Abstract
Poly(ADP-ribose) polymerase inhibitors (PARPi) are a new class of agents with unparalleled clinical achievement for driving synthetic lethality in BRCA-deficient cancers. Recent FDA approval of PARPi has motivated clinical trials centered around the optimization of PARPi-associated therapies in a variety of BRCA-deficient cancers. This review highlights recent advancements in understanding the molecular mechanisms of PARP ‘trapping’ and synthetic lethality. Particular attention is placed on the potential extension of PARPi therapies from BRCA-deficient patients to populations with other homologous recombination-deficient backgrounds, and common characteristics of PARPi and non-homologous end-joining have been elucidated. The synergistic antitumor effect of combining PARPi with various immune checkpoint blockades has been explored to evaluate the potential of combination therapy in attaining greater therapeutic outcome. This has shed light onto the differing classifications of PARPi as well as the factors that result in altered PARPi activity. Lastly, acquired chemoresistance is a crucial issue for clinical application of PARPi. The molecular mechanisms underlying PARPi resistance and potential overcoming strategies are discussed.
Keywords: PARP1, classifications of PARP inhibitors, BRCA1/2, synthetic lethality, acquired chemoresistance to PARP inhibitors, immunotherapy
Introduction
Poly(ADP-ribose) polymerase (PARP) family proteins have attracted attention in the last decade due to the clinical success of PARP inhibitors (PARPi) in cancer treatment. Among the seventeen family members, PARP1 and PARP2 are two key enzymes that mediate DNA damage response (DDR) by serving as DNA damage sensors and signal transducers.1,2 To respond DNA damage such as nicks and double-strand breaks (DSB), PARP1 is rapidly recruited to the sites of damaged DNA, and its catalytic activity increases 10- to 500-fold through an allosteric activation mechanism. This results in the synthesis of protein-conjugated poly(ADP-ribose) (PAR) chains using NAD+ as a critical substrate.3,4 The negatively charged PAR functions as a high-density protein-binding scaffold and recruits components of the DNA damage repair machinery.1,5 PARP2, the less abundant homologous protein of PARP1, probably has a less dominant role due to the absence of N-terminal zinc-finger (Zn) domains for DNA binding.6–8 PARP1 is thus an attractive target for cancer therapy, with four PARPi being licensed in clinic to date: olaparib, talazoparib, niraparib, and rucaparib. All these PARPi compounds share a nicotinamide moiety competing with NAD+ for binding to PARP1, and thus inhibit the catalytic activity of PARP1. Hundreds of clinical trials are currently ongoing to optimize the use of each PARPi and to test effective combination treatment strategies aiming to improve response, overcome resistance, or minimize overlapping toxicity. For example, administration of PARPi in combination with immune checkpoint inhibitors represents a novel therapeutic strategy. In this review, we will discuss the molecular mechanisms of PARPi-based targeted therapy and classification of PARPi, acquired PARPi resistance, and potential biomarkers of PARPi response.
PARPi-associated synthetic lethality
PARPi are developed in breast and ovarian cancers with BRCA gene mutations using synthetic lethal screening.9,10BRCA1 and BRCA2 are two human tumor suppressor genes that play an important role in DNA repair, and their mutations play a critical role in the development of several cancers including breast and ovarian cancer. Byrant et al.9 and Farmer et al.10 have first demonstrated synthetic lethality of BRCA1- and BRCA2-deficient tumor cells by PARP inhibition. Two models are proposed to explain underlying mechanisms. The first model stands on the notion that PARP1, the major target of PARPi, is primarily associated with base excision repair (BER) and single stranded break (SSB) repair by recruiting DNA repair effectors such as XRCC1, DNA polymerase-β and DNA ligase III.10 When PARP1 is inhibited, SSBs cannot be repaired and are switched to DSBs during DNA replication. This forces defective homologous recombination (HR) and cytotoxicity in the absence of BRCA1/2.11,12 In contrast, the second model describes a process by inhibiting PARP1 autoPARylation13 and inducing allosteric changes in the PARP1 structure. The cytotoxicity of PARPi relies on sufficient trapping of PARP1 on DNA lesions.14,15 The trapped PARP1 forms a toxic lesion and stalls the progress of replication forks. This can be repaired by HR in a BRCA1- and BRCA2-dependent manner.16,17 However, the replication fork collapses in BRCA1/2-deficient cancer cells that is paired with an increased genomic instability, ultimately resulting in cell death. The second model is currently favored as the preferred mechanism of synthetic lethality for two reasons. 1) PARP inhibition by inhibitors is more cytotoxic than the genetic deletion of PARP;18 and 2) The cytotoxicity of PARPi is correlated with its ability to trap PARP on DNA rather than its catalytic inhibitory properties. This is evident upon comparison of three PARPis: talazoparib, olaparib, and rucaparib. These PARPis are comparable in their catalytic PARP inhibition. However, talazoparib results in higher levels of cytotoxicity compared to olaparib and rucaparib due to its superior PARP-trapping potency.19 This logic is further supported by recent evidence suggesting that PARPi modulates PARP-1 trapping through effects on a critical allosteric regulatory domain of PARP-1.14 In addition to synthetic lethality in cells with the BRCA1/2-defect, PARPi also shows similar mechanistic pathologies when reactive oxidative species (ROS) accumulate, which cause oxidative DNA damage and are likely to increase replication stress. Alantolactone, a ROS inducer, is shown to synergize with olaparib and induce lethality irrespective of HR status.20
Classification of PARPi based on their allosteric effects on PARP1
PARPi exist in clinical and non-clinical varieties. There are currently five clinical inhibitors. Four of them, namely olaparib, talazoparib, niraparib, and rucaparib have been approved for treating BRCA1/2-deficient ovarian and breast cancers (Table 1).21,22 Two recent trials (NCT02184195 and NCT02987543) have shown that olaparib also benefits patients with either metastatic pancreatic cancer or metastatic castration-resistant prostate cancer.23,24 Notably, niraparib is the first drug granted approval irrespective of the BRCA1/2 status. The fifth clinical inhibitor, veliparib, is still in Phase III clinical trials after two failed Phase III attempts.25 Benzamide adenine dinucleotide (BAD) and EB-47 are two inhibitors in preclinical studies. BAD is an analog of the PARP1 substrate nicotinamide adenine dinucleotide (NAD+),26 and EB-47 can mimic NAD+ binding with an affinity comparable to that of clinical PARPi.27,28 Although all different PARPi compounds bind at the catalytic center to block binding of NAD+ and prevent PAR production, they exhibit different potencies in inducing cancer cell death. This is presumably due to their varying capabilities to induce catalytic inhibition and trap PARP1 on DNA breaks.
Table 1.
The list of approved PARP inhibitors and prescribing information*.
| Drug | Approval date | Single/Combination | Recommended dose | Based clinical trials | Indications |
|---|---|---|---|---|---|
| Olaparib (LYNPARZA, AstraZeneca) | May 19, 2020 | single | 300 mg orally twice daily | NCT02987543 | adult patients with deleterious or suspected deleterious germline or somatic HRR1 gene-mutated mCRPC2, who have progressed following prior treatment with enzalutamide or abiraterone |
| May 8, 2020 | in combination with bevacizumab | olaparib, 300 mg orally twice daily; bevacizumab, 15 mg/kg intravenously every three weeks | NCT03737643 | adult patients with advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer who are in complete or partial response to first-line platinum-based chemotherapy and whose cancer is associated with HRD3-positive status | |
| Dec 27, 2019 | single | 300 mg orally twice daily | NCT02184195 | adult patients with deleterious or suspected deleterious gBRCAm4 metastatic pancreatic adenocarcinoma whose disease has not progressed on at least 16 weeks of a first-line platinum-based chemotherapy regimen | |
| Dec 19, 2018 | single | 300 mg orally twice daily | NCT01844986 | adult patients with deleterious or suspected deleterious gBRCAm4 or sBRCAm5 advanced epithelial ovarian, fallopian tube or primary peritoneal cancer who are in complete or partial response to first-line platinum-based chemotherapy | |
| Jan 12, 2018 | single | 300 mg orally twice daily | NCT02000622 | patients with deleterious or suspected deleterious gBRCAm4, HER2-negative metastatic breast cancer who have been treated with chemotherapy either in the neoadjuvant, adjuvant, or metastatic setting | |
| Aug 17, 2017 | single | 300 mg orally twice daily | NCT01874353 NCT00753545 | adult patients with recurrent epithelial ovarian, fallopian tube, or primary peritoneal cancer who are in a complete or partial response to platinum-based chemotherapy | |
| Dec 19, 2014 | single | 400 mg (capsules) orally twice daily | NCT00494442 | patients with deleterious or suspected deleterious gBRCAm4 advanced ovarian cancer who have been treated with three or more prior lines of chemotherapy | |
| Talazoparib (TALZENNA, Pfizer) | Oct 16, 2018 | single | 1 mg orally once daily | NCT01945775 | patients with deleterious or suspected deleterious gBRCAm4, HER2negative locally advanced or metastatic breast cancer |
| Rucaparib (RUBRACA, Clovis Oncology) | May 15, 2020 | single | 600 mg orally twice daily | NCT02952534 | patients with deleterious gBRCAm4 or sBRCAm5-associated mCRPC2 who have been treated with androgen receptor-directed therapy and a taxane-based chemotherapy |
| Apr 6, 2018 | single | 600 mg orally twice daily | NCT01968213 | patients with recurrent epithelial ovarian, fallopian tube, or primary peritoneal cancer who are in a complete or partial response to platinum-based chemotherapy | |
| Dec 19, 2016 | single | 600 mg orally twice daily | NCT00664781 | patients with deleterious gBRCAm4 or sBRCAm5-associated advanced ovarian cancer who have been treated with two or more chemotherapies | |
| Niraparib (ZEJULA, GlaxoSmithKline) | Apr 29, 2020 | single | 200 or 300 mg orally once daily6 | NCT02655016 | adult patients with advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer who are in a complete or partial response to first-line platinum-based chemotherapy |
| Oct 23, 2019 | single | 300 mg orally once daily | NCT02354586 | patients with advanced ovarian, fallopian tube, or primary peritoneal cancer treated with three or more prior chemotherapy regimens and whose cancer is associated with HRD3-positive status | |
| Mar 27, 2017 | single | 300 mg orally once daily | NOVA | adult patients with recurrent epithelial ovarian, fallopian tube, or primary peritoneal cancer who are in complete or partial response to platinum-based chemotherapy |
All the information is obtained from the Drug Approvals and Databases of FDA.
HRR, homologous recombination repair.
mCRPC, metastatic castration-resistant prostate cancer.
HRD, homologous recombination deficiency.
gBRCAm, germline BRCA-mutated.
sBRCAm, somatic BRCA-mutated.
For patients weighing less than 77 kg or with a platelet count of less than 150,000/μL, the recommended dose is 200 mg taken orally once daily. For patients weighing greater than or equal to 77 kg and who have a platelet count greater than or equal to 150,000/μL, the recommended dose is 300 mg taken orally once daily.
The establishment of a classification system would be useful to predict new PARPi activity and to promote PARPi development. According to recent X-ray structure analysis,14 PARP inhibitors affect a critical allosteric regulatory domain of PARP1 as well as in the helical domain (HD).15,29 Based on the varying impact on PARP1 allostery, PARPi molecules are categorized into three types: type I, allosteric pro-retention on DNA; type II, non-allosteric pro-retention on DNA; and type III, allosteric pro-release from DNA (Table 2).
Table 2.
Distinct properties among three types of PARP inhibitors.
| Item | Type I | Type II | Type III |
|---|---|---|---|
| Inhibitors | EB-47, BAD | olaparib, talazoparib | rucaparib, niraparib, veliparib |
| PARP-1 allostery | allosteric | non-allosteric | allosteric |
| HD conformation | destabilization | neutral | more folded |
| PARP-1 affinity for DNA | large increase | small increase | decrease |
| Trapping potency | pro-retention | pro-retention | pro-release |
BAD and EB-47 are designated as type-I inhibitors that contact the helix αF of the HD and have a strong reverse allosteric effect by destabilizing the HD. HD instability results in functional allosteric changes, increases PARP1 affinity for DNA and retains PARP1 on DNA breaks.14 Laboratory results clearly show downstream effects of BAD and EB-47 on trapping PARP1 on DNA breaks. However, they have not been developed as clinical inhibitors, possibly due to BAD's non-selective nature and EB-47’s impermeability in cell lines.27 A cell permeable version of EB-47 may be worth testing as it could have clinical potential as a potent regulator of PARP1 binding. Type II inhibitors (including olaparib and talazoparib) are relatively neutral toward PARP1 allostery and produce minimal increases in affinity for a DNA break. Olaparib does not contact the HD, and thus has no effect on interdomain communication or DNA binding domains in PARP1. Talazoparib is very efficient at trapping PARP1 on DNA breaks, and it does not exert a pronounced reverse allosteric effect on PARP1. Talazoparib is extremely potent at catalytic inhibition of PARP1 and further limits PARP1 autoPARylation which is required for rapid release of PARP1 from the site of DNA damage.19,30 Even subtle differences in PARPi potencies affecting auto-PARylation levels could yield significant differences in PARP1 release from DNA.31 Talazoparib's high trapping efficiency can possibly be attributed to its inhibitory potency combined with its slow dissociation half-life32 and neutral effect on the HD. Type III inhibitors include rucaparib, niraparib, and veliparib. As allosteric pro-release inhibitors, they promote highly folded conformation of the HD by stabilizing αB/αF helices and decreasing PARP1 affinity for DNA. Thus, they act opposite in function to type I inhibitors. Both niraparib and rucaparib boast a strong catalytic inhibition that can limit the auto-PARylation-dependent release from DNA. Combined with their long half-life, this allows both niraparib and rucaparib to compensate for their pro-release allosteric effects.18 Rucaparib and talazoparib exhibit the similar inhibitory potency and binding properties to PARP1, but they have remarkable differences in trapping abilities.33 Rucaparib's propensity to promote stabilization of the HD decreases PARP1 affinity for DNA.14 In contrast, veliparib is considered a poor trapper, probably due to its pro-release allosteric effect, relatively low catalytic inhibition potency, and rapid clearance rates in vivo.34
HD reverse allostery has proven vital to define the potency of an inhibitor to trap PARP1 on DNA breaks.29,35,36 Mutagenesis breaking interdomain communication in PARP1 or disrupting the inhibitor-HD interaction can abolish the reverse allosteric effect of a type I inhibitor and transform them into type II inhibitors. In contrast, inducing HD contact that generates PARP1 reverse allostery transforms type III inhibitors into type I inhibitors.14 Of note, several inhibitors, such as A-966 492,37 3-Aminobenzamide,38 and AG-14 361,39 have been omitted from this classification system because of a lack of data supporting their influence of PARP1 allostery. To develop new PARPi derivatives in the future, studies should emphasize the outstanding drug-like properties, high potency of catalytic inhibition, and allosteric pro-retention of inhibitors on DNA breaks.
PARPi sensitivity and ‘HRDness’
Growing evidence demonstrates that multiple factors can affect PARPi sensitivity in addition to BRCA gene mutations. Homologous recombination defectiveness, or ‘HRDness’, indicates a non-BRCA-related mechanism of PARPi sensitivity.40 Extending the application of PARP inhibition from tumors with BRCA-defects to other HR-deficient backgrounds, such as PTEN- and ATM-mutations,41,42 allow a greater population of patients to benefit from therapy. To this end, silencing critical mediators of DDR potentially enhances cellular sensitivity to PARPi. For example, the histone lysine demethylase PHF2 was recently identified as a novel regulator of DDR. PHF2-depleted cells display increased sensitivity to PARPi. PHF2 deficiency mechanistically results in impaired HR by affecting CtIP-dependent resection of DSBs and decreasing BRCA1 protein levels.43 Similarly, NFBD1 loss could disrupt HR by decreasing the formation of BRCA1, BRCA2 and RAD51 foci and thereby sensitize nasopharyngeal tumor cells to PARPi.44 The HR repressor ZPET (zinc finger protein proximal to RAD18) may play a role in inhibiting MRE11 binding to chromatin and stalling replication forks, and loss of this protein confers PARPi resistance.45 In addition, cellular sensitivity to PARPi can potentially be increased by silencing some important mediators of DDR, such as RPL6,46 which regulates the recruitment of MDC1 and RNF16827, and VRK1 chromatin kinase, which controls the configuration of locally altered chromatin induced by DNA damage47. Cellular sensitivity was also found to increase upon inhibition of the microtubule-depolymerizing kinesin Kif2C that regulates the mobility of DSBs and the generation of DNA damage foci.48 Alternatively, pharmacological inhibition of key mediators in HR also synergistically enhances PARPi sensitivity in HR-proficient or PARPi-resistant cells. Recent studies indicate that the WEE1/PLK1 dual inhibitor AZD1775, the FOXM1 inhibitor thiostrepton, the DNMT inhibitor 5-azacytidine, or the CDK1/2 inhibitor dinaciclib may induce a HRDness phenotype and thereby sensitize tumor cells to PARPi by downregulating HR-related genes, blocking BRCA1 phosphorylation, and reducing RAD51 foci formation.49–53 Therefore, the identification of the potential inhibitors of the HRDness phenotype will promote the optimal usage of PARPi combined with other inhibitors in clinic.
PARPi and non-homologous end-joining (NHEJ)
Although most studies have focused on HR, DSBs are repaired primarily by non-homologous end-joining (NHEJ), likely due to the fact that HR relies on the presence of a sister chromatid while NHEJ can repair DSBs at any cell cycle stage. There are two major types of NHEJ: the classical pathway initiated by the Ku heterodimer (C-NHEJ), and the alternative pathway initiated by PARP1 (Alt-NHEJ). 54–56 Albeit error prone, C-NHEJ is a critical repair pathway for DSBs and directly joins broken ends of DNA with little to no regard for sequence homology. Therefore, C-NHEJ deficiency might be expected to show synthetic lethality with PARPi. Paradoxically, the inhibition of C-NHEJ by depletion of Ku80 or DNA-PK inhibitors reduced the genomic instability and lethality of PARPi in HR-deficient cells rather than aggravating it.57 Similarly, cancer cell cultures with NHEJ competence and HR were sensitive to rucaparib while NHEJ-defective cell cultures were resistant to rucaparib,58 thus indicating that the C-NHEJ repair pathway is required for the sensitivity to PARPi. Furthermore, impaired C-NHEJ induced by deletion of DYNLL1 or ASCIZ (an organizer of the 53BP1 complex) confers BRCA1-deficient tumor cells resistant to PARPi.59 Thus, evidence conclusively supports that the genomic instability resultant from NHEJ is an important promoter of PARP inhibition. While alt-NHEJ mediated by PARP1 and DNA polymerase θ (Pol θ) using microhomology is a crucial back-up repair pathway for blunt DSBs and stressed replication forks in absence of HR, it comes at a cost of promoting chromosomal translocations and genomic instability.54,60,61 Cancer cells deficient in BRCA1 (or its obligate partner BAP1) often repress miR223–3p and allow repairing stressed replication forks through Alt-NHEJ.62 PARP1 initiates the Alt-NHEJ response by displacing the Ku complex from DSBs and is the rate-limiting initial step of Alt-NHEJ. Therefore, PARP1 inhibition with olaparib or rucaparib could suppress Alt-NHEJ and decrease chromosomal translocations,54 indicating that PARPi are negative regulators of the Alt-NHEJ repair pathway.
PARPi and immunotherapy
Immunotherapy using immune checkpoint blockades has obtained a great success in treating human cancers. Unfortunately, the response rate is relatively low. Accumulating evidence suggests that PARPi might induce the anti-tumor immune response and collaborate with checkpoint blockade to inhibit tumor growth.63–71 PARP inhibition by olaparib or talazoparib may promote cytotoxic T lymphocyte infiltration through BRCA1/2-independent mechanisms in several in vivo tumor models, including small cell lung cancer (SCLC), ovarian cancer, and breast cancer.64–67 PARP inhibition has also been shown to upregulate PD-L1, and this may be mediated by Chk1 phosphorylation, IRF3 expression, or through recruitment of myeloid cells into tumor sites, thereby potentiating tumor responsiveness to PD-1 or PD-L1 blockade therapy.63–66 The mechanistic studies suggest that PARP inhibition promotes the accumulation of cytosolic DNA fragments from unrepaired DNA lesions and activates the DNA-sensing cyclic guanosine monophosphate–adenosine monophosphate (cGAMP)-cyclic GMP-AMP synthase (c-GAS) cascade. Subsequently, cGAMP binding to stimulator of interferon genes (STING) induces the phosphorylation of IRF3 through Tank-binding kinase-1 (TBK1). Phospho-IRF3 then translocates to the nucleus and induces the expression of downstream effector genes, such as type I interferon, various chemokines and PD-L1.63,66,67 This is consistent with the mechanisms that account for activation of type I interferon signaling and PD-L1 upregulation in response to DSBs.68,69 Based on these interesting findings, the PARPi and anti–PD-L1 blockade represents a rational therapeutic combination,70,71 and has been widely tested in clinical trials. The combination of PARPi, olaparib, and the PD-L1 inhibitor durvalumab has been the most common strategy to yield desired clinical success rates in patients with metastatic castrate-resistant prostate cancer or recurrent ovarian cancer.72–74 In addition, niraparib in combination with pembrolizumab, a PD-1 inhibitor, has also exhibited promising antitumor activity and tolerance in metastatic triple-negative breast cancer and recurrent ovarian cancer patients.75,76 Ongoing trials combining PARPi and immune checkpoint blockades demonstrate the synergistic antitumor effect and will benefit more patients regardless of platinum-resistance, BRCA mutations, or prior treatments. In addition, PARP1 was reported to repress the expression of NKG2DLs and therefore mediate immune evasion of leukemic stem cells (LSCs) in acute myeloid leukemia. PARP1 inhibition could restore the expression of NKG2DLs on the LSCs surface and promote their clearance by NK cells.77 Thus, the multifactorial functions of PARP1 in immunomodulation and immune escape of cancer stem cells may pave the way for novel therapeutic strategies in clinic.
PARPi resistance and overcoming strategies
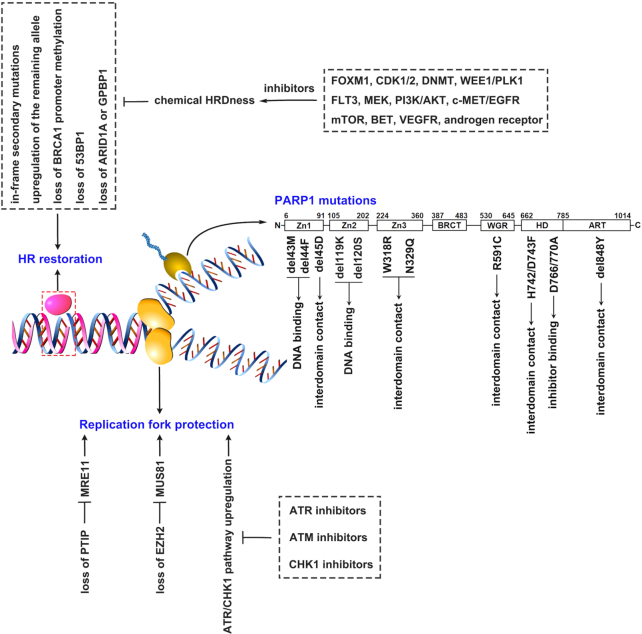
PARPis have boasted unprecedented clinical success for cancer patients with HR-defects. At the same time, acquired resistance to PARPi treatments impedes optimal clinical outcome. Defining both molecular features and elucidating underlying mechanisms of PARPi resistance stand at the foundation of overcoming resistance to PARP treatments. While activation of epithelial-mesenchymal transition (EMT)78,79 and induction of P-glycoprotein expression12,80 have been observed in PARPi-resistant biopsies, they are not sufficient to confer resistance.78 Recent research supports the notion that the primary mechanisms for PARPi resistance are HR restoration and replication fork protection.81 PARP mutations that affect DNA or inhibitor binding have also been explored as a potential mechanism. In addition, combination therapy of PARPi and tyrosine kinase inhibitors might represent a possible avenue to circumvent acquired resistance. This information is summarized in Fig. 1.
Figure 1.
Schematic representation of the mechanisms contributing to PARP inhibitors (PARPi)-resistance and potential overcoming strategies. Homologous recombination (HR) restoration, replication fork protection and PARP1 mutations are three categories of mechanisms underlying acquired PARPi-resistance. HR restoration is usually induced by in-frame secondary mutations restoring BRCA1/2 function, upregulation of the remaining functional allele, loss of BRCA1 promoter methylation, and loss of 53BP1, ARID1A or GPBP1, and can be overcome via combining PARPi with a series of inhibitors that can result in chemical HRDness. Replication fork protection is mediated by loss of PTIP or EZH2 that impairs the recruitment of the MRE11 or MUS81 nuclease to stalled replication forks, and upregulation of the ATR/CHK1 pathway that can be potentially resolved via combining PARPi with inhibitors of ATR, ATM or CHK1. Mutations in PARP1 that can affect interdomain contacts, DNA binding potency or inhibitor binding also have a potential to lead to PARPi-resistance, which might have to be resolved by developing new inhibitors.
HR restoration
HR restoration describes a process in which HR-deficient patients become HR-proficient post-resistance to PARPi; this occurs in about 50% of PARPi-resistant patients with ovarian cancers.81,82 The most common mechanism of HR restoration, genomic reversion of BRCA1/2, has been observed in a subset of post-resistant patients with ovarian cancer,82–84 pancreatic cancer,85 prostate cancer,86 and in about 50% of metastatic breast cancer patients.87 Mechanisms that describe this restoration involve either 1) secondary intragenic mutations; or 2) upregulation of the remaining functional allele; or 3) the loss of BRCA1 promoter methylation. First, secondary intragenic mutations in carcinomas lead to the expression of functional BRCA1/2 proteins by either restoring the open reading frames (ORF) or the inherited mutation.82–86,88,89 Amplification of the mutant BRCA2 allele that expresses the truncated BRCA2 protein90 or the expression of a BRCA1 hypomorphic protein91 also results in resistance. These pathways have been verified in PARPi-resistant PDXs and patients.82,91 Epigenetic silencing of BRCA1 or RAD51C by the promoter hypermethylation can sensitize tumor cells to PARPi. In contrast, the promoter demethylation or de novo gene fusions placing BRCA1 under the transcriptional control of a heterologous promoter will lead to the re-expression of BRCA1 and result in subsequent PARPi resistance.92,93 Heterogeneous tumors treated with PARPi could eliminate tumor cells with BRCA1 methylation, leading to the positive selection of BRCA1-expressing tumor cells.81 Likewise, de novo rearrangements at the BRCA1 locus that skip the BRCA1 promoter hypermethylation could also restore BRCA1 expression.93 In addition, mutant BRCA1 proteins with a loss of the RING domain (185delAG) or that otherwise carry a mutation (C61G) in the RING domain were reported to develop rapid resistance to PARPi.94,95 Loss of 53BP1 has also been shown to confer PARPi-resistance,91 probably due to a shift in DNA repair mechanism from NHEJ to HR.40,80 This might also notably serve as a biomarker to test for PARPi-resistance. Recent patient genomic data enrichment supports the notion that a loss of ARID1A or GPBP1 could drive PARPi-resistance by upregulating the genes involved in HR in ovarian cancer patients.96
Inhibitors targeting DDR or DDR-relative pathway components can pharmacologically induce a HRDness phenotype, known as “chemical HRDness”.40 Any compounds that induces chemical HRDness may overcome PARPi-resistance; common examples can include inhibitors of FOXM1, DNMT, CDK1/2, and WEE1/PLK1. In addition, mTOR, PI3K/AKT, MEK, VEGFR, EGFR, androgen receptors, and BET inhibitors that indirectly regulate DDR might induce a similar HRDness phenotype.40,97,98 Most of these inhibitors have been well discussed in recent reviews40,81,99 and are not covered here. Several studies have reported that receptor tyrosine kinases (RTK) are involved in resistance to PARPi. Inhibition of FLT3, (a member of Class III RTK) by AC220 or AIU2001 could suppress DDR genes (such as BRCA1, BRCA2, PALB2, RAD51, and LIG4) and subsequently result in inhibition of DSB repair pathways. This results in synthetic lethality with PARPi.100,101 In addition, c-MET and EGFR, both similar RTKs, were also found to be hyperactivated in TNBC cells with acquired resistance to PARPi. This is presumably because EGFR and MET heterodimers interact with and phosphorylate the Tyr907 residue of PARP1. While PARP1’s enzymatic activity subsequently increases, PARPi binding slowly decreases.102,103 Combined c-MET and EGFR inhibition by small molecules reversed the acquired resistance of PARPi in TNBC cells.104 Taken together, inhibitors that induce chemical HRDness can contribute to overcoming PARPi-resistance and potentially extend PARPi application to HR-proficient cancer patients.
Replication fork protection
An important alternative mechanism for PARPi resistance is replication fork protection. By trapping PARP1 on chromatin, PARPi forces collapse of the replication fork and increases overall replication stress to induce cell death. Furthermore, BRCA1/2 deficient tumor cells can acquire PARPi resistance through several molecular mechanisms that are independent of BRCA1/2 restoration, including a loss of PTIP or EZH2, which ultimately serves to safeguard their replication forks. More specifically, PTIP deficiency protects replication forks from degradation by impairing the recruitment of the MRE11 nuclease to stalled replication forks, driving PARPi resistance in BRCA1/2-deficient cells.105 Likewise, EZH2 localizes at stalled forks and mediates H3K27 trimethylation and subsequent recruitment of the MUS81 nuclease. A loss of EZH2 confers PARPi resistance by blocking MUS81 recruitment to stalled forks and allowing greater fork stabilization.106
PARPi resistance may also be obtained through increased expression or activity of replication fork stabilizers. Hyperactivation of tousled-like kinases reduces PARPi sensitivity in addition to contributing to chromatin assembly and maintaining replication fork integrity.107 In PARPi-resistant cancer cells, the ATR/CHK1 pathway was often upregulated, thereby inducing the phosphorylation of multiple proteins that stabilize the replication fork.81 Therefore, combination treatment of PARPi with ATR or CHK inhibitors can potentially overcome acquired PARPi-resistance.108–110 Similar to ATR, ATM is another key mediator that resolves DNA replication stress and maintains genomic stability.111,112 ATM inhibition potentially enhances PARPi sensitivity or reverses acquired PARPi-resistance by preventing replication fork protection or by enhancing replication stress. Combination treatment therapy of olaparib and AZD0156, an ATM inhibitor, is currently being tested in patients with advanced-stage solid cancers (NCT02588105).40
PARP mutations
Most clinical PARPis generate cytotoxic lesions by trapping PARP1 at the site of damaged DNA. Therefore, PARPi resistance can be caused through PARP1 mutations that affect trapping potency. The WGR domain-mediated interdomain interactions between regulatory HD and DNA binding domains link DNA binding to catalytic PARP1 activation. These mechanisms are central to PARP1 trapping procedures.29,35 Notably, de novo resistance to olaparib was found to result from the R591C mutation (1771C > T) in the WGR domain and was recently identified in an ovarian cancer patient. More specifically, the R591C mutation affects PARP1 trapping by disrupting the WGR domain at the contact region with the HD and the DNA-binding Zn1 domain.113 Mutations of other residues involved the Zn1-WGR-HD axis, including D45 in the Zn1 domain and H742/D743 in the HD, might also result in PARPi resistance. This has been partially supported in H742/D743F breast cancer cells and p.45delD mouse ES cells in response to talazoparib treatment.113 Similarly, the presence of a W318R mutation in the Zn3 domain that disrupts Zn3/HD interaction also eliminates PARPi pro-retention functions.14 Other mutations that affect interdomain communication, such as N329Q and 848delY, also promote talazoparib resistance through partial PARP1 trapping defects.113
Mutations that directly regulate DNA-binding within the PARP1 Zn domains can also affect PARP1 trapping. More specifically, residues M43 and F44 in the Zn1 domain are conserved and involved in base stacking interactions;114 mutation or deletion of these residues causes talazoparib resistance in BRCA1-deficient SUM149 cells and in mouse ES cells by impairing PARP1 trapping.113 Deletion of residues K119 and S120, both DNA-contacting residues in the Zn2 domain,114 potentially eliminates microirradiation-induced PARP1 recruitment to the sites of DNA damage. This suggests that mutations in K119 and S120 residues result in PARPi resistance by impairing PARP1 DNA-binding functions.113 In addition, mutations affecting PARPi binding to PARP1 can also cause resistance. For example, mutation in the residues that define molecular interaction between EB-47 and HD (D766/770A) truncates EB-47 ability to influence PARP1 allostery and retention on DNA breaks.14
It must be noted that a large number of mutations that affect residues involved in either interdomain communication or direct DNA binding have been identified in cellular or mouse models, but have not been confirmed in PARPi-resistant patients. However, the preliminary data strongly suggest that such mutations can develop in patients that could potentially promote PARPi resistance. Recent progresses in the genomic profiling of from PARPi-resistant tumors86,115 could be used to identify the mutant PARP residues associated with PARPi resistance.
Predictors of clinical response to PARPi treatment
PARPi sensitivity can be predicted utilizing germline BRCA1/2 mutations, somatic BRCA1/2 mutations, or a genomic instability score. However, even with these methods available, identification of predictive biomarkers would allow for treatment regimens to be more effectively tailored to each individual patient. The homologous recombination deficiency loss of heterozygosity (HRD-LOH) assay also serves as an important factor that helps the detection of HRD irrespective of etiology. It is measured by levels of genomic LOH.116,117 Although HRD score or HR gene mutation assessment can be used to augment for PARPi responders, neither test accurately predicts PARPi response. More specifically, objective response rates to these compounds in BRCA1/BRCA2-mutant relapsed platinum sensitive ovarian cancers vary from 30% to 80%.118–120 Attempts to identify predictive biomarkers are ongoing. Potential clinical biomarkers of interest should serve as an indicator of a tumor's HR-defectiveness, PARPi resistance, or ‘PARPness’, which refers to the phenomenon of PARPi benefit despite a lack of HR mutations in the patient.
RAD51 loss as a predictor of HR-defectiveness
Two eukaryotic recombinases, RAD51 and DMC1, mediate homologous DNA pairing and strand exchange during HR. While DMC1 is only expressed in meiosis, RAD51 functions in both mitotic HR and meiotic HR events.121 After recruitment to DNA breaks by BRCA1/2, RAD51 mediates the formation of DNA joints that links homologous DNA molecules.122–124 RAD51 nuclear foci serve as a critical biomarker of intact HR. The low expression of RAD51 has been found to be coupled with an objective response to PARPi. In a recent clinical trial, the presence of RAD51 foci was tested by immunohistochemistry and was detected in all patients with acquired PARPi resistance probably due to reconstitution of HR.87 Interestingly, RAD51 nuclear foci were the common characteristic in PDXs and patient samples with primary or acquired PARPi resistance, regardless of BRCA2 restoration, expression of BRCA1 hypomorphic proteins, or 53BP1 loss. This largely resolves a practical distinction between PARPi sensitivity and resistance which genomic testing alone may not be sufficient to clarify.91,125 Thus, immunostaining of RAD51 foci offers real-time analysis of a tumor's HR proficiency irrespective of mechanism. In addition, results are more rapidly available and economically favorable than in exome sequencing. Taken together, these features favor the use of RAD51 as a biomarker to predict PARPi sensitivity in clinic. The RAD51 score could be used not only to identify PARPi-sensitive cancer patients, but also to expand the optimal patient demographic for PARPi therapies beyond the standard BRCA1/2-mutations.
53BP1 loss as a predictor of PARPi resistance
Recent reports have extensively linked 53BP1 loss to PARPi resistance,126,127 presumably as loss of 53BP1 partially reinstates HR in BRCA1-deficient cells.128 This restoration is made possible because HR and NHEJ contest to repair DNA breaks during DNA replication.129 After DYNLL1 recruits necessary proteins to the damaged site,59 53BP1 protects DNA ends from excessive resection through a shieldin complex;130 the specifics depend on PTIP3 and RIF1 interactions.131 In addition, 53BP1 facilitates NHEJ-related repair of intrachromosomal breaks, immunoglobulin class-switch recombination and fusion of unshielded telomeres.132 53BP1 deletion partially restores the formation of RAD51 filaments at resected DSBs in a PALB2- and BRCA2-dependent manner, and promotes ATM-dependent processing of broken DNA ends to facilitate repair by HR in BRCA1-deficient cells. Moreover, PALB2 chromatin recruitment is facilitated by an interaction between its chromatin associated motif and the nucleosome acidic patch region which is bound by 53BP1’s ubiquitin-directed recruitment domain in 53BP1-proficient cells.133 53BP1 loss attenuates hypersensitivity of BRCA-mutant cells to PARPi and reinstates error-free repair by HR. This suggests that 53BP1 loss may be a predictor of PARPi resistance in patients. In a Phase I open-label clinical trial of PARPi ABT-767 among ovarian cancer patients, a decreased 53BP1 immunostaining score was correlated to a decreased antitumor efficacy of ABT-767 in the HR-deficient subset.120 These results are consistent with those obtained from TNBC patients,134 xenografts,135 and BRCA1-deficient mouse mammary tumors.80 These results unanimously support the use of 53BP1 loss as a predictor of PARPi resistance in clinic.
SMAD4 loss in head and neck squamous cell carcinoma (HNSCC)
A major contributor to HNSCC initiation and progression is a mutation or deletion in the SMAD4 gene and occurs in 35% of primary HNSCC specimens and 41.3% of PDXs.136 This is significant because SMAD4 loss could reduce BRCA1 and RAD51 protein levels, and thus result in genomic instability. In a Phase I clinical trial testing combination treatments of olaparib with radiotherapy and cetuximab for advanced HNSCC, it was found that five of six patients with SMAD4 loss showed prolonged progression-free survival (PFS), while four of eight SMAD4-positive patients exhibited effective response.137 While the sample size leaves much to be desired, this study indicates that SMAD4 loss can possibly sensitize HNSCCs to olaparib, and potentially serve as a biomarker of therapeutic response to PARPi in HNSCC patients. Similarly, SMAD4 FISH assays may function as a new platform for clinical diagnosis of SMAD4 chromosomal loss,136 and promote the application of the biomarker in clinic.
MYC serves as a biomarker of PARPness
A substantial number of patients lacking of HR mutations may still benefit from PARP inhibitors,138 and can be classified on a spectrum of relative ‘PARPness’ to specify their responsiveness to PARPi.40 Usually, PARP1 is the initiator of Alt-NHEJ pathway of DNA repair54 besides for regulating base-excision repair (BER). However, this is not the case in Multiple Myeloma (MM),139 TK-activated leukemia,140 MYC–positive Burkitt lymphoma,141 and neuroendocrine prostate cancer (NEPC).142 A recent analysis of a MM patient dataset (GSE24080) showed significant positive correlation between PARP1 and MYC expression,139 an important oncogene143 and a driver transcription factor hyper-activated in a majority of MM patients.144 MYC directly binds to the PARP1 promoter and mediates transcription; high MYC expression correlates to PARPi sensitivity in MM,139 Burkitt lymphoma140 and neuroblastoma145. Likewise, albeit BRCA-proficient, glioblastoma cells with MYC or MYCN amplification exhibit sensitivity to PARPi, likely due to the repression of CDK18 that facilitates ATR activation.146 In addition, PARPi induced cytotoxic effects are detectable among most MYC-dependent cancer types. All these results strongly suggest that MYC can serve as a potential response biomarker for PARPi-containing treatment protocols irrespective of HR status.
Conclusions and future perspectives
Great strides have been made in PARPi, with several selective and potent inhibitors emerging in the clinic. At present 432 trials of five clinical PARP inhibitors (olaparib, talazoparib, veliparib, niraparib, and rucaparib) are ongoing either alone or in combination with other antitumor agents. These trials aim to extend the license for use beyond BRAC-deficient tumors, expand indications beyond breast and ovarian cancers, and test the efficacy of PARPi combination therapy. To improve predictive accuracy of tumor response to PARPi or PARPi-involved therapies, there remains a need to identify more effective biomarkers. In addition, an emphasis has been placed on understanding underlying mechanisms in preclinical models of acquired PARPi-resistance. These mechanisms are being verified in patients. Moreover, efforts to reveal the relationship between structure and activity and to optimize the chemical structure of PARPi are still in progress. This process involves the development of derivatives of current PARPi as well as the identification of new PARPi compounds. These compounds should ideally contain outstanding drug-like properties and have high potency of both catalytic inhibition and allosteric pro-retention on DNA breaks. Through incremental improvements, PARPi therapeutics can revolutionize patient care by transforming non-responders into responders, and generate optimal outcomes with minimal side effects.
ACKNOWLEDGEMENTS
This work was supported by the National Cancer Institute, the National Institutes of Health (Grant No. R01CA236878).
Contributor Information
Ping Zhou, Laboratory of Molecular Signaling, Division of Oral Biology and Medicine, School of Dentistry, UCLA, Los Angeles, CA 90095, USA; Jonsson Comprehensive Cancer Center, UCLA, Los Angeles, CA 90095, USA.
Justin Wang, Laboratory of Molecular Signaling, Division of Oral Biology and Medicine, School of Dentistry, UCLA, Los Angeles, CA 90095, USA; Jonsson Comprehensive Cancer Center, UCLA, Los Angeles, CA 90095, USA.
Daniel Mishail, Laboratory of Molecular Signaling, Division of Oral Biology and Medicine, School of Dentistry, UCLA, Los Angeles, CA 90095, USA.
Cun-Yu Wang, Laboratory of Molecular Signaling, Division of Oral Biology and Medicine, School of Dentistry, UCLA, Los Angeles, CA 90095, USA; Jonsson Comprehensive Cancer Center, UCLA, Los Angeles, CA 90095, USA; Department of Bioengineering, Henry Samueli School of Engineering and Applied Science, UCLA, Los Angeles, CA 90095, USA.
Author contributions
P.Z., J.W., and D.M. wrote the manuscript. P.Z. and C.Y.W. chose the targeted areas. C.Y.W provided critical revision of the manuscript.
Conflict of interest
All authors declared that they had no conflict of interest.
References
- 1. Vyas S, Chang P. New PARP targets for cancer therapy. Nat Rev Cancer. 2014;14:502–9.. doi: 10.1038/nrc3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sukhanova MV, Abrakhi S, Joshi V, et al. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 2016;44:e60. doi: 10.1093/nar/gkv1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rouleau M, Patel A, Hendzel MJ, et al. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301.. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–24.. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 5. Haince JF, McDonald D, Rodrigue A, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–208.. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 6. Langelier MF, Riccio AA, Pascal JM. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42:7762–75.. doi: 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Langelier MF, Planck JL, Roy Set al. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem. 2011;286:10690–701.. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Menissier DMJ, Ricoul M, Tartier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003;22:2255–63.. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7.. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 10. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21.. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 11. Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–8.. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiarugi A. A snapshot of chemoresistance to PARP inhibitors. Trends Pharmacol Sci. 2012;33:42–8.. doi: 10.1016/j.tips.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 13. Hopkins TA, Shi Y, Rodriguez LE, et al. Mechanistic dissection of PARP1 trapping and the impact on in vivo tolerability and efficacy of PARP inhibitors. Mol Cancer Res. 2015;13:1465–77.. doi: 10.1158/1541-7786.MCR-15-0191-T. [DOI] [PubMed] [Google Scholar]
- 14. Zandarashvili L, Langelier M, Velagapudi UK, et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science. 2020;368:x6367. doi: 10.1126/science.aax6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Langelier MF, Planck JL, Roy S, et al. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–32.. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lomonosov M, Anand S, Sangrithi M, et al. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–22.. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16.. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99.. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murai J, Huang SY, Renaud A, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–43.. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Zhang S, Song L, et al. Synergistic lethality between PARP-trapping and alantolactone-induced oxidative DNA damage in homologous recombination-proficient cancer cells. Oncogene. 2020;39:2905–20.. doi: 10.1038/s41388-020-1191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–40.. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 22. Mateo J, Lord CJ, Serra V, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30:1437–47.. doi: 10.1093/annonc/mdz192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–102.. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 24. Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–27.. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coleman RL, Fleming GF, Brady MF, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. 2019;381:2403–15.. doi: 10.1056/NEJMoa1909707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langelier MF, Zandarashvili L, Aguiar PM, et al. NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun. 2018;9:844. doi: 10.1038/s41467-018-03234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jagtap PG, Southan GJ, Baloglu E, et al. The discovery and synthesis of novel adenosine substituted 2,3-dihydro-1H-isoindol-1-ones: potent inhibitors of poly(ADP-ribose) polymerase-1 (PARP-1). Bioorg Med Chem Lett. 2004;14:81–5.. doi: 10.1016/j.bmcl.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 28. Qiu W, Lam R, Voytyuk O, et al. Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2. Acta Crystallogr D Biol Crystallogr. 2014;70:2740–53.. doi: 10.1107/S1399004714017660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dawicki-McKenna JM, Langelier MF, DeNizio JE, et al. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol Cell. 2015;60:755–68.. doi: 10.1016/j.molcel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pommier Y, O'Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:317–62.. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 31. Liu L, Kong M, Gassman NR, et al. PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic Acids Res. 2017;45:12834–47.. doi: 10.1093/nar/gkx1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen Y, Rehman FL, Feng Y, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19:5003–15.. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hopkins TA, Ainsworth WB, Ellis PA, et al. PARP1 trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol Cancer Res. 2019;17:409–19.. doi: 10.1158/1541-7786.MCR-18-0138. [DOI] [PubMed] [Google Scholar]
- 34. Penning TD, Zhu GD, Gandhi VB, et al. Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem. 2009;52:514–23.. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 35. Eustermann S, Wu WF, Langelier MF, et al. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol Cell. 2015;60:742–54.. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langelier MF, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr Opin Struct Biol. 2013;23:134–43.. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Penning TD, Zhu GD, Gong J, et al. Optimization of phenyl-substituted benzimidazole carboxamide poly(ADP-ribose) polymerase inhibitors: identification of (S)-2-(2-fluoro-4-(pyrrolidin-2-yl)phenyl)-1H-benzimidazole-4-carboxamide (A-966492), a highly potent and efficacious inhibitor. J Med Chem. 2010;53:3142–53.. doi: 10.1021/jm901775y. [DOI] [PubMed] [Google Scholar]
- 38. Brock WA, Milas L, Bergh S, et al. Radiosensitization of human and rodent cell lines by INO-1001, a novel inhibitor of poly(ADP-ribose) polymerase. Cancer Lett. 2004;205:155–60.. doi: 10.1016/j.canlet.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 39. Calabrese CR, Almassy R, Barton S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67.. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 40. Pilie PG, Tang C, Mills GB, et al. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81–104.. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mendes Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. Embo Mol Med. 2009;1:315–22.. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weston VJ, Oldreive CE, Skowronska A, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–87.. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 43. Alonso-de VI, Paz-Cabrera MC, Rother MB, et al. PHF2 regulates homology-directed DNA repair by controlling the resection of DNA double strand breaks. Nucleic Acids Res. 2020;48:4915–27.. doi: 10.1093/nar/gkaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z, Zuo W, Zeng Q, et al. Loss of NFBD1/MDC1 disrupts homologous recombination repair and sensitizes nasopharyngeal carcinoma cells to PARP inhibitors. J Biomed Sci. 2019;26:14. doi: 10.1186/s12929-019-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moquin DM, Genois MM, Zhang JM, et al. Localized protein biotinylation at DNA damage sites identifies ZPET, a repressor of homologous recombination. Genes Dev. 2019;33:75–89.. doi: 10.1101/gad.315978.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang C, Zang W, Ji Y, et al. Ribosomal protein L6 (RPL6) is recruited to DNA damage sites in a poly(ADP-ribose) polymerase-dependent manner and regulates the DNA damage response. J Biol Chem. 2019;294:2827–38.. doi: 10.1074/jbc.RA118.007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Campillo-Marcos I, Lazo PA. Olaparib and ionizing radiation trigger a cooperative DNA-damage repair response that is impaired by depletion of the VRK1 chromatin kinase. J Exp Clin Cancer Res. 2019;38:203. doi: 10.1186/s13046-019-1204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu S, Paydar M, Wang F, et al. Kinesin Kif2C in regulation of DNA double strand break dynamics and repair. Elife. 2020;9:e53402. doi: 10.7554/eLife.53402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin X, Chen D, Zhang C, et al. Augmented antitumor activity by olaparib plus AZD1775 in gastric cancer through disrupting DNA damage repair pathways and DNA damage checkpoint. J Exp Clin Cancer Res. 2018;37:129. doi: 10.1186/s13046-018-0790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Washino S, Rider LC, Romero L, et al. Loss of MAP3K7 sensitizes prostate cancer cells to CDK1/2 inhibition and DNA damage by disrupting homologous recombination. Mol Cancer Res. 2019;17:1985–98.. doi: 10.1158/1541-7786.MCR-18-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fang P, Madden JA, Neums L, et al. Olaparib-induced adaptive response is disrupted by FOXM1 targeting that enhances sensitivity to PARP inhibition. Mol Cancer Res. 2018;16:961–73.. doi: 10.1158/1541-7786.MCR-17-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abbotts R, Topper MJ, Biondi C, et al. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc Natl Acad Sci U S A. 2019;116:22609–18.. doi: 10.1073/pnas.1903765116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Johnson N, Li YC, Walton ZE, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med. 2011;17:875–82.. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wray J, Williamson EA, Singh SB, et al. PARP1 is required for chromosomal translocations. Blood. 2013;121:4359–65.. doi: 10.1182/blood-2012-10-460527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fell VL, Schild-Poulter C. The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res. 2015;763:15–29.. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 56. Wang M, Wu W, Wu W, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–82.. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–11.. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCormick A, Donoghue P, Dixon M, et al. Ovarian cancers harbor defects in nonhomologous end joining resulting in resistance to rucaparib. Clin Cancer Res. 2017;23:2050–60.. doi: 10.1158/1078-0432.CCR-16-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Becker JR, Cuella-Martin R, Barazas M, et al. The ASCIZ-DYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat Commun. 2018;9:5406. doi: 10.1038/s41467-018-07855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang H, Pannunzio NR, Adachi N, et al. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506.. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schimmel J, Kool H, van Schendel R, et al. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. Embo J. 2017;36:3634–49.. doi: 10.15252/embj.201796948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Srinivasan G, Williamson EA, Kong K, et al. MiR223-3p promotes synthetic lethality in BRCA1-deficient cancers. Proc Natl Acad Sci U S A. 2019;116:17438–43.. doi: 10.1073/pnas.1903150116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sen T, Rodriguez BL, Chen L, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9:646–61.. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiao S, Xia W, Yamaguchi H, et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res. 2017;23:3711–20.. doi: 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xue C, Xu Y, Ye W, et al. Expression of PD-L1 in ovarian cancer and its synergistic antitumor effect with PARP inhibitor. Gynecol Oncol. 2020;157:222–33.. doi: 10.1016/j.ygyno.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 66. Shen J, Zhao W, Ju Z, et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 2019;79:311–9.. doi: 10.1158/0008-5472.CAN-18-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ding L, Kim HJ, Wang Q, et al. PARP inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep. 2018;25:2972–80.. doi: 10.1016/j.celrep.2018.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. 2018;215:1287–99.. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sato H, Niimi A, Yasuhara T, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. 2017;8:1751. doi: 10.1038/s41467-017-01883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stewart RA, Pilie PG, Yap TA. Development of PARP and immune-checkpoint inhibitor combinations. Cancer Res. 2018;78:6717–25.. doi: 10.1158/0008-5472.CAN-18-2652. [DOI] [PubMed] [Google Scholar]
- 71. Vikas P, Borcherding N, Chennamadhabani A, et al. Therapeutic potential of combining PARP inhibitor and immunotherapy in solid tumors. Front Oncol. 2020;10:570. doi: 10.3389/fonc.2020.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zimmer AS, Nichols E, Cimino-Mathews A, et al. A phase I study of the PD-L1 inhibitor, durvalumab, in combination with a PARP inhibitor, olaparib, and a VEGFR1-3 inhibitor, cediranib, in recurrent women's cancers with biomarker analyses. J Immunother Cancer. 2019;7:197. doi: 10.1186/s40425-019-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Karzai F, Madan R, Owens H, et al. A phase 2 study of olaparib and durvalumab in metastatic castrate-resistant prostate cancer in an unselected population. J Clin Oncol. 2018;36:163. doi: 10.1200/JCO.2018.36.6_suppl.163. [Google Scholar]
- 74. Drew Y, Jonge M, Hong SH, et al. An open-label, phase II basket study of olaparib and durvalumab (MEDIOLA): Results in germline BRCA-mutated platinum-sensitive relapsed ovarian cancer. Gynecol Oncol. 2018;149:246–7.. doi: 10.1016/j.ygyno.2018.04.555. [Google Scholar]
- 75. Vinayak S, Tolaney S, Schwartzberg L, et al. TOPACIO/Keynote-162: Niraparib + pembrolizumab in patients with metastatic triple-negative breast cancer, a phase 2 trial. J Clin Oncol. 2018;36:1011. doi: 10.1200/JCO.2018.36.15_suppl.1011. [Google Scholar]
- 76. Konstantinopoulos P, Waggoner S, Vidal G, et al. Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. Jama Oncol. 2019;5:1141–9.. doi: 10.1001/jamaoncol.2019.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Paczulla AM, Rothfelder K, Raffel S, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572:254–9.. doi: 10.1038/s41586-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ordonez LD, Hay T, McEwen R, et al. Rapid activation of epithelial-mesenchymal transition drives PARP inhibitor resistance in Brca2-mutant mammary tumours. Oncotarget. 2019;10:2586–606.. doi: 10.18632/oncotarget.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Han Y, Li CW, Hsu JM, et al. Metformin reverses PARP inhibitors-induced epithelial-mesenchymal transition and PD-L1 upregulation in triple-negative breast cancer. Am J Cancer Res. 2019;9:800–15. [PMC free article] [PubMed] [Google Scholar]
- 80. Jaspers JE, Kersbergen A, Boon U, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81.. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. D'Andrea AD. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst). 2018;71:172–6.. doi: 10.1016/j.dnarep.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 82. Christie EL, Fereday S, Doig K, et al. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol. 2017;35:1274–80.. doi: 10.1200/JCO.2016.70.4627. [DOI] [PubMed] [Google Scholar]
- 83. Lheureux S, Bruce JP, Burnier JV, et al. Somatic BRCA1/2 recovery as a resistance mechanism after exceptional response to poly(ADP-ribose) polymerase inhibition. J Clin Oncol. 2017;35:1240–9.. doi: 10.1200/JCO.2016.71.3677. [DOI] [PubMed] [Google Scholar]
- 84. Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94.. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 85. Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5.. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 86. Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 2017;7:999–1005.. doi: 10.1158/2159-8290.CD-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Waks AG, Cohen O, Kochupurakkal B, et al. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann Oncol. 2020;31:590–8.. doi: 10.1016/j.annonc.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20.. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Norquist B, Wurz KA, Pennil CC, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–15.. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Park PH, Yamamoto TM, Li H, et al. Amplification of the mutation-carrying BRCA2 allele promotes RAD51 loading and PARP inhibitor resistance in the absence of reversion mutations. Mol Cancer Ther. 2020;19:602–13.. doi: 10.1158/1535-7163.MCT-17-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cruz C, Castroviejo-Bermejo M, Gutierrez-Enriquez S, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29:1203–10.. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kondrashova O, Topp M, Nesic K, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9:3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ter Brugge P, Kristel P, van der Burg E, et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J Natl Cancer Inst. 2016;108:w148. doi: 10.1093/jnci/djw148. [DOI] [PubMed] [Google Scholar]
- 94. Drost R, Dhillon KK, van der Gulden H, et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Invest. 2016;126:2903–18.. doi: 10.1172/JCI70196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Drost R, Bouwman P, Rottenberg S, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809.. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 96. Hu HM, Zhao X, Kaushik S, et al. A quantitative chemotherapy genetic interaction map reveals factors associated with PARP inhibitor resistance. Cell Rep. 2018;23:918–29.. doi: 10.1016/j.celrep.2018.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yang L, Zhang Y, Shan W, et al. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci Transl Med. 2017;9:l1645. doi: 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ibrahim YH, Garcia-Garcia C, Serra V, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–47.. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bitler BG, Watson ZL, Wheeler LJ, et al. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol Oncol. 2017;147:695–704.. doi: 10.1016/j.ygyno.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ryu H, Choi HK, Kim HJ, et al. Antitumor activity of a novel tyrosine kinase inhibitor AIU2001 due to abrogation of the DNA damage repair in non-small cell lung cancer cells. Int J Mol Sci. 2019;20:4728. doi: 10.3390/ijms20194728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Maifrede S, Nieborowska-Skorska M, Sullivan-Reed K, et al. Tyrosine kinase inhibitor-induced defects in DNA repair sensitize FLT3(ITD)-positive leukemia cells to PARP1 inhibitors. Blood. 2018;132:67–77.. doi: 10.1182/blood-2018-02-834895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Du Y, Yamaguchi H, Wei Y, et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med. 2016;22:194–201.. doi: 10.1038/nm.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dong Q, Du Y, Li H, et al. EGFR and c-MET cooperate to enhance resistance to PARP inhibitors in hepatocellular carcinoma. Cancer Res. 2019;79:819–29.. doi: 10.1158/0008-5472.CAN-18-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chu YY, Yam C, Chen MK, et al. Blocking c-Met and EGFR reverses acquired resistance of PARP inhibitors in triple-negative breast cancer. Am J Cancer Res. 2020;10:648–61. [PMC free article] [PubMed] [Google Scholar]
- 105. Ray CA, Callen E, Ding X, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–7.. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rondinelli B, Gogola E, Yucel H, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19:1371–8.. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- 107. Lee SB, Segura-Bayona S, Villamor-Paya M, et al. Tousled-like kinases stabilize replication forks and show synthetic lethality with checkpoint and PARP inhibitors. Sci Adv. 2018;4:t4985. doi: 10.1126/sciadv.aat4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Southgate H, Chen L, Tweddle DA, et al. ATR inhibition potentiates PARP inhibitor cytotoxicity in high risk neuroblastoma cell lines by multiple mechanisms. Cancers (Basel). 2020;12:E1095. doi: 10.3390/cancers12051095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yazinski SA, Comaills V, Buisson R, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–32.. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kim H, Xu H, George E, et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat Commun. 2020;11:3726. doi: 10.1038/s41467-020-14563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015;149:124–38.. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 112. Lee HJ, Lan L, Peng G, et al. Tyrosine 370 phosphorylation of ATM positively regulates DNA damage response. Cell Res. 2015;25:225–36.. doi: 10.1038/cr.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pettitt SJ, Krastev DB, Brandsma I, et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun. 2018;9:1849. doi: 10.1038/s41467-018-03917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ali A, Timinszky G, Arribas-Bosacoma R, et al. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–92.. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7:1006–17.. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Telli ML, Jensen KC, Vinayak S, et al. Phase II study of gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability: PrECOG 0105. J Clin Oncol. 2015;33:1895–901.. doi: 10.1200/JCO.2014.57.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–82.. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gelmon KA, Tischkowitz M, Mackay H, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–61.. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 119. Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87.. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 120. Hurley RM, Wahner HA, Visscher DW, et al. 53BP1 as a potential predictor of response in PARP inhibitor-treated homologous recombination-deficient ovarian cancer. Gynecol Oncol. 2019;153:127–34.. doi: 10.1016/j.ygyno.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. San FJ, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57.. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 122. Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–83.. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Davies AA, Masson JY, McIlwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–82.. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 124. Tarsounas M, Sung P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol. 2020;21:284–99.. doi: 10.1038/s41580-020-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. Embo Mol Med. 2018;10:e9172. doi: 10.15252/emmm.201809172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Francica P, Rottenberg S. Mechanisms of PARP inhibitor resistance in cancer and insights into the DNA damage response. Genome Med. 2018;10:101. doi: 10.1186/s13073-018-0612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kubalanza K, Konecny GE. Mechanisms of PARP inhibitor resistance in ovarian cancer. Curr Opin Obstet Gynecol. 2020;32:36–41.. doi: 10.1097/GCO.0000000000000600. [DOI] [PubMed] [Google Scholar]
- 128. Bouwman P, Aly A, Escandell JM, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95.. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bunting SF, Callen E, Wong N, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54.. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Noordermeer SM, Adam S, Setiaputra D, et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560:117–21.. doi: 10.1038/s41586-018-0340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Callen E, Zong D, Wu W, et al. 53BP1 enforces distinct pre- and post-resection blocks on homologous recombination. Mol Cell. 2020;77:26–38.. doi: 10.1016/j.molcel.2019.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Gupta R, Somyajit K, Narita T, et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell. 2018;173:972–88.. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Belotserkovskaya R, Raga GE, Lawrence N, et al. PALB2 chromatin recruitment restores homologous recombination in BRCA1-deficient cells depleted of 53BP1. Nat Commun. 2020;11:819. doi: 10.1038/s41467-020-14563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hong R, Ma F, Zhang W, et al. 53BP1 depletion causes PARP inhibitor resistance in ATM-deficient breast cancer cells. BMC Cancer. 2016;16:725. doi: 10.1186/s12885-016-2754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Nacson J, Krais JJ, Bernhardy AJ, et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep. 2018;24:3513–27.. doi: 10.1016/j.celrep.2018.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hernandez AL, Wang Y, Somerset HL, et al. Inter- and intra-tumor heterogeneity of SMAD4 loss in head and neck squamous cell carcinomas. Mol Carcinog. 2019;58:666–73.. doi: 10.1002/mc.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hernandez AL, Young CD, Bian L, et al. PARP inhibition enhances radiotherapy of SMAD4-deficient human head and neck squamous cell carcinomas in experimental models. Clin Cancer Res. 2020;26:3058–70.. doi: 10.1158/1078-0432.CCR-19-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Pilie PG, Gay CM, Byers LA, et al. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin Cancer Res. 2019;25:3759–71.. doi: 10.1158/1078-0432.CCR-18-0968. [DOI] [PubMed] [Google Scholar]
- 139. Caracciolo D, Scionti F, Juli G, et al. Exploiting MYC-induced PARPness to target genomic instability in multiple myeloma. Haematologica. 2020. doi: 10.3324/haematol.2019.240713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Muvarak N, Kelley S, Robert C, et al. c-MYC generates repair errors via increased transcription of alternative-NHEJ factors, LIG3 and PARP1, in tyrosine kinase-activated leukemias. Mol Cancer Res. 2015;13:699–712.. doi: 10.1158/1541-7786.MCR-14-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Maifrede S, Martin K, Podszywalow-Bartnicka P, et al. IGH/MYC translocation associates with BRCA2 deficiency and synthetic lethality to PARP1 inhibitors. Mol Cancer Res. 2017;15:967–72.. doi: 10.1158/1541-7786.MCR-16-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang W, Liu B, Wu W, et al. Targeting the MYCN-PARP-DNA damage response pathway in neuroendocrine prostate cancer. Clin Cancer Res. 2018;24:696–707.. doi: 10.1158/1078-0432.CCR-17-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35.. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Holien T, Vatsveen TK, Hella H, et al. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–3.. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 145. Colicchia V, Petroni M, Guarguaglini G, et al. PARP inhibitors enhance replication stress and cause mitotic catastrophe in MYCN-dependent neuroblastoma. Oncogene. 2017;36:4682–91.. doi: 10.1038/onc.2017.40. [DOI] [PubMed] [Google Scholar]
- 146. Ning JF, Stanciu M, Humphrey MR, et al. Myc targeted CDK18 promotes ATR and homologous recombination to mediate PARP inhibitor resistance in glioblastoma. Nat Commun. 2019;10:2910. doi: 10.1038/s41467-019-10993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]