Abstract
Drug repurposing or repositioning is a technique whereby existing drugs are used to treat emerging and challenging diseases, including COVID-19. Drug repurposing has become a promising approach because of the opportunity for reduced development timelines and overall costs. In the big data era, artificial intelligence (AI) and network medicine offer cutting-edge application of information science to defining disease, medicine, therapeutics, and identifying targets with the least error. In this Review, we introduce guidelines on how to use AI for accelerating drug repurposing or repositioning, for which AI approaches are not just formidable but are also necessary. We discuss how to use AI models in precision medicine, and as an example, how AI models can accelerate COVID-19 drug repurposing. Rapidly developing, powerful, and innovative AI and network medicine technologies can expedite therapeutic development. This Review provides a strong rationale for using AI-based assistive tools for drug repurposing medications for human disease, including during the COVID-19 pandemic.
Introduction
The artificial intelligence (AI) pioneers of the 1950s foresaw building machines that could sense, reason, and think like people—a proof-of-concept known as general AI.1 The rapid growth in computing power and memory storage, an unprecedented wealth of data, and the development of advanced algorithms have led to substantial breakthroughs in AI. AI applications cover diverse fields, such as computer vision, voice recognition, natural language understanding, and digital pathology data analysis. Similarly, AI has been revolutionising drug discovery by extracting hidden patterns and evidence from biomedical data. Pharmaceutical companies and start-ups have used AI for drug discovery and development.2 For example, IBM's Watson Health platform searches for drugs from vast amounts of textual data, including laboratory data, clinical reports, and scientific publications.3
In this Review, we focus on AI technologies for a specific domain in drug discovery—that of drug repurposing—which offers rapid and cost-effective solutions for therapeutic development. These merits are especially clear in the COVID-19 global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), where de-novo drug discovery is almost infeasible (figure 1 ). Thus, the pandemic is a good opportunity for introducing advanced AI algorithms combined with network medicine for drug repurposing.
Figure 1.
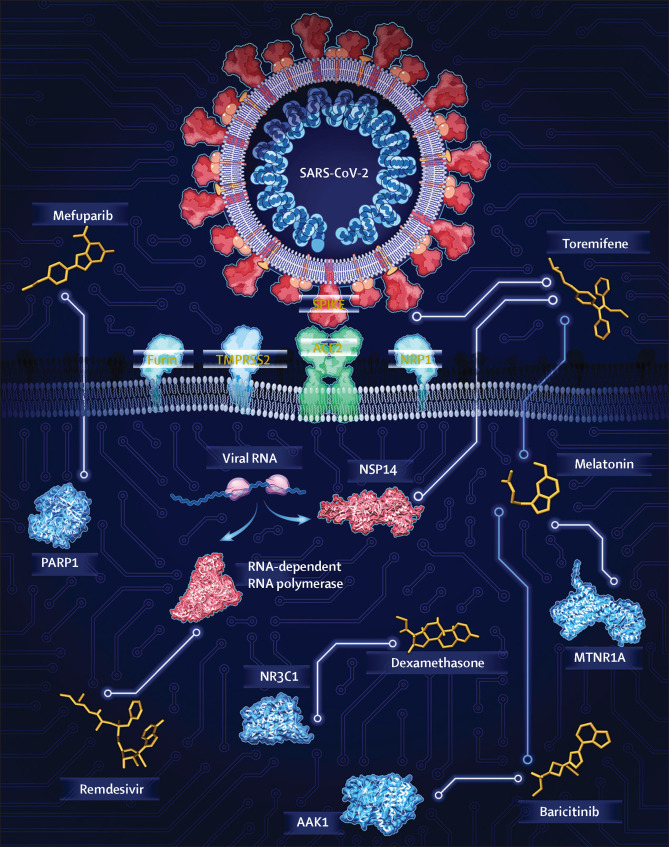
Overview of AI-assisted drug repurposing for COVID-19
AI algorithms can be used for drug repurposing, which is a rapid and cost-effective way to discover new therapy options for emerging diseases. Reproduced by permission of Cleveland Clinic Center for Medical Art and Photography. AI=artificial intelligence. PARP1=poly-ADP-ribose polymerase 1. NR3C1=nuclear receptor subfamily 3 group C member 1. AAK1=AP2-associated protein kinase 1. MTNR1A=melatonin receptor 1A. TMPRSS2=transmembrane serine protease 2. ACE2=angiotensin I converting enzyme 2. NRP1=neuropilin 1. NSP14=non-structural protein 14.
Emerging challenges and opportunities in drug discovery
One study estimated that pharmaceutical companies spent US$2·6 billion in 2015, up from $802 million in 2003, for the development of a new chemical entity approved by the US Food and Drug Administration (FDA).4 The increasing cost of drug development is due to the large volume of compounds to be tested in preclinical stages and the high proportion of randomised controlled trials (RCTs) that do not find clinical benefits or with toxicity issues. Given the high attrition rates, substantial costs, and low pace of de-novo drug discovery, exploiting known drugs can help improve their efficacy while minimising side-effects in clinical trials. As Nobel Prize-winning pharmacologist Sir James Black said, “The most fruitful basis for the discovery of a new drug is to start with an old drug”.5
Drug repurposing, also termed drug repositioning, reprofiling or re-tasking, is a strategy for identifying new indications for approved or investigational (including clinically failed) drugs that have not been approved (panel ). Because the safety of these drugs has already been tested in clinical trials for other applications, repurposing known drugs can bring medications to patients much faster and with less cost than that of developing new drugs. For decades, academic institutions and science funders have championed the idea that screening libraries of existing drugs with various tests could uncover new applications, and have made observations that have led to medicines designed for one disease finding uses in another. Well known examples include sildenafil citrate for erectile dysfunction,6 thalidomide for multiple myeloma,7 and remdesivir for treatment of COVID-19.8 Indeed, the increasing number of repositioned medications led to the idea that a systematic (hypothesis-free) screen of all known drugs might uncover additional compatible targets.
Panel. Terms and concepts.
Drug–target network
A bipartite graph composed of approved drugs and proteins linked by drug–target binary associations.
Drug repurposing
A strategy for identifying new indications for approved or investigational (including clinically failed) drugs that have not been originally approved or dedicated (also termed drug repositioning, reprofiling or re-tasking).
Disease module
Represents a group of nodes (ie, proteins or genes) whose perturbation can be linked to a particular disease (eg, COVID-19) phenotype.
Systems pharmacology
An inter-discipline that applies systems biology principles and data science techniques in pharmacology.
Human interactome
The set of physical protein–protein interactions (the interactome) in human cells.
Network medicine
A discipline that seeks to redefine disease and therapeutics from an integrated perspective using systems biology and network science methodologies, offering important applications to drug design.
Node or vertex
The basic unit of graphs. Usually visualised as circles (or in other shapes), nodes represent basic entities, such as drugs and proteins.
Edge or link
A basic unit of graphs that connects two nodes. Usually visualised as lines (with arrows if directed), edges represent the relationships (eg, protein interaction) between the nodes.
Network proximity
Measures the distances between two modules, such as drug–target and disease–gene modules. Several proximity measures have been defined, such as shortest, closest, separation, kernel, and centre measures.11
Artificial intelligence (AI)
The study of building machines or programmes that exhibit human intelligence in doing specific or general tasks.
Machine learning algorithms
A subset of AI algorithms that can learn from data, therefore removing the need for explicit instructions on how to do certain tasks.
Deep neural networks
A general term referring to multilayer neural networks.
Convolutional neural networks
Neural network architectures specifically designed for analysing image data, which generally include multiple layers of convolutional layers and pooling layers.
Graph representation learning
Specific deep learning techniques that are developed for learning feature representations of graph structure data.
Visible neural network
A new generation of visible approaches that aim to guide the structure of machine learning models with an increasingly extensive knowledge of a biological mechanism.
The strategy of drug repurposing is a powerful solution for emerging diseases,9 such as COVID-19. Yet, without foreknowledge of the complete drug-target network, development of promising and affordable approaches for effective treatment of complex diseases is challenging.10 Because drug targets do not operate in isolation from the complex system of proteins that comprise the molecular machinery of the cells with which they associate, each drug–target interaction (panel) should be examined in an integrative context (figure 2 ).11 Therapeutic interventions need to consider the perturbation of disease system properties (termed network medicine [panel]), and have little to do, functionally speaking, with genetic and genomic events alone.12 Observations and advances in network medicine further indicate that perturbations of cellular systems and the human interactome (panel) underlie the disease, which is the essence of drug discovery and development.12 Knowledge of the interplay between drug targets and human diseases can provide clues for possible drug repurposing because drugs that target one disease might target another through a shared functional protein–protein interaction network.11 For example, SARS-CoV-2 requires host cellular factors (such as angiotensin I converting enzyme 2 [ACE2], transmembrane serine protease 2 [TMPRSS2], and furin; figure 1) for successful replication during infection.13, 14 Systematic targeting of the viral protein and host protein interactions (the SARS-CoV-2 interactome) offers a novel strategy for effective drug repurposing for COVID-19. A SARS-CoV-2 virus–host interactome that contains 332 high-confidence protein–protein interactions between 26 viral proteins and human proteins was structed using affinity purification mass spectrometry.15 69 drug candidates were prioritised that can target the host proteins in the SARS-CoV-2–host interactome.15 Experimental assays validated the antiviral activities of two sets of agents: mRNA translation inhibitors (ie, zotatifin) and sigma-1 and sigma-2 receptor regulators (ie, haloperidol).
Figure 2.
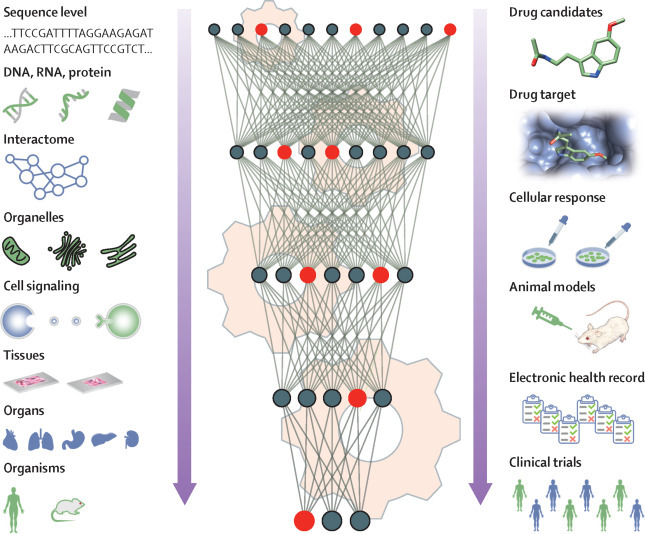
AI for drug repurposing in an integrative context
AI approaches can greatly accelerate drug repurposing by incorporating biological knowledge (eg, human interactome, organelles, tissues, and organs). The cogs indicate computer programs and algorithms. Red and black circles represent neurons in deep neural networks. Red indicates that this neuron carries important information from the biological systems. Green and blue people indicate different subgroups that might have different responses to the treatment. The downward arrows show that AI algorithms can use the information from multi-level biological systems and drug development pipelines to build more powerful models. The left panel shows the biological systems and the right panel shows the drug development pipeline AI=artificial intelligence.
In another study,16 a network-based methodology that quantifies the interplay between the virus–host interactome and drug targets in the human interactome network suggested 16 repurposed drug candidates for potential treatment of COVID-19.16 This finding calls for a detailed approach, including AI and network medicine, and raises the question of not only which protocols to consider, but also which factors to scrutinise, and broadly, how to integrate the disciplines (figure 2).
AI algorithms and recent advancements
Deep learning architecture
Deep learning is a subfield of machine learning that refers to the paradigm of exploring the data with layers of linear and non-linear transformations organised in a hierarchical way.17 The most widely used deep learning model is artificial neural networks, wherein the basic building block is an artificial neuron that non-linearly transforms the weighted sum of input feature variables.
Fully connected feedforward neural network (FNN) is an architecture in which the artificial neurons are connected layer-by-layer from input features to output targets. A weight is associated with each connection and is optimised by minimising the prediction loss of the output targets through backpropagation on training samples.18 FNNs are typically used for data samples represented as vectors. For example, Aliper and colleagues19 used FNN to classify drugs into pharmaceutical therapeutic classes based on the drugs' transcriptomic profile vectors. Lenselink and colleagues20 compared the performance of a diverse set of algorithms on the prediction of molecule and target activity with the ChEMBL database.20 The authors showed that the inclusion of target data can lead to better models. FNN can achieve better performance than that of conventional machine learning methods, such as logistic regression.
In the case of images being the input where each pixel is a feature variable, FNNs become infeasible as the number of weights becomes far too large. However, convolutional neural network (CNN; panel) is particularly suitable for image processing. Instead of fully connecting neurons in adjacent layers, CNN uses filters (small matrices of weights) that apply a convolution operation on local patches of the images, which greatly reduces the number of weights. CNN has been used to analyse chemical images to obtain insight into drug therapeutic functions.21 For example, AtomNet predicts the binding affinity of small molecules to proteins on the basis of the structural information extracted by CNN.22
Biological sequences are another widely explored type of data for drug repurposing. However, neither FNN nor CNN appropriately consider the sequential nature of the data. Recurrent neural networks (RNNs) are specifically designed for sequences in which the main building block is a recurrent cell appearing at each timestamp or sequence location that retains past information while learning new information in a sequence. RNN models have been used for generating focused molecule libraries for drug discovery, with the molecules represented as sequences using simplified molecular input line entry system codes.23 Gao and colleagues24 developed a hybrid approach of graph neural network and RNN to predict drug–target interactions. Beck and colleagues25 developed a hybrid CNN and RNN model called Molecule Transformer-Drug Target Interaction to predict whether any commercially available antiviral drugs could work in SARS-CoV-2. The authors computationally identified several known antiviral drugs, such as atazanavir, remdesivir, efavirenz, ritonavir, and dolutegravir, for the potential treatment of SARS-CoV-2 infection.
Graph representation learning
A classic way to repurpose drugs is through network medicine, which includes the construction of medical knowledge graphs containing relationships between different kinds of medical entities (eg, diseases, drugs, and proteins) and predicts new links between existing approved drugs and diseases (eg, COVID-19). Methods that are based on graph embedding have been gaining attention for link prediction in graphs26 that represent nodes and edges as low-dimensional feature vectors.27, 28, 29, 30 Using the feature vectors of drugs and diseases, we can easily measure their similarities and therefore identify effective drugs for a given disease. One challenge for the graph embedding method is scalability. Real-world (knowledge) graphs are usually large. The number of entities in a medical knowledge graph could be as many as several million. Existing machine learning systems such as TensorFlow and PyTorch are mainly designed for data with regular structures but not for large-scale graphs. Therefore, several systems that are specifically designed for learning representations from large-scale graphs have been developed. For example, Zhu and colleagues31 developed a high-performance system named GraphVite that could be promising for future drug repurposing because the system can efficiently process tens or even hundreds of millions of nodes.
Increasing interest exists in developing graph representation learning techniques for drug repurposing. Sosa and colleagues32 constructed a medical knowledge graph of drugs, diseases, genes, and proteins from the biomedical literature and used graph embedding techniques for predicting the links between drugs and diseases. Gysi and colleagues33 developed a method that was based on graph neural network and presented a case study on SARS-CoV-2 with 81 potential repurposing candidates identified. BenevolentAI's knowledge graph is a large repository of structured medical information, including numerous connections extracted from the scientific literature by machine learning.34 BenevolentAI predicted that baricitinib, a drug used to treat rheumatoid arthritis, could be a potential treatment for COVID-19 through the inhibition of AP2-associated protein kinase 1 (encoded by AAK1; figure 1). A team constructed a comprehensive COVID-19 knowledge graph (termed CoV-KGE) that included 15 million edges across 39 types of relationships connecting drugs, diseases, proteins, genes, pathways, and expressions of genes and proteins35 from a large scientific corpus of 24 million PubMed publications. Using Amazon Web Services' computing resources and graph representation learning techniques, the team identified 41 repurposed drug candidates (including dexamethasone and melatonin) for COVID-19 treatment. To achieve a high prediction performance, the construction of a high-quality medical knowledge graph is essential, which itself is a promising direction for future research.
Repurposed drugs for COVID-19 are under investigation
Several examples exist of repurposed drugs being or having been tested in clinical trials for COVID-19, including antiviral drugs and host-targeting therapies (figure 1). A detailed discussion of repurposed drugs for COVID-19 can be found in a review by Sanders and colleagues.36
Antiviral drugs
Remdesivir, a monophosphate prodrug of an active C-adenosine nucleoside triphosphate analogue, was originally discovered for the potential treatment of Ebola virus disease.36 Remdesivir has shown promise in the treatment of COVID-19, prompting emergency use clearance from the FDA, although indication is limited to severe disease only. The FDA made this decision on the basis of early research showing that the drug might help speed up recovery for hospitalised patients with COVID-19. Mechanistically, remdesivir was shown to inhibit the viral RNA-dependent RNA polymerase (figure 1).37 A double-blind, randomised, placebo-controlled trial of intravenous remdesivir in adults hospitalised with COVID-19 showed that remdesivir significantly shortens the median recovery time to 11 days, compared with 15 days in the placebo group.8 These preliminary findings support the use of remdesivir for patients who are hospitalised with COVID-19 and require supplemental oxygen therapy. However, another randomised, open-label, phase 3 trial involving hospitalised patients not requiring mechanical ventilation did not show a significant difference between a 5-day course and a 10-day course of remdesivir.38 Further investigation of the clinical benefits of remdesivir for patients with COVID-19 in different patient subgroups with or without mechanical ventilation is needed to identify the shortest effective duration of therapy. Additionally, whether remdesivir can shorten the recovery course of individuals with early COVID-19 is unknown. A study using machine learning and statistical analysis approaches discovered that mefuparib (CVL218), a poly-ADP-ribose polymerase 1 inhibitor (figure 1), blocked SARS-CoV-2 replication without obvious toxic effects in vitro.39 The antiviral activity of mefuparib is more potent at viral entry and similar at viral post entry compared with remdesivir, suggesting the drug to be a potential anti-SARS-CoV-2 drug candidate.
Toremifene, a first-generation selective estrogenic receptor modulator that is non-steroidal, was approved for the treatment of breast cancer in 1997.40 A network medicine analysis identified toremifene as a top candidate for the treatment of COVID-19.16 In vitro assays indicated that toremifene blocked various viral infections at micromolar concentration, including Middle East Respiratory Syndrome coronavirus,41 severe acute respiratory syndrome coronavirus,42 and SARS-CoV-2.43 A further computational biophysics study44 suggested that toremifene might block interaction between ACE2 and the spike protein of SARS-CoV-2 and might inhibit non-structural protein 14 of SARS-CoV-2 (figure 1), mechanistically supporting the drug's antiviral activities. The mean plasma concentration of toremifene during administration of 60 mg per day was 0·88 mg/L (2·17 μM) in post-menopausal patients with breast cancer40 and the peak plasma concentration (>10 μM) of toremifene (60 mg per day) was approximately three-times the antiviral effect on SARS-CoV-2 (half-maximal inhibitory concentration of 3·58 μM).43 In summary, toremifene, identified by CoV-KGE35 and network medicine16 approaches, offers a potential drug candidate to be tested in COVID-19 clinical trials.
Host-targeting therapy
SARS-CoV-2 causes up-regulation of systemic inflammation,45 in some cases culminating in a cytokine storm, underscoring the high potential for treatment success by using a drug targeting inflammation and immune response (including baricitinib, dexamethasone, and melatonin; figure 1). By combining findings based on network medicine and large-scale patient data analysis from the COVID-19 patient registry at the Cleveland Clinic, Cleveland, OH, USA, researchers found that melatonin intake was associated with a 50–60% reduced likelihood of a positive laboratory test result for SARS-CoV-2.46 Dexamethasone is a glucocorticoid receptor (figure 1) agonist approved by the US FDA for a variety of inflammatory and autoimmune conditions.47 Dexamethasone was identified as a top repurposed drug candidate by CoV-KGE.35 The randomised trial48 of COVID-19 therapy trial showed that dexamethasone reduced death by a third in patients requiring invasive mechanical ventilation (95 [29%] of 324 patients vs 283 [41%] of 683 patients; rate ratio 0·64 [95% CI 0·51–0·81]) and by a fifth in individuals requiring oxygen without invasive mechanical ventilation (298 [23·3%] of 1279 patients vs 682 [26·2%] of 2604 patients; 0·82 [0·72–0·94]). However, dexamethasone did not reduce mortality in COVID-19 patients not receiving respiratory support. Altogether, these data suggest that targeting excessive host inflammation by immune modulators or anti-inflammatory drugs offers a therapeutic strategy for severe COVID-19, warranting testing in large-scale RCTs.
Drug combinations for COVID-19
Monotherapies, including remdesivir8, 38 and hydroxychloroquine,49 have shown little or no clinical benefit for patients with COVID-19. Because the immune system plays key roles in the worsening health and death of patients with COVID-19,50 combining inflammatory or immune modulators (ie, boosting host immunity) with antiviral drugs might offer an effective treatment for patients with COVID-19. Drug combinations, offering increased therapeutic efficacy and reduced toxicity, play an important role in treating infectious diseases, including COVID-19 (eg, remdesivir plus baricitinib [NCT04401579]). However, our ability to identify and validate effective combinations is limited by a huge increase in the number of possible drug pairs. Using a network-based methodology,51 scientists identified three potential drug combinations for COVID-19,16 including sirolimus plus dactinomycin, mercaptopurine plus melatonin, and toremifene plus emodin. These combinations are based on theoretical analysis using the interactome and have not been tested in preclinical or clinical studies. The same team further observed that combining melatonin and toremifene showed potential for use in the treatment of COVID-19.52 The selective estrogen modulation and melatonin in early COVID-19 (SENTINEL; NCT04531748) trial is being done at the Cleveland Clinic to test the clinical efficacy of combining melatonin and toremifene therapy in patients with early COVID-19.52 Using BenevolentAI's knowledge graph,34 baricitinib was identified as potential treatment for COVID-19. At least two phase 2 randomised, double-blind trials of baricitinib alone or as part of combination therapy with antiviral drugs (eg, remdesivir) are underway for patients with moderate and severe COVID-19 (NCT04373044 and NCT04401579).
Real-world evidence to test drug responsiveness
Another important aspect of using AI for drug repurposing is the use of real-world data, such as electronic health records, in searching for effective repurposed drug candidates. Electronic health records are patient clinical data that are routinely collected, such as demographics, diagnoses, medications, procedures, and laboratory test results, stored in digital form, which can be exchanged and accessed securely.53 Extensive discussions have taken place on leveraging real-world data for drug discovery and development.54 On the one hand, patients in real-world data are more representative of patients who will receive the prescription when the drug is on the market than patients in RCTs are, who are enrolled on the basis of strict inclusion and exclusion criteria. On the other hand, typically treatment and control groups are required to precisely estimate the treatment effects. However, for certain scenarios, such as remdesivir trials for COVID-19, only a single treatment group is possible, which makes estimating the treatment effect difficult.55 In this case, because of the inclusion of many diverse patients, real-world data contain rich information for synthesising a potential control group, which can then be compared with the treatment group in an RCT to help estimate the treatment effects.
Despite the promises of real-world data, deriving insights from real-world data that are similar to those from RCTs is challenging because real-world data have higher dimensionality (including confounding factors), a broader population, and usually lower data quality compared with RCT data. Propensity score, which calculates the likelihood of the patient receiving the treatment from a set of potential confounding factors using logistic regression, is the standard technique to do patient matching.56 However, the calculation of the likelihood of the patient receiving the treatment from such a set is much more complicated in real-world data because of the associated challenges such as high dimensionality, longitudinality, irregularity, and incompleteness. In this case, the advanced machine learning models can estimate propensity scores more accurately than traditional logistic regression-based propensity score matching approaches can.57, 58 Moreover, other types of matching techniques, such as patient similarity analytics,59 also hold promise in these complicated scenarios.
The initiative to build national or international electronic health records repositories for COVID-19 research has been undertaken. One such repository is the international consortium 4CE, which includes the electronic health records of patients from 96 hospitals across five countries. All participants' electronic health records are matched to a common data model with Integrating Biology and the Bedside60 or Observational Medical Outcomes Partnership (OMOP).61 A retrospective cohort study of 1438 patients with laboratory-confirmed COVID-19 admitted to hospital in metropolitan New York, USA, revealed that treatment with hydroxychloroquine, azithromycin, or both, compared with neither treatment, was not significantly associated with differences in in-hospital mortality for patients with COVID-19.49 From a COVID-19 registry of nearly 20 000 patients with 1600 COVID-19-positive patients from the Cleveland Clinic Health System electronic health records, and using a user-active comparator design and propensity score adjustment for confounding, melatonin usage was shown to be associated with a reduced likelihood of a positive SARS-CoV-2 test result by RT-PCR assay.46 Mancia and colleagues62 showed that angiotensin-converting enzyme inhibitors (ACEIs) or angiotensin II receptor blockers (ARBs) were not associated with the risk of COVID-19. An independent study revealed that the use of ACEIs or ARBs were not associated with an increase of the likelihood for a positive COVID-19 test or an increase in the COVID-19 severity using New York University Langone Health electronic health records.63 RCTs are underway to test the clinical benefits of melatonin in patients with COVID-19 (NCT04409522 and NCT04353128).
Discussion, perspective, and future directions
For decades, translational science has faced the challenge of how to translate research findings into new effective medicines and technologies that rapidly deliver the medicines. This challenge has encouraged basic and translational sciences to work together towards this pivotal aim. Generations of scientists have struggled to make headway in de novo drug discovery. In principle, a strategy involving drug repurposing, in which a drug has already been tested and approved by the US FDA, can overcome the barriers of de novo drug discovery. However, the volume of approved or clinically failed drugs is large, emphasising the difficulty of which drug to select that would be highly effective for the disease in question.
Challenges in drug repurposing
Despite the enthusiasm for drug repurposing in treating COVID-19, challenges remain. Cellular or animal assays might not reflect the host environment of the virus infection in humans. Also, repurposed drugs might have been optimised for a particular target, dosing, or tissue in the original indications. Rapid clinical tests of existing antiviral, antimalarial, and immunomodulatory drugs have been done or are underway against COVID-19. Many trials did not optimise the drug's clinical benefits and biological questions because of their expedient design, lack of clinical endpoints, small number of patients enrolled (thus lack of statistical power), and more.64 For example, hydroxychloroquine shows potential anti-SARS-CoV-2 activities in in vitro assays.65 However, hydroxychloroquine has shown very little or no efficiency in preclinical66 and clinical trial studies.49 Few reproducible preclinical animal models and gold-standard clinical outcome measures in COVID-19 trials might also result in some failures to find clinical benefits. Tools and analyses with greater sensitivity are also required to detect differences between drugs and placebos, especially as more mildly affected patients with COVID-19 are included in trials. The presence of heterogeneous populations with different genetic backgrounds might also affect outcomes of clinical results. Possible factors contributing to these clinical trial findings that should be accounted for in future trials include targeting the wrong pathobiological or pathophysiological mechanisms in COVID-19; using drugs that do not engage with the intended target (including virus proteins and virus–host and protein–protein interactome); intervening at the wrong stage of the disease, including early, mild, moderate, and severe illnesses; lacking translatable pharmacodynamic and pharmacokinetic (ie, poor lung penetration) biomarkers; depending on in vitro antiviral activities and not using appropriate animal models with poor predictive efficacy; not addressing the rapid disease progression of COVID-19 in a short period; and not accurately monitoring the complexity of the clinical and biological characteristics to therapeutic intervention.
Although AI-based drug repurposing is in the developmental stage, several examples have shown encouraging results, including baricitinib identified by BenevolentAI,34 dexamethasone48 predicted by CoV-KGE,35 and melatonin from network medicine-based findings (figure 1).46 The development of effective and robust in vitro and in vivo models can reduce the failure rate of drug repurposing between preclinical studies and clinical trials for COVID-19.67, 68 Genotype-informed drug repurposing (termed personalised drug repurposing) might further improve the success rate of clinical trials.69
Challenges in biological interpretation
Given the highly complex and regulated nature of drug development, a long-term vision is needed when developing AI applications in drug repurposing that could increase efficiency and effectiveness in the various processes involved, and reduce the barriers between the numerous research components in the ecosystem to create new therapy options. AI technologies, such as visible neural networks,70 incorporate the AI model's inner workings into real systems of biomedical sciences (eg, human and animal). For example, visible machine learning approaches might guide model structures of data heterogeneity in the life sciences and translate patient data to successful therapies.71 Biological systems are complex and hierarchical (figure 2), composed of multiple levels such as sequences, protein complexes, cells, tissues, organs, and organisms. Drug discovery is a complicated process involving multilevel interactions between chemical compounds and biological systems. Therefore, a potential way of building an effective and interpretable model of drug discovery is to enrich the biologically-inspired visible neural network model72 with drug-related entities such as chemical compounds and diseases. The biomedical knowledge on how different entities interact with each other at different levels can be leveraged to guide the design of the corresponding computing modules.35 Compared to current deep learning models which try to model the entire system with a complex model at once, this divide-and-conquer scheme models the different components in the complex system and how these components interact with each other in an explicit and transparent way. The model parameters can be optimised in an end-to-end way as in other deep learning models.
Challenges in data and model harmonisation
Data harmonisation refers to the process of standardising and integrating information from disparate sources to form a unified database. Data harmonisation is a crucial step for guaranteeing that the machine-learning based models that are developed are widely applicable in different scenarios. Establishing a high quality data model (which is a prerequisite for organising and standardising the data) is the foundation for the harmonisation process. National and international efforts aim to build common data models such as the national patient-centered clinical research network72 and the observational health data science and informatics programme.73 Fast healthcare interoperability resources74 represent another type of standard, which defines how these data should be exchanged.
In addition to data harmonisation, model harmonisation, which defines a unified standard for storing the computational models, is also an important aspect to enhance the generalisability and utility of the computational drug repurposing tools. The open neural network exchange (ONNX) is an example of such efforts aiming to build model exchanging standards that are interoperable. ONNX defines implemented models as an extensible acyclic graph model. Each node on the graph is a call to built-in operators with inputs and outputs defined using standard data types.
Challenges in data sharing and security
With the enhanced availability of health-related data (especially patient data), concerns have been raised regarding data security and privacy.75 For example, demographics and DNA sequencing data have an increased risk of making patients identifiable. Efforts should be made to scrutinise each stage in the data life cycle. For example, questions pertaining to what type of data will be collected; whether the data are necessary; who will collect the data; how the data will be used, stored, and transferred; what the rights are of the person whose data are being collected, and others, should be addressed carefully. Additionally, regulations and transparency are crucial for appropriate data collection and use, and so is an increased public awareness. Towards this goal, federated learning76 could be a promising direction, which trains algorithms across decentralised edge devices (eg, individual mobile phones) or servers hosting different local samples (eg, data owned by different samples). Data samples are not shared or centralised and only the trained models are communicated, which might improve data security and privacy of patient data for drug–disease outcome validation in drug repurposing.
Personalised drug repurposing
Advances in pharmacogenetics and pharmacogenomics indicated that disease treatment would be considerably improved if therapies were guided by individual's genomic profiles. This hypothesis has garnered initial success in some diseases, including cancer.77 Responsiveness to a drug is influenced by genetic, epigenetic, and environmental factors. SARS-CoV-2 infection has shown large inter-individual variabilities, ranging from asymptomatic to severe and lethal disease. One possible hypothesis is that human genetics might determine clinical characteristics and drug responses.69, 78 For example, analysis of approximately 81 000 genomes and exomes from the general population suggested that hydroxychloroquine or chloroquine might only work for TMPRSS2-absent patients who are infected by SARS-CoV-2.69 An international team showed that hydroxychloroquine has antiviral activity in the kidney cells of African green monkeys without TMPRSS2 expression (VeroE6) but not in the model of reconstituted human airway epithelium developed from primary nasal or bronchial cells.67 Additionally, another team showed that chloroquine does not block SARS-CoV-2 infection of the TMPRSS2-positive lung cell line Calu-3.68 These preliminary findings highlight the importance of pharmacogenomics studies in improving clinical benefits and the success rate of drug repurposing. A COVID-19 host genetics initiative is underway to generate, share and analyse data in a search for the genetic determinants of COVID-19 susceptibility, severity, and outcomes, and personalised treatment. Therefore, AI techniques could leverage massive genetic and genomic data to identify human genetic determinants of SARS-CoV-2 pathogenesis, which presents a unique opportunity for drug repurposing in precision medicine and personalised treatment for individuals with COVID-19 (figure 3 ).
Figure 3.
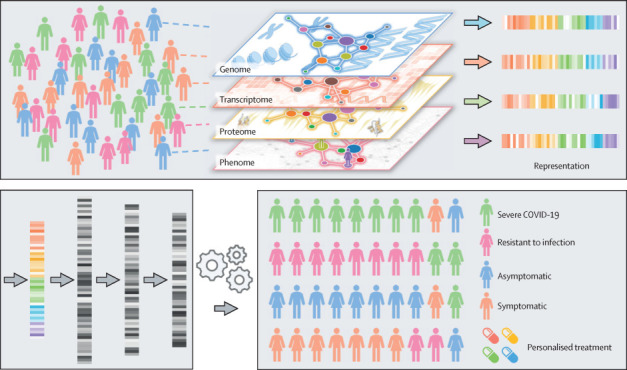
AI for patient stratification and personalised treatment
AI approaches can accelerate precision medicine by the unique integration of genomic, transcriptomic, proteomic, and phenomic profiles from individuals. The bottom left panel represents the feature extraction step using deep learning, the arrows represent successive feature extraction from previous layers, and the shades of grey represent the output (learned features that have high or low values in the output of learned features from previous layers). AI=artificial intelligence.
The future of AI-informed drug repurposing
Drug selection among the many approved ones, while avoiding time-consuming searches, can present uncertainty. To date, AI's potential ability to identify new candidate therapies that can be made available for clinical trials rapidly and, if approved, merged into health care is unparalleled, making AI a centrepiece of advanced technologies. Because of this, AI is a promising method for accelerating drug repurposing for human diseases, especially emerging diseases, such as COVID-19. With the availability of big data, including biological, clinical, and open data (scientific publications and data repositries), novel AI techniques capable of leveraging these large sets of biomedical data are in high demand. Pharmaceutical scientists, computer scientists, statisticians, and physicians are increasingly involved in developing and adopting AI-based technologies for the rapid development of therapeutics. AI approaches, coupled with big data, have the potential to substantially improve the efficiency and effectiveness of drug repurposing and aid medical decision making of therapeutic benefits with real-world evidence for various complex human diseases, such as COVID-19 (figure 1) and Alzheimer's disease.79 However, challenges remain in developing these AI tools, such as data heterogeneity and low quality, and insufficient data sharing by pharmaceutical companies, as well as the security and interpretability of the models. We expect future successful AI models for drug repurposing to be accurate in terms of the generated outcomes, integrative of disparate information types and sources, interoperable in diverse deployment settings, interpretable of internal working mechanisms, and robust to noise and adversarial attacks.
Search strategy and selection criteria
References for this Review were identified through searches of PubMed, Ovid MEDLINE, Scopus, ProQuest, Google Scholar, and Web of Science for papers, including peer-reviewed articles, grey literature, and preprints, published in English from Jan 1, 2010, to July 22, 2020 with “OR/AND” operators. Keywords and the English Medical Subject Headings terms include “drug repurposing”, “drug repositioning”, “network medicine”, “machine learning”, “artificial intelligence”, “convolutional neural networks”, “deep neural networks”, “deep learning”, “COVID-19”, and “SARS-CoV-2”. We also included case reports, case series, and review articles. We independently reviewed the titles and abstracts for inclusion. Additional relevant articles were identified from the review of citations referenced.
Acknowledgments
Acknowledgments
This work was supported by the National Institute of Aging of the US National Institutes of Health (NIH; R01AG066707 and 3R01AG066707-01S1) and the National Heart, Lung, and Blood Institute of the NIH (R00HL138272) to FC. This work was supported by the VeloSano Pilot Program (Cleveland Clinic Taussig Cancer Institute, Cleveland, OH, USA) to FC. This work has been also supported with federal funds from the Frederick National Laboratory for Cancer Research, NIH (HHSN261200800001E) to RN. This research was supported by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research to RN. This work was supported by the US National Science Foundation (1750326 and 2027970) and US Office of Naval Research (N00014-18-1-2585) to FW. We are thankful for all helpful discussions and critical comments regarding this manuscript from the COVID-19 Research Intervention Advisory Committee members at the Cleveland Clinic.
Contributors
FC conceived the study. YZ, FW, JT, and FC, did literature searching and data analysis. FC, YZ, FW, JT, and RN wrote and critically revised the manuscript.
Declaration of interests
RN is principle investigator of Leidos Biomedical Research, National Cancer Institute at Frederick, National Institutes of Health, Frederick, MD, USA. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government. All other authors declare no competing interests.
References
- 1.Turing AM. Computing machinery and intelligence. Mind. 1950;49:433–460. [Google Scholar]
- 2.Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557:S55–S57. doi: 10.1038/d41586-018-05267-x. [DOI] [PubMed] [Google Scholar]
- 3.Smalley E. AI-powered drug discovery captures pharma interest. Nat Biotechnol. 2017;35:604–605. doi: 10.1038/nbt0717-604. [DOI] [PubMed] [Google Scholar]
- 4.Avorn J. The $2·6 billion pill·methodologic and policy considerations. N Engl J Med. 2015;372:1877–1879. doi: 10.1056/NEJMp1500848. [DOI] [PubMed] [Google Scholar]
- 5.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007;448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 6.Ghofrani HA, Osterloh IH, Grimminger F. Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat Rev Drug Discov. 2006;5:689–702. doi: 10.1038/nrd2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laubach JP, Richardson PG, Anderson KC. Hematology: thalidomide maintenance in multiple myeloma. Nat Rev Clin Oncol. 2009;6:565–566. doi: 10.1038/nrclinonc.2009.131. [DOI] [PubMed] [Google Scholar]
- 8.Beigel JH, Tomashek KM, Dodd LE. Remdesivir for the treatment of Covid-19—Preliminary Report. N Engl J Med. 2020;2020:22. doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 9.Cheng F, Murray JL, Zhao J, Sheng J, Zhao Z, Rubin DH. Systems biology-based investigation of cellular antiviral drug targets identified by gene-trap insertional mutagenesis. PLOS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng X, Zhu S, Lu W. Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci (Camb) 2020;11:1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng F, Desai RJ, Handy DE. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene JA, Loscalzo J. Putting the patient back together—social medicine, network medicine, and the limits of reductionism. N Engl J Med. 2017;377:2493–2499. doi: 10.1056/NEJMms1706744. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann M, Kleine-Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779. doi: 10.1016/j.molcel.2020.04.022. 84.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M, Kleine-Weber H, Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271. doi: 10.1016/j.cell.2020.02.052. 80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon DE, Jang GM, Bouhaddou M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 18.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323:533–536. [Google Scholar]
- 19.Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016;13:2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenselink EB, Ten Dijke N, Bongers B. Beyond the hype: deep neural networks outperform established methods using a ChEMBL bioactivity benchmark set. J Cheminform. 2017;9:45. doi: 10.1186/s13321-017-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer JG, Liu S, Miller IJ, Coon JJ, Gitter A. Learning drug functions from chemical structures with convolutional neural networks and random forests. J Chem Inf Model. 2019;59:4438–4449. doi: 10.1021/acs.jcim.9b00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallach I, Dzamba M, Heifets A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv. 2015 https://arxiv.org/abs/1510.02855 published online Oct 10. (preprint) [Google Scholar]
- 23.Segler MHS, Kogej T, Tyrchan C, Waller MP. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci. 2018;4:120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao KY, Fokoue A, Luo H, Iyengar A, Dey S, Zhang P. Interpretable drug target prediction using deep neural representation. IJCAI. 2018:3371–3377. [Google Scholar]
- 25.Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, Qu M, Wang M, Zhang M, Yan J, Mei Q. LINE: large-scale information network embedding. Proceedings of the 24th International Conference on World Wide Web; Florence, Italy: May 2015 (pp 1067–77).
- 27.Bordes A, Usunier N, Garcia-Durán A, Weston J, Yakhnenko O. Translating embeddings for modeling multi-relational data. 26th International Conference on Neural Information Processing Systems; Lake Tahoe, NV: Dec 2013 (pp 1–9).
- 28.Trouillon T, Welbl J, Riedel S, Gaussier E, Bouchard G. Complex embeddings for simple link prediction. In: Maria Florina B, Kilian QW, editors. Proceedings of the 33rd International Conference on Machine Learning. Proceedings of Machine Learning Research; New York, NY, USA: 2016. pp. 2071–2080. [Google Scholar]
- 29.Dettmers T, Minervini P, Stenetorp P, Riedel S. Convolutional 2D knowledge graph embeddings. arXiv. 2017 https://arxiv.org/abs/1707.01476 published online July 5. (preprint) [Google Scholar]
- 30.Sun Z, Deng Z-H, Nie J-Y, Tang J. RotatE: knowledge graph embedding by relational rotation in complex space. arXiv. 2019 https://arxiv.org/abs/1902.10197 published online Feb 26. (preprint). [Google Scholar]
- 31.Zhu Z, Xu S, Qu M, Tang J. GraphVite: a high-performance CPU-GPU hybrid system for node embedding. arXiv. 2019 https://arxiv.org/abs/1903.00757 published online March 2. (preprint). [Google Scholar]
- 32.Sosa DN, Derry A, Guo M, Wei E, Brinton C, Altman RB. A literature-based knowledge graph embedding method for identifying drug repurposing opportunities in rare diseases. Pac Symp Biocomput. 2020;25:463–474. [PMC free article] [PubMed] [Google Scholar]
- 33.Gysi DM, Do Valle I, Zitnik M. Network medicine framework for identifying drug repurposing opportunities for COVID-19. arXiv. 2020 doi: 10.1073/pnas.2025581118. https://arxiv.org/abs/2004.07229 published online April 15. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson P, Griffin I, Tucker C. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X, Song X, Ma T. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00316. published online July 12. [DOI] [PubMed] [Google Scholar]
- 36.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A Review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 37.Yin W, Mao C, Luan X. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman JD, Lye DCB, Hui DS. Remdesivir for 5 or 10 days in patients with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2015301. publish online May 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Y, Tian T, Huang S. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv. 2020 doi: 10.1101/2020.03.11.986836. published online March 12. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiseman LR, Goa KL. Toremifene. A review of its pharmacological properties and clinical efficacy in the management of advanced breast cancer. Drugs. 1997;54:141–160. doi: 10.2165/00003495-199754010-00014. [DOI] [PubMed] [Google Scholar]
- 41.Cong Y, Hart BJ, Gross R. MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyall J, Coleman CM, Hart BJ. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeon S, Ko M, Lee J. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 2020;64:e00819–e00920. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin W, Cheng F. Repurposing of FDA-approved toremifene to treat COVID-19 by blocking the spike glycoprotein and NSP14 of SARS-CoV-2. J Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00397. published online Sept 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A, Madhavan MV, Sehgal K. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y, Hou Y, Shen J. A network medicine approach to prediction and patient-based validation of disease manifestations and drug repurposing for COVID-19. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12579137. published online July 2. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramamoorthy S, Cidlowski JA. Corticosteroids: mechanisms of action in health and disease. Rheum Dis Clin North Am. 2016;42:15–31. doi: 10.1016/j.rdc.2015.08.002. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Group RC, Horby P, Lim WS. Dexamethasone in hospitalised patients with Covid-19—preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. published online July 17. [DOI] [Google Scholar]
- 49.Rosenberg ES, Dufort EM, Udo T. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in new york state. JAMA. 2020;323:2493–2502. doi: 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng F, Kovács IA, Barabási AL. Network-based prediction of drug combinations. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng F, Rao S, Mehra R. COVID-19 treatment: combining anti-inflammatory and antiviral therapeutics using a network-based approach. Cleve Clin J Med. 2020 doi: 10.3949/ccjm.87a.ccc037. published online June 30. [DOI] [PubMed] [Google Scholar]
- 53.Häyrinen K, Saranto K, Nykänen P. Definition, structure, content, use and impacts of electronic health records: a review of the research literature. Int J Med Inform. 2008;77:291–304. doi: 10.1016/j.ijmedinf.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Sherman RE, Anderson SA, Dal Pan GJ. Real-world evidence—what is it and what can it tell us? N Engl J Med. 2016;375:2293–2297. doi: 10.1056/NEJMsb1609216. [DOI] [PubMed] [Google Scholar]
- 55.Bauchner H, Fontanarosa PB. Randomized clinical trials and COVID-19: managing expectations. JAMA. 2020;2020:4. doi: 10.1001/jama.2020.8115. [DOI] [PubMed] [Google Scholar]
- 56.Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 57.Lee BK, Lessler J, Stuart EA. Improving propensity score weighting using machine learning. Stat Med. 2010;29:337–346. doi: 10.1002/sim.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linden A, Yarnold PR. Combining machine learning and propensity score weighting to estimate causal effects in multivalued treatments. J Eval Clin Pract. 2016;22:871–881. doi: 10.1111/jep.12610. [DOI] [PubMed] [Google Scholar]
- 59.Sun J, Wang F, Hu J, Edabollahi S. Supervised patient similarity measure of heterogeneous patient records. SIGKDD Explor. 2012;14:16–24. [Google Scholar]
- 60.Murphy SN, Weber G, Mendis M. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stang PE, Ryan PB, Racoosin JA. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Ann Intern Med. 2010;153:600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 62.Mancia G, Rea F, Ludergnani M, Apolone G, Corrao G. Renin-angiotensin-aldosterone system blockers and the risk of Covid-19. N Engl J Med. 2020;382:2431–2440. doi: 10.1056/NEJMoa2006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reynolds HR, Adhikari S, Pulgarin C. Renin-angiotensin-aldosterone system inhibitors and risk of Covid-19. N Engl J Med. 2020;382:2441–2448. doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta HB, Ehrhardt S, Moore TJ, Segal JB, Alexander GC. Characteristics of registered clinical trials assessing treatments for COVID-19: a cross-sectional analysis. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Cao R, Xu M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park SJ, Yu KM, Kim YI. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. MBio. 2020;11:e01114–e01120. doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maisonnasse P, Guedj J, Contreras V. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;2020:22. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- 68.Hoffmann M, Mösbauer K, Hofmann-Winkler H. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2575-3. published online July 22. [DOI] [PubMed] [Google Scholar]
- 69.Hou Y, Zhao J, Martin W. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma J, Yu MK, Fong S. Using deep learning to model the hierarchical structure and function of a cell. Nat Methods. 2018;15:290–298. doi: 10.1038/nmeth.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu MK, Ma J, Fisher J, Kreisberg JF, Raphael BJ, Ideker T. Visible machine learning for biomedicine. Cell. 2018;173:1562–1565. doi: 10.1016/j.cell.2018.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21:578–582. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hripcsak G, Duke JD, Shah NH. Observational health data sciences and informatics (OHDSI): Opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 74.Braunstein ML. Healthcare in the age of interoperability: the promise of fast healthcare interoperability resources. IEEE Pulse. 2018;9:24–27. doi: 10.1109/MPUL.2018.2869317. [DOI] [PubMed] [Google Scholar]
- 75.Mello MM, Wang CJ. Ethics and governance for digital disease surveillance. Science. 2020;368:951–954. doi: 10.1126/science.abb9045. [DOI] [PubMed] [Google Scholar]
- 76.Xu J, Glicksberg SB, Su C, Walker P, Bian J, Wang W. Federated learning for healthcare informatics. arXiv. 2019 doi: 10.1007/s41666-020-00082-4. https://arxiv.org/abs/1911.06270 published online Nov 13 2019. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nussinov R, Jang H, Tsai CJ, Cheng F. Review: Precision medicine and driver mutations: Computational methods, functional assays and conformational principles for interpreting cancer drivers. PLOS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ellinghaus D, Degenhardt F, Bujanda L. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020 doi: 10.1056/NEJMoa2020283. published online June 17. ht--tps://doi.org/10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang J, Pieper AA, Nussinov R. Harnessing endophenotypes and network medicine for Alzheimer's drug repurposing. Med Res Rev. 2020 doi: 10.1002/med.21709. published online July 13. [DOI] [PMC free article] [PubMed] [Google Scholar]


