Abstract
This article comments on:
Diouf I, Derivot L, Koussevitzky S, Carretero Y, Bitton F, Moreau L, Causse M. 2020. Genetic basis of phenotypic plasticity and genotype×environment interaction in a multi-parental tomato population. Journal of Experimental Botany 71, 5365–5376.
Keywords: Adaptation, breeding, climate change, genotype×environment, multiple stress
The study of plant phenotypic plasticity complements our knowledge of plant response to stresses obtained from controlled single and multiple stress experiments. Diouf et al. (2020) dissect the genetic control of phenotypic plasticity for several traits in tomato. A few loci control both plasticity and mean phenotypes, while most loci are associated only with plasticity or mean phenotypes. The results can be applied to develop new cultivars for different objectives, from stable behavior to those specifically-adapted to different environments, by combining loci with different contributions to plasticity or mean phenotype.
Land plants in natural environments have to respond to different stresses—including heat, cold, salinity, drought, metal toxicity, pests, and diseases, among others—to complete their life cycle. Defense responses for those stresses have evolved in all plant species. These include changes in cuticle (shield), unsaturated fatty acids (membrane modulator), reactive species scavengers (reactive species homeostasis), molecular chaperones (stabilize proteins and subcellular structures), compatible solutes (osmoprotectants), and cellular responses to pathogen attack (Cui et al., 2015; He et al., 2018). Selection pressure has maintained these robust defense responses in natural populations. Domestication of wild species changed the selection pressure towards human needs, and the development of agronomic management techniques reduced the need for adaptation to stressful environments. Modern breeding accelerated the sensitivity of cultivars to stresses by developing high-yielding cultivars with high-input requirements. Biotic and abiotic stress has always been a threat to agricultural production, but the threat is heightened in the current context of climate change, with more frequent extreme heat and drought events (IPPC, 2014; Wu et al., 2018). Furthermore, population growth demands greater agriculture production with minimum ecological impact. The development of high-yielding cultivars capable of responding to changing environmental stresses is today one of the major goals for breeders.
Plant response to abiotic stresses and breeding
Over the past years, an impressive body of knowledge has been generated by the scientific community on plant responses to stresses. The sequences of a large number of genes involved in tolerance to abiotic stresses have been reported (e.g. Gerszberg et al., 2017; Ganie et al., 2019) as well as natural variation and quantitative trait loci (QTLs; Ayean et al., 2019; Morton et al., 2019). Most of these works applied the common scientific reductionist approach: they focused on one single stress and studied the plant response among limited stress levels based on historical values (Arnold et al., 2019). However, combinations of two or more stresses (drought and heat or salinity, for example) are common in many agricultural areas around the world (Suzuki et al., 2014). The effects of combinations of stresses are not additive; for example, combinations of drought with salinity, heat, chilling, pathogens, UV, nutrients, and heavy metals increase the negative effects of each individual stress, while combinations of drought with ozone or high CO2 mitigate the effect of the single stress (Suzuki et al., 2014). Each stress combination imposes specific requirements on the plant and therefore different responses are found when comparing single and multiple stress responses (Zandalinas et al., 2018). Thus, all the previous knowledge should be re-evaluated under multiple stress conditions. Moreover, the shape of the phenotypic response to certain stresses is not linear for most phenotypic traits; in fact, it is typically curved (Arnold et al., 2019), which means it is also appropriate to reconsider conclusions based on limited stress levels. The limitations of common experimental designs may explain the difficulties in extrapolating the research results to applied breeding.
Plasticity
Phenotypic plasticity is defined as the ability of a genotype to display different phenotypes as a response to different environments (Kusmec et al., 2018). The degree of plasticity may vary from zero (phenotype is stable) to far from zero (phenotype is plastic). Variation in plasticity among genotypes is classically known as genotype×environment interaction (Box 1). From the applied point of view, the appropriate degree of plasticity depends on the expected environmental conditions. Reduced plasticity is selected for producing stable yields when environments are relatively homogeneous (e.g. intensive agriculture or greenhouses), although adaptation to future environments would be constrained due the genetic homogeneity of the selected cultivars (Gage et al., 2017). High plasticity is needed to obtain cultivars adapted to specific environments, although lower yields would be obtained in non-target environments (Bernardo, 2010). Climate change forecasts predict that unstable weather will become more frequent, so high-yielding cultivars that provide predictable yields in a broader range of environments are one of the most important challenges facing breeders (Kusmec et al., 2018). In order to exploit the plasticity in breeding programs, genetic control should be elucidated or environments developed for testing in a manner that maximizes the opportunity to identify plastic genotypes. The study of plasticity needs to include a wide range of environments, an appropriate mapping population, and powerful statistical frameworks (Arnold et al., 2019). A few previous works have demonstrated that variation in plasticity has a genetic basis and is therefore amenable to selection (Mangin et al., 2017).
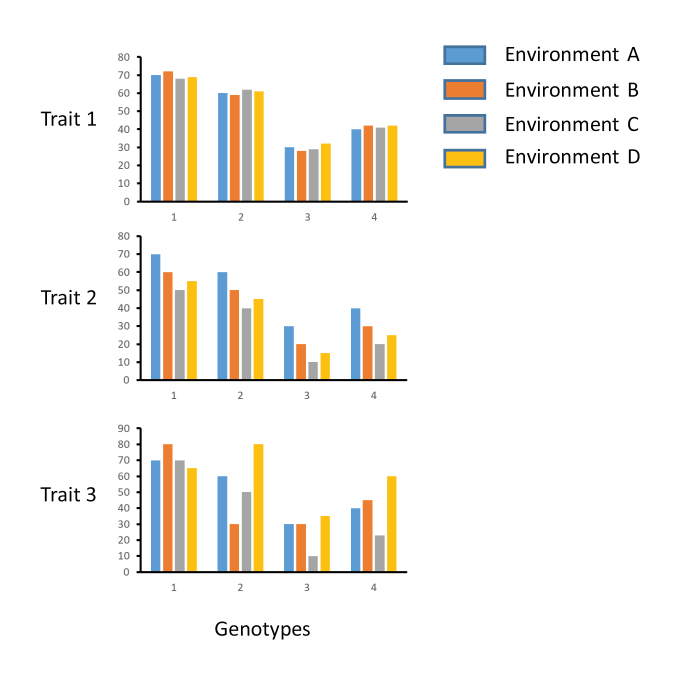
Box 1. Differences in phenotypic plasticity among genotypes.
Several hypothetical traits have been recorded in some genotypes across different environments. Genotypes showed different phenotypes for trait 1, but no differences among environments: no plasticity. For trait 2, genotypes responded to the different environments, but the response is similar in all genotypes: plasticity. Finally, for trait 3, the phenotypic response across environments is different among genotypes: variability in plasticity, genotype×environment interaction.
Diouf et al. (2020) used a multiparental advanced intercross (MAGIC) population, developed from intercrossing between eight tomato genotypes (Pascual et al., 2015) to investigate plasticity for several phenological and agronomic traits. The MAGIC population was evaluated in 12 environments in three different experimental stations, with different levels of heat, water, and salinity. Each combination of experiment and stress treatment could be considered as a single environment in order to allow a simultaneous study of the impact of a single stress, or they could all be studied together for a suitable estimation of the general plasticity accounting for the variations between the different environments. The choice of a MAGIC population deserves to be highlighted. The genetic structure of MAGIC populations offers a good compromise between genetic variability (up to eight alleles may segregate) and balanced allele frequency (a minimum of 0.125 per allele), compared with biparental populations (two alleles) or genome-wide association panels (where rare alleles are common and their effects cannot be detected). The authors observed that the best average performing genotypes were usually the most plastic in their response. This association between higher plasticity and better agronomic performance was also observed by Mangin et al. (2017). Diouf et al. (2020) identified QTLs associated with phenotype means and plasticity. Twenty-one percent of them were involved in both phenotype mean and plasticity, which would explain the association between plasticity and agronomic performance. Interestingly, a hub of plasticity QTLs for several traits were identified on chromosome 11. Most plasticity QTLs were located within domestication and improvement sweep regions previously defined by Zhu et al. (2018), suggesting that selection by breeding of plasticity alleles confers good adaptability in high-quality environments. Their analysis also allowed them to obtain insight into the underlying genetic model (i.e. overdominance, allelic sensitivity, or gene regulatory) that better fits the plasticity in the current research. Given the homozygosity of the tested MAGIC lines, overdominance can be ignored. Of the other two genetic models, the gene regulatory model seems most reasonable because a large proportion of plasticity QTLs did not co-localize with main effect QTLs.
Perspectives
We are changing our paradigm for studying crop responses to stresses and designing new crop varieties for future unpredictable environments. Powerful mapping populations, extensive trials, high-throughput phenotyping, new statistical models (Arnold et al., 2019), and the inclusion of biotic stresses will allow the next generation of breeders to obtain a better understanding of the genetic control of plasticity and its exploitation in breeding programs. Diouf et al. (2020) and previous pioneering works (Gage et al., 2017; Kusmec et al., 2018) have shown that genetic main effects and plasticity have both common and independent genetic control. The determination of these common and specific loci will allow the design of more efficient breeding strategies for different objectives, from varieties adapted to local conditions to varieties with high-yield stability over contrasting environments. The identification of the causal genes underlying plasticity will help us to better understand the complex responses of plants to stresses.
Acknowledgements
Research in my laboratory is kindly funded by the Spanish Ministry of Science, Innovation and University and FEDER, grant RTI2018-097665-B-C22 and the European Commission 510 H2020 research and innovation programme through the TOMGEM project agreement no. 679796.
References
- Arnold PA, Kruuk LEB, Nicotra AB. 2019. How to analyse plant phenotypic plasticity in response to a changing climate. New Phytologist 222, 1235–1241. [DOI] [PubMed] [Google Scholar]
- Ayenan MTA, Danquah A, Hanson P, Ampomah-Dwamena C, Sodedji FAK, Asante IK, Danquah EY. 2019. Accelerating breeding for heat tolerance in tomato (Solanum lycopersicum L.): an integrated approach. Agronomy 9, 720. [Google Scholar]
- Bernardo R. 2010. Breeding for quantitative traits in plants. Woodbury, MN: Stemma Press. [Google Scholar]
- Cui H, Tsuda K, Parker JE. 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Diouf I, Derivot L, Koussevitzky S, Carretero Y, Bitton F, Moreau L, Causse M. 2020. Genetic basis of phenotypic plasticity and genotype×environment interaction in a multi-parental tomato population. Journal of Experimental Botany 71, 5365–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, Jarquin D, Romay C, et al. . 2017. The effect of artificial selection on phenotypic plasticity in maize. Nature Communications 8, 1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganie SA, Molla KA, Henry RJ, Bhat KV, Mondal TK. 2019. Advances in understanding salt tolerance in rice. Theoretical and Applied Genetics 132, 851–870. [DOI] [PubMed] [Google Scholar]
- Gerszberg A, Hnatuszko-Konka K. 2017. Tomato tolerance to abiotic stress: a review of most often engineered target sequences. Plant Growth and Regulation 83, 175–198. [Google Scholar]
- He M, He CQ, Ding NZ. 2018. Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Frontiers in Plant Science 9, 1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC. 2014. Climate Change 2014. Synthesis Report. Contribution of working groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (Core Writing Team, Pachauri RK, Meyer LA, eds). Geneva, Switzerland: IPCC. [Google Scholar]
- Kusmec A, de Leon N, Schnable PS. 2018. Harnessing phenotypic plasticity to improve maize yields. Frontiers in Plant Science 9, 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin B, Casadebaig P, Cadic E, et al. . 2017. Genetic control of plasticity of oil yield for combined abiotic stresses using a joint approach of crop modelling and genome-wide association. Plant, Cell & Environment 40, 2276–2291. [DOI] [PubMed] [Google Scholar]
- Morton MJL, Awlia M, Al-Tamimi N, Saade S, Pailles Y, Negrão S, Tester M. 2019. Salt stress under the scalpel - dissecting the genetics of salt tolerance. The Plant Journal 97, 148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L, Desplat N, Huang BE, Desgroux A, Bruguier L, Bouchet JP, Le QH, Chauchard B, Verschave P, Causse M. 2015. Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnology Journal 13, 565–577. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. 2014. Abiotic and biotic stress combinations. New Phytologist 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Wu W, Ma BL, Whalen JK. 2018. Enhancing rapeseed tolerance to heat and drought stresses in a changing climate: perspectives for stress adaptation from root system architecture. Advances in Agronomy 151, 87–157. [Google Scholar]
- Zandalinas SI, Mittler R, Balfagón D, Arbona V, Gómez-Cadenas A. 2018. Plant adaptations to the combination of drought and high temperatures. Physiologia Plantarum 162, 2–12. [DOI] [PubMed] [Google Scholar]
- Zhu G, Wang S, Huang Z, et al. . 2018. Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261.e12. [DOI] [PubMed] [Google Scholar]