Abstract
Background
This study explored the mechanisms underlying altered neurobehavioural development of female offspring born to mothers with polycystic ovary syndrome (PCOS).
Methods
In total, 20 women with PCOS and 32 healthy women who underwent caesarean deliveries with a single female foetus were recruited. Infants were assessed with Dubowitz scoring. Swan71 cell line with stable FOS overexpression was used to verify the regulatory effects of FOS on brain-derived neurotrophic factor (BDNF) and nerve growth factor (NGF) expression. Learning and memory in female first-generation (F1) and second-generation (F2) offspring in a rat model of PCOS was tested using the Morris water maze at puberty and adulthood. Transcriptome analysis of pubertal hippocampi and hypothalami of female F1 offspring was conducted.
Findings
Total score and behaviour subscales of Dubowitz scoring were significantly lower in female infants of women with PCOS. FOS and NGF protein levels were downregulated in placental villi of the PCOS group. FOS played a key role in BDNF inhibition and enhancing NGF in Swan71 cells. PCOS female F1 rats exhibited lower target crossing times during puberty when compared to controls. Transcriptome analysis revealed significant changes in hippocampal and hypothalamic neuronal pathways in female F1 rats at puberty.
Interpretation
FOS regulation of neurotrophins in the placenta negatively affects neurobehavioural development of female offspring of PCOS mothers.
Funding
This study was funded by the National Key R&D Program of China (2018YFC1004900 to F.Q. and F.W.) and the National Natural Science Foundation of China (81874480 to F.Q.; 81873837 to F.W.).
Keywords: Polycystic ovary syndrome, Offspring, Neurobehaviour, FOS, Neurotrophin, Placenta
Research in context.
Evidence before this study
As the most common endocrine disorder observed in women of reproductive age, polycystic ovarian syndrome (PCOS) can affect multiple aspects of women's health and even alter the foetal endocrine environment, leading to adverse outcomes in offspring including altered cardiometabolic health as well as higher risk for developmental delay and neurobehavioural disorders. FOS, as a marker of stimulation-related neural activation, belongs to the activator protein 1 superfamily of transcription factors. Brain derived neurotrophic factor (BDNF) and nerve growth factor (NGF) are essential for brain development and plasticity, which have been widely implicated in neurobehavioural disorders. BDNF and NGF in umbilical cord blood, placenta and amniotic fluid may have impact on foetal neurodevelopment. Despite the well-established association between maternal PCOS and the altered neurobehavioural development in female offspring of women with PCOS, the underlying mechanisms remain obscure.
Added value of this study
The total score and behaviour subscales of Dubowitz scoring were significantly lower in female infants of women with PCOS. We found negative alternations of proliferation and apoptosis of trophoblasts, vascularization of villi, endocrine functions and nutrient transfer in placenta of women with PCOS. In trophoblasts of PCOS group, the protein levels of FOS and NGF were downregulated, while BDNF unchanged. A cluster of neural function related genes was identified with integrative analysis of methylome and transcriptome. With respect to neural function, the enriched Gene Ontology (GO) terms in differential expressed gene cluster of PCOS group involved brain development, the proliferation and differentiation of neural precursor cell, neuron and neuroblast. Consistently, the enriched GO terms in differential methylated gene cluster of PCOS group were associated with neuropeptide and synaptic transmission, as well as cognition and learning or memory. We found that FOS gene probably plays a core role and predicted the potential sites in neurotrophin genes of FOS transcriptional modification. FOS could inhibit the mRNA expression of BDNF and enhance the mRNA expression of NGF in SWAN71 cells. In Morris water maze test of PCOS rat model, the target crossing times of female F1 offspring in puberty were greatly lower than the controls, however, no significant difference existed in adulthood, neither in puberty nor in adulthood of female F2 offspring. The transcriptome analysis showed that there were significant changes in the neuronal life processes and pathways in the puberty hippocampus and hypothalamus of the female F1 offspring of PCOS model rats.
Implications of all the available evidence
Our study demonstrates that FOS-mediated regulation of neurotrophins in the placenta may underpin the negatively effects of maternal PCOS on neurobehavioural development of female offspring.
Alt-text: Unlabelled box
1. Introduction
Polycystic ovarian syndrome (PCOS) is characterised by hyperandrogenism, irregular ovulation, and polycystic ovaries. It is the most common endocrine disorder observed in women of reproductive age. This heterogeneous condition affects multiple aspects of women's health including reproductive, metabolic, and psychological functions [1]. Moreover, maternal PCOS can alter the foetal endocrine environment, leading to adverse outcomes in offspring [[2], [3], [4]], which are more closely associated with maternal PCOS rather than pregnancy complications [5] Offspring of PCOS mothers exhibit altered cardiometabolic health relative to that of controls; these differences are predominantly observed in female offspring aged between 1 and 18 years [6]. Female offspring of women with PCOS have a higher risk for developmental delay [7] PCOS-exposed offspring also have an increased risk of neurobehavioural disorders, involving attention-deficit/hyperactivity disorder (ADHD), autism spectrum disorders (ASD), Tourette's disorder, chronic tic disorders (TD/CTD). In particular, the risk of ADHD and ASD is higher in female offspring born to PCOS women [[8], [9], [10], [11]].
FOS belongs to the activator protein 1 (AP-1) superfamily of transcription factors and is responsible for diverse cellular processes, including proliferation, differentiation, apoptosis, hypoxia, angiogenesis, and steroidogenesis,[12, 13], which has been confirmed in trophoblast cells [14]. FOS is a marker of stimulation-related neural activation [15]. As a switch that converts short-term stimuli into long-term responses, FOS activity in the brain is closely associated with learning, memory, hyperactivity, locomotion, anxiety-related behaviours, and impaired social behaviour [[16], [17], [18]]. In PCOS, FOS is decreased in adipose tissue and the hypothalamus, which may be associated with insulin resistance and neuroendocrine dysfunction [19, 20]. As FOS protein in ovarian cells regulates androgen production, impaired FOS activity may underpin the pathogenesis of hyperandrogenism in PCOS [21, 22]
Brain-derived neurotrophic factor (BDNF) and nerve growth factor (NGF) are essential for brain development and plasticity, and have been widely implicated in neurobehavioural disorders [23, 24]. BDNF levels in umbilical cord blood reflect the degree of neural maturity in infants [25]. Reduced placental BDNF negatively affects foetal neurodevelopment, as placental BDNF enters the foetal brain to contribute to its development [26]. As a possible source of neurotrophins to the foetus, the placenta plays a more prominent role than maternal passage and endogenous foetal synthesis [25, [27], [28], [29], [30], [31]]. BDNF and NGF are expressed in the placenta and serve diverse roles in differentiation, proliferation, survival, angiogenesis, and transplacental nutrient transport, thus impacting on the development of the materno-foetal-placental unit [[32], [33], [34], [35], [36], [37], [38]]. BDNF has been implicated in “developmental programming” via the regulation of metabolism and energy balance from early life to adulthood [39]. BDNF and NGF in amniotic fluid are markers for the presence of foetal central nervous system abnormalities in utero [40]. With regards to PCOS, neurotrophins in peripheral blood or follicular fluid may affect follicular and embryonic development [41, 42].
Despite the well-established association between maternal PCOS and altered neurobehavioural development in female offspring of women with PCOS, the underlying mechanisms remain obscure. The present study aimed to address this gap in knowledge using RNA sequencing (RNA-seq), reduced representation bisulphite sequencing (RRBS), and subsequent bioinformatics analysis, involving Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, protein-protein interaction (PPI) network, and in silico analysis.
2. Methods
2.1. Subjects and sampling
In total, 20 women with PCOS and 32 healthy control women who underwent caesarean deliveries with a single female foetus in Women's Hospital, School of Medicine, Zhejiang University between June 2016 and June 2017 were recruited. Patients were diagnosed with PCOS according to the Rotterdam Consensus (European Society for Human Reproduction and Embryology/American Society for Reproductive Medicine criteria) [43]. The control women had regular menstrual cycles and normal sex hormone levels prior to pregnancy. Uteri and ovaries of all women were structurally normal. Exclusion criteria were other disorders with a similar clinical presentation, such as congenital adrenal hyperplasias, (ovarian or adrenal) androgen-secreting tumours, Cushing's syndrome, thyroid dysfunction, hyperprolactinemia, primary premature ovarian insufficiency, premature ovarian failure, and/or functional hypothalamic amenorrhea. The study was approved by the Ethics Committee of Women's Hospital, School of Medicine, Zhejiang University. Peripheral blood, umbilical cord blood, and placenta were collected during caesarean section. Anthropometric parameters including age, cycle length, pregestational body mass index (BMI), BMI at delivery, and foetal digit ratio of the right hand were measured using questionnaires and standard equipment. Serum biochemical indexes including, total testosterone (TT) (80979, 0.05 ng/mL, Crystal Chem Inc, USA), sex hormone-binding globulin (SHBG) (RAB0734, 1.2 pmol/L, Sigma-Aldrich, USA), BDNF (RAB0026, 80 pg/mL, Sigma-Aldrich, USA), NGF (CSB-E04683h, 6.86pg/mL, Cusabio, USA), interleukin 6 (IL6) (BMS213-2, 0.92 pg/mL, Invitrogen, USA), and interleukin 15 (IL15) (BMS2106, 3.4 pg/mL, Invitrogen, USA) were measured with enzyme-linked immunosorbent assay (ELISA) in maternal peripheral blood and umbilical cord blood. Free androgen index (FAI) was calculated as TT (nmol/l) divided by SHBG (nmol/l) × 100.
2.2. Neurological examination of the infants
Infants were examined using the Dubowitz neurologic examination method on the day after birth [44]. Trained researchers performed the neurological examinations, and results were recorded. In total, 34 items were classified into six categories, including tone, tone patterns, reflexes, movements, abnormal signs, and behaviour. Written informed consent was obtained from all women included in the present study.
2.3. Human tissue collection
After obtaining informed consent, the term placentas (38–40 weeks of gestation) from non-labour women who underwent elective caesarean section were collected. Placental tissue and villous biopsies were gently separated from the foetal side as described previously [45]. Samples of placental tissue were collected and stored at −80°C after flash freezing in liquid nitrogen. Samples of placental tissue were fixed in 4% paraformaldehyde for haematoxylin and eosin (H&E) staining and immunofluorescence.
2.4. Immunofluorescence
Placental villi were immunostained with primary antibodies against BDNF (ab205067, Abcam, UK, RRID: AB_205067), NGF (ab52918, Abcam, UK, RRID: AB_881254), FOS (GTX25794, GeneTex, USA, RRID: AB_369382), beta-human chorionic gonadotropin (β-hCG) (11615-1-AP, Proteintech, China), glucose transporter 1 (GLUT-1) (1:200, 21829-1-AP, Proteintech, China RRID: AB_10837075), and glucose transporter 3 (GLUT-3) (20403-1-AP, Proteintech, China RRID: AB_10694437) using a standard protocol as previously described [46]. Cell nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI) (32670-5MG-F, Merck, Germany). Staining was visualised with a laser-scanning confocal microscope at × 400 magnification (LSM 780, Zeiss, Germany). The integrated optical density (IOD) and area of trophoblasts were quantified using Image-pro plus 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
2.5. Cell proliferation and apoptosis detection
Ki67 staining and terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay were performed to evaluate proliferation and apoptosis using an anti-Ki67 primary antibody (GB14102, Servicebio, China) and fluorescein-based cell death detection kit (11684795910, Roche, Switzerland). The number of Ki67-positive cells, TUNEL-positive cells, total number of DAPI-stained cells, and IOD were measured with Image-pro plus 6.0 (Media Cybernetics, Inc., Rockville, MD, USA). The proliferation and apoptotic indexes were calculated as the percentage of Ki67-positive and TUNEL-positive nuclei divided by the total number of DAPI-stained cells in each section, respectively.
2.6. RNA extraction and real-time PCR
Vascular endothelial growth factor (VEGF), BDNF, NGF, FOS, Cyclin Dependent Kinase Inhibitor 1A (CDKN1A), Cyclin A2 (CCNA2), lin-7 homolog A (LIN7A), microtubule associated protein 1B (MAP1B), and transcription factor 4 (TCF4) mRNA expression levels in the foetal side of placentae from women with PCOS and controls were analysed with real-time polymerase chain reaction (real-time PCR). Isolation of total RNA was performed using Trizol Reagent (15596026, Invitrogen, USA), and cDNA was synthesised using the RevertAid First Strand cDNA Synthesis Kit (K1621, Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Real-time PCR was performed using the Step One Plus Real-Time PCR System (4376600, Applied Biosystems, USA). The relative quantification of gene expression was performed using the comparative ΔΔCt method. The primer sequences used for RT‑PCR are listed in Table S1.
2.7. Bisulphite sequencing PCR
A methylation assay was performed at the promoter region of BDNF, NGF, and FOS genes in the foetal side of placentae from PCOS and control women using the bisulphite genomic sequencing (BSP) method as previously described [47]. Total genomic DNA was extracted using the TIANamp Genomic DNA Kit (DP304-02, Tiangen Biotech, China). Bisulphite treatment was conducted using EZ DNA Methylation-Gold Kit (D5005, ZYMO Research, USA). The methylation status of cytosine phosphate guanine (CpG) sites located on gene promoters were analysed by cloning and sequencing bisulphite-treated DNA. The modified DNA was amplified by PCR, and the products were purified and cloned into a pMD 19-T vector (3271, Takara, Japan). Ten colonies of each subject were randomly selected for the plasmid DNA and sequenced with 3730 DNA Analyzer Polymers (3730S, Applied Biosystems, USA, RRID: SCR_018052).
2.8. RRBS
RRBS libraries of genomic DNA were prepared using TruSeq DNA LT Sample Prep Kit v2 (Illumina, USA). The libraries were quantified with Quant-iT PicoGreen dsDNA Assay Kit (P7589, Life technologies, USA). All libraries were sequenced with Illumina HiSeq 2500 platform. Clean reads were obtained by removing the reads containing sequencing adaptors and primers or low quality reads. Contaminated and short sequences were filtered. Differentially methylated promoters (DMPs) were obtained with a P-value threshold of 0.05 and two-fold change.
2.9. RNA-seq
Total RNA was extracted using Trizol reagent (15596026, Invitrogen, USA) according to the manufacturer's instructions. RNA quantity and purity were measured with Bioanalyzer 2100 (G2939BA, Agilent, USA) and RNA 6000 Nano LabChip Kit (5065-4476, Agilent, USA). RNA-seq libraries were obtained using TruSeq RNA LT Sample Prep Kit v2 (Illumina, USA). Paired-end sequencing was performed on the Illumina HiSeq 2500 platform (Genergy Biotech, China). The expression values were obtained by cuffnorm based on fragments per kilobase of exon per million reads mapped (FPKM) value [48]. Differentially expressed genes (DEGs) were screened out based on the criteria of two-fold change and P-value less than 0•05 using DESeq2 [49]. The raw data of RRBS and RNA-seq have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE154274 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154274) [50].
2.10. Differential alternative splicing analysis
Replicate multivariate analysis of transcript splicing (rMATS) v3.2.5 was used to detect differential alternative splicing (DAS) genes [51]. Five alternative splicing events were obtained, including skipped exons, retained introns, alternative 5′ splice sites, alternative 3′ splice sites, and mutually exclusive exons.
2.11. GO, KEGG enrichment analysis, and PPI network analysis
For RNA-seq of placentae, significantly enriched GO terms were achieved with DEGs and DMP-related genes by TopGO at the significance level set (P < 0.05) [52]. PPI network analysis was performed with DEGs using the Search Tool for the Retrieval of Interacting Genes (STRING) v.9.05 [53]. Cytoscape software was used to visualise these associations [54]. For RNA-seq of the hippocampus and hypothalamus, GO and KEGG enrichment analysis were conducted with DAS genes using KOBAS v 2.0 software [55].
2.12. In silico analysis
Putative motifs and potential transcription factor binding sites in genomic DNA 5 kb upstream and downstream were analysed using Genomatix MatInspector (Genomatix Software GmbH, Germany).
2.13. Overexpression of FOS in stable clonal lines
The first-trimester human trophoblast cell line, Swan71, was originally provided by Professor Charles H Graham, Queen's University, Kingston, Ontario, Canada and maintained in the Zhejiang Province Key Laboratory of Female Reproductive Health Research. Swan71 cells were cultured in Dulbecco's Modified Eagle's Medium/Ham's Nutrient Mixture F-12 (DMEM/F-12) medium (11320033, Gibco, USA) supplemented with 10% foetal bovine serum (FBS) (10099141C, Gibco, USA) and 1% penicillin–streptomycin (15140148, Invitrogen, USA) at 37°C and 5% CO2.
To establish stable cell lines that overexpressed FOS, we transfected Swan71 cells with the lentiviral gene expression vector of FOS (pRLenti-EF1a-EGFP-P2A-Puro-CMV-FOS-3Flag, H13612) and a control vector (pRLenti-EF1a-EGFP-P2A-Puro-CMV-MCS-3Flag, GL107) using polybrene (H9268, Sigma, USA). Swan71 cells were infected with lentiviral particles at a multiplicity of infection of 60. After infection for 72 hours, transfected cells were selected by culturing with puromycin (P8833, Sigma, USA) for 2 weeks.
RNA samples were isolated from Swan71-GL107 and Swan71-H13612 to validate BDNF and NGF expression levels using real-time PCR. FOS protein expression levels in Swan71 cells were detected with western blot using a standard protocol as previously described [56]. Monoclonal anti-flag M2 antibody (F1804S, Sigma, USA, RRID: AB_262044), goat anti-mouse IgG (A0216, Beyotime, China, RRID:AB_2860575), GAPDH polyclonal antibody (AP0063, Bioworld, USA, RRID: AB_2651132), and goat anti-rabbit IgG (A0208, Beyotime, China) were used.
2.14. Establishment of PCOS rat model
A total of 20 neonatal female Sprague-Dawley rats were provided by the Laboratory Animal Research Center, Zhejiang Chinese Medical University, Hangzhou, China. Animals were randomly divided into PCOS model and control groups (n=10 per group). On postnatal day nine, a PCOS rat model was established as previously described;[47, 57] The study was performed according to the Care and Use of Laboratory Animals protocol of National Research Council of China, following the ARRIVE guidelines, and was approved by Zhejiang University Ethics Committee.
On postnatal day nine, a PCOS rat model was induced by subcutaneous injection of testosterone propionate (57-85-2, Solarbio, China) at a dose of 0.1 mg/0.004 mL olive oil per g of body weight [57]. The control group received olive oil only. Animals were raised and housed in a temperature-controlled room (25°C) under a 12-hour light/dark cycle at a constant humidity of 55% humidity, and were provided ad libitum access to food and water. At 8 week of age, oestrous cycles were determined by analysing the cell types in vaginal smears for 14 consecutive days [58]. In the PCOS model group, only rats with irregular vaginal smears were used for subsequent experiments. Oestrus cycle, serum level of FAI, and ovarian histology were assessed to validate the PCOS rat model [47]. All rats were sacrificed in the dioestrus stage. Serum samples were collected after blood centrifugation and stored for hormone assays. Ovaries were removed from sacrificed rats and fixed in 4% paraformaldehyde. After embedding in paraffin, 4-μm sections were mounted on glass slides and stained with H&E for morphological observation under light microscopy.
2.15. Offspring acquisition
Rats in the PCOS model and control groups were designated as the parental generation (F0). At 12 weeks of age, F0 dams were injected intraperitoneally with 20 IU pregnant mare serum (PMSG, Ningbo Second Hormone Factory, China) at 5:00 pm followed by 20 IU of human chorionic gonadotropin (HCG, Ningbo Second Hormone Factory) 48 hours later and immediately paired with proven male breeders. Vaginal sperm plugs were assessed the following morning [59]. Pups born to F0 dams were designated as the F1 generation. At 12 weeks of age, female F1 rats were used to generate F2 by breeding them with proven male breeders.
2.16. Learning and memory testing with the Morris water maze in female offspring
The Morris water maze test was performed at 5 weeks (puberty) and 13 weeks of age (adulthood) to evaluate learning and memory in female offspring. The test was conducted and recorded between 8:00 am and 2:00 pm under poor lighting conditions. The water maze apparatus consisted of a circular water tank 1.5 m in diameter and was filled to a depth of 60 cm with water (22 ± 2°C). The tank was divided into four virtual quadrants. A circular platform, 8 cm in diameter, was placed in one of the quadrants (target quadrant) 2 cm below the water surface. Rats were subjected to four training sessions per day for 2 days and a memory retention test (probe test) on the third day. In the training sessions, rats were placed at the same starting position in each quadrant and were allowed to swim freely to find the hidden platform (target) for 60 s. The time spent from being placed in the water to reaching the platform was recorded as latency to target. If the rat did not locate the platform within 60 s, it was guided to the platform and allowed to stay there for 10 s, and its latency to target was recorded as 60 s. On the third day of the probe test, the platform was removed, and rats were allowed to swim freely for 60 s. The distance and latency to target in the training sessions; distance, time, and entries in the target zone; and distance, time, and entries in the target quadrant during the probe test were recorded using a camera linked to a computerized video tracking system (SMART v3.0, Panlab SL, Spain).
2.17. Brain tissue collection
After completion of the Morris water maze test, female offspring were deeply an aesthetised with sodium pentobarbital (50 mg/kg, Sigma, P3761, USA) in the dioestrus stage. The hippocampus and hypothalamus were collected and stored at −80°C after flash freezing in liquid nitrogen.
2.18. Electron microscopy
Samples were post-fixed in 2.5% glutaraldehyde for 2 hours at room temperature. After fixation, samples were rinsed with BP buffer, post-fixed with 2% osmium tetroxide for 1-2 hours, rinsed again with BP buffer, dehydrated in an ethanol series, infiltrated in a mixture of Spurr and acetone, and embedded in 100% Spurr embedding agent overnight and polymerised for 24 hours. The specimens were cut into blocks and serially sliced into 70-nm thick slices for transmission electron microscope (H-7650, Hitachi, Japan) analysis. Micrographs of neurons and synapses were obtained using a side-mounted CCD camera (SC200, Gatan, USA).
2.19. Statistical analysis
Data are presented as mean ± SEM (n indicates the number of tissue preparations, cells, or separate experiments, as indicated in the figure legends). Statistical analysis was performed using unpaired two-tailed Student's t-tests, one-way analysis of variance with post-hoc tests, or their equivalent nonparametric tests for continuous variables. Chi-squared test or Fisher's exact test was performed for categorical variables (version 21.0; SPSS). P < 0.05 was considered statistically significant.
3. Results
3.1. Clinical characteristics
Of 20 PCOS patients, 17 presented with all three diagnostic criteria items (oligo- or anovulation, clinical or biochemical signs of hyperandrogenism, and PCO morphology); two did not present with signs of hyperandrogenism, and one did not present with PCO morphology. No significant differences were observed in maternal age at delivery, pregestational BMI, and BMI at delivery between PCOS patients and controls, with the exception of a longer cycle length in PCOS patients (Table 1). Relative to serum FAI levels in the control group at delivery, serum FAI levels in maternal peripheral blood were higher in the PCOS group, but those in umbilical cord blood of the corresponding newborns were not. The birth weights of female newborns were comparable in women with PCOS. Foetal digit ratios (2D:4D) of the right hand were similar in female newborns of women with PCOS and controls, corroborating the data from umbilical cord blood. Compared to controls, women with PCOS had higher IL6 levels and comparable levels of IL15, BDNF, and NGF in peripheral blood. Female newborns of women with PCOS had comparable IL6 and IL15 levels but significantly lower levels of BDNF and NGF in umbilical cord blood compared to those of control women, which is in accordance with a previous study that reported that neural factors are unlikely to traffic between foetus and mother [60].
Table 1.
Clinical characteristics of subjects.
| Characteristics | Control (n = 32) | PCOS (n = 20) |
|---|---|---|
| Pregestational BMI (kg/m2) | 20.81 ± 0.32 | 21.44 ± 0.58 |
| Cycle length (days) | 29.16 ± 0.20 | 55.25 ± 3.56* |
| Maternal age at delivery (years) | 31.78 ± 0.57 | 32.55 ± 0.89 |
| BMI at delivery (kg/m2) | 26.53 ± 0.51 | 27.18 ± 0.51 |
| Serum FAI in maternal peripheral blood at delivery (nmol/L) | 0.73 ± 0.06 | 1.34 ± 0.15* |
| Serum BDNF in maternal peripheral blood at delivery (ng/mL) | 178.52 ± 10.53 | 173.02 ± 13.92 |
| Serum NGF in maternal peripheral blood at delivery (pg/mL) | 20.28 ± 1.24 | 20.58 ± 1.18 |
| Serum IL-6 in maternal peripheral blood at delivery (pg/ml) | 6.59 ± 0.58 | 9.25 ± 0.98* |
| Serum IL-15 in maternal peripheral blood at delivery (pg/ml) | 16.25 ± 1.43 | 19.21 ± 2.20 |
| Gestational age at delivery (weeks) | 38.34 ± 0.10 | 38.45 ± 0.11 |
| Newborn digit ratio of the right hand | 0.930 ± 0.005 | 0.923 ± 0.005 |
| Serum FAI in umbilical cord blood | 1.33 ± 0.09 | 1.79 ± 0.21 |
| Serum BDNF in umbilical cord blood (ng/mL) | 137.64 ± 11.82 | 96.42 ± 7.18* |
| Serum NGF in umbilical cord blood (pg/mL) | 13.91 ± 0.87 | 9.41 ± 0.65* |
| Serum IL-6 in umbilical cord blood (pg/ml) | 7.24 ± 0.74 | 9.14 ± 0.95 |
| Serum IL-15 in umbilical cord blood (pg/ml) | 10.18 ± 1.16 | 9.87 ± 1.40 |
| Birth weight (kg) | 3.56 ± 0.07 | 3.37 ± 0.09 |
Note: PCOS, Polycystic ovarian syndrome; BMI, body mass index; FAI, free androgen index; BDNF, brain-derived neurotrophic factor; NGF, nerve growth factor; IL-6, interleukin 6; IL-15, interleukin 15. Data were presented as mean ± SEM.
P < 0.05.
3.2. Dubowitz neurological examination of female newborns
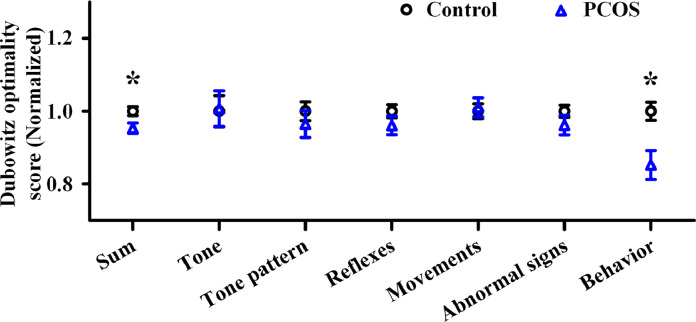
The total score and behaviour subscale of the Dubowitz scoring system were significantly lower in female infants of women with PCOS compared to those of controls. All remaining subscales (including tone, tone pattern, reflexes, movements, and abnormal signs) were comparable between these two populations (Fig. 1).
Fig. 1.
Neurological examination of female offspring in polycystic ovary syndrome (PCOS) and control group with Dubowitz optimality score. PCOS: n = 20; control: n = 32. Data were presented as mean ± SEM. *P < 0.05. PCOS, polycystic ovary syndrome.
3.3. Altered placental function in PCOS patients
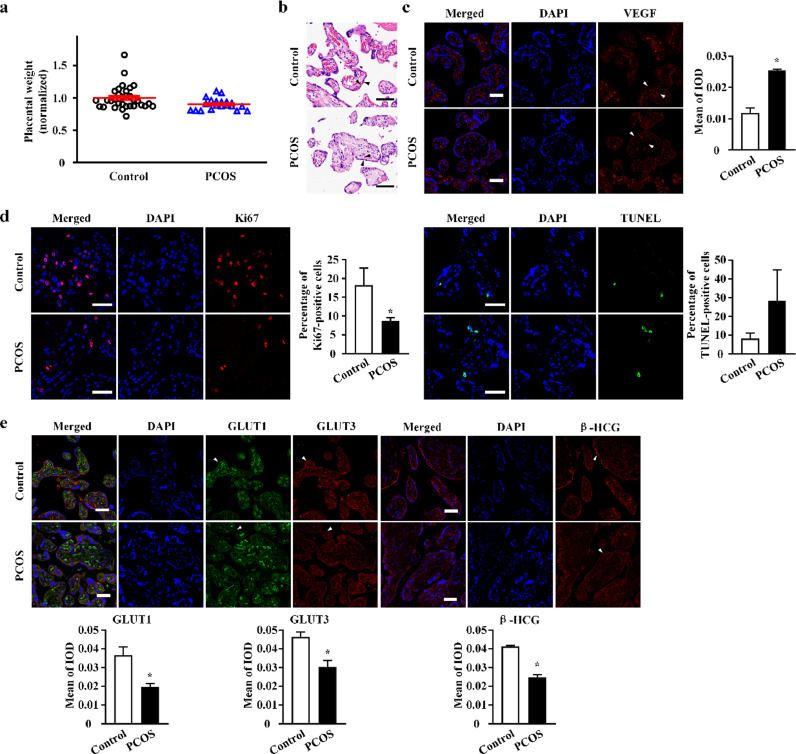
No significant difference in placental weight was noted between PCOS and control groups (Fig. 2a). The morphological features of placental villi in PCOS and control groups are shown in Fig. 2a. With regards to placental vascularisation, we observed substantially decreased vascularisation of villi and elevated levels of vascular endothelial growth factor (VEGF) in placental trophoblasts of PCOS women (Fig. 2c). Ki67 and TUNEL staining revealed suppressed cell proliferation and slightly enhanced apoptosis of trophoblasts in placental villi of the PCOS group (Fig. 2d). With regards to cell-cycle related gene expression, CCNA2 expression was significantly decreased and CDKN1A expression was increased in placentae of the PCOS group (Fig. S1). With regards to placental endocrine function and nutrient transfer in PCOS, reduced levels of β-HCG, GLUT1, and GLUT3 were observed in placentae of the PCOS group (Fig. 2e), suggesting attenuated hormone secretion and carrier-mediated glucose transport in pathological trophoblasts of PCOS.
Fig. 2.
The alterations of placental functions in polycystic ovary syndrome (PCOS). (a) Placental weight (all placental lobes) of female offspring of women with PCOS (n = 20) compared to controls (n = 32). Data were presented as mean ± SEM. (b) Hematoxylin and eosin (H&E) staining of placental tissue. Black arrows point to syncytiotrophoblasts layer and fetal blood vessels. (c) Vascular endothelial growth factor (VEGF) protein expression in human placental fetal side of the women with PCOS and controls, as assessed by immunofluorescence with VEGF (red). White arrows point to syncytiotrophoblasts layer and fetal blood vessels. Bar plot shows the mean of integrated optical density (IOD) (n = 3 in each group). (d) Immunofluorescence staining of Ki-67 (red) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (green), the markers for cellular proliferation and apoptosis, respectively, was performed on trophoblasts in placental villi of PCOS and control group. Bar plot shows the percentage of Ki67-positive cells and percentage of TUNEL-positive cells, respectively (n = 5 in each group). (e) Immunofluorescence staining of glucose transporters 1 (GLUT1) (green), glucose transporters 3 (GLUT3) (red), and beta-human chorionic gonadotropin (β-HCG) (red) were performed on trophoblasts in placental villi of PCOS and control group. The syncytiotrophoblasts layers are labeled with white arrows, and the results of protein quantification are shown by bar plots (n = 5 in each group). All scale bars are 50 μm. DAPI, 4’,6-diamidino-2-phenylindole. Bar plots in (c) and (e) represent the protein expression level on syncytiotrophoblasts. Data were presented as mean ± SEM. *P < 0.05, unpaired two-tailed Student's t tests.
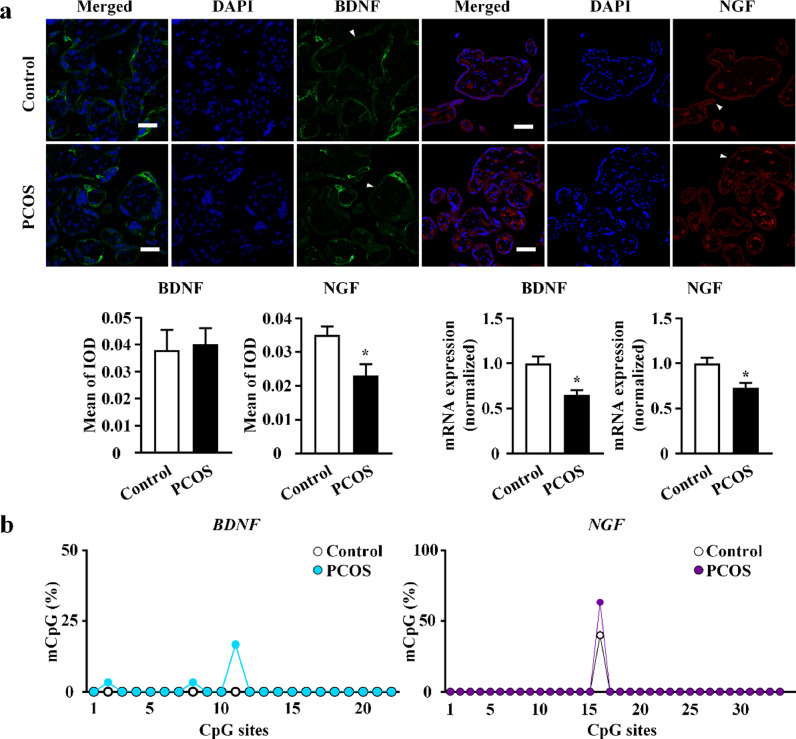
To investigate the placental production of neurotrophins, including BDNF and NGF, real-time PCR and immunofluorescence were performed. In the PCOS group, NGF mRNA levels were significantly downregulated in placental villi, and NGF protein expression in trophoblasts was reduced. BDNF protein expression in trophoblasts remained unchanged, while BDNF mRNA levels were significantly reduced in placental villi of the PCOS group (Fig. 3a). These results were consistent with the decreased levels of NGF (but not BDNF), in umbilical cord blood of the PCOS group (Table 1), underscoring the potential involvement of neurotrophins in PCOS offspring development.
Fig. 3.
Brain derived neurotrophic factor (BDNF) and nerve growth factor (NGF) in placental villi. (a) Visualization of BDNF and NGF in placental villi of polycystic ovarian syndrome (PCOS) and control group by immunofluorescence with BDNF (green) and NGF (red). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue fluorescence). The syncytiotrophoblasts layers are labeled with white arrows, and the results of protein quantification are shown by bar plots (n = 5 in each group). All scale bars are 50 μm. BDNF and NGF mRNA expression level were quantified by real-time polymerase chain reaction (real-time PCR). PCOS: n = 20; control: n = 32. Data were presented as mean ± SEM. *P < 0.05, unpaired two-tailed Student's t tests. (b) The methylation status at the promoter region of BDNF (blue) and NGF (purple) genes in the placental villi from women with PCOS and controls using bisulfite genomic sequencing (BSP) (n = 3 in each group, significance was determined by Chi-square test).
To identify the potential role of DNA methylation in transcriptional regulation, we analysed CpG islands of BDNF and NGF genes in placentae using bisulphite genomic sequencing PCR. No significant differences between PCOS individuals and controls were identified. However, the examined promoter region of all examined genes was hypomethylated (Fig. 3b and S2).
3.4. Genome-wide DNA methylation and expression patterns in placentae of PCOS patients
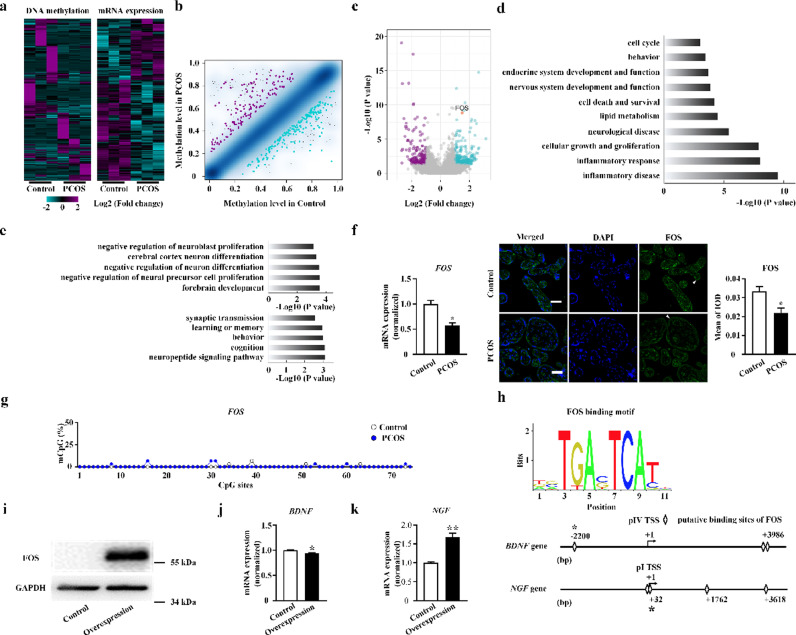
Global analysis of methylomic and transcriptomic data revealed an altered pattern of DNA methylation of promoters(Fig. 4a, b) and gene expression (Fig. 4a, c) in placentae of PCOS patients. A cluster of neural function-related genes, including LIN7A and MAP1B, were identified with integrative analysis of the methylome and transcriptome. mRNA expression levels were confirmed with real-time PCR (Fig. S3).
Fig. 4.
Identification of FOS and its regulation of neurotrophins in placenta of polycystic ovary syndrome (PCOS). (a) Heatmaps of the global DNA methylation in the promoter and gene expression data for placentae from women with PCOS and controls. (n = 3 in each group) (b) Scatter plot of global DNA methylation levels in placentae in controls (x-axis) compared to women with PCOS (y-axis). Point density is shown as blue shading; Peacock blue and purple dots indicate significant down-regulated and up-regulated cytosine phosphate guanine (CpG) sites associated with a methylation difference > 0.20. (c) Volcano plot to inspect differentially expressed genes in placentae from women with PCOS and controls, with fold change as the abscissa and -log10 (P value) as the ordinate. Peacock blue and purple splashes represent genes that significantly up or down regulated respectively. Gray splashes mean genes without significantly different expression. FOS gene is marked as red. (d) Gene Ontology (GO) terms enriched in differentially expressed genes in placentae from women with PCOS. (e) Neural GO terms enriched in differentially expressed gene (top) and methylated DNA (bottom) in placentae from women with PCOS. (f) FOS gene mRNA and protein expression levels in human placentae, as assessed by real-time polymerase chain reaction (real-time PCR) (PCOS: n = 20; control: n = 32) and immunofluorescence with FOS (green), and nuclear DNA (blue) labeling (n = 5 in each group). All scale bars are 50 μm. (g) The summary data of methylation status at the promoter region of FOS gene in the placental villi using bisulfite genomic sequencing (BSP) (n = 3 in each group, significance was determined by Chi-square test). (h) In silico analysis of the genomic DNA 5 kb upstream and downstream of BDNF (pIV) and NGF (pI) transcription start site (TSS). Detection of FOS (i) protein expression levels, BDNF (j) and NGF (k) mRNA expression levels in overexpression group (n = 3) and control group (n = 3). Data were presented as mean ± SEM. *P < 0.05, unpaired two-tailed Student's t tests.
Inflammatory disease and response processes were identified using GO enrichment analysis of differentially transcribed gene clusters in PCOS. Cellular growth, and development and function of nervous and endocrine systems were also closely related to PCOS (Fig. 4d). Enriched GO terms in differentially expressed gene clusters of the PCOS group involved brain development, and proliferation and differentiation of neural precursor cells, neurons, and neuroblast. The enriched GO terms in differentially methylated gene clusters of the PCOS group were consistently associated with neuropeptide and synaptic transmission, cognition, learning, and, memory (Fig. 4e).
To investigate possible protein-protein interactions among these genes, we performed transcriptome analysis with STRING, which identified the FOS gene as having a core role (Fig. S4). mRNA and protein expression levels were significantly downregulated in women with PCOS (Fig. 4f). The methylation level of FOS was detected by bisulphite genomic sequencing PCR (Fig. 4g, S2). However, no significant differences between PCOS individuals and controls were identified.
Next, we employed bioinformatic analysis to elucidate the molecular basis of transcriptional modification of BDNF and NGF in the placenta. The putative motifs of transcriptional factor FOS were identified through in silico analysis of genomic DNA 5 kb upstream and downstream of BDNF and NGF placenta-specific transcription start sites (TSS). The predicted proximal binding sites of FOS were 2.2 kb upstream and 32 bp downstream of BDNF and NGF TSS, respectively (Fig. 4h).
3.5. Expression levels of NGF and BDNF in FOS-overexpressing Swan71 cells
In order to further verify the regulatory effects of FOS on BDNF and NGF expression, we constructed a human placental trophoblast cell line Swan71 with stable overexpression of FOS. Western blot analysis indicated that FOS protein expression levels were higher in the overexpression group (Fig. 4i, S5). BDNF mRNA levels in the overexpression group were lower, while NGF levels were significantly higher than those of controls (Fig. 4j, k), indicating that FOS inhibited BDNF expression and enhanced NGF expression.
3.6. Morris water maze test in female F1 and F2 offspring of PCOS model rats
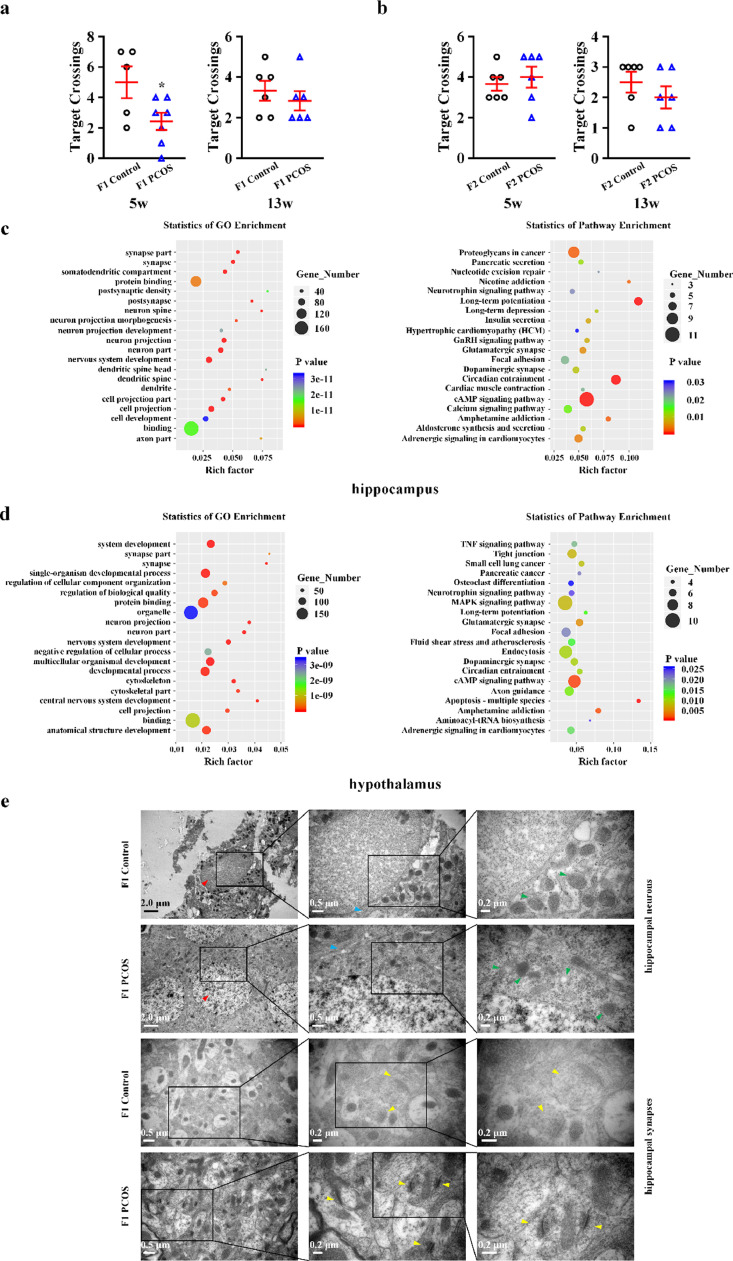
The PCOS rat model was successfully established, indicated by irregular oestrous cycle, elevated serum FAI levels (Fig. S6a), multiple cystic follicles, and decreased numbers of GCs and corpora lutea observed with H&E staining (Fig. S6b). The target crossing times of PCOS female F1 animals at puberty were lower than those of controls. No significant differences between groups were observed in adulthood (Fig. 5a). No significant differences were observed between PCOS and control groups at puberty or in adulthood in female F2 animals (Fig. 5b).
Fig. 5.
Neurobehavioural and cerebral changes in female offspring of polycystic ovary syndrome (PCOS) model rats. (a) Morris water maze test in PCOS female F1 (n = 5 in 5 week, n = 6 in 13 week) and controls (n = 7 in 5 week, n = 6 in 13 week). (b) Morris water maze test in PCOS female F2 (n = 6 in 5 week and 13 week) and controls (n = 6 in 5 week and 13 week). Data are presented as mean ± SEM. *P < 0.05, unpaired two-tailed Student's t tests. (c, d) The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment scatter plot of differential alternative splicing (DAS) genes in hippocampus (c) and hypothalamus (d) of F1 pubertal female offspring. The x-axis indicates the rich factor, and the y-axis indicates the name of the term or pathway. The dot size means the gene number, and the dot color represents the P value. The top 20 GO terms and KEGG pathways were shown. (e) The morphological and ultrastructure study of hippocampus neurons and synapses of F1 pubertal female offspring with electron microscopy. The nuclear chromatin (red arrows), rough endoplasmic reticulum (blue arrows), mitochondria (green arrows) and synapses (yellow arrows) in hippocampus were observed.
3.7. Transcriptomic analysis of the hippocampus and hypothalamus of F1 pubertal female rats
As hypothalamic and hippocampal gene expression changes lead to persistent neurobehavioural consequences in rodents [61], we conducted a global analysis of transcriptomic data of F1 pubertal female rats. The top 20 GO terms and KEGG pathways were identified in brains of pubertal female offspring of PCOS model rats. Synapse- and neuron-related terms were significantly enriched in the hippocampus and hypothalamus (Fig. 5c, d). Nervous system-related pathways including long-term potentiation, amphetamine addiction, glutamatergic synapses, dopaminergic synapses, long-term depression, and neurotrophin signalling were enriched in the hippocampus (Fig. 5c). Pathways involving amphetamine addiction, glutamatergic synapses, dopaminergic synapses, long-term potentiation, and neurotrophin signalling were enriched in the hypothalamus (Fig. 5d). We identified nine significantly enriched DNA stability-related GO terms in analysis of the DEGs in the hypothalamus of F1 pubertal female offspring (Fig. S7); however, none were identified in the hippocampus (data not shown).
3.8. Electron microscopic examination of the pubertal hippocampus of female F1 rats
In the pubertal hippocampus of female F1 rats, nuclear chromatin was uneven. Part of the cytoplasm was dissolved and presented as vacuole-like denaturation. Mitochondria were swollen and dilated with vacuolisation and ruptured mitochondrial cristae. A decrease in rough endoplasmic reticulum was also identified in neurons. Synaptic structures were partially dissolved, and synaptic number was decreased. Aberrant synaptic vesicles and blurred synaptic clefts were observed (Fig. 5e).
4. Discussion
A major finding of this study was impaired neurobehavioural development of female infants of women with PCOS. In trophoblasts of the PCOS group, FOS and NGF protein levels were downregulated. A cluster of neural function-related genes was identified using integrative analysis of the methylome and transcriptome. The FOS gene plays a core role in inhibiting BDNF and enhancing NGF in Swan71 cells. The target crossing times in the Morris water maze test were lower in female F1 offspring of PCOS model rats at puberty than in controls; moreover, significant changes were noted in neuronal processes and pathways in the pubertal hippocampus and hypothalamus based on transcriptomic analysis.
FOS inhibits placental trophoblast proliferation, migration, and invasion [14]. FOS is also critical for regulating placental development and controlling trophoblast function. Dysfunction in FOS may lead to placental-related pregnancy complications involving preeclampsia and abortion [[62], [63], [64]]. As placental FOS expression is diminished and FOS inhibits placental cell proliferation, diminished FOS expression in PCOS may be the result of a compensatory placental response to a sub-optimal maternal environment. BDNF potentiates placental development and plays an important role in cytotrophoblast differentiation, proliferation, and survival [65]. Neurotrophins may also modulate metabolism or nutrient flux to the foetus [66]. BDNF signalling impacts placental responses to adverse maternal effects [67, 68]. The down-regulation of placental BDNF may be implicated in altered foetal programming in these infants [69, 70]. Adequate NGF levels at the fetomaternal interface are essential for the progression of pregnancy [71]. Nevertheless, evidence for the interaction between FOS and neurotrophins in human placenta is limited. Here, we observed that the expression of the neurotrophins NGF and BDNF were regulated by FOS in both human placental trophoblasts and trophoblast cell lines, which may contribute to the maternal impact to the foetus. FOS may regulate BDNF in zebrafish brain [56]. Attenuated FOS expression indirectly decreases NGF in stromal cells from patients with endometriosis [72]. AP-1 transcription factors, including FOS family members, participate in the induction of BDNF exon I, III, and VI transcripts in rat neurons [73]. In addition, we predicted potential sites of FOS transcriptional modification in neurotrophin genes. Although the methylation levels of NGF and BDNF promoters did not significantly differ according to PCOS status, other pathways may underpin FOS-mediated regulation of NGF and BDNF.
A significant reduction in placental weight, thickness, density, and volume; and a higher foetal–placental weight ratio are observed in the PCOS population compared to that in healthy controls. Further, irregular placental shape is more frequent in PCOS patients [74]. Placental pathophysiological examination has revealed that women with PCOS have significantly more inflammatory and thrombotic lesions as well as increased villous immaturity of the placenta when compared with women from a reference group [75]. To date, the clinical evidence on birth weights of offspring of mothers with PCOS remains controversial [75]. The present study demonstrates altered proliferation and apoptosis of trophoblasts, vascularisation of villi, endocrine function, and nutrient transfer in placenta of PCOS women, which may affect foetal development.
The influence of maternal PCOS on offspring has received considerable attention in recent years [[76], [77], [78]]. Environmental factors during foetal development may induce an epigenetic transgenerational inheritance of adult-onset disease [[79], [80], [81]]. Female F0 mice with PCOS-like traits induced by late-gestation injection of dihydrotestosterone resulted in female F1-F3 offspring with PCOS-like reproductive and metabolic phenotypes. The sequencing of single metaphase II oocytes from F1-F3 offspring revealed common and unique altered gene expression across all generations [82]. Female F1 and F2 offspring of rats with ancestral dehydroepiandrosterone exposure exhibited PCOS-like reproductive and metabolic phenotypes, including disrupted oestrous cycles and polycystic ovaries, increased serum levels of testosterone, impaired glucose tolerance, and widespread metabolic abnormalities [83].
In the present study, we evaluated the neurobehavioural development of female first and second filial generations of PCOS model rats at puberty and adulthood using the Morris water maze, which provides insight into the effects of maternal PCOS on offspring. Maternal PCOS significantly affected the neurobehavioural development of the female first filial generation of PCOS model rats at puberty. Based on these results, subsequent GO and KEGG enrichment analysis were conducted. DAS genes were identified in the hippocampus and hypothalamus of pubertal female offspring of PCOS model rats. As alternative splicing occurs at high frequency in brain tissue and contributes to multiple aspects of nervous system development including cell-fate decisions, neuronal migration, axon guidance, and synaptogenesis [67,72], these findings were consistent with those of the Morris water maze test.
Epidemiologic, clinical, and experimental data indicate that the maternal intrauterine environment plays a critical role in tissue and organ development, and may induce aberrant responses later in life by triggering a cascade of events [84]. PCOS induces gestational hyperandrogenism [85], which was also observed in our study, constituting a potential source of androgen excess for the foetus. To address this issue, the present study assessed in utero androgen exposure conditions in both gestational and perinatal periods in an indirect manner, but no significant effects of intrauterine androgen exposure on PCOS offspring were noted. Similar phenomena were observed IL6 changes, although the maternal levels of inflammatory cytokines may compromise brain phenotypes [86]. Based on these findings, other factors inducing placental alterations in PCOS may be involved. Future work should explore other intrauterine variables with higher-powered studies.
The present study has several limitations. First, the sample size of women with PCOS and controls was relatively small which have increased sampling bias in the study. More samples are needed to confirm the present conclusions given the clinical heterogeneity of PCOS across patients and populations. Second, we were unable to conduct a long-term follow-up of the offspring. As our intention was to compare PCOS offspring with controls to provide clinicians insight into relevant neurobehavioural differences, neurological examination of newborns was performed in our study. Third, the Swan71 cell line likely originated from females and is now tetraploid with chromosomal and microsatellite instability, which may compromise the relevance of any in vitro findings. Further investigations are underway to clarify the mechanisms underlying FOS-driven modifications of neurotrophin genes. Fourth, the PCOS rat model used here reflects a restricted range of PCOS phenotypes in women. As F1 and F2 rats were randomly selected from the F0 generation, and six F0 PCOS rats delivered, the number of F1 and F2 rats for neurobehavioural testing was limited. A previous study reported that both mothers and fathers exposed to chronic social instability during adolescence and early adulthood transmit altered behaviours, including enhanced anxiety and social deficits, to their F1 offspring; however, only F1 fathers transmit all behaviours to their F2 and F3 daughters, indicating that maternal exposure may skip a generation and manifest only in F2 and F3 [87]. The low sample size of the study may have obscured differences in FAI. Future studies should follow offspring until F3 with larger sample sizes to investigate the transgenerational effects of PCOS. Finally, the role of placental FOS in neurotrophin regulation in the context of maternal impact on neurobehavioural alterations in PCOS offspring should be examined in the future.
In conclusion, our findings demonstrate that FOS-mediated regulation of neurotrophins in the placenta at least partly underpins the negative effects of maternal PCOS on neurobehavioural development of female offspring.
Data sharing
The raw data of RRBS and RNA-seq have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE154274 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154274).
Declaration of Competing Interests
The authors have no conflict of interest to declare.
Acknowledgments
This study was funded by the National Key R&D Program of China (2018YFC1004900 to F.Q. and F.W.) and the National Natural Science Foundation of China (81874480 to F.Q.; 81873837 to F.W.). The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
The corresponding author has full access to all the data in the study and has final responsibility for the decision to submit for publication.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102993.
Appendix. Supplementary materials
References
- 1.Norman RJ, Dewailly D, Legro RS, Hickey TE. Polycystic ovary syndrome. Lancet. 2007;370(9588):685–697. doi: 10.1016/S0140-6736(07)61345-2. [DOI] [PubMed] [Google Scholar]
- 2.Puttabyatappa M, Cardoso RC, Padmanabhan V. Effect of maternal PCOS and PCOS-like phenotype on the offspring's health. Mol Cell Endocrinol. 2016;435:29–39. doi: 10.1016/j.mce.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sha T, Wang X, Cheng W, Yan Y. A meta-analysis of pregnancy-related outcomes and complications in women with polycystic ovary syndrome undergoing IVF. Reprod Biomed Online. 2019 doi: 10.1016/j.rbmo.2019.03.203. [DOI] [PubMed] [Google Scholar]
- 4.Boomsma CM, Eijkemans MJ, Hughes EG, Visser GH, Fauser BC, Macklon NS. A meta-analysis of pregnancy outcomes in women with polycystic ovary syndrome. Hum Reprod Update. 2006;12(6):673–683. doi: 10.1093/humupd/dml036. [DOI] [PubMed] [Google Scholar]
- 5.Sir-Petermann T, Hitchsfeld C, Maliqueo M, Codner E, Echiburu B, Gazitua R. Birth weight in offspring of mothers with polycystic ovarian syndrome. Hum Reprod. 2005;20(8):2122–2126. doi: 10.1093/humrep/dei009. [DOI] [PubMed] [Google Scholar]
- 6.Gunning MN, Sir Petermann T, Crisosto N, van Rijn BB, de Wilde MA, Christ JP. Cardiometabolic health in offspring of women with PCOS compared to healthy controls: a systematic review and individual participant data meta-analysis. Hum Reprod. 2020;26(1):103–117. doi: 10.1093/humupd/dmz036. Update. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell GA, Sundaram R, Mumford SL, Park H, Mills J, Bell EM. Maternal polycystic ovarian syndrome and early offspring development. Hum Reprod. 2018;33(7):1307–1315. doi: 10.1093/humrep/dey087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsigianni M, Karageorgiou V, Lambrinoudaki I, Siristatidis C. Maternal polycystic ovarian syndrome in autism spectrum disorder: a systematic review and meta-analysis. Mol Psychiatry. 2019;24(12):1787–1797. doi: 10.1038/s41380-019-0398-0. [DOI] [PubMed] [Google Scholar]
- 9.Cesta CE, Oberg AS, Ibrahimson A, Yusuf I, Larsson H, Almqvist C. Maternal polycystic ovary syndrome and risk of neuropsychiatric disorders in offspring: prenatal androgen exposure or genetic confounding? Psychol Med. 2019:1–9. doi: 10.1017/S0033291719000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosidou K, Dalman C, Widman L, Arver S, Lee BK, Magnusson C. Maternal polycystic ovary syndrome and risk for attention-deficit/hyperactivity disorder in the offspring. Biol Psychiatry. 2017;82(9):651–659. doi: 10.1016/j.biopsych.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Kosidou K, Dalman C, Widman L, Arver S, Lee BK, Magnusson C. Maternal polycystic ovary syndrome and the risk of autism spectrum disorders in the offspring: a population-based nationwide study in Sweden. Mol Psychiatry. 2016;21(10):1441–1448. doi: 10.1038/mp.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y, Rosewell KL, Brännström M, Akin JW, Curry TE, Jo M. FOS, a critical downstream mediator of PGR and EGF signaling necessary for ovulatory prostaglandins in the human ovary. J Clin Endocrinol Meta. 2018;103(11):4241–4252. doi: 10.1210/jc.2017-02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preston GA, Lyon TT, Yin Y, Lang JE, Solomon G, Annab L. Induction of apoptosis by c-Fos protein. Mol Cell Biol. 1996;16(1):211–218. doi: 10.1128/mcb.16.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renaud SJ, Kubota K, Rumi MA, Soares MJ. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem. 2014;289(8):5025–5039. doi: 10.1074/jbc.M113.523746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp A, Tischmeyer W, Manahan-Vaughan D. Learning-facilitated long-term depression requires activation of the immediate early gene, c-fos, and is transcription dependent. Behav Brain Res. 2013;254:83–91. doi: 10.1016/j.bbr.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Zhang L, Jiao H, Zhang Q, Zhang D, Lou D. c-Fos facilitates the acquisition and extinction of cocaine-induced persistent changes. J Neurosci. 2006;26(51):13287–13296. doi: 10.1523/JNEUROSCI.3795-06.2006. the official journal of the Society for Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velazquez FN, Caputto BL, Boussin FD. c-Fos importance for brain development. Aging (Albany NY) 2015;7(12):1028–1029. doi: 10.18632/aging.100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ. Microbes & neurodevelopment–Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun. 2015;50:209–220. doi: 10.1016/j.bbi.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Jones MR, Chazenbalk G, Xu N, Chua AK, Eigler T, Mengesha E. Steroidogenic regulatory factor FOS is underexpressed in polycystic ovary syndrome (PCOS) adipose tissue and genetically associated with PCOS susceptibility. J Clin Endocrinol Metab. 2012;97(9):E1750–E17E7. doi: 10.1210/jc.2011-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Johansson J, Shao R, Mannerås L, Fernandez-Rodriguez J, Billig H. Hypothalamic neuroendocrine functions in rats with dihydrotestosterone-induced polycystic ovary syndrome: effects of low-frequency electro-acupuncture. PloS One. 2009;4(8):e6638. doi: 10.1371/journal.pone.0006638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beshay VE, Havelock JC, Sirianni R, Ye P, Suzuki T, Rainey WE. The mechanism for protein kinase C inhibition of androgen production and 17alpha-hydroxylase expression in a theca cell tumor model. J Clin Endocrinol Metab. 2007;92(12):4802–4809. doi: 10.1210/jc.2007-1394. [DOI] [PubMed] [Google Scholar]
- 22.Wickenheisser JK, Quinn PG, Nelson VL, Legro RS, Strauss JF, McAllister JM. Differential activity of the cytochrome P450 17alpha-hydroxylase and steroidogenic acute regulatory protein gene promoters in normal and polycystic ovary syndrome theca cells. J Clin Endocrinol Metab. 2000;85(6):2304–2311. doi: 10.1210/jcem.85.6.6631. [DOI] [PubMed] [Google Scholar]
- 23.Berry A, Bindocci E, Alleva E. NGF, brain and behavioural plasticity. Neural Plast. 2012;2012:1–9. doi: 10.1155/2012/784040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Autry AE, Monteggia LM. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol Rev. 2012;64(2):238–258. doi: 10.1124/pr.111.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chouthai NS, Sampers J, Desai N, Smith GM. Changes in neurotrophin levels in umbilical cord blood from infants with different gestational ages and clinical conditions. Pediatr Res. 2003;53(6):965–969. doi: 10.1203/01.PDR.0000061588.39652.26. [DOI] [PubMed] [Google Scholar]
- 26.Kodomari I, Wada E, Nakamura S, Wada K. Maternal supply of BDNF to mouse fetal brain through the placenta. Neurochem Int. 2009;54(2):95–98. doi: 10.1016/j.neuint.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore JH, Jarskog LF, Vadlamudi S. Maternal infection regulates BDNF and NGF expression in fetal and neonatal brain and maternal–fetal unit of the rat. J Neuroimmunol. 2003;138(1-2):49–55. doi: 10.1016/s0165-5728(03)00095-x. [DOI] [PubMed] [Google Scholar]
- 28.Prusa AR, Marton E, Rosner M, Bettelheim D, Lubec G, Pollack A. Neurogenic cells in human amniotic fluid. Am J Obstetr Gynecol. 2004;191(1):309–314. doi: 10.1016/j.ajog.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Lommatzsch M, Hornych K, Zingler C, Schuff-Werner P, Hoppner J, Virchow JC. Maternal serum concentrations of BDNF and depression in the perinatal period. Psychoneuroendocrinology. 2006;31(3):388–394. doi: 10.1016/j.psyneuen.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Uchida S, Inanaga Y, Kobayashi M, Hurukawa S, Araie M, Sakuragawa N. Neurotrophic function of conditioned medium from human amniotic epithelial cells. J Neurosci Res. 2000;62(4):585–590. doi: 10.1002/1097-4547(20001115)62:4<585::AID-JNR13>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Fan CG, Zhang QJ, Tang FW, Han ZB, Wang GS, Han ZC. Human umbilical cord blood cells express neurotrophic factors. Neurosci Lett. 2005;380(3):322–325. doi: 10.1016/j.neulet.2005.01.070. [DOI] [PubMed] [Google Scholar]
- 32.Toti P, Ciarmela P, Florio P, Volpi N, Occhini R, Petraglia F. Human placenta and fetal membranes express nerve growth factor mRNA and protein. J Endocrinol Invest. 2006;29(4):337–341. doi: 10.1007/BF03344105. [DOI] [PubMed] [Google Scholar]
- 33.Mayeur S, Silhol M, Moitrot E, Barbaux S, Breton C, Gabory A. Placental BDNF/TrkB signaling system is modulated by fetal growth disturbances in rat and human. Placenta. 2010;31(9):785–791. doi: 10.1016/j.placenta.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Fujita K, Tatsumi K, Kondoh E, Chigusa Y, Mogami H, Fujii T. Differential expression and the anti-apoptotic effect of human placental neurotrophins and their receptors. Placenta. 2011;32(10):737–744. doi: 10.1016/j.placenta.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Garcés MF, Sanchez E, Torres-Sierra AL, Ruíz-Parra AI, Angel-Müller E, Alzate JP. Brain-derived neurotrophic factor is expressed in rat and human placenta and its serum levels are similarly regulated throughout pregnancy in both species. Clin Endocrinol (Oxf) 2014;81(1):141–151. doi: 10.1111/cen.12391. [DOI] [PubMed] [Google Scholar]
- 36.Kertes DA, Bhatt SS, Kamin HS, Hughes DA, Rodney NC, Mulligan CJ. BNDF methylation in mothers and newborns is associated with maternal exposure to war trauma. Clin Epigenetics. 2017;9(1) doi: 10.1186/s13148-017-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toti P, Ciarmela P, Florio P, Volpi N, Occhini R, Petraglia F. Human placenta and fetal membranes express nerve growth factor mRNA and protein. J Endocrinol Invest. 2006;29(4):337–341. doi: 10.1007/BF03344105. [DOI] [PubMed] [Google Scholar]
- 38.Mayeur S, Lukaszewski MA, Breton C, Storme L, Vieau D, Lesage J. Do neurotrophins regulate the feto-placental development? Med Hypotheses. 2011;76(5):726–728. doi: 10.1016/j.mehy.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Briana DD, Malamitsi-Puchner A. Developmental origins of adult health and disease: The metabolic role of BDNF from early life to adulthood. Metab Clin Exp. 2018;81:45–51. doi: 10.1016/j.metabol.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Marx CE, Vance BJ, Jarskog LF, Chescheir NC, Gilmore JH. Nerve growth factor, brain-derived neurotrophic factor, and neurotrophin-3 levels in human amniotic fluid. Am J Obstetr Gynecol. 1999;181(5 Pt 1):1225–1230. doi: 10.1016/s0002-9378(99)70113-4. [DOI] [PubMed] [Google Scholar]
- 41.Zangeneh FZ, Naghizadeh MM, Bagheri M, Jafarabadi M. Are CRH & NGF as psychoneuroimmune regulators in women with polycystic ovary syndrome? Gynecol Endocrinol. 2017;33(3):227–233. doi: 10.1080/09513590.2016.1250152. [DOI] [PubMed] [Google Scholar]
- 42.Russo N, Russo M, Daino D, Bucci F, Pluchino N, Casarosa E. Polycystic ovary syndrome: brain-derived neurotrophic factor (BDNF) plasma and follicular fluid levels. Gynecol Endocrinol. 2012;28(4):241–244. doi: 10.3109/09513590.2011.613969. [DOI] [PubMed] [Google Scholar]
- 43.Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19(1):41–47. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 44.Dubowitz L, Mercuri E, Dubowitz V. An optimality score for the neurologic examination of the term newborn. J Pediatr. 1998;133(3):406–416. doi: 10.1016/s0022-3476(98)70279-3. [DOI] [PubMed] [Google Scholar]
- 45.Aban C, Martinez N, Carou C, Albamonte I, Toro A, Seyahian A. Endocannabinoids participate in placental apoptosis induced by hypoxia inducible factor-1. Apoptosis. 2016;21(10):1094–1105. doi: 10.1007/s10495-016-1274-x. [DOI] [PubMed] [Google Scholar]
- 46.Qu F, Wang F-F, Lu X-E, Dong M-Y, Sheng J-Z, Lv P-P. Altered aquaporin expression in women with polycystic ovary syndrome: hyperandrogenism in follicular fluid inhibits aquaporin-9 in granulosa cells through the phosphatidylinositol 3-kinase pathway. Hum Reprod (Oxford, Engl) 2010;25(6):1441–1450. doi: 10.1093/humrep/deq078. [DOI] [PubMed] [Google Scholar]
- 47.Qu F, Wang F-F, Yin R, Ding G-L, El-Prince M, Gao Q. A molecular mechanism underlying ovarian dysfunction of polycystic ovary syndrome: hyperandrogenism induces epigenetic alterations in the granulosa cells. J Mol Med. 2012;90(8):911–923. doi: 10.1007/s00109-012-0881-4. [DOI] [PubMed] [Google Scholar]
- 48.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen S, Park JW, Z-x Lu, Lin L, Henry MD, Wu YN. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111(51):E5593–EE601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R Package Version. 2010;2(0):2010. [Google Scholar]
- 53.STRING. Availabe online: http://string-db.org/ (accessed on 20 March 2017).
- 54.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–WW22. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comakli S, Kokturk M, Topal A, Ozkaraca M, Ceyhun SB. Immunofluorescence/fluorescence assessment of brain-derived neurotrophic factor, c-Fos activation, and apoptosis in the brain of zebrafish (Danio rerio) larvae exposed to glufosinate. Neurotoxicology. 2018;69:60–67. doi: 10.1016/j.neuro.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Tamura N, Kurabayashi T, Nagata H, Matsushita H, Yahata T, Tanaka K. Effects of testosterone on cancellous bone, marrow adipocytes, and ovarian phenotype in a young female rat model of polycystic ovary syndrome. Fertil Steril. 2005;84:1277–1284. doi: 10.1016/j.fertnstert.2005.06.017. Suppl 2. [DOI] [PubMed] [Google Scholar]
- 58.Marcondes FK, Bianchi FJ, Tanno AP. Determination of the estrous cycle phases of rats: some helpful considerations. Braz J Biol = Revista brasleira de biologia. 2002;62(4a):609–614. doi: 10.1590/s1519-69842002000400008. [DOI] [PubMed] [Google Scholar]
- 59.Dong H, Zhong Z, Chen W, Wu X, Zhang Q, Huang G. Effect of acupuncture on endometrial angiogenesis and uterus dendritic cells in COH Rats during peri-implantation period. Evid Based Compl Alternat Med eCAM. 2017;2017 doi: 10.1155/2017/3647080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flöck A, Weber SK, Ferrari N, Fietz C, Graf C, Fimmers R. Determinants of brain-derived neurotrophic factor (BDNF) in umbilical cord and maternal serum. Psychoneuroendocrinology. 2016;63:191–197. doi: 10.1016/j.psyneuen.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Butler MC, Long CN, Kinkade JA, Green MT, Martin RE, Marshall BL. Endocrine disruption of gene expression and microRNA profiles in hippocampus and hypothalamus of California mice: Association of gene expression changes with behavioural outcomes. J Neuroendocrinol. 2020:e12847. doi: 10.1111/jne.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mackenzie RM, Sandrim VC, Carty DM, McClure JD, Freeman DJ, Dominiczak AF. Endothelial FOS expression and pre-eclampsia. BJOG. 2012;119(13):1564–1571. doi: 10.1111/1471-0528.12016. [DOI] [PubMed] [Google Scholar]
- 63.Marzioni D, Todros T, Cardaropoli S, Rolfo A, Lorenzi T, Ciarmela P. Activating protein-1 family of transcription factors in the human placenta complicated by preeclampsia with and without fetal growth restriction. Placenta. 2010;31(10):919–927. doi: 10.1016/j.placenta.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Q, Yang Y, Cui X, Zhang D, Liu S, Yan Q. AP1 mediates uPA/uPAR induced FUT4 expression and trophoblast invasion. J Cell Biochem. 2018;119(8):6442–6451. doi: 10.1002/jcb.26648. [DOI] [PubMed] [Google Scholar]
- 65.Kawamura K, Kawamura N, Kumazawa Y, Kumagai J, Fujimoto T, Tanaka T. Brain-derived neurotrophic factor/tyrosine kinase B signaling regulates human trophoblast growth in an in vivo animal model of ectopic pregnancy. Endocrinology. 2011;152(3):1090–1100. doi: 10.1210/en.2010-1124. [DOI] [PubMed] [Google Scholar]
- 66.Yamanaka M, Tsuchida A, Nakagawa T, Nonomura T, Ono-Kishino M, Sugaru E. Brain-derived neurotrophic factor enhances glucose utilization in peripheral tissues of diabetic mice. Diabetes Obes Metab. 2007;9(1):59–64. doi: 10.1111/j.1463-1326.2006.00572.x. [DOI] [PubMed] [Google Scholar]
- 67.Prince CS, Maloyan A, Myatt L. Maternal obesity alters brain derived neurotrophic factor (BDNF) signaling in the placenta in a sexually dimorphic manner. Placenta. 2017;49:55–63. doi: 10.1016/j.placenta.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saenen ND, Plusquin M, Bijnens E, Janssen BG, Gyselaers W, Cox B. In utero fine particle air pollution and placental expression of genes in the brain-derived neurotrophic factor signaling pathway: an ENVIRONAGE birth cohort study. Environ Health Perspect. 2015;123(8):834–840. doi: 10.1289/ehp.1408549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhobale M. Neurotrophins: Role in adverse pregnancy outcome. Int J Dev Neurosci. 2014:37. doi: 10.1016/j.ijdevneu.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 70.Dhobale MV, Pisal HR, Mehendale SS, Joshi SR. Differential expression of human placental neurotrophic factors in preterm and term deliveries. Int J Dev Neurosci. 2013;31(8):719–723. doi: 10.1016/j.ijdevneu.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Tometten M, Blois S, Arck PC. Nerve growth factor in reproductive biology: link between the immune, endocrine and nervous system? Chem Immunol Allergy. 2005;89:135–148. doi: 10.1159/000087962. [DOI] [PubMed] [Google Scholar]
- 72.Peng B, Alotaibi FT, Sediqi S, Bedaiwy MA, Yong PJ. Role of interleukin-1beta in nerve growth factor expression, neurogenesis and deep dyspareunia in endometriosis. Hum Reprod. 2020 doi: 10.1093/humrep/deaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuvikene J, Pruunsild P, Orav E, Esvald E-E, Timmusk T. AP-1 transcription factors mediate BDNF-positive feedback loop in cortical neurons. J Neurosci. 2016;36(4):1290–1305. doi: 10.1523/JNEUROSCI.3360-15.2016. the official journal of the Society for Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palomba S, Russo T, Falbo A, Di Cello A, Tolino A, Tucci L. Macroscopic and microscopic findings of the placenta in women with polycystic ovary syndrome. Hum Reprod (Oxford, Engl) 2013;28(10):2838–2847. doi: 10.1093/humrep/det250. [DOI] [PubMed] [Google Scholar]
- 75.Koster MPH, de Wilde MA, Veltman-Verhulst SM, Houben ML, Nikkels PGJ, van Rijn BB. Placental characteristics in women with polycystic ovary syndrome. Hum Reprod (Oxford, Engl) 2015;30(12):2829–2837. doi: 10.1093/humrep/dev265. [DOI] [PubMed] [Google Scholar]
- 76.Shen D, Wang F, Jiang Z, Qu F. [Long-term effects of polycystic ovary syndrome on the offspring] Zhejiang da xue xue bao Yi xue ban = J Zhejiang Univ Med Sci. 2017;46(3):300–304. doi: 10.3785/j.issn.1008-9292.2017.06.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Zhang Q, Zhang F, Wang F, Qu F. Effects of prenatal androgen exposure on PCOS offspring. J Huazhong Univ Sci Technol(Health Sciences) 2019;48(6):742–746. [Google Scholar]
- 78.Vanky E, Engen Hanem LG, Abbott DH. Children born to women with polycystic ovary syndrome-short- and long-term impacts on health and development. Fertil Steril. 2019;111(6):1065–1075. doi: 10.1016/j.fertnstert.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 79.Guerrero-Bosagna C, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol Cell Endocrinol. 2012;354(1-2):3–8. doi: 10.1016/j.mce.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PloS One. 2012;7(2):e31901. doi: 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova MI, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PloS One. 2012;7(5):e36129. doi: 10.1371/journal.pone.0036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Risal S, Pei Y, Lu H, Manti M, Fornes R, Pui HP. Prenatal androgen exposure and transgenerational susceptibility to polycystic ovary syndrome. Nat Med. 2019;25(12):1894–1904. doi: 10.1038/s41591-019-0666-1. [DOI] [PubMed] [Google Scholar]
- 83.Zhang HL, Yi M, Li D, Li R, Zhao Y, Qiao J. Transgenerational Inheritance of Reproductive and Metabolic Phenotypes in PCOS Rats. Front Endocrinol. 2020;11:144. doi: 10.3389/fendo.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359(1):61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sir-Petermann T, Maliqueo M, Angel B, Lara HE, Perez-Bravo F, Recabarren SE. Maternal serum androgens in pregnant women with polycystic ovarian syndrome: possible implications in prenatal androgenization. Hum Reprod. 2002;17(10):2573–2579. doi: 10.1093/humrep/17.10.2573. [DOI] [PubMed] [Google Scholar]
- 86.Ramirez JSB, Graham AM, Thompson JR, Zhu JY, Sturgeon D, Bagley JL. Maternal interleukin-6 is associated with macaque offspring amygdala development and behaviour. Cereb Cortex. 2020;30(3):1573–1585. doi: 10.1093/cercor/bhz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saavedra-Rodríguez L, Feig LA. Chronic social instability induces anxiety and defective social interactions across generations. Biol Psychiatry. 2013;73(1):44–53. doi: 10.1016/j.biopsych.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.