Abstract
Background
Advances in prostate cancer (PCa) lag behind other tumor types partly due to the paucity of models reflecting key milestones in PCa progression.
Objective
To develop clinically relevant PCa models.
Design
Since 1996 we have generated clinically annotated patient-derived xenografts (PDXs) (the MDA PCa PDX series) linked to specific phenotypes reflecting all aspects of clinical PCa.
Results
We studied two cell line–derived xenografts and the first 80 PDXs derived from 47 human PCa donors. Of these, 47 PDXs derived from 22 donors are working models and can be expanded either as cell lines (MDA PCa 2a and 2b) or PDXs. The histopathologic, genomic, and molecular characteristics (AR, ERG, and PTEN loss) maintain fidelity with the human tumor and correlate with published findings. PDX growth response to mouse castration and targeted therapy illustrate their clinical utility. Comparative genomic hybridization and sequencing show significant differences in oncogenic pathways in pairs of PDXs derived from different areas of the same tumor. We also identified a recurrent focal deletion in an area that includes the SPOPL gene in PDXs derived from 7 human donors out of 28 studied (25%). SPOPL is a SPOP paralog, and SPOP mutations define a molecular subclass of PCa. SPOPL deletions are found in 7% of TCGA PCas, which suggests that our cohort is a reliable platform for targeted drug development.
Conclusions
The MDA PCa PDX series is a dynamic resource that captures the molecular landscape of PCas progressing under novel treatments and enables optimization of PCa-specific, marker-driven therapy.
Keywords: Patient-derived xenografts, metastatic prostate cancer, gene fusion, preclinical studies, comparative genomic hybridization
INTRODUCTION
Metastatic prostate cancer (PCa) that progresses after androgen ablation therapy (i.e., castration-resistant PCa [CRPC]) remains incurable. PCa consist of clinical subsets including the typical prostate adenocarcinoma with a predictable pattern of progression, as well as other subsets with atypical clinical behavior similar to that of prostatic small cell neuroendocrine carcinoma (aggressive variant PCa [AVPC]), which accounts for about 40% of lethal PCa (1, 2). PCa is also heterogeneous at the morphological and molecular level (3). Thus, responses to targeted therapies differ between subpopulations of PCa patients, and it is essential to have a spectrum of biological models that reflect the diverse clinical, morphological, and molecular PCa phenotypes to further research and therapy development.
PCa research has historically suffered from a lack of patient-derived models. Efforts to develop PCa patient-derived xenografts (PDXs) have been undertaken in several institutions (4). However, few PCa PDX banks with substantial number of models have been established. Some contain PDXs derived from autopsy samples (the LuCaP series) with no information on tumor progression after the samples have been acquired. Others are mainly derived from primary tumors (the Living Tumor Laboratory [LTL] series and the PC series developed in Erasmus Medical Center, Rotterdam, The Netherlands) (4). Therefore, despite these efforts, we still lack a collection of clinically annotated PCa PDXs that reflects the full spectrum of potentially lethal disease, namely therapy-naïve and therapy-resistant PCas reflecting clinical and morphological variants derived from primary sites as well as metastases, and can be related to the tumor donor’s progression to therapy, which is monitored by expert genitourinary (GU) oncologists. We have approached this challenge by establishing PDXs using tissue specimens taken from patients with potentially lethal PCa undergoing surgical resection or biopsy of primary tumors and/or metastatic sites demonstrating clinical progression. This is an ongoing program that provides a diverse repository of well-annotated tissue samples and PDXs that can be linked prospectively with specific stages and states of PCa progression and also reflects the clinical and molecular evolution of therapy resistant PCa.
Here, we report the morphological and molecular characterization of two cell line–derived xenografts and the first 80 PDXs derived from 47 human PCa donors developed in our program. Of these, 47 PDXs derived from 22 donors are working models and can be expanded either as cell lines (MDA PCa 2a and 2b) or PDXs. The present study constitutes the foundation for the development and use of PDXs for precision oncology. Furthermore, as this is a dynamic repository, we also established PDXs from human PCa that are not described in this report because they are currently being characterized and will be the subject of a follow up publication. These new PDXs include those derived from PCas progressing on new hormonal agents, enzalutamide and abiraterone. Enquiries about their availability to the scientific community can be sent to the corresponding author of this publication.
Materials and Methods
The PCa PDX Program at MD Anderson Cancer Center
To efficiently process all aspects of PDX development, the MDA PCa PDX program operates within a highly integrated network of physicians, scientists, laboratory staff, and resources within the Tissue Biospecimen and Pathology Resource at MD Anderson. These individuals include urologists, oncologists, interventional radiologists, and pathologists, as well as staff who provide regulatory compliance support for archived biospecimen requests, consent validation, material transfer agreements, data management, and sample distribution.
PDX development
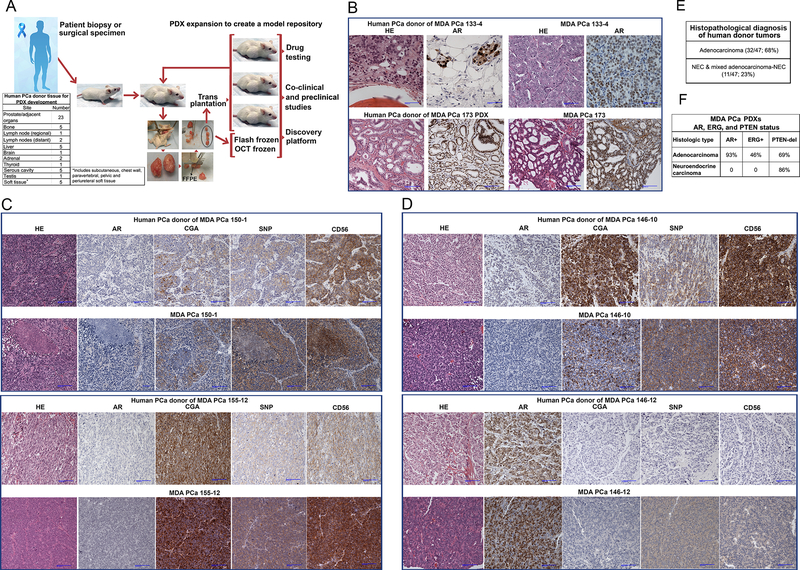
In our program, PCa tissue samples used for PDX development were derived from therapeutic or diagnostic procedures, namely, radical prostatectomies, orthopedic and neurosurgical procedures to palliate complications, and biopsies of metastatic lesions. Written informed consent was obtained from patients before sample acquisition, and all samples were processed according to a protocol approved by the Institutional Review Board of the University of Texas MD Anderson Cancer Center. The studies were conducted in accordance with the Belmont Report and the US Common Rule. On collection, the specimens were placed in cold (4°C), sterile α-MEM (GIBCO; Invitrogen), and small pieces were then implanted into subcutaneous pockets of 6- to 8-week-old male CB17 SCID mice (Charles River Laboratories) (Fig 1A). Tissue adjacent to the implanted samples (mirror image) were formalin-fixed and paraffin-embedded (FFPE), and tissue sections were used for quality control. Mice were monitored weekly for tumor growth. Once the initial implanted tumor grew in the mouse (passage 0 [P0]), it was harvested and sequentially passaged to five mice (Fig 1A). At each mouse-to-mouse passage, representative samples of each PDX were frozen in a DMSO solution for later implantation and PDX expansion, FFPE, flash-frozen, and frozen in OCT medium for histopathological and molecular studies (Fig 1A).
Fig 1.
(A) Schematic diagram outlining the strategy for MDA PCa PDX development, expansion, and storage. PCa tissue samples are implanted subcutaneously into mice. Once the tumor grows, PDXs are expanded in mice to passage 5. At each passage, representative samples of PDX tissue are stored in various forms (e.g., fresh-frozen, formalin-fixed and paraffin-embedded) to create a PDX model repository. The table “Human PCa Donor for PDX Development” indicates the origin of the samples that developed into the MDA PCa PDXs used in this work. (B) Human donor adenocarcinomas and corresponding PDXs have the same morphological and immunohistochemical profile. Representative photomicrographs of HE sections and immunohistochemical stains for AR. PCa donor tumor of MDA PCa 133 was a bone metastasis and PCa donor tumor of MDA PCa 173 was a primary PCa. (C) Human donor neuroendocrine carcinomas and corresponding PDXs have the same morphological and immunohistochemical profile. Representative photomicrographs of HE sections and immunohistochemical stains for AR, and markers of neuroendocrine differentiation CGA, SNP, and CD56. PCa donor tumor of MDA PCa 150 was a bone metastasis, and PCa donor tumor of MDA PCa 155 was a primary PCa. (D) PDXs derived from a mixed adenocarcinoma/neuroendocrine human PCa. Representative photomicrographs showing the adenocarcinoma and neuroendocrine components of human PCa reflected in different PDXs. PCa donor tumor of MDA PCa 146 was a primary PCa. (E) Morphological distribution (adenocarcinoma and neuroendocrine carcinoma) of MDA PCa PDXs reported in this work. (F) AR, ERG, and PTEN status of MDA PCa PDXs reported in this work. HE, hematoxylin and eosin; AR, androgen receptor; CGA, chromogranin; SNP, synaptophysin.
To achieve a reliable and reproducible method for PDX development, we established a standard operating procedure (SOP) with an optimized process for tissue samples, including shortening the time from sample acquisition to implantation in mice and selecting tissue samples with the highest percentage of viable cells. We do not maintain PDXs in mice unless they are being utilized by investigators to perform experiments. Historically, once initial growth was observed and the PDX underwent five or more serial passages in mice, the likelihood that the model would continue to propagate in mice is high (80%−90%). Also, in our experience most PDXs will regrow after cryopreservation either in the first or second attempt, provided that the tissue used for cryopreservation was obtained from a PDX that reached passage 5. Every time we harvested tissue for cryopreservation, a mirror tissue sample for quality control was obtained. The viability of tissue at the time of cryopreservation is essential for regrowth. One caveat is that we have not tested the viability of cryopreserved tissue for more than one year as we have consistently generated new vials at shorter intervals.
All animal experiments were conducted in accordance with accepted standards of animal care and were approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center.
PDX growth in intact and castrated mice
All studies were performed using 6- to 8-week-old male CB17 SCID mice. Twenty-three mice were subcutaneously implanted with MDA PCa 183-A. Nine of these mice were monitored for tumor growth over time (intact mice). Seven were castrated when tumor volume was equal or higher than 500 mm3 (castrated mice). The remaining 7 mice were monitored for tumor volume before and ten days after castration (tumor volume before and after castration).
Twenty-one mice were subcutaneously implanted with MDA PCa 180–30. Seven of these mice were monitored for tumor growth (intact mice), and fourteen were castrated when tumor volume was equal or higher than 500 mm3. Values were used for the plots of castrated mice and tumor volume before and after castration.
PDX growth intrabone in mice treated with vehicle or FGFR inhibitor
Twenty-three male CB17 SCID mice were injected with cells derived from MDA PCa 118b into the distal ends of the femurs according to published protocols (5). Twelve days after cell injection, ten mice were treated with vehicle and thirteen with erdafitinib (Janssen Pharmaceuticals) by oral gavage. Using the same protocol, twenty male CB17 SCID mice were injected with cells derived from MDA PCa 183-A into the distal ends of their femurs. Twenty-three days after cell injection, ten mice were treated with vehicle and ten with erdafitinib by oral gavage. After three weeks of treatment of MDA PCa 118b–bearing mice and five weeks of treatment of MDA PCa 183-A–bearing mice, tumor volume was measured from serial sagittal MR images in vehicle- and erdafitinib-treated mice as previously reported (6).
Statistical Methods
For the studies on tumor size response to mice castration, the change of tumor volume over time was explored using spaghetti plots. Linear mixed models were fit to assess the change of tumor volume over time in all groups, while taking into account the intra-mouse correlations. All statistical analyses were performed using SAS and Splus.
Two-sample t tests were used to study tumor volume response to erdafitinib treatment.
RESULTS
The PCa PDX Program at MD Anderson Cancer Center
In 1996, we initiated efforts to develop patient-derived models of PCa and established two cell lines (MDA PCa 2a and MDA PCa 2b) (7). Subsequent attempts (n = 25) at establishing in vitro models were unsuccessful. These attempts include PCa samples obtained at radical prostatectomy and samples from different metastatic sites (bone, lymph node, skin, and ascites). The methods used during these attempts have been previously published (8). In the cases derived from primary PCa, we saw initial cell growth in most cases, but normal prostatic epithelial cell overgrowth was the most common outcome. In the cases derived from metastases, we frequently obtained short-term cultures, but the cells eventually stopped growing or underwent cell death. Methods and alternatives used during these attempts are discussed in a previous publication (8).
From 1996 to April 2010, we processed PCa tissue samples from 163 patients for PDX development, including samples from different areas of the same tumor in order to understand tumor heterogeneity. In total, during that period, we established 80 PDXs derived from 47 human PCa donors; of these, 47 PDXs derived from 22 donors are working models and can be expanded either as cell lines or PDXs (Table 1, Table S1, and Supplementary Results). These PDXs are named: MDA PCa followed by a number unique to the donor tumor, tumor site, and procedure date. Currently, the success rate for PDX development is 30% to 40%, irrespective of the site of origin of the tissue. The variability probably depends on the amount of viable tissue submitted for PDX development. It takes between 3 months and 3 years (depending on the rate of growth) for a tissue sample implanted in mice to be passed through 5 mice sequentially. The PDXs developed are derived from primary PCas or areas of direct extension to adjacent organs (bladder, rectum) and from metastases, and include therapy-naïve and -resistant adenocarcinomas, as well as clinical and histopathological subtypes (Fig 1A, Tables 1 and S1). For brevity, from now on the term ‘PDXs’ will be inclusive of MDA PCa 2a-T and 2b-T xenografts (T indicates a cell line–derived xenograft).
Table 1.
Morphological and IHC findings in human donor tumors and PDXs. IHC for neuroendocrine markers was performed only on AR– cases with neuroendocrine morphology
| Human PCa of Origin | PDX or Cell Line Derived Xenograft | ||||||
|---|---|---|---|---|---|---|---|
| Specimen number | Tumor Site | Diagnosis | Gleason Grades | Clinical State | Treatment history at the time of tissue acquisition | PDX / Cell line name | Diagnosis@ |
| 2 | Bone | Adenocarcinoma | N/A | CRPC | Bilateral orchiectomy; suramine+doxorubicin; vertebral resection | MDA PCa 2a-T# | Adenocarcinoma |
| MDA PCa 2b-T# | Adenocarcinoma | ||||||
| 117 | Prostate | Adenocarcinoma | 4+5 | CRPC | Leuprolide+bicalutamide; autologous dendritic vaccine; gefitinib; docetaxel | MDA PCa 117–9 | Adenocarcinoma |
| 118 | Bone | Adenocarcinoma (SNP−, CGA−) | N/A | CRPC | Leuprolide+bicalutamide; paclitaxel+carboplatin+ DES; skull/shoulder radiation | MDA PCa-118b | Adenocarcinoma (AR−, SNP−, CGA−, CD56+) |
| 133 | Bone | Adenocarcinoma | N/A | CRPC | Local radiation+leuprolide +bicalutamide +ketoconazole; docetaxel+estramustine; thalidomide+paclitaxel +estramustine; docetaxel+imatinib; ifosfamide+mitoxantrone+gemcitabine; etoposide+ mitoxantrone; gefitinib+ mitoxantrone; vertebral resection | MDA PCa 133–4 | Adenocarcinoma |
| 144 | Prostate, Bladder, Rectum | Mixed adenocarcinoma (AR+) and neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56 +) | N/A | CRPC | Local radiation+hormonal therapy; leuprolide; docetaxel+carboplatin; etoposide+cisplatin | MDA PCa 144–4 | Neuroendocrine carcinoma (AR−, SNP+, CGA +) |
| MDA PCa 144–6 | Neuroendocrine carcinoma (AR−, SNP+, CGA +) | ||||||
| MDA PCa 144–11 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) | ||||||
| MDA PCa 144–13 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) | ||||||
| MDA PCa 144–20 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) | ||||||
| 146 | Bladder | Mixed adenocarcinoma (AR+) and neuroendocrine carcinoma (SNP+, CGA+, CD56+) | 5+4 | CRPC | Brachytherapy+ Leuprolide+ bicalutamide; docetaxel | MDA PCa 146–10 | Neuroendocrine carcinoma (AR−, SNP+, CGA+) |
| MDA PCa 146–12 | Adenocarcinoma (AR+, SNP−, CGA−) | ||||||
| MDA PCa 146–17 | Neuroendocrine carcinoma (AR−, SNP+, CGA+) | ||||||
| MDA PCa 146–20 | Mixed adenocarcinoma and neuroendocrine carcinoma (AR−, SNP+, CGA+) | ||||||
| 149 | Bladder | Adenocarcinoma | 4+5 | CRPC | RPx+leuprolide+local radiation; bicalutamide; docetaxel | MDA PCa 149–1 | Adenocarcinoma |
| 150 | Bone | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) | N/A | CRPC | RPx+leuprolide+CCI-779 | MDA PCa 150–1 | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) |
| MDA PCa 150–3 | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) | ||||||
| MDA PCa 150–5 | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) | ||||||
| MDA PCa 150–7 | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) | ||||||
| MDA PCa 150–10 | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) | ||||||
| 152 | Brain | Adenocarcinoma (ductal) | N/A | CRPC | RPx+leuprolide+local radiation | MDA PCa 152–1 | Adenocarcinoma |
| MDA PCa 152–5 | Adenocarcinoma | ||||||
| 153 | Thyroid Gland | Adenocarcinoma | N/A | CRPC | Leuprolide+ bicalutamide; goserelin; DES; Immunotherapy (NY-ESO-1 plasmid DNA vaccine) | MDA PCa 153–7 | Adenocarcinoma |
| MDA PCa 153–14 | Adenocarcinoma | ||||||
| 155 | Prostate, Bladder | Neuroendocrine carcinoma (AR−, SNP weak, CGA+, CD56+) | N/A | CRPC | Leuprolide+bicalutamide +docetacel+carboplatin; cisplatin+etoposide | MDA PCa 155–2 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) |
| MDA PCa 155–9 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) | ||||||
| MDA PCa 155–12 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) | ||||||
| MDA PCa 155–16 | Neuroendocrine carcinoma (AR−, SNP+, CGA+, CD56+) | ||||||
| 166 | Bladder | Adenocarcinoma | 5+4 | CRPC | Ketoconazol+leuprolide+ bicalutamide+carboplatin+docetaxel | MDA PCa 166–1 | Adenocarcinoma |
| 170 | Prostate | Adenocarcinoma | 5+4 | CRPC | Leuprolide+bicalutamide+docetaxel;
cyclophosphamide+ vincristine |
MDA PCa 170–1 | Adenocarcinoma |
| MDA PCa 170–4 | Adenocarcinoma | ||||||
| 173 | Prostate | Adenocarcinoma | 3+4 | Naïve | Therapy-naïve | MDA PCa 173–2 | Adenocarcinoma |
| 175 | Testis | Adenocarcinoma | N/A | CRPC | Leuprolide+bicalutamide; ketoconazole; paclitaxel +DES; samarium+doxorubicin) | MDA PCa 175–2 | Adenocarcinoma |
| MDA PCa 175–6 | Adenocarcinoma | ||||||
| MDA PCa 175–10 | Adenocarcinoma | ||||||
| 177 | Prostate | Poorly differentiated
carcinoma (SNP−, CGA−) |
N/A | CRPC | Goserelin+bicalutamide | MDA PCa 177-B | Poorly differentiated carcinoma with morphology suggestive of neuroendocrine features (AR−, SNP−, CGA−, CD56+) |
| 178 | Prostate | Adenocarcinoma | 4+5 | Hormone Responsive | Leuprolide+bicalutamide | MDA PCa 178–11 | Adenocarcinoma |
| 180 | Prostate, Bladder | Adenocarcinoma | 5+4 | CRPC | Leuprolide+bicalutamide +docetaxel+carboplatin | MDA PCa 180–11 | Adenocarcinoma |
| MDA PCa 180–14 | Adenocarcinoma | ||||||
| MDA PCa 180–18 | Adenocarcinoma | ||||||
| MDA PCa 180–21 | Adenocarcinoma | ||||||
| MDA PCa 180–30 | Adenocarcinoma | ||||||
| 181 | Bladder | Mixed adenocarcinoma (NE-) and neuroendocrine carcinoma (SNP+, CGA+, CD56+) | N/A | CRPC | Brachytherapy; goserelin; paclitaxel+carboplatin; etoposide+cisplatin; paclitaxel+ doxorubicin) | MDA PCa 181 | Neuroendocrine carcinoma (AR−, SNP weak, CGA−, CD56+) |
| 182 | Prostate | Adenocarcinoma* | 5+4 | CRPC | Local radiation; leuprolide+bicalutamide +DES; docetaxel, carboplatin, etoposide+ cisplatin.) | MDA PCa 182–7 | Adenocarcinoma |
| MDA PCa 182–11 | Adenocarcinoma | ||||||
| 183 | Bone | Adenocarcinoma | N/A | Naïve | Therapy-Naïve | MDA PCa 183-A | Adenocarcinoma |
| 188 | Bladder | Adenocarcinoma | 5+4 | CRPC | RPx; leuprolide+ bicalutamide+docetaxel; carboplatin+paclitaxel | MDA PCa 188 | Adenocarcinoma |
Diagnosis per report, no material was available to review
Indicates cell line–derived xenograft (for simplicity, when we say PDXs this will be inclusive of MDA PCa 2a-T and 2b-T)
Morphology of PDX was the same at early (passage 1–3) and late passages (passage 5–6); N/A, not applicable; IHC, immunohistochemistry; CRPC, castration resistant prostate cancer; AR, androgen receptor; SNP, synaptophysin; CGA, chromogranin; DES, diethylstilbestrol; RPx: radical prostatectomy.
As previously mentioned, this is a dynamic repository and, to date, in addition to the PDXs described in this report, we have also established PDXs from 52 PCa patients, which are currently being characterized. These PDXs derived from primary sites, bone or soft tissue metastases include those derived from treatment naïve PCas and PCas progressing on first and second generation androgen deprivation therapy, chemotherapy and other therapies.
Morphological and immunohistochemical features of human donor tumors and PDXs
Early and late passage PDXs (passage 1–3 and 5–6, respectively) retain the morphology of the human PCa donor (Table 1). PDXs also have the same immunohistochemical (IHC) profile (androgen receptor [AR], synaptophysin [SNP], chromogranin A [CGA], or CD56 [NCAM]) as their human PCa donor and are either adenocarcinomas (Fig 1B) or neuroendocrine carcinomas (NEC), which includes small cell morphology (Fig 1C). PDXs established from mixed adenocarcinoma and NEC, reflect one or both morphological components present in the human PCa (Fig 1D and S1, Supplementary Results). NEC PDXs are all AR-negative and positive for one or more neuroendocrine markers (SNP, CGA, or CD56) (9) (Fig 1C, Table 1, Supplementary Results). Adenocarcinomas with negative or low expression of AR and negative expression of SNP and CGA (e.g., MDA PCa 83 and MDA PCa 118b) fit the recently published definition of double-negative PCas (10).
Of the 47 donor tumors used to establish the PDXs, 32 are adenocarcinomas (68%), and 11 are NEC or mixed adenocarcinoma and NEC (23%) (Fig 1E and Table 1). This distribution recapitulates the morphological landscape of potentially lethal human PCa (1, 2). In the other 4 cases (9%), either there is no material available for review (e.g., serous fluids), there are no malignant cells based on the pathology report, or they are unusual PCa morphologies (Table 1 and S1, Supplementary Results).
AR, ERG, and PTEN status in PDXs and a subgroup of 15 PDX–human donor tumor pairs
Previous reports indicate that 50% to 60% of human PCas have recurrent rearrangements involving ERG, ETV1, ETV4, or ETV5. AR signaling status, a critical determinant of PCa behavior, is a therapeutic target for PCa, and aberrant ERG expression cooperates with PTEN deletions to promote PCa progression (11). IHC and fluorescent in situ hybridization (FISH) analyses of these genes demonstrated that adenocarcinoma PDXs derived from 27 of the 29 human donors available for study are AR-positive (93%), and ERG expression is positive in 12 of 26 of the AR-positive samples available for study (46%) (Fig 1F). MDA PCa 2a-T and 2b-T and MDA PCa 177 have ETV1 rearrangement as confirmed by FISH and RNA-ISH (12) (Fig S2). PDXs derived from NECs, which do not express AR, also do not express ERG in all but one case (Table S2). PDXs derived from 18 of 26 adenocarcinomas (69%) available for evaluation have homo- or heterozygous PTEN deletion (Fig 1F, Table S2, Supplementary Results). PDXs derived from 6 of 7 NECs (86%) have homo- or heterozygous PTEN deletion (Fig 1F). PTEN status is variable in PDXs derived from mixed adenocarcinoma and NEC (Table S1). In summary, the morphological distribution and molecular features of the PDXs established are consistent with the reports on human PCa in the general population (13).
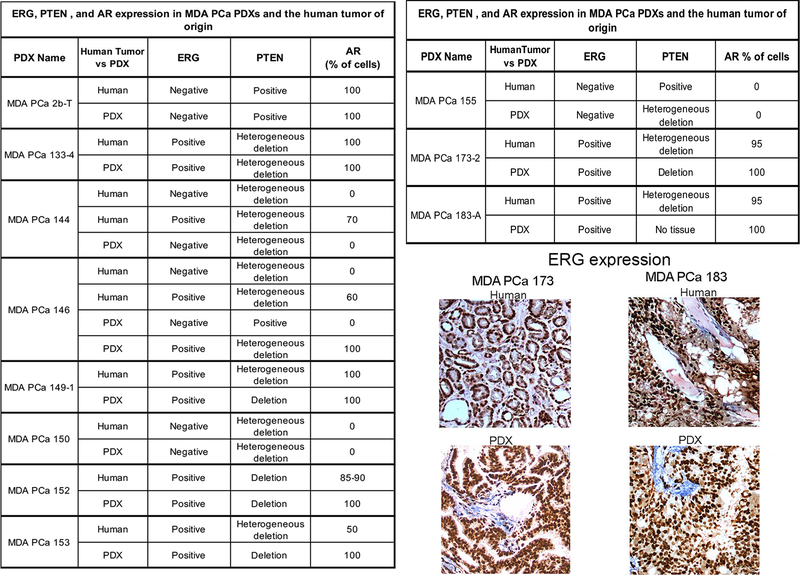
Finally, to further understand how AR, ERG and PTEN status in PCa PDXs recapitulate human PCa donors, we studied 15 PDX–human donor pairs and found concordance in most samples. Fig 2 shows results of the 11 pairs that are not archived, more details are in Supplementary Results.
Fig 2.
Table outlines the status of ERG, PTEN, and AR in 11 pairs of human PCa (Tumor) and corresponding PDX (PDX). Representative photomicrographs show examples of ERG staining in the human prostate donor tumor and PDX in two different pairs. Note that in those cases in which multiple PDXs were generated from different areas of the same tumor (e.g., MDA PCa 144–4, −13, etc; and MDA PCa 146–10; −12, etc) we did not include a suffix that uniquely identifies each individual PDX because this is only an illustration of the different phenotypes found. Table S2 lists the specific phenotype identified in each of these unique PDXs.
MDA PCa PDX growth in intact and castrated male mice
Androgen ablation is the standard first-line treatment for metastatic PCa. The response duration is highly variable and second-generation androgen ablation therapeutics (abiraterone and enzalutamide) have significant activity in patients with CRPC. Nevertheless, the emergence of resistance remains inevitable in most cases. Thus, to develop effective treatment strategies aimed at improving patient management, there is an urgent need to further understand the mechanisms that account for treatment response and resistance.
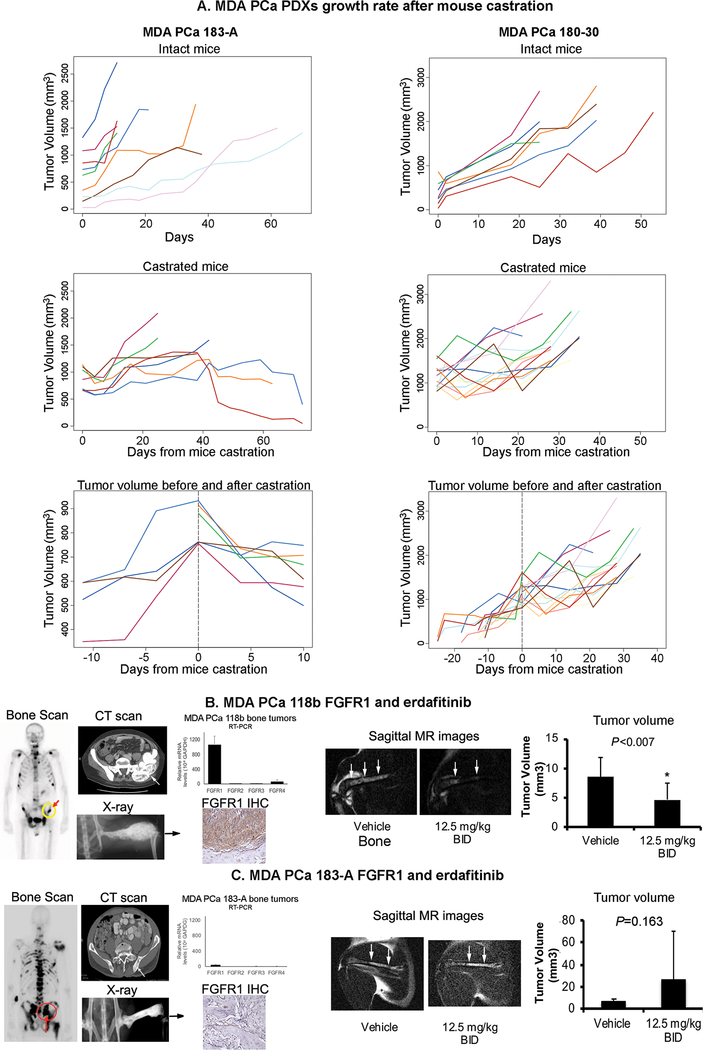
To assess the potential utility of our models in furthering our understanding of the mechanisms underlying CRPC progression and to develop effective therapies, we studied the effect of surgical castration of PDX-bearing mice on the growth of the tumors. Testosterone levels in intact male mice were shown to mimic standard androgen ablation, and in castrated mice mimic abiraterone-treated patients (14). Castration was performed as previously published (7). For these studies we selected two models, MDA PCa 183-A, derived from a treatment-naïve PCa, and MDA PCa 180–30, derived from a PCa that progressed on androgen deprivation and chemotherapy.
Change of tumor volume over time in the intact and castrated groups was assessed by a linear mixed model, while considering the intra-mouse correlations. We found a statistically significant difference between the slopes of the tumor volumes in subcutaneous MDA PCa 183-A growing in intact and castrated mice (P < 0.0001) (Fig 3A). Accordingly, when we analyzed the growth slope of MDA PCa 183-A before and after castration in the same mouse, we found a statistically significant increase in tumor volume before castration (slope = 26.8, P < 0.0001) and a statistically significant decrease in tumor volume after castration (slope = −18.0, P < 0.0001). Of interest, in the castrated group, tumors in three out of seven mice grew fast and reached a volume above what is allowed by our animal facilities guidelines and had to be killed. The tumor volume in the other four mice either plateaued and/or showed a subsequent decline (Fig 3A). Although the small number of mice does not have enough power for a statistically significant conclusion, these results suggest that two different populations of cells exist in MDA PCa 183-A PDXs, which is in alignment with the fact that the cells were derived from an untreated human tumor and therefore there was no prior treatment selection. By contrast, although the growth rate of MDA PCa 180–30 was slowed down after castration (P = 0.008), there was a more uniform response to castration in this tumor with a clear relapse occurring in all tumors over time (Fig 3A). This is in alignment with the fact that MDA PCa 180–30 was derived from a human PCa that underwent multiple therapies, and therefore there was a selection. These results indicate that the MDA PCa PDXs are useful models to study CRPC progression.
Fig 3.
(A) Effect of mouse castration on the growth of two MDA PCa PDXs. MDA PCa 183-A. Intact mice. Tumor volume monitoring detected a significant increase over time (n = 9, slope = 18.0; P < 0.0001, linear mixed models). Castrated mice. Tumor volume monitoring after mouse castration did not detect any significant change over time (n = 7, slope = −0.4; P = 0.85). There was a statistically significant difference between the slopes of tumor volume between the intact and castrated mice (P < 0.0001). Tumor volume before and after castration. A piecewise linear mixed model, with castration day as the cutoff, suggests that there was a statistically significant increase in tumor volume before castration (n = 4, slope = 26.8, P < 0.0001), and there was a statistically significant decrease in tumor volume after castration (n = 6, slope = −18.0, P < 0.0001). MDA PCa 180–30. Intact mice. Tumor volume monitoring detected a significant increase over time (n = 7, slope = 41.7; P < 0.0001, linear mixed models). Castrated mice. Tumor volume monitoring after mouse castration detected a significant increase over time, although the rate of increase (i.e., slope) was smaller than in intact mice (n = 14, slope = 26.4; P < 0.0001). There was a statistically significant difference between the two slopes (P = 0.008). Tumor volume before and after castration. A piecewise linear mixed model, with castration day as the cutoff, suggests that there was a statistically significant increase in tumor volume before castration (n = 14, slope = 36.3, P < 0.0001). Also, there was a statistically significant increase (although at a lower rate) in tumor volume after castration (n = 14, slope = 30.9, P < 0.0001). Tumor volumes based on caliper measurements were calculated by the modified ellipsoidal formula: 1/2(length × width2). (B) Effect of erdafitinib in MDA PCa 118b PDXs growing in the bone of immunodeficient mice. Bone scan (front view) and contrast enhanced CT scan show the lesion involving the left ilium (arrow) that was the source of MDA PCa 118b PDX. X-ray of a mouse pelvis and rear limbs 5 weeks after intrafemoral implantation of MDA PCa 118b–derived cells. mRNA expression by RT-PCR of FGFR1, FGFR2, FGFR3, FGFR4 in mouse femurs using human-specific primers. FGFR1 immunohistochemistry (IHC) of an MDA PCa 118b–bearing femur shows high FGFR1 expression. An example of MR images of MDA PCa 118b tumor–bearing femur in erdafitinib and vehicle-treated mice. The graph shows the quantification of tumor volume. (C) Effect of erdafitinib in MDA PCa 183-A PDXs growing in the bone of immunodeficient mice. Bone scan (rear view) and CT scan show the bone lesion involving the sacrum that was the source of the MDA PCa 183 cells (arrow). X-ray of a mouse pelvis and rear limbs 9 weeks after intrafemoral implantation of MDA PCa 183-A–derived cells. mRNA expression by RT-PCR of FGFR1, FGFR2, FGFR3, FGFR4 in mouse femurs using human-specific primers. FGFR1 IHC of MDA PCa 183-A–bearing femur shows no FGFR1 expression. An example of MR images of MDA PCa 183-A–bearing femur in erdafitinib and vehicle-treated mice. The graph shows the quantification of tumor volume. T, tumor; B, bone.
Although, further analysis of these tumors is beyond the scope of this report, in a technical note, when we find a statistically significant difference in tumor volume between drug vs vehicle treated mice, we perform morphological, IHC and molecular analysis of tumors. Briefly, we cut a longitudinal section in the middle of the harvested tumor and prepare FFPE blocks for morphological and IHC analysis. Adjacent tissue pieces are cut and flash freeze for molecular studies. Alternatively, fresh tissue can be used to perform single cell sequencing. One benefit of performing preclinical studies using PDXs in mice is that certain immunohistochemical and molecular studies can be species specific.
Effect of a specific pan-FGFR inhibitor on two bone-derived PDXs
Bone-forming metastases dominate the clinical picture of men with CRPC (15, 16). Studies by our group and others have implicated the fibroblast growth factor (FGF) axis in the pathogenesis of PCa bone progression (5, 17), and we showed that blockade of FGFRs with the receptor tyrosine kinase inhibitor dovitinib has clinical activity in a subset of men with CRPC and bone metastases (6). More recent studies implicate the FGF axis in the progression to androgen ablation and other therapies (10, 18). We therefore assessed the antitumor activity of erdafitinib, a novel and selective pan-FGFR tyrosine kinase inhibitor, preclinically. For these studies we selected two bone-derived PDXs that recapitulate the bone-forming phenotype observed in human PCa, MDA PCa 118b and MDA PCa 183-A (Fig 3B and C). RT-PCR analysis indicated that FGFR1 was high in MDA PCa 118b and about 20-fold lower in MDA PCa 183-A. All other receptors were expressed at very low levels (Fig 3B and C). Accordingly, FGFR1 expression was high in MDA PCa 118b and not detectable in MDA PCa 183-A at the IHC analysis (Fig 3B and C). Cells derived from these PDXs were injected into the distal ends of the femurs of intact male SCID mice according to published protocols (5) and mice subjected to erdafitinib (12.5 mg/kg body weight BID) or vehicle administration by oral gavage. Potent antitumor effect of erdafitinib against PCa cells derived from MDA PCa 118b (P < 0.007), but not MDA PCa 183-A, was observed by MR analysis (Fig 3). These results indicate that erdafitinib is active in controlling the growth of FGFR1-expressing PCa cells in bone and that our PDXs are informative in the preclinical setting.
The same technical note that was outlined at the end of the previous section applies here, only that in tumor bearing bones it is challenging to cut each tumor in equally representative pieces. Therefore, we use a set of bone tumor (one per each mice) for morphological, IHC and molecular studies. Of course, the flash frozen material will have a significant contribution of mouse (bone) cells. Alternatively, the femoral shafts of the tumors bearing femur can be flashed to obtain a product enriched for tumor cells.
Copy number alteration in PDXs derived from different areas of the same human PCa
We performed array comparative genomic hybridization (aCGH) to assess copy number changes in different PDXs derived from the same human PCa donor.
NEC PDXs MDA PCa 144–13 and 144–4 lost cyclin-dependent kinase inhibitor 2A (CDKN2C) (1p32.3) and PTEN (10q23.31), and had a partial loss of RB (13q14) (Fig S3). However, notable differences in copy number changes in specific regions were also found (Fig S4, Supplementary Results).
Common copy number alterations among MDA PCa 146–10, 146–12, and 146–20 were found despite morphological differences, including loss of PTEN and loss of MAP3K1 via a focal homozygous deletion in chromosome 5 (Fig S5), which increases sensitivity to MEK inhibition (19). However, many marked differences in specific regions were identified (Fig S5).
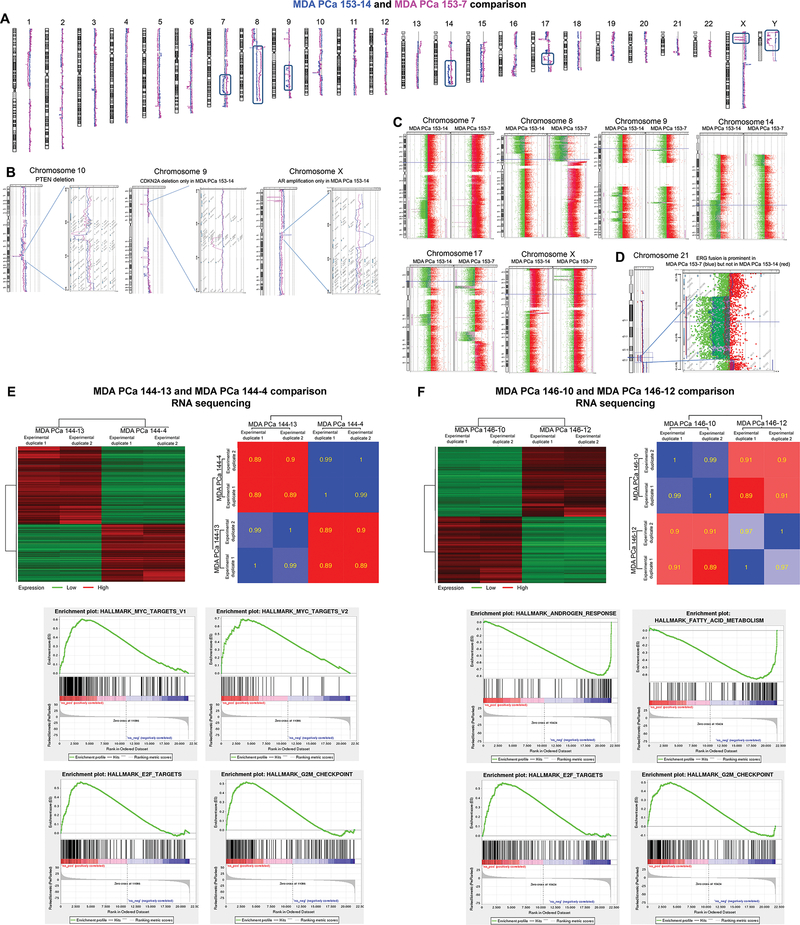
MDA PCa 153 PDXs are adenocarcinomas that share many gains and losses, including PTEN loss (Fig 4A–C). However, MDA PCa 153–14 has AR amplification and a deletion in the area encompassing the CDKN2A (Fig 4B), suggesting alternative mechanisms of progression within cells derived from the same tumor. In this context, it is noticeable that ERG fusion by deletion is very prominent in MDA PCa 153–7 but not in 153–14, showing further evidence of the intra-tumor heterogeneity typical of PCa (Fig 4D). Other cases are presented in Supplementary Results (Fig S6 and S7).
Fig 4. A-C, whole-genome analysis of MDA PCa 153 PDXs.
(A) Comparison of copy number changes at whole genome level in MDA PCa 153–7 (red) and MDA PCa 153–14 (blue) identified many common losses and gains, as well as notable differences in copy number between the two PDXs (boxed areas). (B) Both MDA PCa 153–7 (red) and MDA PCa 153–14 (blue) have a PTEN deletion. However, MDA PCa 153–14 (but not −7) has AR amplification and a deletion in the area encompassing the cyclin-dependent kinase inhibitor 2A (CDKN2A). (C) Notable differences in copy number between the two PDXs can be appreciated at higher resolution. (D) Chromosome view and gene view of ERG fusion by deletion, which is very prominent in MDA PCa 153–7 but not in 153–14. E-F, RNA sequencing analysis of MDA PCa 144 and MDA PCa 146 pairs. (E) Heat maps illustrate correlation and differential expression of genes between MDA PCa 144 and MDA PCa 146 PDX pairs. For the differential expression analysis, significant genes are defined using FDR 0.01 and fold change of 2. (F) Four hallmark gene sets among the most significantly enriched in MDA PCa 144–13 vs MDA PCa 144–4 (left panels) and MDA PCa 146–10 vs MDA PCa 146–12 (right panels).
These results illustrate the high degree of heterogeneity in PCa which may underlie the diverse mechanisms of progression to targeted therapies.
Gene expression analysis by next-generation RNA sequencing
We subsequently focused on MDA PCa 144 and MDA PCa 146 pairs to better understand human PCa heterogeneity at the gene expression level. We found a strong correlation in gene expression between MDA PCa 144–4 and 144–13 PDXs (r = 0.89 to 0.90) (Fig 4E, upper panels). However, 3351 genes are differentially regulated between these PDXs (Table S3), and gene set enrichment analysis (GSEA) identified hallmark MYC, E2F, and G2M checkpoints as the most significantly enriched target gene sets in MDA PCa 144–13 (Fig 4E, lower panels). This is in alignment with the 47-fold higher expression of MYC in MDA PCa 144–13 compared with 144–4 (Table S4). New therapeutic approaches are emerging to target this oncogene (20).
We also found a strong correlation in gene expression between MDA PCa 146–10 and 146–12 (Fig 4F, upper panels). However, 3022 genes are differentially regulated between these PDXs (Table S5), and GSEA showed that hallmark androgen response genes and fatty acid metabolism target genes are among the most significantly enriched in MDA PCa 146–12 compared with 146–10 (Fig 4F, lower panels and Table S6). This is in alignment with our previous finding that MDA PCa 146–12, but not 146–10, expresses AR (Fig 1D). Recent studies demonstrate that AR is tightly linked with lipogenesis in PCa and suggest that targeting fatty acid metabolism will inhibit AR signaling (21). Our studies emphasize the clinical relevance of MDA PCa PDX models in reflecting the heterogeneity of PCa and identifying different pathways (e.g., MYC, AR signals) that drive the growth of heterogeneous cell populations within a single cancer.
Copy number alteration and SPOPL mutational analyses
Results of aCGH analysis of 37 PDXs derived from 28 human tissues identified gains and losses previously reported for PCa (13) (Fig S8). Table S7 outlines specific rearrangements (TMPRRS2-ERG/DSCAM/PRDM15/ETV1), amplifications (AR, MYC), and deletions (PTEN, SPOP, SPOPL, and P53) identified in these PDXs.
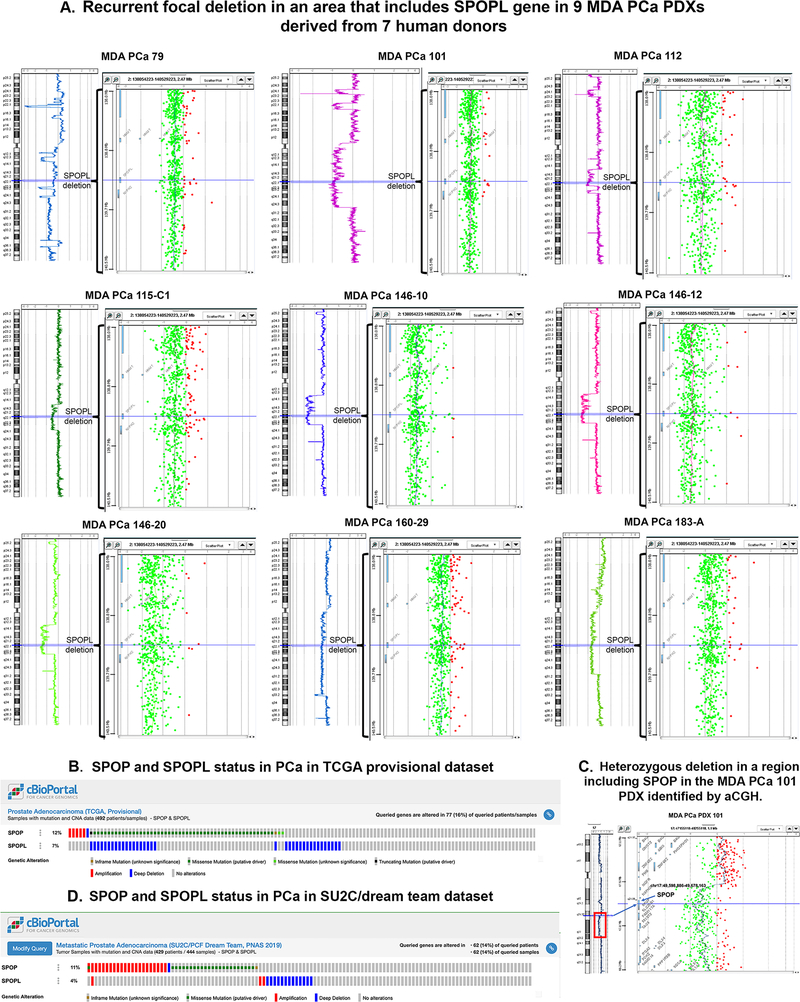
Importantly, by aCGH, we observed that 9 PDXs derived from 7 human PCa donors displayed a focal deletion in the area encompassing speckle-type POZ protein-like (SPOPL) gene (7/28, 25%) (Fig 5A, Table S7). Deletions in SPOPL have been previously reported in PCa (22) and comprise about 7% of the PCas in TCGA dataset (22) (Fig 5B). SPOPL is a MATH-BTB protein that shares an overall 85% sequence identity with SPOP (a SPOPL paralog). SPOP was recently reported to be mutated in about 8% of PCas and defines a molecular subclass (23). No mutations were found in SPOP in our cohort, but we found a heterozygous deletion in a region including SPOP (17q21.33) in MDA PCa 101 (Fig 5C). Other focal deletions identified in our PDXs include those spanning tumor suppressors TP53 (17p13.1), CDKN1B (12p13.1), MAP3K1 (5q11.2), and FANCD2 (3p26) (MDA PCa 79; 101; 115-C1; 146–10, −12, −20; 160–29; and 183-A). These genomic alterations have been reported in human PCa (22, 23), which further supports the clinical relevance of our models.
Fig 5.
(A) Recurrent focal deletion in areas that include the SPOPL gene in MDA PCa PDXs derived from 7 human donors. High resolution aCGH analysis identified focal deletion in the area encompassing SPOPL gene in 9 PDXs derived from 7 human PCa donors. B-D, SPOP and SPOPL status in PCa. (B) SPOP and SPOPL status in TCGA provisional dataset. (C) Heterozygous deletion in a region including SPOP (17q21.33) in the MDA PCa 101 PDX identified by aCGH. (D) SPOP and SPOPL status in SU2C/PCF dream team dataset. Data from TCGA and SU2C/PCF dream team were obtained from cBioPortal (39, 40).
DISCUSSION
The MDA PCa PDX series includes models derived from PCas encompassing the entire clinical spectrum, including hormone-naïve and CRPC, primary tumors and metastases, typical and AVPC. It also includes PCa morphological variants (e.g., adenocarcinoma, NEC). As the MDA PCa PDX program is constantly accruing samples for PDX development, it captures the evolving molecular landscape of PCa progressing under novel therapies. To date, we have 154 PDXs derived from 99 PCa patients, including the non-archived models from the 47 human donor tumors described here that can be expanded either as cell lines (MDA PCa 2a and 2b) or PDXs. The racial distribution of patient donors of the established PCa models reflects the patient population treated at our institution (88 Caucasian, 6 African American, and 5 Hispanic).
Previous sequencing studies of multiple metastatic sites at different times provided unique insights into tumor evolution (24, 25). However, tissue samples obtained at certain time points during therapy are snapshots of progression. The MDA PCa PDX series provides a biological tool to study the role of these alterations in the progression of the disease experimentally. Further, current clinical trials in metastatic CPRC focus on targeting pathways that are altered in this advanced disease state. Clinical trials are the benchmark for establishing the therapeutic activity of drugs, and the value of biomarkers of response and/or resistance to drugs should be established in prospective studies. But studies involving patients and patients’ biological material are limited owing to feasibility, cost, and ethical constraints. Therefore, PDXs are particularly important for establishing the preclinical antitumor activity and tolerability of new drugs or drug combinations prior to clinical studies. The use of PDXs is also important for determining the contribution of the tumor microenvironment (e.g., bone) to PCa progression and resistance to therapy, the antitumor effect of drugs, the identification of predictors of treatment response and the emergence of drug-resistant clones (biomarker identification). This evidence underscores the importance of the availability of PDXs in the preclinical setting for drug selection, therapy development, selection of markers of treatment response, and identification of promising combination treatment strategies. Further, advances in therapy development can be achieved by integrated analysis of clinical and co-clinical studies with PDXs.
One of the most important subjects of PCa research is the identification of mediators of progression and resistance to therapy (relapse), which can be accelerated by using preclinical approaches. A deep understanding of the molecular mechanisms underlying the results of these preclinical studies requires the ability to introduce genetic alterations into PCa cells. However, as it happens with human PCa cells, PCa cells derived from PDXs do not establish as cell lines, which is a significant obstacle to genetically editing these cells. The recent establishment of organoid technology (26, 27) provides a biological platform for PCa cell propagation in vitro, that can be manipulated (e.g., genetic manipulation, in vitro selection). This new methodology complements PDXs as preclinical models of PCa. However, 3D organoid growth conditions do not recapitulate the tumor microenvironment. Thus, findings using 3D organoids need to be complemented by studies using PDXs to translate those findings to the clinic (28). One final point that has not been successfully addressed by PDXs is the study of the immune system in cancer (28).
SPOPL is a component of a cullin-based ubiquitin ligase complex and its clinical significance is reported for medulloblastoma (29). In TCGA provisional dataset, SPOP mutation was reported in 11% and SPOPL was deleted in 7% of primary PCa with a tendency to co-occur (Fig 5B). Interestingly, in the SU2C/PCF dream team dataset (23), this co-occurrence is absent. There is an increase in SPOP amplification and a reduction in the incidence of both SPOP mutation and SPOPL deletion, but the combination of these alterations totaled 8% (Fig 5D). The enriched incidence of SPOP mutations in earlier disease relative to metastatic CRPC has been reported by others (22, 30). Together these results suggest that there is a selection for mutually exclusive SPOP mutation or SPOPL loss in metastatic CRPC. Our cohort is enriched for SPOPL deletions, and therefore constitutes a unique resource to study the role of SPOPL deletion in CRPC.
Finally, we show here that the response of the PDX to castration is different depending on the source of the PDX (e.g., tumor donor prior therapy) and as expected, PDXs derived from a treatment-naïve tumor have a better response to castration than those derived from PCas that progress after therapy. Further, our models also reflect the bone phenotype typical of PCa bone metastases and predict tumor response to targeted therapies.
In summary, The MDA PCa PDX collection presented here include adenocarcinoma and NEC PDXs derived from primary prostate cancers and bone metastases, being prostate and bone the two most frequent sites of progression. These were obtained from therapy naïve and prostate cancers progressing on androgen deprivation and chemotherapy. Therefore, constitutes a unique, clinically relevant, resource for preclinical studies to understand mechanisms of treatment response and resistant to standard as well as less common therapies (e.g., gefitinib). In this context, of particular relevance is the availability of PDXs derived from different areas of the same tumor that would enable the identification of divergent mechanism of progression to therapy in the same tumor due to molecular heterogeneity. Also, the MDA PCa PDXs cohort presented here is unique in that is enriched for SPOPL deletions”
Finally, our studies are in line with those of others (31, 32), demonstrating that MDA PCa PDXs reflect the human donor tumor and are useful for drug testing (33). Furthermore, MDA PCa PDXs have provided unique insights into the biology of PCa. For example, a study of MDA PCa 118b implicated the FGF axis in the pathogenesis of PCa bone metastasis (5), which led to the initiation of a study that demonstrated clinical activity (6). Subsequent studies using our MDA PCa PDXs implicated the FGF axis in progression to therapy (18). A recent study confirmed our reports of the role of the FGF axis in the pathogenesis of advanced PCa (10) and reported a new subgroup that utilizes the FGF axis in progression. MDA PCa 118b fell within this subgroup, further attesting the clinical relevance of our models. Furthermore, the MDA PCa PDXs contributed to the discovery of distinct classes of chromosomal rearrangements in PCa cells (34), the identification of new therapeutic approaches for combination therapy targeting DNA damage response genes (35, 36), the elucidation of new biological roles of genes in PCa (37), and the identification of new mechanisms underlying neuroendocrine differentiation (38). A list of studies that have used the MDA PCa PDXs developed in our program is outlined in Table S1-Addendum.
Conclusions
The MDA PCa PDX series provides insight into the biological basis that accounts for PCa heterogeneity and serves as an invaluable resource for discovery, therapy development, and optimization of personalized therapy targeting PCa-specific drivers of progression. PCa lag behind other tumor types in the marker-informed classification for treatments. The transition to such classification can be achieved by linking detailed characterization of human cancers to “driver mechanisms” in model systems.
Supplementary Material
Statement of translational relevance.
We report the generation of a large number of patient-derived xenografts (PDXs) (MDA PCa PDX series) representative of the clinical spectrum of prostate cancer (PCa). The MDA PCa PDX series is a dynamic resource that captures the molecular landscape of PCas over time. It provides insight into the biological basis that accounts for heterogeneity and serves as an invaluable resource for discovery, therapy development, and optimization of personalized therapy targeting PCa-specific molecular markers. The models developed thus far have led to the identification of clinically relevant therapy targets and have proven valuable for drug testing.
Acknowledgments
We thank Sarah E. Townsend for editing the manuscript. We thank Agilent Technologies for the “Agilent University Relations Grant Award to N.Palanisamy”
Financial support. Prostate Cancer Foundation, The University of Texas MD Anderson Moon Shot Program, NCI Cancer Center Support Grant (P30CA16672), Cancer Center Prostate Cancer SPORE (NIH/NCI P50 CA140388), David H. Koch Center for Applied Research in Genitourinary Cancers at MD Anderson, Houston, TX, Janssen Research and Development, NIH/NCI U01 CA224044
Footnotes
Financial disclosure. None of the contributing authors have any conflict of interest relevant to this manuscript
REFERENCES
- 1.Beltran H, Tomlins S, Aparicio A, Arora V, Rickman D, Ayala G, et al. Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res. 2014;20(11):2846–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logothetis CJ, Gallick GE, Maity SN, Kim J, Aparicio A, Efstathiou E, et al. Molecular classification of prostate cancer progression: foundation for marker-driven treatment of prostate cancer. Cancer discovery. 2013;3(8):849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlachostergios PJ, Puca L, and Beltran H. Emerging Variants of Castration-Resistant Prostate Cancer. Current oncology reports. 2017;19(5):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navone NM, van Weerden WM, Vessella RL, Williams ED, Wang Y, Isaacs JT, et al. Movember GAP1 PDX project: An international collection of serially transplantable prostate cancer patient-derived xenograft (PDX) models. Prostate. 2018;78(16):1262–82. [DOI] [PubMed] [Google Scholar]
- 5.Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J Clin Invest. 2008;118(8):2697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan X, Corn PG, Yang J, Palanisamy N, Starbuck MW, Efstathiou E, et al. Prostate cancer cell-stromal cell crosstalk via FGFR1 mediates antitumor activity of dovitinib in bone metastases. Sci Transl Med. 2014;6(252):252ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navone NM, Olive M, Ozen M, Davis R, Troncoso P, Tu SM, et al. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin Cancer Res. 1997;3(12 Pt 1):2493–500. [PubMed] [Google Scholar]
- 8.Navone NM, Olive M, and Troncoso P. Isolation and culture of prostate cancer cell lines. Methods in molecular medicine. 2004;88:121–32. [DOI] [PubMed] [Google Scholar]
- 9.Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera JM, Reuter VE, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38(6):756–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell. 2017;32(4):474–89 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choucair K, Ejdelman J, Brimo F, Aprikian A, Chevalier S, and Lapointe J. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. BMC Cancer. 2012;12:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–9. [DOI] [PubMed] [Google Scholar]
- 13.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michiel Sedelaar JP, Dalrymple SS, and Isaacs JT. Of mice and men--warning: intact versus castrated adult male mice as xenograft hosts are equivalent to hypogonadal versus abiraterone treated aging human males, respectively. Prostate. 2013;73(12):1316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlin BI, and Andriole GL. The natural history, skeletal complications, and management of bone metastases in patients with prostate carcinoma. Cancer. 2000;88(S12):2989–94. [DOI] [PubMed] [Google Scholar]
- 16.Loberg RD, Logothetis CJ, Keller ET, and Pienta KJ. Pathogenesis and Treatment of Prostate Cancer Bone Metastases: Targeting the Lethal Phenotype. j clin oncol. 2005;23(32):8232–41. [DOI] [PubMed] [Google Scholar]
- 17.Valta MP, Tuomela J, Bjartell A, Valve E, Vaananen HK, and Harkonen P. FGF-8 is involved in bone metastasis of prostate cancer. Int J Cancer. 2008;123(1):22–31. [DOI] [PubMed] [Google Scholar]
- 18.Varkaris A, Corn PG, Parikh NU, Efstathiou E, Song JH, Lee YC, et al. Integrating Murine and Clinical Trials with Cabozantinib to Understand Roles of MET and VEGFR2 as Targets for Growth Inhibition of Prostate Cancer. Clin Cancer Res. 2016;22(1):107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Z, Vis DJ, Bruna A, Sustic T, van Wageningen S, Batra AS, et al. MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models. Cell Res. 2018;28(7):719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowak DG, Katsenelson KC, Watrud KE, Chen M, Mathew G, D’Andrea VD, et al. The PHLPP2 phosphatase is a druggable driver of prostate cancer progression. J Cell Biol. 2019;218(6):1943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zadra G, and Loda M. When fat goes down, prostate cancer is on the ropes. Mol Cell Oncol. 2019;6(3):1595308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beshiri ML, Tice CM, Tran C, Nguyen HM, Sowalsky AG, Agarwal S, et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin Cancer Res. 2018;24(17):4332–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risbridger GP, Toivanen R, and Taylor RA. Preclinical Models of Prostate Cancer: Patient-Derived Xenografts, Organoids, and Other Explant Models. Cold Spring Harbor perspectives in medicine. 2018;8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Yang L, Zhang M, Huang Z, Lin J, and Zhang N. Expression and clinical relevance of SPOPL in medulloblastoma. Oncol Lett. 2017;14(3):3051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzelepi V, Zhang J, Lu JF, Kleb B, Wu G, Wan X, et al. Modeling a lethal prostate cancer variant with small-cell carcinoma features. Clin Cancer Res. 2012;18(3):666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aparicio A, Tzelepi V, Araujo JC, Guo CC, Liang S, Troncoso P, et al. Neuroendocrine prostate cancer xenografts with large-cell and small-cell features derived from a single patient’s tumor: morphological, immunohistochemical, and gene expression profiles. Prostate. 2011;71(8):846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19(5):664–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–9. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Chang W, Yang G, Ren C, Park S, Karantanos T, et al. Targeting poly(ADP-ribose) polymerase and the c-Myb-regulated DNA damage response pathway in castration-resistant prostate cancer. Science signaling. 2014;7(326):ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Liu B, Wu W, Li L, Broom BM, Basourakos SP, et al. Targeting the MYCN-PARP-DNA Damage Response Pathway in Neuroendocrine Prostate Cancer. Clin Cancer Res. 2018;24(3):696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salameh A, Lee AK, Cardo-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A. 2015;112(27):8403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zheng D, Zhou T, Song H, Hulsurkar M, Su N, et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nature communications. 2018;9(1):4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.