Highlights
-
•
Prototype dose-response model for initial infection by respiratory coronaviruses developed based on molecular parameters;
-
•
Number of free virus particles not bound to mucus in the lung modelled as a function of virus dose in mucus and thermodynamic association constant;
-
•
Binding of MERS-CoV to human mucin may contribute to lower human-to-human transmission compared to SARS-CoV-2;
-
•
Binding of MERS-CoV and SARS-CoV-2 to cellular receptors is not a limiting factor in infection;
-
•
This model may help evaluate the impact of distance in the transmission of SARS-CoV-2.
Keywords: SARS-CoV-2, Infection, Dose-response, Mucin, Risk
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Middle East respiratory syndrome coronavirus (MERS-CoV) infect the human respiratory tract. A prototype thermodynamic equilibrium model is presented here for the probability of the virions getting through the mucus barrier and infecting epithelial cells based on the binding affinity (Kmucin) of the virions to mucin molecules in the mucus and parameters for binding and infection of the epithelial cell. Both MERS-CoV and SARS-CoV-2 bind strongly to their cellular receptors, DDP4 and ACE2, respectively, and infect very efficiently both bronchus and lung ex vivo cell cultures which are not protected by a mucus barrier. According to the model, mucin binding could reduce the infectivity for MERS-CoV compared to SARS-CoV-2 by at least 100-fold depending on the magnitude of Kmucin. Specifically Kmucin values up to 106 M−1 have little protective effect and thus the mucus barrier would not remove SARS-CoV-2 which does not bind to sialic acids (SA) and hence would have a very low Kmucin. Depending on the viability of individual virions, the ID50 for SARS-CoV-2 is estimated to be ~500 virions (viral RNA genomic copies) representing 1 to 2 pfu. In contrast MERS-CoV binds both SA and human mucin and a Kmucin of 5 × 109 M−1 as reported for lectins would mop up 99.83% of the virus according to the model with the ID50 for MERS-CoV estimated to be ~295,000 virions (viral RNA genomic copies) representing 819 pfu. This could in part explain why MERS-CoV is poorly transmitted from human to human compared to SARS-CoV-2. Some coronaviruses use an esterase to escape the mucin, although MERS-CoV does not. Instead, it is shown here that “clustering” of virions into single aerosol particles as recently reported for rotavirus in extracellular vesicles could provide a co-operative mechanism whereby MERS-CoV could theoretically overcome the mucin barrier locally and a small proportion of 10 μm diameter aerosol particles could contain ~70 virions based on reported maximum levels in saliva. Although recent evidence suggests SARS-CoV-2 initiates infection in the nasal epithelium, the thermodynamic equilibrium models presented here could complement published approaches for modelling the physical entry of pathogens to the lung based on the fate and transport of the pathogen particles (as for anthrax spores) to develop a dose-response model for aerosol exposure to respiratory viruses. This would enable the infectivity through aerosols to be defined based on molecular parameters as well as physical parameters. The role of the spike proteins of MERS-CoV and SARS-CoV-2 binding to SA and heparan sulphate, respectively, may be to aid non-specific attachment to the host cell. It is proposed that a high Kmucin is the cost for subsequent binding of MERS-CoV to SAs on the cell surface to partially overcome the unfavourable entropy of immobilisation as the virus adopts the correct orientation for spike protein interactions with its protein cellular receptor DPP4.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019, causing a respiratory disease (coronavirus disease 2019, COVID-19) of varying severity in Wuhan, China, subsequently leading to a pandemic (Hui et al 2020). By 8 Aug 2020 there had been some 19.4 million cases of SARS-CoV-2 worldwide with 721,906 deaths globally (Anon 2020). Like SARS-CoV-2 other beta coronaviruses (CoVs) such as severe acute respiratory syndrome virus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) also infect the human respiratory tract and causes severe zoonotic respiratory disease. MERS-CoV is transmitted sporadically to humans from its dromedary camel (Camelus dromedarius) reservoir, causing severe respiratory disease with ~35% fatality rate (Li et al. 2017). A major difference is that both SARS-CoV and SARS-CoV-2 are readily transmitted between humans by close contact, while MERS-CoV is not (Rabaan et al. 2020). Person-to-person community transmission of SARS-CoV-2 is highly efficient while MERS-CoV is transmitted occasionally by human-to-human contact (Killerby et al.2020) through infectious aerosolized particles often in hospital settings (Adhikawi et al. 2019). Thus, humans are considered transient or terminal hosts for MERS-CoV, with no evidence for sustained human-to-human community transmission. In naturally infected camels, MERS-CoV RNA has been recovered most commonly from nasal swabs but also from faecal swabs, rectal swabs, and lung tissue (Killerby et al. 2020). Levels of MERS-CoV RNA and SARS-CoV-2 RNA copies per cm3 of sputum or tracheal aspirate in infected humans are similar with median MERS-CoV levels of 105.5 copies (mild group) to 107.2 copies (severe group) (Oh et al. 2016) and median SARS-CoV-2 levels of 105.9 to 106.7 copies (Pan et al. 2020;To et al. 2020). This suggests these two viruses differ fundamentally in their infectiousness to humans. All three viruses infect bronchus and lung cells with high efficiency and indeed MERS-CoV seems to be the most efficient. Specifically in bronchus cell cultures, MERS-CoV replication competence is similar to that for SARS-CoV-2 and higher than SARS-CoV while in lung cell cultures, MERS-CoV-2 replication is higher than SARS-CoV and SARS-CoV-2 (Hui et al. 2020). While SARS-CoV-2 spike protein may not bind to sialic acid (SA) glycans (Hao et al. 2020) and hence to human mucins, that MERS-CoV spike protein does bind human mucin is well-documented (Li et al. 2017). Since both SARS-CoV-2 and MERS-CoV spike proteins bind strongly to their respective cellular receptors, it may be other factors such as differences in mucin binding affinity that affect infectivity. This is addressed here.
Previously a thermodynamic mechanistic approach was developed to use molecular biology data measured using biochemical methods to calibrate a dose-response model for a virus infecting the human intestine (Gale 2018). Here that model is developed for respiratory viruses such as MERS-CoV and SARS-CoV-2 by focusing on the removal of virus by mucus in the lung through entrapment (Zanin et al. 2016). The objective of this paper is to demonstrate a method to calibrate part of a dose-response model for respiratory viruses such as MERS-CoV and SARS-CoV-2 using available thermodynamic data for the key molecular interactions between the human host and viral factors during each stage of the infection process. The physical barriers and transport of the virions in the airways also have to be included in the model as for inhaled anthrax (Weir and Haas 2011) depending on where in the respiratory system the virus initiates infection, which may be in the nasal epithelium in the case of SARS-CoV-2 (Hou et al. 2020). A key physical barrier is the mucus gel layer which consists of 97% water and 3% solids, representing polymeric mucins. In its protective role, mucus entraps microbes and particles removing them from the lungs via the co-ordinated beating of motile cilia (Ridley and Thornton 2018). The major mucins produced in the airways of the human lung are the secreted polymeric mucins MUC5AC and MUC5B (Ridley and Thornton 2018) and present a barrier to respiratory viruses including influenza A virus (IAV) and adenovirus (De Graaf and Fouchier, 2014; Zanin et al. 2016; McAuley et al. 2017; Stonebraker et al. 2004). Mucins contain SA glycans to which proteins called lectins bind with high affinity, and in this respect viral surface proteins such as IAV haemagglutinin (HA) and MERS-CoV spike protein can be considered as lectins. Lectins show great specificity in the SAs to which they bind (De Graaf and Fouchier, 2014). The spike S1A protein of MERS-CoV is particularly suited to bind human respiratory mucin but not to bovine submaxillary mucin (BSM) reflecting differences in the molecular structure of the SAs (Li et al. 2017). Similarly the mouse hepatitis coronavirus (MHV) and SARS-CoV do not bind BSM (Peng et al. 2012) and to the author's knowledge it is not known if they bind human mucin. In contrast to MERS-CoV, the spike protein of SARS-CoV-2 does not bind to SA (Hao et al. 2020) suggesting it does not bind to mucins. This may represent a fundamental difference between SARS-CoV-2 and MERS-CoV and it is suggested here that this may enhance the ability of SARS-CoV-2 to diffuse through the mucus barrier to reach the lung epithelial cells compared to MERS-CoV which, it is proposed here, becomes trapped in the mucus and removed. Indeed SARS-CoV, SARS-CoV-2 and MERS-CoV are all able to replicate well in ex-vivo cultures of both human bronchus cells and lung cells (Hui et al. 2020). Unlike in the lung in vivo, cell cultures do not have a protective mucous barrier, suggesting that once through the mucus barrier, the three viruses readily infect bronchus and lung cells. Of course, there are other differences too, and the presence of a furin-like cleavage site on the spike protein in SARS-CoV-2 facilitates the spike protein priming and might also increase the efficiency of the spread of SARS-CoV-2 compared to SARS-CoV and MERS-CoV (Rabaan et al. 2020). The fact that SARS-CoV-2 may initiate infection in the nasal epithelium would also increase its human-to-human transmission efficiency.
A dose-response model has been fitted to death data for MHV in mice and also for SARS-CoV in transgenic mice expressing human angiotensin-converting enzyme 2 (hACE2) which is the receptor for SARS-CoV (Watanabe et al. 2010). The ID50 (death in mice through intranasal challenge) was estimated to be 280 plaque-forming units (pfu) (95% CI = 130 to 530 pfu) with p1, the probability of death from a single virion, being 0.00246 (Huang 2013). Dose-response models such as that for MHV and SARS-CoV in mice (Watanabe et al. 2010) assume that the individual virions in a dose act independently and therefore that the risk of infection increases linearly (at low doses at least) with exposure dose, Vexposure. This may not necessarily be the case if virions in a high dose “cooperate” through overwhelming a host defence mechanism, in this case the mucins in the mucus in the lung. It is shown here for a virus that binds mucins that dispersion of the virus in the lung mucus could greatly decrease the fraction of free virus remaining to go on to infect the cells of the lung epithelium compared to the same dose concentrated (or clustered) in a single particle. Thus it is suggested here that rare, high-virion load aerosol particles may be important for MERS-CoV to get through the mucus barrier such that the virions comprising a dispersed dose may be overwhelmed by the mucin defences while the same number of virions if “acting together” may locally overwhelm the mucin defence such that more manage to get through the mucus to the lung epithelial cells. Conceptually this “co-operative action” is analogous to a team of 10 persons forming a human triangle such that one person successfully scales a high wall while acting individually none of the 10 persons can clear the wall. In contrast to MERS-CoV, aerosol particles containing a single virion of SARS-CoV-2 would be more infectious. The actual ID50 in terms of number of virions and the value of p1 depend on the virion (genomic RNA copy number) to pfu ratio. It is not suggested that all those virions comprising a pfu have to co-operate to initiate infection. Instead only a small proportion of virions are viable in terms of being able to initiate infection. It is assumed that all virions whether viable or not are able to bind SA and hence bind mucin through the spike protein in the case of MERS-CoV.
The co-operative effect is only relevant if single aerosol particles small enough to get into the bronchus i.e. <10 μm in diameter (Van Leuken et al. 2016) contain multiple virions. Otherwise the virions must be considered to be fully dispersed and acting independently. A small proportion of 10 μm diameter aerosol particles as produced in coughs from IAV-infected persons (Lindsley et al. 2012) could contain multiple SARS-CoV-2 or MERS-CoV virions. Viral RNA loads for MERS-CoV and SARS-CoV-2 in infected persons of 109 copies per cm3 of sputum have been reported (Oh et al. 2016; To et al. 2020) and even up to 1011 copies per cm3 saliva (Adhikari et al. 2019; Pan et al. 2020). Thus Pan et al. (2020) reported a maximum viral load of SARS-CoV-2 from 80 COVID-19 patients of 1.34 × 1011 copies per cm3 of sputum such that a 10 μm diameter particle of volume of 5.24 × 10−7 mm3 would contain some 70 virions. It should be emphasized these high load particles would represent only a very small proportion and most inhaled aerosol particles would contain either zero or just one virion because the median was 7.5 × 105 to 106.7 copies of SARS-CoV-2 RNA per cm3 of sputum (Pan et al. 2020;To et al. 2020). The mean for MERS-CoV was 6.3 × 108 copies per cm3 saliva (Adhikari et al 2019). However, the thermodynamic dose-response model developed here does need to be able to accommodate inhalation of both particles with a single virion and particles with a cluster of virions in the case of those respiratory viruses such as MERS-CoV that bind to human mucin. Clustering mechanisms such as the secretion in stools of ~0.5 μm diameter extracellular vesicles reported by Santiana et al. (2018) as containing ~15 rotavirus virions could result in some <1 μm diameter particles which are produced in coughs in abundance (Lindsley et al. 2012) containing high virion doses although this has not been demonstrated for coronaviruses (CoVs). Lim et al. (2016) merely say that the assembly of CoV particles is completed through budding of the CoV nucleocapsid through membranes early in the secretory pathway from the endoplasmic reticulum to the Golgi. Bhar and Jones (2019) propose an alternate role (in addition to the role of virion cell binding) for the histo-blood group antigens in norovirus infection as a means of viral aggregation. Another mechanism of aggregation is through multiple viruses binding to a bacterium as in the case of human norovirus and poliovirus attaching to commensal Enterobacter bacteria (Bhar and Jones 2019).
2. Methods
The abbreviations are listed in Table 1 .
Table 1.
List of abbreviations
| (h)ACE2 | (Human) Angiotensin-converting enzyme 2 |
| BSM | Bovine submaxillary mucin, |
| CBD | Carbohydrate binding domain on a lectin or virus spike protein |
| CoV | Coronavirus |
| Cr | Host cell receptor |
| Ctotal | Total number of cells in human lung that have ACE2 receptors and can bind SARS-CoV-2 |
| C.VT | Number of host cells with bound virus at temperature T |
| dpi | Days post infection |
| DPP4 | dipeptidyl peptidase 4 (also called CD26), protein receptor for MERS-CoV |
| Fc | Fraction of virus dose bound to lung cells |
| Fv | Fraction of virus in lung mucus not bound to mucin, i.e. free |
| Ftrans | Fraction of the challenge dose, Vexposure, that is transported through the nasal airways to reach the mucus in the lung-lining fluid |
| GP | Viral (glyco)protein on virus surface that binds to Cr |
| HA | Haemagglutinin |
| HE | Haemagglutinin-esterase |
| HIV | Human immunodeficiency virus |
| IAV | Influenza A virus |
| L | Avogadro number = 6.022 × 1023 molecules per mol |
| Kmucin | Association constant for binding of virus to mucin at temperature T |
| Ka_virus_T | Association constant for binding of virus to host cells at temperature T |
| Kd_mucin | Dissociation constant for each CBD/SA interaction |
| Kd_receptor_T | Dissociation constant for GP from Cr at temperature T |
| LLF | Lung lining fluid |
| M | Molar (moles dm−3) |
| Nv | Number of GP/Cr contacts made on virus binding to cell |
| Nm | Number of GP/SA contacts made on virus binding to mucin molecule |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| MHV | Mouse hepatitis coronavirus |
| Mucfree | Number of free mucin molecules, i.e., not bound to virus |
| Muctotal | Total number of mucin molecules in given volume of mucus |
| NA | Neuraminidase |
| p1 | Probability of infection of host by exposure to a single virion |
| pbudding | Probability progeny virions exit the infected cell |
| pentry | Probability that a virion bound to cell surface enters that cell |
| pcell | Probability, given a virion has bound to the surface of a lung epithelial cell, that that cell becomes infected and releases its progeny viruses |
| phost | Probability of successful infection of the host; |
| ppfu | Probability that a given virion (represented in the exposure as a viral RNA copy) is itself capable of initiating infection in a cell |
| pvirogenesis | Probability virus replicates within cell after entry and progeny virions are assembled. |
| pfu | Plaque-forming unit |
| R | Ideal gas constant = 8.31 J/mol/K |
| S1A | Head of CoV spike protein that binds SA |
| S1B | Head of CoV spike protein that binds protein Cr |
| S2 | Stalk of CoV spike protein involved in membrane fusion and virus entry |
| SARS-CoV | Severe acute respiratory syndrome coronavirus |
| ΔSa_immob | Change in entropy on immobilization of whole virus on binding to cell surface |
| ΔSa_mucin | Change in entropy on immobilization of whole virus on binding to mucin |
| SA | Sialic acid |
| TMPRSS | Transmembrane protease/serine protease that cleaves spike protein into S1 and S2 |
| Vexpsoure | Airborne exposure to a given dose of virions |
| Vfree | Virus not bound to mucin molecules |
| Vmucus | Virus dose entering mucus in lung lining fluid |
| V.Muc | Number of viruses with bound mucin |
2.1. Basic lung model
The human lung surface area was determined as 24–69 m2 with some studies indicating the internal lung surface area to be about 130 m2 (Fröhlich et al. 2016). Determination of the volume of the lining fluid in the lung (LLF), also called epithelial lining fluid or airway surface liquid, is more complicated than for the gastrointestinal fluids because lung volumes are much smaller. In the literature different volumes of 12 cm3, 20–40 cm3, 25 cm3, 10–30 cm3 and 17–20 cm3 have been indicated in different studies reviewed by Fröhlich et al. (2016). Based on the body weight-dependent data obtained in sheep (0.37 ± 0.15 cm3/kg), a 70 kg human would possess 26 cm3 of LLF. The total number of cells in the human lung is 230 × 109 of which 30% are endothelial and 24% are alveolar cells (Crapo et al 1982). Lukassen et al. (2020) investigated ACE2, transmembrane protease/serine protease (TMPRSS2) and furin expression levels in the transcriptome across cell types in lung tissue. Of 39,778 lung cells studied, only 104 expressed ACE2 and TMPRSS2 and/or furin, suggesting that only 0.26% of lung cells are susceptible to infection by SARS-CoV-2. The number of cells expressing ACE2 was 206 accounting for 0.52% of the lung cells (Lukassen et al. 2020). Therefore the total number of lung cells that can bind SARS-CoV-2, Ctotal, is estimated to be 0.52% of 230 × 109 = 1.2 × 109 cells.
2.2. Thermodynamic approach for calibrating a dose-response model for infection of the human lung by a virus
Infection of the host is defined here as the infection of one or more cells in the host such that virogenesis and viral egress successfully take place in at least one cell of the host. The progeny viruses may then go on to infect other host cells leading to disease progression, which is not considered here. The response considered here, namely initial infection, happens before the innate immune response and the acquired immune response both of which are important for stopping progression of infection in the host (Lim et al. 2016). The probability, phost, of being infected by airborne exposure to a given dose of virions, Vexpsoure, may be described mathematically as
| (1) |
where p1 is the probability of infection of the susceptible person by a single virion in an aerosol particle. Although Eq. 1 represents a dose-response for a set of equally susceptible individuals it is important to note that p1 itself may vary between individuals within a population depending for example on genetic factors (Plante et al. 2020) and overall health of the individuals. This is addressed here for individuals with differing concentrations of mucin in the LLF.
At a cellular level, the probability of infection of the host, phost, equals the probability of successful infection of at least one bronchus or lung cell and is related to the number of bronchus/lung cells (C.VT) with bound virus by:-
| (2) |
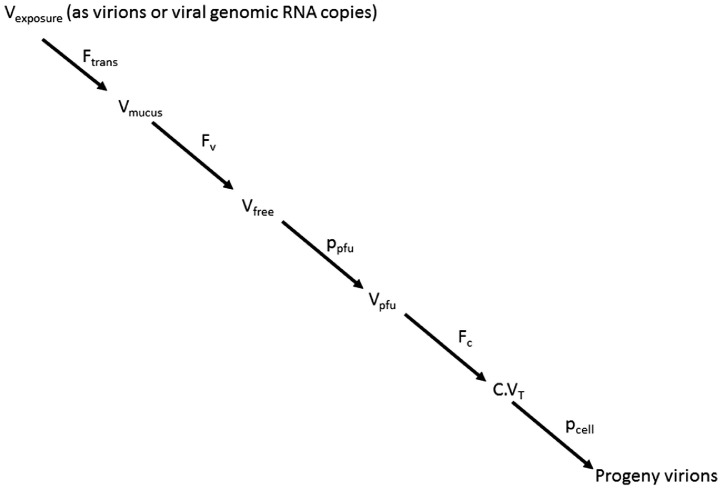
where pcell is the probability of successful infection of a host bronchus/lung cell given a virus has bound to its surface. Thus the more bronchus/lung cells with bound virus then the greater the chance that infection will be successful in at least one of them. Based on the thermodynamic approach described previously (Gale 2018) and accommodating the fate and transport of the virions through the airways (Weir and Haas 2011) the development of the dose-response is broken down into five steps as shown in Fig. 1: -
-
1
Estimation of the fraction, Ftrans, of the exposure dose, Vexposure, that is transported through the nasal airways to reach the mucus in the lung-lining fluid of the nasal epithelium/bronchus/lung;
-
2
Calculation of the number, Vfree, of viruses getting through the host defences e.g. mucins and pathogen pattern recognition receptors to the lung epithelium. This is dependent on the fraction, Fv, of the total virions (Vmucus) in the mucus which are not bound to mucin;
-
3
Estimation of the probability, ppfu that a given virion (represented in the exposure as a viral RNA genomic copy) is fit and actually capable at the molecular level of binding to and initiating infection in a susceptible host cell such that it could be detected as a pfu;
-
4
Calculation of C.VT as the fraction, Fc, of viruses bound to cells based on the thermodynamics of virus/host cell binding and in particular the number and strength of the virus glycoprotein (GP)/cell receptor (Cr) interactions together with the parameter ΔSa_immob, which is the change in entropy on immobilization of whole virus on binding to the cell surface (Gale 2019; Gale, 2020); and
-
5
Estimation of the probability pcell which depends on the ability of the bound virus to enter the cell, replicate and bud releasing progeny virions (Gale, 2017).
Fig. 1.
Pathway from virus exposure to infection of cell.
The total number, Vfree, of viruses surviving the mucin barrier, and getting through to the lung epithelium from Fig. 1 is given by
| (3) |
However, the C.VT term in Eq. 2 cannot be replaced by Vfree calculated as Eq. 3 because C.VT should be an integer to represent a whole number of cells. As an alternative approach from Fig. 1, the probability, p1, of infection of the host by exposure to a single virion in Eq. 1 may be expressed as
| (4) |
since the Ftrans, Fv and Fc terms may be interpreted as probabilities. Thus a prototype thermodynamic dose-response for a respiratory virus in humans is given by:-
| (5) |
It should be noted that the ppfu term represents the fitness of the virion itself, while the pcell term represents the ability of a host cell with bound (viable) virus to undertake and complete virogenesis. The two terms are completely unrelated.
2.3. Estimation of Ftrans, the fraction of inhaled virions which are transported through the airways to reach the mucus in the lung lining fluid
Weir and Haas (2011) model anthrax spore transport through three regions of the respiratory system namely i) the nasal cavity and larynx, ii) the trachea and first branches of the bronchioles and iii) the respiratory bronchioles and alveoli. The model here for SARS-CoV-2 is based on cells within the lung and bronchioles expressing hACE2 being able to bind SARS-CoV-2 and on the basis of data from Hui et al. (2020) and Lukassen et al. (2020), SARS-CoV-2 only needs to get to the bronchus and does not need to reach the alveoli, unlike spores of B. anthracis, to initiate infection in a host. In the case of MERS-CoV which uses a different cell surface receptor from SARS-CoV-2, namely dipeptidyl peptidase 4 (DPP4 or CD26) and possibly certain glycans, cells of the bronchial submucosal glands and bronchus are targeted (Hui et al. 2020; Li et al. 2017). The prototype model presented here does not formally assess the fate and transport of the virus through the respiratory system as described by Weir and Haas (2011) for anthrax spores, and Ftrans is set to 1 such that all the SARS-CoV-2 and MERS-CoV virions in the exposure are assumed to get to the mucus in the bronchus. This is justified for SARS-CoV-2 because recent evidence suggests that the nasal surfaces might be the dominant initial site for respiratory tract infection with likely subsequent aspiration-mediated virus seeding to the lung in SARS-CoV-2 pathogenesis (Hou et al. 2020). It is suggested here that determining Ftrans is much more important for anthrax because once inhaled anthrax spores are infectious only after being transported to the alveoli (Weir and Haas 2011). According to Hou et al. (2020) the nose contains the highest percentage of ACE2-expressing ciliated cells in the proximal airways with high SARS-CoV-2 infectivity of nasal epithelium and a gradient in infectivity characterized by a marked reduction in the distal lung (bronchioles and alveoli). As Hou et al. (2020) point out, aerosol deposition and fomite mechanical delivery deposition modelling suggest that aerosols containing virus inhaled by subjects achieve the highest density of deposition, i.e., highest viral loading per unit surface area, in the nose. This is completely different to the anthrax model and on the basis of the data of Hou et al. (2020) the prototype SARS-CoV-2 model here only needs to focus on the nasal epithelium.
2.4. Prediction of Fv, the fraction of viruses in the mucus layer which are free to bind to receptors on the lung epithelium
It is assumed that one virus binds to one mucin molecule to give a virus/mucin complex (V.Muc) in a dynamic equilibrium (Gale 2018).
The strength of the binding is reflected by the association constant, Kmucin, between virus and mucin and is expressed as:-
| (6) |
The fraction of free virus (Fv) is given by:-
| (7) |
Replacing [V.Muc] with Eq. 6 and rearranging gives Fv in terms of the free mucin concentration [Mucfree] and Kmucin in Eq. 8.
| (8) |
2.4.1. Estimation of [Mucfree] in non-smokers
Kesimer et al. (2017) reported mean (±SE) concentrations of 108±20 pmol per cm3 for MUC5B and 10±4 pmol per cm3 for MUC5AC in lung mucus from persons who had never smoked. The mean concentration for MUC5AC plus MUC5B in non-smokers is therefore 118 pmol per cm3, which is 1.18 × 10−7 M. Using the Avogadro number, L = 6.022 × 1023 molecules per mol, 118 pmol equates to 7.1 × 1010 molecules of MUC5AC and MUC5B in total per mm3 volume of lung mucus.
2.4.2. Variation in lung mucin concentrations between individual non-smokers
It is important for a dose-response model to accommodate variation between indivduals in their susceptibility to infection. Kesimer et al. (2017) measured absolute concentrations of MUC5B and MUC5AC in 19 non-smoker participants and reported large variation between individuals. Thus although the mean [MUC5B] absolute concentration was 108 pmol/cm3 the minimum was 20 pmol/cm3 and the maximum was 300 pmol/cm3.
2.4.3. Calculation of Fv for dispersed virus using Equation 8
The degree of localised dispersion of the virus in the mucus is an important consideration in the methodology in relation to calculation of [Mucfree] in Eq. 8. Where the virus is well dispersed in the lung mucus such that everywhere [Vmucus] << [Muctotal], then Fv can be calculated from Eq. 8 using [Muctotal] to approximate [Mucfree]. The fact that with Muctotal >> Vmucus everywhere in the lung means that even with all the viruses in the initial challenge dose bound to mucin, Mucfree ~ Muctotal and therefore [Muctotal] can be used to approximate [Mucfree] in Eq. 8. Thus [Mucfree] is set to 1.18 × 10−7 M in Eq. 8 for non-smokers.
2.4.4. Calculation of Fv for locally aggregated virus using the difference equation approach
When Kmucin is large and where [Vmucus] approaches or exceeds [Muctotal] locally, [Mucfree] becomes much smaller than [Muctotal] as the virus overwhelms and mops up all the free mucin molecules. In the extreme all the local mucins molecules are bound to virus and [Mucfree] tends to 0 locally. Therefore the difference equation approach has to be used as described previously (Gale 2018). Briefly Kmucin values are calculated using Eq. 6 for the range of Vfree, Mucfree and V.Muc combinations from V.Muc = 1 to V.Muc = Vmucus (or to V.Muc = Muctotal depending on which is the lesser) assuming Muctotal = 7.1 × 1010 molecules of MUC5AC and MUC5B in total per mm3 and converted to their respective concentrations [Vfree], [Mucfree] and [V.Muc] by dividing by 10−6 dm3/mm3 and by L. The fraction of free virus, Fv, is then calculated as Vfree/Vmucus.
2.4.5. Values of Kmucin
For the purpose of demonstration of the model, Kmucin values from 1 M−1 to 1022 M−1 are used. Kmucin is related to the number Nm of interactions between carbohydrate binding domains (CBDs) on the lectin (virus) and the SA glycans on the mucin according to:-
| (9) |
where Kd_mucin is the dissociation constant for each CBD/SA interaction, ΔSa_mucin is the change in entropy on immobilization of whole virus on binding to mucin and R is the ideal gas constant (Gale 2018).
2.5. Modelling the effect of the virus:mucin ratio on the fraction, Fv, of free virus
Two scenarios are modelled for each virion:mucin ratio.
2.5.1. Scenario 1: Inhaled aerosol particles with different viral loads are each engulfed in 1 mm3 of mucus
For the purpose of undertaking the difference equation approach a 1 mm3 volume (10−6 dm3) is chosen to represent the volume of mucus into which the virus dose (Vmucus) is dispersed as the airborne particle(s) enter(s) the LLF of the lung. This volume of mucus contains 7.1 × 1010 mucin molecules. It is assumed that virus doses of 2x, 1x, 0.5x and 0.1 × 7.1 × 1010 virions respectively (Table 2 ) are effectively exposed to the 7.1 × 1010 mucins in 1 mm3 of mucus as they pass through the mucus to the lung epithelium. In effect the 1 mm3 volume of mucus represents the limit of the aerosol dispersion in the mucus, such that within that 1 mm3 volume as many virions as possible (given the dose) are exposed to mucins. It can be envisaged as providing the thinnest layer of mucus film that maximises the number of viruses in the dose that are in direct contact with mucus and hence exposed to a mucin. Eq. 6 is based on concentrations and Kmucin will be same whether it's calculated using 7.1 × 104 viruses and 7.1 × 104 mucins in a volume of 10−6 mm3 for example or using 7.1 × 1010 virus and 7.1 × 1010 mucins in 1 mm3 as performed here in the model. However, although a dose of 7.1 × 104 SARS-CoV-2 virions is perhaps more realistic as an inhaled dose based on 108.94 virions per cm3 of saliva (To et al. 2020), it is preferable to use values of 7.1 × 1010 virus and 7.1 × 1010 mucins in the difference equation approach to achieve the greater range of simulated Kmucin values and greater incremental sensitivity. So with 7.1 × 104 viruses and 7.1 × 104 mucins the highest Kmucin that can be achieved with the difference equation approach is 1016.6 M−1. This compares to 1028.6 M−1 which can be achieved with 7.1 × 1010 viruses and 7.1 × 1010 mucins in 1 mm3.
Table 2.
Fraction, Fv, of free virus in lung mucus calculated by difference equation approach for range of virion to mucin molecule ratios representing exposure of virion to mucins at the aerosol particle/mucus interface.
| Scenario 1: Number of virions to number of mucin molecules in 1 mm3 volume of mucus | Scenario 2: Number of virions to number of mucin molecules (volume of mucus) | Local ratio virion:mucin at aerosol particle/mucus interface | Fv at K mucin = 1010 M−1 (Reduction in risk by mucus) | Fv at K mucin = 1022 M−1. |
| 1.42 × 1011 virions to 7.1 × 1010 mucins | 7.1 × 1010 virions to 3.55 × 1010 mucins in 0.5 mm3. | 2:1 | 0.5004 (2-fold) | *0.5000 |
| 1.07 × 1011 virions to 7.1 × 1010 mucins | 7.1 × 1010 virions to 4.73 × 1010 mucins in 0.666 mm3. | 1.5:1 | 0.334 (3-fold) | *0.333 |
| 7.1 × 1010 virions to 7.1 × 1010 mucins | 7.1 × 1010 virions to 7.1 × 1010 mucins in 1 mm3. | 1:1 | 0.0286 (35-fold) | 0.3 × 10−7 |
| 3.55 × 1010 virions to 7.1 × 1010 mucins | 7.1 × 1010 virions to 1.42 × 1011 mucins in 2 mm3. | 1:2 | 0.00169 (588-fold) | 1.4 × 10−11 |
| <0.71 × 1010 virions to 7.1 × 1010 mucins | 7.1 × 1010 virions to >7.1 × 1011 mucins in >10 mm3. | <0.1:1 (Eq. 8) | 0.00085 (1,176-fold) | <10−15 |
*limit of what is achievable given ratio
2.5.2. Scenario 2: Aerosol particles with the same viral load are each engulfed in different volumes of mucus
A dose of 7.1 × 1010 virions is locally dispersed into 0.5 mm3 (representing 2 virions per mucin), 1 mm3 (representing 1 virion per mucin), 2 mm3 (representing 1 virion per 2 mucins) and 10 mm3 (representing 1 virion per 10 mucins) of lung mucus with [Muctotal] = 1.18 × 10−7 M.
2.6. Estimation of ppfu, the probability that a virion (as represented by a viral RNA genomic copy) is capable of initiating infection in a cell
Not all virions produced during infection are capable of initiating infection in a subsequent host. Thus Bull et al. (2012) showed only 0.01% of norovirus particles could initiate infection in humans because each of the virions is different due to the mutant spectrum. According to Vicenzi et al. (2004) approximately 360 viral genomes of SARS-CoV are required to generate a pfu. This can be interpreted as 359 of every 360 virions being unable to initiate infection (non-infectious). Thus ppfu is calculated as 1/360 = 0.0028.
2.7. Calculation of the fraction, Fc, of virus bound to cells from the association constant, Ka_virus_T representing the strength of virus binding to host cells
Central to calculation of the number of lung epithelial cells with bound virus is the association constant, Ka_virus_T which represents the strength of binding of whole virus to a lung cell at temperature T (Gale 2019, Gale, 2020). This is calculated as
| (10) |
where Kd_receptor_T is the dissociation constant for the CoV spike trimer protein from its proteinaceous cellular receptor, Cr, at temperature T, Nv is the number of spike protein trimer/Cr interactions on virus/host cell binding and ΔSa_immob is the change in entropy on immobilization of whole virus on binding to the cell surface (Gale 2019, Gale, 2020).
2.7.1. Values of Kd_receptor_T for SARS-CoV, SARS-CoV-2 and MERS-CoV
The dissociation constant, Kd_receptor_T represents the strength of the interaction between the CoV spike protein and Cr with smaller values representing stronger binding. Wrapp et al. (2020) demonstrate that each SARS-CoV-2 trimer binds just one ACE2 with a K d_receptor_T of 1.47 × 10−8 M. This is much higher affinity binding than the K d_receptor_T of 1.85 × 10−7 M for the SARS-CoV spike trimer binding to a single ACE2 reported by Kirchdoerfer et al. (2018). The value of Kd_receptor_T for each MERS spike protein binding to its CD26 (DPP4) cellular receptor is 1.67 × 10−8 M (Lu et al., 2013).
2.7.2. Values of Nv
There are on average 74 spike trimers per SARS-CoV virion (Neumann et al. 2011) each of which could in theory make at least one interaction with ACE2. According to Guo et al. (2018) 10% of the IAV virus surface could be in contact with a surface, such that approximately 7 IAV HA trimers can interact with receptor-loaded SA molecules. Thus Nv could be as high as 7 spike trimer protein/receptor contacts in the case of MERS-CoV, SARS-CoV and SARS-CoV-2 which are of similar size to IAV. Yuan et al. (2017) hypothesize that for MERS-CoV bound on the cell surface, one DPP4 receptor may crosslink two S trimers by binding to receptor-binding domains, one from each trimer thus giving large values for Nv involving multiple spike proteins linked together with DPP4 receptors.
2.7.3. Values of ΔSa_immob
The parameter ΔSa_immob was first identified in a thermodynamic analysis of the effect of temperature on virus binding to host cells (Gale 2019) and not surprisingly there are no data for its magnitude as yet although it is likely to be large and negative depending on the degree of immobilisation of the whole virus on binding to the host cell. The translational entropy of a particle of molecular weight 2.5 × 108 Da as for Rous sarcoma virus (Vogt and Simon 1999) is estimated to be +350 J/mol/K using the Sackur-Tetrode equation (P. Gale unpublished results). Assuming half of this entropy is lost on cell binding and that the rotational entropy lost is similar (Finkelstein and Janin 1989) then ΔSa_immob would approximate -350 J/mol/K although estimates based on Eq. 10 suggest values as low as -1,091 J/mol/K (Gale 2020). This large negative entropy change may be offset to some degree by non-specific attachment of the virus to the cell surface through SA and heparan sulphate binding as proposed for HIV such that much of the entropy loss of ΔSa_immob on virus binding is taken prior to specific GP/Cr binding (Gale 2020). For the purpose of the prototype model developed here a value of -350 J/mol/K is used for ΔSa_immob.
2.7.4. Demonstration that Fc ~ 1 in the lung model for Ka_virus_T >~1014 M−1
The previous thermodynamic dose response models for virus binding in the human intestine (Gale 2018) and in the arthropod midgut (Gale 2019) have both shown that when Ka_virus_T is >~1015 M−1 then its actual value is not important because at high virus doses all the host cells have bound virus while at low virus doses all the viruses are bound to host cells. This is therefore investigated for the lung model. Using the difference equation approach (Gale 2018) for binding of 1,000 virions in 25 cm3 of LLF, the fraction, Fc, of virus dose bound to lung cells as a function of Ka_virus_T was calculated for Ctotal = 1.2 × 109 cells and for C.VT = 1 to 1,000 (representing all viruses bound).
2.8. Estimation of pcell
The parameter pcell is the probability of viral entry (through fusion of the virus envelope with the host cell membrane) combined with the probabilities of each of the subsequent steps of virogenesis within the cell being completed, including viral polyprotein synthesis and cleavage by the virus-coded main protease (Zhang et al. 2020), viral RNA replication and capsid assembly followed by budding (viral egress) of progeny virions from the infected cell. Thus pcell can be broken down into three conditional probabilities:-
| (11) |
where pentry, pvirogenesis and pbudding are the probabilities of entry of a bound virus into the cell interior, synthesis of the viral protein and RNAs within the cell and egress of the virus from the cell, respectively. Entry of the bound SARS-CoV-2 virion to the cell through fusion of the viral envelope with the host cell membrane is mediated by the spike protein S2 subunit which is conserved among coronaviruses (Walls et al. 2017). Cleavage at S1/S2 occurs upon biogenesis and viral egress for some coronaviruses, such as MHV and MERS-CoV (Walls et al. 2017). TMPRSS processing of SARS-CoV and MERS-CoV spike protein occurs at the cell membrane (Walls et al. 2017). Hoffmann et al. (2020) demonstrated efficient proteolytic processing of SARS-CoV-2 spike protein by TMPRSS2 in human kidney cell lines suggesting that fusion and entry should occur with high efficiency in human kidney cells at least, such that pentry is high. The values for the probabilities pvirogenesis and pbudding for SARS-CoV-2 are also assumed to be high on the basis of ex vivo studies of SARS-CoV-2 infection in bronchus and lung cell cultures (Hui et al. 2020). It is assumed here that overall pcell is highly efficient with a probability of 0.5 for the purpose of this prototype model.
3. Results
3.1. Increasing Kmucin decreases the fraction of free virus particularly at lower virus to mucin ratios
The predicted fractions of free virus and the efficacy of virus removal by the mucus barrier are presented for a range of virus:mucin ratios in Table 2 and plotted in Fig. 2 as a function of Kmucin. The results for Scenario 2 are identical to those for Scenario 1 in Table 2 which is not surprising since the values of [Vfree] and [Mucfree] in the denominator of Eq. 6 are simply interchanged with the same [V.Muc] in the numerator. It is concluded that the results in Table 2 may be interpreted either in terms of challenge of a varying virus dose in a given fixed volume of mucus, or in terms of dispersion of a given fixed dose, Vmucus, of virions carried on an inhaled aerosol within differing “local microenvironment” volumes of the mucus in the LLF in the lung.
Fig. 2.
The fraction, Fv, of free virus in mucus calculated by the difference equation approach decreases with increasing magnitude of the association constant, Kmucin, for binding of virus to mucin and is strongly affected by the local virus to mucin molecule ratio which is set to 2:1 (dash-dotted line), 1:1 (dashed line), 1:2 (dotted line) and <0.1:1 (solid line). The solid line is also represented by Eq. 8 for fully dispersed virus with [Mucfree] = 1.18 × 10−7 M. In a) the y-axis is linear to visualise the effect for the 2:1 virus:mucin ratio. In b) the y-axis is logarithm transformed to visualise small fractions of Fv as the virus becomes more dispersed in the mucus.
Low values of Kmucin have relatively little impact with Fv greater than 0.9 up to values of Kmucin ~106 M−1 (Fig. 2a) as expected for weak binding. At values of Kmucin >106 M−1 values of Fv decrease significantly with increasing Kmucin (Fig. 2b). The decrease in Fv is enhanced with decreasing local virus:mucin ratio. For the highest virus:mucin ratio of 2:1, the value of Fv can never fall below 0.5 (Fig. 2a) irrespective of Kmucin because there is only sufficient mucin to mop up half of the virus dose, the other half of the virus dose remaining free to go on and infect lung epithelial cells. For virus:mucin ratios of 1:1 or less, Fig. 2b shows that above Kmucin of ~106 M−1 the fraction, Fv, of free virus decreases linearly with increasing Kmucin. The minimum value of Fv which is achievable at the 1:1 ratio is 1.4 × 10−11 reflecting just 1 in the 7.1 × 1010 viruses being free. This is not achieved for the 1:1 ratio even with a Kmucin as high as 1022 M−1 (dashed line in Fig. 2b). The minimum value of Fv which is achievable at the 1 virus:2 mucin ratio is 2.8 × 10−11 reflecting just 1 in the 3.55 × 1010 viruses being free. This is achieved at a Kmucin of 1018 M−1 and therefore the dotted line terminates at Kmucin ~1018 M−1 (Fig. 2b). With the difference method approach it was not possible to achieve a Kmucin of >1018 M−1 for the 2 virus:1 mucin ratio simulation which is why the dash-dotted line in Fig. 2 also terminates at Kmucin ~1018 M−1. Using Eq. 8, values of Fv can be calculated for all values of Kmucin (solid line in Fig. 2b) and superimpose those for the 0.1 virion:1 mucin ratio.
3.2. Reducing the local virus:mucin ratio increases the efficiency of the mucus barrier
A Kmucin of 1010 M−1 is typical for lectins binding to mucins (see below). Table 2 presents the values of Fv over the range of local virion:mucin ratios for Kmucin = 1010 M−1. At a local concentration of 2 virus to 1 mucin molecule, 50% of the virus is free such that the mucus barrier only reduces the risk by 2-fold. At a local ratio of 1:1, 2.9% of the virus is free representing a 35-fold reduction in risk by the mucus barrier, while decreasing the local ratio to 1 virus to 2 mucins reduced the free virus fraction to 0.17% representing a 588-fold reduction in risk by the mucus barrier. Fully dispersing the virus dose in the lung mucin such that the number of mucin molecules greatly exceeds the number of viruses reduces the fraction of free virus to 0.085% such that the risk is reduced almost 1,200-fold by the mucus barrier. These effects are even more marked for Kmucin of 1022 M−1 such that all the virus is bound at a ratio of 1 virus to 2 mucins (Table 2).
3.3. Modelling the number, C.VT, of lung epithelial cells with bound virion
3.3.1. Most of the virus in the lung is predicted to be bound to cells at Ka_virus_T >1014 M−1
The fraction, Fc, of virus bound to cells in the lung as a function of Ka_virus_T is plotted in Fig. 3 and compared with the fractions of bound virus in the human intestine model (Gale 2018) and the mosquito midgut model (Gale 2019). Due to the higher concentration of target cells in the lung, lower values of Ka_virus_T achieve greater virus binding than for the intestine and midgut models. Indeed 90% of the virus is bound to lung cells when Ka_virus_T >1014 M−1. Thus Fc may be assumed to approximate 1 in Equation 5 if Ka_virus_T >1014 M−1.
Fig. 3.
The major proportion of virions is predicted to be bound to host cells in the human lung when Ka_virus_T > ~1014 M−1. Fraction, Fc, of virus dose of 1,000 virions predicted to be bound to host cells as a function of Ka_virus_T for the human lung (solid line, Ctotal = 1.2 × 109 cells in 0.025 dm3 of lung lining fluid, [Ctotal] = 8.0 × 10−14 M); human intestine (dashed line, Ctotal = 4.15 × 108 cells in 0.314 dm3 of gut contents, [Ctotal] = 2.2 × 10−15 M) and mosquito midgut (dotted line, Ctotal = 1 × 103 cells in 10−6 dm3 of midgut contents, [Ctotal] = 1.7 × 10−15 M) as calculated by the difference equation approach (Gale 2018).
3.3.2. Ka_virus_T is >1014 M−1 for the three CoVs if the number of trimer/receptor contacts is at least five
Values of Ka_virus_T are calculated for SARS-CoV, SARS-CoV-2 and MERS-CoV using the reported Kd_receptor_T values for the spike trimer binding and assuming ΔSa_immob = -350 J/mol/K in Eq. 10 for four and five spike trimer/receptor interactions (Table 3 ). With Nv = 4 spike trimer/receptor contacts, values of Ka_virus_T are well below 1014 M−1 while with Nv = 5 spike trimer/receptor contacts values of Ka_virus_T are >1014 M−1 for all three viruses.
Table 3.
Values of Ka_virus_T calculated for SARS-CoV, SARS-CoV-2 and MERS-CoV using the reported Kd_receptor_T values (see text) for the spike trimer binding and assuming ΔSa_immob = -350 J/mol/K (see text) in Eq. 10 for four and five spike trimer/receptor interactions.
| SARS-CoV | SARS-CoV-2 | MERS-CoV | |
|---|---|---|---|
| Kd_receptor_T (M) | 1.85 × 10−7 | 1.50 × 10−8 | 1.67 × 10−8 |
| Ka_virus_T (M−1) Nv = 4 | 4.36 × 108 | 1.01 × 1013 | 6.57 × 1012 |
| Ka_virus_T (M−1) Nv = 5 | 2.36 × 1015 | 6.73 × 1020 | 3.93 × 1020 |
3.4. Prototype dose-response model for initial infection in the lung by dispersed coronavirus
Fv values were calculated as 0.999, 0.894, 8.4 × 10−3, and 8.47 × 10−6 for virus fully dispersed in lung mucus according to Equation 8 using [Muctotal] = 1.18 × 10−7 M and Kmucin values of 103 M−1, 106 M−1, 109 M−1 and 1012 M−1 to represent Nm = 1, 2, 3, and 4 contacts of Kd_mucin = 10−3 M, respectively (Eq. 9). These represent virus removals of 0.012%, 10.5%, 99.1% and 99.999% respectively. The probabilities, phost, of infection over a range of virion exposures according to Equation 5 are compared for the four Fv values in Fig. 4 . As expected from Fig. 2, increasing Kmucin from 103 M−1 to 106 M−1 representing a second additional spike protein/SA contact has little effect on reducing the infectivity. For these low values of Kmucin, the infectivity reflects the proportion of virions in the dose that are actually capable of initiating infection as represented by the value of ppfu with predicted ID50s of 498 virions (1.4 pfu) and 558 virions (1.6 pfu) for Kmucin values of 103 M−1 and 106 M−1 respectively (based on the assumed value of 0.5 for pcell). However increasing Kmucin to 109 M−1 representing a third additional spike protein/SA contact increases the ID50 by ~100-fold to 59,243 virions (165 pfu) (Fig. 4). Increasing Kmucin by a further 1,000-fold to 1012 M−1 representing a forth additional spike protein/SA contact on binding with mucin increases the ID50 by 1,000-fold to ~5.87 × 107 virions (163,000 pfu) (Fig. 4). With a Kmucin value of 5 × 109 M−1 as for a lectin, Fv = 1.7 × 10−3 representing a 99.8% removal of the virus by the mucus giving an ID50 of 2.95 × 105 virions (819 pfu). Validation of the model in Fig. 4 for SARS-CoV-2 and MERS-CoV is not possible currently because there are no data for infectivity in humans. However, data for the proportion of dead mice after challenge by intranasal inoculation with increasing doses of SARS-CoV and MHV from Watanabe et al. (2010) are plotted as filled squares and open squares, respectively, in Fig. 4. The doses in units of pfu from Watanabe et al. (2010) have been converted to virions (or RNA genomic copies) by multiplying by 1/ppfu for comparison with the model output here.
Fig. 4.
Prototype dose-response models according to Equation 5 for respiratory coronavirus virion exposure with Ftrans = 1, ppfu = 0.0028, Fc = 1 and pcell = 0.5. Fv values calculated with Equation 8 using [Mucfree] ~ [Muctotal] = 1.18 × 10−7 M assuming Kmucin values of 103 M−1 (dashed line), 106 M−1 (solid line), 109 M−1 (dash-dot line), and 1012 M−1 (dotted line) to represent Nm = 1, 2, 3, and 4 spike protein/mucin SA contacts, respectively, of Kd_mucin = 10−3 M (Eq. 9). Also Kmucin = 5 × 109 M−1 (dash-dot-dot line) as for the lectin soy bean agglutinin binding to porcine submaxillary mucin (Dam and Brewer 2010). Squares represent proportion of mice which died after intranasal challenge of mouse hepatitis coronavirus (open) or SARS-CoV (filled) from Watanabe et al. (2010) assuming 1 pfu = 360 virions or genomic copies. In b) the y-axis is logarithm transformed to visualise risks from low doses of dispersed virus.
3.5. Estimation of values for p1
From Equation 4, values for p1, the risk of initial infection from exposure to a single virion, are 1.39 × 10−3, 1.24 × 10−3, 1.17 × 10−5, 2.35 × 10−6 and 1.18 × 10−8 for Kmucin values of 103 M−1, 106 M−1, 109 M−1, 5 × 109 M−1 and 1012 M−1 respectively with [Muctotal] = 1.18 × 10−7 M, thus completing the dose-response in Equation 1.
3.6. Predicted effect on efficiency of mucus barrier of variation between individuals in the concentrations of mucins in lung mucus
Assuming full dispersion in the mucus, the virus removals by the mucus barrier with a Kmucin value of 1010 M−1 ranged from 200-fold to 3,000-fold as calculated by Equation 8 using minimum and maximum [MUC5B] reported by Keismer et al. (2017) for 19 non-smokers (Table 4 ). The variation between individuals in concentrations of MUC5B in lung mucus for non-smokers (Table 4) affects the dose-response with predicted ID50s varying from 1.0 × 105 virions (278 pfu) up to 1.5 × 106 virions (4,160 pfu) with Kmucin at 1010 M−1 (Fig. 5 ).
Table 4.
Variation in absolute concentration of MUC5B in lung mucus from 19 non-smokers affects predicted efficiency of mucus barrier at removing respiratory viruses. MUC5B concentrations from Kesimer et al. (2017).
| Participant | [MUC5B] (M) | Fv from Equation 8 (Kmucin = 1010 M−1) | Overall removal of virus by mucus |
|---|---|---|---|
| Minimum | 2.0 × 10−8 | 0.005 | 201-fold |
| Mean | 1.08 × 10−7 | 0.000925 | 1,081-fold |
| Maximum | 3.0 × 10−7 | 0.000333 | 3,001-fold |
Fig. 5.
Dose-response for respiratory coronavirus virion exposure varies between individuals depending on MUC5B concentration in the mucus (Table 4) according to Equation 5 (with Ftrans = 1, ppfu = 0.0028, Fc = 1 and pcell = 0.5) for SARS-CoV-2 fully dispersed in lung mucus with Kmucin = 1010 M−1 and Fv values calculated with Equation 8 using [Mucfree] ~ [Muctotal] = 0.2 × 10−7 M (dotted line), 1.08 × 10−7 M (solid line) and 3.0 × 10−7 M (dash-dot line).
4. Discussion
The work here builds on the thermodynamic approach developed previously for faecal/oral viruses in the human intestine (Gale 2018) and focuses on the fraction, Fv, of exposure dose, Vexposure, which is not bound to mucins in the lung mucus in the LLF and is therefore free to bind to and infect lung epithelial cells. Prototype dose-response models are plotted in Fig. 4 for a range of Kmucin values and in Fig. 5 to demonstrate the effect of varying [Muctotal] in different individual non-smokers.
The models in Fig. 4 and Fig. 5 represent the risk according to Equation 5 from virions which are fully dispersed in the mucus using Fv calculated with Equation 8. Equation 5 cannot be used with Fv values from Table 2 for aggregated virus doses (i.e. where local Vmucus ≥ Muctotal) because Fv calculated by the difference equation approach is dependent on both the aggregation state of the virions and the number of virions in the inhaled aerosol particle, both of which affect the local virus:mucin ratio and hence [Mucfree] (in Equation 8) where the aerosol interfaces with the mucus barrier.
The key conclusion of the prototype dose-response models is that dispersed SARS-CoV-2 virions are highly infectious to human without an effective mucus barrier in the lung with a predicted ID50 ~500 virions which equates to 1 to 2 pfu. This ID50 is dependent on the value of ppfu which is given a point value of 0.0028 here based on 360 genomic copies per pfu. Adhikari et al. (2019) use a point value of 1,239 genomic copies per pfu for MERS-CoV based on SARS-CoV data and note the reported values ranged from 1:1 to 300:1 to 1,200-1,600:1. Using different point values would proportionately affect the number of virions comprising an ID50 and is a source of uncertainty addressed by Adhikari et al. (2019). The low ID50 values of 1 to 2 pfu for SARS-CoV-2 in the absence of effective mucin binding reflect both the tight binding of the virion to the host cell through the high affinity spike protein/hACE2 interactions such that Ka_virus_T is high (Table 3) and the efficient priming of the spike protein by proteolytic cleavage for fusion, such that pentry is high. Increasing the magnitude of Kmucin as suggested here for MERS-CoV directly reduces the infectivity, such that with Kmucin =1012 M−1, the predicted ID50 ~ 5.87 × 107 virions or 163,000 pfu. In terms of other applications, this prototype dose-response model may be used to test the effect of antiviral drugs on the infectivity. For example from drug inhibition data on the viral main protease (Zhang et al. 2020) it may be possible to model the effect of the drug on pvirogenesis in Eq. 11 and hence assess the shift in dose response (Fig. 4) in the presence of the drug.
4.1. Fate and transport of the virions to the lung mucus
The dose response developed here is based on a virion dose being deposited in the mucus in the LLF in the bronchus and with Ftrans = 1 in effect assumes 100% of the inhaled dose is deposited there. Deposition is a critical step in the initiation of infection by anthrax and varies between 9% and 30% of the inhaled dose depending on the host species (Gutting et al 2015). Specifically SARS-CoV-2 infects the nasal epithelium (Hou et al. 2020), the ciliated, mucus-secreting, and club cells of bronchial epithelium and the type 1 pneumocytes in the lung (Hui et al 2020). On the basis of the recent data of Hou et al. (2020) the model for SARS-CoV-2 only needs to focus on the nasal epithelium such that assuming Ftrans = 1 may be justified. The prototype model here does not include lung air flow and pathogen transport principles as used in the anthrax model of Weir and Haas (2011). Instead, it is assumed that the fate of the virions deposited in the mucus in the LLF depends on whether each virion becomes bound subsequently to the mucins in the mucus and hence removed from the lung or remains free to bind to a host epithelial cell and initiate infection. This is accommodated in the Fv term, which is determined by both Kmucin and the mucin concentration according to Equation 8. To develop the model it may be possible to apply the fate and transport approaches developed by Weir and Haas (2011) to viruses using the Kmucin and mucin concentrations and also other diffusion approaches such as that developed for viruses moving through mucus (Erickson et al. 2015). A further difference between anthrax spores and SARS-CoV-2 is that cell tropism for the latter is dependent on the presence of the hACE2 receptor and the TMPRSS that cleaves spike protein into S1 and S2 (Lukassen et al. 2020; Hou et al. 2020). Although hACE2 and TMPRSS are expressed in cells from the lung and epithelial cells from the subsegmental bronchial branches, only a small proportion of cells express both (Lukassen et al. 2020) and are hence susceptible to SARS-CoV-2 infection. This is accommodated in the Ctotal term in this model which affects how Fc varies with Ka_virus_T as shown for the human lung, human intestine and arthropod midgut models in Fig. 3.
4.2. The mucus barrier in the human lung may be more effective against MERS-CoV which binds human mucin than against SARS-CoV-2 which does not bind SA
It might be thought that the similarity of the dose-response data for MHV and SARS-CoV in the mouse data of Watanabe et al. (2010) presented as symbols in Fig. 4 argues against the importance of the mucus barrier in the thermodynamic dose-response model because the spike protein of MHV binds SA (Li et al 2017) while the spike protein of SARS-CoV-2 does not (Hao et al 2010). However, MHV differs from other CoVs in coding for an additional spike protein, the haemagglutinin-esterase (HE), which allows virions to elute from SA surfaces and hence provides the virions with a means of escape from irreversible attachment to non-cell-associated sialoglycoconjugates such as mucins (Langereis et al., 2011). Thus although MHV spike protein itself binds to SAs (Li et al. 2017) while SARS-CoV does not, the ability of the MHV to escape the mucin barrier would explain its similar dose-response in mice to SARS-CoV in the data of Watanabe et al. (2010) plotted in Fig. 4.
Adhikari et al. (2019) adopt the SARS-CoV dose-response model of Watanabe et al. (2010) as a surrogate for MERS-CoV in humans. However MERS-CoV may be different from SARS-CoV and SARS-CoV-2 in that it binds to human mucin through SAs (Li et al. 2017). More significantly it does not encode an HE protein (Du et al. 2017) with which to release itself from the SAs on the mucins (Langereis et al., 2011). Some beta-coronaviruses (as discussed for MHV above) do produce the HE protein which helps in the viral spread through the mucosa (Rabaan et al. 2020).Thus MERS-CoV may be effectively trapped by mucus in the human lung and according to the model here, this would increase the ID50 for MERS-CoV relative to that for SARS-CoV-2 and may contribute to explaining why MERS-CoV is less efficiently transmitted from person to person in the community.
4.3. Obtaining data on Kmucin for MERS-CoV and SARS-CoV-2 binding to mucins in human lung mucus
The results in Fig. 4 emphasise the potential of the mucus as a major barrier depending on the magnitude of the parameter Kmucin. Although SARS-CoV-2 does not bind to SAs (Hao et al. 2020), spike protein interactions with SA have been reported for several CoVs including MERS-CoV (Li et al. 2017). These interactions are mediated by the N-terminal domain (S1A) of the spike protein while proteinaceous receptors (e.g. hACE2) are generally bound via S1B (Li et al. 2017). From Fig. 2a it can be seen that for the mucus to have any effect the magnitude of Kmucin must exceed 106 M−1 and that Kmucin values of 1010 M−1 are required to achieve >1,000-fold removal of dispersed virus (Fig. 2b). Dam and Brewer (2010) report a Kmucin value of 5.0 × 109 M−1 for the lectin soy bean agglutinin binding to porcine submaxillary mucin which has multiple GalNAc sugars to bind. Similarly Vataira macrocarpa lectin has a Kmucin of 1.0 × 1010 M−1 with porcine submaxillary mucin. Thus Kmucin values of 1010 M−1 used in the prototype dose response model (Fig. 4) may be realistic for MERS-CoV. Interactions between carbohydrate binding domains (CBDs) on proteins and monomeric SAs are generally weak with Kd_mucin values ~ 10−3 M (Taylor and Drickamer, 2006; Fei et al. 2015) and indeed Li et al. (2017) describe the S1A/SA interaction as weak for MERS-CoV. However, by making multiple contacts with the virus the affinity can be hugely increased according to Eq. 9 and Li et al. (2017) only detected MERS-CoV S1A binding to mucin when presented on nanoparticles such that multivalent (i.e. Nm > 1) CBD/SA interactions could be made thus enhancing binding affinity as expected from Eq. 9. For MERS-CoV the three S1A subunits of the spike trimer could each bind SA (i.e. one SA bound per S monomer). There are also 74 trimer spikes on each MERS-CoV virion (Neuman et al. 2011) so in theory there is potential for the virion to make multiple interactions with the multiple SAs on a mucin molecule such that Kmucin could become very large. Thus Nm = 4 CBD/SA interactions each of Kd_mucin = 10−3 M would give a Kmucin of ~1012 M−1 according to Eq. 9. The magnitude of ΔSa_mucin for immobilisation of the virus on the mucin surface in Eq. 9 is likely to be small because, unlike ΔSa_immob for virus binding to a cell surface, binding to mucus is random.
Most mucins have a high SA content which, along with the high sulphate content, results in a strongly negative surface charge (Zanin et al. 2016). Hao et al. (2020) have shown that SARS-CoV-2 spike protein binds to heparan sulphate which also contains multiple sulphate groups. Moreover, basic amino acid residues (i.e. positively charged), known to constitute heparin binding domains are solvent accessible on the SARS-CoV-2 spike protein surface and form a continuous patch that is suitable for heparin binding (Mycroft-West et al. 2020). Human CoV NL63 also binds heparan sulphate which is used by this coronavirus for its attachment to target cells (Milewska et al 2014). This suggests SARS-CoV-2 and other human CoVs could bind to negatively charged groups on mucins despite not binding to SAs (Hao et al. 2020). Thus binding of SARS-CoV and SARS-CoV-2 to human mucins will only be determined by experiment with human mucin itself and cannot be ruled out at this stage even though Hao et al. (2020) report it does not bind SA.
4.4. The degree of local dispersion/clustering of the virions in the lung mucus greatly affects the removal efficacy of the mucus
According to Scenario 2 in Table 2 inhalation of a fixed dose of SARS-CoV-2 virions dispersed across many aerosol particles may pose a greatly diminished risk compared to the same dose inhaled as a “cluster” in a single aerosol particle (providing Kmucin ≥1010 M−1). The virus:mucin ratio in Table 2 represents the local virion:mucin contact ratio and not the virus to mucin ratio across the whole 25 cm3 of LLF which will be very low and representing an average. As the virus dose in the aerosol becomes more dispersed in the mucus then the local Muctotal begins to exceed Vmucus such that each virus is in effect surrounded by mucin molecules and has no contact or protective shielding effect from other viruses in the challenge dose from the aerosol particle. The model here suggests that the “local dilution” of a clustered virus dose from an aerosol only has to be ~>2-fold (Fig. 2) to almost maximise the effect of the protective mucus barrier. Thus according to the model each virus only needs to be surrounded by 2 mucin molecules of Kmucin ≥ 1010 M−1 in order to achieve a 588-fold reduction in risk of the virus getting to the epithelium and initiating infection. Infinite dilution as represented by the output of Equation 8 halves the risk again giving at the limit a 1,176-fold reduction in risk with <0.1% of the initial virus dose now free to bind to lung cells and initiate infection (Table 2).
4.5. The greatest change in risk occurs at the local virion/mucin ratio of 1:1 akin to a threshold
A four-fold increase in the local virus concentration in the mucus from 1 virion per 2 mucins to 2 virions per mucin gives ~300-fold increase in Fv (Table 2) and hence the risk of infection. This represents a co-operative effect within a virus “cluster” and breaks away from the traditional concept of independent action of pathogen infection although there is no absolute threshold effect in the model here. Bhar and Jones (2019) also challenge the “free independent virus particle” idea of viral transmission with the discovery of mechanisms by which multiple viral particles can infect the same cell simultaneously.
4.6. Impact of cell-tethered mucins is not considered here
The model here only considered the secreted mucins, MUC5B and MUC5AC, in the mucus and does not consider the cell-tethered mucins which may present an additional barrier to respiratory viruses. The cell-tethered mucins such as MUC1 and MUC4 form the basis of a gel-like layer surrounding the cilia (periciliary layer) that is essential for normal ciliary action to move mucus out of the airways (Ridley and Thornton 2018). The cell-tethered mucins provide a barrier function to mucosal pathogens by displaying sites for pathogen binding, acting as a releasable decoy, and sterically blocking binding of pathogens to underlying cellular receptors (McAuley et al. 2017). The cell-tethered mucin MUC1 gives protection against IAV H1N1 (McAuley et al. 2017). There is some evidence in mice that MUC4 gives protection to mice against SARS-CoV with Muc4−/− mice showing a modest 62% higher titre at 2 days post infection (dpi) and a 51% higher titre at 4 dpi as compared to wild type mice, although the increases were not statically significant (Plante et al. 2020). In contrast, Muc1−/− mice displayed a more rapid statistically significant rise in pulmonary IAV H1N1 loads compared to wild type mice at 1 to 3 dpi, although levels were comparable at 5 to 7 dpi (McAuley et al. 2017). Overall MUC4 offered protection to female mice against SARS-CoV and chikungunya virus pathogenesis although it was concluded this may not reflect a direct role such as the physical barrier function of mucin (Plante et al. 2020).
4.7. Uncertainty in the assumption that Fc = 1 i.e. all virus not bound to mucin is bound to lung epithelial cells
It is shown in Fig. 3 that >90% of the virus is bound to lung cells in the human lung model when Ka_virus_T exceeds 1014 M−1, thus greatly simplifying the model because Fc in Equation 5 does not have to be calculated for each dose using the difference equation approach (Gale 2018) but can be assumed to approximate 1. While there is a published value of Kd_receptor_T for SARS-CoV-2 spike trimer protein binding to a single hACE2 (Wrapp et al. 2020), there is no information on the number of contacts, Nv which must be at least five (Table 3) depending on ΔSa_immob to ensure Ka_virus_T >1014 M−1 according to Eq. 10. According to Yang et al. (2005) HIV virions bearing IAV HA required 8 or 9 HA trimers for virus entry and Guo et al. (2018) suggest that approximately 7 IAV HA trimers can interact with receptor-loaded SA molecules. It is concluded that although there is considerable uncertainty in the exact value of Nv for SARS-CoV-2 and MERS-CoV it is likely to be greater than 4 for both and on the basis that Nv could be as high as 7 spike trimer protein/receptor interactions it is concluded that Ka_virus_T is >1014 M−1 according to Eq. 10 such that Fc can be assumed to approximate 1. This is in agreement with Walls et al. (2020) who suggest the observed tight binding of SARS-CoV-2 S1B to hACE2 could partially explain the efficient transmission of SARS-CoV-2 in humans.
It should also be noted that ΔSa_immob in Eq. 10 is an unknown parameter for any virus and depends on the size of the virus (Gale 2019; Gale, 2020) together with how restricted virion mobility is on attachment to a host cell. The ΔSa_immob of –350 J/mol/K used in Table 3 is estimated using the Sackur-Tetrode equation. A less negative ΔSa_immob at –170 J/mol/K for example would require just three GP/Cr contacts to achieve Ka_virus_T of >1014 M−1 for SARS-CoV-2 according to Eq. 10 and ultimately it would be better to measure Ka_virus_T experimentally if possible.
4.8. The host cell infection component of the prototype dose-response for SARS-CoV-2 is defined solely by cell binding and entry
As a probability, pcell, can take a value between 0 (cell is refractory) and 1 (cell is fully permissive) and a value of 0.5 is assigned here on the basis of the efficiency of cleavage of SARS-CoV-2 spike protein by human cells (Hoffmann et al. 2020). The values of pvirogenesis and pbudding are assumed to ~1 in the calculation of pcell in Eq. 11. In effect therefore the component of the dose-response for SARS-CoV-2 infection of human lungs is based solely on Kd_receptor_T, Nv and pentry. This appears to be acceptable for assessing the zoonotic potential of bat CoVs. Thus Menachery et al. (2020) argue that the combination of receptor binding and proteolytic activation by endogenous proteases permits zoonotic CoV infection as with MERS-CoV and SARS-CoV, and can be used to evaluate zoonotic virus populations in bats for emergence threats. There are several human host cell proteases that can cleave the spike protein. Thus SARS-CoV can use the endosomal cysteine proteases cathepsin B and L and the serine protease TMPRSS2 for S protein priming in cell lines, and inhibition of both proteases is required for robust blockade of viral entry (Hoffmann et al. 2020). However, only TMPRSS2 activity is essential for SARS-CoV viral spread and pathogenesis in the infected host whereas CatB/L activity is dispensable (Hoffmann et al. 2020). Therefore pentry should be based on the probability of cleavage by TMPRSS2 in order to model for spread of the virus in the body.
4.9. Comparing the prototype thermodynamic dose-response model for initial infection with published dose-response data for coronavirus death in mice
Although the Kmucin value from Dam and Brewer (2010) of 5 × 109 M−1 for lectin binding to porcine submaxillary mucin gives a good approximations to the MHV and SARS-CoV mouse death data (symbols in Fig. 4), this is not evidence of validation of the model here because MHV escapes mucin and SARS-CoV does not bind mucin and thus Kmucin values would be expected to be very small (certainly <106 M−1) such that the predicted dose-response curve for initial infection by MHV and SARS-CoV in mice (solid line in Fig. 4) would have ID50s of ~500 virions (1 to 2 pfu) according to the model here. It should be stressed that the data of Watanabe et al. (2010) as represented by the symbols in Fig. 4 are not comparable with the model outputs here, the former representing death while the latter represents initial infection of one or more epithelial cells in the nose or bronchus, with no inclusion of the effect of the innate immune response which is important in disease progression and illness in CoV infection (Lim et al. 2016). It is therefore not surprising that the ID50 for death (symbols) is higher than the ID50 for infection with low Kmucin (solid line) in Fig. 4.
4.10. Is the dose-response for SARS-CoV a good surrogate for SARS-CoV-2?
Although similar, there are some notable differences between the SARS-CoV-2 and SARS-CoV spike proteins (Rabaan et al. 2020; Wrapp et al. 2020). The key amino acid residues involved in ACE2 binding are largely different and the SARS-CoV-2 trimer binds to ACE2 with a ~12-fold lower Kd_receptor_T than that of the SARS-CoV trimer (Table 3). The binding of SARS-CoV-2 is predicted to be much stronger than for SARS-CoV according to Ka_virus_T values as the number of spike trimer/ACE2 contacts increase (Table 3). According to the model here in Fig. 3 this difference could be overcome by a sufficient number, Nv, of spike trimer/hACE2 contacts such that Ka_virus_T exceeds 1014 M−1 (Table 3). However, the TMPRSS cleavage site in the spike protein sequence of SARS-CoV-2 may also account for the increased infectivity of SARS-CoV-2 relative to SARS-CoV (Hao et al 2020; Rabaan et al. 2020) such that pcell may be lower for SARS-CoV than for SARS-CoV-2. Thus the dose-response of Watanabe et al. (2010) based on SARS-CoV and MHV in mice may not be appropriate for SARS-CoV-2 although in the absence of data it is used as a surrogate dose-response in airborne risk assessment (Buonanno et al. 2020).
4.11. Both SARS-CoV and MHV bind to host cells with high affinity consistent with similar dose-response models in mice
Although each SARS-CoV spike trimer could bind three ACE2 receptors, the stoichiometry of spike trimers binding ACE2 is complicated for SARS-CoV (Kirchdoerfer et al. 2018) with most trimers just binding one ACE2. This is because the three copies of SARS-CoV S1A protein which bind ACE2 are all located on the top of the spike trimer and are near each other, leading to steric clashes between bound ACE2 molecules (Shang et al. 2020). This contrasts with mouse hepatitis coronavirus (MHV) for which three receptor molecules bind to the sides of the spike trimer, where there is no steric clash (Shang et al. 2020). Thus each MHV trimer binds three receptors (Kirchdoerfer et al. 2018) and depending on the number of receptor molecules on host cell membranes, Shang et al. (2020) conclude that the high stoichiometry of receptor binding by MHV spike potentially allows efficient viral attachment to target cells. Thus in addition to not being bound by mucins, both MHV and SARS-CoV could attach to host cells with high affinity consistent with their similar dose-response curves for death in mice (symbols in Fig. 4).
4.12. Limitations of the model developed here
In addition to lack of data on the magnitude of Kmucin for viruses associating with mucins, current understanding of mucin functions is limited. This is partly due to the complexity of mucus and its interactions (Zanin et al. 2016). The interaction between mucins and respiratory pathogens is also more complicated than mere entrapment (Zanin et al. 2016). The model here only considers entrapment of the virus, and does not consider mechanisms for the virus to evade the mucus which do not appear to be known for CoVs. Although mucus is an important host barrier to IAV, the neuraminidase (NA) protein of IAV is important in evading this barrier, such that viruses with low NA activity, or viruses treated with NA inhibitors, are severely impeded by mucus in vitro (Zanin et al. 2016). Another limitation of the model is that it is possible that more than one virus can bind a mucin molecule since each mucin molecule has multiple SAs (Zanin et al. 2016).
According to Guo et al. (2018) equilibrium binding models (such as those developed here) are poorly applicable to the IAV-receptor interaction because the virus rolls over a surface (such as a host cell) coated with receptors enabled by NA activity. Migration of attached IAV particles over a receptor coated-surface depends on the very high Kd_receptor_T (0.0003 to 0.003 M) values (i.e. low affinity binding) of monovalent HA/SA interactions resulting in their rapid formation and dissociation. Guo et al. (2018) hypothesize that NA, in combination with the highly dynamic formation and release of individual HA-SA interactions, drives virus rolling by the generation of a receptor gradient due to the receptor destroying activity of NA. This is a different strategy from SARS-CoV-2 cell binding for which Kd_receptor_T for the trimer is very low at 1.5 × 10−8 M (Wrapp et al. 2020) and SARS-CoV-2 and MERS-CoV do not appear to utilize enzymes for viral release with only group 2a coronaviruses (human CoV-HKU1, MHV) having haemagglutinin esterases (de Groot 2006). It is concluded that the equilibrium binding models (such as those used here and previously (Gale 2018)) are applicable to coronaviruses, although not for IAV virions which have both HA and NA present in their viral envelopes. It may also not be applicable to MHV which also has HE.
4.13. The role of sialic acids: ΔSa_immob vs Kmucin
Unlike IAV which only binds SA receptors, the spike protein trimers of CoV bind to protein Cr receptors with very high affinity. This raises the question of why some CoV spike proteins e.g. MERS-CoV, also bind to SAs, which according to the model here would hinder their movement through the mucus barrier. Other viruses also use non-specific attachment factors that bind the virus on the cell surface without initiating endocytosis and viral entry, for which specific receptors are required. In the case of tick-borne encephalitis virus, HIV, pseudorabies virus, sindbis virus, Semliki Forest virus, hepatitis E virus and Rift Valley fever virus the attachment factor is heparan sulphate, a negatively-charged glycosaminoglycan (Pulkkinen et al. 2018). SARS-CoV-2 spike protein also binds heparan sulphate (Hao et al., 2020). It was previously proposed that the attachment factors including cell surface SAs and heparan sulphate serve to take some of the entropy loss of ΔSa_immob on virus binding (Gale 2020). Specifically for MERS-CoV, Li et al. (2017) provide evidence that SAs may serve as low-affinity attachment receptors and thereby aid viral entry through increasing the likelihood of MERS-CoV spike engagement with the DPP4 entry receptor. Thus the potential loss of the virus through binding to SAs on mucins in the mucus as suggested by the model here may be a trade-off for non-specific binding of viral spike proteins to SAs on the cell surface in overcoming the unfavourable ΔSa_immob. In this respect, the magnitude of Kmucin in the Fv term plays off against that of ΔSa_immob in the Fc term in Fig. 1.
5. Conclusions
The conclusions are:-
-
1
The outline of a thermodynamic equilibrium model for a dose-response for SARS-CoV-2 and MERS-CoV is set out here based on molecular parameters. The available parameters for the spike trimer/cell receptor interactions suggest the association constant for virus binding to a human bronchus/lung cell will be so high that any virion getting through the mucus barrier will bind to these cells resulting in entry through spike protein fusion which is also known to be efficient for both viruses.
-
2
It is known that mucin proteins in lung mucus present a barrier to respiratory viruses. According to the model outlined here, the mucus in the 25 cm3 volume of lung lining fluid could present a significant barrier to infection of lung cells in humans by coronaviruses if the magnitude of the association constant (Kmucin) defining the strength of binding of the virion to human mucin is at least 109 M−1 (as has been reported for lectins' binding to mucins).
-
3
The magnitude of Kmucin is currently unknown for any virus to the author's knowledge. While SARS-CoV-2 spike protein does not bind sialic acid, the spike protein of MERS-CoV does and indeed MERS-CoV has been shown to bind human mucin. A Kmucin of at least 1010 M−1 could theoretically be achieved for MERS-CoV with four low affinity contacts between virus spike protein and monomeric sialic acids on the mucin each with dissociation constants of ~10−3 M (which is typical for carbohydrate binding domains on proteins binding to monomeric sialic acids). This could contribute to more efficient removal of MERS-CoV by the mucus barrier in the lung and hence its reduced human-to-human transmission compared to SARS-CoV-2. However SARS-CoV-2 spike protein has positively charged groups which could interact with negatively charged acidic groups on the mucins and its binding to human mucin cannot be ruled out without definitive data.
-
4
The mucus barrier may be more effective against fully dispersed virions in the case of MERS-CoV that binds human mucin. Some coronaviruses use an esterase to escape the mucin, although MERS-CoV does not. Instead, it is shown here that “clustering” of virions into single aerosol particles as recently reported for rotavirus in extracellular vesicles could provide a co-operative mechanism whereby MERS-CoV could theoretically overcomes the mucin barrier locally. In this respect the rare, high virion-load aerosol particles may be more important for transmission of MERS-CoV.
-
5
Assuming efficient transport to the mucus in the lung bronchus (or nasal epithelium in the case of SARS-CoV-2) and depending on the viability of individual virions, the ID50 for SARS-CoV-2 initial infection is estimated to be ~500 virions (viral RNA genomic copies) representing 1 to 2 pfu. Assuming a Kmucin of 5 × 109 M−1 as for lectin/mucin binding, the ID50 for MERS-CoV initial infection is estimated to be ~295,000 virions (viral RNA genomic copies) representing 819 pfu.
-
6
The thermodynamic equilibrium approach developed here based on virus binding to mucins according to Kmucin and the mucin concentration in mucus could complement inhalation models based on fate and transport as for anthrax spores in the lungs.
-
7
It is proposed that Kmucin is the cost for binding of viruses to sialic acids on the cell surface to partially overcome the unfavourable entropy of immobilisation of a virus.
Author Statement
I wrote this paper based on my previous thermodynamic dose-response models with no funding. It is my contribution to the global response against COVID-19.
Disclaimer
The views expressed in this paper are those of the author and not necessarily those of any organisations.
Declaration of Competing Interest
None declared.
References
- Adhikari U., Chabrelie A., Weir M., Boehnke K., McKenzie E., Ikner L., Wang M., Wang Q., Young K., Haas C.N., Rose J., Mitchell J. A case study evaluating the risk of infection from Middle Eastern Respiratory syndrome coronavirus (MERS-CoV) in a hospital setting through bioaerosols. Risk Analysis. 2019;39:2608–2624. doi: 10.1111/risa.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon (2020). Coronavirus resource center (https://coronavirus.jhu.edu/map.html) Accessed 8 August 2020.
- Bhar S., Jones M.K. In Vitro Replication of Human Norovirus. Viruses 11. 2019:547. doi: 10.3390/v11060547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R.A., Eden J.-S., Luciani F., McElroy K., Rawlinson W.D., White P.A. Contribution of intra- and interhost dynamics to norovirus evolution. Journal of Virology. 2012;86:3219–3229. doi: 10.1128/JVI.06712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno G.L, Stabile L, Morawska L. Estimation of airborne viral emission: Quanta emission rate of SARS-CoV-2 for infection risk assessment. Environment International. 2020;141 doi: 10.1016/j.envint.2020.105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapo J.D., Barry B.E., Gehr P., Bachofen M., Weibel E.R. Cell number and cell characteristics of the normal human lung. The American review of respiratory disease. 1982;126:332–337. doi: 10.1164/arrd.1982.126.2.332. [DOI] [PubMed] [Google Scholar]
- Dam T.K., Brewer C.F. Oligosaccharide multivalent lectin–carbohydrate interactions: Energetics and mechanism of binding. Advances in Carbohydrate Chemistry and Biochemistry. 2010;63:139–164. doi: 10.1016/S0065-2318(10)63005-3. [DOI] [PubMed] [Google Scholar]
- De Graaf M., Fouchier R.A.M. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. The EMBO Journal. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson A.M., Henry B.I., Murray J.M., Klasse P.J., Angstmann C.N. Predicting first traversal times for virions and nanoparticles in mucus with slowed diffusion. Biophysical Journal. 2015;109:164–172. doi: 10.1016/j.bpj.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Y., Sun Y.-S., Li Y., Yu H., Lau K., Landry J.P., Luo Z., Baumgarth N., Chen X., Zhu X. Characterization of receptor binding profiles of influenza A viruses using an ellipsometry-based label-free glycan microarray assay platform. Biomolecules. 2015;5:1480–1498. doi: 10.3390/biom5031480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A.V., Janin J. The price of lost freedom: entropy of bimolecular complex formation. Protein Eng. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- Fröhlich E., Mercuri A., Wu S., Salar-Behzadi S. Measurements of Deposition, Lung Surface Area and Lung Fluid for Simulation of Inhaled Compounds. Front. Pharmacol. 2016;7:181. doi: 10.3389/fphar.2016.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Using thermodynamic parameters to calibrate a mechanistic dose-response for infection of a host by a virus. Microbial Risk Analysis. 2018;8:1–13. doi: 10.1016/j.mran.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Towards a thermodynamic mechanistic model for the effect of temperature on arthropod vector competence for transmission of arboviruses. Microbial Risk Analysis. 2019;12:27–43. doi: 10.1016/j.mran.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. How virus size and attachment parameters affect the temperature sensitivity of virus binding to host cells: Predictions of a thermodynamic model for arboviruses and HIV. Microbial Risk Analysis. 2020 doi: 10.1016/j.mran.2020.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Rabouw H., Slomp A., Dai M., van der Vegt F., van Lent J.W.M., McBride R., Paulson J.C., de Groot R.J., van Kuppeveld F.J.M., de Vries E., de Haan C.A.M. Kinetic analysis of the influenza A virus HA/NA balance reveals contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces. PloS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutting B.W., Rukhin A., Mackie R.S., Marchette D., Thran B. Evaluation of inhaled versus deposited dose using the exponential dose‐response model for inhalational anthrax in nonhuman primate, rabbit, and guinea pig. Risk Analysis. 2015;35:811–827. doi: 10.1111/risa.12326. [DOI] [PubMed] [Google Scholar]
- Hao, W., Ma, B., Li, Z., Wang, X., Gao, X., Li, Y., Qin, B., Shang, s., Cui, S. and Tan, Z. (2020) Binding of the SARS-CoV-2 spike protein to glycans. [DOI] [PMC free article] [PubMed]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schäfer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O'Neal W.K., Randell S.H., Boucher R.C., Baric R.S. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. (2013). SARS: Dose response models. Retrieved from http://qmrawiki.canr.msu.edu/index.php/SARS:_Dose_Response_Models#_9f6e9f18777d09a3e4569b0d5784d92f.
- Hui K.P.Y., Cheung M.-C., Perera R.A.P.M., Ng K.-C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.-W., Poon L.L.M., Peiris M., Nicholls J.M., Chan M.C.W. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. The Lancet. 2020 doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesimer M., Ford A.A., Ceppe A., Radicioni G., Cao R., Davis C.W., Doerschuk C.M., Alexis N.E., Anderson W.H., Henderson A.G., Barr R.G., Bleecker E.R., Christenson S.A., Cooper C.B., Han M.K., Hansel N.N., Hastie A.T., Hoffman E.A., Kanner R.E., Martinez F., Paine, III R, Woodruff P.G, O'Neal W.K, Richard C. Boucher R.C. Airway Mucin Concentration as a Marker of Chronic Bronchitis. N Engl J Med 2017. 2017;377:911–922. doi: 10.1056/NEJMoa1701632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Midgley C.M., Gerber S.I., Watson J.T. Middle East respiratory syndrome coronavirus transmission. Emerging Infectious Diseases. 2020;26:191–198. doi: 10.3201/eid2602.190697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep. 2018;8(1):15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., van Vliet A.L., Boot W., de Groot R.J. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. Journal of Virology. 2011;84:8970–8974. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Widjaja I., Rajb V.S., McBride R.Pengc,W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y., Lent J.W.M., Paulson J.C., de Haan C.A.M., de Groot R.J., van Kuppeveld F.J.M., Haagmans B.L., Bosch B.-J. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA. 2017;114(40):E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: A review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley W.G., Pearce T.A., Hudnall J.B., Davis K.A., Davis S.M., Fisher M.A., Khakoo R., Palmer J.E., Clark K.E., Celik I., Coffey C.C., Blachere F.M., Beezhold D.H. Quantity and size distribution of cough-generated aerosol particles produced by influenza patients during and after illness. J. Occup. Environ. Hyg. 2012;9:443–449. doi: 10.1080/15459624.2012.684582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its cellular receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. The EMBO journal. 2020;39(10) doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley J.L., Corcilius L., Tan H.-X., Payne R.J., McGuckin M.A., Brown L.E. The cell surface mucin MUC1 limits the severity of influenza A virus infection. Mucosal Immunol. 2017;10:1581–1593. doi: 10.1038/mi.2017.16. [DOI] [PubMed] [Google Scholar]
- Menachery VD, Dinnon KH, III, Yount BL, Jr McAnarney ET, Gralinski LE, Hale A, Graham RL, Scobey T, Anthony SJ, Wang L, Graham B, Randell SH, Lipkin WI, Baric RS. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J Virol. 2020;94 doi: 10.1128/JVI.01774-19. e01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A, Zarebski M, Nowak P, Stozek K, Potempa J, Pyrc K. Human Coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. Journal of Virology. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West, C., Su, D., Stefano Elli, S., Guimond, S., Miller, G., Turnbull, J., Yates, E., Guerrini, M., Fernig, D., Lima, M. and Skidmore, M. (2020) The 2019 coronavirus (SARS-CoV-2) surface protein (Spike) S1 Receptor Binding Domain undergoes conformational change upon heparin binding.
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. Journal of Structural Biology. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.D., Park W.B., Choe P.G. Viral Load Kinetics of MERS Coronavirus Infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. The Lancet. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Lia F. Crystal structure of mouse coronavirus receptor binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U.S.A. 2012;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante, J.A., Plante, K.S., Gralinski, L.E., Beall, A., Ferris, M.T., Bottomly, D., Green, R., McWeeney, S.K., Heise, M.T., Baric, R.S. and Menachery, V.D. (2020). Mucin 4 protects female mice from coronavirus pathogenesis. ( 10.1101/2020.02.19.957118). [DOI]
- Pulkkinen L.I.A., Butcher S.J., Anastasina M. Tick-borne encephalitis virus: A structural view. Viruses. 2018;10:350. doi: 10.3390/v10070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley C., Thornton D.J. Mucins: the frontline defence of the lung. Biochemical Society Transactions (2018) 2018;46:1099–1106. doi: 10.1042/BST20170402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiana M., Ghosh S., Ho B.A., Rajasekaran V., Du W.L., Mutsafi Y., De Jésus-Diaz D.A., Sosnovtsev S.V., Levenson E.A., Parra G.I., Takvorian P.M., Cali A., Bleck C., Vlasova A.N., Saif L.J., Patton J.T., Lopalco P., Corcelli A., Green K.Y., Altan-Bonnet N. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell host & microbe. 2018;24(2):208–220. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PloS Pathog. 2020;16(3) doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebraker J.R., Wagner D., Lefensty R.W., Burns K., Gendler S.J., Bergelson J.M. Glycocalyx restricts adenoviral vector access to apical receptors expressed on respiratory epithelium in vitro and in vivo: role for tethered mucins as barriers to lumenal infection. J. Virol. 2004;78:13755–13768. doi: 10.1128/JVI.78.24.13755-13768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.E., Drickamer K. Carbohydrate recognition in cell adhesion and signalling. In: Taylor M.E., Drickamer K., editors. Introduction to glycobiology. Oxford University Press; Oxford: 2006. pp. 122–154. [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuken J.G.P., Swart A.N., Havelaar A.H., Van Pul A., Van der Hoek W., Heederik D. Atmospheric dispersion modelling of bioaerosols that are pathogenic to humans and livestock – A review to inform risk assessment studies. Microbial Risk Analysis. 2016;1:19–39. doi: 10.1016/j.mran.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicenzi E., Canducci F., Pinna D., Mancini N., Carletti S., Lazzarin A., Bordignon C., Poli G., Clementi M. Coronaviridae and SARS-associated coronavirus strain HSR1. Emerging infectious diseases. 2004;10:413–418. doi: 10.3201/eid1003.030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V.M., Simon M.N. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. PNAS. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARSCoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Bartrand T.A., Weir M.N., Omura T., Haas C.N. Development of a dose-response model for SARS coronavirus. Risk Analysis. 2010;30:1129–1138. doi: 10.1111/j.1539-6924.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir M.H., Haas C.N. A model for in-vivo delivered dose estimation for inhaled Bacillus anthracis spores in humans with interspecies extrapolation. Environmental Science and Technology. 2011;45:5828–5833. doi: 10.1021/es200901e. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kurteva S., Ren X., Lee S., Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Communications. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin M., Baviskar P., Robert Webster R, Richard Webby R. The interaction between respiratory pathogens and mucus. Cell Host & Microbe. 2016;19:159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun1 X., Curth U., Drosten X., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]