Abstract
A hallmark of severe COVID-19 pneumonia is SARS-CoV-2 infection of the facultative progenitors of lung alveoli, the alveolar epithelial type 2 cells (AT2s). However, inability to access these cells from patients, particularly at early stages of disease, limits an understanding of disease inception. Here, we present an in vitro human model that simulates the initial apical infection of alveolar epithelium with SARS-CoV-2 by using induced pluripotent stem cell-derived AT2s that have been adapted to air-liquid interface culture. We find a rapid transcriptomic change in infected cells, characterized by a shift to an inflammatory phenotype with upregulation of NF-κB signaling and loss of the mature alveolar program. Drug testing confirms the efficacy of remdesivir as well as TMPRSS2 protease inhibition, validating a putative mechanism used for viral entry in alveolar cells. Our model system reveals cell-intrinsic responses of a key lung target cell to SARS-CoV-2 infection and should facilitate drug development.
Keywords: SARS-CoV-2, COVID-19, alveolar epithelial cell, alveolar type 2 cell, human induced pluripotent stem cells, lung, iPSCs, inflammation
Graphical Abstract
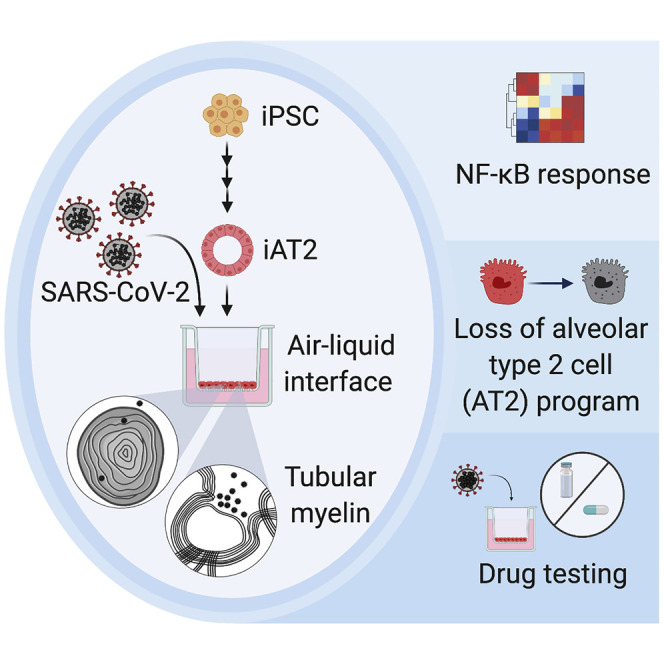
Highlights
-
•
iPSC-derived alveolar type 2 cells (iAT2s) are permissive to SARS-CoV-2 infection
-
•
SARS-CoV-2 induces an iAT2-intrinsic cytotoxicity and inflammatory response
-
•
iAT2s effectively model antiviral drug response and may be used for further drug development
Huang et al. show that human iPSC-derived alveolar type 2 cells (iAT2s) can be used to model COVID-19. They find that iAT2s in air-liquid interface culture are permissive to SARS-CoV-2 infection and show that SARS-CoV-2 induces a rapid inflammatory phenotype predominated by NF-κB signaling.
Introduction
Responding to the COVID-19 pandemic caused by the novel coronavirus, SARS-CoV-2, requires access to human model systems that can recapitulate disease pathogenesis, identify potential targets, and enable drug testing. Access to primary human lung in vitro model systems is a particular priority because a variety of respiratory epithelial cells are the proposed targets of viral entry (Hoffmann et al., 2020; Hou et al., 2020; Zhu et al., 2020). A rapidly emerging literature now indicates that a diversity of epithelial cells of the respiratory tract from the nasal sinuses and proximal conducting airways through the distal lung alveoli express the cell surface receptor for SARS-CoV-2, angiotensin-converting enzyme 2 (ACE2), and appear permissive to infection with SARS-CoV-2 in vivo and, in some cases, in vitro (Bradley et al., 2020; Hou et al., 2020; Leung et al., 2020; Schaefer et al., 2020; Sun et al., 2020; Sungnak et al., 2020; Ziegler et al., 2020). The most severe infections with SARS-CoV-2 result in acute respiratory distress syndrome (ARDS), a clinical phenotype that is thought to arise in the setting of COVID-19 pneumonia as the virus progressively targets the epithelium of the distal lung, particularly the facultative progenitors of this region, alveolar epithelial type 2 cells (AT2s) (Hou et al., 2020). Although small animal models such as Syrian hamsters (Imai et al., 2020; Sia et al., 2020) and humanized ACE2 transgenic mice (Bao et al., 2020; Jiang et al., 2020) have shown changes in the alveolar epithelium after SARS-CoV-2 infection, little is known about the initial responses of human lung alveoli to SARS-CoV-2 infection, in part, because of to the inability to access these cells from patients, particularly at early stages of disease.
To date, primary human AT2s that are harvested from explanted lung tissues require 3D co-culture with supporting fibroblasts, cannot be maintained in culture for more than three passages, and tend to rapidly lose their AT2 phenotype ex vivo (Jacob et al., 2019). Thus, SARS-CoV-2 infection modeling has to this point been predominantly performed with either human airway (non-alveolar) cells in air-liquid interface (ALI) cultures, non-human cell lines that naturally express the ACE2 viral receptor, such as the African Green Monkey Vero E6 cell line (Harcourt et al., 2020), or transformed human cell lines with or without forced overexpression of ACE2. Although some of these cell lines, such as A549 and Calu-3 cells, were originally generated from transformed cancerous lung epithelial cells, they no longer express NKX2-1, the master transcriptional regulator required for differentiated lung epithelial gene expression (Abo et al., 2020) and, thus, are limited in their capacity to simulate an accurate lung cellular response to most perturbations, including viral infections.
To provide alternative sources of self-renewing human lung epithelial lineages, our group and others have recently developed human lung epithelial organoids and 2D ALI lung cultures through the directed differentiation of induced pluripotent stem cells (iPSCs) in vitro (Abo et al., 2020; Hawkins et al., 2017; Huang et al., 2014; Hurley et al., 2020; Jacob et al., 2017; Longmire et al., 2012; McCauley et al., 2017, 2018a, 2018b; Serra et al., 2017; Yamamoto et al., 2017). Here, we report the successful infection of a pure population of human iPSC-derived AT2-like cells (iAT2s) with SARS-CoV-2, providing a reductionist model that reveals the cell-intrinsic distal lung epithelial global transcriptomic responses to infection. By 1 day post-infection (dpi), SARS-CoV-2 induced a rapid global transcriptomic change in infected iAT2s characterized by a shift to an inflammatory phenotype associated with the secretion of cytokines encoded by NF-κB target genes. By 4 dpi, there were time-dependent epithelial interferon responses and progressive loss of the mature lung alveolar epithelial program, exemplified by significant downregulation of surfactant encoding genes—transcriptomic changes that were not predicted by prior human airway or cell line models. Our model system thus reveals the cell-intrinsic responses of a key lung target cell to infection, providing a novel, physiologically relevant platform for further drug development and facilitating a deeper understanding of COVID-19 pathogenesis.
Results
iAT2s in ALI Culture Are Permissive to SARS-CoV-2 Infection and Replication
In order to develop a human model system, we used the technique of directed differentiation (Jacob et al., 2017, 2019) to generate iAT2s from either human embryonic stem cells or iPSCs engineered to carry a tdTomato reporter targeted to the endogenous SFTPC locus (Hurley et al., 2020; Jacob et al., 2017). In 3D Matrigel cultures, we established self-renewing epithelial spheres composed of purified iAT2s, >90% of which expressed surfactant protein-C (SFTPC), the canonical AT2 marker, as monitored by flow cytometry assessment of the SFTPCtdTomato reporter at each passage in culture (Figures 1A and 1B). Serially passaging these epithelial spheres generated >1030 iAT2s per starting sorted tdTomato+ cell over 225 days in culture (Hurley et al., 2020), generating cells that maintained expression of AT2 marker genes, including surfactants as shown by single cell RNA sequencing (scRNA-seq) (Figures 1C, 1D, and S1), and providing a stable source of human primary-like AT2 cells for viral infection disease modeling.
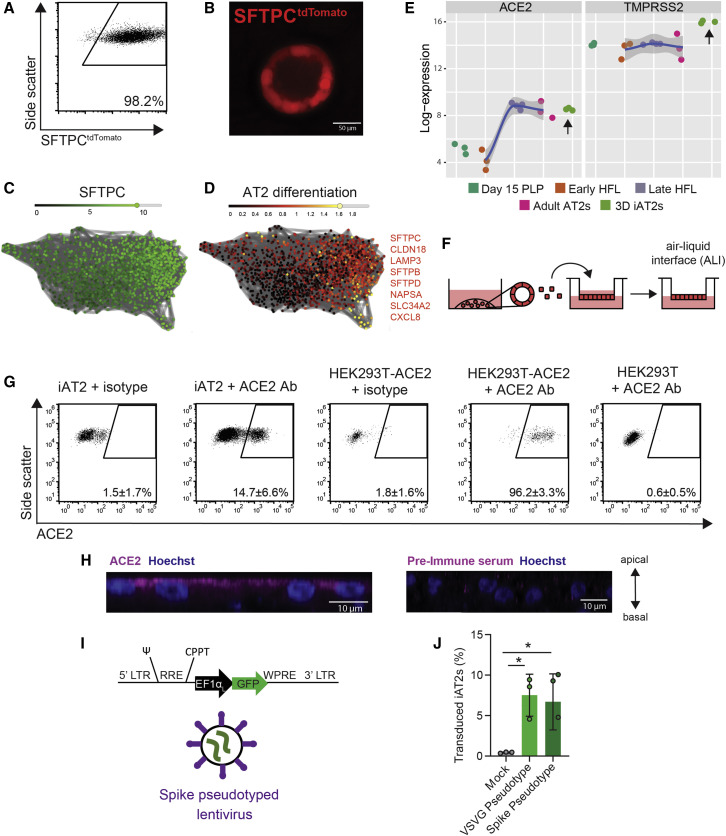
Figure 1.
iPSC-Derived Alveolar Epithelial Type 2 Cells (iAT2s) Express Functional SARS-CoV-2 Entry Factors ACE2 and TMPRSS2
(A and B) iAT2s, carrying a tdTomato reporter targeted to the endogenous SFTPC locus by gene editing (SPC2 line), can be serially passaged while maintaining >90% SFTPCtdTomato+ expression in 3D sphere cultures (day 160 of differentiation, passage 8 shown; scRNA-seq profile provided in Figure S1 and compared to Habermann et al., 2020).
(C and D) scRNA-seq data of iAT2s (SPC2 line at day 114 of differentiation) visualized in SPRING plots (Weinreb et al., 2018) based on reanalysis of a dataset previously published in (Hurley et al., 2020) showing expression of (C) SFTPC as well as (D) an eight-gene benchmark of AT2 cell differentiation (SFTPC, CLDN18, LAMP3, SFTPB, SFTPD, NAPSA, SLC34A2, and CXCL8 as characterized in [Hurley et al., 2020]).
(E) iAT2s (RUES2 line) express ACE2 and TMPRSS2 transcripts at comparable levels to purified primary adult human lung AT2s (day 15 PLP, primordial lung progenitors derived from pluripotent stem cells at day 15 of differentiation; early HFL, primary early human fetal lung alveolar epithelium at 16–17.5 weeks gestation; late HFL, alveolar epithelium at 20–21 weeks gestation; and adult AT2s, adult alveolar epithelial type 2 cells from 3 different individuals freshly sorted using the antibody against HTII-280. Primary adult and fetal sample procurement described in detail in Hurley et al., [2020]).
(F–H) iAT2s (SPC2 line) cultured at air-liquid interface (ALI) (F) express ACE2 protein, as observed by flow cytometry, n = 9 (G; additional scRNA-seq profiling in Figure S1), which is apically localized, as observed by immunofluorescence staining (scale bar, 10 μm) (H).
(I and J) iAT2s infected with a GFP-expressing lentivirus pseudotyped with either VSVG or SARS-CoV-2 Spike envelopes, n = 3. ∗p < 0.05, one-way ANOVA. All bars represent mean ± standard deviation. See also Figure S1.
Because the directed differentiation of human iPSCs in vitro is designed to recapitulate the sequence of developmental milestones that accompanies in vivo human fetal organogenesis, we first sought to understand the ontogeny of expression of the coronavirus receptor, ACE2, during human fetal development from early endodermal progenitors through increasingly more mature stages of alveolar development. By analyzing our previously published transcriptomic time series profiles of developing human fetal and adult primary AT2s (Hurley et al., 2020), we found that ACE2 expression increases from early to late canalicular stages of distal human lung development, and expression levels were similar to adult AT2 levels present by week 21 of gestation in developing alveolar epithelial cells (Figure 1E). We found the directed differentiation of human pluripotent stem cells (RUES2 embryonic stem cells and SPC2 iPSCs) in vitro into purified distal lung AT2-like cells resulted in cells expressing similar levels of ACE2 to those of adult primary cell controls in head-to-head comparisons (Figures 1E and S1, respectively). We and others (Abo et al., 2020; Ziegler et al., 2020) have recently profiled the frequency of ACE2 mRNA-expressing primary adult and iPSC-derived AT2-like cells by scRNA-seq (Figure S1), finding that mRNA expression occurs in a minority of cells (1%–3%) at any given time; similar frequencies were observed in primary AT2s compared to iAT2s. In contrast, the gene encoding the protease utilized for viral entry, TMPRSS2, is expressed more robustly in both AT2s and iAT2s (Figure S1 and [Abo et al., 2020]) and is less developmentally variable, being stably expressed by week 16 of distal fetal lung development (Figure 1E).
Because lung epithelial infection by SARS-CoV-2 occurs at the apical membrane of cells facing the air-filled lumens of airways and alveoli, submerged cultures of 3D epithelial spheres with apical membranes oriented interiorly are unlikely to faithfully recapitulate infection physiology. Therefore, we adapted our SFTPCtdTomato+ iAT2s (SPC2-ST-B2 iPSC line) to 2D ALI cultures (Figure 1F), generating monolayered epithelial cultures of pure iAT2s with apical-basal polarization and barrier integrity (transepithelial electrical resistance = 454 ± 73 ohms × cm2) while preserving or augmenting expression of AT2-specific genes (e.g., SFTPC, SFTPB, SFTPA1, SFTPA2, SFTPD, NAPSA, and PGC) as detailed in our recent preprint (Abo et al., 2020) as well as in Figure S1.
To quantify protein-level expression frequency of ACE2 in iAT2s in ALI cultures, we employed flow cytometry, observing that 14.7 ± 6.6% of live iAT2s demonstrated cell surface expression of ACE2 (Figure 1G) and indicating more frequent expression at the protein level than had been predicted from analysis of published primary AT2 or ALI-cultured iAT2 scRNA-seq profiles (Figure S1 and [Abo et al., 2020]). We validated the sensitivity and specificity of the ACE2 antibody by staining controls consisting of human 293T cells, which lack ACE2, and 293T cells lentivirally transduced to overexpress ACE2 (293T-ACE2; Figure 1G) (Mou et al., 2020). Apical localization of ACE2 in iAT2s was confirmed by immunofluorescence staining (Figure 1H). To test functionality of ACE2 as the viral receptor, we used a GFP-expressing lentivirus pseudotyped with either viral spike protein (S) or VSV-G envelope to infect iAT2s, 293T cells, and 293T-ACE2 cells, finding that both pseudotypes infected iAT2s and 293T-ACE2 cells, whereas the Spike pseudotype did not infect 293T cells (Figures 1I, 1J, and S1C; Crawford et al., 2020).
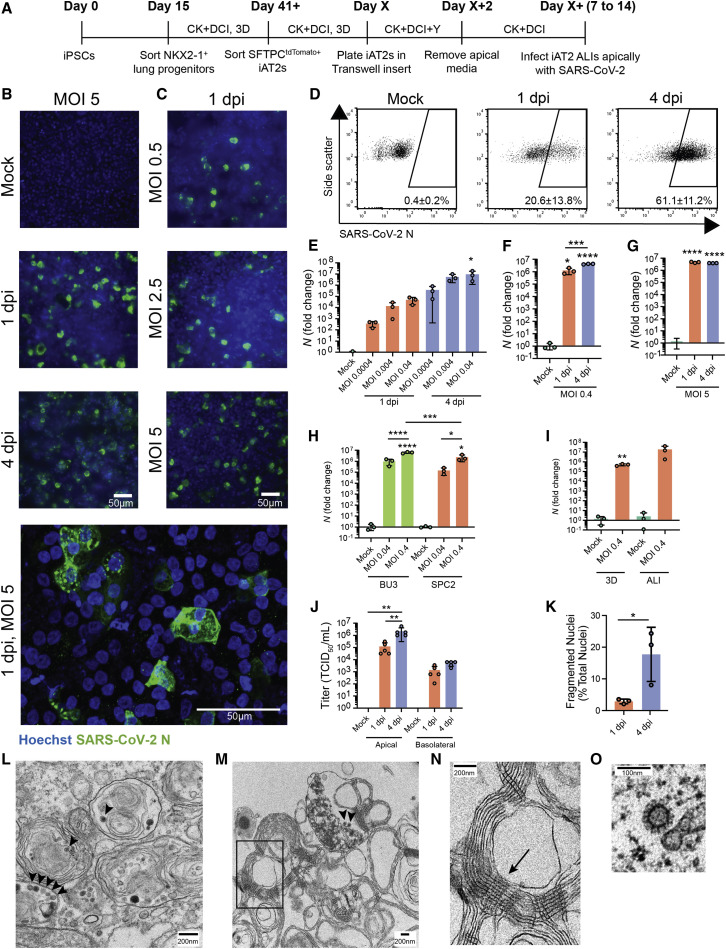
To test whether ALI cultures of iAT2s are permissive to SARS-CoV-2 infection, we employed escalating viral doses representing a broad range of multiplicities of infection (MOIs). iAT2s in ALI culture were apically exposed to SARS-CoV-2 for 1 h (Figure 2 A), and viral infection was visualized by immunofluorescence analysis using an antibody against the viral nucleoprotein (N). The number of N-positive cells increased over time (Figure 2B) and with increasing MOIs (Figure 2C), indicative of viral replication and spread, and the majority of cells remained viable at 4 dpi (Figure S1C). Quantification of N-positive cells by flow cytometry revealed an infection level of about 20% at 1 dpi and 60% at 4 dpi (Figure 2D). The time- and dose-dependent increases in infection efficiencies were observed at both low MOIs (0.0004–0.04; unpurified virus) and high MOIs (up to 5; purified virus); MOI 5 saturated N transcript levels by 1 dpi (Figures 2E–2G). Additionally, similar infection efficiencies were observed for iAT2 ALI cultures derived from multiple iPSC lines generated from distinct donors of different genetic backgrounds (Figure 2H), and iAT2s could be infected while growing either in ALI cultures or when grown as 3D spheres that were mechanically disrupted at the time of infection (Figure 2I). In iAT2 ALI cultures, infectious virus was predominantly released from the apical side at increasing titers over time (4 dpi versus 1 dpi; Figure 2J), providing further evidence of ongoing viral replication in iAT2s in this model system. Although initial viral infection efficiency was similar to the percentage of ACE2-expressing iAT2s, the discrepancy between the number of ACE2+ iAT2s (13%) and N+ cells at 4 dpi (60%) may suggest alternative mechanisms of viral entry or spread. It is also conceivable that ACE2 surface expression of some SARS-CoV-2 target cells was below our flow cytometry-based detection limit. Immunofluorescence analysis of infected iAT2s suggests time-dependent cytopathogenicity, as indicated by increasing numbers of fragmented nuclei from 1 to 4 dpi (Figure 2K). Transmission electron microscopy (TEM) confirmed the successful infection of mature, functional iAT2s (Figures 2L–2O and S2). For example, viral particles were visible both intracellularly in lamellar body-containing iAT2s (Figures 2L and S2H) as well as in the apical extracellular space (Figure 2M) adjacent to tubular myelin (Figures 2M and 2N), an ultrastructure specific for secreted surfactant.
Figure 2.
SARS-CoV-2 Infects iAT2s in a Dose- and Time-Dependent Manner
(A) Schematic of the iAT2 directed differentiation protocol in which robustly self-renewing iAT2s can be plated at ALI for SARS-CoV-2 infections. “CK+DCI,” distal lung medium components detailed in the STAR★Methods.
(B and C) Immunofluorescence images of viral nucleoprotein (N, green) of iAT2s infected with SARS-CoV-2 (MOI = 5) at 1 and 4 days post infection (dpi) (B) or with increasing MOIs (0.5, 2.5, 5) shown at 1 dpi (C) (20×, scale bar, 50 μm).
(D) Efficiency of iAT2 infections scored by representative FACS plots of SARS-CoV-2 N at 1 and 4 dpi (MOI 5) compared to mock; mean gated percentages ± standard deviation for n = 3 replicates are shown; results representative of three independent experiments.
(E) RT-qPCR of viral N gene expression at 1 and 4 dpi via a range of low MOIs of an unpurified SARS-CoV-2 virus stock, n = 3.
(F and G) RT-qPCR of N gene expression at 1 and 4 dpi via a purified virus stock to infect with an MOI of 0.4 (F) or an MOI of 5 (G), n = 3. Fold change expression compared to Mock [2-ΔΔCt] after 18S normalization is shown.
(H) RT-qPCR of N gene expression of BU3 and SPC2 iAT2s at MOIs 0.04 and 0.4 at 1 dpi.
(I) RT-qPCR of N gene expression at an MOI of 0.4 in iAT2s in alveolospheres and iAT2s at ALI at 2 dpi.
(J) Viral titers were determined in apical washes and basolateral media at 1 and 4 dpi (n = 5).
(K) Mean percent fragmented nuclei in immunofluorescence images of infected iAT2s at 1 and 4 dpi (MOI 5; n = 3).
(L–N) Electron micrograph of infected iAT2s showing virions (arrow heads) intracellularly, including in a lamellar body and (M) extracellular virions around tubular myelin (N, arrow). Tubular myelin meshwork (inset from M) that forms upon secretion of pulmonary surfactant (scale bar, 200 nm).
(O) Higher magnification of a coronavirus virion at 1 dpi (MOI 5) (scale bar, 100 nm).
All bars represent mean ± standard deviation with biological replicates indicated for each panel. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, unpaired, two-tailed Student’s t test (I and K) or one-way ANOVA with multiple comparisons (E, F, G, H, and J) were performed.
RNA Sequencing Profiles of Infected iAT2s Indicate an Epithelial-Intrinsic Innate Immune Response
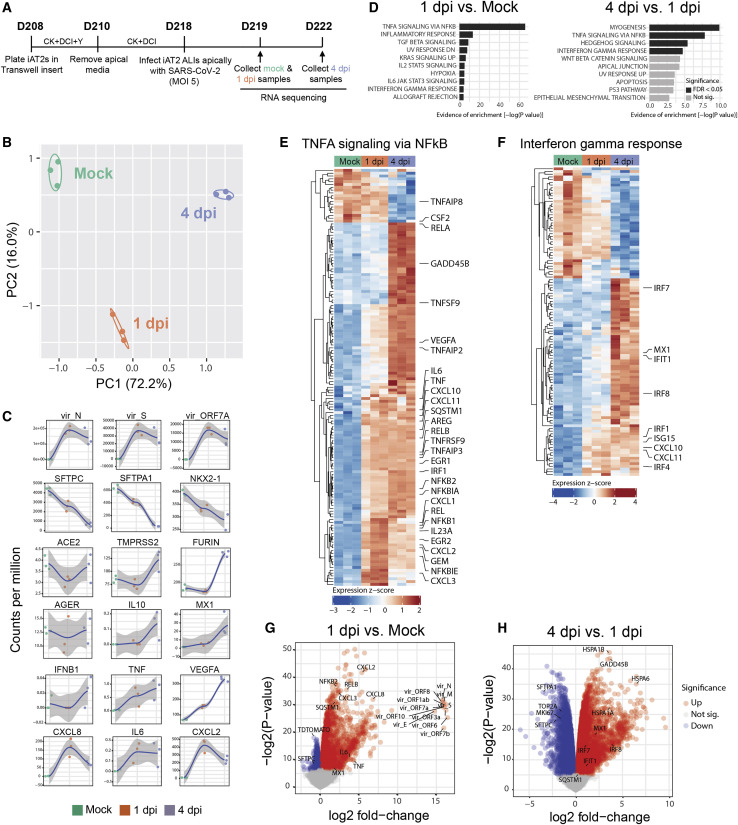
Having established a putative human model system for SARS-CoV-2 infection of AT2-like cells, we next sought to define the global, time-dependent transcriptomic responses of cells to infection. We performed bulk RNA sequencing of SARS-CoV-2-infected iAT2s at 1 dpi or 4 dpi compared to mock-infected iAT2 controls (n = 3 biological replicates at each time point, Figure 3 A). Profound and progressive changes in the global transcriptomes of infected iAT2s were observed based on principal-component analysis and differential gene expression (Figure 3B and Table S1). For example, 4,519 genes were differentially expressed between mock-infected and SARS-CoV-2-infected cells at 1 dpi, 10,725 genes between SARS-CoV-2-infected cells at 1 dpi and 4 dpi, and 10,462 genes between the infected samples (1 and 4 dpi combined) and mock (FDR < 0.05; empirical Bayes moderated t test; Table S1). Viral transcripts, including the viral genome, were among the top differentially expressed transcripts at both 1 dpi and 4 dpi, representing 28% to 33% of all reads mapped at 1 dpi (Figure 3C). AT2-specific genes, such as SFTPC, were among the top downregulated genes by 1 dpi, and progressive loss of the AT2 program continued to 4 dpi with significant continued loss of SFTPC-, SFTPA1-, SFTPD-, and SFTPC tdTomato-encoding transcripts (FDR < 0.05; Table S1; Figure 3C). This loss of AT2 marker gene expression was not accompanied by any detectable emergence of alternate lung fates as there was no transcriptomic evidence of any upregulation of airway (SCGB1A1, FOXJ1, FOXI1, TP63, MUC5B, or MUC5AC) or alveolar type 1 cell (AGER; Figure 3C) markers. Gene set enrichment analysis (GSEA) revealed significant upregulation of inflammatory pathways both at 1 dpi (FDR < 0.05; Figure 3D) and 4 dpi: NF-κB-mediated inflammatory signaling represented the first and second most-upregulated pathways at 1 dpi and 4 dpi, respectively, and the most upregulated pathway over the entire time course (Figures 3D–3G). Interferon (IFN) signaling was in the top ten pathways significantly upregulated at both time points, and pathways reported to be activated by interferon signaling were also significantly upregulated (FDR < 0.05), including KRAS (MAPK) signaling, IL6-JAK-STAT3, and IL2-STAT5 signaling (Figure 3D). Despite the GSEA findings, the global transcriptomic analysis showed no significant induction of individual type I and III IFN genes (e.g., IFNB1, IFNL1, or IFNL2) at any time point after infection, and upregulation of multiple IFN signaling-related genes and targets (IFNAR2, IRF1/4/7/8/9, IFIT1, MX1, CXCL10, CXCL11, SOCS3, and ISG15) was observed mainly at 4 dpi rather than 1 dpi, consistent with delayed signaling (Figures 3F–3H). These results were confirmed by RT-qPCR (Figures 4 and S4) and indicate that SARS-CoV-2 infection of iAT2s elicits a delayed, modest IFN response compared to treatment of iAT2s with either IFNβ or transfection with the synthetic double-stranded RNA analog, poly(I:C), which we found stimulate stronger interferon responses (Figure S4). Compared to other published datasets of SARS-CoV-2 infection models in Calu-3, A549-ACE2, and primary (non-alveolar) normal human bronchial epithelium (NHBE) (Blanco-Melo et al., 2020), iAT2s were able to uniquely capture the downregulation of AT2-specific programs, such as decreased surfactant gene expression and loss of lamellar body Gene Ontology (GO) terms (comparative gene set enrichment based on lung-related GO biological processes; Figure S3).
Figure 3.
SARS-CoV-2 Elicits Transcriptomic Changes in iAT2s That Highlight Epithelial-Intrinsic Inflammatory Responses to Infection
(A) Schematic of the iAT2 ALI samples (starting with day 208 iAT2s) infected with SARS-CoV-2 (MOI 5) and collected at 1 and 4 dpi (mock collected 1 dpi) for bulk RNA sequencing (RNA-seq).
(B) Principal-component analysis (PCA) of iAT2 samples (n = 3 biological replicates per condition) showing global transcriptomic variance (%) of PC1 and PC2 components.
(C) Local regression (LOESS) plots of viral, AT2, NF-κB, and interferon (IFN) gene expression levels quantified by RNA-seq normalized expression (counts per million reads).
(D) Gene set enrichment analysis (GSEA, Camera using Hallmark gene sets) of the top ten upregulated gene sets in 1 dpi versus mock or 4 dpi versus 1 dpi conditions (black color indicates statistical significance; FDR < 0.05).
(E and F) Unsupervised hierarchical clustered heatmaps of differentially expressed genes (DEGs; FDR < 0.05) in the Hallmark gene sets “TNFA signaling in NFkB” (E) and “interferon gamma response” (F) as plotted with row normalized Z score; a selected subset of these DEGs are highlighted with large font.
(G and H) Volcano plots of differentially expressed genes in 1 dpi versus mock (G) and 4 dpi versus 1 dpi (H).
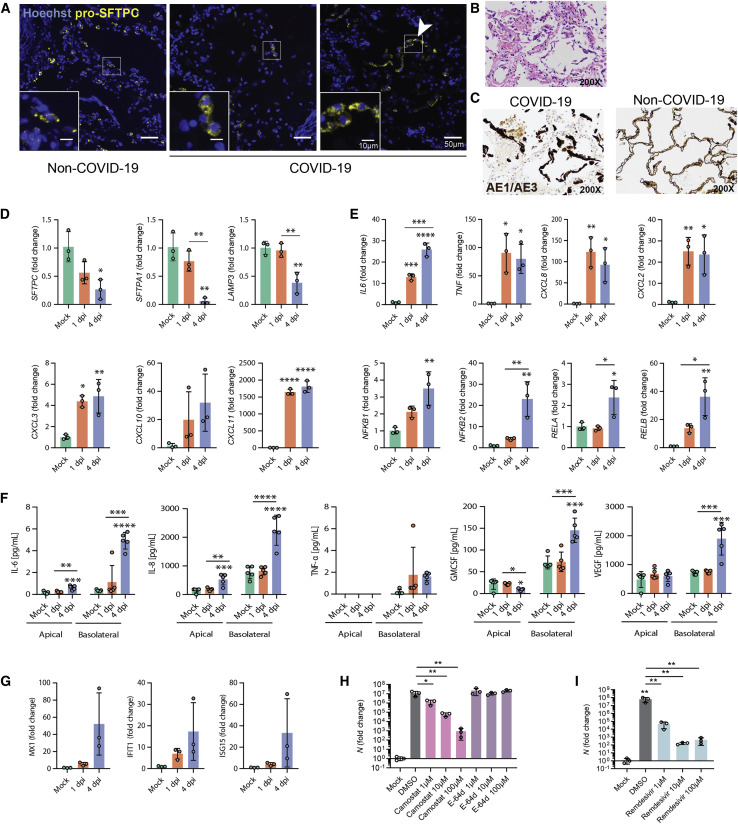
Figure 4.
Infection of iAT2s with SARS-CoV-2 Prompts the Loss of the Lung AT2 Program, Activation of the NF-κB Pathway, and Delayed Activation of IFN Signaling
(A) Immunofluorescence staining of pro-surfactant protein C (pro-SFTPC) in tissue sections of non-COVID-19 and COVID-19 lungs with zoomed insets showing the typical cytoplasmic punctate appearance of lamellar body-localized pro-SFTPC. The arrow indicates AT2 hyperplasia (right panel), a typical and non-specific response to injury, juxtaposed with regions that have a paucity of pro-SFTPC (middle panel).
(B) COVID-19 decedents’ autopsied lung tissue sections, stained with H&E.
(C) Non-COVID-19 and COVID-19 decedents’ sections stained with cytokeratin AE1/AE3 (brown) showing early acute phase of diffuse alveolar damage with sloughed alveolar epithelium (200× magnification).
(D and E) RT-qPCR of AT2 (D) and NF-κB-related (E) transcripts in iAT2s infected with SARS-CoV-2 (MOI 5; n = 3; Fold change expression over “Mock” = 2-ΔΔCt) at 1 and 4 dpi.
(F) Luminex analysis of apical washes and basolateral media collected from iAT2 ALI cultures (n = 5).
(G) RT-qPCR of interferon-stimulated genes (ISGs) in infected (MOI 5) iAT2s at 1 and 4 dpi (n = 3).
(H and I) RT-qPCR of N gene expression at 2 dpi (MOI 0.04) with (H) vehicle control, camostat (TMPRSS2 inhibitor), E-64d (cathepsin B/L inhibitor), or (I) remdesivir treatment, n = 3.
All bars represent mean ± standard deviation.∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA with multiple comparisons were performed.
Consistent with the cytopathogenicity suggested by our microscopy studies, cell death-related genes were significantly upregulated over the entire time course after infection, and stress-related signaling was evident by 4 dpi because multiple heat-shock proteins were in the top most-upregulated transcripts when comparing 4 dpi to 1 dpi (Figure 3H; e.g., HSPA1A, HSPA1B, HSPA6, and HSP90AB1). Significant downregulation of proliferation markers (TOP2A and MKI67; Figure 3H) was evident by 4 dpi, and there was a significant decrease in cell viability (Figure S1). Taken together, these results suggest that SARS-CoV-2 infection of human iAT2s results in a cell-intrinsic shift away from an AT2 program toward an inflammatory program with NF-κB-mediated inflammatory responses significantly upregulated and limited induction of interferon signaling.
To compare our findings to changes in AT2s in vivo, we performed pro-SFTPC immunostaining of lung tissue sections from the autopsies of two individuals who died from SARS-CoV-2-induced respiratory failure (clinical information provided in STARMethods). In contrast to the typical, frequent pattern of pro-SFTPC immunostaining in control lung sections, COVID-19 decedent lungs exhibited regions of reduced and sporadic pro-SFTPC staining interspersed with regions of AT2 cell hyperplasia, evident in our samples as rows of cuboidal pro-SFTPC+ alveolar cells with additional histopathologic findings of diffuse alveolar damage, such as hyaline membrane formation, as has been described in prior COVID-19 patient autopsies (Chen et al., 2020). Sloughing of cells that stained positively for cytokeratin AE1/AE3 further confirmed regional injury to the alveolar epithelium, consistent with injury observed in the iAT2 in vitro model. (Figures 4A–4C).
We validated downregulation of iAT2-specific genes by RT-qPCR and observed significantly diminished expression of SFTPC, SFTPA1, and LAMP3 at 4 dpi (Figure 4D). Moreover, we demonstrated functional activation of NF-κB signaling in infected iAT2s, as predicted by our bioinformatics analysis, by quantifying expression of NF-κB-modulated target mRNAs and proteins. Upregulation of NF-κB target transcripts IL6, CXCL8, CXCL2, CXCL3, CXCL10, and CXCL11, as well as NF-κB-related mRNA NFKB1, NFKB2, RELA, and RELB, was validated by RT-qPCR (Figure 4E). Secretion of NF-κB target proteins by infected iAT2s was determined by Luminex analysis of apical washes and basolateral media. IL-6 and IL-8 were increased both apically and basolaterally, whereas GM-CSF and VEGF were secreted into the basolateral media, as has been shown previously in other models of AT2 injury (Pham et al., 2002) (Figure 4F).
Finally, to assess the potential of iAT2s to screen for COVID-19 therapeutics that might target the alveolar epithelium, we tested the effect of a TMPRSS2 inhibitor, camostat mesylate, that was recently shown to block SARS-CoV-2 infection in Vero cells, Calu-3 cells, and human airway epithelial cells (Hoffmann et al., 2020) but has not been tested previously in human alveolar cells. Camostat significantly reduced the levels of detectable viral N transcript at 2 dpi (Figure 4H), indicating its potential as an antiviral drug and suggesting that SARS-CoV-2 infection of iAT2s relies on priming by the protease TMPRSS2, which is expressed in both iAT2s and primary adult human AT2s (Figures 1 and S1). Conversely, the cathepsin B/L inhibitor E-64d, which blocks SARS-CoV-2 infection of Vero, HeLa-ACE2, Calu-3, and 293T-ACE2 cells (Figure S4C; Hoffmann et al., 2020; Ou et al., 2020; Shang et al., 2020), had no effect in iAT2s (Figure 4H), suggesting that SARS-CoV-2 infection in the alveolar epithelium is independent of the endosomal cysteine proteases cathepsin B and L even though they are expressed in both iAT2s and primary AT2s (Figure S1). In addition, the FDA-approved broad-spectrum antiviral drug remdesivir (GS-5734) that is used to treat COVID-19 patients and was shown to inhibit SARS-CoV-2 in cell culture and mouse models (Pruijssers et al., 2020; Wang et al., 2020a) significantly reduced viral N transcript (Figure 4I). Altogether, these data highlight the importance of using a physiologically relevant cell model to study SARS-CoV-2.
Discussion
Taken together, our approach provides a new human model of SARS-CoV-2 infection of AT2s, a key lung target cell that is otherwise difficult to access in vivo and hard to maintain in vitro. Because iAT2s can be propagated indefinitely in 3D culture in a form that is easily shareable between labs (Hurley et al., 2020; Jacob et al., 2019), our adaptation of these cells to 2D ALI cultures now allows straight-forward simulations of apical viral respiratory infections of a self-renewing cell type that can be scaled up nearly inexhaustibly and studied in a highly pure form, thus simulating cell-autonomous or “epithelial-only” host responses to pathogens. Our results implicate AT2s as inflammatory signaling centers that respond to SARS-CoV-2 infection within 24 h and NF-κB signaling predominates this response; these findings are independently replicated in freshly purified adult human primary AT2 cells infected with SARS-CoV-2, as reported by Mulay et al. (2020). The significant loss of surfactant gene expression and cellular stress, toxicity, and death of iAT2s observed in our model are likely to be clinically relevant because similar observations were evident in vivo in the lung autopsies of multiple COVID-19 decedents. Others have previously demonstrated that primary adult human AT2s are infectible with SARS-CoV (SARS1) in vitro (Qian et al., 2013) and more recently have shown that AT2s are either infected in vivo with SARS-CoV-2 in COVID-19 autopsy lungs (Bradley et al., 2020; Hou et al., 2020; Schaefer et al., 2020) or can contribute to lung regeneration in COVID-19 ARDS survivors (Chen et al., 2020), further highlighting the relevance of AT2s and their study in SARS-CoV-2 infection.
Importantly, IFN responses were found to be moderate in our model: only a subset of ISGs and no canonical type I or III IFN ligands (IFNB1, IFNL1, or IFNL2) were significantly differentially expressed in SARS-CoV-2-infected samples compared to noninfected cells. SARS-CoV-2 has been shown to be sensitive to IFNλ and IFNβ treatment, so the absence of a robust IFN response in AT2s, if verified in vivo, would have significant clinical implications and might suggest pathways to augment therapeutically in COVID-19 patients before they progress to ARDS (Blanco-Melo et al., 2020; Broggi et al., 2020; Clementi et al., 2020). Indeed, some clinical studies suggest a beneficial effect of early type I IFN treatment on COVID-19 progression (Wang et al., 2020b). In prior studies of cell lines that lacked AT2-specific gene expression, the absence of a robust IFN response either was overcome by using a high MOI for infection or IFN induction occurred at late time points after infection when using a low MOI (Lei et al., 2020). This is in contrast to our results that indicate a moderate and delayed (4 dpi) IFN response at low and high MOIs (MOI 0.4 and MOI 5). Interestingly, a delayed IFN response was also observed in SARS-CoV- and MERS-CoV-infected human airway epithelial cells and is a determinant of SARS disease severity (Channappanavar et al., 2016; Menachery et al., 2014).
Recent studies have shown that host cell factors, such as ACE2, TMPRSS2, cathepsins, and furin, are key to SARS-CoV-2 entry and, thus, may be potential therapeutic targets (Hoffmann et al., 2020; Ou et al., 2020). We found that TMPRSS2 is used for viral entry in our system, whereas cathepsins are not. In the iAT2 model, ACE2 is heterogeneously expressed (Figure 1), concordant with the level of heterogeneity in adult human AT2s in vivo (Abo et al., 2020). Although the frequency of ACE2 expression in our model system at the time of infection is comparable to the initial frequency of infection of iAT2s, this does not necessarily prove that initial iAT2 infection is ACE2 dependent, and proving this requires further study potentially with ACE2 knockout iPSC lines.
An important caveat of our study is the well-published observation that most human lineages derived in vitro from iPSCs are immature or fetal in phenotype, possibly confounding disease modeling. In addition, the robust self-renewing capabilities of iAT2s also distinguish them from primary AT2s that proliferate poorly ex vivo, suggesting potential differences in how they may respond to SARS-CoV-2. However, our iAT2s show expression of maturation genes, including surfactant proteins (Figure 1D, [Hurley et al., 2020]). Furthermore, iAT2s after adaptation to ALI culture augment expression of AT2 maturation markers and are less proliferative than those in 3D culture and are thus more similar to primary AT2s (Figure S1; Abo et al., 2020). This, together with the observation of SARS-CoV-2 virions intracellularly in lamellar bodies and extracellularly in the vicinity of tubular myelin, confirms that surfactant-secreting, functionally mature AT2-like cells were the targets of infection in our studies. The presence of virions within lamellar bodies also implies that this surfactant-packaging organelle, specific to mature AT2 cells within the lung epithelium and absent in lung cell lines, may be a site directly utilized for and potentially dysregulated by SARS-CoV-2 infection. Thus, our model system reveals the cell-intrinsic responses of a key lung target cell to infection, facilitating a deeper understanding of COVID-19 pathogenesis and providing a platform for drug discovery.
Limitations of the Study
A limitation of our study is the observation that iAT2s in our culture conditions do not generate alveolar type 1 (AT1)-like cells, an ability that is similarly lacking in other published reports of in vitro cultures of both iPSC-derived and primary human AT2s to date (Barkauskas et al., 2013). Although the basis for this divergence from mouse AT2 cell culture behavior remains unclear, it most likely either reflects differences between human and mouse alveolar epithelial systems or missing factors in human in vitro culture conditions that are present in vivo in the AT2 niche, shortcomings that could potentially be addressed with future cell culture modifications. Regardless, our findings should not detract from published observations that SARS-CoV-2 infection has also been observed in AT1 cells in COVID-19 autopsy specimens (Bradley et al., 2020) in addition to several airway epithelial cell types. Additionally, because we do not have mesenchymal support cells present in our cell cultures, we ensure that epithelial-intrinsic responses can be studied without any confounding effects that can arise in culture systems that depend on mesenchymal cell co-cultures. Although there are differences between our in vitro model and other ex vivo or in vivo models, human in vitro models recapitulating infections of those lineages or introducing further complexity by adding immune lineages are likely to similarly advance our understanding of COVID-19 disease pathogenesis in concert with our iAT2 model.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal to CD26, PE conjugated (clone BA5b) | Biolegend | Cat# 302705; RRID: AB_314289 |
| Mouse monoclonal to CD47, PerCP-Cy5.5 conjugated (clone CC2C6) | Biolegend | Cat# 323110; RRID: AB_940461 |
| Anti-SARS-CoV nucleoprotein (N) | Rockland Immunochemicals | Cat# 200-401-A50; RRID: AB_828403 |
| AlexaFluor 488 AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat# 711-545-152; RRID: AB_2313584 |
| Anti-ACE2 primary antibody | R&D Systems | Cat# AF933; RRID: AB_355722 |
| AlexaFluor 647 Donkey Anti-Goat IgG (H+L) | Invitrogen | Cat# A-21447; RRID: AB_2535864 |
| Anti-AE1/AE3 antibody | Ventana | Cat# 760-2135; RRID: AB_2810237 |
| Anti-pro-SFTPC antibody | Santa Cruz | Cat# sc-518029 |
| AlexaFluor 647 Donkey Anti-Mouse IgG (H+L) | Invitrogen | Cat# A32787; RRID: AB_2762830 |
| Hoechst 33342 | Life Technologies | Cat# H3570 |
| ProLong Diamond Antifade Mountant | Invitrogen | Cat# P36965 |
| Virus Strains | ||
| SARS-CoV-2 USA_WA1/2020 | Kindly provided by CDC’s Principal Investigator Natalie Thornburg and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Growth Factor Reduced Matrigel | Corning | Cat# 356230 |
| SB431542 | Tocris | Cat# 1614 |
| Dorsomorphin | Stemgent | Cat# 04-0024 |
| CHIR99021 | Tocris | Cat# 4423 |
| Recombinant human FGF10 | R&D Systems | Cat# 345-FG-025 |
| Recombinant human KGF | R&D Systems | Cat# 251-KG-010 |
| Recombinant human BMP4 | R&D Systems | Cat# 314-B |
| Retinoic acid | Sigma | Cat# R2625 |
| Y-27632 dihydrochloride | Tocris | Cat# 1254 |
| Dexamethasone | Sigma | Cat# D4902 |
| 8-bromoadenosine 30,50-cyclic monophosphate sodium salt (cAMP) | Sigma | Cat# B7880 |
| 3-Isobutyl-1-methylxanthine (IBMX) | Sigma | Cat# I5879 |
| 0.05% trypsin-EDTA | Invitrogen | Cat# 25300-120 |
| Defined Fetal Bovine Serum | Thermo Fisher | Cat# NC0652331 |
| Calcein blue | Life Technologies | Cat# C1429 |
| Dispase | Thermo Fisher | Cat# 354235 |
| Recombinant human IFNβ | PeproTech | Cat# 300-02BC |
| Poly(I:C) | InvivoGen | Cat# TLRL-PIC |
| Oligofectamine Transfection Reagent | Invitrogen | Cat# 12252011 |
| Camostat mesylate | Tocris | Cat# 59721-29-5 |
| E-64d | Selleck Chem | Cat# S7393 |
| Remdesivir | Selleck Chem | Cat# S8932 |
| Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| QIAzol Lysis Reagent QIAGEN | QIAGEN | Cat# 79306 |
| TaqMan Fast Universal PCR Master Mix (2X) | Thermo Fisher | Cat# 4364103 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| Magnetic Luminex® Performance Assay Human HighSensitivity Cytokine Base Kit A | R&D Systems | Cat# LHSCM000 |
| Deposited Data | ||
| Bulk RNA-seq series | This paper | GEO: GSE153277, https://crem.shinyapps.io/iAEC2infection/ https://dx.doi.org/10.17632/p58k2bd6t5.1 |
| Experimental Models: Cell Lines | ||
| Human: Normal donor iPSC line targeted with NKX2-1-GFP SFTPC-tdTomato (BU3 NGST) | Kotton Lab (Jacob et al., 2017) | http://stemcellbank.bu.edu |
| Human: Donor iPSC line targeted with SFTPC-tdTomato (SPC2) | Kotton Lab (Hurley et al., 2020) | http://stemcellbank.bu.edu |
| Human: RUES2 ST embryonic stem cell line targeted with SFTPC-tdTomato | Original line is a gift from Dr. Ali H. Brivanlou, Rockefeller University; modified to include SFTPCtdTomato by the Kotton Lab (Jacob et al., 2017) | http://stemcellbank.bu.edu |
| Oligonucleotides | ||
| TaqMan Gene Expression Assay Primer/Probe Sets | N/A | N/A |
| CXCL2 | Thermo Fisher | Hs00601975_m1 |
| CXCL3 | Thermo Fisher | Hs00171061_m1 |
| CXCL8 | Thermo Fisher | Hs00174103_m1 |
| CXCL10 | Thermo Fisher | Hs00171042_m1 |
| CXCL11 | Thermo Fisher | Hs00171138_m1 |
| IFNB1 | Thermo Fisher | Hs01077958_s1 |
| IFNL1 (IL29) | Thermo Fisher | Hs00601677_g1 |
| IFNL2 (IL28A) | Thermo Fisher | Hs00820125_g1 |
| IL6 | Thermo Fisher | Hs00174131_m1 |
| IFIT1 | Thermo Fisher | Hs00356631_g1 |
| ISG15 | Thermo Fisher | Hs00192713_m1 |
| LAMP3 | Thermo Fisher | Hs01111316_m1 |
| MX1 | Thermo Fisher | Hs00895608_m1 |
| NFKB1 | Thermo Fisher | Hs00765730_m1 |
| NFKB2 | Thermo Fisher | Hs01028890_g1 |
| RELA | Thermo Fisher | Hs00153294_m1 |
| RELB | Thermo Fisher | Hs00232399_m1 |
| SARS-CoV-2 N | IDT | 10006606 |
| SFTPC | Thermo Fisher | Hs00161628_m1 |
| SFTPA1 | Thermo Fisher | Hs00831305_s1 |
| TNF | Thermo Fisher | Hs00174128_m1 |
| 18S | Thermo Fisher | 4318839 |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Prism | GraphPad | https://www.graphpad.com/ |
| Other | ||
| StemDiff Definitive Endoderm Kit | StemCell Technologies | Cat# 05110 |
| mTeSR1 | StemCell Technologies | Cat# 05850 |
| Glutamax | Life Technologies | Cat# 35050-061 |
| Gentle Cell Dissociation Reagent | StemCell Technologies | Cat# 07174 |
| Accutase | Sigma | Cat# A6964 |
| 6.5mm Transwell with 0.4 μm Pore Polyester Membrane Inserts, Sterile | Corning | Cat# 3470 |
Resource Availability
Lead Contact
Further information and requests for other reagents may be directed to, and will be fulfilled by, the Lead Contact, Darrell Kotton (dkotton@bu.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. Pluripotent stem cell lines used in this study are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://www.bu.edu/dbin/stemcells/.
Data and Code Availability
The accession number for the RNA-seq data reported in this paper is GEO: GSE153277. Interactive time series visualization of gene expression available at: https://crem.shinyapps.io/iAEC2infection/. Original data have been deposited to Mendeley Data: https://dx.doi.org/10.17632/p58k2bd6t5.1.
Experimental Model Details
iPSC lines and maintenance
All experiments involving the differentiation of human pluripotent stem cell (PSC) lines were performed with the approval of the Institutional Review Board of Boston University (protocol H33122). The SPC2 induced pluripotent stem cell (iPSC) line carrying a SFTPC-tdTomato reporter and the BU3 iPSC line carrying NKX2-1-GFP and SFTPC-tdTomato reporters were obtained from our previous studies (Hawkins et al., 2017, Jacob et al., 2017, Hurley et al., 2020). The human embryonic stem cell line RUES2 was a kind gift from Dr. Ali H. Brivanlou of The Rockefeller University and we previously targeted this line with a SFTPC-tdTomato reporter (Jacob et al., 2017). All PSC lines used in this study displayed a normal karyotype when analyzed by G-banding (Cell Line Genetics). All PSC lines were maintained in feeder-free conditions, on growth factor reduced Matrigel (Corning) in 6-well tissue culture dishes (Corning), in mTeSR1 medium (StemCell Technologies) using Gentle Cell Dissociation Reagent for passaging. Further details of iPSC derivation, characterization, and culture are available for free download at http://www.bu.edu/dbin/stemcells/protocols.php.
Method Details
iPSC directed differentiation into alveolar epithelial type 2 cells (iAT2s) and air-liquid interface (ALI) culture
Human iPSC lines, clones SPC-ST-B2 and BU3 NGST, underwent directed differentiation to generate iPSC-derived alveolar epithelial type II like cells (iAT2s) in 3D Matrigel cultures using methods we have previously published (Jacob et al., 2019). As previously described (Jacob et al., 2019), we performed PSC directed differentiation via definitive endoderm into NKX2-1 lung progenitors as follows. In short, cells maintained in mTeSR1 media were differentiated into definitive endoderm using the STEMdiff Definitive Endoderm Kit (StemCell Technologies) and after the endoderm-induction stage, cells were dissociated with Gentle Cell Dissociation Reagent (GCDR) and passaged into 6 well plates pre-coated with growth factor reduced Matrigel in ‘‘DS/SB’’ anteriorization media, consisting of complete serum-free differentiation medium (cSFDM) base as previously described (Jacob et al., 2017) supplemented with 10 μM SB431542 (‘‘SB’’; Tocris) and 2 μM Dorsomorphin (‘‘DS’’; Stemgent). For the first 24 h after passaging, 10 μM Y-27632 was added to the media. After anteriorization in DS/SB media for 3 days (72 h), cells were cultured in ‘‘CBRa’’ lung progenitor-induction media for 9-11 days. ‘‘CBRa’’ media consists of cSFDM containing 3 μM CHIR99021 (Tocris), 10 ng/mL recombinant human BMP4 (rhBMP4, R&D Systems), and 100 nM retinoic acid (RA, Sigma), as previously described (Jacob et al., 2017). On day 15 of differentiation, live cells were sorted on a high-speed cell sorted (MoFlo Legacy or MoFlo Astrios EQ) to isolate NKX2-1+ lung progenitors based on CD47hi/CD26neg gating (Hawkins et al., 2017). Sorted day 15 progenitors were then resuspended in undiluted growth factor-reduced Matrigel (Corning) and distal/alveolar differentiation of cells was performed in ‘‘CK+DCI’’ medium, consisting of cSFDM base, with 3 μm CHIR99021, 10 ng/mL rhKGF, and 50 nM dexamethasone (Sigma), 0.1 mM 8-Bromoadenosine 30,50-cyclic monophosphate sodium salt (Sigma) and 0.1 mM 3-Isobutyl-1-methylxanthine (IBMX; Sigma) (DCI) with a brief period of CHIR99021 withdrawal on days 31-35 to achieve iAT2 maturation. To establish pure cultures of iAT2s, cells were sorted by flow cytometry to isolate SFTPCtdTomato+ cells on days 41 and 69 of differentiation. iAT2s were then maintained through serial passaging as self-renewing monolayered epithelial spheres (“alveolospheres”) by plating in Matrigel (Corning) droplets at a density of 400 cells/μl with refeeding every other day in CK+DCI medium according to our previously published protocol (Jacob et al., 2019). iAT2 culture quality and purity was monitored at each passage by flow cytometry, with > 80% of cells expressing SFTPCtdTomato over time, as we have previously detailed (Jacob et al., 2019, Hurley et al., 2020).
To establish air-liquid interface (ALI) cultures, single cell suspensions of iAT2s were prepared as we have recently detailed (Abo et al., 2020). Briefly, Matrigel droplets containing iAT2s as 3D sphere cultures were dissolved in 2 mg/mL dispase (Sigma) and alveolospheres were dissociated in 0.05% trypsin (GIBCO) to generate a single-cell suspension. 6.5mm Transwell inserts (Corning) were coated with dilute Matrigel (Corning) according to the manufacturer’s instructions. Single-cell iAT2s were plated on Transwells at a density of 520,000 live cells/cm2 in 100 μL of CK+DCI with 10 μM Rho-associated kinase inhibitor (“Y”; Sigma Y-27632). 600 μL of CK+DCI+Y was added to the basolateral compartment. 24 h after plating, basolateral media was refreshed to CK+DCI+Y. 48 h after plating, apical media was aspirated to initiate air-liquid interface culture. 72 h after plating, basolateral media was changed to CK+DCI to remove the rho-associated kinase inhibitor. Basolateral media was changed 3 times per week thereafter.
SARS-CoV-2 propagation and titration
SARS-CoV-2 stocks (isolate USA_WA1/2020, kindly provided by CDC’s Principal Investigator Natalie Thornburg and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA)) were grown in Vero E6 cells (ATCC CRL-1586) cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2% fetal calf serum (FCS), penicillin (50 U/mL), and streptomycin (50 mg/mL). To remove confounding cytokines and other factors, viral stocks were purified by ultracentrifugation through a 20% sucrose cushion at 80,000xg for 2 h at 4°C (25). SARS-CoV-2 titer was determined in Vero E6 cells by tissue culture infectious dose 50 (TCID50) assay. All work with SARS-CoV-2 was performed in the biosafety level 4 (BSL4) facility of the National Emerging Infectious Diseases Laboratories at Boston University, Boston, MA following approved SOPs.
SARS-CoV-2 infection of iAT2s
iAT2s plated in ALI culture were infected with purified SARS-CoV-2 stock at the indicated multiplicity of infection (MOI). 100 μL inoculum was prepared in CK+DCI media (or mock-infected with medium-only). Inoculum was added to the apical chamber of each Transwell and incubated for 1 h at 37°C and 5% CO2. After the adsorption period, the inoculum was removed and cells were incubated at 37°C for 1 or 4 days. At the time of harvest, basolateral media was collected for further analysis and apical washes were performed by adding 100 μL CK+DCI media to the apical chamber, incubated for 15 min at room temperature before collection for further analysis. Both the apical washes and basolateral media were used for viral titration and Luminex assays as described below. Infected and mock-infected iAT2s were fixed in 10% formalin and used for immunofluorescence analysis or electron microscopy as described below. For flow cytometry, infected and mock-infected iAT2 cells were first detached by adding 0.2 mL Accutase (A6964, Sigma) apically and incubated at room temperature for 15 min. Detached cells were pelleted by low speed centrifugation, resuspended in 10% formalin, and used for flow cytometry as described below. Cells were lysed in TRIzol for RNA-seq and RT-qPCR analysis.
To infect iAT2 alveolospheres in 3D culture, cells were dissolved in 2 mg/mL dispase (Sigma) for 1 h at 37°C, and alveolospheres were mechanically dissociated. Cells were washed with PBS and resuspended in inoculum (MOI 0.4) for 1 h at 37°C. Cells were subsequently replated in matrigel droplets and collected at 2 dpi in TRIzol for RT-qPCR analysis.
For determining cell viability, infected and mock-infected iAT2s at ALI were first detached by adding 0.2 mL Accutase apically and incubated at room temperature for 15 min. Detached cells were pelleted by low speed centrifugation, resuspended in PBS, diluted 1:1 in trypan blue, and counted using a LUNA-II Automated Cell Counter (Logos Biosystems).
Immunofluorescence microscopy of iAT2s
For nucleoprotein immunofluorescence, infected or control iAT2s on Transwell inserts were fixed in 10% formalin for 6 h, washed twice in PBS (10 min, room temperature), permeabilized with PBS containing 0.25% Triton X-100 and 2.5% normal donkey serum (30 min, room temperature), and blocked with PBS containing 2.5% normal donkey serum (20 min, room temperature). Subsequently, cells were incubated with primary antibody diluted in anti-SARS-CoV nucleoprotein (N) antibody (rabbit polyclonal, 1:2500, Rockland Immunochemicals, Cat #200-401-A50) diluted in 4% normal donkey serum overnight at 4°C. This antibody cross-reacts with the SARS-CoV-2 nucleoprotein (Thi Nhu Thao et al., 2020). Next, cells were washed with PBS three times (5 min, room temperature), and incubated with secondary antibody (AlexaFluor 488 AffiniPure Donkey Anti-Rabbit IgG (H+L), 1:500, Jackson ImmunoResearch 711-545-252) for 2 h at room temperature. Cells were washed with PBS three times (5 min, room temperature), incubated with Hoechst (1:500, Life Technologies) for 30 min, and washed again. Transwell inserts were then cut out with a scalpel and mounted with Prolong Diamond Mounting Reagent (Life Technologies). Slides were imaged with an upright fluorescence microscope (Nikon Eclipse Ni-E) and a confocal microscope (Zeiss LSM 700).
For ACE2 immunofluorescence, never-infected iAT2s at ALI were fixed for 10 min at RT in 4% PFA. After fixation, the same protocol was followed as above, using an anti-ACE2 primary antibody (R&D, AF933, 1:100) or pre-immune serum for overnight incubation, and an appropriate secondary (AlexaFluor 647 Donkey Anti-Goat IgG (H+L), 1:500, Invitrogen A21447).
Pseudotyped lentiviral entry experiments
Pseudotyped particles carrying the pHAGE-EF1αL-GFP lentiviral vector were packaged using a 5-plasmid transfection protocol (27). In brief, 293T cells were transfected using Trans-IT (Mirus) with a plasmid carrying the lentiviral backbone (pHAGE-EF1αL-GFP; plasmid map downloadable from www.kottonlab.com), 3 helper plasmids encoding Rev, tat, and gag/pol genes, in addition to plasmids encoding either the VSV-G or the SARS-CoV-2 Spike envelope (plasmid HDM-IDTSpike-FixK, cloned by Jesse Bloom and a kind gift from Alex Balazs (Crawford et al., 2020)). Supernatants carrying packaged lentivirus were collected at 48, 60 and 72 h and then concentrated by ultracentrifugation. Lentiviral titers were determined by infecting FG293 with VSV-G pseudotype or 293T cells overexpressing ACE2 (created by Michael Farzan and a kind gift from Alex Balazs (Crawford et al., 2020; Mou et al., 2020)) with Spike pseudotype and then quantifying GFP+ cells by flow cytometry. To transduce iAT2s growing in ALI cultures, VSV-G (MOI 50) or Spike (MOI 30) pseudotyped lentiviruses, diluted in CK+DCI medium supplemented with Polybrene (5 μg/mL; EMD Millipore), were applied to the apical surface for 4 h. GFP expression was assessed 48-72 h after transduction by microscopy (Keyence) or flow cytometry.
Flow cytometry
For post infection flow cytometry, fixed iAT2s were either stained for cell surface expression of ACE2 (R&D, #AF933, 4-8 μg/2.5x106 cells) followed by donkey anti-goat IgG-AF647 (Invitrogen, #A21447) or were permeabilized with saponin buffer (Biolegend) then stained with SARS-CoV nucleoprotein (N) antibody (Rockland, #200-401-A50, 1:1000), followed by donkey anti-rabbit IgG-AF488 (Jackson ImmunoResearch, #711-545-152). Gating was based on either mock infected stained controls or infected, isotype-stained controls. Flow cytometry staining was quantified using a Stratedigm S1000EXI and analyzed with FlowJo v10.6.2 (FlowJo, Tree Star Inc). FACS plots shown represent single-cells based on forward-scatter/side-scatter gating.
Reverse Transcription Quantitative PCR (RT-qPCR)
iAT2 cells were collected in Qiazol (QIAGEN) or TRIzol (ThermoFisher) then RNA was extracted using the RNAeasy mini kit (QIAGEN) or following the manufacturer’s protocol, respectively. Complementary DNA (cDNA) was generated by reverse transcription using MultiScribe Reverse Transcriptase (Applied Biosystems). PCR was then run for 40 cycles using an Applied Biosystems QuantStudio 384-well system. Predesigned TaqMan probes were from Applied Biosystems or IDT (see Key Resources Table). Relative gene expression was calculated based on the average Ct value for technical triplicates, normalized to 18S control, and fold change over mock-infected cells was calculated using 2-ΔΔCt. If transcripts were undetected, they were assigned a Ct value of 40 to allow for fold change calculations, and biological replicates, as indicated in each figure legend, were run for statistical analyses.
Transmission electron microscopy
iAT2 ALI cultures on Transwell inserts were infected with SARS-CoV-2 at an MOI of 5 or mock-infected. At 1 dpi, cells were fixed and inactivated in 10% formalin for 6 h at 4°C and removed from the BSL-4 laboratory. The cells were washed with PBS and then post-fixed in 1.5% osmium tetroxide (Polysciences) overnight at 4°C. The membrane was excised from the insert, block stained in 1.5% uranyl acetate (Electron Microscopy Sciences, EMS) for 1 h at room temperature (RT). The samples were dehydrated quickly through acetone on ice, from 70% to 80% to 90%. The samples were then incubated 2 times in 100% acetone at RT for 10 min each, and in propylene oxide at RT for 15 min each. Finally, the samples were changed into EMbed 812 (EMS), left for 2 h at RT, changed into fresh EMbed 812 and left overnight at RT, after which they were embedded in fresh EMbed 812 and polymerized overnight at 60°C. Embedded samples were thin sectioned (70 nm) and grids were stained in 4% aqueous uranyl acetate for 10 min at RT followed by lead citrate for 10 min at RT. Electron microscopy was performed on a Philips CM12 EM operated at 100kV, and images were recorded on a TVIPS F216 CMOS camera with a pixel size of 0.85-3.80 nm per pixel.
Drug efficacy testing in iAT2 and Vero E6 cells
iAT2s plated in ALI culture were pre-treated apically (100 μL) and basolaterally (600 μL) with the indicated concentrations of camostat mesylate (Tocris, #59721-29-5), E-64d (Selleckchem, #S7393), remdesivir (Selleckchem, #S8932), or DMSO control for 30 min at 37°C. Following pre-treatment, all apical media were aspirated and SARS-CoV-2 (MOI 0.04) was added for 1 h without any drugs apically, after which the inoculum was removed. iAT2s were exposed to the compounds basolaterally for the entire duration of the experiment. Cells were harvested in TRIzol after 2 dpi and processed for RT-qPCR.
Vero E6 cells were treated with the indicated concentrations of camostat mesylate, E-64d, remdesivir, or DMSO control for 30 min at 37°C. After 30 min, SARS-CoV-2 (MOI 0.1) was added. Cells were harvested in TRIzol after 2 dpi and processed for RT-qPCR.
For immune stimulation treatments, iAT2s at ALI were treated with poly(I:C) delivered with Oligofectamine Transfection Reagent (Invitrogen) or treated with recombinant human IFNβ (rhIFNβ). Prior to treatment, 10 μL poly(I:C) was mixed with 10 μL Oligofectamine and incubated at RT for 15 min. After the incubation period, the poly(I:C) and Oligofectamine mixture was added to 80 μL of CK+DCI media, for a total of 100 μL per well. iAT2s at ALI were treated apically (100 μL) with poly(I:C) (10 μg/mL) and Oligofectamine, or apically and basolaterally (600 μL) with IFNβ (10 ng/mL) for 24 h at 37°C. Cells were subsequently harvested and processed for RT-qPCR.
Luminex analysis
Apical washes and basolateral media samples were clarified by centrifugation and analyzed using the Magnetic Luminex® Performance Assay Human High Sensitivity Cytokine Base Kit A (R&D Systems, Inc). Apical washes were diluted 1:2, basolateral media was undiluted, except to detect IL-8 and VEGF, where it was diluted 1:10. Limit of detection: GM-CSF = 0.13 pg/mL, IL-6 = 0.31 pg/mL, IL-8 = 0.07 pg/mL, TNF-α = 0.54 pg/mL and VEGF = 1.35 pg/mL. Mean fluorescence intensity was measured to calculate final concentration in pg/mL using Bioplex200 and Bioplex Manager 5 software (Biorad).
RNA sequencing and bioinformatic analyses
For bulk RNA sequencing (RNA-Seq), biological triplicate (n = 3) samples of purified RNA extracts were harvested from each group of samples prepared as follows. After 208 days of total time in culture, iAT2s cultured as serially passaged 3D spheres were single-cell passaged onto Transwell inserts. Apical media was removed on day 210 to initiate air-liquid interface (ALI) culture. On day 218, 6 replicate wells of iAT2s were exposed to SARS-CoV-2 in an apical inoculum and 3 replicate wells were exposed to mock. On day 219, 3 mock and 3 post-infection samples (1 dpi) were collected. Three additional post-infection samples (4 dpi) were collected on day 222. mRNA was isolated from each sample using magnetic bead-based poly(A) selection, followed by synthesis of cDNA fragments. The products were end-paired and PCR-amplified to create each final cDNA library. Sequencing of pooled libraries was done using a NextSeq 500 (Illumina). The quality of the raw sequencing data was assessed using FastQC v.0.11.7. Sequence reads were aligned to a combination of the human and SARS-CoV-2 genome reference (GRCh38 and Wuhan Hu-1 isolate) and the TdTomato reporter sequence, using STAR v.2.5.2b (Dobin et al., 2013). Counts per gene were summarized using the featureCounts function from the subread package v.1.6.2. The edgeR package v.3.25.10 was used to import, organize, filter and normalize the data. Genes that were not expressed in at least one of the experimental groups were filtered out (keeping only genes that had at least 10 reads in at least 3 libraries, that is, a worthwhile number of samples as determined by the replicate number in the design matrix). The TMM method was used for normalization. Principal Component Analysis (PCA) and Multidimensional Scaling (MDS) were used for exploratory analysis, to assess sample similarities and potential batch effects. Subsequently, the limma package v3.39.19 (Law et al., 2016) with its voom method, namely, linear modeling and empirical Bayes moderation was used to test differential expression (moderated t tests). P values were adjusted for multiple hypothesis testing using Benjamini-Hochberg correction (false discovery rate-adjusted p value; FDR). Differentially expressed genes between the groups in each experiment were visualized using Glimma v1.11.1, and FDR < 0.05 was set as the threshold for determining significant differential gene expression. Gene set analysis was performed with Hallmark gene sets using the Camera package (Wu and Smyth, 2012). For the comparisons between different infection models (Figure S3), we put our analyses above in context with other publicly available datasets (Riva et al., 2020, Blanco-Melo et al., 2020). Specifically, for the SARS-CoV-2 infections (versus mock-treated samples) of normal human bronchial epithelial cells (NHBE), A549 with forced ACE2 overexpression (A549-ACE2) and Calu-3, we used Series 1, Series 16 and Series 7 respectively from the GEO dataset (GEO: GSE147507) mapped to the human genome reference (GRCh38). For the Vero E6 infection model, we used the samples from GSE153940 mapped to the African Green Monkey (Chlorecebus sabaeus) genome reference (Ensembl, ChlSab1.1). For these contrasts, log-fold-change ranked gene lists were generated using the same procedure described above. We then used FGSEA (v.1.9.7, (Korotkevich et al., 2019)) to test for enrichment in lung-related Gene Ontology sets in the pre-ranked gene lists. The inclusion criteria for gene sets to test was: any GO in the Molecular Signature Database (MSigDB, https://www.gsea-msigdb.org/gsea/msigdb) with any of the following terms in its name: “LUNG,” “SURFACTANT,” “ALVEO,” “LAMEL,” “MULTIVESICULAR.” Normalized enrichment scores (NES) for tests that resulted in FDR < 0.2 were then plotted in a heatmap (Figure S3).
Human COVID-19 autopsy specimens and lung tissue sections
This study was reviewed by the IRB of Boston University and found not to constitute human subjects research. With consent from next-of-kin, human lung tissues from decedents with COVID-19 were collected at the time of autopsies performed at Boston Medical Center. Samples from individuals with shorter duration between symptom onset and death were prioritized for analysis. Samples were fixed in formalin and embedded in paraffin. Healthy tissue adjacent to a lung tumor resected prior to the emergence of COVID-19 was collected with IRB approval under protocol H-37859 and utilized as a control.
| Patient | Age | Sex | Symptom onset to death (Days) | Hospitalization to death (Days) | O2 requirement at death |
|---|---|---|---|---|---|
| COVID 1 | 91 | M | 8 | 7 | 7L nasal cannula |
| COVID 2 | 72 | M | 9 | 4 | 15L nasal cannula |
Immunohistochemistry on autopsy lung tissue was performed using freshly cut 5 μm thick FFPE tissue sections and stained with AE1/AE3 (Ventana, 760-2135) on a Ventana Benchmark Ultra (Ventana Medical Systems, Tucson, AZ, USA) after mild protease 3 (Ventana, 760-2020) digestion and heat induced epitope retrieval with alkaline CC1 buffer (Ventana, 950-124). Slides were visualized with DAB using Optiview detection (Ventana, 760-700). Parallel sections stained with H&E and AE1/AE3 were performed to identify sloughed pneumocytes from admixed inflammatory cells. Immunofluorescent staining was performed on additional FFPE sections from each patient. After deparaffinization in xylene, hydration, and antigen retrieval in citrate-based unmasking solution (Vector, H-3300), sections were blocked and permeabilized in 4% normal donkey serum and 0.1% Triton X-100 (Sigma) for 1 h and incubated overnight with anti-pro-SFTPC antibody (Santa Cruz, sc-518029) in blocking solution. Sections were washed with 0.1% Triton X-100 in PBS and incubated with secondary antibody (anti-mouse IgG AlexaFluor647, Invitrogen A32787, 1:500) for 2 h at RT. Nuclei were counterstained with Hoechst and sections were mounted with Prolong Diamond Anti-Fade Mounting Reagent (ThermoFisher) and coverslipped. Immunofluorescent imaging was performed on a Zeiss LSM 700 confocal microscope.
Quantification and Statistical Analysis
Statistical methods relevant to each figure are outlined in the figure legend. In short, unpaired, two-tailed Student’s t tests were used to compare quantitative analyses comprising two groups of n = 3 or more samples, or one-way ANOVAs with multiple comparisons were used to compare three or more groups. Further specifics about the replicates used in each experiment are available in the figure legends. In these cases, a Gaussian distribution and equal variance between samples was assumed as the experiments represent random samples of the measured variable. The p value threshold to determine significance was set at p = 0.05. p value annotations on graphs are as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Data for quantitative experiments is typically represented as the mean with error bars representing the standard deviation or standard error of the mean, as specified in the figure legends.
Acknowledgments
The authors wish to thank all members of the Boston University COVID-19 affinity research collaborative (ARC) as well as the Wilson, Hawkins, Mühlberger, Saeed, and Kotton Labs for helpful discussions. We thank Olivia Hix, Michael Herriges, Andrea Alber, Nora Lee, Marall Vedaie, Mitchell White, and Baylee Heiden for excellent technical assistance and histologic scoring; George Murphy for donation of the N RT-qPCR probe; Alejandro Balazs for donation of the spike pseudotype plasmid and guidance; Michael Farzan, Huihui Mou, and Alejandro Balazs for donation of the 293T-ACE2 cell line; CDC’s Principal Investigator Natalie Thornburg and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) for donation of the SARS-CoV-2 USA_WA1/2020; Suryaram Gummuluru for donation of camostat and E-64d; Vickery Trinkaus-Randall for help with confocal microscopy; Yuriy Aleksyeyev for RNA sequencing; Brian Tilton for cell sorting; and Ronald Corley, Katya Ravid, and Jay Mizgerd for critical review of results. The graphical abstract was created with BioRender. This work was supported by Evergrande MassCPR awards to D.N.K., E.M., and M.S.; an NHLBI Progenitor Cell Translational Consortium (PCTC) Jump Start Award to J.H.; NIH grant F30HL147426 to K.M.A.; a CJ Martin Early Career Fellowship from the Australian National Health and Medical Research Council to R.B.W.; the I.M. Rosenzweig Junior Investigator Award from The Pulmonary Fibrosis Foundation to K.D.A.; a Harry Shwachman Cystic Fibrosis Clinical Investigator Award, Gilead Research Scholars, Gilda and Alfred Slifka and Gail and Adam Slifka funds, and CFMS fund to R.W.; a Cystic Fibrosis Foundation (CFF) grant (HAWKIN19XX0), Fast Grants award to E.M.; NIH grants U01HL148692 and R01HL139799 to F.H.; the C3.ai Digital Transformation Institute Award and NIH grants U01HL148692, U01HL134745, U01HL134766, and R01HL095993 to D.N.K.; NIH grants U01TR001810, UL1TR001430, R01DK101501, and R01DK117940 to A.A.W.; UL1TR001430 to M.B.; and R21AI135912 to E.M. iPSC distribution and disease modeling is supported by NIH grants U01TR001810 and N01 75N92020C00005. This work was partially supported by the Evans Center for Interdisciplinary Biomedical Research ARC on “Respiratory Viruses: A Focus on COVID-19” at Boston University.
Author Contributions
D.N.K., E.M., and A.A.W. conceptualized the study; J.H., A.J.H., K.M.A., and R.B.W. designed experiments; J.H., A.J.H., K.M.A., R.B.W., J.O., E.L.S., and A.S. performed experiments and analyzed data; C.V.-M. and J.L.-V. performed bioinformatics analysis; E.B., A.H., and E.J.B. contributed to sample preparation; D.N.K. prepared the original draft; and D.N.K., E.M., A.A.W., J.H., A.J.H., K.M.A., R.B.W., K.-D.A., M.L.B., C.S.-R., M.S., and M.B. reviewed and edited the manuscript. All authors contributed to the interpretation of the results.
Declaration of Interests
The authors declare no competing interests.
Published: September 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2020.09.013.
Supplemental Information
Listed are names and statistics (fold-change and false-discovery rate-adjusted values; FDR) for all genes tested through bulk RNA-seq analysis of iAT2s infected with SARS-CoV-2 (MOI 5) at 1 and 4 dpi, along with mock-infected cells at 1 day. Biostatistical comparisons were performed between 1 dpi and mock-infected, 4 dpi and 1 dpi, and between all infected samples versus mock-infected samples, with an expression cut-off for significant differential expression of FDR < 0.05.
References
- Abo K.M., Ma L., Matte T., Huang J., Alysandratos K.D., Werder R.B., Mithal A., Beermann M.L., Lindstrom-Vautrin J., Mostoslavsky G. Human iPSC-derived alveolar and airway epithelial cells can be cultured at air-liquid interface and express SARS-CoV-2 host factors. bioRxiv. 2020 2020.2006.2003.132639. [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., Hogan B.L. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu H., Yu Y., Tang N. Pulmonary alveolar regeneration in adult COVID-19 patients. Cell Res. 2020;30:708–710. doi: 10.1038/s41422-020-0369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., Ferrarese R., Criscuolo E., Diotti R.A., Castelli M., Scagnolari C., Burioni R., Antonelli G., Clementi M., Mancini N. Interferon-β-1a Inhibition of Severe Acute Respiratory Syndrome–Coronavirus 2 In Vitro When Administered After Virus Infection. The Journal of Infectious Diseases. 2020;222:722–725. doi: 10.1093/infdis/jiaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A.C., Gutierrez A.J., Bui L.T., Yahn S.L., Winters N.I., Calvi C.L., Peter L., Chung M.-I., Taylor C.J., Jetter C. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020;6:eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins F., Kramer P., Jacob A., Driver I., Thomas D.C., McCauley K.B., Skvir N., Crane A.M., Kurmann A.A., Hollenberg A.N. Prospective isolation of NKX2-1-expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Invest. 2017;127:2277–2294. doi: 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X., Islam M.N., O’Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., Snoeck H.W. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley K., Ding J., Villacorta-Martin C., Herriges M.J., Jacob A., Vedaie M., Alysandratos K.D., Sun Y.L., Lin C., Werder R.B. Reconstructed Single-Cell Fate Trajectories Define Lineage Plasticity Windows during Differentiation of Human PSC-Derived Distal Lung Progenitors. Cell Stem Cell. 2020;26:593–608.e8. doi: 10.1016/j.stem.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H., Na C.L., Weaver T.E., Vedaie M., Hurley K. Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell. 2017;21:472–488.e410. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Vedaie M., Roberts D.A., Thomas D.C., Villacorta-Martin C., Alysandratos K.D., Hawkins F., Kotton D.N. Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 2019;14:3303–3332. doi: 10.1038/s41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182:50–58.e58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019 doi: 10.1101/060012. [DOI] [Google Scholar]
- Law C.W., Alhamdoosh M., Su S., Dong X., Tian L., Smyth G.K., Ritchie M.E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Research. 2016;5 doi: 10.12688/f1000research.9005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J.M., Yang C.X., Tam A., Shaipanich T., Hackett T.-L., Singhera G.K., Dorscheid D.R., Sin D.D. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur. Respir. J. 2020;55:2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire T.A., Ikonomou L., Hawkins F., Christodoulou C., Cao Y., Jean J.C., Kwok L.W., Mou H., Rajagopal J., Shen S.S. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell. 2017;20:844–857.e846. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Alysandratos K.D., Jacob A., Hawkins F., Caballero I.S., Vedaie M., Yang W., Slovik K.J., Morley M., Carraro G. Single-Cell Transcriptomic Profiling of Pluripotent Stem Cell-Derived SCGB3A2+ Airway Epithelium. Stem Cell Reports. 2018;10:1579–1595. doi: 10.1016/j.stemcr.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Hawkins F., Kotton D.N. Derivation of Epithelial-Only Airway Organoids from Human Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2018;45:e51. doi: 10.1002/cpsc.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schäfer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. MBio. 2014;5 doi: 10.1128/mBio.01174-14. e01174–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Quinlan B.D., Peng H., Guo Y., Peng S., Zhang L., Davis-Gardner M.E., Gardner M.R., Crynen G., Voo Z.X. Mutations from bat ACE2 orthologs markedly enhance ACE2-Fc neutralization of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.06.29.178459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay A., Konda B., Garcia G., Yao C., Beil S., Sen C., Purkayastha A., Kolls J.K., Pociask D.A., Pessina P., Sainz de Aja J., Garcia-de-Alba C., Kim C.F., Gomperts B., Arumugaswami V., Stripp B.R. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. bioRxiv. 2020 doi: 10.1101/2020.06.29.174623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham I., Uchida T., Planes C., Ware L.B., Kaner R., Matthay M.A., Clerici C. Hypoxia upregulates VEGF expression in alveolar epithelial cells in vitro and in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;283:L1133–L1142. doi: 10.1152/ajplung.00464.2001. [DOI] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H., 3rd, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J., Ito Y., Holmes K.V., Mason R.J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020:1–7. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer I.-M., Padera R.F., Solomon I.H., Kanjilal S., Hammer M.M., Hornick J.L., Sholl L.M. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod. Pathol. 2020 doi: 10.1038/s41379-020-0595-z. Published online June 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra M., Alysandratos K.D., Hawkins F., McCauley K.B., Jacob A., Choi J., Caballero I.S., Vedaie M., Kurmann A.A., Ikonomou L. Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung- versus thyroid-lineage specification. Development. 2017;144:3879–3893. doi: 10.1242/dev.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Nhu Thao T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe. 2020;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C., Wolock S., Klein A.M. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics. 2018;34:1246–1248. doi: 10.1093/bioinformatics/btx792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Research. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Gotoh S., Korogi Y., Seki M., Konishi S., Ikeo S., Sone N., Nagasaki T., Matsumoto H., Muro S. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods. 2017;14:1097–1106. doi: 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., HCA Lung Biological Network. Electronic address. lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Listed are names and statistics (fold-change and false-discovery rate-adjusted values; FDR) for all genes tested through bulk RNA-seq analysis of iAT2s infected with SARS-CoV-2 (MOI 5) at 1 and 4 dpi, along with mock-infected cells at 1 day. Biostatistical comparisons were performed between 1 dpi and mock-infected, 4 dpi and 1 dpi, and between all infected samples versus mock-infected samples, with an expression cut-off for significant differential expression of FDR < 0.05.
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GEO: GSE153277. Interactive time series visualization of gene expression available at: https://crem.shinyapps.io/iAEC2infection/. Original data have been deposited to Mendeley Data: https://dx.doi.org/10.17632/p58k2bd6t5.1.