The sequencing of plant genomes is challenging due to their large size and abundant repetitive DNA, but third-generation platforms such as Oxford Nanopore Technologies can now generate high-quality assemblies.
Keywords: Basecalling, de novo assembly, gene annotation, MinION flow cell, Oxford Nanopore, third-generation sequencing
Abstract
DNA sequencing was dominated by Sanger’s chain termination method until the mid-2000s, when it was progressively supplanted by new sequencing technologies that can generate much larger quantities of data in a shorter time. At the forefront of these developments, long-read sequencing technologies (third-generation sequencing) can produce reads that are several kilobases in length. This greatly improves the accuracy of genome assemblies by spanning the highly repetitive segments that cause difficulty for second-generation short-read technologies. Third-generation sequencing is especially appealing for plant genomes, which can be extremely large with long stretches of highly repetitive DNA. Until recently, the low basecalling accuracy of third-generation technologies meant that accurate genome assembly required expensive, high-coverage sequencing followed by computational analysis to correct for errors. However, today’s long-read technologies are more accurate and less expensive, making them the method of choice for the assembly of complex genomes. Oxford Nanopore Technologies (ONT), a third-generation platform for the sequencing of native DNA strands, is particularly suitable for the generation of high-quality assemblies of highly repetitive plant genomes. Here we discuss the benefits of ONT, especially for the plant science community, and describe the issues that remain to be addressed when using ONT for plant genome sequencing.
Introduction
DNA sequencing technology was introduced more than four decades ago and has evolved over time to produce data at ever-increasing rates. First-generation sequencing was established in 1977 when Sanger and Coulson published the first virus genome sequence, bacteriophage ϕX174 (Sanger et al., 1977). First-generation sequencing dominated the field until the mid-2000s when high-throughput sequencing technologies, dubbed second-generation sequencing, emerged. The maximum read length of second-generation sequencing methods was typically shorter than for Sanger sequencing, but the higher throughput and relatively low cost made them competitive choices for large-scale sequencing projects (Lu et al., 2016; Bolger et al., 2019). These second-generation sequencing technologies remain popular for the analysis of simple genomes, resequencing, and RNA sequencing (RNA-seq), but the short reads they generate often lead to suboptimal assemblies, especially for de novo assemblies of large, highly repetitive genomes (Lu et al., 2016).
The most recent developments in sequencing technology make it possible to obtain significantly longer reads while still generating data at faster rates than first-generation methods. These third-generation technologies sequence single DNA molecules in real time, and the reads can be many kilobases in length. Such reads can span the large repetitive regions of complex genomes, thus improving sequence assemblies (Lu et al., 2016). Third-generation sequencing was spearheaded by Pacific Biosciences (PacBio) with their single-molecule real-time (SMRT) technology and was soon applied to plant genomes (VanBuren et al., 2015). This was followed by the launch of Oxford Nanopore Technologies (ONT) in 2014 (Box 1). Here we discuss the current advantages and challenges of the third-generation ONT sequencing platform and its potential as a method of choice for the plant genome sequencing community.
Box 1. Key developments in Oxford Nanopore Technologies application for plants.
• One MinION flow cell can generate enough data to assemble a small plant genome
Michael et al. (2018) report the assembly of a highly contiguous Arabidopsis genome using only one MinION flow cell. This study demonstrated that ONT technology can be used to assemble small plant genomes (i.e. <200 Mb) to an early draft stage using a single flow cell and with minimal effort.
• Medium size plant genome assemblies are possible and competitive using ONT technology
Schmidt et al. (2017) used ~135 Gb of ONT long-read data generated from 31 flow cells to assemble the genome of a wild tomato species to a high contiguity. This assembled genome was then compared with a related accession that had been sequenced and assembled using short reads. Given the higher output that can be obtained per flow cell and better read lengths using improved protocols, even quicker turnarounds may be possible today.
• Medium to small plant genomes can be assembled and brought to chromosome scale using additional techniques
Belser et al. (2018) showed that ONT data can be used to assemble a genome that can then be subsequently brought to chromosome scale using their case optical mapping. It can be expected that simpler techniques such as Hi-C (Feng et al., 2014) would produce similar results.
• Long reads generated from ONT flow cells are found to be useful for validating heterozygous genome assemblies
Wang et al. (2020) sequenced and assembled a highly heterozygous eucalyptus genome using a combination of long read data generated from ONT and short read Illumina data. They demonstrate how ONT long read sequencing provides important information for de novo assemblies and use a 10% hold out strategy to assess different assembly pipelines that incorporate long read data.
The potential of Oxford Nanopore Technologies sequencing for plant genomics
The release of the MinION platform in 2014 established ONT at the forefront of low-cost third-generation sequencing platforms. The MinION features a flow cell containing 2048 pores divided into four groups of 512, which are monitored by ONT software (Jain et al., 2016). The MinION was quickly followed by the GridION (designed to run five MinION flowcells) and PromethION (designed to run 24 or 48 larger capacity flow cells), which utilize the same core technology as the MinION but are designed for larger sequencing loads.
Unlike PacBio, which is a ‘sequencing by synthesis’ platform, ONT uses a novel approach where native DNA molecules are pulled through nanoscale pores (nanopores) that accept only one DNA molecule at a time. As the DNA moves through the pore, sensors detect changes in the ionic current corresponding to the characteristics of each passing nucleotide. This information can be visualized in a ‘squiggle plot’ and provides the signal used for basecalling (Deamer et al., 2016). Theoretically, sequencing continues until the end of the DNA fragment or until the pore becomes physically blocked, allowing for unprecedented read lengths that have the potential to significantly improve de novo genome assemblies and the detection of structural variations in large genomes. This is especially important in plant genomes, which contain highly repetitive regions derived from transposons and tandem repeats (Bolger et al., 2019).
ONT has been used to sequence small genomes such as that of the bacterium Escherichia coli (Loman et al., 2015), as well as large and repetitive plant and animal genomes. Examples include the human genome (Jain et al., 2018) and plant genomes, ranging from the ~119.5 Mbp genome of Arabidopsis thaliana (Michael et al., 2018) to the 2.53 Gbp genome of Chrysanthemum nankingense (Song et al., 2018) (Table 1). ONT has also been used to improve the accuracy of single nucleotide polymorphism (SNP) genotyping in complex polyploid plant genomes, where low-coverage long-read sequencing achieves superior genome alignments (Malmberg et al., 2019).
Table 1.
Plant species sequenced using the ONT platform
| Plant species | Genome size/N50 | Sequencing technology | Assembler | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | 119.5 Mbp/N50 12.3 Mbp (contig) | Illumina, ONT | Canu, Miniasm, Pilon | Michael et al. (2018) |
| Anthoceros agrestis (field hornwort) | 116.9 Mbp/N50 155.5 kbp (contig) 17.3 Mbp (scaffold) (Bonn strain); 122.9 Mbp/N50 1.8 Mbp (contig) (Oxford strain) | ONT, Hi-C, Illumina (Bonn strain); ONT, Illumina (Oxford strain) | MaSuRCA, Pilon, HiRise (Bonn strain); Miniasm, Racon, Pilon (Oxford strain) | F.W. Li et al. (2020) |
| Anthoceros punctatus | 132.8 Mbp/N50 1.7 Mb (contig) | ONT, Illumina | Miniasm, Racon, Pilon | |
| Spirodela polyrhiza (common duckweed) | 138.49 Mbp/N50 3.34 Mbp (contig), 7.68 (scaffold) | ONT, Hi-C | Miniasm; Proximo (for Hi-C data) | Harkness et al. (2020, Preprint) |
| 139.7 Mbp/N50 2.9 Mbp (contig) | Illumina, ONT | Miniasm, Racon, Pilon | Hoang et al. (2018) | |
| Tectona grandis (teak) | 317 Mbp/N50 357 kbp (scaffold), 277 kbp (contig) | Illumina, Illumina Mate Pairs, ONT | MaSuRCA, SSPACE, GapCloser, | Yasodha et al. (2018 |
| Oryza sativa L. (rice) IR64 | 367 Mbp/N50 1.6 Mbp (scaffold) | ONT, 10× Genomics | Supernova, Canu | Tanaka et al. (2020) |
| Corylus avellana L. (European hazel) | 370 Mbp/N50 36.65 Mbp (scaffold) | Illumina, ONT, Hi-C | MaSuRCA, HiRise | Lucas et al. (2019, Preprint) |
| Oryza sativa (rice) Carolina Gold Select | 377 Mbp/N50 1.72 Mbp (scaffold), N50 1.63 Mbp (contig) | ONT, Illumina | MaSuRCA, Flye | Read et al. (2020) |
| Oryza sativa (rice) | 386.5 Mbp N50 6.32 Mbp (contig) (Basmati 334); 383.6 Mbp/N50 10.53 Mbp (contig) (Dom Sufid) | ONT, Illumina | Canu, Fly, Medaka, Pilon | Choi et al. (2020) |
| Lupinus albus (white lupin) | 451 Mbp/N50 9.88 Mbp (scaffold), 7.11 Mbp (contig) | ONT, PacBio, Illumina, Bionano optical mapping | Canu, Falcon (for PacBio data only), Pilon, Bionano Solve | Hufnagel et al. (2019) |
| Dioscorea dumetorum (yam) | 485 Mbp/N50 3.2 Mbp (contig) | ONT, Illumina | Canu, Racon, Pilon | Siadjeu et al. (2020) |
| Juglans sigillata (iron walnut) | 536.5 Mbp/N50 16.43 Mbp (scaffold), N50 4.34 Mbp (contig) | ONT, Illumina, Bionano, Hi-C | Canu, wtdbg, Pilon | Ning et al. (2020) |
| Juglans regia (walnut) | 547 Mbp/N50 31.49 Mbp (scaffold), 1.36 Mbp (contig) | ONT, Illumina short read, Hi-C | MaSuRCA, HiRise | Marrano et al. (2019 Preprint) |
| Eucalyptus pauciflora (snow gum) | 594.87 Mbp/N50 3.23 Mb | ONT, Illumina | MaSuRCA | Wang et al. (2020) |
| Brassica oleracea | 630 Mbp N50 29.5 Mbp (scaffold), 7.3 Mbp (contig) | Illumina, ONT, Bionano | Ra, (SMARTdenovo, wtdbg), Racon, Pilon, Bionano Solve and Access | Belser et al. (2018) |
| Brassica rapa | 529 Mbp/N50 15.4 Mbp (scaffold), 3.8 Mbp (contig) | |||
| Musa schizocarpa | 587 Mbp/N50 36.8 Mbp (scaffold), 4.0 Mbp (contig) | |||
| Oryza coarctata (wild rice) | 665 Mbp/N50 1.86 Mbp (scaffold), 15.13 kbp (contig) | Illumina, ONT, Illumina Mate-Pair | PLATANUS, SSPACE, GapCloser | Mondal et al. (2018) |
| Asparagus setaceus (asparagus fern) | 710.15 Mbp/N50 2.19 Mbp (scaffold) | ONT, Illumina, 10× Genomics, Hi-C | Canu, Pilon; LACHESIS (for Hi-C data) | S.F. Li et al. (2020) |
| Euryale ferox (prickly waterlily) | 725.2 Mbp/N50 4.75 Mbp (contig) | ONT, Illumina, Hi-C | Canu, Pilon; LACHESIS (for Hi-C data) | Yang et al. (2020) |
| Ceratophyllum demersum (rigid hornwort) | 733.3 Mbp/N50 1.56 Mbp (contig) | |||
| Sorghum bicolor (sorghum) | 732 Mbp/N50 33.28 Mbp (scaffold), 3.05 Mbp (contigs) | Illumina, ONT, Bionano | Canu, SMARTdenovo, Pilon, Nanopolish, Bionano | Deschamps et al. (2018) |
| Cannabis sativa (cannabis) | 748 Mbp (1.39 Gbp F1 hybrid)/N50 742 kbp (contig) (172 kbp for F1 hybrid) | Illumina, PacBio, ONT | Miniasm, Racon, Pilon | Grassa et al. (2018, Preprint) |
| Eriobotrya japonica (loquat) | 760.1 Mbp/N50 39.7 (scaffold) | ONT, Illumina, Hi-C | Canu, SMARTdenovo, Racon, Pilon; BWA and LACHESIS (for Hi-C data) | Jiang et al. (2020) |
| Lonicera japonica (Japanese honeysuckle) | 843.2 Mbp N50 84.4 Mbp (scaffold) | ONT, Illumina, Hi-C | Canu, SMARTdenovo, Pilon; LACHESIS, SLR, SALSA (for Hi-C data) | Pu et al. (2020) |
| Solanum pennellii (wild tomato) | 1.0 Gbp/N50 2.45 Mbp (contig) | Illumina, ONT | Canu, SMARTdenovo, Pilon | Schmidt et al. (2017) |
| Chrysanthemum nankingense (chrysanthemum) | 2.53 Gbp/N50 130.7 kbp (contig) | Illumina, ONT | Canu, SMARTdenovo, Pilon | Song et al. (2018) |
Additional benefits of the MinION include its low investment cost and portability. Currently, an ONT MinION starter pack is available for US$1000 (https://nanoporetech.com/products/minion). The MinION plugs into a normal laptop via USB 3.0 and the entire system weighs only 103 g, making it possible to sequence at any location with access to power and an internet connection. Sequencing has been carried out on the International Space Station (Castro-Wallace et al., 2017), in the field to identify closely related plants in Snowdonia National Park (Parker et al., 2017), on site in West Africa to analyse Ebolavirus samples (Quick et al., 2016), and on farms in East Africa to identify strains of Cassava virus (Boykin et al., 2018).
Even the larger ONT systems such as the GridION X5 and PromethION 24 (rental costs of US$49 995 and US$165 000, respectively) are significantly less expensive than competing platforms. For small-scale projects, costs can be further reduced by multiplexing samples on one MinION flow cell using a barcoding kit, or by using a Flongle adaptor that plugs into a MinION or GridION system, allowing for sequencing on even smaller flow cells. These contain 126 channels (compared with MinION’s 512) that can produce up to 2 Gb output in a run. The significantly lower start-up costs of ONT compared with its competitors mean that even smaller laboratories have the opportunity to generate their own third-generation sequencing data (Maestri et al., 2019).
One unique advantage of ONT is the ability to detect epigenetic modifications in native DNA (Jain et al., 2016). DNA methylation detection (Rand et al., 2017; Simpson et al., 2017) was originally limited to methylated CpG dinucleotides (Shim et al., 2013), but the technology has improved to include other DNA methylation states such as isolated 5mC and 6mA (Ni et al., 2019). Additionally, Parker et al. (2019, Preprint) demonstrated that ONT can detect N6-methyladenosine in native A. thaliana RNA. ONT’s basecaller Guppy (from v3.2.1 onward) also allows certain DNA methylation sites to be called, such as 5mA, and 6mC in a CpG context, although it has currently only been trained on human and microbial data. A basecalling augmentation tool by ONT called Megalodon (https://github.com/nanoporetech/megalodon) can be combined with Taiyaki to train machine-learning algorithms (neural networks) for detecting plant-specific modifications. However, this requires additional data and significant computational resources such as graphics processing units (GPUs). Since DNA methylation plays a key role in the regulation of gene expression and in other cellular processes such as responses to stimuli (Law and Jacobsen, 2010), detecting these modifications during DNA sequencing provides valuable additional data (Simpson et al., 2017). The investigation of CHG and CHH context-dependent methylation (Law and Jacobson, 2010) remains important, especially in plants. Whole-genome bisulfide sequencing is a widely adopted method for investigating these methylations. However, different approaches, which range from the experimental conditions to the downstream bioinformatics pipelines, make it difficult to compare studies between research groups (Zhang et al., 2018), highlighting the potential advantages of ONT as a standardized method for detecting native DNA methylation (Fig. 1).
Fig. 1.
ONT offers a variety of important advantages to the wider plant genomics community.
The challenges of Oxford Nanopore Technologies sequencing for plant genomics
Although ONT is already established at the forefront of third-generation sequencing, several limitations of the technology remain, especially for sequencing highly repetitive plant genomes (Jiao and Schneeerger, 2017). Large amounts of high-quality DNA are required for a successful ONT sequencing run, defined as a high yield run with long reads (Schmidt et al., 2017). However, extracting intact high molecular weight DNA from plants is hindered by cell walls and secondary metabolites, with residual metabolites also remaining bound to the DNA, reducing sequencing yields (Schalamun et al., 2019; Vaillancourt and Buell, 2019, Preprint). There is often an inverse correlation between the quality and quantity of extracted DNA (Schalamun et al., 2019), and multiple DNA extraction protocols should be tested and optimized before sequencing a new plant species (Fig. 2; Table 2).
Fig. 2.
From plant tissue to genome assembly: the main steps in ONT sequencing. Optimizing each step can significantly increase the sequencing output and assembly quality.
Table 2.
Current challenges and solutions when using ONT to sequence plant genomes
| Challenge | Potential solutions |
|---|---|
| Low DNA quality and quantity | Test multiple extraction protocols and optimize for each plant species. |
| Short read contamination | Removal of short and medium-sized fragments using BluePippin Prep or Circulomics Short Read Eliminator kits, the latter being easier to use. |
| Basecalling speed and computational requirements | PromethION includes the hardware needed for fast basecalling. MinION basecalling time can be significantly reduced by using GPUs. |
| Long assembly computation time | Newer assemblers can significantly reduce computational time (e.g. wtdbg2). |
| Remaining uncorrectable base errors | Additional Illumina sequencing and polishing is currently required (Watson and Warr, 2019). This might be addressed with newer pore versions or basecalling models trained for particular species. Useful software includes Racon and Pilon. |
| Assembly is not (near) chromosome scale | Additional techniques such as optical mapping or Hi-C can be used to order and place contigs and obtain (near) chromosome-scale assemblies, at least for small and medium-sized plant genomes. |
| Genome structural and functional annotation | For structural annotation, long-read technology can be used with programs such as Stringtie2 (Kovaka et al., 2019). For functional annotation, free online tools relying on specific plant expertise are available, such as Mercator (Schwacke et al., 2019), TRAPID (Van Bel et al., 2013), or Hayai (Ghelfi et al., 2019), in addition to general tools such as Blast2GO (Götz et al., 2008). The plant repeat database (Nussbaumer et al., 2013) can be used to analyse repetitive DNA, and structural variations can be analysed using NGMLR/sniffles (Sedlazeck et al., 2018). |
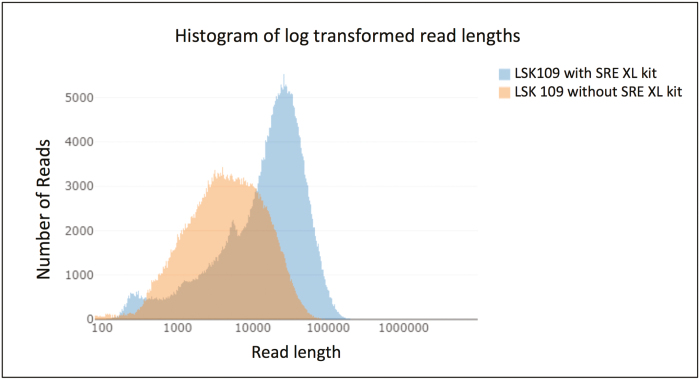
It is important to generate read lengths that span complex, repetitive DNA segments. Various protocols can be used to remove short DNA fragments, the easiest of which involves an adjustment to the quantity of NaCl and polyethylene glycol (PEG) used during bead clean-up steps (Schalamun and Schwessinger, 2017). An alternative is nuclear extraction followed by electrophoretic size selection, using equipment such as the Sage Science BluePippin Prep method (Schmidt et al., 2017). Although BluePippin achieves a clean size cut-off, sample recovery can be <50%, meaning that large quantities of input DNA are required. Furthermore, this method involves a substantial capital investment and recurring costs for consumables. A newer method for depleting short fragments is the Short Read Eliminator kit from Circulomics. Adopting a similar approach to bead clean-up, this kit relies on the precipitation of large DNA fragments, which are pelleted by centrifugation, while the shorter fragments remain in solution and are discarded (Fig. 3).
Fig. 3.
Difference in read lengths between an untreated sample and a sample treated with the Circulomics Short Read Eliminator kit. DNA was extracted from rapeseed (Brassica napus) and sequenced on an ONT MinION (image created using NanoComp by De Coster et al. (2018).
The correction of random read errors in the PacBio system is achieved using the circular consensus read technology that re-reads circularized DNA molecules multiple times, which are combined to produce high-fidelity results (Vollger et al., 2020). Because ONT reads are not circularized, an analogous read consensus option is not available beyond 1D2 sequencing, which aims to sequence both strands. Therefore, ONT sequences still have markedly higher error rates compared with second-generation sequencing platforms. This reflects the low signal-to-noise ratio of ONT sequencing, which remains a key challenge (Rang et al., 2018). Several factors contribute to this, including structural similarities between nucleotides and multiple nucleotides concurrently influencing the signal (Rang et al., 2018). ONT therefore developed the flip–flop basecalling model, which uses two overlapping windows to interpret the raw signal. Nucleotides containing methyl groups or other modifications will also modify the signal, making basecalling more difficult.
An additional factor that significantly influences signal quality is the speed at which the DNA strand moves through the pore, as signal strength depends on the time each nucleotide resides within the sensing region. ONT chemistry therefore includes the attachment of a motor protein to the DNA, which slows the translocation of the nucleotides through the pore signalling region, improving signal quality and robustness (Rang et al., 2018). However, the translocation speed of the motor protein can be sequence dependent, generating inconsistent signals especially in atypical segments such as homopolymer runs and multiple short repeats.
A comprehensive study on the basecalling accuracy of different sequencing platforms was performed using sequencing data from the bacterium Klebsiella pneumoniae (Wick et al., 2019). Even with the best standard basecallers, read identity was just below 90%, whereas consensus accuracy was 99.4%. This can make the assembly of plant genomes more difficult than animal genomes, because the former tend to contain more repetitive DNA and are more likely to be polyploid (Jiao and Schneeberger, 2017). In part, this reflects the fact that ONT’s basecaller Guppy is only trained on PCR, human and bacterial data, resulting in a lack of optimization for native plant DNA containing side chain modifications. This contributes to the significantly lower quality scores of plant ONT data compared with data from other domains, and hinders downstream alignment and assembly pipelines.
As discussed above, an alternative approach that could address this challenge is the development of plant-specific basecalling models generated using the ONT tool Taiyaki. Wick et al. (2019) achieved consensus accuracy >99.9% with K. pneumoniae after training Taiyaki using Klebsiella-specific models. A major improvement was that the self-trained models accounted for base read errors caused by DNA methylation. From a hardware perspective, the new R10 pore, which facilitates a longer read-head design, promises higher raw read accuracy. Improvements to the accuracy of ONT basecallers rely solely on software improvement and can be applied retrospectively to existing ONT sequencing data.
From Oxford Nanopore Technologies reads to genomes and useful data
As ONT sequencing technology continues to improve, the computational tools used to analyse raw sequencing data must also be optimized (Rang et al., 2018). One key post-sequencing step is the translation of the electrical current output signal into the nucleotide sequence, which is the technological principle of basecalling. The latest improvements in ONT basecallers require GPU computing for the rapid processing of raw data (Nobile et al., 2017), significantly improving basecalling speed compared with CPU-based workstations (Wick et al., 2019). Although such GPU resources are made available through national/international service providers such as iPLANT/CyVerse or ELXIR/de.NBI (Tauch and Al-Dilaimi, 2019), it may nevertheless be advisable to invest in standard NVIDIA graphics cards, which are known to support high basecalling speeds. Consequently, the PromethION comes with enterprise-grade GPU computing installed. For MinION and Flongle, ONT has developed the MinIT and Mk1C for data acquisition and basecalling, eliminating the need for any external hardware. The alternative basecaller Chiron, developed by Teng et al. (2018), achieves throughput of only a few thousand bases per second despite running on GPUs, making it too slow for typical plant sequencing projects.
Assembly
Several toolkits and pipelines are available for genome assembly (Fig. 2). One example, Canu, is based on the overlap layout consensus (OLC) principle (Koren et al., 2017). Canu uses a ‘correction then assembly’ strategy, making it also useful as a pre-processing tool before switching to another assembler. One consideration when assembling larger plant genomes is that Canu needs to run on computer clusters and still requires significant run time (Schmidt et al., 2017).
Similarly, MECAT (Xiao et al., 2017) first corrects reads and then uses the basic Canu engine for genome assembly, although Canu was replaced with a string graph assembler in the more recent version, MECAT2. A string graph assembler is also used in NECAT (Chen et al., 2020, Preprint), which has been adopted by ONT. However, both MECAT2 and NECAT still require initial read error correction as part of their assembly pipeline. Alternative OLC assemblers such as Ra (Vaser and Šikić, 2019, Preprint) and Miniasm (Li, 2016) directly assemble raw, uncorrected reads.
A number of alternative long-read assemblers have also been successfully applied to plant genomes (Schmidt et al., 2017; Belser et al., 2018; Wang et al., 2020). These include SMARTdenovo and its successor wtdbg2/Redbean (Ruan and Li, 2020), the latter using fuzzy de Bruijn graphs as a more error-tolerant extension of the de Bruijn graph data structure typically used to assemble Illumina sequencing data. Another example, Flye, relies on a repeat graph data structure that also tolerates more sequencing errors (Kolmogorov et al., 2019). In addition to these long-read assemblers, hybrid assemblers that use short, low-error sequences coupled with more error-prone long-read data are also available. One example is MaSuRCA (Zimin et al., 2013), which can be slow when applied to complex plant genomes but has nevertheless been tested successfully in plant species, including the annual grass Aegilops tauschii (Zimin et al., 2017).
Polishing and consensus
Although recent advances in assembly algorithms have improved consensus handling, it is often still necessary to post-process the assembly before biological analysis (Fig. 2). Typically, ONT reads are used to correct the assembly as an additional consensus step. This can be achieved rapidly using Racon, which realigns the reads and should therefore provide good consensus accuracy (Vaser et al., 2017). Racon is currently undergoing modifications to increase its speed by making it GPU compatible. However, Nanopolish can usually achieve superior accuracy by utilizing the original signal level traces rather than basecalled reads (Loman et al., 2015). Even so, at least in the case of bacteria (Wick et al., 2019), a custom-trained basecaller provided such high consensus accuracy after Racon-based polishing (>99.9%) that additional Nanopolish processing actually reduced the accuracy. Machine learning can also be used to correct errors. The ONT program Medaka (https://nanoporetech.github.io/medaka/benchmarks.html#evaluation-across-samples-and-depths) promises to outperform Racon and Nanopolish in terms of speed and accuracy for bacterial sequences, although it is currently trained only on bacterial and human data. Alternatively, the community-developed tool HELEN uses a similar approach, but is currently only trained on human data (Shafin et al., 2019, Preprint).
It is also necessary to correct assemblies using an orthogonal technology, such as Illumina sequencing, to remove remaining small-scale sequence errors. The Pilon polisher is often used for this purpose (Walker et al., 2014), following autocorrection of the assembly using ONT reads. This is because the best consensus accuracy of ≥99.9% is still not sufficient to achieve the minimum 99.99% base accuracy benchmark defined for a ‘finished human genome assembly’ or the actual accuracy of ~99.999% achieved by the International Human Genome Sequencing Consortium (2004). This level of accuracy is necessary because errors can significantly affect downstream protein prediction and subsequent interpretations (Watson and Warr, 2019). However, the technology is developing rapidly and it may not be appropriate to test old results against such benchmarks (Koren et al., 2019). Nevertheless, efficient error correction is important, and even high-quality reference genomes may lack genes due to assembly problems, regardless of which sequencing technology was used.
Assembly pipeline, improvement, and quality control
Researchers have a variety of options for data processing and de novo genome assembly, and some combinations are better than others depending on parameters such as data volume, genome size, and the heterozygosity and ploidy of the plant species. One approach, used by Schmidt et al. (2017) and Belser et al. (2018), is to first correct reads using Canu (Koren et al., 2017) followed by assembly using SMARTdenovo (J. Ruan, unpublished github) and polishing with Illumina data using Pilon (Walker et al., 2014). If available computational resources are not sufficient for Canu, Deschamps et al. (2018) showed that, at least for medium-sized genomes, the Canu correction step can be omitted.
The resulting assemblies can be scaffolded to near chromosome scale using Bionano optical mapping technology (Belser et al., 2018; Deschamps et al., 2018). The latter also carried out post-scaffolding polishing with ONT data using Racon (Vaser et al., 2017) and 10× genomics data using the Long Ranger ALIGN pipeline to resolve medium-sized structural errors that Pilon could not fix before scaffolding
The need for polishing and overall assembly quality can be assessed using BUSCO, a tool that provides quantitative measures for genome completeness based on the anticipated gene content (Waterhouse et al., 2018). Unpolished long-read assemblies often contain large numbers of small indels; hence many genes are not detected during BUSCO analysis. Polishing with tools such as Racon, Nanopolish, or Pilon will resolve these indels and increase the completeness score in BUSCO. Another approach for quality assessment is the LTR Assembly Index (LAI), which checks for the presence and integrity of long terminal repeats (LTRs) in the genome assembly (Ou et al., 2018). LAI is therefore complementary to BUSCO because it uses the non-genic parts of the assembly, further evaluating the quality of genomes (Ou et al., 2018).
Gene calling and other forms of downstream analysis
As the ONT platform and associated gene assembly tools continue to develop, there will be a shift towards the downstream analysis of gene platforms, especially for gene calling. Pipelines such as MAKER-P (Campbell et al., 2014) and BRAKER2 (Hoff et al., 2016) are already available, but require computational resources and effort in model training. However, given ongoing developments in ONT for RNA-seq analysis (both full-length cDNA and native RNA), and more widespread adoption of PacBio’s full-length self-corrected RNA-seq analysis (dubbed ‘isoseq’), we are likely to see a move towards evidence-only-based gene finders, such as Stringtie2 (Kovaka et al., 2019), which rely on long-read RNA/cDNAs. One limitation of Stringtie2 is that only genes corresponding to RNAs expressed with high enough coverage are detected. Unlike gene finding, gene functional annotation has already made the switch to high-throughput automated analysis using tools such as Mercator, TRAPID, or Hayai (Van Bel et al., 2013; Ghelfi et al., 2019; Schwacke et al., 2019) as well as generalists such as Blast2GO (Götz et al., 2008) to allow for the coming wave of ultra-large genome projects encompassing thousands of species (Lewin et al., 2018).
Conclusions and future directions
Many plant genomes are large and complex with highly repetitive regions, making it difficult to generate high-quality assemblies using first-generation or even second-generation sequencing methods (Bolger et al., 2014; Jiao and Schneeberger, 2017). The increasing quantity and quality of long-read sequence data from low-cost ONT platforms therefore provide confidence for the success of future plant genome sequencing projects, which will lead to significant advances in plant genome and pangenome assemblies. Current challenges in areas such as read error rates will be overcome by the rapid advances of third-generation technologies, and the advantages of ONT already outweigh the shortcomings. In the future, ONT is set to provide unprecedented insight into the complexities of plant genomes, while ongoing developments for modified basecalling will also provide a sound basis for epigenomic and transcriptomic analysis.
Acknowledgements
This work was supported by the German Ministry of Education and Research (FKZ 031B0293A to KD, FKZ 031A536C to BU, and 031B0187 to RS and BU). BU acknowledges the support of the Deutsche Forschungsgemeinschaft through EXC 2048/1. The authors would like to thank Rainer Schwacke (www.plabipd.de) for the assistance in completing the list of ONT-sequenced plant genomes. The authors also thank Dr Richard M. Twyman for editing the manuscript.
References
- Belser C, Istace B, Denis E, et al. 2018. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nature Plants 4, 879–887. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Poorter H, Dumschott K, et al. 2019. Computational aspects underlying genome to phenome analysis in plants. The Plant Journal 97, 182–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger ME, Weisshaar B, Scholz U, Stein N, Usadel B, Mayer KF. 2014. Plant genome sequencing—applications for crop improvement. Current Opinion in Biotechnology 26, 31–37. [DOI] [PubMed] [Google Scholar]
- Boykin LM, Ghalab A, De Marchi BR, et al. 2018. Real time portable genome sequencing for global food security [version 1; peer review: 1 approved, 1 approved with reservations]. F1000Research 7, 1101. [Google Scholar]
- Campbell MS, Law M, Holt C, et al. 2014. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiology 164, 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Wallace SL, Chiu CY, John KK, et al. 2017. Nanopore DNA sequencing and genome assembly on the international space station. Scientific Reports 7, 18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Nie F, Xie S-Q, et al. 2020. Fast and accurate assembly of Nanopore reads via progressive error correction and adaptive read selection. bioRxiv doi:10.1101/2020.02.01.930107 [Preprint]. [Google Scholar]
- Choi JY, Lye ZN, Groen SC, Dai X, Rughani P, Zaaijer S, Harrington ED, Juul S, Purugganan MD. 2020. Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biology 21, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D, Akeson M, Branton D. 2016. Three decades of nanopore sequencing. Nature Biotechnology 34, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, King M, May G, Lin H. 2018. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nature Communications 9, 4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE. 2014. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Molecular Cell 55, 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelfi A, Shirasawa K, Hirakawa H, Isobe S. 2019. Hayai-annotation plants: an ultra-fast and comprehensive functional gene annotation system in plants. Bioinformatics 35, 4427–4429. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Research 36, 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassa CJ, Wenger JP, Dabney C, Poplawski SG, Timothy Motley ST, Michael TP, Schwartz CJ, Weiblen GD. 2018. A complete Cannabis chromosome assembly and adaptive admixture for elevated cannabidiol (CBD) content. bioRxiv 458083. doi: 10.1101/458083. [Preprint]. [DOI] [Google Scholar]
- Harkness A, McLoughin F, Bilkey N, et al. 2020. A new Spirodela polyrhiza genome and proteome reveal a conserved chromosomal structure with high abundances of proteins favoring energy production. bioRxiv doi: 10.1101/2020.01.23.909457. [Preprint]. [DOI] [PubMed] [Google Scholar]
- Hoang PNT, Michael TP, Gilbert S, Chu P, Motley ST, Appenroth KJ, Schubert I, Lam E. 2018. Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. The Plant Journal 96, 670–684. [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel B, Marques A, Soriano A, et al. 2019. Genome sequence of the cluster root forming white lupin. Nature Communications 11, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431, 931–45. [DOI] [PubMed] [Google Scholar]
- Jain M, Koren S, Miga KH, et al. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnology 36, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M. 2016. The Oxford nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biology 17, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, An H, Xu F, Zhang X. 2020. Chromosome-level genome assembly and annotation of the loquat (Eriobotrya japonica) genome. GigaScience. 9, doi: 10.1093/gigascience/giaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao WB, Schneeberger K. 2017. The impact of third generation genomic technologies on plant genome assembly. Current Opinion in Plant Biology 36, 64–70. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology 37, 540–546. [DOI] [PubMed] [Google Scholar]
- Koren S, Phillippy AM, Simpson JT, Loman NJ, Loose M. 2019. Reply to ‘Errors in long-read assemblies can critically affect protein prediction’. Nature Biotechnology 37, 127–128. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Research 27, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biology 20, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews. Genetics 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Robinson GE, Kress WJ, et al. 2018. Earth BioGenome Project: sequencing life for the future of life. Proceedings of the National Academy of Sciences, USA 115, 4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Nishiyama T, Waller M, et al. 2020. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nature Plants 6, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Wang J, Dong R, Zhu HW, Lan LN, Zhang YL, Li N, Deng CL, Gao WJ. 2020. Chromosome-level genome assembly, annotation and evolutionary analysis of the ornamental plant Asparagus setaceus. Horticulture Research 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature Methods 12, 733–735. [DOI] [PubMed] [Google Scholar]
- Lu H, Giordano F, Ning Z. 2016. Oxford nanopore MinION sequencing and genome assembly. Genomics, Proteomics & Bioinformatics 14, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas SJ, Kahraman K, Avşar B, Buggs RJA, Bilge I. 2019. A chromosome-scale genome assembly of European Hazel (Corylus avellana L.) reveals targets for crop improvement. bioRxiv doi: 10.1101/817577. [Preprint]. [DOI] [PubMed] [Google Scholar]
- Maestri S, Cosentino E, Paterno M, Freitag H, Garces JM, Marcolungo L, Alfano M, Njunjić I, Schilthuizen M, Slik F. 2019. A rapid and accurate MinION-based workflow for tracking species biodiversity in the field. Genes 10, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg MM, Spangenberg GC, Daetwyler HD, Cogan NOI. 2019. Assessment of low-coverage nanopore long read sequencing for SNP genotyping in doubled haploid canola (Brassica napus L.). Scientific Reports 9, 8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrano A, Britton M, Zaini PA, et al. 2019. High-quality chromosome-scale assembly of the walnut (Juglans regia L) reference genome. bioRxiv doi: 10.1101/809798. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Jupe F, Bemm F, Motley ST, Sandoval JP, Lanz C, Loudet O, Weigel D, Ecker JR. 2018. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nature Communications 9, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal TK, Rawal HC, Chowrasia S, Varshney D, Panda AK, Mazumdar A, Kaur H, Gaikwad K, Sharma TR, Singh NK. 2018. Draft genome sequence of first monocot-halophytic species Oryza coarctata reveals stress-specific genes. Scientific Reports 8, 13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni P, Huang N, Zhang Z, Wang DP, Liang F, Miao Y, Xiao CL, Luo F, Wang J. 2019. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 35, 4586–4595. [DOI] [PubMed] [Google Scholar]
- Ning DL, Wu T, Xiao LJ, Ma T, Fang WL, Dong RQ, Cao FL. 2020. Chromosomal-level assembly of Juglans sigillata genome using Nanopore, BioNano, and Hi-C analysis. GigaScience 9, doi: 10.1093/gigascience/giaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile MS, Cazzaniga P, Tangherloni A, Besozzi D. 2017. Graphics processing units in bioinformatics, computational biology and systems biology. Briefings in Bioinformatics 18, 870–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaumer T, Martis MM, Roessner SK, Pfeifer M, Bader KC, Sharma S, Gundlach H, Spannagl M. 2013. MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Research 41, D1144–D1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Chen J, Jiang N. 2018. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Research 46, e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Helmstetter AJ, Devey D, Wilkinson T, Papadopulos AST. 2017. Field-based species identification of closely-related plants using real-time nanopore sequencing. Scientific Reports 7, 8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall A, Barton GJ, Simpson GG. 2019. Nanopore direct RNA sequencing maps an arabidopsis N6 methyladenosine epitranscriptome. bioRxiv doi: 10.1101/706002. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu X, Li Z, Tian Y, et al. 2020. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytologist 10.1111/nph.16552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Loman NJ, Duraffour S, et al. 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand AC, Jain M, Eizenga JM, Musselman-Brown A, Olsen HE, Akeson M, Paten B. 2017. Mapping DNA methylation with high-throughput nanopore sequencing. Nature Methods 14, 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rang FJ, Kloosterman WP, de Ridder J. 2018. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biology 19, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read AC, Moscou MJ, Zimin AV, Pertea G, Meyer RS, Purugganan MD, Leach JE, Triplett LR, Salzberg SL, Bogdanove AJ. 2020. Genome assembly and characterization of a complex zfBED-NLR gene-containing disease resistance locus in Carolina Gold Select rice with Nanopore sequencing. PLoS Genetics 16, e1008571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nature Methods 17, 155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. 1977. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265, 687–695. [DOI] [PubMed] [Google Scholar]
- Schalamun M, Nagar R, Kainer D, Beavan E, Eccles D, Rathjen JP, Lanfear R, Schwessinger B. 2019. Harnessing the MinION: an example of how to establish long-read sequencing in a laboratory using challenging plant tissue from Eucalyptus pauciflora. Molecular Ecology Resources 19, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalamun M, Schwessinger B. 2017. DNA size selection (>1 kb) and clean up using an optimized SPRI beads mixture V.1 10.17504/protocols.io.idmca46. [DOI]
- Schmidt MH, Vogel A, Denton AK, et al. 2017. De novo assembly of a new Solanum pennellii accession using nanopore sequencing. The Plant Cell 29, 2336–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Ponce-Soto GY, Krause K, Bolger AM, Arsova B, Hallab A, Gruden K, Stitt M, Bolger ME, Usadel B. 2019. MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Molecular Plant 12, 879–892. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC. 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nature Methods 15, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafin K, Pesout T, Lorig-Roach R, et al. 2019. Efficient de novo assembly of eleven human genomes using PromethION sequencing and a novel nanopore toolkit. bioRxiv doi: 10.1101/715722. [Preprint]. [DOI] [Google Scholar]
- Shim J, Humphreys GI, Venkatesan BM, et al. 2013. Detection and quantification of methylation in DNA using solid-state nanopores. Scientific Reports 3, 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siadjeu C, Pucker B, Viehöver P, Albach DC, Weisshaar B. 2020. High contiguity de novo genome sequence assembly of trifoliate yam (Dioscorea dumetorum) using long read sequencing. Genes 11, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. 2017. Detecting DNA cytosine methylation using nanopore sequencing. Nature Methods 14, 407–410. [DOI] [PubMed] [Google Scholar]
- Song C, Liu Y, Song A, et al. 2018. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Molecular Plant 11, 1482–1491. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nishijima R, Teramoto S, Kitomi Y, Hayashi T, Uga Y, Kawakatsu T. 2020. De novo genome assembly of the indica rice variety ir64 using linked-read sequencing and nanopore sequencing. G3 10, 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Al-Dilaimi A. 2019. Bioinformatics in Germany: toward a national-level infrastructure. Briefings in Bioinformatics 20, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng H, Cao MD, Hall MB, Duarte T, Wang S, Coin LJM. 2018. Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. Gigascience 7, doi: 10.1093/gigascience/giy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt B, Buell CR. 2019. High molecular weight DNA isolation method from diverse plant species for use with Oxford nanopore sequencing. bioRxiv 783159; doi: 10.1101/783159. [Preprint]. [DOI] [Google Scholar]
- Van Bel M, Proost S, Van Neste C, Deforce D, Van de Peer Y, Vandepoele K. 2013. TRAPID: an efficient online tool for the functional and comparative analysis of de novo RNA-Seq transcriptomes. Genome Biology 14, R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, Bryant D, Edger PP, et al. 2015. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527, 508–511. [DOI] [PubMed] [Google Scholar]
- Vaser R, Šikić M. 2019. Yet another de novo genome assembler. bioRxiv doi: 10.1101/656306. [Preprint]. [DOI] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Research 27, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollger MR, Logsdon GA, Audano PA, et al. 2020. Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. Annals of Human Genetics 84, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Das A, Kainer D, Schalamun M, Morales-Suarez A, Schwessinger B, Lanfear R. 2020. The draft nuclear genome of assembly of Eucalyptus pauciflora: new approaches to comparing de novo assemblies. GigaScience 9, doi: 10.1093/gigascience/giz160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Molecular Biology and Evolution 35, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M, Warr A. 2019. Errors in long-read assemblies can critically affect protein prediction. Nature Biotechnology 37, 124–126. [DOI] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Holt KE. 2019. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biology 20, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao CL, Chen Y, Xie SQ, Chen KN, Wang Y, Han Y, Luo F, Xie Z. 2017. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nature Methods 14, 1072–1074. [DOI] [PubMed] [Google Scholar]
- Yang Y, Sun P, Lv L, et al. 2020. Prickly waterlily and rigid hornwort genomes shed light on early angiosperm evolution. Nature Plants 6, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasodha R, Vasudeva R, Balakrishnan S, Sakthi AR, Abel N, Binai N, Rajashekar B, Bachpai VKW, Pillai C, Dev SA. 2018. Draft genome of a high value tropical timber tree, Teak (Tectona grandis L. f): insights into SSR diversity, phylogeny and conservation. DNA Research 25, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Harris CJ, Liu Q, et al. 2018. Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 115, E1069–E1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29, 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Puiu D, Luo MC, Zhu T, Koren S, Yorke JA, Dvorak J, Salzberg S. 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the mega-reads algorithm. Genome Research 1, 066100. [DOI] [PMC free article] [PubMed] [Google Scholar]