Abstract
The study described here was undertaken to extend the observation that some transcription factors can either stimulate or suppress gene expression depending on the local environment of their DNA binding site. It is suggested that if such transcription factors also had a mechanism to sense the expression level of the gene they control, then they could create a feedback loop able to keep expression of a gene within a limited range. The transcription factor would be activating if gene expression were determined to be too low and repressing if it were too high. To test the above idea, I have examined the effect of gene expression on the ability of the transcription factor binding areas, the promoter/enhancers, to stimulate or attenuate gene expression depending on the existing expression level of a gene. Studies were carried out with a population of 61 human genes expressed selectively in liver. A similar study was carried out with thyroid genes. The total length of all promoter/enhancers in each gene sequence was determined and compared in weakly and strongly expressed genes. The results showed that the level of expression was stimulated by promoter/enhancers in weakly expressed genes and antagonized in strongly expressed ones. The results are interpreted to indicate that promoter/enhancers act to keep expression of a gene within a defined range that is appropriate for the gene's function.
Keywords: Molecular biology, Cell biology, Genetics, Gene expression, Genomics, Human genetics, Promoter, Transcription factor, Transcription, Feedback loop, Promoter/enhancer
Molecular biology; Cell biology; Genetics; Gene expression; Genomics; Human genetics; Promoter; Transcription factor; Transcription; Feedback loop; Promoter/enhancer
1. Introduction
Among the most intriguing features of transcription factors is their ability to activate or suppress expression of a target gene depending on the DNA sequence and other components near the transcription factor binding site. Transcription factors are not always activating or always suppressing; some can do either one depending on the situation. This feature allows transcription factors to act flexibly to regulate gene expression by responding to signals in the DNA sequence rather than only to the properties of the transcription factor itself. An example is the rat Pax-8 transcription factor [1]. In the presence of TTF-2, Pax-8 activates the Slc5a5 (NIS) enhancer, but not the thyroglobulin promoter, a site it is able to activate in the absence of TTF-2. Similar context-dependent gene activation or repression has been documented in other transcription factors including the glucocorticoid receptor and mouse USF2 [2,3].
The ability to act either positively or negatively raises the possibility that transcription factors may be involved in a feedback loop to keep expression of their gene within a limited range. Apart from the known functions of transcription factors, the only novel feature needed would be the ability of the transcription factor to sense the current expression level of its gene. With such a sensing ability a transcription factor would be able to suppress gene expression if was too high and activate expression that was too low.
Here I provide evidence that such feedback loops exist in the case of human liver genes and in genes expressed in thyroid. Liver gene studies were carried out with a population 61 genes all of which contain annotated promoter/enhancer regions. In all 61, liver is the tissue where expression is highest. For each gene, I downloaded the aggregate length of all annotated promoter/enhancer regions within the gene boundary. Total promoter/enhancer lengths were then compared with the level of gene expression in gene sub-populations having low and high expression levels. The results were interpreted to support the existence of a feedback loop if greater promoter/enhancer length was found to correlate with gene activation among weakly expressed genes and suppression among highly expressed ones.
2. Materials and methods
2.1. Gene databases
Genes employed in this study were all obtained from a database of 2413 human genes each with selective expression in a sub-set of tissues [4]. Broadly expressed genes are not present in the database. For each gene, the tissue with the highest expression is recorded in the data base, and genes for liver (117 genes) and thyroid (56 genes) were used for this study (see Supplementary Tables 1 and 2, respectively).
As only genes with promoter/enhancer regions were required, genes lacking them were deleted from the liver and thyroid datasets described above. Of the 117 liver database genes, 61 (52%) were found to have annotated promoter/enhancer sequences and were accepted for the study. Similarly, 39 (70%) of 56 thyroid genes were accepted. Information about genes in the two promoter/enhancer-containing populations are shown in Tables 1 and 2.
Table 1.
Liver promoter/enhancer containing genes employed in the study.
| Gene | Chr | Expa | Pr/Enh Lengthb |
Gene | Chr | Exp | Prom/Enh length |
|---|---|---|---|---|---|---|---|
| NR0B2 | 1 | 40.9 | 7024 | SLCO1B3 | 12 | 32.6 | 367 |
| CYP4A11 | 1 | 155.0 | 2942 | AMDHD1 | 12 | 46.0 | 2355 |
| C8A | 1 | 175.8 | 4210 | NR1H4 | 12 | 24.3 | 13271 |
| F5 | 1 | 21.5 | 9132 | MIA2 | 14 | 1.1 | 3267 |
| C4BPA | 1 | 262.8 | 848 | LINC01146 | 14 | 5.3 | 113 |
| PROX1 | 1 | 18.8 | 10345 | ASPG | 14 | 59.8 | 5005 |
| AGT | 1 | 630.3 | 2560 | PCSK6 | 15 | 64.1 | 16783 |
| APOB | 2 | 206.5 | 2623 | ACSM5 | 16 | 64.4 | 3426 |
| FABP1 | 2 | 469.8 | 6721 | CES1 | 16 | 223.0 | 1371 |
| ABCB11 | 2 | 6.5 | 6310 | CA5A | 16 | 15.5 | 3170 |
| MOGAT1 | 2 | 1.4 | 3313 | SERPINF2 | 17 | 669.0 | 3747 |
| C2orf72 | 2 | 60.5 | 4072 | TM4SF5 | 17 | 66.9 | 3086 |
| RAB17 | 2 | 59.4 | 10699 | SLC13A5 | 17 | 163.4 | 3825 |
| ITIH1 | 3 | 329.0 | 2342 | ASGR2 | 17 | 232.2 | 4103 |
| NR1I2 | 3 | 23.8 | 8604 | PIPOX | 17 | 81.8 | 25025 |
| UROC1 | 3 | 47.4 | 1171 | ENPP7 | 17 | 2.3 | 9837 |
| SLC2A2 | 3 | 88.8 | 16823 | TMEM105 | 17 | 0.7 | 11323 |
| FETUB | 3 | 29.9 | 1100 | TTR | 18 | 1516.9 | 706 |
| KLB | 4 | 5.1 | 1682 | ONECUT2 | 18 | 2.6 | 16026 |
| F11 | 4 | 47.8 | 2781 | LRG1 | 19 | 463.4 | 6659 |
| AGXT2 | 5 | 34.9 | 3262 | TNFSF14 | 19 | 27.8 | 3870 |
| ACOT12 | 5 | 23.2 | 1200 | C3 | 19 | 1009.1 | 6411 |
| IGFBP1 | 7 | 198.9 | 2430 | PALM3 | 19 | 21.0 | 4458 |
| MLXIPL | 7 | 244.9 | 2644 | CYP4F3 | 19 | 76.1 | 3318 |
| ABCB4 | 7 | 18.1 | 3044 | HPN | 19 | 275.0 | 3243 |
| PON3 | 7 | 83.7 | 2259 | FOXA3 | 19 | 19.1 | 5686 |
| AKR1D1 | 7 | 31.3 | 21240 | SULT2A1 | 19 | 241.9 | 2677 |
| FGL1 | 8 | 848.6 | 5244 | LINC00261 | 20 | 38.7 | 6145 |
| ADRA1A | 8 | 8.4 | 1952 | HNF4A | 20 | 43.9 | 50902 |
| GAS2 | 11 | 5.0 | 11464 | SEC14L4 | 22 | 8.7 | 1522 |
| OTC | 23 | 34.4 | 8346 |
Gene expression in RPKM.
Total Promoter/Enhancer length in gene (bp).
Table 2.
Thyroid promoter/enhancer containing genes employed in the study.
| Gene | Chr | Expa | Pr/Enh lengthb | Gene | Chr | Exp | Pr/Enh length |
|---|---|---|---|---|---|---|---|
| BMP8A | 1 | 7.1 | 4391 | PREX2 | 8 | 6.6 | 9502 |
| TPO | 2 | 499.9 | 3459 | PKHD1L1 | 8 | 28.5 | 3741 |
| IQCA1 | 2 | 14.1 | 2915 | FAM189A2 | 9 | 56.2 | 7691 |
| ITGA9 | 3 | 15.7 | 37584 | AFAP1L2 | 10 | 49.9 | 21550 |
| FRMD4B | 3 | 11.8 | 22466 | TCERG1L | 10 | 9.7 | 2451 |
| ATP13A4 | 3 | 23.2 | 4789 | RMST | 12 | 37.2 | 13555 |
| ATP8A1 | 4 | 26.6 | 19234 | LINC00571 | 13 | 0.3 | 2873 |
| KDR | 4 | 31.9 | 2738 | SFTA3 | 14 | 77.5 | 738 |
| FRAS1 | 4 | 5.5 | 12453 | TSHR | 14 | 84.4 | 11846 |
| MMRN1 | 4 | 19.6 | 9406 | OCA2 | 15 | 3.6 | 3743 |
| UNC5C | 4 | 4.6 | 4357 | DAPK2 | 15 | 21.8 | 47601 |
| NPNT | 4 | 108.6 | 8506 | C16orf89 | 16 | 94.2 | 1348 |
| EPB41L4A | 5 | 4.5 | 8141 | KCNJ16 | 17 | 51.8 | 4897 |
| COL23A1 | 5 | 42.7 | 23633 | KLHL14 | 18 | 7.7 | 4870 |
| IYD | 6 | 147.2 | 1080 | TLE6 | 19 | 4.3 | 4235 |
| FNDC1 | 6 | 12.3 | 3098 | PLVAP | 19 | 340.8 | 556 |
| ABCA13 | 7 | 0.1 | 2790 | ISM1 | 20 | 35.7 | 4142 |
| WDR86 | 7 | 21.9 | 6292 | SALL4 | 20 | 3.9 | 6014 |
| GFRA2 | 8 | 9.3 | 2673 | LONRF3 | 23 | 4.8 | 2325 |
Gene expression in RPKM.
Total promoter/enhancer length in gene (bp).
2.2. Promoter/enhancers and gene expression
Promoter/enhancers were those identified by GeneHancer [5] and were downloaded from the UCSC Genome browser (https://genome.ucsc.edu/). For all genes, the value of the promoter/enhancer length was the sum of the lengths of the promoter and all enhancers present in the gene. Nearly all genes have a promoter near the transcription start site plus between one and several annotated enhancers, and all were used. No effort was made to include enhancers outside the gene sequence. Enhancer numbers varied from 0-8 in the case of liver genes and 0–15 in thyroid. Gene expression values were downloaded from the UCSC Genome Browser human reference genome (hg38).
2.3. Randomization of promoter/enhancer lengths
Control studies were performed beginning with promoter/enhancer and gene expression results shown in Tables 1 and 2. For each gene in the control analysis, the promoter/enhancer length result was assigned at random to the expression value of a different gene using Python random.randint(). Randomized and authentic data were thereafter treated identically.
2.4. Data handling
SigmaPlot v13.0 was employed to render results graphically, compute linear regression plots and compute statistical parameters.
3. Results
3.1. Experimental strategy
The strategy is described here for liver genes although the same approach was employed for both liver and thyroid populations. The 61 liver genes were first divided into three groups based on their level of expression. For each of the three (low, high and very high expression groups), the expression level of each gene was plotted against its total promoter/enhancer length and a linear regression was calculated. If the hypothesis to be tested is correct, then the regression line was expected to have a positive slope in the case of weakly expressed genes and a negative slope with strongly expressed ones. The expected result would indicate that each additional increment of promoter/enhancer length produces an increase in the expression of weakly expressed genes, but a decreased expression in strongly expressed ones. A control experiment was carried out in which the promoter/enhancer length for each gene was associated with the expression level of a different, randomly chosen one of the 61 liver genes. Regression analysis of the randomized population was expected to differ from that of the authentic data.
3.2. Liver genes
3.2.1. Gene populations that differ in expression level
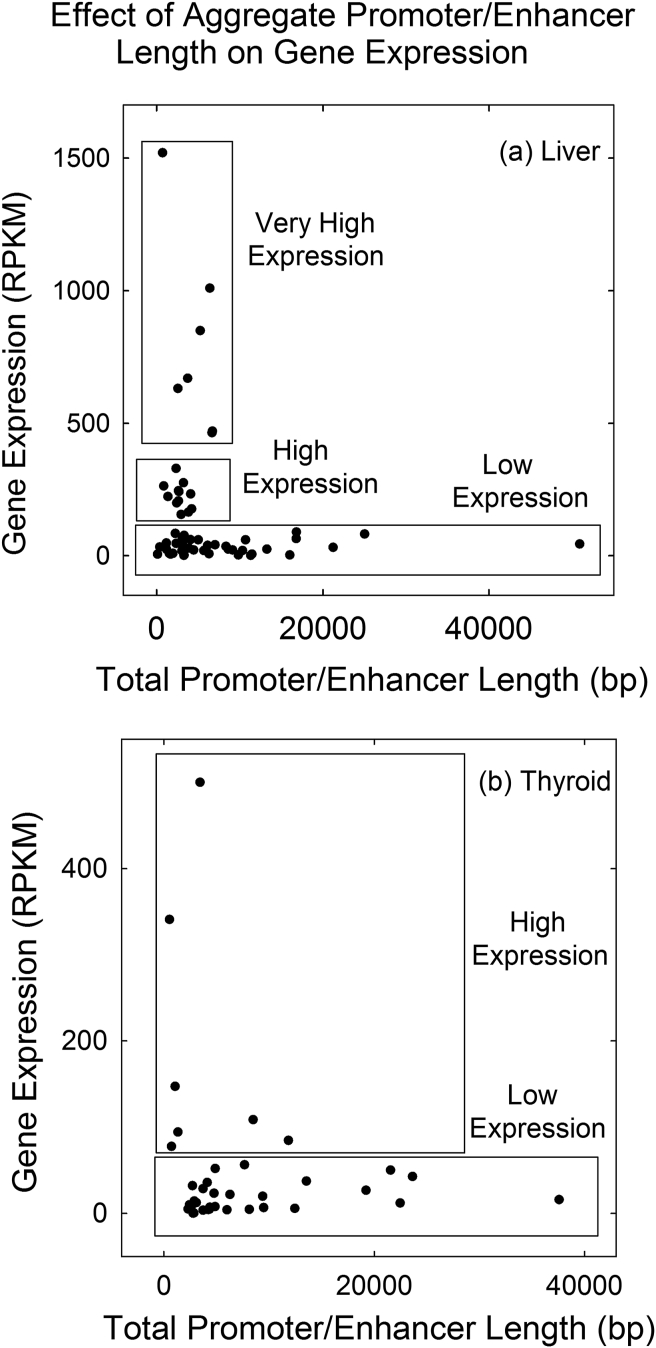
Figure 1a shows a plot of the expression level of the 61 promoter/enhancer-containing liver genes against the promoter/enhancer length. The plot indicates that although there are some genes with very high expression levels and lengths of promoter/enhancer sequence, most genes have expression levels of <400 RPKM and promoter/enhancer lengths of <10,000bp. Groups were chosen to conform to data clusters indicated in Figure 1a. Genes in the low expression group had expression values of <100 RPKM, high 100-40 RPKM and very high >400 RPKM. A gap in expression level was observed between the low and high expression groups (Figures 1a and 2). The observed gap between the two groups suggests it may correlate with other features of gene regulatory control.
Figure 1.
Plot of gene expression against total promoter/enhancer length for database liver (a) and thyroid (b) genes. Boxes indicate genes that were used for the low, high and very high (liver only) expression groups.
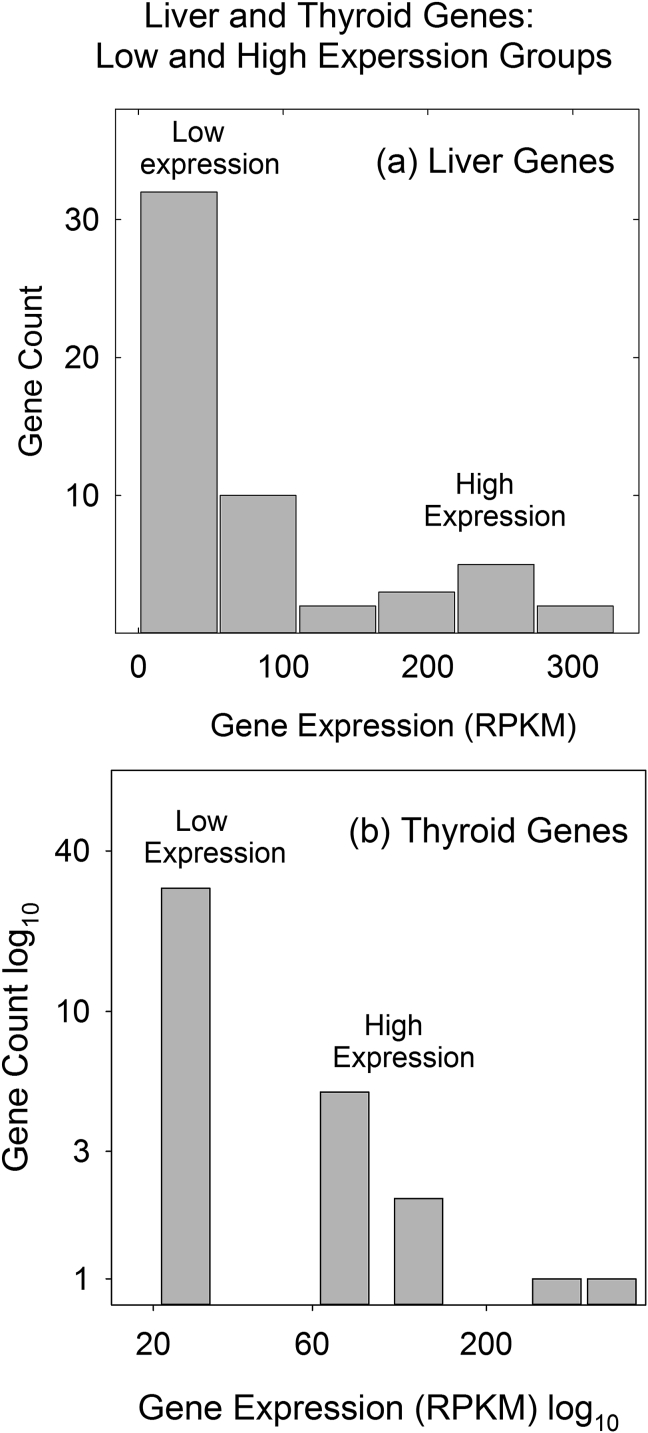
Figure 2.
Plot of number of genes against expression level for liver (a) and thyroid (b) genes. High and low expression groups are indicated. Note that high and low expression groups are distinct in both liver and thyroid genes.
3.2.2. Dependence of promoter/enhancer length on expression gene group
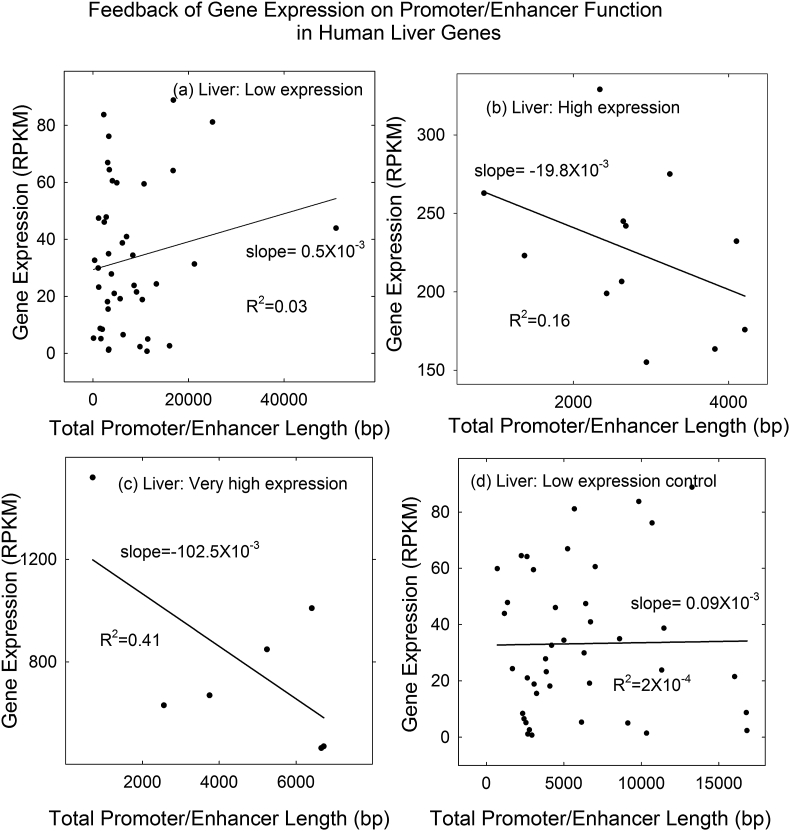
The expression level of genes in the three groups was plotted separately against promoter/enhancer length as shown in Figure 3a-c. A regression line was calculated for each plot. The results demonstrated the expected outcome. The regression line was found to have a positive slope in the case of genes in the low expression group and negative in the high and very high groups. The outcome is interpreted to support the hypothesis that promoter/enhancer length can activate weakly expressed genes and suppress expression of highly expressed ones.
Figure 3.
Plot of gene expression against total promoter/enhancer length for liver genes in the low (a), high (b) and very high (c) expression groups. Panel (d) shows a control plot in which low expression group genes were randomized among promoter/enhancer length values. Note that gene expression correlates positively with promoter/enhancer length in low expression group, but negatively in the high and very high groups.
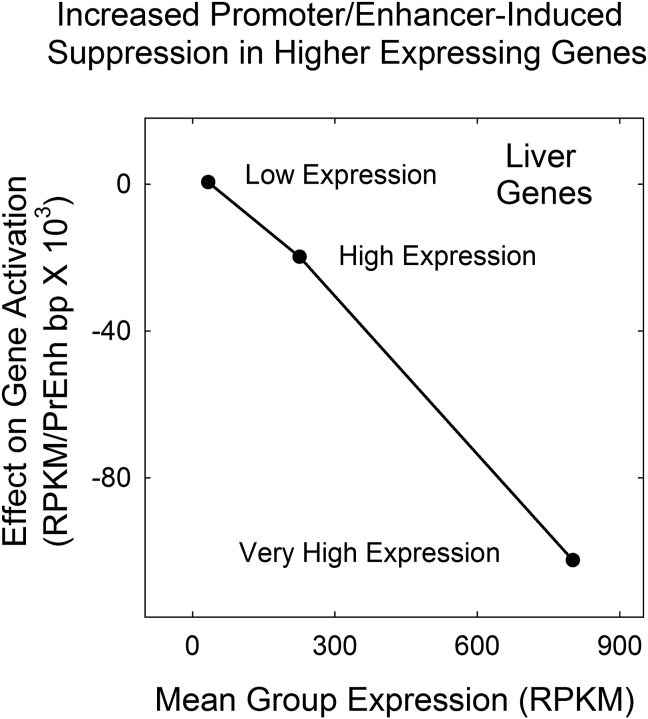
Slopes of the gene expression vs. promoter/enhancer length curves were found to span a substantial range, from +0.5 to -102.5 × 10−3 RPKM/bp (Figures 3a-c and 4). The range of values is interpreted to be revealing about the operation of the proposed feedback loop. In particular, I suggest it is noteworthy that most of the slope values in the observed range are negative. This indicates that the feedback loop must act most often to suppress the activity of over expressing genes rather than to activate under expressing ones (at least among the genes examined here).
Figure 4.
Plot of gene activation or suppression in liver gene groups differing in their level of expression. Note that positive activation is observed with low expression genes but negative (i.e. suppressive) in the case of high expressing genes as expected in a feedback loop.
R2 values were computed for the results shown in Figure 3a-c as they provide a measure of how closely the data points match the linear regression shown. Although the values support the regression slopes shown, they are modest in all three cases (range: 0.03–0.41). The modest values are interpreted to indicate that promoter/enhancers are not the only things that influence the expression level of the genes examined. Other influencing factors that do not correlate with promoter/enhancer length would be expected to obscure the relationship involving promoter/enhancers and contribute to the observed departure of data points from the regression line.
3.2.3. Control with randomized promoter/enhancer length values
As described above, a control experiment was performed with the low expression gene population. Promoter/enhancer lengths were associated at random with gene expression values and randomized data pairs were plotted in the same way as authentic length observations (Figure 3d). The result demonstrated little dependence of gene expression on promoter/enhancer length in the randomized dataset, a result that supports the view that a positive relationship exists in the case of the authentic values.
3.3. Thyroid genes
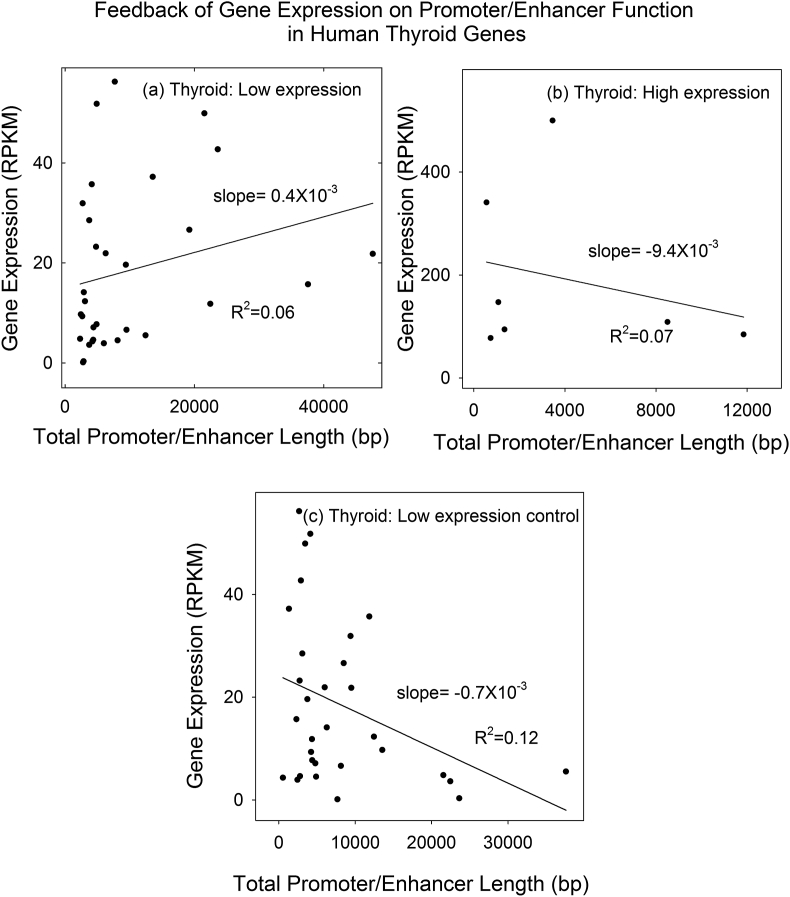
Analysis of 38 thyroid promoter/enhancer-containing genes was similar to that carried out with liver genes. Thyroid genes were separated into weakly and strongly expressed groups containing 31 and 7 genes, respectively (Figure 1b). For each group, gene expression values were plotted against promoter/enhancer length and a regression line was computed. Slopes of the regression lines were found to be positive in the case of low expression genes a negative with the highly expressed ones (Figure 5a and 5b). As in the case of liver genes, the results are interpreted to support the view that transcription factors bound to thyroid promoter/enhancer sequences have the potential to be involved in a feedback loop that maintains gene expression within a circumscribed range.
Figure 5.
Plot of gene expression against total promoter/enhancer length for thyroid genes in the low (a) and high (b) expression groups. Panel (c) shows a control plot in which low expression group genes were randomized among promoter/enhancer length values. Note that gene expression correlates positively with promoter/enhancer length in the low expression group, but negatively in the high group.
The same control study described above for liver genes was also carried out with thyroid. Gene expression values were randomized with respect to the promoter/enhancer lengths and the results were plotted as a function of promoter/enhancer length. A negative slope was observed in contrast to the positive slope expected and observed with the unrandomized input data (Figure 5c). The results support the view that the authentic data (Figure 5a) did not produce an outcome that could have arisen from random input information.
The range of slope values for thyroid genes contrasts with the much higher range observed with liver (i.e. 0.4 to -9.4 × 10−3 for thyroid compared with 0.5 to -102.5 × 10−3 RPKM/bp for liver). It is suggested that this difference in range of slope values may be related to the generally higher expression levels of liver genes compared to thyroid (compare Figure 1a and 1b). The higher level of liver gene expression may require a greater input from transcription factors to control its level of synthesis.
4. Discussion and conclusions
4.1. Feedback loop
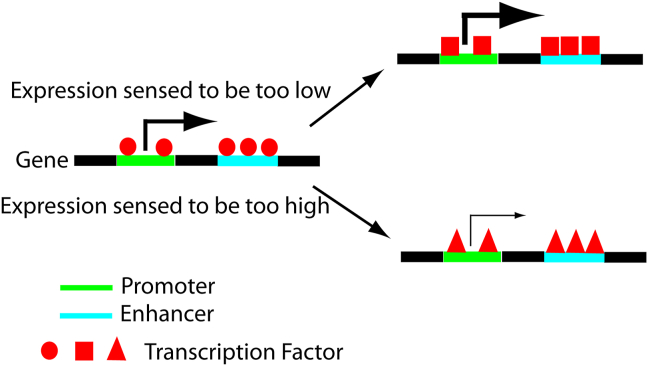
For a transcription factor to be able to function in a feedback loop as described here, it is of central importance that the factor be able to activate or suppress gene expression in a context-dependent manner. While a transcription factor or other regulatory element only able to activate or only suppress its target gene might be able to establish a level of expression well suited for one set of conditions, it would not be able to respond to change due to unexpected factors such as mutation in the genome or environmental conditions. With the ability to maintain its level of expression in a wider range, a gene would acquire a degree of protection against things that could go wrong. In view of the value such protection might provide, it is not at all surprising to find feedback in the human genome. In fact, it is attractive to speculate that such feedback loops might be widespread in the genomes of humans and other species. Figure 6 shows a graphical representation of the proposed feedback loop controlling gene expression.
Figure 6.
Graphical representation of the proposed feedback loop to control human gene expression. Note that a transcription factor is proposed to activate gene expression when expression is sensed to be too low (red squares) and suppress transcription when expression is sensed to be too high (red triangles).
It is a strength of the feedback loop mechanism proposed here that many transcription factors have been demonstrated to activate or suppress transcription depending on the molecular context. Among the best known is the glucocorticoid receptor (GR). Its ability to activate or repress gene expression can depend on a variety of context factors including hormone binding, receptor dimerization and receptor binding to other proteins [2, 3, 6, 7]. Other transcription factors in which binding to a modification protein can alter gene activation/repression include thyroid transcription factor 1, a bacterial transcription factor (AggR) and certain bHLH transcription factors [1, 8, 9]. Unusual cases have been reported in which an intramolecular rearrangement of the transcription factor protein itself can influence whether the factor is activating or repressing [10, 11]. Interactions between different transcription factors have also been reported to influence activation versus suppression [12].
Although it is well documented that some transcription factors can activate or suppress gene expression in a context dependent manner, it is likely that many cannot. Thus, for flexible transcription factors to function in a feedback loop as proposed here, they would need to be able to act in an environment of non-flexible ones. Studies with model systems have the potential to clarify this issue, but the required studies have not yet been done. In the meantime, I suggest two possibilities: (1) as only flexible transcription factors are able to sense the current level of gene expression, their activity may predominate over that of non-flexible transcription factors; and (2) promoter and enhancer regions may be enriched in the presence of binding sites for flexible compared to non-flexible transcription factors.
Operation of a feedback loop as described here depends critically on the ability of the transcription machinery to sense the current level of a gene's transcription. Without that information, promoter/enhancers would not be able to adjust the transcription rate in the appropriate direction. The studies with liver and thyroid genes reported here indicate that the required sensing mechanisms must exist, but there is no suggestion regarding the details of the sensing mechanism or what features of the transcription process may be sensed. There is no shortage, however, of possible processes that could be sensed. Anything that would suffice as an overall measure of the transcription rate would be appropriate. Possibilities include open compared to closed chromatin, epigenetic marks such as histone acetylation or methylation, un-methylated CpG islands, the density of RNA polymerase molecules along a gene and others [13, 14, 15, 16]. The results reported here suggest tests of such possibilities may be a productive area of future research.
As some genes lack annotated promoter/enhancers entirely (see for example Supplementary Tables 1 and 2), it is reasonable to ask how such genes could have feedback loops of the type described here. Two possibilities suggest themselves: (1) candidate genes may have promoter/enhancers that are not identified by current methods for promoter/enhancer detection; (2) some genes may not require feedback control of their expression level. For instance, a gene on/gene off only mechanism may be sufficient for the role of such a gene.
4.2. Experimental design
The experimental design employed here is a demanding one. To support the existence of a feedback loop, the slope of the expression vs. promoter/enhancer length curve needs to be positive with weakly expressed genes and negative with highly expressed ones as observed here. Any other result would invalidate the hypothesis. For example, lack of a dependence of gene expression on promoter/enhancer length would be produced by transcription factors that only activate or only suppress their target gene expression. Such an outcome would not support the idea of feedback. Similarly, the idea of feedback would be invalidated by expression curves with slopes that are opposite from the ones reported here. For instance, further activation of genes already expressed at a high level would be expected to be useful in only a restricted number of genes.
Operation of a feedback loop as described here provides some important advantages for regulation of gene expression. Instead of specifying only an expression level, a feedback loop specifies a range of levels and also a mechanism for keeping expression within that range. This “guardrail” function is expected to contribute to keeping the level of a gene's expression from becoming toxic.
The experimental results reported here demonstrate that feedback loops can be detected despite the extended length of promoter/enhancer regions employed. Control regions used for the analyses described here were tens of thousands of base pairs in some cases. As such long control regions would provide the genome with a level of protection against mutagenic change, it would be of interest in the future to know how much of this length may be required or tolerated.
Declarations
Author contribution statement
J. Brown: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2020.e04934.
Acknowledgements
I gratefully acknowledge Forde Upshur, Ava Roth and Karsten Siller for advice on computational aspects of this project.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Perrone L., Pasca di Magliano M., Zannini M., Di Lauro R. The thyroid transcription factor 2 (TTF-2) is a promoter-specific DNA-binding independent transcriptional repressor. Biochem. Biophys. Res. Commun. 2000;275(1):203–208. doi: 10.1006/bbrc.2000.3232. [DOI] [PubMed] [Google Scholar]
- 2.Cato A.C., Wade E. Molecular mechanisms of anti-inflammatory action of glucocorticoids. Bioessays. 1996;18(5):371–378. doi: 10.1002/bies.950180507. [DOI] [PubMed] [Google Scholar]
- 3.Lefstin J.A., Yamamoto K.R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392(6679):885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 4.Brown J.C. 2020. Role of Gene Length in Control of Human Gene Expression: Chromosome-specific and Tissue-specific Effects. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T. 2017. GeneHancer: Genome-wide Integration of Enhancers and Target Genes in GeneCards. Database (Oxford). 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhen T., Cidlowski J.A. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. N. Engl. J. Med. 2005;353(16):1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 7.Heck S., Kullmann M., Gast A., Ponta H., Rahmsdorf H.J., Herrlich P. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13(17):4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckner R., Yao T.P., Oldread E., Livingston D.M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10(19):2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 9.Mickey A.S., Nataro J.P. Dual function of aar, a member of the new AraC negative regulator family, in Escherichia coli gene expression. Infect. Immun. 2020;88(6) doi: 10.1128/IAI.00100-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X.Y., Green M.R. Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev. 1996;10(5):517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- 11.Skalicky J.J., Donaldson L.W., Petersen J.M., Graves B.J., McIntosh L.P. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;5(2):296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miner J.N., Yamamoto K.R. Regulatory crosstalk at composite response elements. Trends Biochem. Sci. 1991;16(11):423–426. doi: 10.1016/0968-0004(91)90168-u. [DOI] [PubMed] [Google Scholar]
- 13.Chen T., Dent S.Y. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014;15(2):93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu. Rev. Genom. Hum. Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veloso A., Kirkconnell K.S., Magnuson B., Biewen B., Paulsen M.T., Wilson T.E. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24(6):896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.