Abstract
Coronaviruses infect many animals, including humans, due to interspecies transmission. Three of the known human coronaviruses: MERS, SARS-CoV-1, and SARS-CoV-2, the pathogen for the COVID-19 pandemic, cause severe disease. Improved methods to predict host specificity of coronaviruses will be valuable for identifying and controlling future outbreaks. The coronavirus S protein plays a key role in host specificity by attaching the virus to receptors on the cell membrane. We analyzed 1238 spike sequences for their host specificity. Spike sequences readily segregate in t-SNE embeddings into clusters of similar hosts and/or virus species. Machine learning with SVM, Logistic Regression, Decision Tree, Random Forest gave high average accuracies, scores, sensitivities and specificities of 0.95–0.99. Importantly, sites identified by Decision Tree correspond to protein regions with known biological importance. These results demonstrate that spike sequences alone can be used to predict host specificity.
Keywords: coronaviruses, Spike protein, Machine learning, Sequence clustering, Viral host specificity, t-SNE
Highlights
-
•
Coronavirus spike protein sequences can predict host specificity.
-
•
Machine learning had accuracies of >0.98 for classification of human vs non-human hosts.
-
•
Decision Tree classifier identified spike regions with important biological roles .
-
•
Clustering with t-SNE embeddings correctly segregated sequences by virus genus and host .
1. Introduction
The COVID-19 pandemic has heightened public awareness of coronaviruses (CoVs) and our vulnerability to highly contagious infections. CoVs are positive-sense enveloped RNA viruses comprising 2 subfamilies (Orthocoronavirinae and Letovirinae) and 5 genera: (Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Deltacoronavirus, and Alphaletovirus) [1,2]. CoVs infect a wide range of species, including humans, due to a high level of interspecies transmission. Alpha and beta CoVs only infect mammals, including humans, while delta and gamma CoVs are known to infect birds and some mammals, but have not yet been shown to infect humans [3]. Seven species are known to infect humans. HCoV-NL63, HCoV-229E, HCoV-HKU1, HCoV-OC43, and BCoV-1, cause mild respiratory disease. In contrast, SARS-CoV-1, MERS and SARS-CoV-2, the pathogen causing the COVID-19 pandemic, have emerged since 2003 as severe pathogens with significant rates of mortality. These outbreaks are presumed to result from interspecies transmission from bats via intermediate hosts of civets or camels to humans [2]. The SARS-CoV-1 outbreak in China in 2003 had a 10% mortality rate. Ten years later, MERS appeared in the Middle East with a staggering mortality rate of 35%. SARS-CoV-2 is the latest CoV species that was first identified in late 2019, and induced an unprecedented outbreak that continues to affect millions worldwide.
The large genomes of CoVs code for 4 structural proteins and 16 non-structural proteins. The non-structural S protein or spike is critical for virus attachment and fusion to the host cell membrane and is an important determinant of host specificity [[4], [5], [6]]. S protein sequences are highly variable among CoVs and may bind to different host cell receptors. For example, the human pathogen, MERS, uses DPP4 as its host cell receptor, while SARS-CoV-1 and SARS-CoV-2 attach to ACE2 receptor [5,6]. After binding to the host cell receptor, S protein is cleaved into S1 and S2 subunits for cell fusion and entry. These critical stages in the viral life cycle have been proposed as targets for development of antiviral drugs [7].
Due to the lack of a vaccine for human CoVs, it is critical to better understand the potential infectivity of different CoVs. Therefore, we have applied several machine learning algorithms to sequences of S proteins to assess predictions of host cell specificity.
2. Methods
Data collection and preprocessing. Data were obtained from the Virus Pathogen Database https://www.viprbrc.org on April 11, 2020. It included 1253 unaligned S protein sequences of various lengths, which belonged to 67 species of CoVs. We used a standard preprocessing pipeline consisting of: (i) sequence cleaning; (ii) sequence alignment; (iii) sequence encoding; and (iv) dimensionality reduction performed by truncated singular value decomposition (tSVD). This method is very close to principal component analysis (PCA), a more traditional approach for dimensionality reduction. We preferred tSVD since it works more efficiently with sparse vectors as it does not need to center the data [8].
As a part of sequence cleaning, we used known accession numbers to identify viral and/or host species for more than a hundred “unknown” sequences. Also, we removed 15 sequences in the Torovirinae, Piscanivirinae, and Serpentovirinae subfamilies which were previously part of the Coronaviridae family but were reassigned to the Tobaniviridae family in 2018 [1]. The overall distribution of the data with respect to viral species and hosts is shown in Fig. 1 . We used MEGA X software https://www.megasoftware.net to align the sequences. After alignment, all sequences were represented with an identical length of 2396 residues. We applied a well-known one-hot encoding to convert the sequences into numerical vectors for input to machine learning algorithms. The amino acid sequences contained 25 letters ABCDEFGHIJKLMNPQRSTUVWXYZ which represent the canonical 20 amino acids plus 5 codes for ambiguous amino acid assignments [9]. The additional 5 codes are: B for aspartic acid or asparagine, J for leucine or isoleucine, U for selenocysteine, X for any amino acid, and Z for glutamic acid or glutamine. Each amino acid was encoded as a corresponding unit vector, and the gap sign - was encoded as a zero vector. This encoding produced 0–1 sparse vectors of length . Thus, in this mapping, Alanine (A) was encoded as a 25-dimensional vector , and Cysteine (C), for example, was encoded as a 25-dimensional vector , etc. The gap was encoded as a 25-dimensional zero vector . Finally, we performed tSVD retaining the 50 leading principal components.
Fig. 1.
The distribution of the cleaned dataset (1238 sequences). a The top 6 genera of CoVs: avian CoV, porcine epidemic diarrhea virus (PEDV), betacoronavirus 1 (BCoV-1), MERS, porcine CoV HKU15, SARS-CoV-2. b The top 5 of CoVs’ hosts: swine, avians, human, bats, and camels.
We experimented with omitting step (iv) in the pipeline as well as sequence removal in step (i). Our experiments showed that SVD significantly improves computational efficiency since it dramatically reduces the dimensionality of the data set. Even reducing to only 50 components has little influence on the results of both t-SNE and the tested classifiers (e.g., the change in the average scores was in the range of to +2%). The only disadvantage of SVD is that it is a challenge to interpret the results of the DT classifier.
Deletion of the 15 sequences in Tobaniviridae family had little or no effect on the average scores (accuracy, -score, sensitivity, and specificity) of the classifiers and t-SNE embeddings. However, it affected the identification of important sites in S proteins as described later.
t -SNE visualisation of the data. We used t-SNE (t-distributed Stochastic Neighbor Embedding) [10] which is known to reveal local structure of high-dimensional data and yield excellent results on RNA sequences [11]. t-SNE uses Student’s t-distribution as the output kernel and Kullback–Leibler divergence as the loss function. t-SNE’s main advantage over traditional clustering methods is that it is prone to produce a two-dimensional visualisation with distinctly isolated clusters if there are hidden clusters in the data. As an alternative, we tried another classical approach to visualise the data: first cluster the data with one of the clustering algorithms and then embed it into two-dimensional space with multidimensional scaling (MDS). However, the latter method demonstrated slightly worse results as compared to t-SNE.
The most important parameter of t-SNE, perplexity, controls the width of the kernel that measures similarities between points. Thus, for each point it determines the number of the nearest neighbours to which it is attracted. It is possible to choose any perplexity from 1 to , where n is the number of data points, however, the recommended range is from 5 to 100 [11]. For all t-SNE embeddings, we used a perplexity equal to 30, which is the default value in the majority of t-SNE implementations. Our experiments demonstrated that perplexity values in the recommended range yield similar results, confirming that t-SNE is not very sensitive to the exact value of perplexity in that range.
Since t-SNE uses a non-convex objective, it may yield different results depending on its initialisation (random state). We experimented with choosing various values of random state and found that even though the location of clusters may change, their content does not. For reproducibility, we chose the random state 1 to perform t-SNE embedding.
Classifications. Three classification were considered:
: the human related CoVs (463 entries) vs other CoVs (775 entries);
: the CoVs whose hosts are avians (300 entries) vs the CoVs whose hosts are swines (367 entries);
: the CoVs whose hosts are mammals (938 entries) vs the CoVs with all other hosts, which in fact are all avians (300 entries);
Note that in , only the species of virus matter, not the hosts. Thus, human related CoVs (i.e., HCoV-NL63, HCoV-229E, HCoV-HKU1, HCoV-OC43, BCoV-1, MERS, SARS-CoV-1, and SARS-CoV-2) do not necessarily have human hosts. However, if a host is human, then the virus belongs to the set of the human related CoVs.
Classifiers. Four well-known classifiers: Support Vector Machine (SVM), Logistic Regression (LR), Decision Tree (DT), and Random Forest (RF) were used to perform the classifications , , and . The performance was assessed by computing means and standard deviations of 4 scores: accuracy, -score, sensitivity, and specificity for each of 2-, 3-, 5-, 7-, 9-, and 10-fold cross-validations.
Identification of important sites. Since the DT classifier is one of the best for exploratory analysis [12], we used it to identify important features (sites) in the classification. Since our classifications are binary, the feature importance was calculated as Gini importance of a split [13]. To ensure that the sites do not depend on the choice of training and test subsets, we performed 10 times each of 2-, 3-, 5-, 7-, 9-, and 10-fold splits of the data. Thus, the DT classifier was run 20, 30, 50, 70, 90, and 100 times respectively.
Code and data availability. We prepared a self-contained Jupyter notebook in Python that shows the methods and results presented here. The code and the dataset are available at https://github.com/kuzminkg/CoVs-S-pr.
3. Results
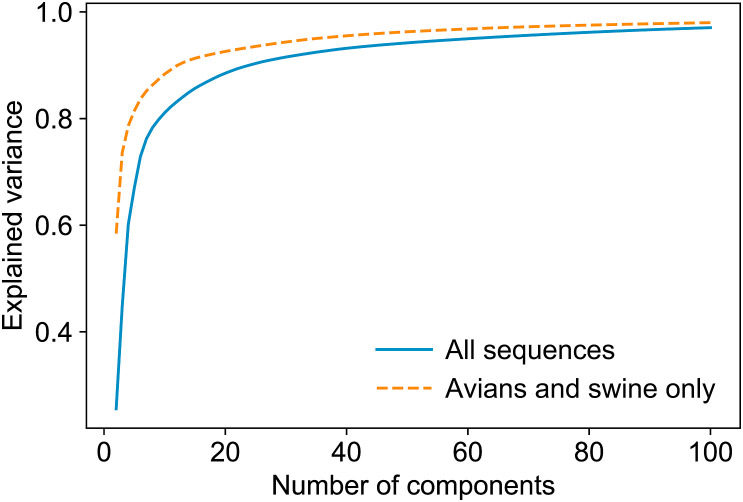
Dimensionality reduction. Since the protein sequences were transformed into high dimensional vectors with 59,900 components, we wanted to decrease the number of components to make the programs run faster and decrease the level of noise in the data. Dimensionality reduction requires choosing the final number of components. We used calculated explained variance to find the number of components. Fig. 2 demonstrates the dependence between the number of components left and the level of explained variance (1 corresponds to 100% of explained variance) for (i) all data, and for (ii) the sequences whose hosts are avians or swine only. The former is used in the and classification, while the latter is used in . Starting with 2 components which give the levels of 0.2557/0.5843 for (i)/(ii) respectively, the explained variance grows quickly reaching the level 0.9 with 24/13 or more components, and level 0.95 with 61/35 or more components. We chose 50 components to further perform SVD, which preserves 94.20% and 96.28% of explained variance for (i) and (ii) respectively.
Fig. 2.
Explained variance vs number of components for (i) all 1238 sequences and (ii) for the sequences whose hosts are either avian or swine (667 entries). The 0.9 threshold is and for (i) and (ii) respectively. The 0.95 threshold is and for (i) and (ii) respectively. We chose 50 components to perform SVD which provided 94.20% and 96.28% of variance for (i) and (ii) respectively.
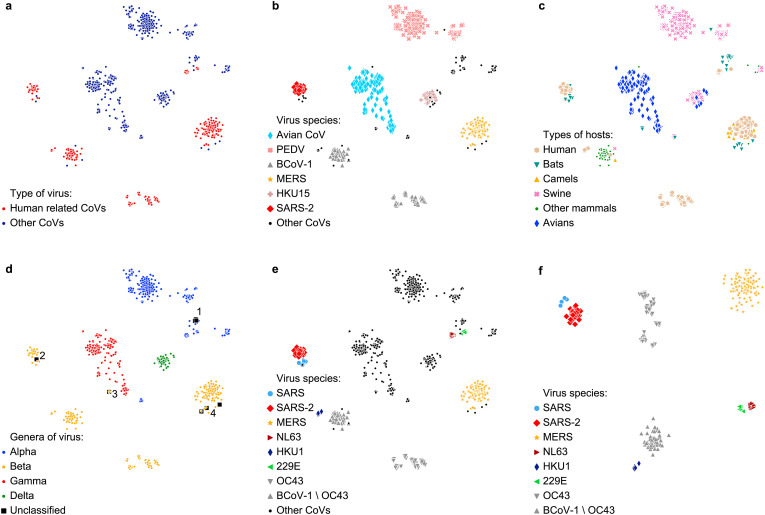
Clusters in the data. In order to demonstrate that vectors readily segregate into clusters with respect to genetic similarities in S proteins, we applied t-SNE, which mapped the input vectors to 2-dimensional vectors, see Fig. 3 . In all figures but 3f, we use the same embedding of all data. Known human related CoVs form distinct clusters as shown in 3f.
Fig. 3.
t-SNE embeddings of all sequences (a–e), and human related CoV sequences only (f). a Human related CoVs vs the other CoVs. b The most represented virus species in the dataset. c Major types of hosts in the dataset. d Relative locations of different genera of CoVs. e Human related CoVs embedded with all other CoV sequences (1238 entries in total). f Embeddings of human related CoVs only (463 entries in total).
Fig. 3a demonstrates the overall data distribution and relative locations of human related CoVs with respect to other CoVs. Fig. 3b and c shows relative locations of CoV species and their hosts. We note that t-SNE without special adjustments may distort distances between far apart clusters [11]. Therefore, the fact that a pair of clusters are further apart than another pair does not necessarily imply that the original (not t-SNE embedded) points have the same relationship. However, t-SNE preserves distances very well locally as seen in Fig. 3c (compare 3e), where humans are close to bats, which are known [14] to be the natural hosts of 5 human related CoVs: SARS-CoV-1, SARS-CoV-2, MERS, HCoV-229E, and HCoV-NL63. In case of MERS, humans cluster is close to camels which are intermediate hosts for this species of CoV [14,15]. Moreover, the sequences near HCoV-OC43 and HCoV-HKU1 belong to cattle or rodents, both known to be either intermediate or natural hosts for these viruses [14,16].
Interesting results are shown in Fig. 3d where the sequences are grouped by genera. All 4 genera segregate into non-overlapping clusters, which makes it possible to accurately predict the genus of a new or unknown virus in the Orthocoronavirinae subfamily. As our dataset contained a few unclassified sequences (marked as black boxes in Fig. 3d), we used this t-SNE embedding to predict their genera. The unclassified sequences in the area 1 are YN2012_Rs3376, YN2012_Rs4125, YN2012_Rs4259, and YN2012_Ra13591 that have been recently identified as alphacoronaviruses [17]. The unclassified sequences in the area 2 are JTMC15, and 16BO133 of SARS related CoVs (and, thus, are betacoronaviruses) and are hypothesized to be the origin of SARSs [18]. The unclassified sequence in the area 3 is BtCoV92 which was identified as a Nobecovirus [19] which is a subgenus of betacoronaviruses. Finally, the 3 unclassified sequences JPDB144, PREDICT/PDF_2180, Vs_CoV_1 in the area 4 are likely to belong to betacoronaviruses.
In Fig. 3e, the CoVs that are located next to HCoV-NL63 and HCoV-229E are related to those species. For instance, the sequence with GenBank accession number MN611517, was recently identified as 229E-related CoV [20], while the sequences BtKYNL63_9a and BtKYNL63_9b were identified as NL63-related CoVs [21]. CoVs located close to MERS include Hedgehog CoV1, Erinaceus hedgehog CoV HKU31, as well as HKU4 and HKU5, which are known to be closely genetically related to MERS [22,23]. The 3 unclassified sequences JPDB144, Vs_CoV_1, and PREDICT/PDF_2180 are also close to MERS. The CoVs located close to BCoV-1HCoV-OC43 (i.e, BCoV-1 without HCoV-OC43) are Murine CoV, HKU14, HKU23, and HKU24, which are all closely related to BCoV-1 [[24], [25], [26]].
Classifications. In order to see how well the standard machine learning classifiers recognize cluster structure intrinsic to the data and revealed by t-SNE embedding, we ran 3 classifications (, , and ) with 4 classifiers (SVM, LR, DT, and RF) each. We compared the performances of the classifiers with 2-, 3-, 5-, 7-, 9-, and 10-fold cross-validations and calculated means and standard deviations of accuracy, -score, sensitivity, and specificity. We also compared the influence of the dimensionality reduction in the input data on the classifier performance. Table S1 demonstrates results of 3-fold cross-validation for classification.
Table S1.
3-fold cross-validation of different classifiers for . The results are presented as mean (μ) standard deviation (σ) of 4 measures of performance: accuracy (Ac), -score (), sensitivity (Sn), and specificity (Sp). The best performances (with the greatest value ) are shown in bold.
| SVM | LR | DT | RF | |
|---|---|---|---|---|
| Inputs with 59,900 components | ||||
| Ac | .983 .007 | .985 .005 | .986.009 | .984.006 |
| .978 .009 | .980 .006 | 982.012 | .979.008 | |
| Sn | .981 .014 | .987 .011 | .996.003 | .985.011 |
| Sp | .985 .011 | .983 .012 | .981.016 | .983.012 |
| Inputs with 50 components | ||||
| Ac | .986 .004 | .974 .018 | .969.026 | .977.016 |
| .981 .005 | .966 .024 | .960.034 | .969.016 | |
| Sn | .989 .011 | .970 .029 | .968.021 | .961.029 |
| Sp | .983 .012 | .977 .016 | .970.032 | .987.010 |
Similar results (within to range) were obtained for all other folds of cross-validation and other classifiers as seen in Table S2, Table S3. Dimensionality reduction sightly improved the performance of SVM and marginally worsened the performances of the other classifiers.
Table S2.
k-fold cross-validations for performed by DT and SVM classifiers run on non-reduced and SVD-reduced inputs respectively. The results are presented as mean (μ) standard deviation (σ) of 4 measures of performance: accuracy (Ac), -score (), sensitivity (Sn), and specificity (Sp). The best performances (with the greatest value ) are shown in bold.
| 2-fold | 3-fold | 5-fold | 10-fold | |
|---|---|---|---|---|
| DT, inputs with 59,900 components | ||||
| Ac | .983 .011 | .986 .009 | .973 .029 | .974 .047 |
| .978 .014 | .982 .012 | .967 .034 | .970 .053 | |
| Sn | .991 .000 | .996 .003 | .989 .007 | .994 .014 |
| Sp | .978 .017 | .981 .016 | .964 .046 | .963 .075 |
| SVM, inputs with 50 components | ||||
| Ac | .972 .022 | .986 .004 | .986 .013 | .989 .023 |
| .962 .030 | .981 .005 | .981 .017 | .986 .029 | |
| Sn | .950 .032 | .989 .011 | .989 .017 | .998 .006 |
| Sp | .985 .016 | .983 .012 | .983 .021 | .983 .034 |
Table S3.
3-fold cross-validation of DT and SVM classifier run on inputs without and with dimensionality reduction respectively. The results are presented as mean (μ standard deviation (σ) of 4 measures of performance: accuracy (Ac), -score (), sensitivity (Sn), and specificity (Sp).
| DT, inputs with 59,900 components | |||
| Ac | .986 .009 | .977 .023 | .978 .012 |
| .982 .016 | .974 .027 | .986 .008 | |
| Sn | .996 .003 | .957 .048 | .984 .018 |
| Sp | .980 .016 | .995 .004 | .960 .057 |
| SVM, inputs with 50 components | |||
| Ac | .986 .004 | .976 .025 | .987 .018 |
| .981 .005 | .972 .030 | .992 .012 | |
| Sn | .989 .011 | .957 .061 | 1.000 .000 |
| Sp | .983 .012 | .992 .012 | .947 .075 |
Table S3 demonstrates decent results for all performed classifications (referring to the 4 statistical metrics used – accuracy, -score, sensitivity, and specificity) reaching more than 98% for , 95% for , and 94% for Important sites. We used DT to identify important sites in classification, see Table S4. Only two sites (1483 and 2258) had high importance of greater than 0.80. Remarkably, they appeared in each run of DT classifier independently of the number of splits k. All other sites used in DT had importance of less than 0.13. As k increases, the proportion of occurrences of the two sites changes in favor of 2258, reaching 100% in the 10-fold split.
Since DT consistently demonstrated the best performance for non-reduced inputs (binary vectors with 59,900 components), while SVM exhibited the best behaviour on SVD-reduced inputs (vectors with 50 components only), to save space only the results of these two classifiers in the corresponding cases are shown in Table S2, Table S3. Table S2, Table S3 demonstrates that 3-fold cross-validation gives the best results among other k-fold cross-validations (; the results for were similar to and not shown in Table S2). Notably, on SVD-reduced inputs, the means of accuracy, -score, sensitivity, and specificity tend to increase with the growth of k, while on non-reduced inputs, they vary with no particular dependence. This regularity was consistently observed not only in classification, but also in the other two and , and may be considered as evidence that SVD successfully decreased the level of noise in the data.
We experimented with adding or deleting sequences in the dataset to assess the robustness of site selection. The sequence addition/deletion had minimal effect unless it altered the composition of species in the dataset. Deletion of the species that have genetic analogs (e.g., deletion of all SARS-CoV-1 sequences while SARS-CoV-2 sequences remain untouched) has no significant effect. However, addition and deletion of “unique” species may alter results dramatically. For instance, when we included the 15 Tobaniviridae sequences deleted from the original dataset during the prepossessing, the sites 1483 and 2258 were identified, but their proportion gradually changed from 11:8 for a 2-fold split to 35:64 for a 10-fold split. Additionally, another important site 2213 appeared among the sites with high (0.7) importance. In another example, deletion of all BCoVs drastically altered the ranking of important sites identifying 1634 as the only site with high importance.
4. Discussion
Our analysis shows that host specificity of CoVs can be predicted with high accuracy using only S protein sequences. A total of 1238 sequences were aligned and SVD used for dimensionality reduction. Machine learning classifiers SVM, LR, DT, and RF gave excellent results with high accuracies of 0.98 for 3-fold cross-validation. Similar results were obtained by another group using RF models applied to a different database of spike sequences [27].
Clustering of data with t-SNE embeddings confirms the reliability of the analysis. S protein sequences segregate correctly by virus genus and type of host species. The observed overlap of human with bats or camel hosts is likely due to the hypothesized origin of SARS-CoV-1 in bats and MERS from bats via the intermediate host of camels [2]. As expected, the two SARS-CoVs comprise two regions of the same cluster. A recent alternate approach applied supervised machine learning on whole genome sequences of CoV for rapid classification of COVID-19 [28]. Our approach achieves excellent classification and clustering results using only CoV spike sequences.
The DT classifier for identifies two sites (1483 and 2258) with an average importance of 0.81. Other sites were more variable and less significant with importance of 0.13 or less. We examined the location of the two key sites in S protein and their known biological roles. Site 1483 is equivalent to the conserved arginine (R685 in SARS-CoV2) in the S1/S2 protease cleavage site of the S protein. Protease cleavage at this site is required for viral cell entry [7]. Site 2258 corresponds to Q1201 in the heptad repeat 2 (HR2) of SARS-CoV2 S protein. HR2 is critical for viral fusion to the host cell membrane. This region has been targeted in development of antibodies and peptide inhibitors as antiviral agents for SARS CoVs [29,30]. Therefore, DT has identified key amino acid residues in biologically important regions of S protein.
Accurate predictions of CoV host specificity are essential in light of the COVID19 pandemic and the potential for new trans-species infections in future. Our analysis relies only on the S protein sequences rather than using the entire viral genome. Similar analyses might be valuable to predict the host specificity of novel CoV sequences.
Funding
This research was supported in part by a Georgia State University Molecular Basis of Disease fellowship (KK).
Declaration of competing interest
The authors declared no conflict of interest.
Table S4.
The average importance and number of occurrences (in parentheses) for deciding sites identified by DT classifier in . The classifier was run times, where k is the number of folds.
| Site | 2-fold | 3-fold | 5-fold | 10-fold |
|---|---|---|---|---|
| 1483 | .811 (3) | .803 (2) | .800 (1) | NA (0) |
| S2258 | .807 (17) | .806 (28) | .806 (49) | 0.806 (100) |
Acknowledgments
KK thanks his scientific advisor, Dr. Pavel Skums, who has been kind and infinitely patient with his student, and Dr. Sergey Plis, who showed how beautiful machine learning could be.
References
- 1.de Groot R., Baker S., Baric R. Virus Taxonomy: 2019 Release. July 2019. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/222/coronaviridae ec 51, berlin, germany, accessed on July 13, 2020.
- 2.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. https://www.ncbi.nlm.nih.gov/pubmed/30531947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo P.C., Lau S.K., Lam C.S. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. https://www.ncbi.nlm.nih.gov/pubmed/22278237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. https://www.ncbi.nlm.nih.gov/pubmed/27578435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls A.C., Park Y.J., Tortorici M.A. Structure, function, and antigenicity of the sars-cov-2 spike glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. https://www.ncbi.nlm.nih.gov/pubmed/32155444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang T., Bidon M., Jaimes J.A. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. https://www.ncbi.nlm.nih.gov/pubmed/32272173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franceschini A., Lin J., von Mering C., Jensen L.J. SVD-phy: improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics. 2015;32(7):1085–1087. doi: 10.1093/bioinformatics/btv696. https://academic.oup.com/bioinformatics/article-pdf/32/7/1085/19568709/btv696.pdf arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotyk A. Iupac-iubmb joint commission on biochemical nomenclature (jcbn) and nomenclature committee of iubmb (nc-iubmb), Newsletter 1999. Folia Microbiol. 1999;44(3):243–246. doi: 10.1007/BF02818542. https://europepmc.org/articles/PMC6882829 [DOI] [PubMed] [Google Scholar]
- 10.van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. http://www.jmlr.org/papers/v9/vandermaaten08a.html [Google Scholar]
- 11.Kobak D., Berens P. The art of using t-sne for single-cell transcriptomics. Nat. Commun. 2019;10(1):5416. doi: 10.1038/s41467-019-13056-x. https://europepmc.org/articles/PMC6882829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Wang M., Zhang H. The use of classification trees for bioinformatics. Wiley interdisciplinary reviews. Data mining and knowledge discovery. 2011;1(1):55–63. doi: 10.1002/widm.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nembrini S., König I.R., Wright M.N. The revival of the gini importance? Bioinformatics. 2018;34(21):3711–3718. doi: 10.1093/bioinformatics/bty373. https://academic.oup.com/bioinformatics/article-pdf/34/21/3711/26146978/bty373.pdf arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Z.-W., Yuan S., Yuen K.-S. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. http://www.ijbs.com/v16p1686.htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paden C.R., Yusof M.F.B.M., Al Hammadi Z.M. Zoonotic origin and transmission of middle east respiratory syndrome coronavirus in the uae. Zoonoses and Public Health. 2018;65(3):322–333. doi: 10.1111/zph.12435. https://onlinelibrary.wiley.com/doi/pdf/10.1111/zph.12435 https://onlinelibrary.wiley.com/doi/abs/10.1111/zph.12435 arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi S., Koma T., Noopdoi N. Commentary: origin and evolution of pathogenic coronaviruses. Front. Immunol. 2020;11:811. doi: 10.3389/fimmu.2020.00811. https://www.frontiersin.org/article/10.3389/fimmu.2020.00811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y. Tan, T. Schneider, M. Leong, L. Aravind, D. Zhang, Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of sars-cov-2-related viruses, mBio 11 (3). arXiv:https://mbio.asm.org/content/11/3/e00760-20.full.pdf, doi:10.1128/mBio.00760-20. URL https://mbio.asm.org/content/11/3/e00760-20. [DOI] [PMC free article] [PubMed]
- 18.Lau S.K., Luk H.K., Wong A.C. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(7):1542–1547. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassenaar T.M., Zou Y. 2019_ncov/sars-cov-2: rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses. Lett. Appl. Microbiol. 2020;70(5):342–348. doi: 10.1111/lam.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Si H.R., Zhu Y. Discovery of bat coronaviruses through surveillance and probe capture-based next-generation sequencing. mSphere. 2020;5(1) doi: 10.1128/mSphere.00807-19. e00807–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao Y., Shi M., Chommanard C. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses nl63 and 229e and their recombination history. J. Virol. 2017;91(6) doi: 10.1128/JVI.01953-16. e01953–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau S., Luk H., Wong A. Identification of a novel betacoronavirus (merbecovirus) in amur hedgehogs from China. Viruses. 2019;11(11):980. doi: 10.3390/v11110980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y., Du L., Liu C. Receptor usage and cell entry of bat coronavirus hku 4 provide insight into bat-to-human transmission of mers coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2014;111(34):12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau S.K.P., Woo P.C.Y., Yip C.C.Y. Isolation and characterization of a novel betacoronavirus subgroup a coronavirus, rabbit coronavirus hku 14, from domestic rabbits. J. Virol. 2012;86(10):5481–5496. doi: 10.1128/JVI.06927-11. https://jvi.asm.org/content/86/10/5481.full.pdf https://jvi.asm.org/content/86/10/5481 arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. T. Y. So, D. K. W. Chu, E. Miguel, et al., Diversity of dromedary camel coronavirus hku 23 in african camels revealed multiple recombination events among closely related betacoronaviruses of the subgenus embecovirus, J. Virol. 93 (23). arXiv:https://jvi.asm.org/content/93/23/e01236-19.full.pdf, doi:10.1128/JVI.01236-19. URL https://jvi.asm.org/content/93/23/e01236-19. [DOI] [PMC free article] [PubMed]
- 26.Lau S.K.P., Woo P.C.Y., Li K.S.M. Discovery of a novel coronavirus, China rattus coronavirus hku 24, from Norway rats supports the murine origin of betacoronavirus 1 and has implications for the ancestor of betacoronavirus lineage a. J. Virol. 2015;89(6):3076–3092. doi: 10.1128/JVI.02420-14. https://jvi.asm.org/content/89/6/3076.full.pdf https://jvi.asm.org/content/89/6/3076 arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiang X.-L., Xu P., Fang G., Liu W.-B., Kou Z. Using the spike protein feature to predict infection risk and monitor the evolutionary dynamic of coronavirus. Infectious Diseases of Poverty. 2020;9(1):33. doi: 10.1186/s40249-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randhawa G.S., Soltysiak M.P.M., El Roz H. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: covid-19 case study. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0232391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elshabrawy H.A., Coughlin M.M., Baker S.C., Prabhakar B.S. Human monoclonal antibodies against highly conserved hr1 and hr2 domains of the sars-cov spike protein are more broadly neutralizing. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0050366. https://www.ncbi.nlm.nih.gov/pubmed/23185609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia S., Liu M., Wang C. Inhibition of sars-cov-2 (previously 2019-ncov) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]