Abstract
Purpose
Clear cell renal cell carcinoma (ccRCC) is among the most common malignant tumors worldwide, with a high incidence rate and poor prognosis. Currently, there are no biomarkers that can accurately guide prognostic evaluation and therapeutic strategy for ccRCC. The prognostic value and potential biological function of claudin-8 (CLDN8), a critical component of tight junctions in ccRCC, remain unclear.
Methods
Sequencing data were obtained from The Cancer Genome Atlas, International Cancer Genome Consortium, and Gene Expression Omnibus databases. R packages were used to explore CLDN8 mRNA expression levels and analyze differentially expressed genes. Results were validated in clinical specimens and cell lines, and bioinformatics analyses were conducted to explore the potential biological functions of CLDN8. Finally, functional analyses were carried out using 786–O ccRCC cell line.
Results
Both CLDN8 mRNA and protein expression levels were significantly lower in ccRCC compared with the normal control tissues. Kaplan–Meier analyses showed that low CLDN8 expression levels were associated with the poor overall survival, while univariate and multivariate Cox regression indicated that CLDN8 could serve as an independent prognostic factor in patient with ccRCC. Bioinformatic and Western blot analyses showed that CLDN8 suppressed proliferation, migration, and invasion of 786–O ccRCC cells through the epithelial–mesenchymal transition and AKT pathways.
Conclusion
CLDN8 could serve as an independent prognostic factor in ccRCC, in which it suppresses 786–O proliferation, migration, and invasion through EMT and AKT pathways.
Keywords: CLDN8, clear cell renal cell carcinoma, biomarker, prognosis, potential biological functions
Introduction
Renal cell carcinoma (RCC) is one of the most common malignant tumors of the genitourinary system with a high incidence rate and poor patient prognosis.1,2 Clear cell RCC (ccRCC) is the major histologic subtype of RCC, accounting for 70–80% cases.3 Although advances in imaging technology mean that patients increasingly being detected when the disease is at an early stage 30% of those with ccRCC still have metastasis at initial diagnosis.4 More importantly, despite the implementation of nephrectomy or targeted therapies, the 5-year survival rate for patients with metastatic ccRCC is less than 10%.5 Further, the TNM staging system currently applied in clinical practice is considered relatively inaccurate for prognostic evaluation.6 Hence, exploring new biomarkers to guide diagnosis and prognostic evaluation in patient with ccRCC remains of great significance.
Claudins are integral membrane proteins of 20–37 kDa in size that are critical components of tight junctions (TJs).7 TJs appear to have vital roles in the process of metastasis, during which they are loosened or dismantled in cancer cells,8 and claudins are crucial modulators of carcinogenesis and metastasis. Many studies have shown altered expression of claudins in numerous epithelial-derived tumors, including lung, prostate, and ovarian cancers.9–11 As a member of the claudin family, CLDN8 shows significantly altered expression between ccRCC and normal kidney tissues in GEPIA database. Previous studies demonstrated that CLDN8 expression was significantly correlated with prognosis in patients with breast and prostate cancers,12,13 however, there have been few literature about the prognostic value and potential biological functions of CLDN8 in ccRCC to date.
In this work, CLDN8 mRNA and protein expression levels were examined using the data from online databases, cell lines, and our clinical specimens. Then, the prognostic value of CLDN8 was evaluated in patients with ccRCC. Following this, the potential biological functions of CLDN8 were explored using bioinformatic analyses and experiments with ccRCC cell lines.
Materials and Methods
Bioinformatics and Data Processing
Data including mRNA transcript profiles (HTSeq-FPKM) and corresponding clinical information were obtained from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/), including 539 ccRCC and 72 normal tissue samples. In addition, nine microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/). More than 700 specimens were obtained from nine GEO datasets: GSE4282, GSE46699, GSE53757, GSE15641, GSE68417, GSE14994, GSE40435, GSE71963, and GSE76351. Moreover, a total of 91 RCC and 45 normal tissue samples were downloaded from the International Cancer Genome Consortium (ICGC) database (https://icgc.org). Normalization was conducted by log2 transformation after adding a 1 pseudocount, based on the pre-processed data. The sangerbox online tool (http://sangerbox.com/) and HPA database (https://www.proteinatlas.org/) were used to further explore CLDN8 mRNA and protein levels.
Patients and Specimens
A total of 148 paraffin-embedded samples from patients diagnosed with ccRCC were selected from the Peking University First Hospital (PUFH) between June 2008 and January 2011 and the clinical information from those patients obtained from medical records. Moreover, 20 paired ccRCC and adjacent normal tissues were obtained from patients with ccRCC undergoing surgical resection at PUFH. This study was approved by the Ethics Committee of PUFH, and all patients signed informed consent. All procedures were performed according to the World Medical Association Declaration of Helsinki.
Immunohistochemistry (IHC)
Sections were prepared from the paraffin-embedded tissue samples. Immunostaining was performed using a two-step detection kit (Zsbio PV-9000, China). Sections were dewaxed, rehydrated, and then boiled in citrate buffer (pH 6.0) for 30 min. Endogenous peroxidase was blocked by incubation in 3% H2O2 for 30 min at room temperature. Sections were then washed in PBS solution and subsequently and blocked with 10% goat serum (Zsbio, China) for 1 h and incubated with anti-CLDN8 (ab211439,1:20,000, Abcam) at 4°C overnight, followed by HRP-conjugated secondary antibody for 30 min and counterstaining with diaminobenzidine (DAB) solution. Two experienced independent investigators assessed all tumor slides by examining at five random views and observing 100 cells per view at ×400 magnification. Staining intensity was classified as 0 (no staining), 1 (weak), 2 (moderate), or 3 (strong), and the proportion of stained tumor cells was scored as 0 (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), or 4 (>75%). The multiplication of these two variables was calculated as a final score: 0 (score 0–3); 1 (score 4–6); 2 (score 7–9); or 3 (score 10–12).14
Cell Culture and Transfection
Seven human kidney and ccRCC cell lines (HK-2, 786-O, 769-P, ACHN, Caki-1, OSCR2, and A498) were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM or RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) in a 5% CO2 humidified atmosphere at 37°C.
We amplified a target mRNA CLDN8 product using PCR, incorporating the enzyme sites XhoI and BamHI for cloning into the pLVX vector, using the following primers: forward CLDN8 primer: 5ʹ- CCGCTCGAGGCCACCATGGCAACCCATGCCTTAG-3ʹ; The reverse CLDN8 primer: 5ʹ- CGGGATCCCTACACATACTGACTTCTGGAGTAGAC-3ʹ. Following this, we transformed the recombinant plasmid into Escherichia.coli and picked a single monoclonal colony. Then, the plasmid was sent for sequencing by a commercial company (TIANYI HUIYUAN, Ltd), to ensure its accuracy, followed by packaging into a lentivirus. Next, the lentivirus was transformed into 1 × 104 786-O cells at a multiplicity of infection (MOI) of = 50. Finally, a stable strain was screened using blasticidin (5 µg/mL).
Real-Time Quantitative PCR (qPCR)
Total RNA was extracted from 20 paired clinical samples and six cell lines using a Takara kit, according to the manufacturer’s protocol. Then, the RNA was reverse transcribed to generate cDNA in a 20µL reaction system. Gene transcripts were quantified by qPCR using a SYBR Premix ExTaq kit, with α-Tubulin as a control for normalization. Primer sequences used for qPCR were shown as follows: forward CLDN8 primer: 5ʹ-TGAATGTTGCCCAAAAACGTG-3ʹ, reverse CLDN8 primer: 5ʹ- GCGATGGGAAGGTATCGAGTATC-3ʹ. Each reaction was performed four times, and the 2−ΔΔCT method used to calculate relative mRNA expression level.
Western Blot Analysis
Total proteins were extracted from cells using NP-40 lysis buffer and quantified using the bicinchonininc acid (BCA) method. Samples were transferred to polyvinylidene fluoride membranes and incubated overnight at 4°C with antibody against CLDN8 (1:1000) or β-actin (1:1000). After incubation with peroxidase-coupled anti-rabbit IgG (1:1000) at 37°C for 2 h, bound proteins were visualized using ECL (Pierce) and detected using a DNR Bioimaging System (DNR, Jerusalem, Israel). Relative protein levels were quantified using β-actin as the loading control. Detailed information regarding the antibodies used is presented in supplementary Table S1.
Bioinformatics Analyses
X-tile was used to divide ccRCC samples from the TCGA database into low (n = 385) and high (n = 145), according to CLDN8 expression. The “Limma” R package was then used to screen for differentially expressed genes (DEGs) between the groups. DEGs with absolute log2foldchange>3, and false discovery rate (FDR) <0.001 were considered statistically significant.
Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis were performed using the “clusterprofiler” R package (version 3.6.1). P < 0.05 was considered as statistically significant.15
Gene set enrichment analysis (GSEA) was performed to identify potential biological pathways. GSEA software (http://software.broadinstitute.org/gsea/index.jsp) was run on the JAVA 8.0 platform. Using information from TCGA, samples were classified into high and low groups, according to average CLDN8 mRNA expression. For each analysis, gene set permutations were implemented 1000 times to identify significantly enriched gene sets, defined as those with FDR q-value <0.05.16
Wound Healing Assay
A total of 5 × 105 786-O cells in logarithmic growth were inoculated in each wells of 6-well plates. The cells were cultured in RPMI 1640 medium containing 10% FBS for 6 h. Subsequently, wounds were created in each well and growth medium replaced with serum-free medium. Based on the growth characteristics of 786-O cells, observations and photography were conducted at 0, 6, and 24 h.
Clone Formation Experiment
A total of 1 × 103 786-O cells in logarithmic growth were inoculated in each well of 6-well plates. Cells were then cultured in RPMI 1640 medium containing 10% FBS for 2 weeks, fixed, and stained with 0.5% crystal violet. Colonies were counted under a microscope.
Migration and Invasion Experiment
Cells (786-O) in logarithmic growth transformed with vector only (control) or plasmid expressing CLDN8 were harvested by trypsinization, then 1 × 104 added to each insert of the upper chambers in 24-well transwell chambers, with RPMI 1640 containing no FBS. For the invasion experiment, before preparing the cells, a dried layer of Matrigel matrix (Corning, USA) was rehydrated with RPMI 1640 medium for 2 h at room temperature. Plates were incubated for 24 h, and then the cells fixed and stained with 0.5% crystal violet.
Cell Proliferation Assay
Cells were seeded in 96-well assay plates and subjected MTS analysis using a CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega). For MTS analysis, 100μL of cells in culture medium were added to each well of the 96-well plate, 20μL reagent pipetted into each well, and the plates incubated for 1 h. A standard curve was created according to the OD values corresponding to different numbers of cells. Finally, optical density (OD) values at 490 nm were used to evaluate cell viability.
Statistical Analyses
The Student’s t-test was used to calculate mRNA and protein expression levels in ccRCC and adjacent normal renal tissues, using SPSS 26.0 (IBM). One-way ANOVA was conducted to evaluate differences among multiple groups, and Bonferroni’s multiple comparisons test was used as a post hoc test. Patients with NX, MX, GX, or missing values were excluded from subsequent analyses. The Kaplan–Meier method and Cox regression were used to compare the impact of CLDN8 expression on patient overall survival (OS), alongside other clinical variables from both TCGA and our clinical database. All p-values are based on two-sided statistical analysis, and p < 0.05 was considered to indicate statistical significance.
Results
CLDN8 is Downregulated in ccRCC in Data from Inline Databases
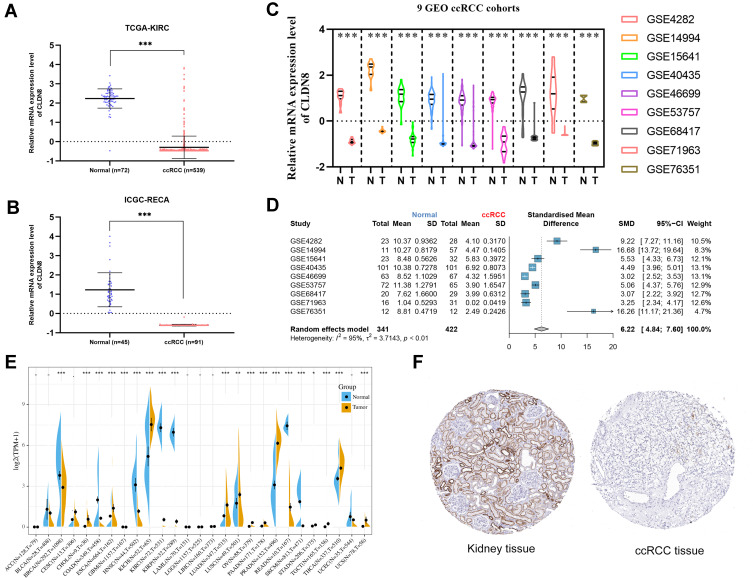
We first examined the CLDN8 mRNA expression level in TCGA, GEO, and ICGC databases. In TCGA and ICGC data, CLDN8 was observed significantly downregulated in ccRCC compared with normal controls (Figure 1A and B). Further, CLDN8 mRNA levels were remarkable decreased ccRCC samples in nine GEO datasets (Figure 1C). We also constructed a forest plot for meta-analysis (Figure 1D).
Figure 1.
CLDN8 expression levels in the online database. CLDN8 mRNA expression level. (A and B) Relative CLDN8 mRNA expression levels in ccRCC according to the TCGA and ICGC RNA sequencing data. (C) Relative CLDN8 mRNA expression levels in nine GEO datasets. (D) A meta-analysis of nine GEO datasets. (E) CLDN8 mRNA expression levels between cancer and normal controls among 27 types of tumors. (F) Representative images of ccRCC and paracancerous immunohistochemistry in the HPA database (*p<0.05, **p<0.01, ***p<0.001).
Next, we used the sangerbox online tool to compare CLDN8 mRNA expression levels between normal and cancerous tissues from 27 types of tumor (Figure 1E). Significantly altered CLDN8 expression was detected in multiple cancers. Additionally, representative protein expression images for CLDN8 in ccRCC and normal tissues from the HPA database were evaluated (Figure 1F).
CLDN8 is Downregulated in ccRCC Clinical Specimens and Various Cell Lines
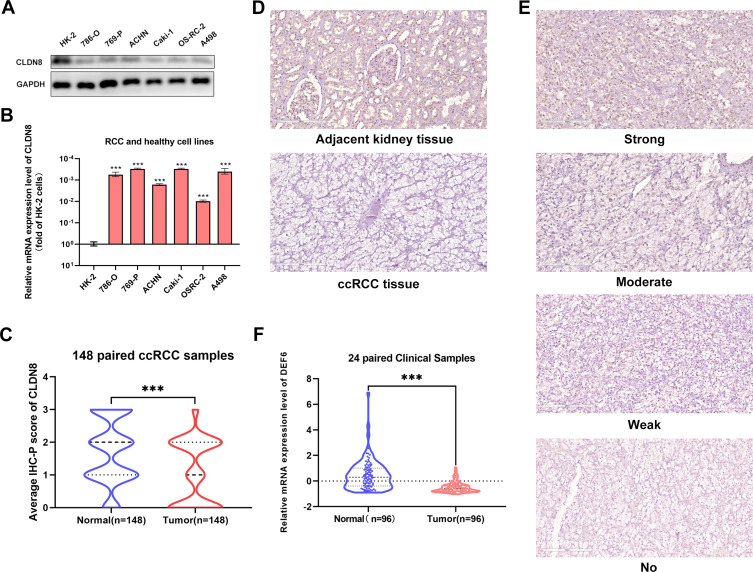
CLDN8 expression levels were assessed in clinical specimens and cell lines. Western blot analysis revealed lower CLDN8 protein levels in RCC cell lines compared with normal kidney cell lines (Figure 2A). Further, qPCR analysis showed lower CLDN8 mRNA levels in RCC cell lines compared to normal kidney cell lines (Figure 2B). Following this, qPCR was conducted using 24 paired randomly selected ccRCC specimens and confirmed that CLDN8 mRNA levels were lower in ccRCC tissues compared with the normal controls (Figure 2C). CLDN8 mRNA expression levels in normal and ccRCC tissues for each specimen are presented in Supplementary Figure S1.
Figure 2.
CLDN8 expression levels in the specimens and cell lines. (A and B) Western blot and qPCR analysis of ccRCC cell lines and normal cell lines. (C) The CLDN8 mRNA expression in ccRCC tissues and paired normal tissues in 20 paired clinical samples. (D) Representative adjacent normal renal tissue staining and ccRCC tissue staining are shown. (E) Four representative images of CLDN8 expression were interpreted by immunohistochemistry as no, weak, moderate, and strong. (F) The average scores of immunohistochemical staining were shown (***p<0.001).
Subsequently, IHC staining of 148 paired clinical specimens was performed. Representative IHC staining images (200 μm) of CLDN8 in normal and ccRCC specimens are shown in Figure 2D. Further, as shown in Figure 2E, the representative IHC staining in ccRCC specimens revealed no, weak, moderate, and strong staining (200 μm). The results indicated that CLDN8 protein expression was lower in ccRCC tissues than that in normal control specimens (Figure 2F).
Association of CLDN8 Expression with ccRCC Patient Survival
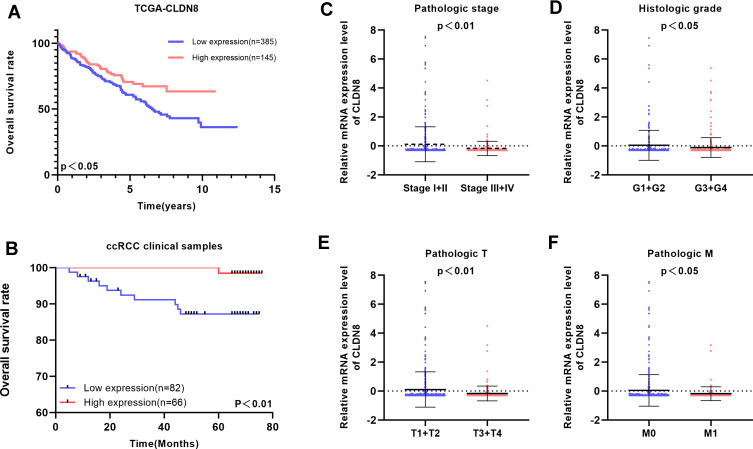
To explore the relationship between CLDN8 expression and the prognosis of patients with ccRCC, we analyzed samples from TCGA. The baseline information for patients with ccRCC is presented in Table 1. Kaplan–Meier analyses indicated that patients with low CLDN8 expression levels had inferior OS (Figure 3A). Moreover, lower CLDN8 expression was significantly associated with higher AJCC stage, ISUP grade, pT stage, and pM stage (Figure 3C–F). Further, univariate Cox regression analyses showed that CLDN8 expression, age, grade, T, M, and stage were all remarkable risk factors (Table 2), while, multivariate Cox regression analyses confirmed that CLDN8 expression, age, grade, T, M, and stage were all independent factors influencing prognosis in patients with ccRCC (Table 2).
Table 1.
Clinicopathological Parameters Baseline in TCGA-KIRC Cohort
| Parameters | TCGA-KIRC Cohort (n=530) | N (%) |
|---|---|---|
| Age | ||
| ≤60 | 264 | 49.81 |
| >60 | 266 | 50.19 |
| Gender | ||
| Male | 344 | 64.91 |
| Female | 186 | 35.09 |
| pT stage | ||
| T1 | 271 | 51.13 |
| T2 | 69 | 13.02 |
| T3 | 179 | 33.77 |
| T4 | 11 | 2.08 |
| pN stage | ||
| N0 | 239 | 45.09 |
| N1 | 16 | 3.02 |
| NX | 275 | 51.89 |
| pM stage | ||
| M0 | 420 | 79.25 |
| M1 | 78 | 14.72 |
| MX | 32 | 6.04 |
| AJCC stage | ||
| Stage I | 265 | 50.00 |
| Stage II | 57 | 10.75 |
| Stage III | 123 | 23.21 |
| Stage IV | 83 | 15.66 |
| Unknown | 2 | 0.38 |
| ISUP grade | ||
| G1 | 14 | 2.64 |
| G2 | 227 | 42.83 |
| G3 | 206 | 38.87 |
| G4 | 75 | 14.15 |
| GX | 8 | 1.51 |
Figure 3.
Clinical significance of CLDN8 expression in ccRCC patients. (A and B) Kaplan–Meier survival curves for OS of CLDN8 in TCGA and our clinical database are shown. (C–F) The correlation between CLDN8 expression and clinicopathological characteristics in ccRCC patients was explored in the TCGA database, including pathologic stage (p<0.01), histologic grade (p<0.05), pathologic T stage (p<0.01), pathologic M stage (p<0.05).
Table 2.
Univariate and Multivariate Analyses in TCGA Database
| Variables | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| CLDN8 (High vs Low) | 0.538 (0.369,0.784) | 0.001 | 0.674 (0.460,0.986) | 0.042 |
| Age (>60 vs ≤60) | 1.751 (1.282,2.391) | <0.001 | 1.636 (1.195,2.240) | 0.002 |
| Gender (Male vs Female) | 0.940 (0.686,1.288) | 0.701 | ||
| Grade (G3+G4 vs G1+G2) | 2.588 (1.834,3.652) | <0.001 | 1.647 (1.143,2.373) | 0.007 |
| T (T3+T4 vs T1+T2) | 3.040 (2.233,4.139) | <0.001 | 1.799 (1.137,2.847) | 0.012 |
| M (M1 vs M0) | 4.258 (3.109,5.832) | <0.001 | 2.373 (1.628,3.459) | <0.001 |
| Stage (III+IV vs I+II) | 3.661 (2.655,5.048) | <0.001 | 1.974 (1.331,2.929) | 0.001 |
Subsequently, we validated TCGA outcome data in 148 paired specimens. Consistent with previous results, patients with low CLDN8 expression had worse prognosis (Figure 3B). Univariate and multivariate Cox analyses showed that CLDN8 expression was an independent indicator of poor prognosis in patients with ccRCC after adjustment for age, gender, Fuhrman score, T, M, N, and stage (Table 3).
Table 3.
Univariate and Multivariate Analyses in Clinical Samples
| Variables | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR(95% CI) | p-value | HR(95% CI) | p-value | |
| CLDN8 (High vs Low) | 0.108 (0.014,0.846) | 0.034 | 0.099 (0.012,0.798) | 0.030 |
| Age (>60 vs ≤60) | 2.510 (0.735,8.577) | 0.142 | ||
| Gender (Male vs Female) | 4.605 (0.589,35.975) | 0.145 | ||
| Fuhrman (III+IV vs I+II) | 9.930 (3.003,32.829) | <0.001 | 6.336 (1.755,22.875) | 0.005 |
| T (T3+T4 vs T1+T2) | 7.173 (2.083,24.702) | 0.002 | ||
| M (M1 vs M0) | 146.499 (9.163,2342.166) | <0.001 | 57.879 (2.799,1196.940) | 0.009 |
| N (N1 vs N0) | 35.736 (3.994,319.754) | 0.001 | ||
| Stage (III+IV vs I+II) | 7.173 (2.083,24.702) | 0.002 | ||
Functional Enrichment Analyses of CLDN8
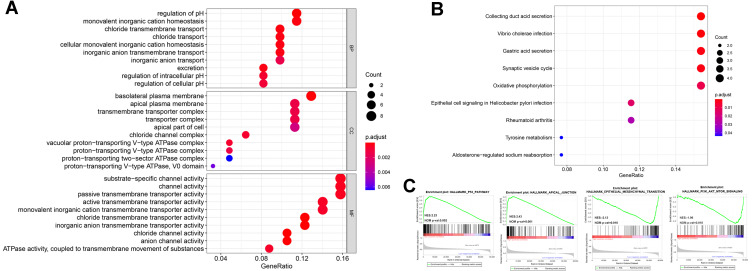
GO, KEGG, and GESA functional enrichment analyses were used to explore the potential biological functions of CLDN8. First, DEGs were classified into three functional groups, based on GO analysis, including biological processes (BP), molecular function (MF), and cell composition (CC). As shown in Figure 4A, DEGs were primarily enriched for BP including regulation of pH and ion transport; For CC comprising, ion-exchange-related plasma membranes and ATPase-related complex; and for several MFs involved in channel and transporter activities. KEGG analysis indicated that DEGs were mainly enriched in pathways involved in acid secretion and metabolism (Figure 4B), while, GESE enrichment analysis identified several representative pathways related to ccRCC, such as AKT and P53 signalings (Figure 4C).
Figure 4.
Bioinformatics analyses of the potential biological pathway. (A and B) GO and KEGG enrichment analyses were constructed, where larger dot size is correlated with higher counts and darker red color is related to the lower p-value. (C) Four representative GSEA results were shown.
Exploration of CLDN8 Biological Functions Using 786-O Cells
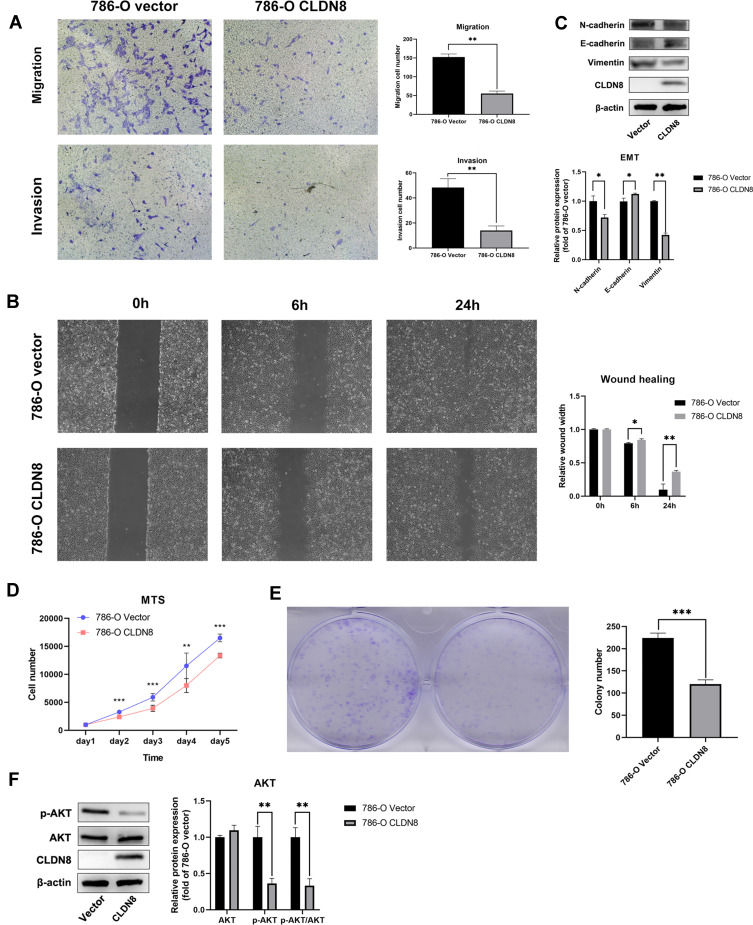
Based on the results of bioinformatics analyses, we further explored the biological functions of CLDN8 in 786-O cells. Transwell assays indicated that overexpression of CLDN8 decreased the migration and invasion abilities of ccRCC cells (Figure 5A), while wound healing was also reduced in ccRCC cells overexpressing CLDN8 (Figure 5B). To further investigate the relevant molecular mechanisms, we performed Western blot analysis and found that N-cadherin and vimentin were downregulated in 786-O CLDN8 cells while E-cadherin was upregulated, suggesting that CLDN8 may regulate the process of epithelial to mesenchymal transition (EMT) to influence the ccRCC cell migration and invasion (Figure 5C). Furthermore, MTS and colony formation experiments indicated that overexpression of CLDN8 could impair the proliferation ability of ccRCC cells (Figure 5D and 5E), while p-AKT and p-AKT/AKT were significantly downregulated in 786-O CLDN8 cells (Figure 5F), indicating that CLDN8 may regulate ccRCC cell proliferation via the AKT pathway. These findings were consistent with those of GSEA.
Figure 5.
CLDN8 regulates ccRCC cells in vitro. (A and B) Overexpression of CLDN8 inhibited the migration and invasion of ccRCC cells. (C) The effect of overexpression of CLDN8 on the EMT pathway was detected by Western blot. (D and E) Overexpression of CLDN8 inhibited the proliferation of ccRCC cells. (F) The effect of overexpression of CLDN8 on the AKT pathway was detected by Western blot (*p<0.05, **p<0.01, ***p<0.001).
Discussion
Among kidney cancers, ccRCC is the most common subtype and is often diagnosed with metastasis, resulting in poor patient prognosis.1,17 Currently, diagnosing ccRCC mainly depends on computed tomography, magnetic resonance imaging, and pathologic examination, with no specific diagnostic biomarkers.2,18 Under these circumstances, patients with ccRCC are often at an advanced stage when initially diagnosed. Further, the treatment of metastatic ccRCC is one of the thorniest problems in urology.17,19
The emergence of bioinformatics has contributed to addressing the above issues. As a reliable established bioinformatics tool, TCGA database is an invaluable resource that provides the foundation for the development of diagnostic and therapeutic methods for ccRCC.20 Through the use of TCGA data, many different prognostic biomarkers have been identified for ccRCC, including miRNAs, lncRNAs, and ceRNAs,21–23 however, few of these biomarkers are suitable for use in clinical practice. Hence, identification of new biomarkers for diagnostic and therapeutic purposes continues to be a research focus.
Claudin family members are strongly associated with TJs.7 More importantly, TJs must be loosened in cancer cells during the metastatic process, to enable migration and invasion.24 As a member of the claudin family, CLDN8 has been associated with basic kidney functions.25 Previous studies have shown that CLND8 expression levels are significantly altered in some tumors.12,26 GEIPA data show large differences in CLND8 expression levels between ccRCC and normal kidney tissue, hence exploration of the prognostic value and potential biological function of CLDN8 were evaluated.
In this study, both CLDN8 mRNA and protein expression levels were downregulated in ccRCC tissues relative to normal control specimens and cell lines, which is consistent with the results found in online databases. Furthermore, CLDN8 was strongly correlated with several clinical parameters, such as pathologic stage and histologic grade, while univariate and multivariate Cox regression analysis indicated that CLDN8 expression could serve as an independent prognostic factor for OS in patients with ccRCC.
We also conducted bioinformatic analyses to explore the potential biological functions of CLDN8 in regulating ccRCC. As noted in previous reports, GO and KEGG analyses showed that CLDN8 was strongly associated with numerous basic functions of the kidney, including regulation of pH and control of the transport of substances.25,27 Moreover, ccRCC is a metabolic disease, characterized by metabolic reprogramming.28,29 The metabolic flux through glycolysis is partitioned and mitochondrial bioenergetics and oxidative phosphorylation are impaired in ccRCC, as well as lipid metabolism,30 as supported by the results of GO and KEGG analyses in this study, which indicated a role for CLDN8 in oxidative phosphorylation. GSEA analyses showed enrichment in several tumor-related pathways. The results of our experiments using a ccRCC cell line overexpressing CLDN8 showed that CLDN8 could suppress the proliferation, migration, and invasion of ccRCC cells via AKT and EMT pathways, corroborating the results of the bioinformatics analyses.
This study has several limitations. Firstly, we are limited by the sample size in TCGA and clinical specimens, which may have resulted slight bias. Second, more additional clinical indicators should be included to optimize the prognostic value of CLDN8. Third, more in-depth analysis of the molecular biological mechanisms underlying the role of CLDN8 in regulating ccRCC should be specifically explored in future studies.
In conclusion, this study is the first to confirm the prognostic value and potential biological function of CLDN8 in ccRCC. Our data indicate that CLDN8 could serve as an independent prognostic factor in ccRCC, and that suppresses proliferation, migration, and invasion of ccRCC cells via EMT and AKT pathways.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.81670617). The authors also would like to thank Xin Gao and Anbang He, who thoroughly copyedited the manuscript for language use, spelling, and grammar.
Data Sharing Statement
The datasets used in the current study are available from TCGA (http://cancergenome.nih.gov/), and the GEO (https://www.ncbi.nlm.nih.gov/), ICGC (https://icgc.org), and HPA (https://www.proteinatlas.org/).
Ethics Statement
This study was approved by the Ethics committee of Peking University First Hospital, and patients’ informed consent was acquired before the study. All procedures were consistent with the declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1). [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67(5):913–924. doi: 10.1016/j.eururo.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 3.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029 [DOI] [PubMed] [Google Scholar]
- 4.Gray RE, Harris GT. Renal cell carcinoma: diagnosis and management. Am Fam Physician. 2019;99(3):179–184. [PubMed] [Google Scholar]
- 5.Mitsui Y, Shiina H, Kato T, et al. Versican promotes tumor progression, metastasis and predicts poor prognosis in renal carcinoma. Mol Cancer Res. 2017;15(7):884–895. doi: 10.1158/1541-7786.MCR-16-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verine J, Colin D, Nheb M, et al. Architectural patterns are a relevant morphologic grading system for clear cell renal cell carcinoma prognosis assessment: comparisons with WHO/ISUP grade and integrated staging systems. Am J Surg Pathol. 2018;42(4):423–441. doi: 10.1097/PAS.0000000000001025 [DOI] [PubMed] [Google Scholar]
- 7.Tsukita S, Tanaka H, Tamura A. The claudins: from tight junctions to biological systems. Trends Biochem Sci. 2019;44(2):141–152. doi: 10.1016/j.tibs.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 8.Tabariès S, Siegel PM. The role of claudins in cancer metastasis. Oncogene. 2017;36(9):1176–1190. doi: 10.1038/onc.2016.289 [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto T, Oshima T, Yoshihara K, et al. Reduced expression of claudin-7 is associated with poor outcome in non-small cell lung cancer. Oncol Lett. 2010;1(3):501–505. doi: 10.3892/ol_00000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landers KA, Samaratunga H, Teng L, et al. Identification of claudin-4 as a marker highly overexpressed in both primary and metastatic prostate cancer. Br J Cancer. 2008;99(3):491–501. doi: 10.1038/sj.bjc.6604486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahiya N, Becker KG, Wood WH, Zhang Y, Morin PJ, Campbell M. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS One. 2011;6(7):e22119. doi: 10.1371/journal.pone.0022119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Zheng A, Lu H, Jin Z, Peng Z, Jin F. The expression and prognostic significance of claudin-8 and androgen receptor in breast cancer. Onco Targets Ther. 2020;13:3437–3448. doi: 10.2147/OTT.S242406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashikari D, Takayama K-I, Obinata D, Takahashi S, Inoue S. CLDN8, an androgen-regulated gene, promotes prostate cancer cell proliferation and migration. Cancer Sci. 2017;108(7):1386–1393. doi: 10.1111/cas.13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, He A, Lv T, Xu C, Lin L, Lin J. Overexpression of P4HB is correlated with poor prognosis in human clear cell renal cell carcinoma. Cancer Biomark. 2019;26(4):431–439. doi: 10.3233/CBM-190450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angulo JC, Shapiro O. The changing therapeutic landscape of metastatic renal cancer. Cancers (Basel). 2019;11(9):1227. doi: 10.3390/cancers11091227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedke J, Gauler T, Grünwald V, et al. Systemic therapy in metastatic renal cell carcinoma. World J Urol. 2017;35(2):179–188. doi: 10.1007/s00345-016-1868-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linehan WM, Ricketts CJ. The cancer genome atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. 2019;16(9):539–552. doi: 10.1038/s41585-019-0211-5 [DOI] [PubMed] [Google Scholar]
- 21.Luo Y, Chen L, Wang G, Xiao Y, Ju L, Wang X. Identification of a three-miRNA signature as a novel potential prognostic biomarker in patients with clear cell renal cell carcinoma. J Cell Biochem. 2019;120(8):13751–13764. doi: 10.1002/jcb.28648 [DOI] [PubMed] [Google Scholar]
- 22.Zeng J-H, Lu W, Liang L, et al. Prognosis of clear cell renal cell carcinoma (ccRCC) based on a six-lncRNA-based risk score: an investigation based on RNA-sequencing data. J Transl Med. 2019;17(1):281. doi: 10.1186/s12967-019-2032-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao K, Zhang Q, Wang Y, et al. The construction and analysis of competitive endogenous RNA (ceRNA) networks in metastatic renal cell carcinoma: a study based on the cancer genome atlas. Transl Androl Urol. 2020;9(2):303–311. doi: 10.21037/tau.2020.02.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh AB, Dhawan P. Claudins and cancer: fall of the soldiers entrusted to protect the gate and keep the barrier intact. Semin Cell Dev Biol. 2015;42:58–65. doi: 10.1016/j.semcdb.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 25.Leiz J, Schmidt-Ott KM. Claudins in the renal collecting duct. Int J Mol Sci. 2019;21(1):221. doi: 10.3390/ijms21010221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Yang Y, Hao P, Ding X. Claudin 8 contributes to malignant proliferation in human osteosarcoma U2OS cells. Cancer Biother Radiopharm. 2015;30(9):400–404. doi: 10.1089/cbr.2015.1815 [DOI] [PubMed] [Google Scholar]
- 27.Kielgast F, Schmidt H, Braubach P, et al. Glucocorticoids regulate tight junction permeability of lung epithelia by modulating claudin 8. Am J Respir Cell Mol Biol. 2016;54(5):707–717. doi: 10.1165/rcmb.2015-0071OC [DOI] [PubMed] [Google Scholar]
- 28.Lucarelli G, Loizzo D, Franzin R, et al. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev Mol Diagn. 2019;19(5):397–407. doi: 10.1080/14737159.2019.1607729 [DOI] [PubMed] [Google Scholar]
- 29.Bianchi C, Meregalli C, Bombelli S, et al. The glucose and lipid metabolism reprogramming is grade-dependent in clear cell renal cell carcinoma primary cultures and is targetable to modulate cell viability and proliferation. Oncotarget. 2017;8(69):113502–113515. doi: 10.18632/oncotarget.23056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucarelli G, Rutigliano M, Sallustio F, et al. Integrated multi-omics characterization reveals a distinctive metabolic signature and the role of NDUFA4L2 in promoting angiogenesis, chemoresistance, and mitochondrial dysfunction in clear cell renal cell carcinoma. Aging (Albany NY). 2018;10(12):3957–3985. doi: 10.18632/aging.101685 [DOI] [PMC free article] [PubMed] [Google Scholar]