Abstract
The glycoprotein nonmetastatic melanoma protein B (GPNMB, also known as osteoactivin) is highly expressed in many cell types and regulates the homeostasis in various tissues. In different physiological contexts, it functions as a melanosome-associated protein, membrane-bound surface receptor, soluble ligand, or adhesion molecule. It is therefore involved in cell differentiation, migration, inflammation, metabolism, and neuroprotection. Because of its various involvement in different physiological conditions, GPNMB has been implicated in many diseases, including cancer, neurological disorders, and more recently immune-mediated diseases. This review summarizes the regulation and function of GPNMB in normal physiology, and discusses the involvement of GPNMB in disease conditions with a particular focus on its potential role and therapeutic implications in autoimmunity.
Keywords: GPNMB, autoimmunity, osteoactivin, lupus, vasculitis
1. Introduction
The relatively recently described glycoprotein nonmetastatic melanoma protein B (GPNMB) is also known as dendritic cell-heparin integrin ligand (DC-HIL, mouse ortholog), osteoactivin (OA, rat ortholog), or hematopoietic growth factor inducible neurokinin-1 type (HGFIN). The gene encoding GPNMB was first identified from a subtractive cDNA screen of human melanoma cells with different metastatic potential (1). It acquired its name from preferentially being expressed at high levels in a melanoma cell line with low metastatic potential. GPNMB shares high sequence homology with a common melanocytic protein called premelanosome protein (PMEL17/gp100), which is critical for pigment biogenesis in melanocytes. It also shares homology, to a lesser extent, with lysosome-associated membrane proteins (LAMPs) which are glycosylated proteins found abundant in lysosomal membranes (2). Since the discovery of GPNMB, studies have shown that this protein is in fact highly expressed in a variety of tumors, including breast cancer, glioblastoma, lung cancer, to name a few (3–5). In addition, this protein promotes tumor growth as well as tumor invasion and metastasis (6–8).
Although GPNMB was recently identified, significant research on the function and regulation of GPNMB has been performed to dissect the molecular complexities of this protein in various diseases. Furthermore, recent evidence suggests a role for GPNMB in immune regulation and in immune-mediated diseases. This review comprehensively discusses current knowledge of GPNMB structure and function, as well as implications of GPNMB dysregulation in autoimmunity and as potential therapeutic target.
2. GPNMB Structure
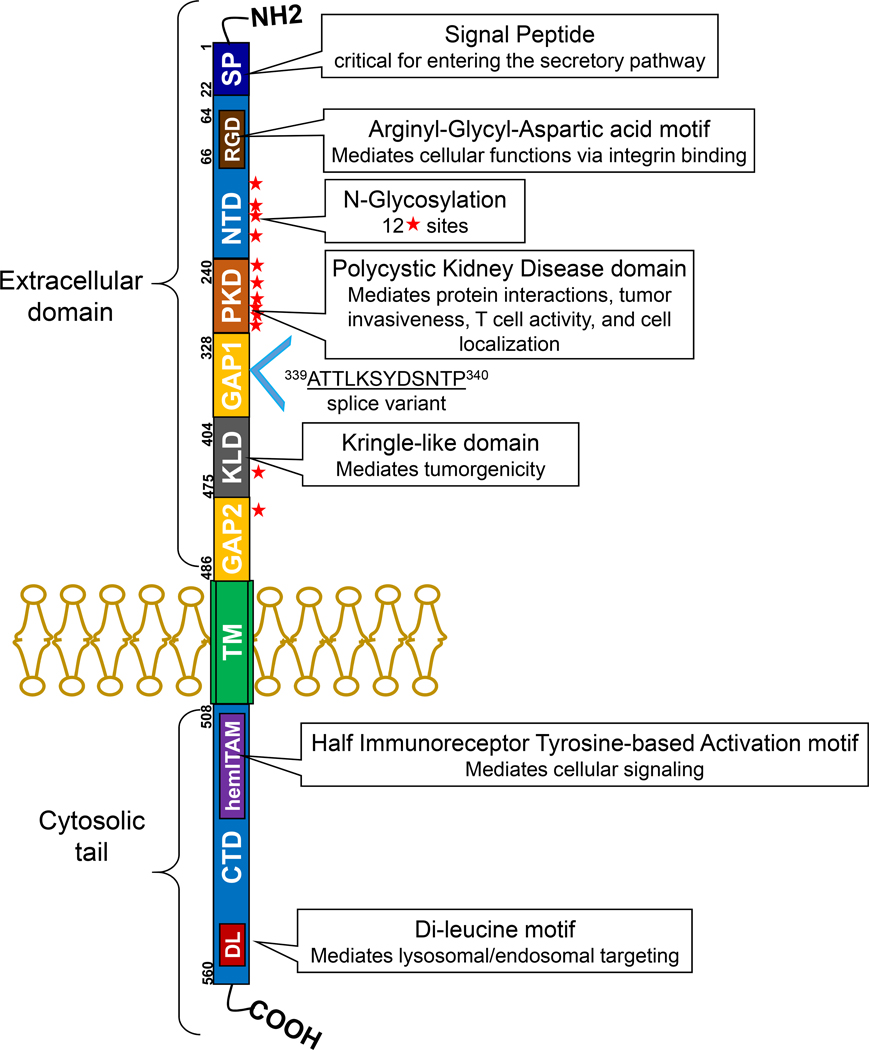
The GPNMB gene, mapped to chromosome 7p15.1, encodes a type I transmembrane glycoprotein that has two isoforms due to alternative splicing. The length of the protein is 572 vs. 560 amino acids (9); the longer variant contains a 12 amino acid insertion within the extracellular GAP1 domain (Figure 1). There are no reports suggesting that the short and long isoforms possess different functions.
Figure 1. Structure of GPNMB.
This membrane bound protein contains a signaling domain (SP), an RGD motif in the N-terminal domain (NTD), a polycystic kidney disease (PKD) domain, and a Kringle-like domain (KLD) in the extracellular portion. The cytosolic domain, which is separated by the transmembrane domain (TM), consists of a half immunoreceptor tyrosine-based activation motif (hemITAM), and a lysosomal/endosomal targeting dileucine motif (DL) in the C-terminal domain (CTD). Solid stars indicate potential N-glycosylation sites. A GPNMB isoform is identified where 12 amino acids are inserted between amino acid 339 and 340 in GAP1, most likely due to alternative splicing.
By homology modeling prediction, GPNMB contains an extracellular domain, a single pass transmembrane domain, and a 53 amino acid cytoplasmic tail (10). The extracellular domain has an N-terminal signal peptide (SIG), an integrin-binding RGD motif embedded in the N-terminal domain (NTD), a polycystic kidney disease (PKD) domain, and GAP1 and GAP2 domain separated by a Kringle-like domain (KLD, Figure 1). There are 12 potential N-glycosylation sites in the extracellular domain and these N-glycans are linked to asparagine residues. In immunoblots, human GPNMB is detected at around 90 and 115 kDa as the precursor and mature forms, while the un-glycosylated form is detected around 65 kDa. The SIG domain is believed to send GPNMB into a secretory pathway (10). The RGD motif is well characterized as an integrin-binding motif, critical for GPNMB-mediated cell adhesion, migration, and proliferation (11–14). The PKD domain is an immunoglobulin-like folding structure with possible roles in mediating protein interactions and cell adhesion (15). It is suggested that the invasive capacity of GPNMB in breast cancer cells is dependent on this domain (16). The PKD domain also distinguishes the trafficking and amyloidogenic properties between GPNMB and its homologue PMEL; the differences of these properties is due to extensive glycosylation of the PKD domain of GPNMB (17). In addition, GPNMB interacts with syndecan-4 on activated T cells through the PKD, which leads to suppression of T cell activation and proliferation (18). The KLD is conserved across species and recently shown to be critical in controlling the tumorgenic potential of GPNMB, as the KLD deletion mutant not only affected the molecular functions of GPNMB, but also reduced its ability to promote cell migration (19).
The half immunoreceptor tyrosine-based activation (hemITAM) motif located in the C-terminal domain (CTD) contains a highly conserved YxxI sequence. The tyrosine residue in hemITAM can be phosphorylated thereby leads to activation of several downstream pathways. For instance, the epidermal growth factor receptor interacts with GPNMB and this induces tyrosine phosphorylation in the hemITAM, which correlates with poor prognosis of breast cancer patients (20). The hemITAM is also essential for the induction of epithelial-mesenchymal transition, tumorigenic activities, and stem-like properties of GPNMB (21, 22). In addition to the hemITAM domain, GPNMB also has a dileucine (DL) motif in the cytoplasmic tail. The D/ExxxLL sequence is believed to be associated with receptor internalization and lysosomal/endosomal targeting (23).
Interestingly, GPNMB can undergo proteolytic processing to form a 20 kDa C-terminal fragment (24). This only occurs with the mature form of GPNMB (115 kDa protein) (10). In breast cancer cells, ADAM10 was identified as a sheddase that can release the GPNMB ectodomain, which induces endothelial cell migration (25). In fibroblasts, the ectodomain of GPNMB induces MMP-3 production via ERK activation (26).
3. GPNMB Regulation
Epigenetics
GPNMB is regulated by epigenetic mechanisms. Methylation at the promoter region of GPNMB has been documented in colorectal cancer (27, 28). Hypermethylation of GPNMB was found in 95% of the patients with colorectal cancer while only 20% of healthy subjects (27). In addition, GPNMB methylation status correlates with cancer progression, as it is lower in early colorectal cancer stages and higher in advanced cancer. An inverse relationship between methylation status and protein expression was also noted. Interestingly GPNMB showed tumor suppressive effects in human colorectal cancer cells.
In addition to DNA methylation, GPNMB might be controlled by histone acetylation. Treating gastric cancer cells BGC-823 with a histone deacetylase inhibitor trichostatin A (TSA) significantly downregulated GPNMB expression (29). Since GPNMB is elevated in advanced gastric carcinoma tissue and TSA significantly induced cell apoptosis, the authors suggested that GPNMB might be involved in TSA-induced apoptosis in gastric cancer. In breast cancer cells, GPNMB has been shown to be one of the target genes of enhancer of zeste homolog 2 (EZH2), a histone methyltransferase (30). Using gene expression profiling arrays in EZH2-knockdown breast cancer cells coupled with analyzing ChiP-seq datasets, GPNMB was identified to be an EZH2-target gene. Since EZH2-mediated H3K27me3 is a repressor mark, GPNMB was upregulated when EZH2 was silenced in these cells.
The expression of GPNMB is also regulated by miRNAs. Screening of differentially expressed miRNAs in a hyperoxia rodent model revealed Gpnmb as a potential target of miR-150 (31). This was further validated experimentally, as upregulation of GPNMB under hyperoxia exposure in lungs of miR-150 knockout mice was observed (32). In human dental pulp stem cells, GPNMB is suppressed by miR-508–5p (33).
Transcription Factors
Melanogenesis associated transcription factor (MITF) belongs to the Microphthalmia family of basic helix-loop-helix leucine zipper transcription factors (MiTF/TFE), which regulate genes involved in lysosome and autophagy biogenesis (34). In melanocytes, MITF is a master regulator of cell development and a key transcription factor in melanoma progression; it controls genes that govern cell proliferation, survival, and differentiation (35). The involvement of MITF in GPNMB regulation has been studied extensively. Overexpression of MITF in macrophage-like cell line RAW/C4 cells upregulated GPNMB expression (36). Using EMSA and luciferase transfection assays the authors confirmed that MITF binds and transactivates the Gpnmb promoter. This was further supported by work by Loftus et al, where they identified a conserved element of MITF binding site on GPNMB that can drive GPNMB expression in melanoblasts both in a luciferase assay and in vivo (37). An interesting study by Gabriel et al showed that GPNMB is critical for macrophage infiltration in obese adipose tissue (38). In these cells, GPNMB was induced in the presence of lysosomal stress inducers in a MITF-dependent manner. In addition to macrophages and melanocytes, the involvement of MITF in GPNMB expression was also shown in human monocyte-derived dendritic cells (39); treating these cells with a MITF inhibitor ML329 dose-dependently reduced GPNMB expression. A mechanistic study was conducted in the context of central nervous system development. The intracellular domain of teneurin-1 regulated MITF-dependent GPNMB expression by competing for histidine triad nucleotide binding protein 1 (40). In a recent study, the anti-cancer effect of inhibiting chromosomal segregation 1-like protein in gastric cancer cells was due to reduction in MITF levels thereby suppressing GPNMB expression, further confirming the regulatory effect of MITF on GPNMB expression and function (41).
Other members of the MiTF/TFE family include TFE3, TFEB, and TFEC. Among them, TFE3 has been shown to regulate GPNMB. The critical involvement of tumor suppressor gene FLCN in renal cell carcinoma development was found to be mediated via the TFE3/GPNMB axis (42). Through a series of in vitro and in vivo studies, the authors showed that GPNMB is a downstream target of TFE3, whose expression was strictly dependent on FLCN inactivation. Interestingly MITF seems to play a minor role in the effect of FLCN, as MITF-induced GPNMB promoter activity was not reduced by FLCN in luciferase promoter assays. This shows the complexity of GPNMB regulation in different cell types and different disease context. A recent study by Baba et al. also confirmed that GPNMB is a direct transcriptional target gene of chimeric TFE3, a fusion protein of oncogene PRCC-TFE3 responsible for a subtype of renal cell carcinoma (43).
In addition to MITF and TFE3, the musculoaponeurotic fibrosarcoma (MAF) oncogene family protein K (MAFK) transcription factor also regulates GPNMB expression (22). In breast cancer cells, MAFK induced epithelial-to-mesenchymal transition and promoted invasive tumor formation by upregulating GPNMB. Since MAFK lacks canonical transcriptional activation domains (44), its effect on GPNMB transcription could rely on its dimeric partner or other indirect mechanisms. In hepatic stellate cells, GPNMB appears to be regulated by transcription factor SOX9, and plays a critical role in SOX-9 mediated liver fibrosis (45).
Cytokines and Growth Factors
GPNMB can be stimulated by a variety of growth factors and cytokines, including IFNγ (in bone marrow-derived macrophages, myelogenous leukemia cell line k562, lung epithelial cell line CCL-64, and CD11b+Gr1+ cells) (46–48), IL1β (in CD11b+Gr1+ cells) (47), IL10 (in human monocyte-derived dendritic cells) (39), TGFβ (CD14+ monocytes) (49), and granulocyte-macrophage-colony stimulating factors (GM-CSF, in T47D breast cancer cells) (48). Other stimulants include LPS (microglia cells, RAW264.7 macrophages, and bone marrow-derived macrophages) (46, 50, 51) and α-melanocyte-stimulating hormone (in melanoma cells) (52). UVA upregulated GPNMB expression in a dose-dependent manner in murine melanoma cells while UVB had no effect (52). In addition to regulating GPNMB expression, cytokines also affect cellular localization of GPNMB. Indeed, IFNγ and TNFα upregulated surface expression of GPNMB in melanocytes but had no effect on intracellular GPNMB expression (52).
The downstream signaling pathways involved in cytokine stimulation of GPNMB were elucidated in some studies. Metz et al. showed that cytokines such as IFNγ and GM-CSF regulated GPNMB by p53 and other transcription factors induced by cytokines (48). In hepatocellular carcinoma cells, epithelial cell adhesion molecule (EpCAM) and CSF-1 significantly enhanced GPNMB levels in part by c-myc (53). Bone morphogenic protein-2 induced GPNMB in osteoblast culture via activation of Smad1 (54). In addition, bone morphogenic protein-2 acts through homeodomain transcription factors Dlx3, Dlx5, and Msx2 during osteoblast differentiation (55). Endothelin-1 upregulated GPNMB via MITF to enhance melanogenesis (56). The laminin-derived peptide C16 interacts with integrin β1 which leads to Src phosphorylation and in turn upregulates GPNMB, resulting in the invasive phenotype of breast cancer cells (57).
4. GPNMB Partners and Receptors
As mentioned earlier, GPNMB is a membrane-bound protein and it also exists in a soluble form (24, 26). In either form, GPNMB has been shown to interact with different partners, including integrins, heparan sulfate proteoglycan, tyrosine kinase receptors, transporters, and others.
Integrins
Since GPNMB, or osteoactivin, was identified in an osteoporotic rat model (58), it has been studied extensively in regulating bone formation. In MC3T3-E1 osteoblast-like cells, GPNMB acted as a matricellular protein that stimulated osteoblast adhesion through binding to integrin αvβ1 and cell surface heparan sulfated proteoglycans (59). This interaction is believed to lead to increased cell spreading, actin reorganization, and osteoblast differentiation. In osteoclasts, GPNMB co-immunoprecipitated with integrin β1 or β3, however, the functional consequence of this interaction is not known (60). Another reported integrin partner for GPNMB is integrin α5β1 (14). The RGD integrin-binding domain in GPNMB can promote cell invasion. Indeed, Maric et al. showed that GPNMB binds to α5β1 fibronectin receptor in tumor cells through its RGD domain to enhance downstream signaling events essential for tumor invasion and metastasis. Interestingly, the RGD motif is essential for GPNMB to induce α2, α5, β1 and β3 integrin subunits in breast cancer cells (14). This possibly provides a positive feedback loop to augment the metastatic potential of GPNMB.
Syndecan-4
In the immune system, GPNMB modulates adaptive immunity by acting through its ligand heparin sulfate proteoglycan syndecan-4. The extracellular domain of GPNMB on antigen presenting cells suppresses T-cell activation and proliferation by binding to heparan sulphate-like structures on syndecan-4 on activated T cells (49, 61–67). The critical role of GPNMB-syndecan-4 interaction between antigen presenting cells and T cells has been suggested in different disease models, such as graft-versus-host disease, experimental autoimmune encephalomyelitis (EAE), and cancer (47, 63–65, 67). The antigen presenting cells that express high levels of GPNMB include CD14+ monocytes and dendritic cells. In particular, the GPNMB-positive CD11b+Gr1+ myeloid-derived suppressor cells (MDSCs) were the most expanded and most potent in suppressing T cells in both EAE and melanoma models (47, 64). Blockade of GPNMB worsened autoimmune response and enhanced antitumor immunity in melanoma models. These data suggest that the GPNMB-syndecan-4 pathway has the potential to regulate autoimmune responses by mediating the T cell suppressor function of MDSCs. It is also a potential target for immunotherapy. Indeed, toxin-conjugated GPNMB was shown to suppress an activated T cell-driven response by depleting syndecan-4-positive T cells, and deemed to be therapeutically beneficial in diseases in which these cells play critical pathogenic roles (61, 63). A recent study by Kobayashi et al. showed in vitro and in vivo anti-cancer efficacy using anti-GPNMB monocloncal antibodies to reduce MDSCs while increasing IFNγ-secreting T cells in the tumor microenvironment (66).
Tyrosine Kinase Receptors
In the investigation of how long non-coding RNA LINK-A is involved in triple negative breast cancer, Lin et al. showed that transmembrane proteins GPNMB and epidermal growth factor receptor (EGFR) interact with each other upon HB-EGF stimulation (20). The kinase domain in EGFR intracellular domain binds to the hemITAM domain of GPNMB, resulting in phosphorylation of tyrosine (Tyr) 525 in GPNMB. This interaction is critical for LINK-A mediated recruitment of BRK and LRRK2, and subsequent activation of HIFα-mediated signaling in breast cancer cells.
The effect of soluble GPNMB in promoting angiogenesis and osteogenesis appears to be mediated through the fibroblast growth factor receptor-1 (FGFR-1) (68). The effect of GPNMB in human bone marrow stromal cells and endothelial cells was blocked by knocking down FGFR1 or FGFR1 inhibitor SU5402.
Na+/K+-ATPase
In amyotrophic lateral sclerosis (ALS) patients, elevated GPNMB protein levels were observed in the cerebrospinal fluid and sera (69). Tanaka et al. showed that the ectodomain of GPNMB was protective against mutant superoxide dismutase 1- induced neurotoxicity via the activation of the PI3K/Akt and MEK/ERK pathways (69). In a follow up study the group utilized Membrane Protein Library/BLOTCHIP-MS technology to perform a comprehensive analysis of cell surface proteins/receptors, and eventually identified the alpha subunits of Na+/K+-ATPase as binding partners of the soluble GPNMB (70). This was further confirmed by co-immunoprecipitation as well as functional assays incorporating Na+/K+-ATPase inhibitor ouabain and knocking down the alpha subunits of Na+/K+-ATPase. Both inhibitor and siRNA of Na+/K+-ATPase effectively blocked the protective effect of GPNMB against stress-induced cell death and PI3K/Akt and MEK/ERK activation. Although Na+/K+-ATPase is a transporter, it has been reported to be able to function as a non-canonical receptor to activate the PI3K/Akt and MEK/ERK pathways (71).
CD44
The soluble form of GPNMB has been shown to mediate its effect on mesenchymal stem cells, osteoclasts, astrocytes, and adipocytes through acting on CD44. Also referred as P-glycoprotein 1, CD44 is a transmembrane glycoprotein that is widely expressed on various cell types. It is involved in cell-matrix or cell-cell interactions to modulate various cellular events including cell proliferation, adhesion, migration, as well as lymphocyte activation and extravasation (72). CD44 has multiple variants and it also undergoes post-translational modifications such as N- and O-glycosylation. These unique qualities and its abundance on cell surfaces are likely the reason for its diverse cellular activity. The first study that identified CD44 as a GPNMB receptor was conducted by Yu et al. They showed that M2 macrophages secreted higher levels of GPNMB than M1 macrophages, and this subsequently promoted the proliferative and migratory properties of mesenchymal stem cells (73). They further demonstrated that CD44 was the receptor for macrophage-released GPNMB on these stem cells. Via interacting with CD44, GPNMB stimulated ERK and AKT signaling pathways in these cells. This study provided the framework for how macrophages, through GPNMB, regulates mesenchymal stem cells.
To dissect the function and signaling mechanisms of GPNMB in osteoclasts, Sondag et al. showed that GPNMB dose dependently inhibited receptor activator of nuclear factor-k B ligand (RANKL)-induced osteoblast differentiation in both bone marrow-derived osteoclast progenitor cells and in mice (74). In addition, the inhibitory effect of GPNMB on RANKL-induced ERK phosphorylation was mediated by CD44. These data suggest that GPNMB negatively regulates osteoclastogenesis, and that CD44/ERK activation is critical for this process. The CD44/GPNMB axis is also critical in neurodegenerative diseases including Parkinson’s disease. Elevated levels of GPNMB and CD44 were observed in the substantia nigra of human Parkinson’s disease brains and in astrocytes of an animal model for Parkinson’s disease (75). GPNMB treatment showed neuroprotective effect in an astrocyte cell line and primary mouse astrocytes. Using primary mouse astrocytes isolated from CD44 knockout mice, it was confirmed that the anti-inflammatory effects of GPNMB were CD44-mediated. Interestingly, GPNMB was recently shown to promote fatty acid synthesis in adipocytes through CD44-AKT-SREBP1c in adipocytes (76).
Others
Chung et al. showed that GPNMB is a pathogen recognition receptor for dermatophytic fungi (77). GPNMB on dendritic cells can directly bind to the cell wall of Trichophyton rubrum and Microsporum audouinii to transduce signals via its membrane-proximal YxxI sequence thereby resulting in activation of the dendritic cells.
In hepatic macrophages and stellate cells, GPNMB was found to interact with calnexin (78). Located in the endoplasmic reticulum, calnexin is a chaperone protein that assists folding and assembly of N-linked glycoproteins. Although the mechanism of the direct interaction between GPNMB and calnexin is not known, the authors posited that GPNMB, overexpressed in hepatic stellate cells, binds to calnexin thereby exerts anti-oxidative properties and subsequently inhibits liver fibrosis.
5. Physiological Functions
GPNMB is expressed in a variety of tissues with the highest expression in the adipose tissue and skin (79, 80). It is also expressed in multiple cell types, including osteoclasts, osteoblasts, immune cells, melanocytes, to name a few. The wide expression of GPNMB points to the involvement of this protein in many physiological processes.
5.1. The Bones
GPNMB is involved in bone development. It controls the differentiation and activities of both osteoclasts and osteoblasts (60, 81–83). In osteoblasts, blockade of GPNMB inhibited differentiation, while overexpression of GPNMB enhanced osteoblast differentiation and function (81). The osteogenic activity of GPNMB was also confirmed in animal models (82, 84). Also expressed in osteoclasts, the role of GPNMB in osteoclasts is less clear. Mutation of Gpnmb led to increased osteoclastogenesis but impaired bone resorption (83). Recombinant GPNMB inhibited RANKL-mediated osteoclast differentiation via activation of the CD44/ERK pathway (74). In contrast, other studies have suggested that GPNMB stimulates osteoclast differentiation and function (60, 85). More studies are needed to investigate the involvement of GPNMB in osteoclast function.
5.2. The Skin
GPNMB is highly expressed in the skin. It is found to be predominantly expressed in melanocytes, which are located in the basal layer (10, 80). It is also expressed in other cell types of the skin, including keratinocytes and Langerhans cells (49, 52). Melanosomes are intracellular vesicles that produce, package, and store melanin in melanocytes. GPNMB is localized in early, stage III, and stage IV melanosomes, with relatively higher amounts detected in mature melanosomes (10). GPNMB is critical for the biogenesis of early melanosomes, as knockdown of GPNMB led to significant decrease of the number of melanosomes in melanocytes (86). GPNMB also mediates melanocyte adhesion to keratinocytes in a RGD-dependent manner (52). The significance of GPNMB in the skin is highlighted by the study by Yang et al., where they showed that GPNMB truncating alleles are the cause of autosomal-recessive amyloidosis cutis dyschromica, a rare variant of primary cutaneous amyloidosis that causes localized hyperpigmentation and hypopigmentation (87).
GPNMB is also involved in wound repair. Yu et al. elegantly showed that GPNMB, secreted from macrophages, signals mesenchymal stem cells to be recruited to the site of skin injury (88). These mesenchymal stem cells can in turn promote M2 polarization of macrophages for rapid wound repair. The authors further showed that macrophages in diabetic wounds showed diminished levels of GPNMB, and topical treatment of GPNMB not only restored mesenchymal stem cell recruitment, but also increased M2 macrophages and enhanced wound closure.
5.3. The Central Nervous System
GPNMB is widely expressed in normal central nervous system (CNS) of adult rats, and mainly colocalized with microglia/macrophages (89). Various studies have suggested that GPNMB is neuroprotective. Transgenic mice with GPNMB overexpressed demonstrate significantly ameliorated infarct volume after ischemia–reperfusion injury compared to wild-type mice (90). This effect appears to be mediated by the ectodomain of GPNMB, which increases phosphorylated-Akt and ERK. In addition, GPNMB shows neuroprotective effects in NSC-34 motor neurons via the Na+/K+-ATPase/PI3K/Akt and MAPK/ERK axis (70). GPNMB also improves both spatial and non-spatial memory (91). In both GPNMB transgenic mice and mice treated with the soluble form of GPNMB, the memory improvement was associated with increased levels of the α-amino-3-hydroxy-5-methylisoxazole-4-propionate receptor subunit GluA1.
GPNMB is also involved in neuroinflammation. Treating an astrocyte cell line or mouse astrocytes with GPNMB attenuated inflammatory responses in these cells, and this process appears to be CD44-dependent (75). Interestingly elevated levels of GPNMB is observed in the CNS in patients with ALS, Parkinson’s disease, and Alzheimer’s disease (69, 75, 92, 93). Under these conditions GPNMB might be neuroprotective and promoting neuron survival. Studies exploring GPNMB as a possible therapeutic target or biomarker for these diseases are currently underway (94).
5.4. The Immune System
GPNMB has been implicated in both adaptive and innate immunity (65, 77). In immune cells, GPNMB expression is detected in monocytes, lymphocytes, dendritic cells, and granulocytes (79). In innate immunity, GPNMB is shown to be a pathogen recognition receptor for dermatophytic fungi (77). On the other hand, the anti-inflammatory role of GPNMB was shown in macrophages (46). It was reported that the expression of GPNMB is inhibited in RAW264.7 cells exposed to LPS. In addition, GPNMB reduced the LPS/interferon-γ-induced secretion of IL-6 and IL-12p40. The results indicated that GPNMB suppressed pro-inflammatory cytokine production in macrophages, as was also shown in a model of experimental colitis (51). The notion that GPNMB-positive macrophages negatively regulates inflammation is further supported by the observation that anti-inflammatory M2 macrophages expressed more GPNMB than the pro-inflammatory M1 macrophages (73). Moreover, M2 polarization from bone marrow-derived macrophages was inhibited when GPNMB was knocked down in these cells, while M1 polarization was enhanced (95). The GPNMB-mediated anti-inflammatory effect on macrophages has been demonstrated in wound healing (88), fibrosis (95, 96), kidney injury (97), as well as inflammatory responses in the adipose tissue (38, 78). The involvement of GPNMB in the innate immune response was also implicated in autoimmune pigmentary glaucoma in mice (98).
Various studies support the role of GPNMB in controlling adaptive immunity. As we have discussed previously, the high expression of GPNMB on antigen-presenting cells represses T-cell activation via binding to syndecan-4 (49, 62). The ability of GPNMB to modulate adaptive immunity has been documented in various disease contexts including graft-versus-host disease, EAE, and cancer (47, 63–65, 67).
5.5. Metabolism and Immunometabolism
GPNMB is linked to obesity-driven inflammation in adipose tissue. This is not surprising, as the adipose tissue has high amount of GPNMB (79). Microarray analysis in mice fed with high-fat diet revealed Gpnmb as one of the top upregulated genes (99). Similar results were obtained from transcriptomic analysis of backfat tissue extracted from fat and lean pigs; GPNMB is expressed higher in fat pigs (100). The association of GPNMB with adipose tissue and inflammation is also indicated in Gaucher disease, which is a lysosomal storage disorder characterized by macrophage lipid storage problems and deficiency in lysosomal acid β-glucosidase. Elevated GPNMB levels were found in a mouse model and patients of Gaucher disease (101, 102). GPNMB levels not only correlated with overall disease severity but also decreased when patients were treated.
The exact function of GPNMB in adipose tissue is not known. In a recent study Gong et al. showed that hepatic GPNMB stimulates lipogenesis in adipocytes via binding to CD44, thereby triggering the PI3K-AKT-mTOR-SREBP1c signaling pathway (76). Excessive GPNMB increases fat storage in adipose tissue and exacerbates insulin resistance. In addition, treatment with neutralizing antibodies targeting GPNMB attenuated lipogenesis and adipocyte size, as well as improved metabolic parameters in mice. GPNMB also affects adipose tissue macrophages. It is highly expressed in macrophages in adipose tissue of obese mice and humans (38). GPNMB is induced by lysosomal stress inducers via MITF in macrophages, and potentiates alternative macrophage activation, as indicated by IL-4 mediated arginase-1 induction. These results suggest that GPNMB plays a critical role in the phenotype and function of obesity-induced adipose tissue macrophages. The involvement of macrophage GPNMB in obesity was also shown by Katayama et al., where overexpression of GPNMB in transgenic mice ameliorated fat accumulation and fibrosis in the liver in a diet-induced obesity model (78). GPNMB in hepatic macrophages interacted with calnexin to reduce oxidative stress.
5.6. The Eyes
GPNMB was found in high amounts in the outer layer of the retina during early development, while the expression was restricted in the retinal pigment epithelium and iris later in development (103). It was suggested that based on the sequence homology to melanosomal proteins and endosomal localization, GPNMB might be involved in the development of the retinal pigment epithelium and iris. In addition to the multiple pigmented cells of the eye such as melanocytes of the iris and pigmented cells of the iris epithelium, GPNMB was found to localize to ocular antigen-presenting cells, specifically macrophages and dendritic cells (98). A spontaneous mutation within the Gpnmb locus that creates a truncated form of GPNMB has been identified in DBA/2J mice. Homozygous mice exhibit autoimmune pigmentary glaucoma and compromising ocular immunosuppression (104). It was later shown that GPNMB influenced the glaucoma phenotype of these mice through mechanisms independent of adaptive immunity (98).
5.7. Others
GPNMB is involved in odontogenesis (105). This is not surprising, since odontoblast-like differentiation of dental pulp cells is similar to human bone marrow stromal cells osteogenesis. Knockdown of GPNMB in dental pulp cells inhibited odontoblastic differentiation, suggesting that GPNMB promoted odontoblast differentiation in these cells. Later it was shown that miR-508–5p regulated the osteogenesis of human dental pulp stem cells by targeting GPNMB (33).
The soluble form of GPNMB shows potent angiogenic properties (25, 68, 106). In a mouse model of mammary carcinoma, GPNMB-mediated enhancement of tumor growth was accompanied with increased vascular density (25). In addition, the ectodomain of GPNMB, shed by ADAM10, promoted endothelial cell migration. In human umbilical vascular endothelial cells soluble GPNMB induced angiogenesis by activating FGFR-1 and the corresponding downstream molecule ERK1/2 (68). Similar results were shown by Narasaraju et al. where the protective effect against hyperoxia induced-impaired vascular growth impairment and increase in cellular infiltrates in miR-150 knockout mice was mediated through elevated levels of GPNMB (106). The pro-angiogenic property of GPNMB provides a mechanistic tool for cancer cells to promote tumorigenesis. Indeed, GPNMB silencing in glioma cells inhibited cell growth. In addition, conditioned media from these transfected cells significantly reduced tube formation in endothelial cells (107).
6. Implications in Autoimmunity
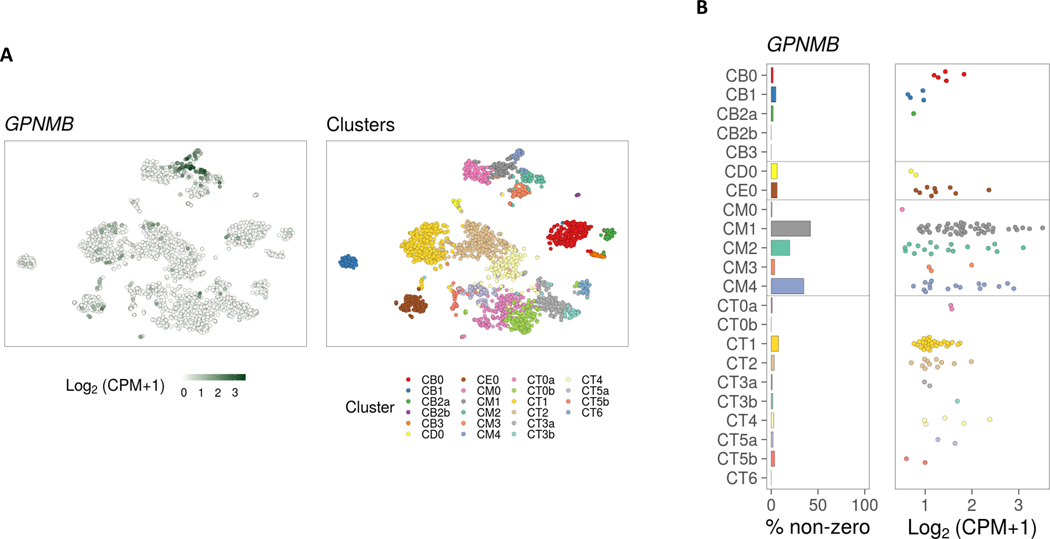
GPNMB has been associated with kidney disease (108, 109) and promotes repair in kidney injury (97). It is therefore not surprising that there are reports associating GPNMB with lupus nephritis. Indeed, elevated GPNMB levels were found in the urine and exosomes in a lupus patient with acute lupus nephritis (109). In addition, immunostaining of GPNMB showed cortical tubular expression in kidney biopsies from a patient with lupus nephritis. A recent study profiling the immune cell landscape in kidney samples from lupus nephritis patients using single-cell RNA sequencing revealed upregulation of GPNMB in myeloid cells (110). In particular, GPNMB appears to be primarily expressed in phagocytic CD16+ macrophages, M2-like CD16+ macrophages, and to a lesser extent in tissue-resident macrophages (Figure 2). This echoes what was found in animal models of lupus. Characterization of renal mononuclear phagocyte populations in the lupus mouse model NZB/W showed that phagocytic F4/80hi/CD11cint macrophages were located throughout the kidney interstitium. They were associated with tissue damage and disappeared after remission (111). Using Affymetrix Gene-Chip arrays, confirmed by qPCR, Gpnmb was upregulated in F4/80hi cells from nephritic kidneys compared with young kidneys, and downregulated after disease remission. The underlying mechanisms of how GPNMB is involved in lupus nephritis warrant further examination.
Figure 2. GPNMB expression in kidney biopsy tissue in lupus nephritis.
(A) TSNE plot generated from unsupervised clustering analysis of single cell RNA-seq data from kidney biopsies of lupus patients generated by the Accelerating Medicines Partnership (AMP) project (right panel). Feature plot showing GPNMB expression, revealing 3 myeloid cell clusters with GPNMB expression (CM1, CM4, and CM2) (left panel). (B) Bar plot showing percentage of GPNMB expressing cells in kidney-infiltrating cell clusters and dot pot showing relative GPNMB expression levels. This figure was generated using the viewing browser https://immunogenomics.io/ampsle/. CM0, Inflammatory CD16+ machrophages; CM1: Phagocytic CD16+ macrophages; CM2, Tissue-resident macrophages; CM3, Conventional dendritic cells; CM4, M2-like CD16+ macrophages; CT0a, Effector memory CD4+ T cells; CT0b, Central memory CD4+ T cells; CT1, CD56dim CD16+ NK cells; CT2, Cytotoxic T lymphocytes; CT3a, Tregs; CT3b, TFH-like cells; CT4, GZMK+ CD8+ T cells; CT5a, Resident memory CD8+ T cells; CT5b, CD56bright CD16- NK cells; CT6, ISG-high CD4+ T cells; CB0: Activated B cells; CB1, Plasma cells/Plasmablasts; CB2a, Naive B cells; CB2b, Plasmacytoid dendritic cells; CB3, ISG-high B cells; CD0, Dividing cells; CE0, Epithelial cells.
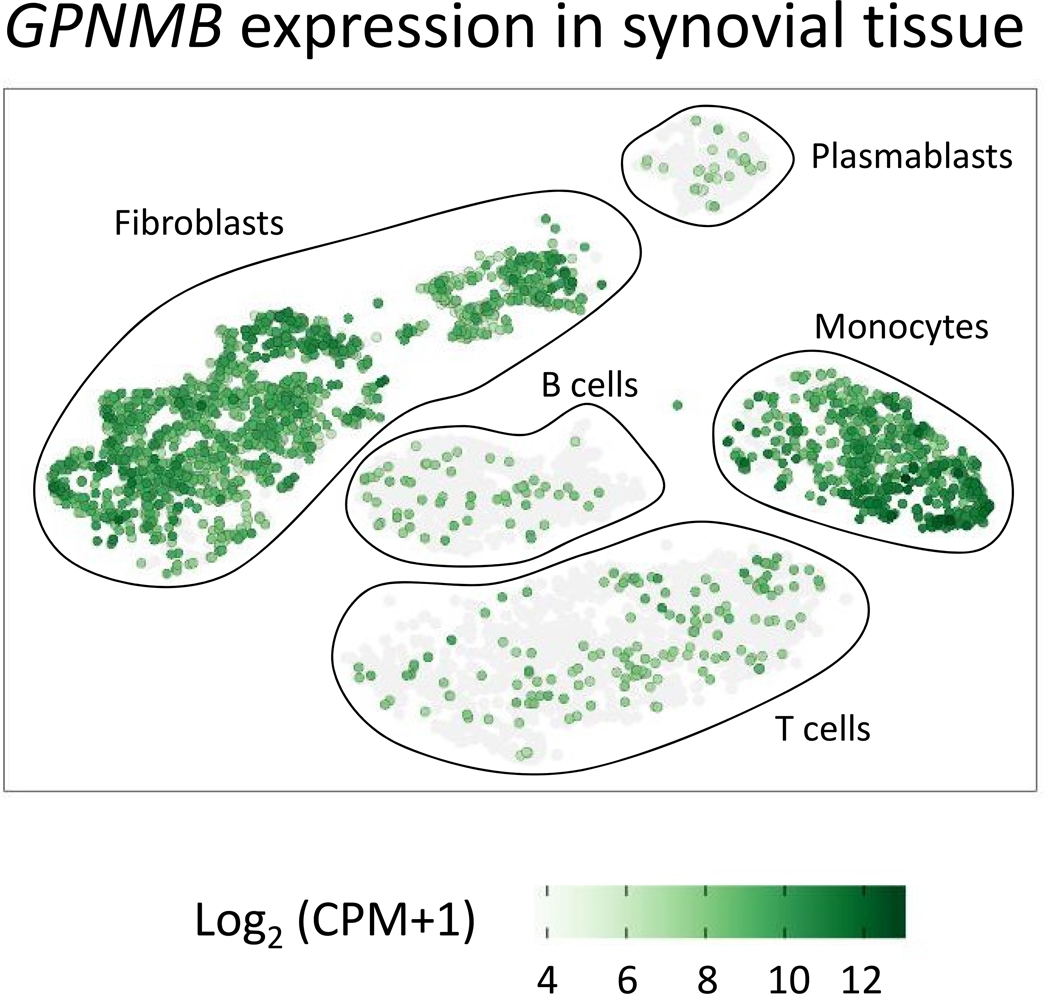
In psoriatic arthritis, GPNMB was upregulated in synovial membranes compared to tissues obtained from healthy subjects (112). GPNMB was among the most highly differentially expressed genes found in the gene array analysis (147 fold change vs. control). It was also elevated in peripheral blood cells from psoriatic arthritis patients. The serum levels of GPNMB was elevated in patients with psoriatic arthritis compared to healthy controls while no difference was observed in patients with rheumatoid arthritis or ankylosing spondylitis, suggesting that GPNMB could be used as a marker for psoriatic arthritis. Analysis of recent single-cell RNA sequencing data derived from the synovial tissue of patients with rheumatoid arthritis and osteoarthritis revealed GPNMB expression in the majority of synovial fibroblasts and tissue-infiltrating monocytes (Figure 3). In monocytes, cells within a NUPR1+ subset which correlated inversely with tissue inflammation demonstrated the highest levels and frequency of GPNMB expression (113).
Figure 3. GPNMB expression in synovial tissue.
Feature plot showing GPNMB expression using single cell RNA-seq data in synovial tissue from rheumatoid arthritis and osteoarthritis patients generated by the Accelerating Medicines Partnership (AMP) project. This figure was generated using the viewing browser https://immunogenomics.io/ampra/
GPNMB has been shown to be possibly implicated in the pathogenic mechanisms underlying Takayasu arteritis in the context of functional genomics (114). Our group revealed a functional effect of a non-coding Takayasu arteritis-associated genetic variant (rs2069837) located within an enhancer region in IL6 on regulating the expression of GPNMB, a gene located more than 500 kb away from this risk locus. By incorporating bioinformatics analysis with experimental assays, they showed that through CTCF-mediated chromatin looping, the Takayasu arteritis risk allele in this genetic variant results in increased recruitment of MEF2 and accumulation of histone deacetylases that repress GPNMB expression. These results were further supported by significantly lower GPNMB mRNA expression levels in monocyte-derived macrophages from individuals with the homozygous risk genotype compared to the heterozygous genotype. This work characterized the functional consequences of this disease-associated risk locus in autoimmunity, and suggested that GPNMB might be involved in the pathogenesis of Takayasu arteritis.
There are no reports suggesting the involvement GPNMB in the pathogenesis of systemic sclerosis, despite data indicating that GPNMB attenuates fibrosis in various disease models. Hepatic fibrosis was reduced in Gpnmb-transgenic rats, accompanied with downregulation of TIMP1, 2, alpha-smooth muscle actin, collagen, and platelet-derived growth factor receptors (115). The protective effect of GPNMB in liver fibrosis has been attributed to its role in macrophages. The CD11Bhi F4/80int Ly-6Clo macrophage subset was found to be most abundant in livers during fibrosis resolution and expressed MMPs, as contrast to profibrotic Ly-6Chi macrophages (116). The gene expression profiles of these Ly-6Clo macrophages is characteristic of favoring scar resolution, including upregulation of Gpnmb. The role of macrophage-mediated protective effect of GPNMB was also suggested in diet-induced obesity model afflicted with liver fibrosis (78). In addition, GPNMB secreted from macrophages was critical for mesenchymal stem cell recruitment in wounds to promote tissue repair (88). In addition to macrophages, GPNMB also exerts its effect on fibroblasts. Stimulating fibroblasts with soluble form of GPNMB activated ERK1/2 and p38 pathway and induced MMP-3 expression in these cells (117). Since GPNMB promotes angiogenesis (25, 68, 106), it could also alleviate the prominent vascular complications seen in patients with systemic sclerosis.
As discussed earlier, GPNMB is critical in the interaction between dendritic cells and T cells. GPNMB on antigen presenting cells inhibits T-cell activation by binding to syndecan-4 on T cells (49, 62). In the EAE model for multiple sclerosis, the GPNMB-syndecan-4 pathway regulates autoimmune responses by mediating the T-cell suppressor function of CD11b+Gr1+ MDSCs (64). The role of GPNMB in regulating T cell activation in autoimmune diseases should be further investigated in the future.
7. Conclusion and Future Perspectives
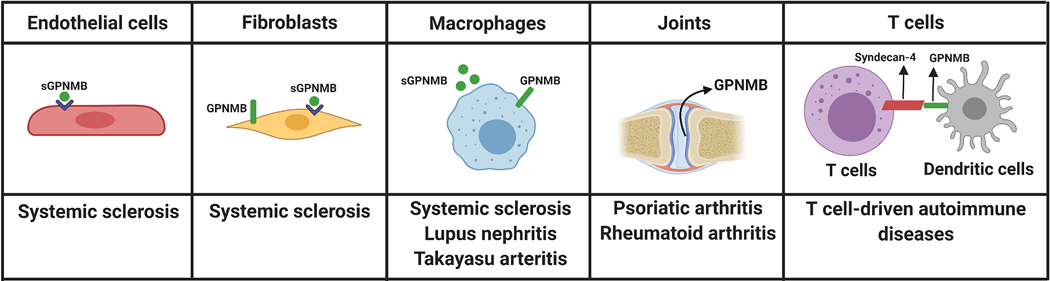
As discussed in this review, GPNMB is widely expressed in many tissues and involved in regulatory roles in various cellular functions. Because of its functions in cell adhesion, migration, proliferation, and differentiation in various cell types, GPNMB has been implicated in a variety of pathological conditions. Aberrant GPNMB expression has been reported in patients with neurodegenerative diseases (69, 75, 92, 93), fatty liver disease (78), Gaucher disease (102), and several types of cancer (3, 118), to name a few. Although the involvement of GPNMB in autoimmune diseases has not been fully elucidated, given the involvement of GPNMB in immune responses, osteogenesis, angiogenesis, and fibrosis, it is possible that GPNMB play an important role in the pathogenesis of autoimmune diseases. Indeed, we discussed current findings related to the involvement of GPNMB in several immune-mediated disease as summarized in Figure 4.
Figure 4. Schematic representation demonstrating potential involvement for GPNMB in multiple autoimmune diseases.
Dysregulation of GPNMB, particularly the soluble form, could play a role in systemic sclerosis, as GPNMB mediates pro-angiogenic effects on endothelial cells, anti-fibrotic potential on fibroblasts, and anti-inflammatory properties in macrophages. GPNMB-expressing macrophages also play a role in lupus nephritis and Takayasu arteritis. The expression of GPNMB in the synovial tissue has been reported in both psoriatic arthritis and rheumatoid arthritis patients. Since GPNMB on antigen presenting cells, such as dendritic cells, inhibits T-cell activation by binding to syndecan-4 on T cells, this interaction might be relevant to T cells-driven autoimmune diseases.
In addition to being involved in disease pathogenesis, GPNMB could be a potential biomarker for autoimmune diseases. Elevated serum levels of GPNMB have been reported in several diseases and its expression correlated with disease severity or prognosis in several conditions (78, 102, 109, 119). In addition, elevated serum levels of GPNMB were found in patients with psoriatic arthritis, but not in patients with rheumatoid arthritis or ankylosing spondylitis (112). Urine GPNMB might be a potential biomarker for lupus nephritis (109). A recent report suggested that the DNA methylation status of GPNMB can be used as a biomarker to assess the progression of colorectal lesions (27). As DNA methylation has been shown to be involved in autoimmune diseases such as systemic sclerosis and lupus (120, 121), it will be interesting to examine the expression and DNA methylation status of GPNMB in these diseases.
Initially found to play multiple roles in the bone, our understanding for the function of GPNMB has since evolved tremendously. Indeed, the anti-inflammatory properties of GPNMB and evidence for dysregulation of GPNMB expression in multiple immune-mediated diseases, suggest that GPNMB regulation or GPNMB-dependent pathways might serve as novel therapeutic target in autoimmunity. However, more research is needed to fully characterize the role of GPNMD upon the pathogenesis and progression of autoimmune diseases.
Acknowledgement
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health grant number R01 AR070148.
Abbreviations
- GPMNB
Glycoprotein nonmetastatic melanoma protein B
- OA
Osteoactivin
- DC-HIL
Dendritic cell-heparin integrin ligand
- SIG
N-terminal signal peptide
- NTD
N-terminal domain (NTD)
- PKD
Polycystic kidney disease
- KLD
Kringle-like domain
- ITAM
Immunoreceptor tyrosine-based activation motif
- CTD
C-terminal domain
- MMP
Matrix metalloproteinase
- ERK
Extracellular signal-regulated kinase
- EZH2
Enhancer of zeste homolog 2
- TSA
Trichostatin A
- MITF
Melanogenesis associated transcription factor
- NUPR1
Nuclear Protein 1, Transcriptional Regulator
- MEF2
Myocyte enhancer factor-2
- EAE
Experimental autoimmune encephalomyelitis
- MDSCs
Myeloid-derived suppressor cells
References
- 1.Weterman MA, Ajubi N, van Dinter IM, Degen WG, van Muijen GN, Ruitter DJ, and Bloemers HP (1995) nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. Int J Cancer 60, 73–81 [DOI] [PubMed] [Google Scholar]
- 2.Terasawa K, Tomabechi Y, Ikeda M, Ehara H, Kukimoto-Niino M, Wakiyama M, Podyma-Inoue KA, Rajapakshe AR, Watabe T, Shirouzu M, and Hara-Yokoyama M (2016) Lysosome-associated membrane proteins-1 and −2 (LAMP-1 and LAMP-2) assemble via distinct modes. Biochem Biophys Res Commun 479, 489–495 [DOI] [PubMed] [Google Scholar]
- 3.Rose AA, Grosset AA, Dong Z, Russo C, Macdonald PA, Bertos NR, St-Pierre Y, Simantov R, Hallett M, Park M, Gaboury L, and Siegel PM (2010) Glycoprotein nonmetastatic B is an independent prognostic indicator of recurrence and a novel therapeutic target in breast cancer. Clin Cancer Res 16, 2147–2156 [DOI] [PubMed] [Google Scholar]
- 4.Kuan C-T, Wakiya K, Dowell JM, Herndon JE, Reardon DA, Graner MW, Riggins GJ, Wikstrand CJ, and Bigner DD (2006) Glycoprotein Nonmetastatic Melanoma Protein B, a Potential Molecular Therapeutic Target in Patients with Glioblastoma Multiforme. Clinical Cancer Research 12, 1970–1982 [DOI] [PubMed] [Google Scholar]
- 5.Li YN, Zhang L, Li XL, Cui DJ, Zheng HD, Yang SY, and Yang WL (2014) Glycoprotein nonmetastatic B as a prognostic indicator in small cell lung cancer. Apmis 122, 140–146 [DOI] [PubMed] [Google Scholar]
- 6.Zhang YX, Qin CP, Zhang XQ, Wang QR, Zhao CB, Yuan YQ, and Yang JG (2017) Knocking down glycoprotein nonmetastatic melanoma protein B suppresses the proliferation, migration, and invasion in bladder cancer cells. Tumour Biol 39, 1010428317699119 [DOI] [PubMed] [Google Scholar]
- 7.Arosarena OA, Barr EW, Thorpe R, Yankey H, Tarr JT, and Safadi FF (2018) Osteoactivin regulates head and neck squamous cell carcinoma invasion by modulating matrix metalloproteases. J Cell Physiol 233, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maric G, Annis MG, MacDonald PA, Russo C, Perkins D, Siwak DR, Mills GB, and Siegel PM (2019) GPNMB augments Wnt-1 mediated breast tumor initiation and growth by enhancing PI3K/AKT/mTOR pathway signaling and beta-catenin activity. Oncogene 38, 5294–5307 [DOI] [PubMed] [Google Scholar]
- 9.Kuan CT, Wakiya K, Dowell JM, Herndon JE 2nd, Reardon DA, Graner MW, Riggins GJ, Wikstrand CJ, and Bigner DD (2006) Glycoprotein nonmetastatic melanoma protein B, a potential molecular therapeutic target in patients with glioblastoma multiforme. Clin Cancer Res 12, 1970–1982 [DOI] [PubMed] [Google Scholar]
- 10.Hoashi T, Sato S, Yamaguchi Y, Passeron T, Tamaki K, and Hearing VJ (2010) Glycoprotein nonmetastatic melanoma protein b, a melanocytic cell marker, is a melanosome-specific and proteolytically released protein. FASEB J 24, 1616–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza SE, Ginsberg MH, and Plow EF (1991) Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem Sci 16, 246–250 [DOI] [PubMed] [Google Scholar]
- 12.Shikano S, Bonkobara M, Zukas PK, and Ariizumi K (2001) Molecular cloning of a dendritic cell-associated transmembrane protein, DC-HIL, that promotes RGD-dependent adhesion of endothelial cells through recognition of heparan sulfate proteoglycans. J Biol Chem 276, 8125–8134 [DOI] [PubMed] [Google Scholar]
- 13.Tomihari M, Hwang SH, Chung JS, Cruz PD Jr., and Ariizumi K (2009) Gpnmb is a melanosome-associated glycoprotein that contributes to melanocyte/keratinocyte adhesion in a RGD-dependent fashion. Exp Dermatol 18, 586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maric G, Annis MG, Dong Z, Rose AAN, Ng S, Perkins D, MacDonald PA, Ouellet V, Russo C, and Siegel PM (2015) GPNMB cooperates with neuropilin-1 to promote mammary tumor growth and engages integrin α5β1 for efficient breast cancer metastasis. Oncogene 34, 5494–5504 [DOI] [PubMed] [Google Scholar]
- 15.Jing H, Takagi J, Liu J. h., Lindgren S, Zhang R. g., Joachimiak A, Wang J. h., and Springer TA (2002) Archaeal Surface Layer Proteins Contain β Propeller, PKD, and β Helix Domains and Are Related to Metazoan Cell Surface Proteins. Structure 10, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 16.Rose A, Russo C, Annis M, and Siegel P (2008) Functional requirement for RGD and PKD domains of GPNMB in breast cancer metastasis. Cancer Res 68, 3681–3681 [Google Scholar]
- 17.Theos AC, Watt B, Harper DC, Janczura KJ, Theos SC, Herman KE, and Marks MS (2013) The PKD domain distinguishes the trafficking and amyloidogenic properties of the pigment cell protein PMEL and its homologue GPNMB. Pigment Cell & Melanoma Research 26, 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung JS, Dougherty I, Cruz PD Jr., and Ariizumi K (2007) Syndecan-4 mediates the coinhibitory function of DC-HIL on T cell activation. J Immunol 179, 5778–5784 [DOI] [PubMed] [Google Scholar]
- 19.Xie R, Okita Y, Ichikawa Y, Fikry MA, Huynh Dam KT, Tran STP, and Kato M (2019) Role of the kringle-like domain in glycoprotein NMB for its tumorigenic potential. Cancer science 110, 2237–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, and Yang L (2016) The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol 18, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Okita Y, Watanabe Y, Abe F, Fikry MA, Ichikawa Y, Suzuki H, Shibuya A, and Kato M (2018) Glycoprotein nmb Is Exposed on the Surface of Dormant Breast Cancer Cells and Induces Stem Cell-like Properties. Cancer Res 78, 6424–6435 [DOI] [PubMed] [Google Scholar]
- 22.Okita Y, Kimura M, Xie R, Chen C, Shen LT-W, Kojima Y, Suzuki H, Muratani M, Saitoh M, Semba K, Heldin C-H, and Kato M (2017) The transcription factor MAFK induces EMT and malignant progression of triple-negative breast cancer cells through its target GPNMB. Science signaling 10, eaak9397 [DOI] [PubMed] [Google Scholar]
- 23.Bonifacino JS, and Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 24.Oyewumi MO, Manickavasagam D, Novak K, Wehrung D, Paulic N, Moussa FM, Sondag GR, and Safadi FF (2016) Osteoactivin (GPNMB) ectodomain protein promotes growth and invasive behavior of human lung cancer cells. Oncotarget 7, 13932–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose AA, Annis MG, Dong Z, Pepin F, Hallett M, Park M, and Siegel PM (2010) ADAM10 releases a soluble form of the GPNMB/Osteoactivin extracellular domain with angiogenic properties. PLoS One 5, e12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furochi H, Tamura S, Mameoka M, Yamada C, Ogawa T, Hirasaka K, Okumura Y, Imagawa T, Oguri S, Ishidoh K, Kishi K, Higashiyama S, and Nikawa T (2007) Osteoactivin fragments produced by ectodomain shedding induce MMP-3 expression via ERK pathway in mouse NIH-3T3 fibroblasts. FEBS letters 581, 5743–5750 [DOI] [PubMed] [Google Scholar]
- 27.Ashktorab H, Rahi H, Nouraie M, Shokrani B, Lee E, Haydari T, Laiyemo AO, Siegel P, and Brim H (2018) GPNMB methylation: a new marker of potentially carcinogenic colon lesions. BMC Cancer 18, 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E, Smoot DT, and Ashktorab H (2009) Distinct High-Profile Methylated Genes in Colorectal Cancer. PLOS ONE 4, e7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruan W-M, Li Y-L, Nie G, Zhou W-X, and Zou X-M (2014) Differential expression of glycoprotein non-metastatic melanoma protein B (GPNMB) involved in trichostatin A-induced apoptosis in gastric cancer. International journal of clinical and experimental medicine 7, 4857–4866 [PMC free article] [PubMed] [Google Scholar]
- 30.Kumari K, Das B, Adhya AK, Rath AK, and Mishra SK (2019) Genome-wide expression analysis reveals six contravened targets of EZH2 associated with breast cancer patient survival. Scientific Reports 9, 1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhaskaran M, Xi D, Wang Y, Huang C, Narasaraju T, Shu W, Zhao C, Xiao X, More S, Breshears M, and Liu L (2012) Identification of microRNAs changed in the neonatal lungs in response to hyperoxia exposure. Physiological Genomics 44, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narasaraju T, Shukla D, More S, Huang C, Zhang L, Xiao X, and Liu L (2015) Role of MicroRNA-150 and Glycoprotein Nonmetastatic Melanoma Protein B in Angiogenesis during Hyperoxia-Induced Neonatal Lung Injury. American Journal of Respiratory Cell and Molecular Biology 52, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F, Wang X, Yang Y, Hu R, Wang W, and Wang Y (2019) The suppressive effects of miR-508–5p on the odontogenic differentiation of human dental pulp stem cells by targeting glycoprotein non-metastatic melanomal protein B. Stem cell research & therapy 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, Liu E, Tang L, Lei Y, Sun X, Hu J, Dong H, Yang S-M, Gao M, and Tang B (2018) Emerging roles and regulation of MiT/TFE transcriptional factors. Cell Communication and Signaling 16, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy C, Khaled M, and Fisher DE (2006) MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12, 406–414 [DOI] [PubMed] [Google Scholar]
- 36.Ripoll VM, Meadows NA, Raggatt L-J, Chang MK, Pettit AR, Cassady AI, and Hume DA (2008) Microphthalmia transcription factor regulates the expression of the novel osteoclast factor GPNMB. Gene 413, 32–41 [DOI] [PubMed] [Google Scholar]
- 37.Loftus SK, Antonellis A, Matera I, Renaud G, Baxter LL, Reid D, Wolfsberg TG, Chen Y, Wang C, Program NCS, Prasad MK, Bessling SL, McCallion AS, Green ED, Bennett DC, and Pavan WJ (2009) Gpnmb is a melanoblast-expressed, MITF-dependent gene. Pigment Cell & Melanoma Research 22, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabriel TL, Tol MJ, Ottenhof R, van Roomen C, Aten J, Claessen N, Hooibrink B, de Weijer B, Serlie MJ, Argmann C, van Elsenburg L, Aerts JM, and van Eijk M (2014) Lysosomal stress in obese adipose tissue macrophages contributes to MITF-dependent Gpnmb induction. Diabetes 63, 3310–3323 [DOI] [PubMed] [Google Scholar]
- 39.Gutknecht M, Geiger J, Joas S, Dörfel D, Salih HR, Müller MR, Grünebach F, and Rittig SM (2015) The transcription factor MITF is a critical regulator of GPNMB expression in dendritic cells. Cell Communication and Signaling 13, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholer J, Ferralli J, Thiry S, and Chiquet-Ehrismann R (2015) The intracellular domain of teneurin-1 induces the activity of microphthalmia-associated transcription factor (MITF) by binding to transcriptional repressor HINT1. J Biol Chem 290, 8154–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Yuan S, Liu J, Wang Y, Zhang Y, Chen X, and Si W (2020) CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. Journal of Cellular Physiology 235, 2071–2079 [DOI] [PubMed] [Google Scholar]
- 42.Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, and Linehan WM (2010) Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 5, e15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba M, Furuya M, Motoshima T, Lang M, Funasaki S, Ma W, Sun H-W, Hasumi H, Huang Y, Kato I, Kadomatsu T, Satou Y, Morris N, Karim BO, Ileva L, Kalen JD, Wilan Krisna LA, Hasumi Y, Sugiyama A, Kurahashi R, Nishimoto K, Oyama M, Nagashima Y, Kuroda N, Araki K, Eto M, Yao M, Kamba T, Suda T, Oike Y, Schmidt LS, and Linehan WM (2019) TFE3 Xp11.2 Translocation Renal Cell Carcinoma Mouse Model Reveals Novel Therapeutic Targets and Identifies GPNMB as a Diagnostic Marker for Human Disease. Molecular Cancer Research 17, 1613–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujiwara KT, Kataoka K, and Nishizawa M (1993) Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene 8, 2371–2380 [PubMed] [Google Scholar]
- 45.Athwal VS, Pritchett J, Martin K, Llewellyn J, Scott J, Harvey E, Zaitoun AM, Mullan AF, Zeef LAH, Friedman SL, Irving WL, Hanley NA, Guha IN, and Piper Hanley K (2018) SOX9 regulated matrix proteins are increased in patients serum and correlate with severity of liver fibrosis. Sci Rep 8, 17905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ripoll VM, Irvine KM, Ravasi T, Sweet MJ, and Hume DA (2007) Gpnmb is induced in macrophages by IFN-gamma and lipopolysaccharide and acts as a feedback regulator of proinflammatory responses. J Immunol 178, 6557–6566 [DOI] [PubMed] [Google Scholar]
- 47.Chung J-S, Tamura K, Cruz PD, and Ariizumi K (2014) DC-HIL-Expressing Myelomonocytic Cells Are Critical Promoters of Melanoma Growth. Journal of Investigative Dermatology 134, 2784–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metz RL, Yehia G, Fernandes H, Donnelly RJ, and Rameshwar P (2005) Cloning and characterization of the 5’ flanking region of the HGFIN gene indicate a cooperative role among p53 and cytokine-mediated transcription factors: relevance to cell cycle regulation. Cell cycle (Georgetown, Tex.) 4, 315–322 [PubMed] [Google Scholar]
- 49.Chung J-S, Bonkobara M, Tomihari M, Cruz PD Jr., and Ariizumi K (2009) The DC-HIL/syndecan-4 pathway inhibits human allogeneic T-cell responses. European Journal of Immunology 39, 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi F, Duan S, Cui J, Yan X, Li H, Wang Y, Chen F, Zhang L, Liu J, and Xie X (2014) Induction of matrix metalloproteinase-3 (MMP-3) expression in the microglia by lipopolysaccharide (LPS) via upregulation of glycoprotein nonmetastatic melanoma B (GPNMB) expression. Journal of molecular neuroscience : MN 54, 234–242 [DOI] [PubMed] [Google Scholar]
- 51.Sasaki F, Kumagai K, Uto H, Takami Y, Kure T, Tabu K, Nasu Y, Hashimoto S, Kanmura S, Numata M, Moriuchi A, Sakiyama T, Tsubouchi H, and Ido A (2015) Expression of glycoprotein nonmetastatic melanoma protein B in macrophages infiltrating injured mucosa is associated with the severity of experimental colitis in mice. Molecular medicine reports 12, 7503–7511 [DOI] [PubMed] [Google Scholar]
- 52.Tomihari M, Hwang S-H, Chung J-S, Cruz PD Jr., and Ariizumi K (2009) Gpnmb is a melanosome-associated glycoprotein that contributes to melanocyte/keratinocyte adhesion in a RGD-dependent fashion. Exp Dermatol 18, 586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian F, Liu C, Wu Q, Qu K, Wang R, Wei J, Meng F, Liu S, and Chang H (2013) Upregulation of glycoprotein nonmetastatic B by colony-stimulating factor-1 and epithelial cell adhesion molecule in hepatocellular carcinoma cells. Oncology research 20, 341–350 [DOI] [PubMed] [Google Scholar]
- 54.Abdelmagid SM, Barbe MF, Arango-Hisijara I, Owen TA, Popoff SN, and Safadi FF (2007) Osteoactivin acts as downstream mediator of BMP-2 effects on osteoblast function. J Cell Physiol 210, 26–37 [DOI] [PubMed] [Google Scholar]
- 55.Singh M, Del Carpio-Cano FE, Monroy MA, Popoff SN, and Safadi FF (2012) Homeodomain transcription factors regulate BMP-2-induced osteoactivin transcription in osteoblasts. J Cell Physiol 227, 390–399 [DOI] [PubMed] [Google Scholar]
- 56.Zhang P, Liu W, Yuan X, Li D, Gu W, and Gao T (2013) Endothelin-1 enhances the melanogenesis via MITF-GPNMB pathway. BMB reports 46, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smuczek B, Santos E. d. S., Siqueira AS, Pinheiro JJV, Freitas VM, and Jaeger RG (2017) The laminin-derived peptide C16 regulates GPNMB expression and function in breast cancer. Experimental Cell Research 358, 323–334 [DOI] [PubMed] [Google Scholar]
- 58.Safadi FF, Xu J, Smock SL, Rico MC, Owen TA, and Popoff SN (2001) Cloning and characterization of osteoactivin, a novel cDNA expressed in osteoblasts. J Cell Biochem 84, 12–26 [DOI] [PubMed] [Google Scholar]
- 59.Moussa FM, Hisijara IA, Sondag GR, Scott EM, Frara N, Abdelmagid SM, and Safadi FF (2014) Osteoactivin promotes osteoblast adhesion through HSPG and alphavbeta1 integrin. J Cell Biochem 115, 1243–1253 [DOI] [PubMed] [Google Scholar]
- 60.Sheng MH, Wergedal JE, Mohan S, and Lau KH (2008) Osteoactivin is a novel osteoclastic protein and plays a key role in osteoclast differentiation and activity. FEBS letters 582, 1451–1458 [DOI] [PubMed] [Google Scholar]
- 61.Akiyoshi H, Chung J-S, Tomihari M, Cruz PD, and Ariizumi K (2010) Depleting Syndecan-4+ T Lymphocytes Using Toxin-Bearing Dendritic Cell-Associated Heparan Sulfate Proteoglycan-Dependent Integrin Ligand: A New Opportunity for Treating Activated T Cell-Driven Disease. The Journal of Immunology 184, 3554–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung J-S, Dougherty I, Cruz PD, and Ariizumi K (2007) Syndecan-4 Mediates the Coinhibitory Function of DC-HIL on T Cell Activation. The Journal of Immunology 179, 5778–5784 [DOI] [PubMed] [Google Scholar]
- 63.Chung J-S, Shiue LH, Duvic M, Pandya A, Cruz PD Jr, and Ariizumi K (2011) Sézary syndrome cells overexpress syndecan-4 bearing distinct heparan sulfate moieties that suppress T-cell activation by binding DC-HIL and trapping TGF-β on the cell surface. Blood 117, 3382–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung J-S, Tamura K, Akiyoshi H, Cruz PD, and Ariizumi K (2014) The DC-HIL/Syndecan-4 Pathway Regulates Autoimmune Responses through Myeloid-Derived Suppressor Cells. The Journal of Immunology 192, 2576–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung J-S, Tomihari M, Tamura K, Kojima T, Cruz PD Jr, and Ariizumi K (2013) The DC-HIL ligand syndecan-4 is a negative regulator of T-cell allo-reactivity responsible for graft-versus-host disease. Immunology 138, 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kobayashi M, Chung JS, Beg M, Arriaga Y, Verma U, Courtney K, Mansour J, Haley B, Khan S, Horiuchi Y, Ramani V, Harker D, Gopal P, Araghizadeh F, Cruz PD Jr., and Ariizumi K (2019) Blocking Monocytic Myeloid-Derived Suppressor Cell Function via Anti-DC-HIL/GPNMB Antibody Restores the In Vitro Integrity of T Cells from Cancer Patients. Clin Cancer Res 25, 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomihari M, Chung J-S, Akiyoshi H, Cruz PD, and Ariizumi K (2010) DC-HIL/Glycoprotein Nmb Promotes Growth of Melanoma in Mice by Inhibiting the Activation of Tumor-Reactive T Cells. Cancer Res 70, 5778–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu X, Zhang P, Xu Z, Chen H, and Xie X (2013) GPNMB enhances bone regeneration by promoting angiogenesis and osteogenesis: potential role for tissue engineering bone. J Cell Biochem 114, 2729–2737 [DOI] [PubMed] [Google Scholar]
- 69.Tanaka H, Shimazawa M, Kimura M, Takata M, Tsuruma K, Yamada M, Takahashi H, Hozumi I, Niwa J, Iguchi Y, Nikawa T, Sobue G, Inuzuka T, and Hara H (2012) The potential of GPNMB as novel neuroprotective factor in amyotrophic lateral sclerosis. Sci Rep 2, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ono Y, Tsuruma K, Takata M, Shimazawa M, and Hara H (2016) Glycoprotein nonmetastatic melanoma protein B extracellular fragment shows neuroprotective effects and activates the PI3K/Akt and MEK/ERK pathways via the Na+/K+-ATPase. Scientific Reports 6, 23241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayed M, Drummond CA, Evans KL, Haller ST, Liu J, Xie Z, and Tian J (2014) Effects of Na/K-ATPase and its ligands on bone marrow stromal cell differentiation. Stem cell research 13, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naor D, Sionov RV, and Ish-Shalom D (1997) CD44: structure, function, and association with the malignant process. Adv Cancer Res 71, 241–319 [DOI] [PubMed] [Google Scholar]
- 73.Yu B, Sondag GR, Malcuit C, Kim MH, and Safadi FF (2016) Macrophage-Associated Osteoactivin/GPNMB Mediates Mesenchymal Stem Cell Survival, Proliferation, and Migration Via a CD44-Dependent Mechanism. J Cell Biochem 117, 1511–1521 [DOI] [PubMed] [Google Scholar]
- 74.Sondag GR, Mbimba TS, Moussa FM, Novak K, Yu B, Jaber FA, Abdelmagid SM, Geldenhuys WJ, and Safadi FF (2016) Osteoactivin inhibition of osteoclastogenesis is mediated through CD44-ERK signaling. Exp Mol Med 48, e257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neal ML, Boyle AM, Budge KM, Safadi FF, and Richardson JR (2018) The glycoprotein GPNMB attenuates astrocyte inflammatory responses through the CD44 receptor. Journal of neuroinflammation 15, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gong X-M, Li Y-F, Luo J, Wang J-Q, Wei J, Wang J-Q, Xiao T, Xie C, Hong J, Ning G, Shi X-J, Li B-L, Qi W, and Song B-L (2019) Gpnmb secreted from liver promotes lipogenesis in white adipose tissue and aggravates obesity and insulin resistance. Nature Metabolism 1, 570–583 [DOI] [PubMed] [Google Scholar]
- 77.Chung J-S, Yudate T, Tomihari M, Akiyoshi H, Cruz PD, and Ariizumi K (2009) Binding of DC-HIL to Dermatophytic Fungi Induces Tyrosine Phosphorylation and Potentiates Antigen Presenting Cell Function. The Journal of Immunology 183, 5190–5198 [DOI] [PubMed] [Google Scholar]
- 78.Katayama A, Nakatsuka A, Eguchi J, Murakami K, Teshigawara S, Kanzaki M, Nunoue T, Hida K, Wada N, Yasunaka T, Ikeda F, Takaki A, Yamamoto K, Kiyonari H, Makino H, and Wada J (2015) Beneficial impact of Gpnmb and its significance as a biomarker in nonalcoholic steatohepatitis. Sci Rep 5, 16920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.The Human Protein Atlas. Vol. 2020 [Google Scholar]
- 80.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, and Pontén F (2015) Tissue-based map of the human proteome. Science 347, 1260419 [DOI] [PubMed] [Google Scholar]
- 81.Abdelmagid SM, Barbe MF, Rico MC, Salihoglu S, Arango-Hisijara I, Selim AH, Anderson MG, Owen TA, Popoff SN, and Safadi FF (2008) Osteoactivin, an anabolic factor that regulates osteoblast differentiation and function. Exp Cell Res 314, 2334–2351 [DOI] [PubMed] [Google Scholar]
- 82.Frara N, Abdelmagid SM, Sondag GR, Moussa FM, Yingling VR, Owen TA, Popoff SN, Barbe MF, and Safadi FF (2016) Transgenic Expression of Osteoactivin/gpnmb Enhances Bone Formation In Vivo and Osteoprogenitor Differentiation Ex Vivo. J Cell Physiol 231, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abdelmagid SM, Sondag GR, Moussa FM, Belcher JY, Yu B, Stinnett H, Novak K, Mbimba T, Khol M, Hankenson KD, Malcuit C, and Safadi FF (2015) Mutation in Osteoactivin Promotes Receptor Activator of NFkappaB Ligand (RANKL)-mediated Osteoclast Differentiation and Survival but Inhibits Osteoclast Function. J Biol Chem 290, 20128–20146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abdelmagid SM, Belcher JY, Moussa FM, Lababidi SL, Sondag GR, Novak KM, Sanyurah AS, Frara NA, Razmpour R, Del Carpio-Cano FE, and Safadi FF (2014) Mutation in osteoactivin decreases bone formation in vivo and osteoblast differentiation in vitro. Am J Pathol 184, 697–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheng MH, Wergedal JE, Mohan S, Amoui M, Baylink DJ, and Lau KH (2012) Targeted overexpression of osteoactivin in cells of osteoclastic lineage promotes osteoclastic resorption and bone loss in mice. PLoS One 7, e35280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang P, Liu W, Zhu C, Yuan X, Li D, Gu W, Ma H, Xie X, and Gao T (2012) Silencing of GPNMB by siRNA inhibits the formation of melanosomes in melanocytes in a MITF-independent fashion. PloS one 7, e42955-e42955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang CF, Lin SP, Chiang CP, Wu YH, H’Ng W S, Chang CP, Chen YT, and Wu JY (2018) Loss of GPNMB Causes Autosomal-Recessive Amyloidosis Cutis Dyschromica in Humans. Am J Hum Genet 102, 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu B, Alboslemy T, Safadi F, and Kim MH (2018) Glycoprotein Nonmelanoma Clone B Regulates the Crosstalk between Macrophages and Mesenchymal Stem Cells toward Wound Repair. J Invest Dermatol 138, 219–227 [DOI] [PubMed] [Google Scholar]
- 89.Huang J-J, Ma W-J, and Yokoyama S (2012) Expression and immunolocalization of Gpnmb, a glioma-associated glycoprotein, in normal and inflamed central nervous systems of adult rats. Brain Behav 2, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakano Y, Suzuki Y, Takagi T, Kitashoji A, Ono Y, Tsuruma K, Yoshimura S, Shimazawa M, Iwama T, and Hara H (2014) Glycoprotein nonmetastatic melanoma protein B (GPNMB) as a novel neuroprotective factor in cerebral ischemia-reperfusion injury. Neuroscience 277, 123–131 [DOI] [PubMed] [Google Scholar]
- 91.Murata K, Yoshino Y, Tsuruma K, Moriguchi S, Oyagi A, Tanaka H, Ishisaka M, Shimazawa M, Fukunaga K, and Hara H (2015) The extracellular fragment of GPNMB (Glycoprotein nonmelanosoma protein B, osteoactivin) improves memory and increases hippocampal GluA1 levels in mice. J Neurochem 132, 583–594 [DOI] [PubMed] [Google Scholar]
- 92.Moloney EB, Moskites A, Ferrari EJ, Isacson O, and Hallett PJ (2018) The glycoprotein GPNMB is selectively elevated in the substantia nigra of Parkinson’s disease patients and increases after lysosomal stress. Neurobiology of disease 120, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huttenrauch M, Ogorek I, Klafki H, Otto M, Stadelmann C, Weggen S, Wiltfang J, and Wirths O (2018) Glycoprotein NMB: a novel Alzheimer’s disease associated marker expressed in a subset of activated microglia. Acta neuropathologica communications 6, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu S, Wuolikainen A, Wu J, Ohman A, Wingsle G, Moritz T, Andersen PM, Forsgren L, and Trupp M (2019) Targeted Multiple Reaction Monitoring Analysis of CSF Identifies UCHL1 and GPNMB as Candidate Biomarkers for ALS. Journal of molecular neuroscience : MN 69, 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou L, Zhuo H, Ouyang H, Liu Y, Yuan F, Sun L, Liu F, and Liu H (2017) Glycoprotein non-metastatic melanoma protein b (Gpnmb) is highly expressed in macrophages of acute injured kidney and promotes M2 macrophages polarization. Cellular Immunology 316, 53–60 [DOI] [PubMed] [Google Scholar]
- 96.Kumagai K, Tabu K, Sasaki F, Takami Y, Morinaga Y, Mawatari S, Hashimoto S, Tanoue S, Kanmura S, Tamai T, Moriuchi A, Uto H, Tsubouchi H, and Ido A (2015) Glycoprotein Nonmetastatic Melanoma B (Gpnmb)-Positive Macrophages Contribute to the Balance between Fibrosis and Fibrolysis during the Repair of Acute Liver Injury in Mice. PLOS ONE 10, e0143413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li B, Castano AP, Hudson TE, Nowlin BT, Lin S-L, Bonventre JV, Swanson KD, and Duffield JS (2010) The melanoma-associated transmembrane glycoprotein Gpnmb controls trafficking of cellular debris for degradation and is essential for tissue repair. The FASEB Journal 24, 4767–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anderson MG, Nair KS, Amonoo LA, Mehalow A, Trantow CM, Masli S, and John SW (2008) GpnmbR150X allele must be present in bone marrow derived cells to mediate DBA/2J glaucoma. BMC genetics 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi MS, Kim YJ, Kwon EY, Ryoo JY, Kim SR, and Jung UJ (2015) High-fat diet decreases energy expenditure and expression of genes controlling lipid metabolism, mitochondrial function and skeletal system development in the adipose tissue, along with increased expression of extracellular matrix remodelling- and inflammation-related genes. Br J Nutr 113, 867–877 [DOI] [PubMed] [Google Scholar]
- 100.Zambonelli P, Gaffo E, Zappaterra M, Bortoluzzi S, and Davoli R (2016) Transcriptional profiling of subcutaneous adipose tissue in Italian Large White pigs divergent for backfat thickness. Anim Genet 47, 306–323 [DOI] [PubMed] [Google Scholar]
- 101.Kramer G, Wegdam W, Donker-Koopman W, Ottenhoff R, Gaspar P, Verhoek M, Nelson J, Gabriel T, Kallemeijn W, Boot RG, Laman JD, Vissers JPC, Cox T, Pavlova E, Moran MT, Aerts JM, and van Eijk M (2016) Elevation of glycoprotein nonmetastatic melanoma protein B in type 1 Gaucher disease patients and mouse models. FEBS Open Bio 6, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murugesan V, Liu J, Yang R, Lin H, Lischuk A, Pastores G, Zhang X, Chuang WL, and Mistry PK (2018) Validating glycoprotein non-metastatic melanoma B (gpNMB, osteoactivin), a new biomarker of Gaucher disease. Blood Cells Mol Dis 68, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bachner D, Schroder D, and Gross G (2002) mRNA expression of the murine glycoprotein (transmembrane) nmb (Gpnmb) gene is linked to the developing retinal pigment epithelium and iris. Brain Res Gene Expr Patterns 1, 159–165 [DOI] [PubMed] [Google Scholar]
- 104.Mo J-S, Anderson MG, Gregory M, Smith RS, Savinova OV, Serreze DV, Ksander BR, Streilein JW, and John SWM (2003) By Altering Ocular Immune Privilege, Bone Marrow–derived Cells Pathogenically Contribute to DBA/2J Pigmentary Glaucoma. Journal of Experimental Medicine 197, 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang YL, Hu YJ, and Zhang FH (2015) Effects of GPNMB on proliferation and odontoblastic differentiation of human dental pulp cells. International journal of clinical and experimental pathology 8, 6498–6504 [PMC free article] [PubMed] [Google Scholar]
- 106.Narasaraju T, Shukla D, More S, Huang C, Zhang L, Xiao X, and Liu L (2015) Role of microRNA-150 and glycoprotein nonmetastatic melanoma protein B in angiogenesis during hyperoxia-induced neonatal lung injury. Am J Respir Cell Mol Biol 52, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bao G, Wang N, Li R, Xu G, Liu P, and He B (2016) Glycoprotein non-metastaticmelanoma protein B promotes glioma motility and angiogenesis through the Wnt/beta-catenin signaling pathway. Experimental biology and medicine (Maywood, N.J.) 241, 1968–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pahl MV, Vaziri ND, Yuan J, and Adler SG (2010) Upregulation of monocyte/macrophage HGFIN (Gpnmb/Osteoactivin) expression in end-stage renal disease. Clin J Am Soc Nephrol 5, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patel-Chamberlin M, Wang Y, Satirapoj B, Phillips LM, Nast CC, Dai T, Watkins RA, Wu X, Natarajan R, Leng A, Ulanday K, Hirschberg RR, Lapage J, Nam EJ, Haq T, and Adler SG (2011) Hematopoietic growth factor inducible neurokinin-1 (Gpnmb/Osteoactivin) is a biomarker of progressive renal injury across species. Kidney international 79, 1138–1148 [DOI] [PubMed] [Google Scholar]
- 110.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, and Diamond B (2019) The immune cell landscape in kidneys of patients with lupus nephritis. Nature immunology 20, 902–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bethunaickan R, Berthier CC, Ramanujam M, Sahu R, Zhang W, Sun Y, Bottinger EP, Ivashkiv L, Kretzler M, and Davidson A (2011) A Unique Hybrid Renal Mononuclear Phagocyte Activation Phenotype in Murine Systemic Lupus Erythematosus Nephritis. The Journal of Immunology 186, 4994–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dolcino M, Ottria A, Barbieri A, Patuzzo G, Tinazzi E, Argentino G, Beri R, Lunardi C, and Puccetti A (2015) Gene Expression Profiling in Peripheral Blood Cells and Synovial Membranes of Patients with Psoriatic Arthritis. PloS one 10, e0128262-e0128262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, Watts GFM, Jonsson AH, Rangel-Moreno J, Meednu N, Rozo C, Apruzzese W, Eisenhaure TM, Lieb DJ, Boyle DL, Mandelin AM 2nd, Accelerating Medicines Partnership Rheumatoid, A., Systemic Lupus Erythematosus, C. Boyce BF, DiCarlo E, Gravallese EM, Gregersen PK, Moreland L, Firestein GS, Hacohen N, Nusbaum C, Lederer JA, Perlman H, Pitzalis C, Filer A, Holers VM, Bykerk VP, Donlin LT, Anolik JH, Brenner MB, and Raychaudhuri S (2019) Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 20, 928–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kong X, and Sawalha AH (2019) Takayasu arteritis risk locus in IL6 represses the anti-inflammatory gene GPNMB through chromatin looping and recruiting MEF2–HDAC complex. Ann Rheum Dis 78, 1388–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Abe H, Uto H, Takami Y, Takahama Y, Hasuike S, Kodama M, Nagata K, Moriuchi A, Numata M, Ido A, and Tsubouchi H (2007) Transgenic expression of osteoactivin in the liver attenuates hepatic fibrosis in rats. Biochemical and Biophysical Research Communications 356, 610–615 [DOI] [PubMed] [Google Scholar]
- 116.Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, Williams MJ, Dunbar DR, Manning JR, van Rooijen N, Fallowfield JA, Forbes SJ, and Iredale JP (2012) Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci U S A 109, E3186–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Furochi H, Tamura S, Mameoka M, Yamada C, Ogawa T, Hirasaka K, Okumura Y, Imagawa T, Oguri S, Ishidoh K, Kishi K, Higashiyama S, and Nikawa T (2007) Osteoactivin fragments produced by ectodomain shedding induce MMP-3 expression via ERK pathway in mouse NIH-3T3 fibroblasts. FEBS letters 581, 5743–5750 [DOI] [PubMed] [Google Scholar]
- 118.Ren F, Zhao Q, Liu B, Sun X, Tang Y, Huang H, Mei L, Yu Y, Mo H, Dong H, Zheng P, and Mi Y (2020) Transcriptome analysis reveals GPNMB as a potential therapeutic target for gastric cancer. J Cell Physiol 235, 2738–2752 [DOI] [PubMed] [Google Scholar]
- 119.Li H, Xiao Y, Wu CC, Yang LL, Cao LY, Chen DR, Zhou JJ, Zhang WF, and Sun ZJ (2019) High expression of GPNMB predicts poor prognosis in head and neck squamous cell carcinoma. Histology and histopathology 34, 803–810 [DOI] [PubMed] [Google Scholar]
- 120.Altorok N, Tsou PS, Coit P, Khanna D, and Sawalha AH (2015) Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis 74, 1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, Merrill JT, McCune WJ, and Sawalha AH (2013) Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. Journal of autoimmunity 43, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]