Abstract
Recent studies have shown how intestinal parasites can modulate gut microbiota. This observation is not surprising since the human intestinal lumen, like any other niche, is a battlefield of microbial competition, and Eukaryotes can affect bacterial populations. Intestinal pathogenic protist has been associated with reshaping the microbial community structure; however, the interactions between the colonic bacterial communities and parasites like Blastocystis spp., Entamoeba coli, and Endolimax nana have been poorly studied. In this work, we studied the distal intestinal bacterial microbiota of 49 children attending 7 public daycare centers in Medellin, Colombia, and compared the bacterial microbiota structure in the presence or absence of the protists Blastocystis spp., E. coli, and E. nana. Parasite colonization was associated with an increase in bacterial richness. Moreover, Blastocystis spp. presented a positive relationship with Prevotella, since this bacterium was selectively enriched in children carrying it. Remarkably, the E. coli colonized children showed a microbial profile that was closer to uninfected controls, although some bacterial taxa displayed to be enriched. This is the case for Akkermansia, which showed to be favored in E. coli colonized individuals, while notably reduced in the Blastocystis spp. parasitized group.
Subject terms: Microbiome, Parasite host response
Introduction
A complex microbial community, mostly bacteria, colonizes human intestines since birth, although it also bears archaea, fungi, viruses and occasionally parasites. Recent research works have confirmed that under normal conditions, human adults can harbor between 500 to 1,000 different species of bacteria that along with its metabolic products can influence the health state of the organism1. These abundant microbes can help us processing food, absorb vitamins, stimulate the immune system2,3 and compete with intestinal pathogens, amid other functions4–6.
In early childhood, the offspring microbiota is strongly associated with the mother’s and with the delivery mode. This will influence if the colonizing bacteria mainly come from skin, vagina, or even stool. As the child grows and solid foods are introduced, new microbes arrive increasing the diversity and favoring bacteria capable of degrading complex polymers. It is estimated that diversity becomes stable, adult-like, at around 2.5 years old7,8. By this age two Phyla dominate the microbiota of healthy individuals: Bacteroidetes and Firmicutes9,10.
It has been extensively described that the main driving force for the intestinal microbial structure is the diet2,6,11–14. Other factors such as host genetics15, sex16, and antibiotic use might also have a significant influence in the community shaping as well17–19.
Other prominent players in the gut microbiome competition are parasites, which gained attention in recent years. Several works have shown how microbiota richness and community structure is influenced by protist and metazoan parasites. Human intestinal nematodes have been described to be associated with increased microbial richness and diversity20–23, whereas intestinal protist have shown a broader panorama, where interactions with the intestinal microbiota depend on the species studied24. One of the most studied models is Entamoeba histolytica, in which specific bacterial taxa are disease predictors. Patients with diarrhea and positive for E. histolytica had higher levels of Prevotella copri, compared to nondiarrheal subjects25. Moreover, it was demonstrated that trophozoites of this parasite phagocyte bacteria like Lactobacillus and Shigella dysenteriae conferring an augmented cytopathic effect to the parasite26,27.
Another studied case is Blastocystis spp. which has been associated in several studies with a bacterial species richness increase in the distal intestine28–30. Blastocystis is an anaerobic Stramenopila that inhabits the gastrointestinal tract of a wide range of animal hosts, including humans. It is one of the most frequent intestinal protists worldwide colonizing around 1 billion people, being associated with an oral-fecal route of transmission. There are reports of a high prevalence of this protist in both developing (up to 100%)31 and developed countries (0.5–30%)32–34. In Colombia, studies from 2009–2019 describe a prevalence between 12.6 and 87.1% in children35,36, which are the age group at a higher risk of intestinal parasite infection34.
Based on the sequence analysis of the SSU rDNA gene, to date 17 Blastocystis subtypes (STs) have already been identified (ST1-ST17), and genetic diversity studies have shown intra-genetic diversity among STs37–39. Subtypes ST1-ST9 and ST12 parasite humans; being STs 1–4 the most prevalent. ST9 has been reported exclusively on human hosts. Differences in the development of symptoms and their severity have been described (abdominal pain, constipation, diarrhea, flatulence, irritable bowel syndrome-like symptoms-IBS, and even skin disorders such as urticaria)34,40,41. One reason that could explain this broad spectrum of reactions can be attributed to the genetic diversity described in this parasite.
Several studies have reported a Blastocystis spp. long-term colonization in asymptomatic carriers, suggesting that it could be a frequent member of the healthy intestinal microbiota42–44. Recent data about the relationship between this protist and the gut bacteria supports the latter hypothesis. Most of the research, performed mainly in the developed world, shows a higher fecal bacterial richness and diversity in individuals carrying the parasite29,30. In the developing world, the same phenomenon was observed, where adults in Mexico (Morelos) positive to Blastocystis spp. showed increased bacterial diversity compared with not infected ones28. Besides, this protist is associated with gut microbiota profiles characterized by low relative abundances of Bacteroides-driven enterotype and high levels of Ruminococcus- and Prevotella-driven enterotype, taxa that are typically associated with gut health28,45–49. A eubiotic condition was also associated with Blastocystis spp. since a significantly higher ratio of beneficial species (Faecalibacterium prausnitzii) versus potentially harmful species (Escherichia coli) was found in individuals positive for this parasite47.
Additionally, some authors described a lower abundance of potentially pathogenic species of the Enterobacteriaceae family in the presence of Blastocystis spp. and Entamoeba compared to negative controls47. On the contrary, a few studies describe a dysbiosis in patients with Blastocystis spp. Nourrisson et al.50 found lower protective bacteria in the fecal microbiota in patients with irritable bowel syndrome and healthy controls. Yason et al. described that mice infected with Blastocystis ST7 had less beneficial Bifidobacterium and Lactobacillus bacteria. Microbial communities are probably differently shaped according to the Blastocystis subtypes51.
Most of the Blastocystis spp. and intestinal microbiota interaction studies have been done in the adult population. Data regarding gut bacteria and Blastocystis spp. interactions in children are scarce. Popovic et al.49 characterized the eukaryotic microbiota of hospitalized Malawian children suffering from Severe Acute Malnutrition (SAM). Blastocystis colonization correlated with bacterial alpha diversity and increased abundance of specific taxa from Firmicutes, particularly those from the clostridia class (Oscillibacter, Sporobacter, Cellulosibacter, and Roseburia). Consistent with the results of previous studies, there was a negative correlation between Blastocystis and the Enterobacteriaceae family. In Colombia, Toro et al.52 evaluated the bacterial gut microbiota composition in children infected with intestinal parasites. Four groups were included according to the parasites identified. Children from the Giardia group (only infected with Giardia) and Helm-pro group (infected with nematode and another protist including Blastocystis) suffer a switch from a type I (Bacteroides-driven enterotype) to a type II enterotype (Prevotella-driven enterotype).
In this work, we study the alterations of the gut microbiota on children from one to five years old colonized by the protists Blatocystis spp., Entamoeba coli, and Endolimax nana. Albeit the intense research of the gut microbiota, still, little information is available on the effect of these parasites in children and, in the case of E. coli and E. nana, information considering this topic is scarce.
Results
The 16S metataxonomic experiment started with 50 k reads per library. After MOTHUR processing, the read number was rarefied with the totalgroup function to an average of 12,249 clean reads per sample, and they ranged between 12,326 and 12,526. The coverage analysis showed that at least 97.4% of the expected OTUs were observed within our analysis individuals and went up to 99% in several samples. The observed OTU (3%) count varied between 193 and 551 within all tested individuals (Supplementary Table 1).
Effect of harboring protists
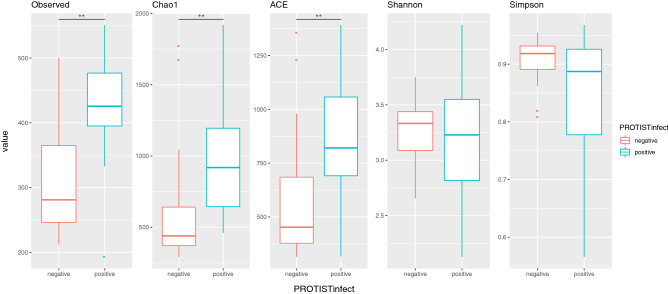
Initially, we tested the differences between children with no detectable parasites on the stool microscopical analysis (NPDM group) and those confirmed to harbor either Blastocystis spp. (Blasto group), Entamoeba coli (E_coli group) or Endolimax nana (E_nana); these last three were grouped in the PROTIST-infected category. As shown in Fig. 1, the Observed OTU median values rise from 281 in NPDM controls (negative) to 426 in the protist-infected ones (positive), with a p-value = 3.785e-05, using the Kruskal–Wallis rank-sum test. Similarly, the Chao1 and ACE indices were significantly higher, using the same test, in infected individuals compared to controls, rising from 439 to 919 (p-value = 0.0001335), and from 453 to 821 (p-value = 0.0003182), respectively. The Shannon and Simpson indices for both groups were similar and showed no statistically significant differences with the Kruskal–Wallis test. Furthermore, sex did not show any significant difference with the richness/diversity indices studied, when the Kruskal–Wallis test was applied.
Figure 1.
Richness and diversity indices of protist infected and control children. Boxplot representation of the median values of the Observed OTUs and the Chao1, ACE, Shannon, and Simpson indices; comparing the control children (no parasites detectable on the stool analysis—negative) and the Protist infected group (positive: colonized either with Blastocystis spp., Entamoeba coli, or Endolimax nana). The observed differences in the median values between the Protist negative and positive groups for the observed OTUs, Chao1 index, and ACE index, were statistically significant with p < 0.001 (Kruskal–Wallis rank-sum test).
Regarding age, there was a weak significative correlation with the Shannon index (Spearman correlation p-value = 0.02515, rho 0.3196735), which was not the case for the Simpson index, that showed no significant results. The ‘lm’ model of Age and Shannon gives shannon = 0.1Age + 2.8 with (p-value = < 2e−16, for the intercept and p-value = 0.0596 for the slope). Our group of selected children, most are in the range of 2 to 5yo, life period in which the intestinal microbiota tends to stabilize.
Finally, Fig. 2 shows the ordination plot using NMDS distance, which mostly separates the individuals of both groups. We determined which variables most strongly affected the structure of the children gut microbiota using a permutational multivariate analysis of variance (PERMANOVA) test of the Bray–Curtis dissimilarities. Again, the ordination analysis plot shows a significant segregation pattern (PERMANOVA R2 = 0.036, p-value < 0.05) of most of the non-parasites controls versus the PROTIST-infect group.
Figure 2.
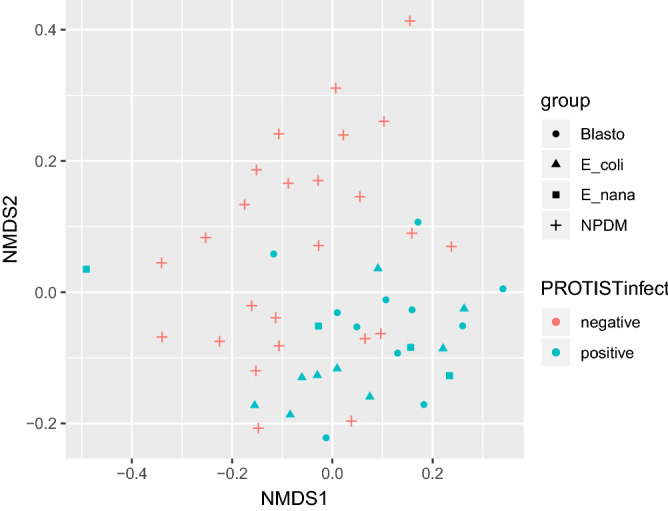
Ordination analysis of the studied children groups. Ordination analysis plot showing the calculated distances (NMDS and Bray) of the studied children in the four tested groups: NPDM (+ controls with no parasites detected on the microscopic stool analysis), Blasto ( • Children parasitized only by Blastocystis spp.). E_coli ( ∆ Children parasitized only by Entamoeba coli). E_nana (□ Children parasitized only by Endolimax nana). Colored in red are the uninfected control children (NPDM), while in blue are the Protist infected children (colonized with either Blastocystis spp., Entamoeba coli, or Endolimax nana).
Bacterial microbiota profiles associated with Blastocystis spp., Entamoeba. coli or Endolimax nana
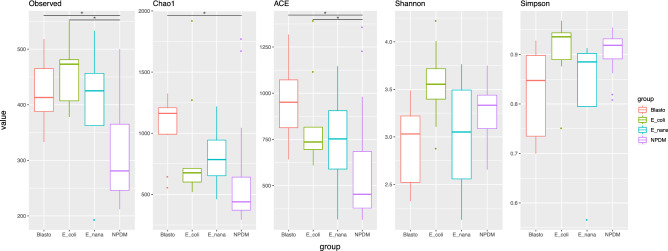
The next step in our analysis was oriented to discriminate if there was any differential relationship between the specific protist harbored by each individual and the microbiota profiles. To do so, we separated the infected children into three groups regarding the detected parasite: Blastocystis spp., E. coli, or E. nana; and compare them with the NPDM controls. Following the previous observations, the median values of the richness indices observed OTUs, Chao1, and ACE were augmented in all the infected groups (Fig. 3). The species richness (Observed OTUs) showed statistical significance, applying the Kruskal–Wallis rank-sum test, among all four tested groups (p-value = 0.0004175) and proved to be significantly higher in pairwise comparisons with the Wilcoxon rank-sum test between the Blastocystis spp. (median 413) and E. coli (median 473) parasitized groups (Blasto and E_coli, p = 0.0043 and p = 0.0010, respectively) compared to the control group (NPDM, median = 281). Additionally, the Kruskal–Wallis test showed statistical differences within the groups in all measured indices (Simpson, p-value = 0.004808; ACE, p-value = 0.002159; Chao1, p-value = 0.001095; Shannon, p-value = 0.03467 (Age-adjusted)). The Pairwise comparisons using the Wilcoxon rank-sum test indicated that the Chao1 index was only significantly different between the controls (median = 439) and the Blasto (median = 1,162, p = 0.0016) groups. The ACE index showed similar results in these two groups (NMPD, median 453; vs. Blasto, median 951; p = 0.0019). Additionally, this index showed significant differences among the NMPD controls vs. E_coli group (p = 0.0313). The Shannon and Simpson indices median values showed slight variations but were not statistically significant with the Wilcoxon rank-sum test when the controls were compared with the parasitized groups (Fig. 3).
Figure 3.
Richness and diversity indices of the control and the colonized children either with Blastocystis spp., Entamoeba coli, or Endolimax nana. Boxplot representation of the median values of the Observed OTUs and the indices Chao1, ACE, Shannon, and Simpson; comparing the control children (no parasites detectable on the stool analysis—NPDM) and the Blasto ( Children parasitized only by Blastocystis spp.), E_coli ( Children parasitized only by Entamoeba coli), and E_nana ( Children parasitized only by Endolimax nana) parasitized groups. The observed differences in the median values of the NPDM controls vs. Blasto or E_coli groups were statistically significant using the Wilcoxon rank-sum test, with p < 0.05 for observed OTUs and ACE index. The Chao1 index was statistically significant between NPDM controls and Blasto groups with a p < 0.01 using the same test.
Again, the ordination analysis plot shows a segregation pattern of most of the non-parasites controls versus the Blastocystis spp., E. coli, or E. nana groups. Among the parasitized groups, it is not possible to observe an apparent clustering of the individual based on each protist species (Fig. 2).
In general, the most copious Phyla were Bacteroidetes and Firmicutes, followed by Proteobacteria, Actinobacteria, and Verrucomicrobia. This last Phylum was observed significatively more abundant in the E_coli (median = 470) group compare to the Blasto group (median = 0) (Wilcoxon rank-sum test, p = 0.019). Remarkably, two individuals of the NMPD control group presented a significant proportion of Fusobacteria (SupplementaryFigure 1). At the family taxonomic category, we observed that the taxa Prevotellaceae, Ruminococcaceae, Bacteroidaceae, and Lachnospiraceae were the most prevalent across all samples. It is noteworthy that the bars of Prevotellaceae are more prominent in the Blasto group samples compare to NPDM controls. Conversely, the control group seems to have a higher relative proportion of Ruminococcaceae compared to Blastocystis spp. parasitized individuals (SupplementaryFigure 2).
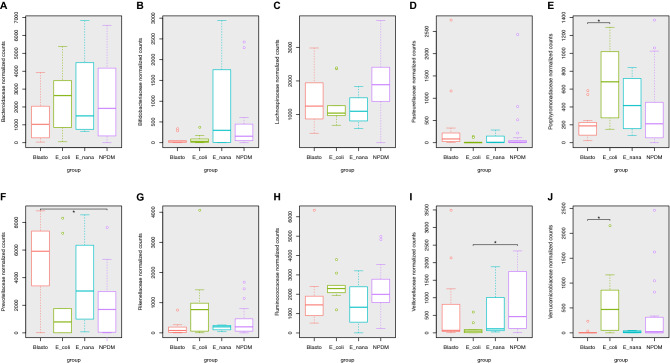
Quantitative comparisons of the relative abundance of the top 10 most abundant classified bacterial families showed significative (Kruskal–Wallis test) changes in Prevotellaceae (p-value = 0.01638), Porphyromonadaceae (p-value = 0.03995), Veillonellaceae (p-value = 0.04452), Verrucomicrobiaceae (p-value = 0.0165), and Pasteurellaceae (p-value = 0.04312) (Fig. 4).
Figure 4.
Relative abundance of the top ten most abundant bacterial families across the tested groups. Box plot graphic representation of the normalized median counts within the tested groups: NPDM (Controls with no parasites detected in the microscopic stool analysis), Blasto (Children parasitized only by Blastocystis spp.). E_coli (Children parasitized only by Entamoeba coli). E_nana (Children parasitized only by Endolimax nana). A Bacteroidaceae. B Bifidobacteriaceae. C Lachnospiraceae. D Pasteurellaceae. E Porphyromonadaceae. The difference in the median values between E_coli and Blasto groups was statistically significant with a p = 0.021. F Prevotellaceae. The difference in the median values between NPDM controls and Blasto groups was statistically significant with a p = 0.013. G Rikenellaceae. H Ruminococcaceae. I Veillonellaceae. The difference in the median values between NPDM controls and E_coli groups was statistically significant with a p = 0.031. J Verrucomicrobiaceae. The difference in the median values between E_coli and Blasto groups was statistically significant with a p = 0.019.
Prevotellaceae and Verrucomicrobiaceae also showed the best statistical significance in the pairwise comparisons of the relative abundances using the Wilcoxon rank-sum test. For Prevotellaceae, the significative difference (p-value = 0.01638) was observed between the Blasto (median = 5,912) and the NMPD controls (1696). In the case of Verrucomicrobiaceae, median values drop from 470 in the E_coli group to 0 in the NPDM controls (p-value = 0.019). Porphyromonadaceae also showed significant results between the Blasto and E_coli groups (p-value = 0.021), while in Veillonellaceae the significative differences were observed between the NPDM controls and the E_coli group (p-value = 0.031).
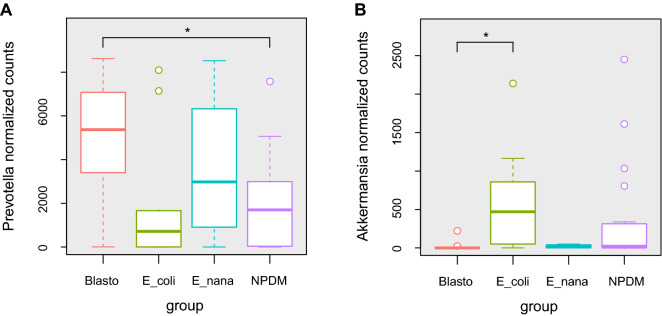
At the genus category, Prevotella, Bacteroides, and Faecalibacterium were, in order, the dominant microoganisms over all individuals (SupplementaryFigure 3). These results indicate changes in Prevotella proportions, which seem to be enriched in the Blasto group. One more detailed and quantitative analysis on Prevotella showed an increase in the median values of normalized counts of this anaerobe in the Blasto (5,367) group compared to the controls (1696) that were statistically significative (p = 0.011) (Fig. 5A).
Figure 5.
A. Relative abundance of Prevotella across tested groups. Box plot graphic representation of the normalized median counts of Prevotella within the tested groups: NPDM (Controls with no parasites detected in the microscopic stool analysis), Blasto (Children parasitized only by Blastocystis spp.). E_coli (Children parasitized only by Entamoeba coli). E_nana (Children parasitized only by Endolimax nana). The difference in the median values between NPDM controls and Blasto groups was statistically significant with a p < 0.05. C. Relative abundance of Akkermansia across tested groups. Box plot graphic representation of the normalized median counts of Akkermansia within the tested groups: NPDM (Controls with no parasites detected in the microscopic stool analysis), Blasto (Children parasitized only by Blastocystis spp.). E_coli (Children parasitized only by Entamoeba coli). E_nana (Children parasitized only by Endolimax nana). The difference in the median values between Blasto and E_coli groups was statistically significant with a p < 0.05.
Prevotella is enriched within the individuals harboring Blastocystis while Akkermansia seems to be favored with Entamoeba coli colonization
To further confirm the previous findings on the changes in the relative abundance of the mentioned microbe families, a LEfSe analysis was performed, aiming to identify marker microorganisms within the tested groups. After filtering the results excluding the unclassified organisms and LDA score above 3, 15 OTUs were kept with significant p values (Table 1). Prevotella (OTU00001) showed the highest abundance shift with an LDA score of 5.32 towards the Blasto group. This group was also enriched with the bacteria Haemophilus, Holdemanella, and Butyricicoccus.
Table 1.
LEfSe analyses of the marker microorganisms of each tested group.
| OTU | LogMaxMean | Class | LDA | pValue | OTUsize | Phylum | Family | Genus |
|---|---|---|---|---|---|---|---|---|
| Otu00015 | 4.11044 | NPDM | 3.75427 | 0.0195828 | 10,821 | Firmicutes | Lachnospiraceae(100) | Blautia(100) |
| Otu00085 | 3.50502 | NPDM | 3.47811 | 0.01566 | 1924 | Firmicutes | Streptococcaceae(100) | Streptococcus(100) |
| Otu00122 | 3.576 | E_nana | 3.16078 | 0.00075462 | 1,020 | Bacteroidetes | Prevotellaceae(100) | Paraprevotella(100) |
| Otu00005 | 4.67852 | E_coli | 4.56941 | 0.0158793 | 21,936 | Verrucomicrobia | Verrucomicrobiaceae(100) | Akkermansia(100) |
| Otu00075 | 3.71921 | E_coli | 3.47399 | 0.0314539 | 2,442 | Firmicutes | Lachnospiraceae(100) | Coprococcus(100) |
| Otu00141 | 3.36365 | E_coli | 3.06432 | 0.0130117 | 741 | Bacteroidetes | Rikenellaceae(100) | Alistipes(100) |
| Otu00001 | 5.48881 | blasto | 5.32462 | 0.0324499 | 181,806 | Bacteroidetes | Prevotellaceae(100) | Prevotella(100) |
| Otu00010 | 4.51725 | blasto | 4.43657 | 0.0386207 | 15,792 | Proteobacteria | Pasteurellaceae(100) | Haemophilus(100) |
| Otu00068 | 3.60055 | blasto | 3.42977 | 0.0354466 | 2,802 | Firmicutes | Erysipelotrichaceae(100) | Holdemanella(100) |
| Otu00088 | 3.4176 | blasto | 3.3222 | 0.0298255 | 1836 | Firmicutes | Ruminococcaceae(100) | Butyricicoccus(100) |
The second highest score was obtained for Akkermansia (LDA 4.57) in the E_coli group, which was also enriched with Coprococcus and Alistipes. The control group had two enriched biomarker organisms, both from the Firmicutes phylum, Blautia, and Streptococcus. The case of Akkermansia is quite impressive since it is not included in the top ten most abundant organisms, although it was the second most abundant biased OTU in the tested groups. To gain insights into this particular group, the normalized counts of Akkermansia were extracted and plotted (Fig. 5B). These results display a reduced number of counts in the Blasto group with a median value of zero, while the control and the E_coli group, 23 and 470, respectively. The Kruskal–Wallis test showed statistical significance in the differences among groups (p-value = 0.0165), and the Pairwise Wilcoxon test showed there only significant differences between the Blasto and the E_coli groups (p = 0.019).
Discussion
Children distal intestinal microbiota tends to stabilize and begin to be more adult-like around 3-yo8. In our selected children group only two were younger than 2-yo, most are in the range of 3 to 5-yo, when intestinal microbiota starts to stabilize. This can explain why, albeit the described association of gut microbiota diversity and age in children, a weak correlation with the Shannon diversity index was observed. Additionally, since these children receive most of their meals in the daycare center, allowing them to have a more similar intestinal microbiota due to the effect of a normalized diet. Nonetheless, despite the influence of age within the intestinal microbiota in the studied children, our results showed that Protist colonization have a relevant impact on the intestinal microbial community in children.
Eukaryotic parasites are major competitors in the microbial world due to bacterivorous activity or direct competition for nutrients27,53,54. The effect of intestinal nematodes and protists on the human gut bacterial microbiota has been proved in several studies, and some of them show, as a common trend, that bacterial richness is increased in individuals that carry intestinal parasites. However, diversity is not always augmented as well20–23,28–30,52.
In the present study, we observed that all the studied protist, Blastocystis spp., Entamoeba coli, and Endolimax nana; were associated with a significant increase in bacterial richness, with median values that almost doubles the control group. We cannot conclude if this is cause or effect, but these findings are in concordance with previous reports for similar studies on the relation of Blastocystis spp. with the intestinal microbiota. For Entamoeba coli and Endolimax nana it was not possible to find previous scientific publications addressing this topic. To the best of our knowledge, this is the first report that observes the relation of the intestinal gut microbiota and these two amoebae using a 16S metataxonomic approach. It is essential to highlight that no definitive conclusion can be drawn from the individuals infected with Endolimax nana due to the low number of children included in this group, only 4. Endolimax nana is an intestinal amoeba of humans that has a cosmopolitan distribution, most likely as a commensal or nonpathogenic protozoon, with an estimated global prevalence in healthy individuals of 13.4%. The scientific evidence to date is inconclusive in terms of its host specificity, epidemiology, morphology, taxonomy and genetic diversity55. Although some authors suggest that E. nana feed exclusively on bacteria and that it could have a pathogenic potential, with case reports of parasitized patients suffering arthritis56, intestinal symptoms57–59, and urticaria60,61, there is not enough evidence that supports this statements. Therefore, studies focused on the parasite genetic variability and crosstalk interaction with the microbiota and the immune system are needed to provide data that clarify the effects of this protozoan in the human intestine. Furthermore, the fecal–oral transmission suggests that Endolimax can be used as a biological marker suitable for the hygiene measures of the population and fecal contamination of food or water62.
Blastocystis spp. has been associated in several studies with increased bacterial diversity in western European adults and Mexicans30,46. In our study in Colombian children, although we found an increased richness in the colonized individuals, no significant differences were observed in the diversity indices. This controversial finding might be attributed to a differential response in the child or adult intestinal microbiota to the Blastocystis challenge. More studies in children need to be performed in order to understand in detail this phenomenon.
Since its first observation in 1849, Blastocystis spp. was initially described as a pathogenic parasite being formally termed in early 20th Century63,64. After several decades of debate about its classification and host preferences, in the second half of this century, it was generally accepted as a pathogenic protist for humans that can cause diarrhea and abdominal pain65. However, several researchers have raised concerns about the evidence that supports the pathogenicity of Blastocystis spp. in humans, and its clinical significance remains controversial66–68. In vitro and in vivo studies demonstrate pathogenic potential but also show considerable inter and intra subtype variation, which provides a possible explanation on the conflicting reports on clinical significance. Blastocystis spp. have intestinal immunomodulatory effects and can release proteases that affect the integrity of the epithelium. This situation might facilitate colonization by other enteric pathogens either directly or by the resultant changes in the gut microbiota69.
Our findings unveil that children carrying Blastocystis spp. display a different microbial community compared to uninfected controls with a tendency to a Prevotella-driven enterotype. This is in concordance with similar studies carried out in adults around the world28–30. Blastocystis spp. showed a significant shift in the Prevotella proportion enriching it, favoring an enterotype switch. Prevotella strains are associated with plant-rich diets, including fibers, simple sugars, and carbohydrates, suggesting that it is a beneficial microbe70. However, Prevotella in the gut has also been linked with inflammatory diseases, which made it difficult to predict its behavior in any given gut ecosystem70. Our results are similar to those found in healthy children from several developing countries, who had a gut microbiome dominated at the genus rank by Prevotella71–73. High species diversity at this genus could be related to its different effects on host health.
Andersen et al. found that Blastocystis spp. colonization was positively associated with species richness, and this parameter was negatively correlated with the Bacteroides-driven enterotype. The authors concluded that Blastocystis spp. establishment in the intestine probably depends on the activity of certain types of bacteria that are generally not present in individuals with low richness colonic microbiota74. Since Blastocystis spp. is an obligated anaerobe, in order to survive, it should favor the predominance of bacterial taxa that maintains a strict anaerobic environment in the gut lumen75.
An interesting finding was that Akkermansia, a bacterium effective in increasing mucus thickness and gut barrier function, was reduced in the Blastocystis spp. infected group, suggesting that this protist poses an unfavorable condition to this beneficial bacterium. This reduction has been previously described, and it was related to specific Blastocystis subtypes, with an inverse correlation of subtypes 3 and 4 with Akkermansia, suggesting differential associations between subtypes and host health29. Other authors have also shown that Blastocystis spp. infection also leads to changes in the abundance of other groups of bacteria, reducing Bacteroides, and increasing Prevotella28,29,75.
Entamoeba coli colonized children showed a bacterial community that closely resembles the control group without a Prevotella-driven enterotype. We also found an increase in the relative abundance of the beneficial bacterium Akkermansia in this group, contrary to the pattern observed in the Blastocystis spp. infected children. This commensal amoeba probably contributes to maintaining gut favorable conditions to beneficial bacteria, like Akkermansia. Although it is challenging to fully interpret the role of any microorganism in a complex community such as the gut microbiota, our results indicate that commensal protists like Entamoeba coli could be related to a healthy status.
Nowadays, the definition of the pathogenicity of intestinal parasites should not only be restricted to its capacity to alter or invade the intestinal mucosa, but the alteration of the healthy gut microbiota might also be a cause of disease1,14,76,77. The alteration profile of the distal microbiota observed in the individuals colonized by Blastocystis spp. have been associated with intestinal bowel disease, and a reduced abundance of Akkermansia is associated with diseases like Atherosclerosis78, ulcerative colitis79, appendicitis80, overweight and obesity81. From this point of view, changes in the intestinal gut microbiota seem to correlate or exacerbate several diseases, so it should be considered at the moment of defining the pathogenic capacity of a parasite. It seems clear with the actual scientific evidence that Blastocystis spp. has the power to promote the displacement of the “healthy intestinal microbiota”, rendering the children more susceptible to other diseases thanks to the increase in Prevotella and the reduction of Akkermansia. By definition, a commensal parasite, like Entamoeba coli, should not affect the normal physiology of the host. However, the evidence shown in this paper add arguments in favor of the pathogenic behavior of Blastocystis spp. in children.
Methods
Population and sample selection
Children attending seven public daycare centers in the three nearby neighborhoods in Medellin, Colombia, were selected for this study. Feces samples were collected in screw-capped containers without any preservatives and then transported to the lab within a maximum period of 3 h. Samples for DNA extraction were frozen at − 70 °C for a period that did not exceed 7 days, and then DNA was extracted. The microscopical parasitological analysis was performed on the same day of the collection by observation of direct (fresh and iodine solution) and modified Ritchie concentrated stool samples. Modified Ziehl–Neelsen slides of the feces samples were also prepared and observed microscopically to detect intestinal apicomplexan parasites. Specific PCR for Cryptosporidium was performed using the protocol described by Xiao et al.82,83. All selected samples were negative with the test mentioned above for intestinal apicomplexan parasites.
Subjects were selected based on being positive for any of the following protists: Blastocystis spp., Entamoeba coli, or Endolimax nana. A control group negative for intestinal parasites was also included. These children received the same food (breakfast and lunch) while assisting the daycare centers and the age ranged from one to five years old (1yo, n = 2; 2yo, n = 11; 3yo, n = 16, 4yo, n = 9, 5yo, n = 11). By sex, the distribution was 17 females and 32 males. Enrolled children were classified into four groups:
NPDM: Control group of children with no positive results for parasites (n = 25). Blasto: Children parasitized only by Blastocystis spp., no other parasites observed (n = 11). E_coli: Children parasitized only by Entamoeba coli, no other parasites observed (n = 9). E_nana: Children parasitized only by Endolimax nana, no other parasites observed (n = 4). The Protist-infect group was set adding all the children of the Blasto, E_coli and E_nana groups (n = 24).
DNA extraction and 16S metataxonomic experiment
DNA extraction was carried with the STOOL DNA kit NORGEN (Canada). Extracted DNA was quantified using UV absorption and the Picogreen fluorescent method. The DNA quality was assessed by gel electrophoresis and control PCR amplifying the complete eDNA 16S gene with universal primers 27F and 1492R. For the metataxonomic experiment, the primers Bakt_341F CCTACGGGNGGCWGCAG and Bakt_805R GACTACHVGGGTATCTAATCC, that amplify the V3/V4 region, were used. The 16S metataxonomic experiment was hired to MACROGEN, Korea. An average of 100.000 reads per library was generated in the MiSeq platform with PE reads of 300 bases. For further bioinformatic analysis 50,000 reads were randomly selected for each sample with the program SEQTK (https://github.com/lh3/seqtk).
Bioinformatic analysis
Amplicon processing was carried out with MOTHUR V1.4284. Following the MiSeq standard operating protocol provided by the authors (https://www.mothur.org/wiki/MiSeq_SOP). Briefly, Amplicon forward and reverse read were merged and those containing Ns or homopolymers larger than 6 bases were removed. Then, the reads were mapped to the SILVA database, and only those that mapped to the V3/V4 region were kept. Chimera removal was performed with VSEARCH85. Reads per library were rarified with the totalgroup strategy to an average of 12,249 clean sequences. OTUs supported by less than 4 reads were excluded. A BIOM file was prepared with the MOTHUR function make.biom, for further statistical analysis in R language.
Statistics analysis
The statistical analysis was performed with package PHYLOSEQ in the R environment. The data was imported as a BIOM file generated with MOTHUR. Alpha diversity statistics were calculated and plotted in boxplots. The ordination plot was generated with NMDS and BRAY distances. Read counts per library were normalized with median sequencing depth. With these normalized counts, we compared taxa abundance at Phylum, family, and genus categories and generated the bar plots of the top ten most abundant taxa, the remaining groups were gathered in the others category. Statistical significance of the differences in the median values was performed with Kruskal–Wallis and pairwise Wilcoxon rank-sum test. The age effect on the Shannon index was assessed using the ‘lm’ function. It was adjusted by subtracting the age contribution to the index.
Permutational multivariate analysis of variance (PERMANOVA) was performed using the Adonis function in the vegan library of R with the Bray–Curtis dissimilarity matrix.
Ethics statement
The ethical clearance of this study was followed by the ethics of the Helsinki declaration and resolution No. 008430 of 1993 from the Ministry of Health from Colombia. The study was approved by the Ethics Committee from Sede de Investigación Universitaria, Universidad de Antioquia, under the official document No. 14-06-564. Parents or legal guardians of all the enrolled individuals signed informed consent.
Supplementary information
Acknowledgements
This project was funded by Colciencias grant (Convocatoria para proyectos de ciencia, tecnología e innovación en salud 2017- Project ID 1115-777-57608), Government of Colombia and Vicerrectoría de Investigación (CODI), Universidad de Antioquia grant number 2017-16171.
Author contributions
J.F.A., G.G.M., and A.G.D. were responsible for the general design of the project and data analysis. F.C. and M.T.L. helped with software implementations and data analysis. All authors reviewed the manuscript.
Data availability
Raw data is available upon publication at SRA website bioproject PRJNA572583. The datasets generated during and/or analyzed during the current study are also available from the corresponding author on reasonable request.
Competing of interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72286-y.
References
- 1.Tuddenham S, Sears CL. The intestinal microbiome and health. Curr. Opin. Infect. Dis. 2015;28:464–470. doi: 10.1097/QCO.0000000000000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonnenburg JL, Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 4.Leung JM, Graham AL, Knowles SCL. Parasite-microbiota interactions with the vertebrate gut: Synthesis through an ecological lens. Front. Microbiol. 2018;9:1–20. doi: 10.3389/fmicb.2018.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The human microbiome and child growth – first 1000 days and beyond. Trends Microbiol. 2019;27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg PB, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science (80-) 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019;16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naito Y, Uchiyama K, Takagi T. A next-generation beneficial microbe: Akkermansia muciniphila. J. Clin. Biochem. Nutr. 2018;63:33–35. doi: 10.3164/jcbn.18-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 18.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C-A, et al. Impact of Enterobius vermicularis infection and mebendazole treatment on intestinal microbiota and host immune response. PLoS Negl. Trop. Dis. 2017;11:e0005963. doi: 10.1371/journal.pntd.0005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosa BA, et al. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome. 2018;6:33. doi: 10.1186/s40168-018-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SC, et al. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl. Trop. Dis. 2014;8:e2880. doi: 10.1371/journal.pntd.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giacomin P, et al. Changes in duodenal tissue-associated microbiota following hookworm infection and consecutive gluten challenges in humans with coeliac disease. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep36797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess SL, Gilchrist CA, Lynn TC, Petri WA. Parasitic protozoa and interactions with the host intestinal microbiota. Infect. Immun. 2017;85:1–12. doi: 10.1128/IAI.00101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilchrist CA, et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. J. Infect. Dis. 2016;213:1579–1585. doi: 10.1093/infdis/jiv772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galván-Moroyoqui JM, del Carmen Domínguez-Robles M, Franco E, Meza I. The interplay between Entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl. Trop. Dis. 2008;2:1–12. doi: 10.1371/journal.pntd.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer LR, Verma AK, Paul J, Bhattacharya A. Phagocytosis of Gut Bacteria by Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2019;9:1–9. doi: 10.3389/fcimb.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieves-Ramírez ME, et al. Asymptomatic intestinal colonization with protist blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems. 2018;3:7–18. doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tito RY, et al. Gut microbiota population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut. 2019;68:1180–1189. doi: 10.1136/gutjnl-2018-316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsell J, et al. The relation between Blastocystis and the intestinal microbiota in Swedish travellers. BMC Microbiol. 2017;17:231. doi: 10.1186/s12866-017-1139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Safadi D, et al. Children of Senegal River Basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect. Dis. 2014;14:164. doi: 10.1186/1471-2334-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark CG, van der Giezen M, Alfellani MA, Stensvold CR. Recent developments in Blastocystis research. Adv. Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- 33.Bart, A. et al. Diagnosis and subtype analysis of Blastocystis sp. in 442 patients in a hospital setting in the Netherlands. BMC Infect. Dis.13, (2013). [DOI] [PMC free article] [PubMed]
- 34.Wawrzyniak I, et al. Blastocystis, an unrecognized parasite: An overview of pathogenesis and diagnosis. Ther. Adv. Infect. Dis. 2013;1:167–178. doi: 10.1177/2049936113504754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramírez JD, Flórez C, Olivera M, Bernal MC, Giraldo JC. Blastocystis subtyping and its association with intestinal parasites in children from different geographical regions of Colombia. PLoS ONE. 2017;12:1–13. doi: 10.1371/journal.pone.0172586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sánchez A, Munoz M, Gómez N, Tabares J, Segura L, Salazar A, Restrepo C, Ruíz M, Reyes P, Qian Y, Xiao L, López MC, Ramírez JD. Molecular epidemiology of Giardia, Blastocystis and Cryptosporidium among Indigenous children from the Colombian Amazon Basin. Front. Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiménez, P. A., Jaimes, J. E. & Ramírez, J. D. A summary of Blastocystis subtypes in North and South America. Parasit. Vectors12, (2019). [DOI] [PMC free article] [PubMed]
- 38.Skotarczak B. Genetic diversity and pathogenicity of blastocystis. Ann. Agric. Environ. Med. 2018;25:411–416. doi: 10.26444/aaem/81315. [DOI] [PubMed] [Google Scholar]
- 39.Stensvold CR, Clark CG. Current status of Blastocystis: a personal view. Parasitol. Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Rostami A, et al. The role of Blastocystis sp. and Dientamoeba fragilis in irritable bowel syndrome: a systematic review and meta-analysis. Parasitol. Res. 2017 doi: 10.1007/s00436-017-5535-6. [DOI] [PubMed] [Google Scholar]
- 41.Iqbal J, Khalid N, Hira PR. Cryptosporidiosis in Kuwaiti children: association of clinical characteristics with cryptosporidium species and subtypes. J. Med. Microbiol. 2011;60:647–652. doi: 10.1099/jmm.0.028001-0. [DOI] [PubMed] [Google Scholar]
- 42.Scanlan PD, et al. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014;90:326–330. doi: 10.1111/1574-6941.12396. [DOI] [PubMed] [Google Scholar]
- 43.Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 44.Petersen AM, et al. Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand. J. Gastroenterol. 2013;48:638–639. doi: 10.3109/00365521.2013.780094. [DOI] [PubMed] [Google Scholar]
- 45.Andersen LO, Stensvold CR. Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J. Clin. Microbiol. 2016;54:524–528. doi: 10.1128/JCM.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Audebert, C. et al. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep.6, (2016). [DOI] [PMC free article] [PubMed]
- 47.Iebba V, et al. Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte D’ivoire. J. Infect. Dev. Ctries. 2016;10:1035–1041. doi: 10.3855/jidc.8179. [DOI] [PubMed] [Google Scholar]
- 48.Beghini F, et al. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11:2848–2863. doi: 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popovic, A. et al. Design and application of a novel two-amplicon approach for defining eukaryotic microbiota. Microbiome6, (2018). [DOI] [PMC free article] [PubMed]
- 50.Nourrisson C, et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS ONE. 2014;9:e111868. doi: 10.1371/journal.pone.0111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yason, J. A., Liang, Y. R., Png, C. W., Zhang, Y. & Tan, K. S. W. Interactions between a pathogenic Blastocystis subtype and gut microbiota: In vitro and in vivo studies. Microbiome7, (2019). [DOI] [PMC free article] [PubMed]
- 52.Toro-Londono MA, Bedoya-Urrego K, Garcia-Montoya GM, Galvan-Diaz AL, Alzate JF. Intestinal parasitic infection alters bacterial gut microbiota in children. PeerJ. 2019;2019:1–10. doi: 10.7717/peerj.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batani G, Pérez G, Martínez de la Escalera G, Piccini C, Fazi S. Competition and protist predation are important regulators of riverine bacterial community composition and size distribution. J. Freshw. Ecol. 2016;31:609–623. [Google Scholar]
- 54.Bauer MA, Kainz K, Carmona-Gutierrez D, Madeo F. Microbial wars: Competition in ecological niches and within the microbiome. Microbial Cell. 2018;5:215–219. doi: 10.15698/mic2018.05.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poulsen CS, Stensvold CR. Systematic review on Endolimax nana: a less well studied intestinal ameba. Trop. Parasitol. 2016;6:8–29. doi: 10.4103/2229-5070.175077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burnstein SL, Liakos S. Parasitic rheumatism presenting as rheumatoid arthritis. J. Rheumatol. 1983;10:514–515. [PubMed] [Google Scholar]
- 57.Graczyk TK, et al. The association of Blastocystis hominis and Endolimax nana with diarrheal stools in Zambian school-age children. Parasitol. Res. 2005;98:38–43. doi: 10.1007/s00436-005-0003-0. [DOI] [PubMed] [Google Scholar]
- 58.Shah M, et al. Blastocystis hominis and Endolimax nana co-infection resulting in chronic diarrhea in an immunocompetent male. Case Rep. Gastroenterol. 2012;6:358–364. doi: 10.1159/000339205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kulik RA, Morais Falavigna DL, Nishi L, Marques Araujo S. Blastocystis sp. and other intestinal parasites in hemodialysis patients. Brazilian J. Infect. Dis. 2008;12:338–341. doi: 10.1590/s1413-86702008000400017. [DOI] [PubMed] [Google Scholar]
- 60.Veraldi S, Schianchi-Veraldi R, Gasparini G. Urticaria probably caused by Endolimax nana. Int. J. Dermatol. 1991;30:376–376. doi: 10.1111/j.1365-4362.1991.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 61.Veraldi S, Angileri L, Rossi LC, Nazzaro G. Endolimax nana and urticaria. J. Infect. Dev. Ctries. 2020;14:321–322. doi: 10.3855/jidc.12389. [DOI] [PubMed] [Google Scholar]
- 62.Sard BG, Navarro RT, Esteban Sanchis JG. Amebas intestinales no patógenas: una visión clinicoanalítica. Enferm. Infecc. Microbiol. Clin. 2011;29:20–28. doi: 10.1016/S0213-005X(11)70023-4. [DOI] [PubMed] [Google Scholar]
- 63.Brumpt E. Blastocystis hominis n sp. et formes voisines. Bull Soc Pathol Exot. 1912;5:725–730. [Google Scholar]
- 64.Alexeieff A. Sur la nature des formations dites kystes de Trichomonas intestinalis. C. R. Soc. Biol. 1911;71:296–298. [Google Scholar]
- 65.Zierdt CH. Blastocystis hominis–past and future. Clin. Microbiol. Rev. 1991;4:61–79. doi: 10.1128/cmr.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scanlan PD. Blastocystis: past pitfalls and future perspectives. Trends Parasitol. 2012;28:327–334. doi: 10.1016/j.pt.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 67.Leder K, Hellard ME, Sinclair MI, Fairley CK, Wolfe R. No correlation between clinical symptoms and Blastocystis hominis in immunocompetent individuals. J. Gastroenterol. Hepatol. 2005;20:1390–1394. doi: 10.1111/j.1440-1746.2005.03868.x. [DOI] [PubMed] [Google Scholar]
- 68.Stensvold CR, van der Giezen M. Associations between gut microbiota and common luminal intestinal parasites. Trends Parasitol. 2018;34:369–377. doi: 10.1016/j.pt.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 69.Ajjampur SSR, Tan KSW. Pathogenic mechanisms in Blastocystis spp.—interpreting results from in vitro and in vivo studies. Parasitol. Int. 2016;65:772–779. doi: 10.1016/j.parint.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016;13:69. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh TS, et al. Gut Microbiomes of Indian Children of Varying Nutritional Status. PLoS ONE. 2014;9:e95547. doi: 10.1371/journal.pone.0095547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monira S, et al. Gut microbiota of healthy and Malnourished children in Bangladesh. Front. Microbiol. 2011;2:228. doi: 10.3389/fmicb.2011.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kisuse J, et al. Urban diets linked to gut microbiome and metabolome alterations in children: a comparative cross-sectional study in Thailand. Front. Microbiol. 2018;9:1345. doi: 10.3389/fmicb.2018.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andersen, L. O. B., Bonde, I., Nielsen, H. B. H. B. & Stensvold, C. R. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol. Ecol.91, (2015). [DOI] [PubMed]
- 75.O’Brien Andersen L, et al. Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1427–1431. doi: 10.1007/s10096-016-2680-2. [DOI] [PubMed] [Google Scholar]
- 76.Almeida A, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Partida-Rodríguez O, et al. Human intestinal microbiota: interaction between parasites and the host immune response. Arch. Med. Res. 2017;48:690–700. doi: 10.1016/j.arcmed.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 78.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe-/- Mice. Circulation. 2016;133:2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- 79.Vigsnæs LK, Brynskov J, Steenholdt C, Wilcks A, Licht TR. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef. Microbes. 2012;3:287–297. doi: 10.3920/BM2012.0018. [DOI] [PubMed] [Google Scholar]
- 80.Swidsinski A, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 81.Depommier C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao L, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiao L, et al. Molecular characterization of cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 2001;67:1097–1101. doi: 10.1128/AEM.67.3.1097-1101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data is available upon publication at SRA website bioproject PRJNA572583. The datasets generated during and/or analyzed during the current study are also available from the corresponding author on reasonable request.