent years, DNA self-assembly has been exploited to produce bulk quantities of nanometric particles of predefined shape and size. The possibility to take control at design time of the particle-particle interactions and built-in functionalities has led to several applications in the assembly of biodegradable and biocompatible DNA-gels with tailored properties (Um et al., 2006; Biffi et al., 2013; Bomboi et al., 2016; Nguyen and Saleh, 2017; Fernandez-Castanon et al., 2018). One important class of DNA constructs is represented by limited-valence DNA nanostars (NSs), that is, DNA nanoparticles with a finite number f of arms departing from a common central junction. These NSs have attracted increasing interest, encouraging the rational design of soft materials with unconventional behavior and tunable properties such as controllable shape (Um et al., 2006), kinetics, and structure (Nguyen and Saleh, 2017), permanent network structure formation via enzymatic reaction (Um et al., 2006; Xiang et al., 2016), re-entrant phase diagram (Bomboi et al., 2016), tunable viscosity (Bomboi et al., 2015), and self-healing (Bomboi et al., 2019). NSs can be experimentally produced by mixing in solution equimolar quantities of f purposely designed single-stranded (ss) DNA sequences, which are able to spontaneously self-assemble into a branched geometry with f double-helical arms. Particle flexibility can be increased by adding a small number of unpaired bases in the sequences at the location of the NS center. Finally, short single-stranded “sticky sequences”, placed at the end of each arm, promote particle-particle attractive interactions via strand hybridization. In the explanatory Figure 1A, we report an example of a tetravalent (f=4) NS with the particular DNA sequence design. The NS structure is composed of four double-stranded (ds) arms with a flexible core of 8 unpaired adenines, positioned at the center of the structure, which provide for particle flexibility. Each arm terminates with a self-complementary ss-sequence that allows NS-NS interactions (see Figure 1B). An additional unpaired adenine, positioned between the ds-regions and the sticky sequences, enhances the sticky-end flexibility facilitating the binding between different NSs. A series of studies focused on the phase behavior of DNA NS suspensions and their gelation properties, aiming at verifying how the functionality of the nanostars and the tunable external parameters (temperature, pH, salt-concentration, cosolute concentration) affect the probability of base paring and, consequently, how they can be exploited to control the (reversible) gel formation, its structure, and its viscoelastic properties. The emerging picture is consistent with recent predictions of the physics of patchy colloidal particles (Bianchi et al., 2011). Specifically, Biffi et al. (2013) studied the phase behavior upon temperature variation of trivalent (f=3) and tetravalent (f=4) DNA NS systems, formed by the DNA strands displayed in Figure 2. They showed that at high temperatures, around T ≈ 90°C, the system is composed of freely diffusing DNA single strands. Upon cooling, still at relatively high temperatures, the strands progressively assemble into star-shaped particles. The NSs do not show any relevant interaction, with the other NSs in solution, except for excluded volume and electrostatic repulsion. In an implicit solvent picture, the system behaves as a gas of weakly interacting particles. At sufficiently low temperatures, below the binding temperature of the sticky ends Tb (the melting temperature of the sticky sequence), the NSs start to bind via the sticky tips on their arms, progressively assembling into larger aggregates up to the formation of a spanning network of bonded particles (gel state). The binding process, differently from a chemical reaction originating covalent bonds, is reversible so that the gel can be melted and reformed multiple times on changing the temperature. The authors also showed that a phase-separation (ps) phenomenon takes place in the system at low enough densities (when the concentration of DNA NSs is not sufficient to give rise to a connected network). When this is the case, particles separate into a NS-poor and NS-rich phase for temperatures below Tps. This process is very similar to the liquid-liquid phase-separation that forms membrane-less compartments in cells (Hyman et al., 2014) and could be fruitfully employed to create biocompatible chemical reaction zones (Nguyen and Saleh, 2017). Outside the phase-separation region, the gelation proceeds via equilibrium states in which the bond breaking and reforming allows stress release. Later, Bomboi et al. (2015) investigated the effect of the solution ionic strength on the phase behavior. In particular, they demonstrated that a higher NaCl salt concentration results in an increase of the temperature below which the network forms. Indeed, the larger concentration of ions screens the electrostatic repulsion of the NSs (due to the negative charge of the DNA phosphate backbone), facilitating the hybridization processes between complementary sequences. In line with experimental results, numerical simulations carried on by Rovigatti et al. (2014) showed that high salt concentrations broaden the phase-separation region to higher temperature and concentration values. The results of these studies can be summarized in the following points: (i) the binding temperature increases with the increasing length of the sticky-end sequence and the ionic strength of the solution. One must be careful to design the DNA sequences to guarantee a well-defined temperature-gap between the self-assembly of the NSs and the temperature at which the sticky sequences start to bind. By playing with the sequence, it becomes possible to generate systems that gel at the desired temperature (higher or lower than the body temperature, for example). (ii) The phase-separation concentration range shifts to higher values with the increase of the ionic strength of the solution, as the electrostatic repulsion between the NSs decreases. (iii) As the valence, f, of the NSs increases, both the temperature and concentration values of the phase-separation curve shift to larger values. This teaches us that gels with very low concentration of DNA can be created when the number of arms is small (i.e. functionality three). This behavior was also found in colloidal systems (for example, in patchy hard spheres), which constitute the simplest model of sticky NSs (Bianchi et al., 2006; Bianchi et al., 2011). Single component mixtures often provide sufficient control over the behavior of a DNA-gel. The addition of one or more components vastly increases their versatility and the range of applicability. By carefully selecting particles with different valence, one is able to assemble DNA-hydrogels with peculiar structural and dynamical properties.
Figure 2.
Experimental realization of limited-valence DNA particles of valence f=3 (A) and f=4 (B), respectively, with the corresponding oligonucleotide composition. The particles are realized by self-assembling in solution three and four complementary DNA strands, respectively, that spontaneously hybridize into the desired arm-structures. (C) Temperature behavior of the two systems. At high temperature, the solutions are composed of freely diffusive DNA single strands; at intermediate temperature, the strands start to hybridize into the star-shaped geometries that, on further cooling, spontaneously assemble via the hybridization of the sticky sequences positioned at the end of each arm. (D) Phase diagram of the selected DNA NSs highlighting how the reduction of the valence shrinks the coexistence region to lower temperature and concentration values. Figure adapted from (Biffi et al., 2013).
Re-Entrant DNA-Gel
Bomboi et al. (2016) used DNA NSs mixed with single DNA strands, which are able to cap the sticky overhangs below a specific temperature. They experimentally proved that one could obtain a suspension of tetravalent A-particles capable of assembling into a gel structure only within a specific range of temperatures, being fluid outside this selected temperature interval. The temperature range of this “circular” (re-entrant) phase diagram (see Figure 3) can be modified at design time by a careful selection of the DNA sequences. As the upper-temperature limit is given by the ability of the NSs to interact with each other, hence from the sequence encoded in the sticky-ends, the lower temperature below which the gel melts can be adjusted by the proper selection of the competitor ss-DNA sequence. Interestingly, the authors have demonstrated that this material can be fluid at room temperature and a viscous gel at body temperature.
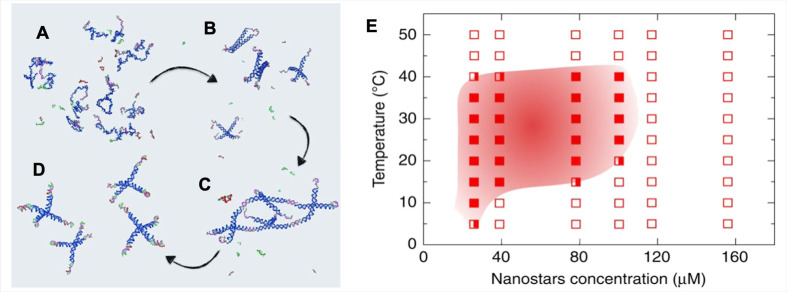
Figure 3.
Re-entrant DNA gel. (A) At very high temperatures, the strands composing the A and B particles are all non-hybridized. (B) High-temperature behavior, the four strands comprising the A particles have now hybridized to form the tetravalent NSs, while the B particle sequences remain in their ss-configuration. (C) Intermediate-temperature behavior, different NSs bind via the sticky sequences to form a gel. The B particles have not yet hybridized. (D) Low-temperature behavior, where the B particles have displaced the AA bonds, originating free-floating AB4 clusters. (E) Phase-diagram of the re-entrant DNA system. Full squares denote phase-separated samples; open squares identify homogeneous solutions; half-full squares represent borderline cases. Figure adapted with permission under Creative Commons license CC BY 4.0 from (Bomboi et al., 2016).
Swappable DNA-Gels
The physics of the bond swapping process (Zhang and Winfree, 2009) makes it possible to realize a reconfigurable all-DNA-gel system (Bomboi et al., 2019). The study provides proof that a smart design of the DNA sequences, able to self-assemble into tetra- and bi-valent nanoparticles, can generate a permanently bonded DNA-gel capable of swapping its bonds in a controlled way via toehold-mediated exchange reactions (Zhang and Winfree, 2009; Smallenburg et al., 2013; Srinivas et al., 2013). This peculiar behavior confers self-restructuring and stress-releasing properties to the gel, making it suitable for the implementation of tailored materials whose mechanical properties can be easily tuned by varying few external parameters (Montarnal et al., 2011). Besides, the non-toxicity and biocompatibility of the DNA nanoparticles make this system a promising platform for in vivo engineering applications.
Stimuli-Responsive DNA-Gels
Temperature is not the only knob one can turn to tune the behavior of DNA-hydrogels. In fact, in biological applications, temperature control may not even be possible in principle since the environment is – to a certain extent – fixed to physiological conditions. The response to other external conditions can be tweaked, however, by smart modifications of the DNA sequences which account for the gel formation. Nucleic acids have been thus exploited as a flexible toolbox for the realization of many different intriguing systems.
Response to Environment pH
DNA-hydrogels consisting of interconnected Y-shaped monomers (Figure 4A), able to pass from a fluid to a gel state in a reversible fashion by varying the pH of the solution, were engineered by Cheng et al. (2009). This peculiar behavior is achieved by properly designing the DNA monomers with ss-terminal interlocking domains, rich in cytosine. In a slightly acidic environment, the partial protonation of some cytosines favors the conjugation with the unprotonated cytosines belonging to a different domain, effectively linking two different monomers. At a proper monomer concentration, this leads to the formation of a spanning three-dimensional DNA-hydrogel. The further variation of the pH back to a basic solution allows restoring the free monomers configuration of the system.
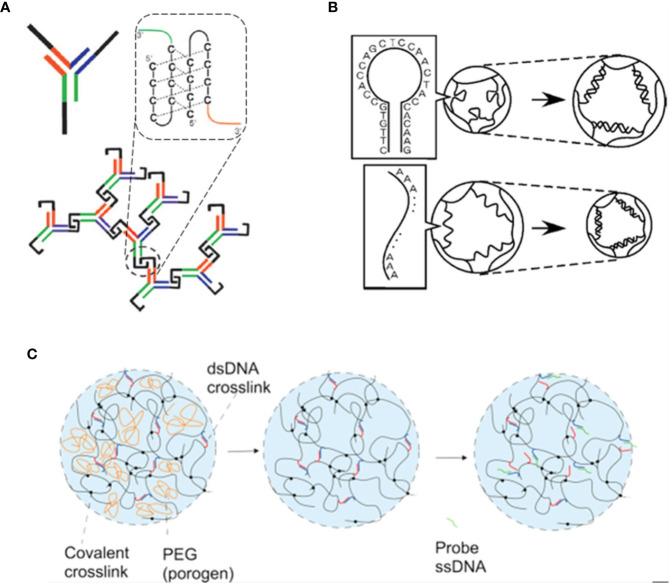
Figure 4.
Examples of stimuli-responsive DNA-gels. (A) Schematics of a pH-responsive DNA system. Y-shaped DNA monomers bind to form a crosslinked hydrogel structure. The network formation is driven by varying the pH of the solution: at acidic pH values, the partial protonation of the C-rich domains (black) favor the hybridization between different monomers, as highlighted in the dashed box. Figure adapted from (Cheng et al., 2009). (B) Example of DNA hydrogels that can swell or shrink depending on the type of DNA crosslinkers. For stem-loop DNA crosslinks, the gel swells (top), while for linear ss-DNA intermolecular links, the gel shrinks (bottom). Figure from (Murakami and Maeda, 2005). (C) A DNA-polyacrylamide hybrid hydrogel with ds-DNA crosslinks able to differently respond to specific target ss-DNA sequences. The addition of polyethylene glycol (PEG) during the hydrogel preparation (left) - which is removed after the polymerization process (center) - allows the system to improve its swelling dynamics after the addition of a ss-DNA probe (right). Figure from (Gao et al., 2015).
Response to Species in Solution
In Figure 4B, we report a gel that can selectively respond to the presence of specific ss-DNA sequences in solution. Murakami and Maeda (Murakami and Maeda, 2005) showed that the network of a polymer gel could be made shrink or swell in the presence of ss-DNA sequences in the solvent if the crosslinkers (the bridges that keep the network glued) consist of purposely designed ss-DNA sequences. More precisely, by using as crosslinkers ss-DNA that hybridize into a stem-loop structure, the gel would swell when the target DNA in solution pairs with the sequence. In contrast, when DNA crosslinks maintain their linear geometry, the gel is found to shrink. Based on the same competitive displacement reactions, Gao et al. (2015) showed the experimental realization of a DNA-polyacrylamide hybrid hydrogel able to swell the network if a polyethylene glycol (PEG) agent is introduced during the preparation of the hydrogel and removed at the end of the synthesis. In such a case, the PEG acts as a pore-forming compound improving the swelling network dynamics that is activated by specific ss-DNA sequences (see Figure 4C). The proper design of hybrid hydrogels with selective responsiveness to sequences in solution could open many new pathways for biomedical applications and DNA-triggered sensing schemes. It might be also applied to load noncoding RNA molecules, such as microRNAs and long noncoding RNAs, active key regulators of cell differentiation and other cellular processes.
Photoresponsivity
An intriguing class of stimuli-responsive systems includes gels that selectively respond to light excitations. Such type of DNA-hydrogels can be realized by adequately modifying the DNA sequences with an azobenzene moiety, a light-responsive molecule that undergoes a trans to cis isomerization after irradiation with UV light and is able to return to the stable trans conformation when illuminated with visible light. The attractive possibility to convert such type of DNA-hydrogels into sol-gel states via reversible reactions, driven by the photo-regulated transition of the azobenzene, has been exploited by Kang et al. (2011) for the controllable encapsulation and release of several loads, including specific drugs, small molecules, nanoparticles, and proteins.
DNA-Gels Joined by Ligation
In 2006, Luo et al. experimentally realized anisotropic DNA nanoparticles with different shapes, able to self-assemble into different DNA-hydrogels (Figure 5) whose crosslinks have been made permanent via the use of the appropriate T4-ligase enzyme (Um et al., 2006). The tailored mechanical properties of the assembled network architectures can be changed at will by varying the type and concentration of the monomers. Besides, the exploitation of appropriate ligase-mediated reactions allows the gelation process to take place at physiological conditions. The intrinsic properties of DNA make these materials promising platforms for large applicability, in particular for in vivo encapsulation and release of specific drugs.
Figure 5.
Examples of different DNA hydrogels realized by the assembly of specific DNA monomers of different shapes: X-DNA (A), Y-DNA (B), and Z-DNA (C). Figure redrawn from (Um et al., 2006).
Stability of DNA-Gels
In the clinical conditions, a ‘perfect’ nano-material should guarantee efficacy of the treatment and safety. In this respect, stability/degradation are important features to be considered when developing DNA-GEL for biomedical applications. Stability has been analyzed only in vitro with different results likely dependent on the type of DNA-gel. Um et al. designed a DNA-gel with tetra- and trimeric NSs bound by ligation (Um et al., 2006) which degradation in vitro, measured as the amount of DNA-gel/day in PBS at 37°C, was linked to the number of NSs branches. Over 70% of tetrameric NSs were still present after 14 days, while trimeric NSs were soon degraded in a few days. Interestingly, they found that the degradation was delayed if DNA-gel was loaded with drugs. DNA-hydrogel stability was also measured after DNase digestion (Nishikawa et al., 2014; Song et al., 2018). Although a variable degradation of DNA-hydrogel was reported (5-60%), higher stability of the DNA network with respect to free components was found. In other experiments, the stability was analyzed in serum to mimic cell culture conditions. However, the degradation of DNA-hydrogels was higher with high disassembly after 12hrs (Nishikawa et al., 2011; Wei et al., 2019).
DNA-Hydrogels and Immune Response
The ability of the immune system to recognize external materials, with activation of the immune response by antigen-presenting cells and phagocytic cells (e.g. neutrophils, monocytes and macrophages) is well known (Chaplin, 2010). Nanomaterials were often found to induce the activation of the immune response at different levels, depending on properties like size, surface charge, among others (Adabi et al., 2017). This variability in response should be considered when designing new nanomaterials. The use of DNA-hydrogels is therefore of interest due to the low immunogenicity described in literature (Nishikawa et al., 2014; Umeki et al., 2017; Shao et al., 2018; Obuobi et al., 2019) and the possibility to functionalize them in order to boost the immune response in a controlled manner, with applications in vaccine and immunological therapy. Many DNA-gels have been designed with the insertion of CpG Oligodeoxynucleotides (ODN), synthetic oligonucleotides derived from bacteria, very effective in activating immune response (Klinman, 2004). Distinct classes of ODNs, composed of unmethylated CG dinucleotides flanked by a phosphorothioate backbone and palindromic regions, have been identified (Klinman, 2004; Scheiermann and Klinman, 2014) and tested, thus showing their high immunogenicity (Nishikawa et al., 2014; Ishii-Mizuno et al., 2017; Umeki et al., 2017; Shao et al., 2018; Komura et al., 2020). Shao et al. developed a vaccine, based on a self-assembled DNA-hydrogel of trimeric NSs and linker DNA containing CpG ODNs, functionalized with an antigen recognized by B and T-helper cells effective in the recruitment and activation of antigen-presenting cells (Shao et al., 2018). In murine macrophage-like cells, CpG DNA-hydrogels activation of cytokines was increased with respect to CpG ODN alone, thus suggesting that immobilization of CpG in the DNA network might improve their efficiency (Shao et al., 2018). Nishikawa et al. developed an injectable DNA-hydrogel (Nishikawa et al., 2014) functionalized with CpG ODNs and tested as a carrier of antigenic drug Ovalbumin (OVA), showing stimulation of immune response and increased activation of antigen-presenting cells in mouse macrophage-like cells. In mice, this DNA-hydrogel induced prolonged expression of cytokines at the injection site and not in serum, suggesting a local activation.
DNA-Hydrogels for Delivery of Drugs and Nucleic Acids
Many efforts in nanomedicine have been made to develop carriers for drug delivery capable of overcoming the limits of free drug administration, such as low stability and insolubility in biological systems, short half-life, and systemic distribution (Yadav et al., 2020). As previously discussed, DNA-hydrogels are biodegradable and, in principle, the versatile nature of DNA sequences and their ability to respond to external stimuli allow their utilization for efficient encapsulation and release of drugs and other molecules. Several systems based on DNA-hydrogels made of NSs have been designed for loading and release of drugs (Um et al., 2006; Nishikawa et al., 2011; Nishikawa et al., 2014; Song et al., 2015b; Ishii-Mizuno et al., 2017; Umeki et al., 2017; Wei et al., 2019; Yue et al., 2019), nucleic acids, (Li et al., 2015; Nishida et al., 2016; Ma et al., 2018; Song et al., 2018; Komura et al., 2020), and proteins (Liu et al., 2018). The DNA-hydrogel can easily trap molecules by varying the stiffness and pore size of the DNA structure, as shown by Yue et al. (2019). The release of molecules can occur passively, through degradation of DNA (Um et al., 2006; Nishikawa et al., 2011; Nishikawa et al., 2014; Obuobi et al., 2019), or, actively, in response to external stimuli (e.g. when gold particles are heated upon irradiation with NIR wavelength) (Song et al., 2015a; Song et al., 2015b). The main achievement in nanomedicine is the development of nanocarriers for tissue and cell targeting reviewed in (Yu et al., 2016; Patra et al., 2018). In this case, the most used ligands are antibodies or peptides, though their selection and conjugation are difficult (Yu et al., 2016). An alternative is the use of DNA-hydrogels functionalized with aptamers (Li et al., 2015; Liu et al., 2018; Wei et al., 2019). In this perspective, DNA-hydrogels are suitable tools in the field of cancer therapy. The major limit in chemotherapy is the high toxicity of the treatment due to the systemic administration and the incapacity of chemo-drug to discriminate between normal and tumor cells (Ahlawat et al., 2018; Schirrmacher, 2019). Tumor environment shows increased permeability and acidity, and selected receptors are overexpressed on cancer cells allowing precise targeting (Ahlawat et al., 2018) and the above-mentioned characteristics of DNA-hydrogels well fit with cancer issues for the targeted delivery of chemo-drugs. Several DNA-hydrogels have been designed for release of Doxorubicin (DOX) (Nishikawa et al., 2011; Song et al., 2015a; Song et al., 2015b; Wei et al., 2019), which can be easily loaded into DNA-hydrogels for its interaction with DNA (Jawad et al., 2019; Mirzaei-Kalar et al., 2020). Among others, Wei et al. developed a sophisticated tumor-targeting DNA-hydrogel with a pH-controlled release of DOX (Wei et al., 2019). Recently, DNA-gels have been developed for the therapeutic release of nucleic acids (Li et al., 2015; Nishida et al., 2016; Song et al., 2018; Komura et al., 2020), such as small interfering RNA (siRNAs) for inhibition of target mRNA expression (Ahmadzada et al., 2018). siRNA systems have elicited interest in cancer therapy to suppress the expression of oncogenes. However, prototypes increasing the stability of siRNA must be developed (Kim et al., 2016). Song et al. designed a DNA-hydrogel producing siRNA (I-Gel) (Song et al., 2018) showing high silencing capability in vitro and a good cellular uptake without cell toxicity.
Biological Effects of DNA-Hydrogels on Cell Cultures and Prospective Applications
The developing of DNA-hydrogels for biomedical applications requires the evaluation of their biological effects in human body tissues. The first step is the characterization of DNA-gels on cell cultures, which allows deep understanding of cellular and molecular mechanisms (Garcia et al., 2019). Moreover, the availability of cell lines derived from several tissues is useful to predict specific organ responses and develop tailoring protocols for different therapeutic applications (Adams and Brantner, 2006). However, while the studies on the physical structure and properties of DNA-gels are advanced, the application on cells is less investigated. Consequently, very little is known about the interaction between DNA-gels and cells and the possible cell specific response. One of the main problems in nanomedicine is cell toxicity caused by chemical and physical characteristics of nanomaterials (Sukhanova et al., 2018). To evaluate this issue and the interplay between DNA-gel and cells, preliminary characterization should consider the ability of DNA-hydrogels to affect basic cell properties like viability, proliferation and migration (Garcia et al., 2019). Currently, cell viability is the most analyzed parameter. It was found that DNA-gels did not cause alteration of viability in human dermal and tumor cell lines, and in mouse macrophage-like cells treated with trimeric or tetrameric NSs DNA-gels (Liu et al., 2018; Obuobi et al., 2019; Wei et al., 2019; Yue et al., 2019), and with NSs complexed with gold nanoparticles (Song et al., 2015a; Song et al., 2015b). The evaluation of cell death pathways is necessary, together with viability, since programmed cell death is a common outcome after nanoparticles exposure (Namvar et al., 2015; Sun et al., 2018; Fritsch-Decker et al., 2019). Wey et al. studied the effects of a DNA-gel composed of tetrameric NSs on cell viability and apoptosis in human breast cancer MCF-7 and lung adenocarcinoma A549 cell lines showing an increase in early apoptotic cells (Wei et al., 2019). Another question concerns the ability of NSs DNA-hydrogels to assembly in well-defined networks and the possibility that DNA-gels can be sensed by cells as physical stimuli, similar to artificial matrix used in cell culture applications (Trepat et al., 2012; Loh and Choong, 2013; Panzetta et al., 2019; Huang et al., 2020). In this view, the study of cell migration could be helpful to understand the interaction between DNA-gels and cells. Migration capability has been analyzed by Li et al. using a tetrameric NSs DNA-hydrogel containing an aptamer targeting lung adenocarcinoma A549 cell line (Li et al., 2015). They found that DNA-gel reduced cell migration on plates and a weak effect was shown even in the absence of the targeting aptamer. The above-mentioned works suggest the ability of DNA-hydrogel to influence basic cell properties. However, further studies are needed to better characterize the effect of DNA-gels on cell toxicity, cell cycle progression, proliferation and cell sensing in different cell models. Novel biological applications may take advantage from a choice of different DNA-hydrogel prototypes, however, their use in cell biology and tumors is at beginning due to their novelty. To this aim, the previously described re-entrant DNA-gel (Bomboi et al., 2016) is a very promising material for novel biomedical applications. Indeed, as above discussed, the re-entrant DNA-gel exhibits unusual phase-behavior, namely a fluid phase at 20°C and a gel state within a temperature range centered around the human body value (Figure 3). These peculiarities suggest that re-entrant DNA-gels might be used to encapsulate cells without detrimental effects. Actually, the possibility to encapsulate cells makes biomaterials promising tools for therapeutic applications, as they make it possible to retain cells in the site of injection and to replace or repair tissue damaged through trauma or disease (Cai et al., 2015). Mesenchymal (MSC), embryonic (ESC), and adult neural (NSC) stem cells have been widely used for this purpose. Their encapsulation into hydrogels was able to sustain cell growth and induce differentiation, with promising effects in tissue regeneration (Lei et al., 2011; Lim et al., 2012; Cheng et al., 2013; Kim et al., 2015; Xu et al., 2015). In addition, biomimetic scaffolds can also be used to reproduce sophisticated 3D culture systems to study cancer cells in more reliable conditions than conventional 2D models, through cell microenvironment engineering. Indeed, the features of the hydrogels, such as elasticity, permeability, adhesion, pore size, and biodegradability, can impact signal transduction pathways of cancer cells, particularly the growth and invasion ability as well as the response to therapies (Loessner et al., 2010; Ananthanarayanan et al., 2011; Liang et al., 2011; Szot et al., 2011; Beck et al., 2013; Del Bufalo et al., 2016). Therefore, in vitro engineered 3D tumors with defined microenvironment would provide novel information concerning the role of microenvironment in directing biological responses. Based on this information, the use of a re-entrant DNA-gel may be a novel opportunity to support medical treatment and therapies. In particular, among various possible applications, the interplay between the re-entrant DNA-gel and cancer stem cells, a small subpopulation of cells found in tumors and considered responsible for tumor onset and recurrence (Reya et al., 2001), could be an interesting field to explore. Cancer stem cells isolated from many types of tumor have the ability to grow as floating spheres in vitro (Salvatori et al., 2012; Favia et al., 2019) offering an additional model to investigate the effects mediated by re-entrant DNA-GEL. Moreover, as tumor spheroids better reflect the in vivo microenvironment than 2D cell cultures, they represent an appropriate model to evaluate signaling pathways and drug delivery (Mehta et al., 2012; Lee et al., 2013). Although Matrigel is often used as a scaffold for cancer cell spheres (Lang et al., 2001; Yeung et al., 2010; Badea et al., 2019), a re-entrant DNA-gel may be a new advance for spheroid growth and applications in cancer research, including cell response to therapies. Furthermore, cell-based therapy for regenerative medicine purposes could also benefit from the use of re-entrant DNA-gels. However, to be validated as a 3D scaffold for biomedical applications, it is mandatory for re-entrant DNA-gel to possess specific properties necessary to sustain the interaction with cells. The most important requirement is biocompatibility, which means that the material is able to support cell growth and that after implantation it does not induce an immune reaction (O’Brien, 2011). Hydrogels also need precise control over bioligand presentation in order to elicit desired cellular responses. Indeed, cell adhesion to ECM provides structural support and dynamic biochemical signalling regulating cell behaviors regarding the form, function, and fate of the cells, critical to tissue morphogenesis, homeostasis, and repair (Hynes, 2009; Rice et al., 2013; Schiller and Fassler, 2013). Biodegradability is another important factor to evaluate as the hydrogel should possess favorable degradation properties which provide the space for cell stretching, proliferation, and release (Kloxin et al., 2010; Lei et al., 2011; Schultz et al., 2015). Polymer concentration and crosslinking density are essential to determine hydrogel stiffness which can affect both stem cell pluripotency and differentiation, as well as cell migration (Leipzig and Shoichet, 2009; Musah et al., 2012; Kim et al., 2015). Adequate porosity of the scaffold is also needed for proper tissue regeneration. Indeed, although large pore size facilitates the transport of nutrients, too large pores may reduce cell infiltration due to the lack of contacts between cells and ECM. However, too small pores may trap cells, decreasing migration speed again (Dadsetan et al., 2008; Keskar et al., 2009; Teng et al., 2014). Following the many examples reported above, the main characteristics of the re-entrant DNA-gels will have to be investigated to validate this 3D scaffold as a novel tool in the integration of biomaterials engineering with the control of the aberrant homeostasis of cancer tissue, including cell reprogramming.
Conclusion
Recent advances in DNA nanotechnology have allowed interdisciplinary cooperation among physical, biomedical, and chemical sciences, unlocking many new pathways for possible diagnostic and detection schemes. In particular, rationally designed DNA-hydrogels have garnered an increasing interest due to their versatility, cost-effectiveness, and biocompatibility, making them potential candidates for tissue engineering applications, self-healing, drug delivery, and cell culture. DNA-gels have been developed by physicists and material scientists. There is a lack of biological applications. DNA-gels might be successfully employed to obtain a targeted delivery inside tumor mass and preliminary ongoing applications on cells are extremely promising. DNA-gels show peculiar differences respect to classical substrates for cell cultures such as Matrix protein components. Among these, controlled gelation properties and the possibility to design sequences to obtain the desired structure with desired bulk behavior. In addition, DNA-gels can be supplied with nucleic acids such as microRNA, siRNA or DNA molecules. These peculiarities make DNA-gels an optimal matrix that might be used as a molecular selective filter to remove dangerous molecules such as oncomiRs or others expressed in the serum of cancer patients. Another application relies on the use of re-entrant gels that could be injected as a fluid at low temperature (20°C) and then become gelified inside the tissue at body temperature (37°C). Finally, the use of DNA-gels as a controlled environment where cancer cells can be included supplemented with desired effectors or molecules might be an additional possibility. In this respect it was interestingly shown that cell senescence, induced by low doses of chemotherapeutic drug, boost anticancer natural immune response in a Multiple Myeloma mouse model (Antonangeli et al., 2016), in this respect treatment of tumor cells with DNA-hydrogel might enhance the natural killer NK cells activity leading to eradication of cancer cells. We believe also that the 3D sponge-like structure of DNA-gels might represent a promising novel substrate for testing the cell behaviour in an intercellular microenvironment. Specifically, it is suitable to assay if selected drugs, hormones or other factors might be able to redifferentiate the cancer phenotype to obtain cells with a more physiological behaviour according to the Anakoinosis concept.
Author Contributions
All authors contributed to the article and approved the submitted version. EL and ML contributed equally to writing and revision.
Funding
The review has been supported by LAZIO INNOVA Regione Lazio, Progetto DNA-GEL Grant No. 85857-0051-0085.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lazio Innova for Post-Doc grants to EL and ML. All images were modified and used in respect to ‘Rights and permissions’ where required.
References
- Adabi M., Naghibzadeh M., Adabi M., Zarrinfard M. A., Esnaashari S. S., Seifalian A. M., et al. (2017). Biocompatibility and nanostructured materials: applications in nanomedicine. Artif. Cells Nanomed. Biotechnol. 45, 833–842. 10.1080/21691401.2016.1178134 [DOI] [PubMed] [Google Scholar]
- Adams C. P., Brantner V. V. (2006). Estimating the cost of new drug development: is it really 802 million dollars? Health Aff. (Millwood) 25, 420–428. 10.1377/hlthaff.25.2.420 [DOI] [PubMed] [Google Scholar]
- Ahlawat J., Henriquez G., Narayan M. (2018). Enhancing the Delivery of Chemotherapeutics: Role of Biodegradable Polymeric Nanoparticles. Molecules 23, 2157. 10.3390/molecules23092157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzada T., Reid G., Mckenzie D. R. (2018). Fundamentals of siRNA and miRNA therapeutics and a review of targeted nanoparticle delivery systems in breast cancer. Biophys. Rev. 10, 69–86. 10.1007/s12551-017-0392-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan B., Kim Y., Kumar S. (2011). Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials 32, 7913–7923. 10.1016/j.biomaterials.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonangeli F., Soriani A., Ricci B., Ponzetta A., Benigni G., Morrone S., et al. (2016). Natural killer cell recognition of in vivo drug-induced senescent multiple myeloma cells. Oncoimmunology 5, e1218105. 10.1080/2162402X.2016.1218105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea M. A., Balas M., Hermenean A., Ciceu A., Herman H., Ionita D., et al. (2019). Influence of Matrigel on Single- and Multiple-Spheroid Cultures in Breast Cancer Research. SLAS Discov. 24, 563–578. 10.1177/2472555219834698 [DOI] [PubMed] [Google Scholar]
- Beck J. N., Singh A., Rothenberg A. R., Elisseeff J. H., Ewald A. J. (2013). The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination. Biomaterials 34, 9486–9495. 10.1016/j.biomaterials.2013.08.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini T., Zanchetta G., Fraccia T. P., Cerbino R., Tsai E., Smith G. P., et al. (2012). Liquid crystal self-assembly of random-sequence DNA oligomers. Proc. Natl. Acad. Sci. 109, 1110. 10.1073/pnas.1117463109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., Largo J., Tartaglia P., Zaccarelli E., Sciortino F. (2006). Phase diagram of patchy colloids: towards empty liquids. Phys. Rev. Lett. 97, 168301. 10.1103/PhysRevLett.97.168301 [DOI] [PubMed] [Google Scholar]
- Bianchi E., Blaak R., Likos C. N. (2011). Patchy colloids: state of the art and perspectives. Phys. Chem. Chem. Phys. 13, 6397–6410. 10.1039/c0cp02296a [DOI] [PubMed] [Google Scholar]
- Biffi S., Cerbino R., Bomboi F., Paraboschi E. M., Asselta R., Sciortino F., et al. (2013). Phase behavior and critical activated dynamics of limited-valence DNA nanostars. Proc. Natl. Acad. Sci. U. S. A. 110, 15633–15637. 10.1073/pnas.1304632110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomboi F., Biffi S., Cerbino R., Bellini T., Bordi F., Sciortino F. (2015). Equilibrium gels of trivalent DNA-nanostars: Effect of the ionic strength on the dynamics. Eur. Phys. J. E. 38, 64. 10.1140/epje/i2015-15064-9 [DOI] [PubMed] [Google Scholar]
- Bomboi F., Romano F., Leo M., Fernandez-Castanon J., Cerbino R., Bellini T., et al. (2016). Re-entrant DNA gels. Nat. Commun. 13191, 1–6. 10.1038/ncomms13191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomboi F., Caprara D., Fernandez-Castanon J., Sciortino F. (2019). Cold-Swappable DNA gels. Nanoscale 11, 9691–9697. 10.1039/C9NR01156K [DOI] [PubMed] [Google Scholar]
- Cai L., Dewi R. E., Heilshorn S. C. (2015). Injectable Hydrogels with In Situ Double Network Formation Enhance Retention of Transplanted Stem Cells. Adv. Funct. Mater. 25, 1344–1351. 10.1002/adfm.201403631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliari S. R., Burdick J. A. (2016). A practical guide to hydrogels for cell culture. Nat. Methods 13, 405–414. 10.1038/nmeth.3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J., Lin Y., Liu H., Wang L., Fan C. (2015). DNA-based plasmonic nanostructures. Elsevier 18, 326–335. 10.1016/j.mattod.2015.01.018 [DOI] [Google Scholar]
- Chaplin D. D. (2010). Overview of the immune response. J. Allergy Clin. Immunol. 125, S3–23. 10.1016/j.jaci.2009.12.980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E., Xing Y., Chen P., Yang Y., Sun Y., Zhou D., et al. (2009). A pH-Triggered, Fast-Responding DNA Hydrogel. Angew. Chem. Int. Ed. 48, 7660–7663. 10.1002/anie.200902538 [DOI] [PubMed] [Google Scholar]
- Cheng T. Y., Chen M. H., Chang W. H., Huang M. Y., Wang T. W. (2013). Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials 34, 2005–2016. 10.1016/j.biomaterials.2012.11.043 [DOI] [PubMed] [Google Scholar]
- Cherkasov V. R., Mochalova E. N., Babenyshev A. V., Vasilyeva A. V., Nikitin P. I., Nikitin M. P. (2020). Nanoparticle Beacons: Supersensitive Smart Materials with On/Off-Switchable Affinity to Biomedical Targets. ACS Nano 14, 1792–1803. 10.1021/acsnano.9b07569 [DOI] [PubMed] [Google Scholar]
- Condon A. (2006). Designed DNA molecules: principles and applications of molecular nanotechnology. Nat. Rev. Genet. 7, 565–575. 10.1038/nrg1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadsetan M., Hefferan T. E., Szatkowski J. P., Mishra P. K., Macura S. I., Lu L., et al. (2008). Effect of hydrogel porosity on marrow stromal cell phenotypic expression. Biomaterials 29, 2193–2202. 10.1016/j.biomaterials.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bufalo F., Manzo T., Hoyos V., Yagyu S., Caruana I., Jacot J., et al. (2016). 3D modeling of human cancer: A PEG-fibrin hydrogel system to study the role of tumor microenvironment and recapitulate the in vivo effect of oncolytic adenovirus. Biomaterials 84, 76–85. 10.1016/j.biomaterials.2016.01.030 [DOI] [PubMed] [Google Scholar]
- Favia A., Salvatori L., Nanni S., Iwamoto-Stohl L. K., Valente S., Mai A., et al. (2019). The Protein Arginine Methyltransferases 1 and 5 affect Myc properties in glioblastoma stem cells. Sci. Rep. 9, 15925. 10.1038/s41598-019-52291-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castanon J., Bomboi F., Sciortino F. (2018). Binding branched and linear DNA structures: From isolated clusters to fully bonded gels. J. Chem. Phys. 148, 025103–025108. 10.1063/1.5011720 [DOI] [PubMed] [Google Scholar]
- Fritsch-Decker S., An Z., Yan J., Hansjosten I., Al-Rawi M., Peravali R., et al. (2019). Silica Nanoparticles Provoke Cell Death Independent of p53 and BAX in Human Colon Cancer Cells. Nanomater. (Basel) 9, 1172 10.3390/nano9081172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Gawel K., Stokke B. T. (2015). Swelling Dynamics of a DNA-Polymer Hybrid Hydrogel Prepared Using Polyethylene Glycol as a Porogen. Gels 1, 219–234. 10.3390/gels1020219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E., Shinde R., Martinez S., Kaushik A., Chand H. S., Nair M., et al. (2019). “Chapter 12 - Cell-Line-Based Studies of Nanotechnology Drug-Delivery Systems: A Brief Review,” in Nanocarriers for Drug Delivery. Eds. Mohapatra S. S., Ranjan S., Dasgupta N., Mishra R. K., Thomas S. (Amsterdam, Netherlands: Elsevier; ), 375–393. [Google Scholar]
- Gu L., Mooney D. J. (2016). Biomaterials and emerging anticancer therapeutics: engineering the microenvironment. Nat. Rev. Cancer 16, 56–66. 10.1038/nrc.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Abdalla A. M. E., Xiao L., Yang G. (2020). Biopolymer-Based Microcarriers for Three-Dimensional Cell Culture and Engineered Tissue Formation. Int. J. Mol. Sci. 21, 1895. 10.3390/ijms21051895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A. A., Weber C. A., Jülicher F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell. Dev. Biol. 30, 39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Hynes R. O. (2009). The Extracellular Matrix: Not Just Pretty Fibrils. Science 326, 1216. 10.1126/science.1176009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii-Mizuno Y., Umeki Y., Onuki Y., Watanabe H., Takahashi Y., Takakura Y., et al. (2017). Improved sustained release of antigen from immunostimulatory DNA hydrogel by electrostatic interaction with chitosan. Int. J. Pharm. 516, 392–400. 10.1016/j.ijpharm.2016.11.048 [DOI] [PubMed] [Google Scholar]
- Jawad B., Poudel L., Podgornik R., Steinmetz N. F., Ching W. Y. (2019). Molecular mechanism and binding free energy of doxorubicin intercalation in DNA. Phys. Chem. Chem. Phys. 21, 3877–3893. 10.1039/C8CP06776G [DOI] [PubMed] [Google Scholar]
- Kang H., Liu H., Zhang X., Yan J., Zhu Z., Peng L., et al. (2011). Photoresponsive DNA-cross-linked hydrogels for controllable release and cancer therapy. Langmuir 27, 399–408. 10.1021/la1037553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskar V., Marion N. W., Mao J. J., Gemeinhart R. A. (2009). In vitro evaluation of macroporous hydrogels to facilitate stem cell infiltration, growth, and mineralization. Tissue Eng. Part A. 15, 1695–1707. 10.1089/ten.tea.2008.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., An D. B., Oh S. H., Kang M. K., Song H. H., Lee J. H. (2015). Creating stiffness gradient polyvinyl alcohol hydrogel using a simple gradual freezing–thawing method to investigate stem cell differentiation behaviors. Biomaterials 40, 51–60. 10.1016/j.biomaterials.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Kim A., Miyata K., Kataoka K. (2016). Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv. Rev. 104, 61–77. 10.1016/j.addr.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Klinman D. M. (2004). Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 4, 249–258. 10.1038/nri1329 [DOI] [PubMed] [Google Scholar]
- Kloxin A. M., Tibbitt M. W., Kasko A. M., Fairbairn J. A., Anseth K. S. (2010). Tunable hydrogels for external manipulation of cellular microenvironments through controlled photodegradation. Adv. Mater. (Deerfield Beach Fla.) 22, 61–66. 10.1002/adma.200900917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura F., Okuzumi K., Takahashi Y., Takakura Y., Nishikawa M. (2020). Development of RNA/DNA Hydrogel Targeting Toll-Like Receptor 7/8 for Sustained RNA Release and Potent Immune Activation. Molecules 25, 728. 10.3390/molecules25030728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S. H., Sharrard R. M., Stark M., Villette J. M., Maitland N. J. (2001). Prostate epithelial cell lines form spheroids with evidence of glandular differentiation in three-dimensional Matrigel cultures. Br. J. Cancer 85, 590–599. 10.1054/bjoc.2001.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Mhawech-Fauceglia P., Lee N., Parsanian L. C., Lin Y. G., Gayther S. A., et al. (2013). A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Invest. 93, 528–542. 10.1038/labinvest.2013.41 [DOI] [PubMed] [Google Scholar]
- Lei Y., Gojgini S., Lam J., Segura T. (2011). The spreading, migration and proliferation of mouse mesenchymal stem cells cultured inside hyaluronic acid hydrogels. Biomaterials 32, 39–47. 10.1016/j.biomaterials.2010.08.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipzig N. D., Shoichet M. S. (2009). The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials 30, 6867–6878. 10.1016/j.biomaterials.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Li J., Zheng C., Cansiz S., Wu C., Xu J., Cui C., et al. (2015). Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 137, 1412–1415. 10.1021/ja512293f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Jeong J., Devolder R. J., Cha C., Wang F., Tong Y. W., et al. (2011). A cell-instructive hydrogel to regulate malignancy of 3D tumor spheroids with matrix rigidity. Biomaterials 32, 9308–9315. 10.1016/j.biomaterials.2011.08.045 [DOI] [PubMed] [Google Scholar]
- Lim T. C., Toh W. S., Wang L. S., Kurisawa M., Spector M. (2012). The effect of injectable gelatin-hydroxyphenylpropionic acid hydrogel matrices on the proliferation, migration, differentiation and oxidative stress resistance of adult neural stem cells. Biomaterials 33, 3446–3455. 10.1016/j.biomaterials.2012.01.037 [DOI] [PubMed] [Google Scholar]
- Liu C., Han J., Pei Y., Du J. (2018). Aptamer Functionalized DNA Hydrogel for Wise-Stage Controlled Protein Release. Appl. Sci. 8, 1941. 10.3390/app8101941 [DOI] [Google Scholar]
- Loessner D., Stok K. S., Lutolf M. P., Hutmacher D. W., Clements J. A., Rizzi S. C. (2010). Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31, 8494–8506. 10.1016/j.biomaterials.2010.07.064 [DOI] [PubMed] [Google Scholar]
- Loh Q. L., Choong C. (2013). Three-dimensional scaffolds for tissue engineering applications: role of porosity and pore size. Tissue Eng. Part B. Rev. 19, 485–502. 10.1089/ten.teb.2012.0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., He C., Cheng Y., Yang Z., Zang J., Liu J., et al. (2015). Localized Co-delivery of Doxorubicin, Cisplatin, and Methotrexate by Thermosensitive Hydrogels for Enhanced Osteosarcoma Treatment. ACS Appl. Mater. Interfaces 7, 27040–27048. 10.1021/acsami.5b09112 [DOI] [PubMed] [Google Scholar]
- Ma Y., Liu H., Mou Q., Yan D., Zhu X., Zhang C. (2018). Floxuridine-containing nucleic acid nanogels for anticancer drug delivery. Nanoscale 10, 8367–8371. 10.1039/C8NR01226A [DOI] [PubMed] [Google Scholar]
- Macfarlane R. J., Lee B., Jones M. R., Harris N., Schatz G. C., Mirkin C. A. (2011). Nanoparticle Superlattice Engineering with DNA. Science 334, 204–208. 10.1126/science.1210493 [DOI] [PubMed] [Google Scholar]
- Mehta G., Hsiao A. Y., Ingram M., Luker G. D., Takayama S. (2012). Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control Release 164, 192–204. 10.1016/j.jconrel.2012.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei-Kalar Z., Yavari A., Jouyban A. (2020). Increasing DNA binding affinity of doxorubicin by loading on Fe3O4 nanoparticles: A multi-spectroscopic study. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 229, 117985. 10.1016/j.saa.2019.117985 [DOI] [PubMed] [Google Scholar]
- Montarnal D., Capelot M., Tournilhac F., Leibler L. (2011). Silica-Like Malleable Materials from Permanent Organic Networks. Science 334, 965–968. 10.1126/science.1212648 [DOI] [PubMed] [Google Scholar]
- Murakami Y., Maeda M. (2005). DNA-Responsive Hydrogels That Can Shrink or Swell. Biomacromolecules 6, 2927–2929. 10.1021/bm0504330 [DOI] [PubMed] [Google Scholar]
- Musah S., Morin S. A., Wrighton P. J., Zwick D. B., Jin S., Kiessling L. L. (2012). Glycosaminoglycan-binding hydrogels enable mechanical control of human pluripotent stem cell self-renewal. ACS Nano 6, 10168–10177. 10.1021/nn3039148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naahidi S., Jafari M., Logan M., Wang Y., Yuan Y., Bae H., et al. (2017). Biocompatibility of hydrogel-based scaffolds for tissue engineering applications. Biotechnol. Adv. 35, 530–544. 10.1016/j.biotechadv.2017.05.006 [DOI] [PubMed] [Google Scholar]
- Namvar F., Rahman H. S., Mohamad R., Azizi S., Tahir P. M., Chartrand M. S., et al. (2015). Cytotoxic Effects of Biosynthesized Zinc Oxide Nanoparticles on Murine Cell Lines. Evidence-Based Complement. Altern. Med. 2015, 593014. 10.1155/2015/593014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. T., Saleh O. A. (2017). Tuning phase and aging of DNA hydrogels through molecular design. Soft. Matter 13, 5421–5427. 10.1039/C7SM00557A [DOI] [PubMed] [Google Scholar]
- Nishida Y., Ohtsuki S., Araie Y., Umeki Y., Endo M., Emura T., et al. (2016). Self-assembling DNA hydrogel-based delivery of immunoinhibitory nucleic acids to immune cells. Nanomedicine 12, 123–130. 10.1016/j.nano.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Nishikawa M., Mizuno Y., Mohri K., Matsuoka N., Rattanakiat S., Takahashi Y., et al. (2011). Biodegradable CpG DNA hydrogels for sustained delivery of doxorubicin and immunostimulatory signals in tumor-bearing mice. Biomaterials 32, 488–494. 10.1016/j.biomaterials.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Nishikawa M., Ogawa K., Umeki Y., Mohri K., Kawasaki Y., Watanabe H., et al. (2014). Injectable, self-gelling, biodegradable, and immunomodulatory DNA hydrogel for antigen delivery. J. Control Release 180, 25–32. 10.1016/j.jconrel.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Obuobi S., Tay H. K., Tram N. D. T., Selvarajan V., Khara J. S., Wang Y., et al. (2019). Facile and efficient encapsulation of antimicrobial peptides via crosslinked DNA nanostructures and their application in wound therapy. J. Control Release 313, 120–130. 10.1016/j.jconrel.2019.10.013 [DOI] [PubMed] [Google Scholar]
- O’Brien F. J. (2011). Biomaterials & scaffolds for tissue engineering. Mater. Today 14, 88–95. 10.1016/S1369-7021(11)70058-X [DOI] [Google Scholar]
- Pan A., Wang Z., Chen B., Dai W., Zhang H., He B., et al. (2018). Localized co-delivery of collagenase and trastuzumab by thermosensitive hydrogels for enhanced antitumor efficacy in human breast xenograft. Drug Deliv. 25, 1495–1503. 10.1080/10717544.2018.1474971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzetta V., Fusco S., Netti P. A. (2019). Cell mechanosensing is regulated by substrate strain energy rather than stiffness. Proc. Natl. Acad. Sci. U. S. A. 116, 22004–22013. 10.1073/pnas.1904660116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N., Ho Um S., Funabashi H., Xu J., Luo D. (2009). A cell-free protein-producing gel. Nat. Mater. 8, 432–437. 10.1038/nmat2419 [DOI] [PubMed] [Google Scholar]
- Parmar P. A., Chow L. W., St-Pierre J. P., Horejs C. M., Peng Y. Y., Werkmeister J. A., et al. (2015). Collagen-mimetic peptide-modifiable hydrogels for articular cartilage regeneration. Biomaterials 54, 213–225. 10.1016/j.biomaterials.2015.02.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra J. K., Das G., Fraceto L. F., Campos E. V. R., Rodriguez-Torres M. D. P., Acosta-Torres L. S., et al. (2018). Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 16, 71. 10.1186/s12951-018-0392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell B. P., Lobb D., Charati M. B., Dorsey S. M., Wade R. J., Zellars K. N., et al. (2014). Injectable and bioresponsive hydrogels for on-demand matrix metalloproteinase inhibition. Nat. Mater. 13, 653–661. 10.1038/nmat3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Morrison S. J., Clarke M. F., Weissman I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111. 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Rice J. J., Martino M. M., De Laporte L., Tortelli F., Briquez P. S., Hubbell J. A. (2013). Engineering the regenerative microenvironment with biomaterials. Adv. Healthc. Mater. 2, 57–71. 10.1002/adhm.201200197 [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. (2006). Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302. 10.1038/nature04586 [DOI] [PubMed] [Google Scholar]
- Rovigatti L., Bomboi F., Sciortino F. (2014). Accurate phase diagram of tetravalent DNA nanostars. J. Chem. Phys. 140, 154903–154910. 10.1063/1.4870467 [DOI] [Google Scholar]
- Salvatori L., Caporuscio F., Verdina A., Starace G., Crispi S., Nicotra M. R., et al. (2012). Cell-to-cell signaling influences the fate of prostate cancer stem cells and their potential to generate more aggressive tumors. PLoS One 7, e31467. 10.1371/journal.pone.0031467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann J., Klinman D. M. (2014). Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine 32, 6377–6389. 10.1016/j.vaccine.2014.06.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H. B., Fassler R. (2013). Mechanosensitivity and compositional dynamics of cell-matrix adhesions. EMBO Rep. 14, 509–519. 10.1038/embor.2013.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirrmacher V. (2019). From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 54, 407–419. 10.3892/ijo.2018.4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz K. M., Kyburz K. A., Anseth K. S. (2015). Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc. Natl. Acad. Sci. U. S. A. 112, E3757–E3764. 10.1073/pnas.1511304112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C. (2003). DNA in a material world. Nature 421, 427–431. 10.1038/nature01406 [DOI] [PubMed] [Google Scholar]
- Seeman N. C. (2016). Structural DNA Nanotechnology (Cambridge: Cambridge University Press; ). [Google Scholar]
- Shao Y., Sun Z. Y., Wang Y., Zhang B. D., Liu D., Li Y. M. (2018). Designable Immune Therapeutical Vaccine System Based on DNA Supramolecular Hydrogels. ACS Appl. Mater. Interfaces 10, 9310–9314. 10.1021/acsami.8b00312 [DOI] [PubMed] [Google Scholar]
- Smallenburg F., Leibler L., Sciortino F. (2013). Patchy Particle Model for Vitrimers. Phys. Rev. Lett. 111, 188002–188005. 10.1103/PhysRevLett.111.188002 [DOI] [PubMed] [Google Scholar]
- Song J., Hwang S., Im K., Hur J., Nam J., Hwang S., et al. (2015. a). Light-responsible DNA hydrogel–gold nanoparticle assembly for synergistic cancer therapy. J. Mater. Chem. B. 3, 1537–1543. 10.1039/C4TB01519C [DOI] [PubMed] [Google Scholar]
- Song J., Im K., Hwang S., Hur J., Nam J., Ahn G. O., et al. (2015. b). DNA hydrogel delivery vehicle for light-triggered and synergistic cancer therapy. Nanoscale 7, 9433–9437. 10.1039/C5NR00858A [DOI] [PubMed] [Google Scholar]
- Song J., Lee M., Kim T., Na J., Jung Y., Jung G. Y., et al. (2018). A RNA producing DNA hydrogel as a platform for a high performance RNA interference system. Nat. Commun. 9, 4331. 10.1038/s41467-018-06864-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas N., Ouldridge T. E., Šulc P., Schaeffer J. M., Yurke B., Louis A. A., et al. (2013). On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 41, 10641–10658. 10.1093/nar/gkt801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova A., Bozrova S., Sokolov P., Berestovoy M., Karaulov A., Nabiev I. (2018). Dependence of Nanoparticle Toxicity on Their Physical and Chemical Properties. Nanoscale Res. Lett. 13, 44–44. 10.1186/s11671-018-2457-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Jia J., Jiang C., Zhai S. (2018). Gold Nanoparticle-Induced Cell Death and Potential Applications in Nanomedicine. Int. J. Mol. Sci. 19, 754. 10.3390/ijms19030754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szot C. S., Buchanan C. F., Freeman J. W., Rylander M. N. (2011). 3D in vitro bioengineered tumors based on collagen I hydrogels. Biomaterials 32, 7905–7912. 10.1016/j.biomaterials.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng W., Long T. J., Zhang Q., Yao K., Shen T. T., Ratner B. D. (2014). A tough, precision-porous hydrogel scaffold: ophthalmologic applications. Biomaterials 35, 8916–8926. 10.1016/j.biomaterials.2014.07.013 [DOI] [PubMed] [Google Scholar]
- Thiele J., Ma Y., Bruekers S. M., Ma S., Huck W. T. (2014). 25th anniversary article: Designer hydrogels for cell cultures: a materials selection guide. Adv. Mater. 26, 125–147. 10.1002/adma.201302958 [DOI] [PubMed] [Google Scholar]
- Tibbitt M. W., Anseth K. S. (2009). Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 103, 655–663. 10.1002/bit.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X., Chen Z., Jacobson K. (2012). Cell migration. Compr. Physiol. 2, 2369–2392. 10.1002/cphy.c110012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou Y.-H., Khoneisser J., Huang P.-C., Xu X. (2016). Hydrogel as a bioactive material to regulate stem cell fate. Bioact. Mater. 1, 39–55. 10.1016/j.bioactmat.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S. H., Lee J. B., Park N., Kwon S. Y., Umbach C. C., Luo D. (2006). Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 5, 797–801. 10.1038/nmat1741 [DOI] [PubMed] [Google Scholar]
- Umeki Y., Saito M., Takahashi Y., Takakura Y., Nishikawa M. (2017). Retardation of Antigen Release from DNA Hydrogel Using Cholesterol-Modified DNA for Increased Antigen-Specific Immune Response. Adv. Healthc. Mater. 6, 1700355. 10.1002/adhm.201700355 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Zheng X., Ducrot É., Yodh J. S., Weck M., et al. (2015). Crystallization of DNA-coated colloids. Nat. Commun. 6, 1–8. 10.1038/ncomms8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Zhao Z., Wang Y., Zou J., Lin Q., Duan Y. (2019). One-Step Self-Assembly of Multifunctional DNA Nanohydrogels: An Enhanced and Harmless Strategy for Guiding Combined Antitumor Therapy. ACS Appl. Mater. Interfaces 11, 46479–46489. 10.1021/acsami.9b15874 [DOI] [PubMed] [Google Scholar]
- Winfree E., Liu F., Wenzler L. A., Seeman N. C. (1998). Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–:544. 10.1038/28998 [DOI] [PubMed] [Google Scholar]
- Xiang B., He K., Zhu R., Liu Z., Zeng S., Huang Y., et al. (2016). Self-Assembled DNA Hydrogel Based on Enzymatically Polymerized DNA for Protein Encapsulation and Enzyme/DNAzyme Hybrid Cascade Reaction. ACS Appl. Mater. Interfaces 8, 22801–22807. 10.1021/acsami.6b03572 [DOI] [PubMed] [Google Scholar]
- Xu K., Narayanan K., Lee F., Bae K. H., Gao S., Kurisawa M. (2015). Enzyme-mediated hyaluronic acid-tyramine hydrogels for the propagation of human embryonic stem cells in 3D. Acta Biomater. 24, 159–171. 10.1016/j.actbio.2015.06.026 [DOI] [PubMed] [Google Scholar]
- Yadav S., Sharma A. K., Kumar P. (2020). Nanoscale Self-Assembly for Therapeutic Delivery. Front. Bioeng. Biotechnol. 8, 127. 10.3389/fbioe.2020.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T. M., Gandhi S. C., Wilding J. L., Muschel R., Bodmer W. F. (2010). Cancer stem cells from colorectal cancer-derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 107, 3722–3727. 10.1073/pnas.0915135107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Trase I., Ren M., Duval K., Guo X., Chen Z. (2016). Design of Nanoparticle-Based Carriers for Targeted Drug Delivery. J. Nanomater. 2016, 1087250. 10.1155/2016/1087250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L., Wang S., Wulf V., Willner I. (2019). Stiffness-switchable DNA-based constitutional dynamic network hydrogels for self-healing and matrix-guided controlled chemical processes. Nat. Commun. 10, 4774. 10.1038/s41467-019-12697-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. Y., Winfree E. (2009). Control of DNA Strand Displacement Kinetics Using Toehold Exchange. J. Am. Chem. Soc 131, 17303–17314. 10.1021/ja906987s [DOI] [PubMed] [Google Scholar]
- Zheng J., Birktoft J. J., Chen Y., Wang T., Sha R., Constantinou P. E., et al. (2009). From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77. 10.1038/nature08274 [DOI] [PMC free article] [PubMed] [Google Scholar]