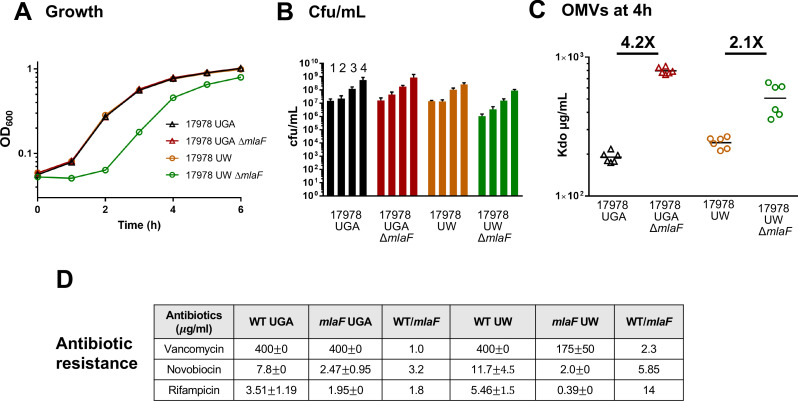
Figure 1. ΔmlaF mutant from the UW group exhibits significant growth defects.
(A) Growth curves of WT and ΔmlaF deletion mutants from the UGA lab (triangles) and UW lab (circles). Standard deviation error bars, if smaller than symbols, are not shown. (B) Colony-forming units (cfu) of cultures at hours 1–4. (C) Outer membrane vesicle (OMV) quantification of WT and ΔmlaF deletion mutants harvested at 4 hr and quantified by measuring 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) levels using the Purpald assay. Growth curves were performed in biological triplicate. OMV assays were performed in biological duplicate and quantified in technical triplicates. (D) MICs for vancomycin, novobiocin, and rifampicin for both UGA and UW strains. MICs were calculated as described in the Materials and methods.
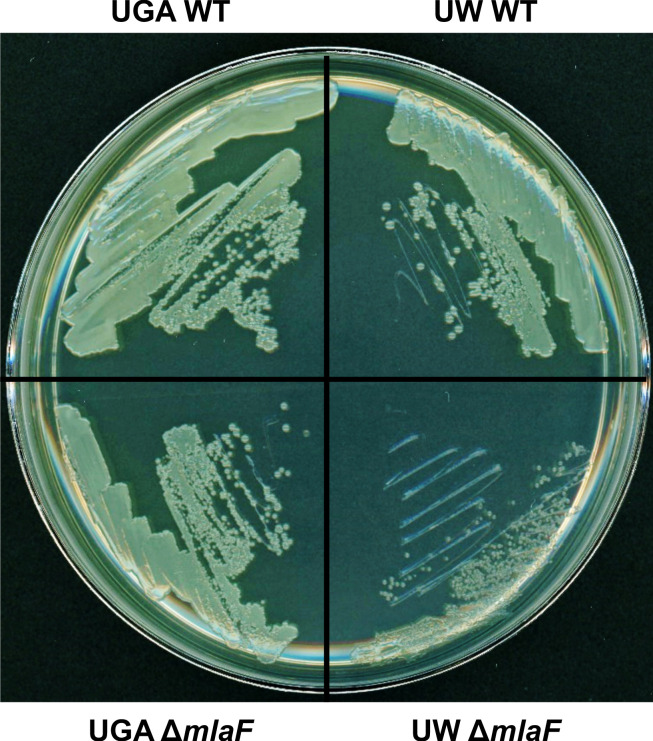
Figure 1—figure supplement 1. UW ΔmlaF has a distinct colony morphology.
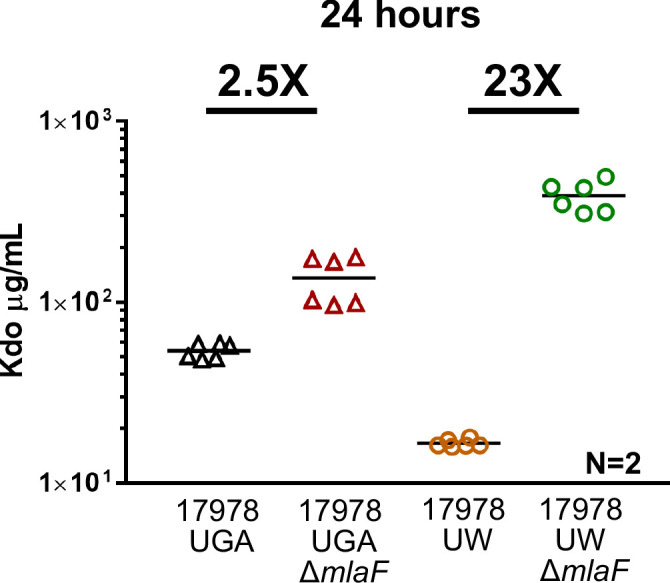
Figure 1—figure supplement 2. OMVs collected from stationary phase cultures at 24 hr.