Abstract
Background
Inflammatory bowel disease (IBD) can manifest both macroscopically and microscopically in the oral cavity; however, little is known about salivary changes in IBD. Therefore, this study aimed to assess salivary and circulatory inflammatory profiles in IBD and to compare their potential to reflect the presence and activity of IBD.
Methods
We measured 92 known inflammatory proteins in serum and in unstimulated and stimulated whole saliva samples from patients with IBD with active intestinal inflammation (n = 21) and matched control patients (n = 22) by proximity extension assay. Fifteen of the patients with IBD returned 10 to 12 weeks after treatment escalation for resampling.
Results
Sixty-seven of the proteins were detected in all 3 sample fluids but formed distinct clusters in serum and saliva. Twenty-one inflammatory proteins were significantly increased and 4 were significantly decreased in the serum of patients with IBD compared with that of the control patients. Two of the increased serum proteins, IL-6 and MMP-10, were also significantly increased in stimulated saliva of patients with IBD and correlated positively to their expressions in serum. None of the investigated proteins in serum or saliva were significantly altered by IBD treatment at follow-up. Overall, inflammatory proteins in serum correlated to biochemical status, and salivary proteins correlated positively to clinical parameters reflecting disease activity.
Conclusions
Saliva and serum inflammatory profiles in IBD share a similar composition but reflect different aspects of disease activity. The oral cavity reflects IBD through elevated IL-6 and MMP-10 in stimulated saliva.
Keywords: saliva, serum, inflammatory bowel disease, proteomics
Saliva and serum inflammatory profiles reflect different aspects of inflammatory bowel disease (IBD) activity. The proteins IL-6 and MMP-10, both associated with extraintestinal manifestations, are elevated in stimulated saliva of patients with IBD. This finding adds new insights to the oral-gut connection in IBD.
INTRODUCTION
Inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC), is a group of chronic immune-mediated inflammatory diseases that affect more than 3.5 million individuals worldwide.1 It is a complex multifactorial disease not exclusive to the gastrointestinal tract, known to manifest itself in other organs in up to 50% of patients, coincident with intestinal manifestations or independently.2, 3
One such organ is the oral cavity, where patients can develop mucocutaneous lesions that resemble those in the bowel as they go into flare and remission and respond to IBD treatment.4 These include CD-specific oral mucosal cobblestoning, orofacial granulomatosis, and mucosal tags, along with an increased prevalence of common oral pathologies such as aphtous ulcers, stomatitis, and glossitis—all recognized as important early signs of disease because they may precede intestinal symptoms and are associated with more severe phenotypes of CD in pediatric patients.4, 5 Oral manifestations of UC have been less studied, although pyostomatitis vegetans, characterized by oral intra- and subepithelial microabscesses similar to those found in colons affected by UC, is considered to be specific to UC.6 Clinically healthy oral mucosa of patients with CD has also been shown to contain granulomas, histopathological hallmarks of intestinal CD.7 Moreover, both adult and pediatric patients with IBD suffer from caries and periodontal disease to a larger extent compared to healthy control patients.8
However, do not seem to be restricted to oral mucosa but have also been described in saliva. Saliva is an easily accessible oral biofluid secreted from the parotid-, submandibular-, sublingual-, and minor salivary glands that reside in the oral mucosa. It can be obtained either by passive drooling or by gustatory/masticatory stimulation and is thus referred to as unstimulated and stimulated saliva, respectively. Unstimulated saliva consists mainly of secretions from the submandibular and sublingual glands with a high mucin component, and stimulated saliva is mainly composed of parotid secretions, which are less viscous and more abundant in volume.9 Saliva contributes to mucous membrane integrity, protects from dryness and ulcer formation, enhances mucosal repair, and serves important immunoregulatory functions including the expression of secretory immunoglobulin A and the secretion of immune-suppressive cytokines and defensins along with mucins.10
Salivary constituents are known to reflect local and systemic diseases.11 Several proinflammatory proteins, such as interleukin (IL)-6, IL-1β, and tumor necrosis factor (TNF)-α have been shown to be significantly elevated in the saliva of patients with IBD compared with that of control patients.12, 13 We have recently shown that calprotectin is significantly elevated in the saliva of patients with IBD, most markedly in naive patients with CD, in whom it tends to decrease following treatment.14 Other described changes in saliva from patients with IBD include altered antioxidant activity, increased oxidative stress, and dysbiosis of the oral microbiome.15-17
Still, investigations involving the saliva of patients with IBD are scarce and limited to singular findings. Our aim was to broadly explore the inflammatory protein profiles in saliva compared to those in serum in patients with IBD and control patients using a high-multiplex immunoassay panel, to relate the findings to clinical data that reflect IBD presence and activity, and further, to assess the impact of IBD treatment on saliva and serum protein profiles.
METHODS
Study Participants
Patients with IBD (n = 21) were recruited at the Division of Gastroenterology at Karolinska University Hospital, Stockholm, Sweden, between October 2015 and March 2018. Inclusion criteria were age ≥18 years, a diagnosis of CD or UC, and active intestinal inflammation verified by endoscopy. The exclusion criteria were pregnancy, current breastfeeding, comorbidities of the gastrointestinal tract (ie, any diagnosis other than IBD), symptoms from the oral cavity, diagnosis of Sjögren’s syndrome or systemic lupus erythematosus, and previous irradiation of the head and neck. Demographic and clinical data, including Montreal classification,18 surgical and medical treatment, and blood counts were retrieved from patient records when available. Endoscopic disease activity at baseline was graded according to the Ulcerative Colitis Endoscopic Index of Severity (UCEIS) or the Simple Endoscopic Score for Crohn’s disease (SES-CD).19, 20 Further, patients’ overall disease activity was graded on a 4-grade scale as a physician global assessment (PGA) by an experienced gastroenterologist and was based on the following parameters; presence of diarrhea, stool frequency, abdominal pain, fatigue, fever, and weight loss. The grades were 0 = clinical remission, 1 = mild disease activity, 2 = moderate disease activity, and 3 = severe disease activity. All 21 patients with IBD were invited for resampling 10 to 12 weeks after treatment escalation, and 15 were seen for follow-up. Reasons for dropouts included lack of treatment compliance (n = 1) or study withdrawal for unknown reasons (n = 5).
Control patients (n = 22) matched for age, sex, body mass index (BMI), and smoking/use of Swedish snuff were recruited at the GHP Stockholm Gastro Center, Stockholm, Sweden, between March and May 2018. The exclusion criteria were the same as for the patients with IBD, with the additional exceptions: inflammatory disease with ongoing medication and antibiotic treatment ≤ 3 months before sampling. All control patients had undergone colonoscopy in the workup of gastrointestinal disorders, and none of them had any macroscopic signs of intestinal inflammation.
Saliva and Serum Sampling
Serum and unstimulated and stimulated whole saliva were sampled from all study participants in conjunction with the endoscopic examination and from 15 of the patients with IBD who returned 10 to 12 weeks after treatment escalation. Blood was drawn into a Z Serum Clot Activator Vacuette tube (Greiner Bio-One, Kremsmünster, Austria) and centrifuged for 15 minutes at 2000 × g, and serum was collected and aliquoted for storage at –80°C until analysis. Unstimulated saliva was collected through passive drooling for 5 to 8 minutes, and stimulated saliva was collected through 5-minute masticatory stimulation by chewing a 0.5 g paraffin tablet (Ivoclar Vivadent, Schaan, Liechtenstein). Samples were placed on ice and centrifuged for 10 minutes at 600 × g, and supernatant was collected and aliquoted for storage at –80°C until analysis. Salivary flow (mL/min) was calculated by dividing sample volume by collection time.
Protein Analysis in Serum and Saliva
Ninety-two inflammation-related proteins were analyzed in the serum and saliva samples using the Olink Inflammation panel (Olink Proteomics, Uppsala, Sweden; Supplementary Table 1). A protease inhibitor was added to the saliva samples before analysis (Sigma-Aldrich, St. Louis, MO). In short, the method was based on proximity extension assay technology: the 92 antibody probe pairs bound to their specific target protein, forming a polymerase chain reaction target sequence through proximity-dependent DNA polymerization, which was detected and quantified using standard real-time polymerase chain reaction.21 The output was normalized in 2 steps and presented in a relative semi-quantitative Normalized Protein eXpression unit. Last, the Normalized Protein eXpression data were transformed into a log2 scale. Samples that did not pass the assay quality control (serum: 5% control patients, 5% patients with IBD baseline; unstimulated saliva: 14% control patients, 10% patients with IBD baseline; stimulated saliva: 14% control patients) were excluded from analysis, and saliva values were adjusted for salivary flow. For this exploratory cohort we set a detection limit of 50%, meaning that analytes detected in < 50% of the serum or unstimulated or stimulated saliva samples were excluded and referred to as undetectable. Total protein concentrations in saliva were measured using the Qubit Protein Assay according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA). Protein interactions and biological functions were investigated using the STRING database.22 Delta values were calculated by subtracting values retrieved from patients with IBD during flare from values obtained after treatment.
Statistical Analysis
Differences between groups in continuous variables were tested by the Mann-Whitney U test. Sex, smoking/Swedish snuff consumption, and protein detectability differences between patients with IBD and control patients were tested using the Pearson χ 2 test. The Wilcoxon signed-rank test was used for all paired analyses. Sample clustering was visually investigated on score plots resulting from principal component analysis (PCA). Significances in fold differences were adjusted for false discovery through the original false discovery rate method of Benjamini and Hochberg, with the false discovery rate set at 5%. Spearman correlation coefficients were calculated for correlations, and analyte sensitivity and specificity were calculated by means of areas under the curve of receiver-operating characteristics (AU-ROC). We used SPSS (version 25.0; IBM Corporation, Armonk, NY), and GraphPad Prism 7 (version 7.04; GraphPad Software Inc., La Jolla, CA) for statistical analysis and the graphical presentation of results. The significance levels were set to P ≤ 0.05 (*), 0.01 (**), or 0.001 (***). All tests were 2-sided.
Ethical Considerations
The study was approved by the Regional Ethical Review Board in Stockholm, Sweden (Dnr 2015/17–31) and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
RESULTS
Demographic and Clinical Characteristics of Study Participants
Demographic and clinical characteristics of the 21 patients with IBD and 22 control patients are presented in Table 1. Eleven of the patients were newly diagnosed and untreated (8 with CD, 3 with UC), and 10 had established and treated disease (4 with CD, 6 with UC). There were no significant differences in age, sex, BMI, or nicotine consumption between patients with IBD and control patients.
TABLE 1.
Demographic and Clinical Characteristics of Patients With IBD and Control Patients
| IBD (n = 21) | Control (n = 22) | P | |
|---|---|---|---|
| Age, y (mean ± SD)* | 43 ± 16 | 47 ± 14 | 0.319 |
| Gender, female/male, n (%)† | 7/14 (33/67) | 8/14 (36/64) | 1.000 |
| BMI, kg/m2 (mean ± SD)* | 25 ± 5 | 27 ± 4 | 0.072 |
| Smoking/snuff habits, n (%)† | 0.755 | ||
| Active | 8 (38) | 7 (32) | |
| Never/former | 13 (62) | 15 (68) | |
| IBD diagnosis, n (%) | |||
| CD | 12 (57) | - | |
| UC | 9 (43) | - | |
| Disease duration, y (mean ± SD) | 3 ± 5 | - | |
| Disease location, CD, n (%)‡ | |||
| Ileum (L1) | 3 (25) | - | |
| Colon (L2) | 6 (50) | - | |
| Ileocolic (L3) | 3 (25) | - | |
| Upper GI (L4) | 0 (0) | - | |
| Disease extent, UC, n (%)‡ | |||
| Proctitis (E1) | 0 (0) | - | |
| Left-sided (E2) | 4 (44) | - | |
| Extensive (E3) | 4 (44) | - | |
| Unknown | 1 (1) | - | |
| Abdominal surgery, n (%) | |||
| Yes | 4 (19) | - | |
| No | 17 (81) | - | |
| Medical treatment, n (%) | |||
| Glucocorticoids | 13 (62) | - | |
| Anti-TNFs/biologics | 4 (19) | - | |
| Thiopurines | 4 (19) | - | |
| 5-ASAs | 3 (14) | - | |
| No treatment | 2 (10) | - | |
| SES-CD (mean ± SD) | 11 ± 5 | - | |
| UCEIS (mean ± SD) | 2 ± 1 | - | |
| Total protein, mg/mL (mean ± SD)* | |||
| Unstimulated saliva | 1.1 ± 0.5 | 0.9 ± 0.3 | 0.392 |
| Stimulated saliva | 0.9 ± 0.3 | 0.8 ± 0.3 | 0.796 |
| Salivary flow, mL/min (mean ± SD)* | |||
| Unstimulated saliva | 0.4 ± 0.2 | 0.3 ± 0.2 | 0.067 |
| Stimulated saliva | 1.8 ± 1.0 | 1.1 ± 0.7 | 0.006 |
*Mann-Whitney U test, control patients vs patients with IBD.
†χ 2 test, control patients vs patients with IBD.
‡According to the Montreal Index.
5-ASA indicates 5-aminosalicylic acid/mesalazine; SD, standard deviation.
Detectability and Interfluid Correlation of Panel Proteins
The 92 inflammatory proteins analyzed are listed in Supplementary Table 1. Altogether, 41 serum samples (20 from patients with IBD, 21 from control patients), 37 unstimulated saliva samples (18 from patients with IBD, 19 from control patients), and 40 stimulated saliva samples (21 from patients with IBD, 19 from control patients) passed the assay quality control and were included in the analysis.
Overall, 80 of the proteins were detected in the serum samples, 72 in the unstimulated saliva, and 69 in the stimulated saliva. Sixty-seven of the proteins were common for all the sample types, and 10 proteins were unique to serum (IL-1RB, SLAMF1, FGF-5, FGF-21, IL-15RA, β-NGF, IL-24, CCL25, NT-3, and IL-5), and IL-1α and LIF were exclusively detected in saliva (Fig. 1A). The proteins MCP-3, GDNF, and IL-10RA were detected in serum and unstimulated saliva but not in stimulated saliva. Another 10 proteins were detectable in less than 50% of the samples in all 3 sample fluids and were thus defined as undetectable (IL-2, TSPL, IL-22RA1, IL-13, TNF, IL-20, IL-33, IFN-γ, IL-4, and NRTN).
FIGURE 1.
Protein panel detectability and fluid correlations. A, Venn diagram depicts the amount of proteins detected in ≥ 50% of all serum, unstimulated, and stimulated saliva samples. B, PCA plot shows distribution of serum, unstimulated, and stimulated saliva for proteins detected in all 3 fluids. C, Correlation between protein expression in serum and saliva for proteins detected in at least 2 of the fluid types. Missing values (because of proteins not being detected in respective fluid) are crossed out. Proteins showing no significant correlation between fluids were excluded from the plot. PC, principal component; S, serum; SS, stimulated saliva; US, unstimulated saliva.
The differences in analyte detection between serum and saliva are shown in Supplementary Figs. 1A-C. In summary, the measured proteins were detected to a higher degree in serum samples than in saliva samples. We detected IL-13 in serum and IL-33 in stimulated saliva in significantly fewer IBD samples compared with control samples (Supplementary Figs. 1A, C). IL-24 was detected in significantly more serum and stimulated saliva samples from patients with IBD than from control patients (Supplementary Figs. 1A, C), and GDNF was detected in more stimulated saliva samples from control patients than from patients with IBD (Supplementary Fig. 1C). There was no significant difference between patients with IBD and control patients regarding the percentage of detectable proteins in unstimulated saliva samples for any protein (Supplementary Fig. 1B).
A PCA of the 67 proteins that were common for all fluids revealed that serum and saliva samples formed distinct clusters (Fig. 1B). Despite certain overlap between unstimulated and stimulated saliva, discrete separation was evident between the two saliva types (Fig. 1B). The expression of most proteins correlated positively between unstimulated and stimulated saliva (Fig. 1C). Eleven proteins common to serum and stimulated saliva correlated positively and one (AXIN1) correlated negatively, whereas only 6 proteins in unstimulated saliva correlated positively to their expression in serum (Fig. 1C). Four proteins correlated positively in all 3 fluids (CXCL9, CCL11, CCL23, and MMP-10).
Significantly Altered Inflammatory Proteins in Serum and Saliva of Patients With IBD
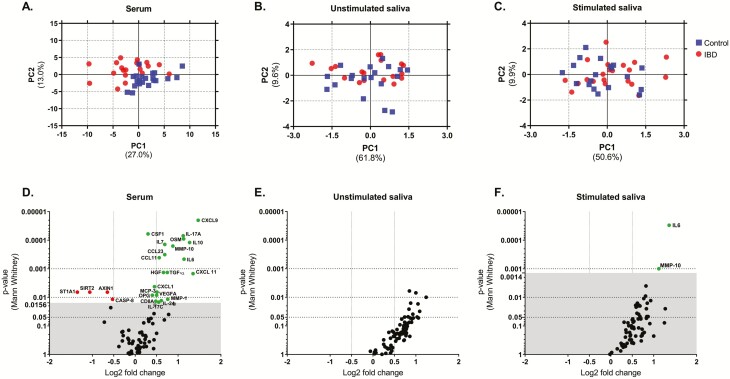
The PCA of detected proteins in serum segregated patients with IBD from control patients, whereas no obvious difference based on the protein expression patterns in either unstimulated or stimulated saliva was observed between patients with IBD and control patients (Figs. 2A-C).
FIGURE 2.
Sample clustering and protein expression in patients with IBD vs control patients. PCA plot shows distribution of IBD and control samples for proteins detected in ≥50% samples, in (A) serum, (B) unstimulated, and (C) stimulated saliva. Volcano plots for statistical significance (Mann-Whitney U test, adjusted for false discovery rate) against mean fold changes of detected proteins between patients with IBD and control patients in (D) serum, (E) unstimulated, and (F) stimulated saliva. The red dots represent significantly reduced proteins in patients with IBD compared with control patients, green dots represent significantly elevated proteins, and black dots represent insignificant proteins. False discovery rate threshold for discoveries in serum was 0.0156, and it was 0.0014 in stimulated saliva.
Out of the detected proteins, 21 were significantly elevated in the serum of patients with IBD with active disease compared with control patients, and 4 were decreased (Fig. 2D). None of the proteins differed significantly between patients with IBD and control patients in unstimulated saliva (Fig. 2E), and IL-6 and MMP-10 were significantly elevated in stimulated saliva of patients with IBD (Fig. 2F). The altered serum proteins were mainly interconnected cytokines and chemokines involved in regulation of the cellular response to stimuli, cell proliferation and function (Supplementary Fig. 2). Certain increased proteins were involved in the IL-17 and Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway. The decreased proteins did not have any known or predicted interactions with each other and were mainly involved in the regulation of proteolysis (Supplementary Fig. 2).
Out of the significantly altered proteins, salivary IL-6 had the highest AU-ROC for distinguishing patients with IBD from control patients, followed by the significantly increased proteins in serum (salivary IL-6 AU-ROC = 0.886; serum CXCL9 AU-ROC = 0.864; Table 2). The sensitivity and specificity of MMP-10 in stimulated saliva corresponded to the AU-ROC of the lowest third of serum proteins. The 4 significantly decreased proteins had an AU-ROC ranging from 0.729 to 0.745 (Table 2).
TABLE 2.
Sensitivity and Specificity of Altered Serum Proteins in IBD: ROC Analysis
| Protein | AUC | 95% CI | P * | |
|---|---|---|---|---|
| Elevated, serum | CXCL9 | 0.864 | 0.744-0.984 | <0.001 |
| CSF1 | 0.848 | 0.720-0.975 | <0.001 | |
| IL-17A | 0.845 | 0.709-0.982 | <0.001 | |
| OSM | 0.840 | 0.723-0.958 | <0.001 | |
| IL-7 | 0.833 | 0.704-0.962 | <0.001 | |
| IL-10 | 0.836 | 0.710-0.961 | <0.001 | |
| MMP-10 | 0.831 | 0.707-0.955 | <0.001 | |
| CCL23 | 0.817 | 0.672-0.961 | 0.001 | |
| CCL11 | 0.812 | 0.675-0.949 | 0.001 | |
| IL-6 | 0.810 | 0.677-0.942 | 0.001 | |
| HGF | 0.786 | 0.638-0.934 | 0.002 | |
| TGF-α | 0.786 | 0.638-0.934 | 0.002 | |
| CXCL11 | 0.783 | 0.638-0.929 | 0.002 | |
| CXCL1 | 0.757 | 0.599-0.915 | 0.005 | |
| MCP-3 | 0.745 | 0.594-0.897 | 0.007 | |
| VEGFA | 0.738 | 0.586-0.890 | 0.009 | |
| OPG | 0.738 | 0.569-0.907 | 0.009 | |
| MMP-1 | 0.729 | 0.576-0.881 | 0.012 | |
| CD8A | 0.724 | 0.564-0.883 | 0.014 | |
| IL-17C | 0.721 | 0.564-0.879 | 0.015 | |
| IL-24 | 0.720 | 0.562-0.878 | 0.016 | |
| Elevated, saliva | IL-6 | 0.886 | 0.776-1.000 | <0.001 |
| MMP-10 | 0.778 | 0.620-0.935 | 0.003 | |
| Decreased, serum | AXIN1 | 0.745 | 0.583-0.907 | 0.007 |
| SIRT1 | 0.745 | 0.589-0.901 | 0.007 | |
| STA1A | 0.745 | 0.595-0.895 | 0.007 | |
| CASP-8 | 0.729 | 0.572-0.885 | 0.012 |
*ROC, asymptotic significance.
AUC indicates area under the curve; ROC, receiver operating characteristics.
Inflammatory Protein Expression in Relation to Clinical Parameters During IBD Flare
Evaluation of correlations between protein expression and clinical parameters revealed more significant correlations in saliva compared with serum (Figs. 3A-C). Serum proteins tended to correlate to biochemical markers of disease activity to a greater extent than saliva proteins. Three proteins in serum correlated positively to C-reactive protein (CRP) and leukocyte concentrations, respectively, and 5 proteins correlated positively and 2 negatively to fecal calprotectin levels (Fig. 3A). Out of the 25 proteins that were altered in the serum of patients with IBD, 9 correlated significantly to at least one of the clinical parameters (IL-17A, IL-17C, IL-10, IL-6, OSM, HGF, CXCL1, VEGFA, and ST1A1).
FIGURE 3.
Correlation between clinical parameters and protein expression in patients with IBD. Correlation between clinical parameters: disease duration, PGA, CRP, fecal calprotectin, blood leukocytes, SES-CD, and the UCEIS and the protein expression in (A) serum, (B) unstimulated saliva, and (C) stimulated saliva. Analyzed by Spearman’s ρ. Only proteins with significant correlation to at least 1 of the clinical parameters are shown. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Salivary inflammatory proteins mainly correlated to clinical parameters reflecting disease activity. Twenty-four proteins in stimulated saliva and 6 in unstimulated saliva correlated positively to SES-CD (Figs. 3B, C). Conversely, IL-8 in unstimulated saliva correlated negatively to SES-CD, and 4 stimulated saliva proteins correlated negatively to the UCEIS (Figs. 3B, C). Overall, the salivary proteins correlated negatively to PGA. In unstimulated saliva, 4 proteins correlated to CRP, and CXCL1 to leukocyte concentrations (Fig. 3B). None of the proteins in stimulated saliva correlated to CRP, and CCL4 correlated negatively to leukocyte concentrations (Fig. 3C). Four proteins in unstimulated saliva and IL-10 in stimulated saliva correlated positively to fecal calprotectin (Figs. 3B, C). Nine of the proteins detected in stimulated saliva correlated positively to disease duration (Fig. 3C). In stimulated saliva, MMP-10 correlated positively with SES-CD, and IL-6 did not correlate significantly to any of the clinical parameters.
Effects of IBD Treatment on the Level of Inflammatory Proteins in Serum and Saliva
Fifteen of the patients with IBD were resampled after 10 to 12 weeks of treatment escalation. Eight had established, previously treated IBD (3 with CD, 5 with UC), and 7 were newly diagnosed and treatment naïve (6 with CD, 1 with UC). Between baseline and resampling, one of the patients with established disease was treated using glucocorticoids, 3 were treated using a combination of glucocorticoids and thiopurines, one was treated using mesalazine, and 3 were treated using TNF-α inhibitors. Five of the newly diagnosed patients had been treated using glucocorticoids, one was treated using glucocorticoids plus mesalazine, and one received no treatment between baseline and resampling.
The PGA and CRP significantly decreased after treatment, whereas no differences were observed in P-albumin, B-leukocytes, or fecal calprotectin (Table 3). No significant differences were observed in flow or total protein concentrations in saliva (data not shown). None of the investigated proteins were significantly altered in serum or in saliva after treatment (Figs. 4A–C). Changes in IL-6 expression after treatment correlated between the 3 fluids (data not shown).
TABLE 3.
Circulatory and Fecal Markers Before and After Treatment of Patients With IBD
| Active Inflammation | After Treatment | P * | |
|---|---|---|---|
| PGA (mean ± SD) | 2 ± 1 | 1 ± 1 | 0.001 |
| CRP, mg/L (mean ± SD) | 14.1 ± 20.6 | 1.3 ± 0.5 | 0.047 |
| Plasma albumin, g/L (mean ± SD) | 34.5 ± 4.6 | 37.4 ± 1.7 | 0.219 |
| Blood leukocytes × 109/L (mean ± SD) | 8.5 ± 4.3 | 6.6 ± 3.2 | 0.147 |
| Fecal calprotectin, mg/kg (mean ± SD) | 1033 ± 959 | 1051 ± 1976 | 0.063 |
*Wilcoxon signed-ranks test, active inflammation vs after treatment (paired samples only).
n = 15.
FIGURE 4.
Alterations in protein expression in patients with IBD following treatment. Volcano plots for statistical significance (Wilcoxon signed-ranks test, adjusted for false discovery rate) against mean fold change of detected proteins in (A) serum, (B) unstimulated, and (C) stimulated saliva after treatment compared with active flare in patients with IBD.
DISCUSSION
In this study, we explored the salivary and circulatory inflammatory profiles of patients with IBD with active intestinal inflammation before and after treatment and related their expression to the presence and activity of IBD. To the best of our knowledge, this is the broadest multiplex analysis of inflammatory proteins in both unstimulated and stimulated whole saliva of patients with IBD with matched serum samples during flare and after treatment. Most of the analyzed proteins were detected in both saliva and in circulation yet followed a distinct expression pattern in saliva compared with serum. Patients with IBD presented significantly elevated levels of IL-6 and MMP-10 in stimulated saliva and several altered serum proteins, none which were affected by treatment at short-term follow-up. Overall, inflammatory proteins in serum correlated to biochemical status and salivary proteins correlated positively to clinical parameters reflecting disease activity.
The majority of the inflammatory proteins, 67 out of 92, were detected in serum and in unstimulated and stimulated saliva samples, which is the highest detection rate of the Olink inflammation panel in saliva to date.23, 24 This result supports the hypothesis that salivary proteins may be analogous to circulatory biomarkers, since approximately 40% of previously suggested disease markers in plasma are detected in saliva as well.25 Yet distinct differences in protein expression patterns were observed between serum and saliva, and the correlation of protein expression between saliva and serum was scarce. Instead, the expression of the majority of the proteins correlated positively between unstimulated and stimulated saliva. It therefore seems that most of the detected proteins in saliva are of local oral origin rather than a transudate of systemic expression.
Interestingly, the protein expression in stimulated saliva was negatively affected by increased salivary flow, implying that parotid secretions dilute the proteins. This result suggests that the potential source of the investigated proteins may be secretions by the submandibular/lingual or minor salivary glands, or mucosal constituents such as oral keratinocytes and leukocytes shed through masticatory stimulation, all of which are present in both saliva types but more abundantly so in stimulated saliva.26
Comparing the expression of inflammatory proteins between patients with IBD and control patients, 21 proteins were significantly elevated and 4 decreased in the serum from patients with IBD. These proteins were mainly cytokines and chemokines related to cellular signaling and migration. The decreased proteins were involved in the positive regulation of proteolysis, while the increased serum proteins were involved in the IL-17 and JAK-STAT signaling pathways, all previously implicated in the pathogenesis of IBD.27-29 Nine of these proteins have been suggested as possible serum markers of IBD when utilizing the same assay; however, there has been no great overlap in findings, which could be due to previous investigations based on patients with IBD without endoscopic evaluation of intestinal inflammation.30
Two of the significantly elevated serum proteins, IL-6 and MMP-10, were also significantly increased in the stimulated saliva of patients with IBD and correlated positively to their respective expressions in serum. The two saliva proteins seemed similarly accurate in distinguishing patients with IBD from control patients as the significantly altered serum proteins. They have both been implicated in the pathogenesis of IBD and have previously been shown to be elevated in serum and in inflamed as well as nonlesional intestinal mucosa of patients with IBD.30-32 Research has also shown IL-6 to be elevated in the unstimulated saliva of patients with IBD, which we could not confirm, possibly because of differences in saliva sampling methodology.12, 13 IL-6 is believed to maintain intestinal inflammation by suppressing the apoptosis of T-cells through signal transducer and activator of transcription 3 and is found in residing macrophages, lymphocytes, and intestinal epithelial cells.33, 34
MMP-10, on the other hand, is a stromelysin whose main function is enzymatic regulation of the extracellular matrix with a hitherto unknown function in IBD. However, overexpression of the enzyme in damaged epithelium results in disordered wound healing, which goes well in hand with the clinical manifestation of IBD in intestinal mucosa.35, 36 Notably, both IL-6 and MMP-10 are speculated to be part of the mechanism generating dermatological manifestations of IBD.35, 37 It would therefore be relevant to validate our findings of elevated IL-6 and MMP-10 in the stimulated saliva of patients with IBD in another cohort and investigate whether they are involved in the pathogenesis of oral mucosal manifestations in IBD as well.
When we related the protein profiles to relevant clinical data that reflected IBD activity, only a few serum proteins correlated with the clinical parameters, and when they did, they mainly reflected the biochemical status of disease activity. In contrast, inflammatory proteins in saliva showed a higher degree of correlation to clinical parameters and predominantly reflected endoscopic activity. Interestingly, MMP-10 expression in stimulated saliva significantly correlated to SES-CD, and IL-6 expression followed a similar but insignificant trend. Our findings suggest that the oral cavity is affected by, and thus reflects, ongoing intestinal inflammation.
We did not observe any significant effect of treatment at short-term follow-up on any of the inflammatory proteins in serum or saliva. An explanation to this could lie within IL-6, which is known not to decrease in saliva or in circulation in several chronic inflammatory conditions after immunomodulatory treatment.38, 39 Given that our STRING-analysis revealed that IL-6 orchestrates the expression of the rest of the significantly altered proteins, it would therefore not be surprising that no other proteins were affected by treatment. Observing this condition from another angle, it would be interesting to assess whether this effect, or rather the lack of it, remains after IL-6 blockade.
This exploratory study comprises a small group of with IBD and control patients; however, it includes an analysis of a large panel of markers measured using a highly specific multiplex immunoassay. Furthermore, the control patients were matched to the patients according to sex, age, BMI, and nicotine consumption to exclude potential confounding factors. We lacked detailed information regarding the amount of consumed nicotine units, but very few proteins (2 in serum, 6 in unstimulated saliva, and 2 in stimulated saliva) were significantly affected by nicotine consumption (data not shown). Another setback of this study is the lack of a detailed oral examination of the study participants. We proceeded from self-reported oral status and can therefore not rule out an effect of undisclosed oral pathology. However, the patient/control patient–matching equalized variables known as predispositions to general oral pathologies such as caries and periodontal disease.40
Another strength of this study is the endoscopic evaluation of the intestinal inflammation in all study participants in direct conjunction to baseline sampling and objective measures of disease activity through blood counts and fecal calprotectin levels. The longitudinal design that encompassed matched sampling after treatment of the patients allowed for investigations on the alterations of the inflammatory profile as an effect of treatment, but the follow-up after treatment escalation was relatively short and patients were no re-assessed endoscopically, which may explain the lack of significant change in inflammatory proteins after treatment.
In conclusion, saliva and serum inflammatory profiles share a similar composition but reflect different aspects of IBD activity. In this exploratory study, patients with IBD presented increased levels of IL-6 and MMP-10 in stimulated saliva; two proteins which have been associated with the pathogenesis and extraintestinal manifestations of IBD. These results add new insights into the oral-gut connection in IBD. If they are proven reliable, the oral cavity may not only provide potential noninvasive disease markers for IBD presence and activity but may also contain clues to the pathophysiology of extraintestinal manifestations of IBD.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Ingrid Ackzell, Lisa Sundin Zarouki, Karin Thourot Nouchi, and Susana Soto Villagran, research nurses at the Department of Gastroenterology at Karolinska Universitetssjukhuset, Stockholm, Sweden, and GHP Stockholm Gastro Center, Stockholm, Sweden, for their help in recruiting and obtaining samples from the patients.
Supported by: This work was supported by the Swedish Research Council (to EAB), the Stockholm County Council (to EAB), Karolinska Institutet Funds (to EAB, SA), the Swedish Dental Society (to MM), and the Ihre Foundation and Thuréus Foundation (to SA). EAB is a recipient of a grant for a half-time position in the clinical research environment from the Swedish Research Council (2012–07110). MM is a recipient of a PhD scholarship through the Clinical Scientist Training Program from the Karolinska Institutet, Sweden. The funders had no role in the study design, data collection/analysis, decision to publish, or manuscript preparation.
Conflicts of interest: The authors declare that this work was carried out without any personal, professional or financial relationships that could potentially be construed as a conflict of interest.
REFERENCES
- 1. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–727. [DOI] [PubMed] [Google Scholar]
- 2. Greuter T, Vavricka SR. Extraintestinal manifestations in inflammatory bowel disease—epidemiology, genetics, and pathogenesis. Expert Rev Gastroenterol Hepatol. 2019;13:307–317. [DOI] [PubMed] [Google Scholar]
- 3. Vavricka SR, Rogler G, Gantenbein C, et al. Chronological order of appearance of extraintestinal manifestations relative to the time of IBD diagnosis in the Swiss inflammatory bowel disease cohort. Inflamm Bowel Dis. 2015;21:1794–1800. [DOI] [PubMed] [Google Scholar]
- 4. Plauth M, Jenss H, Meyle J. Oral manifestations of Crohn’s disease. An analysis of 79 cases. J Clin Gastroenterol. 1991;13:29–37. [DOI] [PubMed] [Google Scholar]
- 5. Pittock S, Drumm B, Fleming P, et al. The oral cavity in Crohn’s disease. J Pediatr. 2001;138:767–771. [DOI] [PubMed] [Google Scholar]
- 6. Ruiz-Roca JA, Berini-Aytés L, Gay-Escoda C. Pyostomatitis vegetans. Report of two cases and review of the literature. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;99:447–454. [DOI] [PubMed] [Google Scholar]
- 7. Sinčić BM, Tomaš MI, Gobić MB, et al. Clinical relevance of CD-68 positive cells in normal buccal mucosa in patients with inflammatory bowel disease. Croat Chem Acta. 2012;85:171–176. [Google Scholar]
- 8. Papageorgiou SN, Hagner M, Nogueira AV, et al. Inflammatory bowel disease and oral health: systematic review and a meta-analysis. J Clin Periodontol. 2017;44:382–393. [DOI] [PubMed] [Google Scholar]
- 9. Edgar WM. Saliva and dental health. Clinical implications of saliva: report of a consensus meeting. Br Dent J. 1990;169:96–98. [DOI] [PubMed] [Google Scholar]
- 10. Mandel ID. The role of saliva in maintaining oral homeostasis. J Am Dent Assoc. 1989;119:298–304. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, Sun J, Lin CC, et al. The emerging landscape of salivary diagnostics. Periodontol 2000. 2016;70:38–52. [DOI] [PubMed] [Google Scholar]
- 12. Nielsen AA, Nielsen JN, Schmedes A, et al. Saliva interleukin-6 in patients with inflammatory bowel disease. Scand J Gastroenterol. 2005;40:1444–1448. [DOI] [PubMed] [Google Scholar]
- 13. Szczeklik K, Owczarek D, Pytko-Polończyk J, et al. Proinflammatory cytokines in the saliva of patients with active and non-active Crohn’s disease. Pol Arch Med Wewn. 2012;122:200–208. [DOI] [PubMed] [Google Scholar]
- 14. Majster M, Almer S, Boström EA. Salivary calprotectin is elevated in patients with active inflammatory bowel disease. Arch Oral Biol. 2019;107:104528. [DOI] [PubMed] [Google Scholar]
- 15. Rezaie A, Ghorbani F, Eshghtork A, et al. Alterations in salivary antioxidants, nitric oxide, and transforming growth factor-beta 1 in relation to disease activity in Crohn’s disease patients. Ann N Y Acad Sci. 2006;1091:110–122. [DOI] [PubMed] [Google Scholar]
- 16. Szczeklik K, Krzyściak W, Cibor D, et al. Markers of lipid peroxidation and antioxidant status in the serum and saliva of patients with active Crohn disease. Pol Arch Intern Med. 2018;128:362–370. [DOI] [PubMed] [Google Scholar]
- 17. Docktor MJ, Paster BJ, Abramowicz S, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5A–36A. [DOI] [PubMed] [Google Scholar]
- 19. Travis SP, Schnell D, Krzeski P, et al. Developing an instrument to assess the endoscopic severity of ulcerative colitis: the Ulcerative Colitis Endoscopic Index of Severity (UCEIS). Gut. 2012;61:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daperno M, D’Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc. 2004;60:505–512. [DOI] [PubMed] [Google Scholar]
- 21. Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Acharya S, Jin C, Bylund J, et al. Reduced sialyl-Lewisx on salivary MUC7 from patients with burning mouth syndrome. Mol Omics. 2019;15:331–339. [DOI] [PubMed] [Google Scholar]
- 24. Di Pietro V, Porto E, Ragusa M, et al. Salivary MicroRNAs: diagnostic markers of mild traumatic brain injury in contact-sport. Front Mol Neurosci. 2018;11:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loo JA, Yan W, Ramachandran P, et al. Comparative human salivary and plasma proteomes. J Dent Res. 2010;89:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Theda C, Hwang SH, Czajko A, et al. Quantitation of the cellular content of saliva and buccal swab samples. Sci Rep. 2018;8:6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pender SLF, MacDonald TT. Proteolytic enzymes in inflammatory bowel disease. Inflamm Bowel Dis. 1998;4:157–164. [DOI] [PubMed] [Google Scholar]
- 28. Moschen AR, Tilg H, Raine T. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol. 2019;16:185–196. [DOI] [PubMed] [Google Scholar]
- 29. De Vries LCS, Wildenberg ME, De Jonge WJ, et al. The future of Janus kinase inhibitors in inflammatory bowel disease. J Crohns Colitis. 2017;11:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersson E, Bergemalm D, Kruse R, et al. Subphenotypes of inflammatory bowel disease are characterized by specific serum protein profiles. PLoS One. 2017;12:e0186142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. León AJ, Gómez E, Garrote JA, et al. High levels of proinflammatory cytokines, but not markers of tissue injury, in unaffected intestinal areas from patients with IBD. Mediators Inflamm. 2009;2009:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dobre M, Milanesi E, Mănuc TE, et al. Differential intestinal mucosa transcriptomic biomarkers for Crohn’s disease and ulcerative colitis. J Immunol Res. 2018;2018:9208274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stevens C, Walz G, Singaram C, et al. Tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-6 expression in inflammatory bowel disease. Dig Dis Sci. 1992;37:818–826. [DOI] [PubMed] [Google Scholar]
- 34. Atreya R, Mudter J, Finotto S, et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in Crohn disease and experimental colitis in vivo. Nat Med. 2000;6:583–588. [DOI] [PubMed] [Google Scholar]
- 35. Bister V, Mäkitalo L, Jeskanen L, et al. Expression of MMP-9, MMP-10 and TNF-alpha and lack of epithelial MMP-1 and MMP-26 characterize pyoderma gangrenosum. J Cutan Pathol. 2007;34:889–898. [DOI] [PubMed] [Google Scholar]
- 36. Shimshoni E, Yablecovitch D, Baram L, et al. ECM remodelling in IBD: innocent bystander or partner in crime? The emerging role of extracellular molecular events in sustaining intestinal inflammation. Gut. 2015;64: 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bernardo D, Vallejo-Díez S, Mann ER, et al. IL-6 promotes immune responses in human ulcerative colitis and induces a skin-homing phenotype in the dendritic cells and T cells they stimulate. Eur J Immunol. 2012;42: 1337–1353. [DOI] [PubMed] [Google Scholar]
- 38. Sikorska D, Orzechowska Z, Rutkowski R, et al. Diagnostic value of salivary CRP and IL-6 in patients undergoing anti-TNF-alpha therapy for rheumatic disease. Inflammopharmacology. 2018;26:1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eng GP, Bouchelouche P, Bartels EM, et al. Anti-drug antibodies, drug levels, interleukin-6 and soluble TNF receptors in rheumatoid arthritis patients during the first 6 months of treatment with adalimumab or infliximab: a descriptive cohort study. PLoS One. 2016;11:e0162316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jepsen S, Blanco J, Buchalla W, et al. Prevention and control of dental caries and periodontal diseases at individual and population level: consensus report of group 3 of joint EFP/ORCA workshop on the boundaries between caries and periodontal diseases. J Clin Periodontol. 2017;44(Suppl 18):S85–S93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.