Abstract
Background and Objectives:
Nowadays, high-level aminoglycosides and ampicillin resistant Enterococcus species are among the most common causes of nosocomial infections. The present study was conducted to determine the prevalence of high-level resistance to aminoglycosides and ampicillin among clinical isolates of Enterococcus species in Ardabil, Iran.
Materials and Methods:
In this cross–sectional study, a total of 111 Enterococcus species were collected from different clinical specimens between 2013 and 2015. Enterococcus species were identified using standard phenotypic and genotypic methods. BHI agar screen and agar dilution methods were used for detection of high-level gentamicin and streptomycin resistance (HLGR and HLSR) and minimal inhibitory concentration (MIC) of ampicillin, respectively.
Results:
Of 111 clinical isolates, 59 (53.2%) and 25 (22.5%) isolates were E. faecalis and E. faecium, respectively, based on the PCR results. Totally, 60.3% and 56.7% of isolates were HLGR and HLSR, respectively, as well as 51.35% were HLGR plus HLSR. Among HLGR isolates, 36 (61.01%), 18 (72%) and 13 (48.14%) were E. faecium, E. faecalis and non-faecalis non-faecium species, respectively. Among HLSR isolates, 33 (55.93%), 16 (64%) and 14 (51.85%) were E. faecalis, E. faecium and non-faecalis non-faecium species, respectively. All HLGR isolates contained aac(6′)Ie-aph(2″)Ia gene. Overall, the prevalence of high-level ampicillin resistance among Enterococcus species was 17.1%. For E. faecalis, E. faecium and non-faecalis non-faecium species, ampicillin resistance rates were as follows: 11 (40.74%), 7 (28%) and 1 (1.69%), respectively. For aminoglycoside antibiotics, the resistance rate was significantly higher in E. faecium isolates and for ampicillin it was higher in E. faecalis isolates.
Conclusion:
The frequency of high-level aminoglycoside resistant enterococcal isolates in our hospital was high and significant ampicillin resistance was noticed. This would require routine testing of enterococcal isolates for HLAR and ampicillin susceptibility.
Keywords: Enterococcus faecalis, Enterococcus faecium, High-level resistance, Gentamicin, Streptomycin, Ampicillin
INTRODUCTION
Enterococcus species have been ranked as the second to third most common organisms responsible for nosocomial infections, especially in critically ill patients or individuals who received multiple antibiotics (1). In clinical practice, most of non-invasive and uncomplicated enterococcal infections are usually cured using a single antibiotic regimen. While, to achieve an efficient bactericidal activity, combination therapy is needed for treating deep-seated enterococcal infections (such as endocarditis) (2). The most common therapy regimen is a combination of a cell wall active agent (e.g., ampicillin and vancomycin) and an aminoglycoside, gentamicin or streptomycin (2). These agents act synergistically to enhance killing of the bacteria, since the beta-lactam agents damage the cell wall and increase the entry of the aminoglycosides into the cell (3). Generally, penicillins (e.g., ampicillin) have the highest activity, carbapenems slightly lower and cephalosporins and aztreonam have the lowest activity (4). Enterococcus spp. show low-level intrinsic resistance against beta-lactam and aminoglycoside antibiotics (3). The major categories of acquired antibiotic resistance in enterococci include high-level penicillin and ampicillin resistance, high-level aminoglycoside resistance (HLAR) and vancomycin resistance (3, 4). The acquisition of these types of resistance eliminates the synergistic effect of combination therapy (2).
The aim of this study was to determine the frequency of high-level resistance to aminoglycosides and ampicillin among enterococcal isolates collected from different clinical specimens in an Iranian hospital.
MATERIALS AND METHODS
Media and chemicals.
DNP™ Genomic DNA Extraction Kit was purchased from Cinagen Company (Cinagen, Iran). AccuPower™ PCR PreMix Kit and oligonucleotide primers were obtained from Bioneer Company (Daejon, South Korea). Gentamicin, streptomycin and ampicillin powders were purchased from Bio Basic Company (Bio Basic Inc. Canada). Brain Hart Infusion Agar (BHI) was obtained from Difco Laboratories (Detroit, MI, USA) and Trypticase Soy Broth (TSB), Blood Agar, Bile Esculin Agar (BEA) and Mueller Hinton Agar (MHA) were purchased from Himedia laboratories (Mumbai, India).
Isolation and identification of bacteria.
From November 2013 to September 2015, a total of 111 enterococci strains were isolated from different clinical specimens collected from patients admitted to a referral teaching hospital affiliated to Ardabil University of Medical Sciences. Isolates were examined by conventional phenotypic methods at the genus level. E. faecium and E. faecalis were identified using PCR analysis of the ddl gene as described previously (1, 5). E. faecalis ATCC 29212 and E. faecium ATCC 51559 were used as controls. Identified isolates were stored at −80ºC in TSB containing 15% glycerol.
Antimicrobial susceptibility testing.
High-level aminoglycosides resistance (HLAR), gentamicin (HLGR) and streptomycin (HLSR), were determined using BHI agar screen method (1). Testing for minimum inhibitory concentration (MIC) of ampicillin was performed by standard agar dilution method (concentration rang: 0.12–1024 μg/ml) (1). Resistance to ampicillin was defined as MIC ≥ 16 (μg/ml).
Antimicrobial susceptibility testing was performed and interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (6). Enterococcus faecalis ATCC 29212 was used as control strain.
PCR amplification of HLGR resistance gene.
The presence of aac(6′)Ie-aph(2″)Ia gene in the genome of HLGR isolates was detected using PCR method as reported earlier (10, 12).
Statistical analysis.
Chi-square test was used to compare the prevalence of HLAR strains between specimen type and hospital wards.
RESULTS
In the present study, a total of 111 Enterococcus species were isolated from different clinical specimens from patients who referred to Imam Khomeini Hospital in Ardabil province. Totally, 61 (54.9%) and 50 (45.1%) isolates were collected from female and male patients, respectively. Enterococci isolates were collected from urine (n=87), blood (n=21), wound (n=2) and sputum (n=1) specimens.
PCR analysis of ddl gene.
Identification of E. faecalis and E. faecium species was performed by amplification of ddl gene using PCR method. According to the PCR results, 59 (53.2%) and 25 (22.5%) isolates were E. faecalis and E. faecium, respectively, as well as 27 (24.3%) isolates belonged to non-faecalis non-faecium Enterococcus species.
Detection of HLGR and HLSR strains.
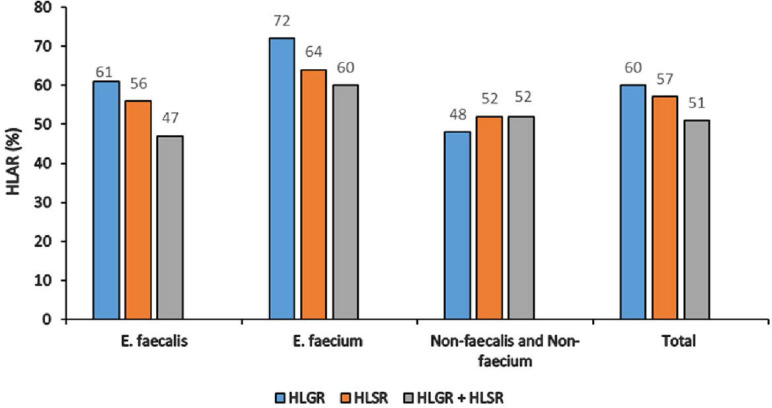
Totally, 60.3% and 56.7% of isolates were HLGR and HLSR, respectively, as well as 51.35% were HLGR plus HLSR. Among HLGR isolates, 36 (61.01%), 18 (72%) and 13 (48.14%) were E. faecium, E. faecalis and non-faecalis non-faecium species, respectively. Among HLSR isolates, 33 (55.93%), 16 (64%) and 14 (51.85%) were E. faecalis, E. faecium and non-faecalis non-faecium species, respectively. Statistically, there was no significant difference between the prevalence of HLGR and HLSR phenotypes in Enterococcus spp. (P > 0.05) (Fig. 1). All HLGR isolates were contained the aac(6′)Ie-aph(2″)Ia gene.
Fig. 1.
Prevalence of HLAR among Enterococcus species isolated from clinical specimens
HLAR; High level aminoglycoside resistance, HLGR; High level gentamicin resistance, HLSR; High level streptomycin resistance
The prevalence of HLGR and HLSR phenotypes was significantly different according to the hospital wards in both E. faecium and E. faecalis species (P < 0.001). The majority of them were isolated from patients who admitted to internal medicine ward, infectious diseases ward and outpatient clinic, respectively (Table 1). However, statistically there was no significant difference in distribution of Enterococcus spp. between wards, except for HLGR E. faecium isolates in infectious disease ward and HLGR and HLSR non-faecalis non-faecium species in internal medicine ward (P < 0.05).
Table 1.
Prevalence of HLAR among Enterococcus species isolated from clinical specimens in different hospital ward
| HLAR | ||||||
|---|---|---|---|---|---|---|
| HLGR | HLSR | |||||
| Hospital ward | E. faecalis (n = 36) n (%) | E. faecium (n = 18) n (%) | non-faecalis non-faecium (n = 13) n (%) | E. faecalis (n = 33) n (%) | E. faecium (n = 16) n (%) | non-faecalis non-faecium (n = 14) n (%) |
| Internal medicine | 13 (36.11) | 9 (50) | 9(69.23 )* | 12 (36.37) | 7 (43.75) | 9 (64.30)* |
| ICU | 3 (8.33) | 1 (5.6) | - | 3 (9.10) | 2 (12.5) | - |
| Infectious disease | 6 (16.70) | 4 (22.20)* | 1(7.7) | 6 (18.2) | 3 (18.75) | 2 (14.30) |
| Outpatient clinic | 6 (16.70) | 2 (11.10) | 2(15.40) | 6 (18.2) | 2 (12.5) | 2 (14.30) |
| Emergency | 3 (8.33) | 1 (5.6) | - | 3 (9.10) | 1 (6.25) | - |
| Cardiology | 2 (5.55) | - | - | 1 (3.03) | - | - |
| Cardiothorax | 1 (2.80) | - | 1(7.7) | 1 (3.03) | - | 1 (7.14) |
| Coronary care unit (CCU) | 1 (2.77) | 1 (5.6) | - | 1 (3.03) | 1 (6.25) | - |
Statistically significant (P < 0.05)
HLAR; High level aminoglycoside resistance, HLGR; High level gentamicin resistance, HLSR; High level streptomycin resistance
The majority of HLAR Enterococcus species were isolated from urine specimens (P < 0.001). However, there was no significant difference between distribution of HLAR Enterococcus species and specimen type (P > 0.05) (Table 2).
Table 2.
Prevalence of HLAR among Enterococcus species isolated from clinical specimens based on specimen type
| HLAR | ||||||
|---|---|---|---|---|---|---|
| HLGR | HLSR | |||||
| Specimen type | E. faecalis (n = 36) n (%) | E. faecium (n = 18) n (%) | non-faecalis non-faecium (n = 13) n (%) | E. faecalis (n = 33) n (%) | E. faecium (n = 16) n (%) | non-faecalis non-faecium (n = 14) n (%) |
| Urine | 31 (86.10) | 15 (83.33) | 11 (84.61) | 27 (81.80) | 12 (75) | 11 (78.60) |
| Blood | 4 (11.11) | 3 (16.66) | 2 (15.40) | 5 (15.15) | 4 (25) | 3 (21.43) |
| Wound | 1 (2.80) | - | - | 1 (3.00) | - | - |
HLAR; High level aminoglycoside resistance, HLGR; High level gentamicin resistance, HLSR; High level streptomycin resistance
Determining MIC for ampicillin.
In this study, MIC of ampicillin was determined using agar dilution method. Totally, 19 (17.1%) out of 111 enterococci isolates showed high-level resistance to ampicillin (MIC ≥ 16 μg/mL). Ampicillin resistance in E. faecalis isolates was found in 11 (18.6%) out of 59 isolates. The MIC values for ampicillin resistant E. faecalis isolates were ranged from 32–512 μg/mL [32; 1 (9.10%), 64; 1 (9.10%), 256; 4 (36.36%) and 512; 5 (45.45%)]. Twenty-eight percent (7/25) of E. faecium isolates with MIC values from 32 to 1024 μg/mL [32; 2 (28.57%), 64; 1 (14.28%), 256; 1 (14.28%), 512; 4 (45.45%) and 1024; 1 (14.28%)] were found to be ampicillin resistant. In non-faecalis and non-faecium enterococci 3.6% of isolates were ampicillin resistant (MIC; 32 μg/mL).
DISCUSSION
Nowadays, acquired high-level ampicillin resistance in Enterococcus species is an increasing global concern. Epidemiology of enterococci resistant to ampicillin varies among hospitals and countries. In the USA and Europe, the majority of nosocomial invasive enterococci isolates are resistant to ampicillin (7). Ampicillin resistance is most commonly seen in E. faecium isolates (7). Accordingly, in this study, resistance to ampicillin in E. faecium (28%) was higher than E. faecalis (19%) isolates. Previously it has been shown that 90%, 84%, 74% and 69% of E. faecium recovered from healthcare-associated infections in the USA, Argentina, Denmark and Iran were ampicillin resistant, respectively (4, 7–9). In Ardabil, the prevalence of ampicillin-resistant E. faecium isolates was significantly lower than the other parts of the world as well as Iran (15). Interestingly, the prevalence of ampicillin-resistant E. faecalis isolates was high (19%) in this study which is similar to Asadollahi et al. report from Iran (17.1%) (15). Given the negligible incidence of ampicillin-resistant E. faecalis isolates in other parts of the world, our findings represent a significant increase in ampicillin resistance rate in E. faecalis isolates in Iran. In a report from Argentina in 2015, ampicillin resistance rate was only 1.8% in E. faecalis isolates (4). While, high-level resistance to penicillin and aminopenicillins have not yet been described in non-faecalis non-faecium Enterococcus species (4), in this study one non-faecalis non-faecium isolate was found to be resistant against ampicillin.
Recently several antibiotics were introduced to be used in combination with ampicillin in the treatment of enterococcal invasive infections (2). However, aminoglycoside antibiotics are still the main component of combination therapy of enterococcal invasive infections (2). With the emergence of high-level resistance, any synergistic effect between aminoglycosides and cell wall active agents is lost. In recent years, acquired high-level resistance to aminoglyco-sides increased significantly by the wide spread of genes encoding aminoglycoside modifying enzymes (AMEs). In a survey conducted in Australia in 2013, 32.4% of E. faecalis and 61.8% of E. faecium isolates showed HLGR phenotype (10). Because of limited use of streptomycin, very few studies have recently studied prevalence of HLSR phenotype among Enterococcus species. In a twelve-year surveillance in Japan, HLSR phenotype was detected in 18% of E. faecalis and 39% of E. faecium isolates in 2016 (11). Unfortunately, there is no a nationwide surveillance data for HLGR and HLSR phenotypes in Iran. However, sporadic studies were reported a HLGR phenotype range of 33 to 77.3% in E. faecium and 25.84% to 89% in E. faecalis isolates. Similarly, HLSR phenotype was reported as 27.27% to 90.10% and 40.44% to 73.10% in E. faecium and E. faecalis isolates, respectively (12–16). In the current study, the prevalence of HLGR was observed among 61.01% of E. faecalis, 72% of E. faecium, and 48.14% of non-faecalis non-faecium species. Additionally, 33 (55.93%) of E. faecalis, 16 (64%) of E. faecium and 14 (51.85%) of non-faecalis non-faecium species had HLSR phenotype. Similar to other reports around the world (11), the proportion of HLGR and HLSR were higher in E. faecium compared to E. faecalis isolates. In this study, 45.47% and 60% of E. faecalis and E. faecium isolates exhibited dually high-level gentamicin and streptomycin resistance, respectively, which is similar to findings reported in other studies (15). As high-level resistance to gentamicin and streptomycin is caused by different mechanisms (3), these antibiotics can be used surrogate to each other in enterococcal infections treatment. Therefore, the emergence of isolates with simultaneous resistance phenotype, HLGR and HLSR, will limit the therapeutic options of enterococcal infections.
High-level aminoglycosides resistance is primarily due to the presence of the AMEs. In Enterococcus species, there are several AMEs responsible for HLGR and HLSR (1). The aac(6′)Ie-aph(2′)Ia gene is the most prevalent gene encoding a AME which causes high-level resistance to aminoglycosides except for streptomycin (1). In the present study, we examined only the presence of the aac(6′)Ie-aph(2′) Ia gene for strains with HLGR. Our findings showed that all HLGR isolates were positive for aac(6′)Ieaph(2′)Ia gene. Similar results were reported from Iran and other countries (11, 14). Genetic basis for HLSR was not further studied in this study.
CONCLUSION
The frequency of high-level aminoglycoside resistant enterococcal isolates in our hospital was high and significant ampicillin resistance was noticed. Therefore, to achieve an optimal therapeutic outcome, continuous monitoring is needed through routine susceptibility testing of enterococcal isolates against aminoglycosides and ampicillin.
ACKNOWLEDGEMENTS
This work was performed in partial fulfilment of the requirements for M.Sc thesis in Medical Bacteriology (Seyed Hossein Mossavi). We gratefully acknowledge Dr. Firouz Amani for his assistance in statistical analysis.
REFERENCES
- 1.Jannati E, Amirmozaffari N, Saadatmand S, Arzanlou M. Faecal carriage of high-level aminoglycoside-resistant and ampicillin-resistant Enterococcus species in healthy Iranian children. J Glob Antimicrob Resist 2020; 20:135–144. [DOI] [PubMed] [Google Scholar]
- 2.Fraser Susan L. Enterococcal infections treatment and management. 2018. Available from https://emedicine.medscape.com/article/216993-treatment
- 3.Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 2012; 3:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagetti P, Bonofiglio L, García Gabarrot G, Kaufman S, Mollerach M, Vigliarolo L, et al. Resistance to β-lactams in enterococci. Rev Argent Microbiol 2019; 51: 179–183. [DOI] [PubMed] [Google Scholar]
- 5.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 1995;33:24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 26th ed CLSI supplement M100s. CLSI; 2016. [Google Scholar]
- 7.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect Control Hosp Epidemiol 2008; 29: 996–1011. [DOI] [PubMed] [Google Scholar]
- 8.Lester CH, Sandvang D, Olsen SS, Schønheyder HC, Jarløv JO, Bangsborg J, et al. Emergence of ampicillin-resistant Enterococcus faecium in Danish hospitals. J Antimicrob Chemother 2008; 62:1203–1206. [DOI] [PubMed] [Google Scholar]
- 9.Asadollahi P, Razavi S, Asadollahi K, Pourshafie MR, Talebi M. Rise of antibiotic resistance in clinical enterococcal isolates during 2001–2016 in Iran: a review. New Microbes New Infect 2018; 26: 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coombs GW, Pearson JC, Daley DA, Le TT, Robinson JO, Gottlieb T, et al. Australian enterococcal sepsis outcome programme annual report, 2013. Commun Dis Intell Q Rep 2014;38:E320–326. [PubMed] [Google Scholar]
- 11.Osuka H, Nakajima J, Oishi T, Funayama Y, Ebihara T, Ishikawa H, et al. High-level aminoglycoside resistance in Enterococcus faecalis and Enterococcus faecium causing invasive infection: Twelve-year surveillance in the Minami Ibaraki Area. J Infect Chemother 2016; 22:61–63. [DOI] [PubMed] [Google Scholar]
- 12.Khodabandeh M, Mohammadi M, Abdolsalehi MR, Hasannejad-Bibalan M, Gholami M, Alvandimanesh A, et al. High-level aminoglycoside resistance in Enterococcus faecalis and Enterococcus faecium; as a serious threat in hospitals. Infect Disord Drug Targets 2020;20:223–228. [DOI] [PubMed] [Google Scholar]
- 13.Khani M, Fatollahzade M, Pajavand H, Bakhtiari S, Abiri R. Increasing prevalence of aminoglycoside-resistant Enterococcus faecalis isolates due to the aac(6′)-aph(2″) gene: A therapeutic problem in Kermanshah, Iran. Jundishapur J Microbiol 2016; 9(3): e28923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dadfarma N, Fooladi AAI, Oskoui M, Mahmoodzadeh Hosseini H. High level of gentamicin resistance (HLGR) among enterococcus strains isolated from clinical specimens. J Infect Public Health 2013;6:202–208. [DOI] [PubMed] [Google Scholar]
- 15.Behnood A, Farajnia S, Moaddab SR, Ahdi-Khosroshahi S, Katayounzadeh A. Prevalence of aac(6′)-Ieaph(2″)-Ia resistance gene and its linkage to Tn5281 in Enterococcus faecalis and Enterococcus faecium isolates from Tabriz hospitals. Iran J Microbiol 2013;5:203–208. [PMC free article] [PubMed] [Google Scholar]
- 16.Emaneini M, Aligholi M, Aminshahi M. Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol J Microbiol 2008; 57:173–178. [PubMed] [Google Scholar]