Abstract
Clinical laboratory diagnostic evaluation of the genomes of children with suspected genetic disorders, including chromosomal microarray and exome sequencing, cannot detect copy number neutral genomic rearrangements such as inversions, balanced translocations and complex chromosomal rearrangements (CCRs). We describe an infant with a clinical diagnosis of Cornelia de Lange syndrome (CdLS) in whom chromosome analysis revealed a de novo complex balanced translocation, 46,XY,t(5;7;6)(q11.2;q32;q13)dn. Subsequent molecular characterization by whole genome sequencing (WGS) identified twenty three breakpoints, delineating segments derived from four chromosomes (5;6;7;21) in ancestral or inverted orientation. One of the breakpoints disrupted a known CdLS gene, NIPBL. Further investigation revealed paternal origin of the CCR allele, clustering of the breakpoint junctions, and molecular repair signatures suggestive of a single catastrophic event. Notably, very short DNA segments (25bp, 41bp) were included in the reassembled chromosomes, lending additional support that the DNA repair machinery can detect and repair such segments. Interestingly, there was an independent paternally-derived miniscule complex rearrangement, possibly predisposing to subsequent genomic instability. In conclusion, we report a CCR causing a monogenic Mendelian disorder, urging WGS analysis of similar unsolved cases with suspected Mendelian disorders. Breakpoint analysis allowed for identification of the underlying molecular diagnosis and implicated chromoanagenesis in CCR formation.
Keywords: Cornelia de Lange syndrome, whole genome sequencing, complex chromosomal rearrangement, breakpoint junction, replicative repair, chromothripsis
INTRODUCTION
Conventional G-banded chromosome analysis has been replaced in the clinical arena with higher-resolution genomic assay techniques, including chromosomal microarray analysis (CMA) and exome sequencing (ES), to detect pathogenic variation (copy number variation, CNV, and single nucleotide variation, SNV) potentially contributing to disease. These have become first-tier diagnostic screens for individuals with developmental disorders and/or congenital malformations. However, these techniques are blind to copy-neutral events such as balanced translocations, inversions, and complex chromosomal rearrangements (CCRs). Whole genome sequencing (WGS), on the contrary, has the potential to detect CCRs and to define breakpoints at nucleotide-pair resolution. In the context of genomic disorders, delineating the breakpoints of copy-neutral events has enabled novel gene discovery, and characterization at the nucleotide resolution has yielded remarkable insights into mechanisms of formation (Carvalho & Lupski, 2016; Liu et al., 2011; Nazaryan-Petersen et al., 2018; Nilsson et al., 2017; Redin et al., 2017).
CCRs can arise as a single catastrophic event. The term chromothripsis (‘chromosome shattering and stitching’) is a phenomenon originally defined from somatic mutagenesis studies and whole genome sequencing of cancer genomes. It is often reserved for an event thought to be due to the shattering of a single chromosome into segments of different lengths followed by stitching, or gluing, together of the segments in random orientation, with minimal overall DNA loss. Another term, chromoplexy (‘chromosome restructuring’) has been used to describe a closed chain of translocations between multiple chromosomes, again with little or no copy number change (Baca et al., 2013; Chiang et al., 2012; Meyerson & Pellman, 2011; Shaikhibrahim, Offermann, & Perner, 2014; C. Z. Zhang, Leibowitz, & Pellman, 2013). Finally, chromoanasynthesis, defined in constitutional mutational studies of patients, results from a chromothripsis-like pattern with a multitude of break-join events followed by chromosome reconstitution with resulting multiple copy number variants (CNVs) including gains such as locus duplication and triplication events (Liu et al., 2011). These terms, thought also to reflect the potential underlying rearrangement mechanisms of non-homologous end joining (NHEJ) versus replicative repair by microhomology-mediated break-induced replication (MMBIR) (Maher & Wilson, 2012; F. Zhang, Carvalho, & Lupski, 2009), have been grouped together under the umbrella term chromoanagenesis, or ‘chromosome rebirth’ (Pellestor, 2019; Zepeda-Mendoza & Morton, 2019).
Initiation of chromothripsis may include various exogenous sources such as ionizing radiation, free radicals, environmental toxins, viral integration with subsequent genomic imbalance, or chemotherapeutic drugs (Kloosterman et al., 2012; Redin et al., 2017). Alternatively, it has been proposed that chromothripsis might be caused by abortive apoptosis (Tubio & Estivill, 2011). Interestingly, DNA fragments involved in CCR formation are more likely to be co-localized in the same or neighbouring sub-compartments of nuclear organization prior to chromosomal reassembly, and Hi-C interaction data has suggested that pairs of loci comprising CCR breakpoint junctions are more likely to interact as compared to random pairs (Redin et al., 2017). Experimental findings have implicated that chromothripsis and chromoplexy may result from lagging chromosomes encapsulated in micronuclei (C. Z. Zhang et al., 2015). Other mechanisms that have been proposed for certain cases include telomere attrition and end-to-end telomere fusion (Lowden, Flibotte, Moerman, & Ahmed, 2011; Stephens et al., 2011).
Repair of chromothripsis and chromothripsis-like events can involve NHEJ, fork stalling and template switching (FoSTeS), and MMBIR. NHEJ is a pathway that repairs double-strand breaks by direct ligation without a homologous template, resulting in blunt ends; micro-deletions or insertion of random nucleotides (i.e., nontemplated insertions) may be observed at the breakpoint junctions. FoSTeS and MMBIR are error-prone DNA-replication mechanisms that utilize microhomologous regions to repair breaks, resulting in microhomology at breakpoint junctions (Beck et al., 2019; Carvalho et al., 2011; Liu et al., 2011; Redin et al., 2017).
Cornelia de Lange syndrome (CdLS, MIM 122470, 300590, 300882, 610759, 614701) is a clinically recognizable genetic disorder characterized by severe growth restriction, developmental delay, a distinctive facial appearance including synophrys, highly arched eyebrows, long eyelashes and a long smooth philtrum, and limb abnormalities. Pathogenic variants in NIPBL, HDAC8, SMC1A, SMC3, and RAD21 are associated with CdLS (Deardorff, Bando, et al., 2012; Deardorff et al., 2007; Deardorff, Wilde, et al., 2012; Krantz et al., 2004; Musio et al., 2006; Tonkin, Wang, Lisgo, Bamshad, & Strachan, 2004; Yuan et al., 2019). The encoded proteins are all components of, or interact with, the cohesin complex, which is responsible for regulating sister chromatid cohesion and segregation, as well as maintaining genomic stability (Cucco & Musio, 2016). Of note, genomic rearrangements involving NIPBL, deletion CNV, have also been reported in some CdLS patients (Pehlivan et al., 2012). Variants in different ‘cohesinopathy-genes’ can have varying severity of clinical phenotypes (Yuan et al., 2019). Despite identification of multiple genes associated with CdLS, over 25% of individuals with a clinical diagnosis have negative genetic testing (Gil-Rodríguez et al., 2015), suggesting the existence of additional yet unidentified genes or yet undefined mechanisms of disease.
We report a child with a clinical diagnosis of CdLS, in whom exome sequencing (ES) and chromosome microarray analysis (CMA) were noncontributory toward establishing a molecular diagnosis. Conventional G-banded chromosome analysis followed by WGS revealed a CCR, 46,XY,t(5;7;6)(q11.2;q32;q13)dn, involving at least 3 chromosomes and over twenty breakpoint junctions, with one of the breakpoints interrupting NIPBL and correlating with the clinical diagnosis. We elaborate on the mechanism of formation and parental origin of the CCR, and highlight the limitations of current first-tier genetic molecular diagnostic testing.
MATERIALS AND METHODS
Short-read whole-genome sequencing
Following informed consent, genomic DNA derived from blood of the patient was sequenced using the Illumina 30X PCR-free protocol at National Genomics Infrastructure (NGI), Stockholm, Sweden. Data was processed using the NGI-piper pipeline (https://github.com/johandahlberg/piper) and structural variants were called using the FindSV pipeline (https://github.com/J35P312/FindSV). Briefly, the FindSV pipeline combines CNVnator V0.3.2 (Abyzov, Urban, Snyder, & Gerstein, 2011) and TIDDIT V2.2.4 (Eisfeldt, Vezzi, Olason, Nilsson, & Lindstrand, 2017). CNVnator detects copy number variants based on read depth, while TIDDIT detects a wide range of structural variants including balanced events based on searches for clusters of discordant read pairs and split reads. Output from FindSV was compiled into a single Variant Calling Format (VCF) file which was subsequently annotated by the variant effect predictor (VEP) (McLaren et al., 2010) and filtered based on VCF quality flag. Finally, the VCF file was sorted based on a local structural variant frequency database consisting of 351 patient samples. The BAM files of variants of interest were manually inspected in the Integrative Genome Viewer (IGV: http://software.broadinstitute.org/software/igv/) (Robinson et al., 2011) and the exact position of the breakpoint could be estimated and sometimes exactly determined after alignment of split reads to the hg19 reference genome in the BLAST-like alignment tool (BLAT:https://genome.ucsc.edu/cgi-bin/hgBlat).
Breakpoint junction PCR
Specific breakpoints, which could not be resolved by WGS or were of particular interest, were investigated by breakpoint junction polymerase chain reaction (PCR). Primer sequences designed to flank estimated breakpoint regions are provided in Table S1. For junction der6_jct1, which disrupted NIPBL, control primers were also designed to amplify the wild-type alleles (Fig. S1). Breakpoint junction PCR was performed by standard methods using DreamTaq DNA Polymerase (Thermo Fisher Scientific). Products were Sanger sequenced, and aligned by BLAT (UCSC Genome Browser; genome.ucsc.edu) to the human genome assembly hg19 (GRCh37) to determine unresolved breakpoint junctions.
Phasing to determine parental origin of alleles
To investigate the parental origin of the rearranged allele, single nucleotide polymorphisms (SNPs) in cis to the rearrangement and within ~200 nucleotides from the estimated breakpoint junction were sequenced in both parents. The SNPs and respective primers are provided as Table S2.
Fluorescent in situ hybridization (FISH)
FISH analyses were performed on metaphase spreads prepared from lymphocyte cultures using RP11–244H11 (Empire Genomics, Buffalo, NY) for the 1p31.2 area; telomere 1q (LPT 01Q) probe, telomere 21q (LPT 21Q) probe and centromere 14/22 (LPE 014) (Cytocell®, Cytocell Ltd, Cambridge, UK) according to standard procedures following respective manufacturer’s protocols.
RESULTS
Clinical Report
The proband was a 15-month-old male, the firstborn child to non-consanguineous parents of Arab Christian descent. Pregnancy was remarkable for oligohydramnios and severe intrauterine growth restriction (IUGR). Delivery was at 36 2/7 weeks via urgent C-section due to nonreassuring monitor and severe oligohydramnios. Birthweight was 1740 grams (Z-score −2.49), length 41 cm (Z-score −2.67), and head circumference 30 cm (Z-score −1.92). Apgar scores were 8 at 1 minute and 8 at 5 minutes. After delivery, the infant was noted to have dysmorphic features, bushy eyebrows, bilateral cleft palate, micrognathia, low-set ears, elbow joint contractures, bilateral upper limb reduction defects, back hirsutism, undescended testes and hypospadias. Based on the recognizable pattern of facial dysmorphology and limb reduction defects, the infant was given a clinical diagnosis of Cornelia de Lange syndrome (CdLS: MIM 122470). At last exam at 15 months of age, development was severely delayed. The child could track, smile, and vocalize yet did not babble nor say specific words. He could support his head, roll over, and sit unsupported for a few seconds, yet could not crawl nor stand. He did not have seizures. Physical exam revealed weight of 5 kg (~25th %tile on CdLS chart), length 64.5 cm (~40th %tile on CdLS chart) and head circumference 39.2 cm (~10th %tile on CdLS chart). Dysmorphic features included thick, high arched eyebrows, synophrys, short nose with anteverted nares, long and smooth philtrum, thin lips, cleft palate, upper and lower lip frenulum, low-set ears with fleshy lobes, retrognathia, and hirsutism. Limb evaluation revealed an absent ulna and short radius bilaterally, with three fingers absent on each hand. Additional medical issues included sensorineural hearing loss (60 dB on right and 70 dB on left), ptosis with an otherwise normal ophthalmology exam, and a small patent foramen ovale with mild left pulmonary branch stenosis. Brain ultrasound showed normal size ventricles and presence of a corpus callosum.
Chromosome analysis revealed a complex rearrangement: 46,XY,t(5;7;6)(q11.2;q32;q13) (Fig. 1A). Parental chromosome analysis was normal, suggesting that the complex rearrangement occurred de novo. Presumed breakpoints on chromosome analysis did not involve any known CdLS gene, and chromosomal microarray analysis (CMA) did not detect disturbances of balance in chromosomal material around the breakpoint junctions or elsewhere in the genome. Exome sequencing from whole blood was noncontributory. Due to the recognized high rate of somatic mosaicism in individuals with CdLS (Huisman, Redeker, Maas, Mannens, & Hennekam, 2013), exome sequencing was repeated on DNA obtained from a buccal swab; no variants of interest in CdLS nor other disease-associated genes were identified.
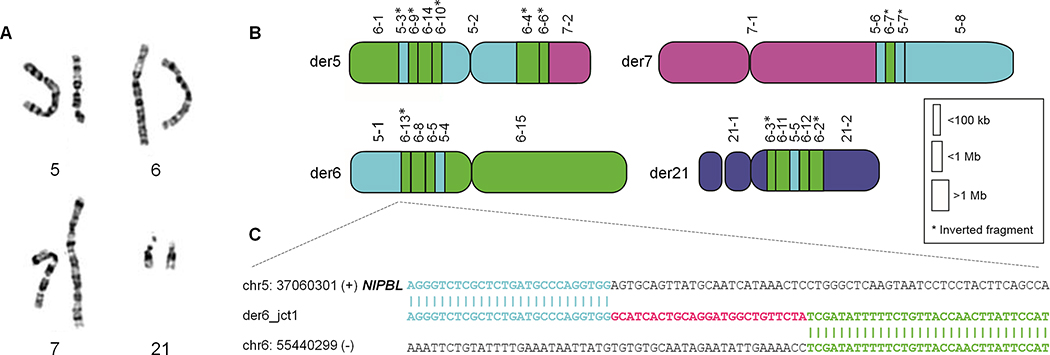
Figure 1. Complex chromosome rearrangement disrupts NIPBL gene.
(A) Karyotype indicated a complex rearrangement involving chromosomes 5, 6, and 7. Both copies of chromosome 21 were considered cytogenetically intact per chromosome analysis. (B) Whole genome sequencing (WGS) indicated a balanced complex rearrangement involving 26 breakpoints on four chromosomes. (C) WGS followed by Sanger sequencing of the breakpoint junction disrupting NIPBL indicated a non-templated insertion of 25 nucleotides.
Short read (paired-end) WGS detects numerous breakpoints on four chromosomes
WGS detected 23 breakpoints (Fig. 1B, S2; Tables 1, S3), far more than were anticipated by conventional chromosome analysis. Four chromosomes were involved – chromosomes 5, 6, and 7, which were seen to be rearranged per conventional chromosome analysis, as well as chromosome 21 which seemed structurally intact on chromosome analysis even when viewed in retrospect. Numerous segments were reincorporated into the reconstructed chromosomes in inverted orientation. Telomeres always remained at the outer ends of the chromosomes, likely due to their unique sequence and structure.
Table 1.
Breakpoint junctions and characterization of molecular signature
| Breakpoint junction | Genomic coordinate (prox end) | Genomic coordinate (distal end) | BL/INS/ MH | Size (bp) | Source |
|---|---|---|---|---|---|
| der5_jct1 | 6:52164816 | 5:66424678 | MH | 4 | NR |
| der5_jct2 | 5:66416578 | 6:54477651 | BL | - | NR |
| der5_jct3 | 6:54468957 | 6:55440339 | BL | - | NR |
| der5_jct4 | 6:55549994 | 6:54532787 | MH | 5 | NR |
| der5_jct5 | 6:54477667 | 5:37060348 | INS | 16 | NR |
| der5_jct6 | 5:66416448 | 6:54144313 | BL | - | NR |
| der5_jct7 | 6:52268476 | 6:54160494 | INS | 91 | NA |
| der5_jct8 | 6:54160401 | 7:123818519 | INS | 71 | chr6:55419517–55419557 [41bp]; chr6:52268424–52268448 [25bp] |
| der6_jct1 | 5:37060326 | 6:55440329 | INS | 25 | NA |
| der6_jct2 | 6:55419612 | 6:54233402 | MH | 2 | NR |
| der6_jct3 | 6:54468932 | 6:54144318 | INS | 8 | NR |
| der6_jct4 | 6:54160398 | 5:66424725 | INS | 32 | NA |
| der6_jct5 | 5:66444130 | 6:55550005 | INS | 83 | chr6:52266027–52266050 [24bp] |
| der7_jct1 | 7:123818492 | 5:66449983 | MH | 1 | NR |
| der7_jct2 | 5:66463171 | 6:54233319 | MH | 2 | NR |
| der7_jct3 | 6:54160547 | 5:68215114 | INS | 117 | NA |
| der7_jct4 | 5:66463179 | 5:68215535 | BL | - | NR |
| der21_jct1 | 21:14598180 | 6:52268420 | INS | 43 | chr6:54160487–54160508 [22bp] |
| der21_jct2 | 6:52266075 | 6:54532789 | MH | 1 | NR |
| der21_jct3 | 6:55416595 | 5:66444415 | INS | 111 | chr6:55419496–55419523 [28bp]; chr6:55419553–55419573 [21bp] |
| der21_jct4 | 5:66449980 | 6:55416610 | BL | - | NR |
| der21_jct5 | 6:55419503 | 6:52266036 | INS | 4 | NR |
| der21_jct6 | 6:52164824 | 21:14598357 | BL | - | NR |
Abbreviations: BL - blunt, INS - insertion, MH - microhomology, NA - not available, NR- not relevant
NIPBL disrupted by the complex rearrangement
To determine the mechanism by which the complex chromosome rearrangement led to a Cornelia de Lange phenotype, breakpoint junctions were investigated for disruption of genes. In total, five genes were disrupted, including HMGCLL1, MAST4, NIPBL, PAQR8, TINAG. Three of these have probability of loss-of-function intolerance (pLI) scores (https://gnomad.broadinstitute.org) equal to zero, and a fourth (PAQR8) has a low pLI score of 0.16. Thus, it was presumed that those four genes would not cause disease by mechanism of haploinsufficiency. Conversely, NIBPL, disrupted by der6_jct1 (Fig. 1C, 2A), has a pLI score of 1 and is a known dosage-sensitive gene associated with CdLS. Breakpoint junction analysis by PCR and Sanger sequencing confirmed this junction in the proband, and its absence in the parents (Fig. S1). Numerous variants resulting in premature termination codons in NIPBL lead to CdLS (Yan et al., 2006), as do heterozygous deletion CNV consistent with a haploinsufficiency phenotype (Pehlivan et al., 2012). The breakpoint junction disrupting NIBPL (located in intron 44 of 46 introns) is within an AluJr element, yet the junction has a non-templated insertion and was not dependent on Alu-Alu recombination (Song et al., 2018).
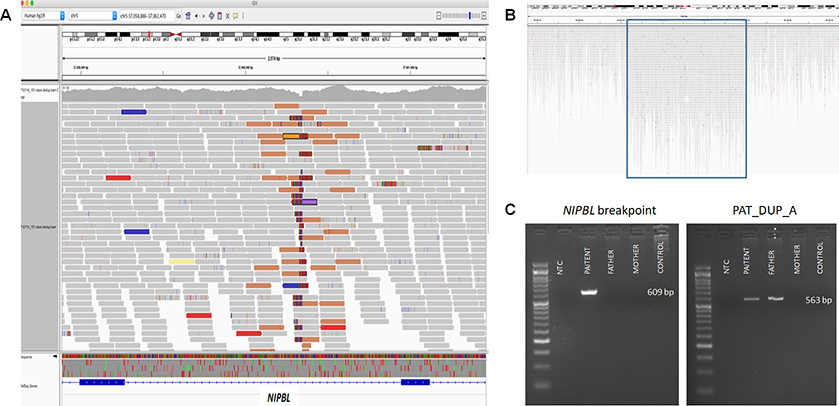
Figure 2. Whole genome sequencing data and PCR amplification of NIBPL breakpoint and of paternally inherited duplication.
(A) Screenshot of whole genome sequencing (WGS) data showing breakpoint junction region disrupting NIPBL. (B) Screenshot of WGS data indicating duplication of chromosome 1: 69,063,596–69,119,765 [hg19]. (C) Breakpoint junction analysis of der6_jct1, interrupting NIPBL, indicated that the rearrangement occurred de novo in the proband (left panel). Amplification of der7_jct1 (between segments 6–16 and the duplication on chromosome 1) indicated that the duplication on chromosome 1 was inherited from the father, and presumably moved en bloc with segment 6–16 during formation of the complex rearrangement (right panel).
Parental origin and mechanism of rearrangement formation
Analysis of SNPs in close proximity to the breakpoint junction and in cis to the rearranged allele (Table S2) revealed that the complex rearrangement occurred on the paternal allele. This is consistent with previous reports showing a paternal bias for CCR formation, possibly attributed to the larger number of mitotic divisions during gametogenesis in males as compared to females (Campbell et al., 2014; Kloosterman et al., 2012).
In order to surmise the potential mechanism underlying formation of the CCR, 21 of the 23 breakpoints were defined at the nucleotide level by WGS split-read analysis. The remaining junctions (der5_jct7, der5_jct8, and der7_jct1) were resolved by breakpoint junction PCR analysis and Sanger sequencing. The majority of the breakpoints clustered within 1.8Mb and 3.4Mb on chromosomes 5 and 6, respectively (Table S3), suggesting chromothripsis as the underlying catastrophic event. Among the breakpoint junctions delineated at base-pair resolution, seven were precisely joined as blunt ends, consistent with non-homologous end joining (NHEJ) as an underlying repair mechanism. Microhomology ranging from 1 to 5 nucleotides was observed in six junctions, implicating microhomology-mediated break-induced replication (MMBIR) as a repair mechanism. The other breakpoints had insertions varying from 4 to 117 nucleotides (Fig. S2, Table 1). Low-copy repeats and repetitive elements were present at some of the junctions (Table S3), yet did not seem to mediate non-allelic homologous recombination (NAHR) derived DNA rearrangements. Overall, less than 1000 kilobases of total genomic imbalance was appreciated.
Many of the larger insertions at breakpoint junctions were non-templated insertions; yet, in others, runs of 21 to 41 nucleotides could be mapped back to the regions of chromosomal shattering, mostly to chromosome 6 (Tables 1, S3). Some of these likely represent very short DNA segments that were incorporated into the reassembled chromosomes by the repair machinery (Slamova et al., 2018). As an example, 41bp in junctions der5_jct7&8 mapped to the gap between segment 6–12 and 6–13, and 25bp at the same junction mapped to the segment between 6–3 and 6–4 (Table S3). Thus, there seemed to be several cryptic breakpoint junctions in addition to those initially detected by WGS analysis.
Detection of an independent complex rearrangement inherited from the father
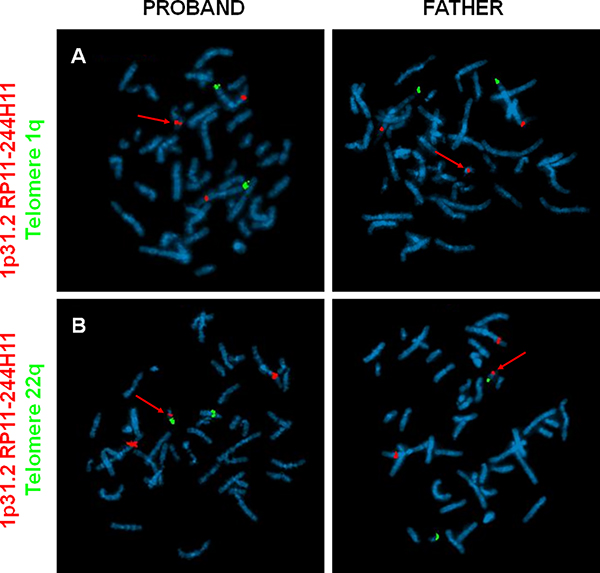
Whilst delineating the extensive number of breakpoint junctions by PCR, we noted a 56kb duplication originating from chromosome 1 (coordinates: chr1:69,063,596–69,119,765 [hg19]) (Fig. 2B–C). One end of this duplication bordered chromosome 6 (position: chr6:166,668,890), and surprisingly could be amplified not only from the proband DNA but from paternal DNA as well. Further analysis and Sanger sequencing confirmed a paternally-inherited contig consisting of fragments derived from chromosomes 1, 6 and 7. However, the contig could not reliably be placed in the context of a derivative chromosome, since repetitive elements flanked it on the genome sequencing data. Therefore, we turned to fluorescent in situ hybridization (FISH) utilizing a probe targeting the 56kb duplication of chromosome 1p31.2. FISH confirmed the duplication in both the father and proband, and revealed that the duplication was located on one of the smaller chromosomes (Fig. 3A). Subsequent analyses (Fig. 3B, Fig. S3) showed that the duplication, and thus the paternally-inherited second complex rearrangement, was inserted onto chromosome 22. Details of the paternally inherited duplication are provided in Fig. S4 and Table S4. It remains to be determined whether this miniature complex rearrangement might promote genomic instability and ignite additional catastrophic events.
Figure 3. Fluorescent in situ hybridization (FISH) localizes paternally-inherited duplication of chromosome 1p31.2 to chromosome 22.
(A) Chromosomes from the proband (left panels) and father (right panels) were hybridized with a probe targeting telomere 1q (green fluorescent label) and a probe targeting the paternal duplication originating from 1p31.2 (red fluorescent label). A third signal of the 1p31.2 probe can be seen on one of the smaller chromosomes (red arrow). (B) Utilizing probes targeting 1p31.2 (red) and chromosome 22 (telomere 22q, green), the duplication was localized to chromosome 22 (red arrow) in both the proband and his father.
DISCUSSION
In this study, we report a cytogenetically balanced complex chromosomal rearrangement masquerading as a monogenic disorder. Resolution of the breakpoints at the nucleotide level by WGS revealed that the phenotype of CdLS was due to disruption of the NIPBL gene by one of 23 identified breakpoints (Fig. 1C, S2). The disruption was intronic, and was thus invisible to exome sequencing. Additional genes were interrupted by the CCR (Table S3), yet these are unlikely to be haploinsufficient genes based on their pLI scores and/or presumed function. Since the phenotype of CdLS is quite severe, the pathogenic contribution of a minor disrupted gene may have been overlooked. Moreover, genes mapping within the ~1.5kb of genomic imbalance may contribute to the severity of the phenotype. Nevertheless, we thus expand the molecular mechanisms by which a monogenic disorder such as CdLS can occur to include CCRs, and highlight the limitations of conventional cytogenetic approaches.
The severe presentation of the affected individual was consistent with previously reported genotype-phenotype correlations for NIPBL variants, wherein individuals with truncating variants have more severe presentations as compared to individuals with missense variants (Yan et al., 2006). Chromosomal rearrangements have been previously reported in CdLS; in fact, a de novo translocation t(5;13)(p13.1;q12.1) with a deleted fragment encompassing NIPBL aided in identification of the gene-disease association (Hulinsky, Byrne, Lowichik, & Viskochil, 2005; Krantz et al., 2004) and confirmed haploinsufficiency as the mechanism. Additionally, several individuals with exonic deletions in NIBPL have been described (Pehlivan et al., 2012). The current report of a CCR leading to a very severe phenotype of CdLS lends further support to this being a ‘transcriptomopathy’ and suggests that CCRs may perturb gene regulation and expression to a greater degree than point mutations, perhaps by position effects (Yuan et al., 2015). Albeit, further studies involving RNA expression data would be necessary to confirm or refute this hypothesis.
In the studied case, the paternal origin of the CCR allele, the clustering of the majority of breakpoints within a few megabases of nucleotides, and the molecular signature of the breakpoint junctions (i.e., blunt ends, microhomology, and non-templated insertions) supported a single catastrophic event as the mechanism of formation. Similar to a recent report by Slamova et al. (2018), we observed very small fragments from shattered chromosomes that were apparently captured and incorporated into derivative chromosomes by the repair machinery (Slamova et al., 2018), providing further support that copy number variants can be very short. We conclude that differentiation between copy number variants and indels should be defined by the mechanism by which they arise rather than by size alone.
Complex rearrangements have been reported to segregate stably through multiple generations (Bertelsen et al., 2016). Other reports show unbalanced but stable transmission of a subset of the derivative chromosomes leading to a phenotype in the offspring, and yet other cases document de novo rearrangements complicating the parentally inherited complex rearrangement (de Pagter et al., 2015; Di Gregorio et al., 2014; Gu et al., 2016). In the reported case, the paternally-inherited contig consisting of duplicated short fragments of chromosomes 1, 6, and 7 translocated onto chromosome 22 (Fig. 3, Fig. S4), posed a major challenge to its ultimate molecular unravelling. Notably, the regions involved on chromosomes 6 and 7 in the paternal duplication (6q27 and 7p14.1) were isolated and distinct from those regions involved in the chromothripsis event (6p12 and 7q31.33). This further supported the conclusion that there were two independent events – a paternally inherited duplication, and a de novo chromothriptic event. Reversion to FISH technology and, perhaps more so, the dialogue between WGS, FISH, and Sanger breakpoint sequencing, proved to be critical to the understanding of the case. Long read WGS would be expected to be of high utility in such complex cases. Future studies may be directed at understanding whether, and by which mechanism, parentally-derived chromosome rearrangements predispose to catastrophic events in offspring.
CONCLUSIONS
It has been noted that in chromosomal abnormalities, prediction of the resultant phenotype depends heavily on the specific genes and regions that are altered, rather than the mere presence of a chromosomal abnormality or the number of genes disrupted (Redin et al., 2017). WGS has an added benefit of unraveling cryptic complexity at breakpoint junctions (Baptista et al., 2008; Higgins et al., 2008). This case underscores the urgent need to implement genomic technologies capable of detecting both balanced and unbalanced rearrangements in routine diagnostic use.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank the family for their participation in this study, and Ms. Terry Scher, Ms. Marion Werner and Ms. Orly Gafni-Weinstein for their invaluable input regarding chromosome and FISH analyses. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project SNIC Sens2017130. AL was supported by the Swedish Research Council (2017–02936), the Stockholm County Council and the Swedish Brain Foundation. JRL is supported in part by the National Institute of Neurological Disorders and Stroke NINDS (R35NS105078).
Footnotes
CONFLICT OF INTEREST
Baylor College of Medicine and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), formerly the Baylor Miraca Genetics Laboratories, which performs chromosomal microarray analysis and clinical exome sequencing. JRL serves on the Scientific Advisory Board of the BG. JRL has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The other authors declare that they have no conflict of interest.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abyzov A, Urban AE, Snyder M, & Gerstein M (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res, 21(6), 974–984. doi: 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, … Garraway LA (2013). Punctuated evolution of prostate cancer genomes. Cell, 153(3), 666–677. doi: 10.1016/j.cell.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, … Crolla JA (2008). Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet, 82(4), 927–936. doi: 10.1016/j.ajhg.2008.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Carvalho CMB, Akdemir ZC, Sedlazeck FJ, Song X, Meng Q, … Lupski JR (2019). Megabase Length Hypermutation Accompanies Human Structural Variation at 17p11.2. Cell, 176(6), 1310–1324.e1310. doi: 10.1016/j.cell.2019.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen B, Nazaryan-Petersen L, Sun W, Mehrjouy MM, Xie G, Chen W, … Tümer Z (2016). A germline chromothripsis event stably segregating in 11 individuals through three generations. Genet Med, 18(5), 494–500. doi: 10.1038/gim.2015.112 [DOI] [PubMed] [Google Scholar]
- Campbell IM, Stewart JR, James RA, Lupski JR, Stankiewicz P, Olofsson P, & Shaw CA (2014). Parent of origin, mosaicism, and recurrence risk: probabilistic modeling explains the broken symmetry of transmission genetics. Am J Hum Genet, 95(4), 345–359. doi: 10.1016/j.ajhg.2014.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, & Lupski JR (2016). Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet, 17(4), 224–238. doi: 10.1038/nrg.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, … Lupski JR (2011). Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet, 43(11), 1074–1081. doi: 10.1038/ng.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, … Talkowski ME (2012). Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet, 44(4), 390–397, S391. doi: 10.1038/ng.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucco F, & Musio A (2016). Genome stability: What we have learned from cohesinopathies. Am J Med Genet C Semin Med Genet, 172(2), 171–178. doi: 10.1002/ajmg.c.31492 [DOI] [PubMed] [Google Scholar]
- de Pagter MS, van Roosmalen MJ, Baas AF, Renkens I, Duran KJ, van Binsbergen E, … Kloosterman WP (2015). Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am J Hum Genet, 96(4), 651–656. doi: 10.1016/j.ajhg.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, … Shirahige K (2012). HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature, 489(7415), 313–317. doi: 10.1038/nature11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, … Krantz ID (2007). Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet, 80(3), 485–494. doi: 10.1086/511888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, … Kaiser FJ (2012). RAD21 mutations cause a human cohesinopathy. Am J Hum Genet, 90(6), 1014–1027. doi: 10.1016/j.ajhg.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio E, Savin E, Biamino E, Belligni EF, Naretto VG, D’Alessandro G, … Brusco A (2014). Large cryptic genomic rearrangements with apparently normal karyotypes detected by array-CGH. Mol Cytogenet, 7(1), 82. doi: 10.1186/s13039-014-0082-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeldt J, Vezzi F, Olason P, Nilsson D, & Lindstrand A (2017). an efficient and comprehensive structural variant caller for massive parallel sequencing data. F1000Res, 6, 664. doi: 10.12688/f1000research.11168.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Rodríguez MC, Deardorff MA, Ansari M, Tan CA, Parenti I, Baquero-Montoya C, … Pié J (2015). De novo heterozygous mutations in SMC3 cause a range of Cornelia de Lange syndrome-overlapping phenotypes. Hum Mutat, 36(4), 454–462. doi: 10.1002/humu.22761 [DOI] [PubMed] [Google Scholar]
- Gu S, Szafranski P, Akdemir ZC, Yuan B, Cooper ML, Magriñá MA, … Lupski JR (2016). Mechanisms for Complex Chromosomal Insertions. PLoS Genet, 12(11), e1006446. doi: 10.1371/journal.pgen.1006446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, … Morton CC (2008). Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet, 82(3), 712–722. doi: 10.1016/j.ajhg.2008.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman SA, Redeker EJ, Maas SM, Mannens MM, & Hennekam RC (2013). High rate of mosaicism in individuals with Cornelia de Lange syndrome. J Med Genet, 50(5), 339–344. doi: 10.1136/jmedgenet-2012-101477 [DOI] [PubMed] [Google Scholar]
- Hulinsky R, Byrne JL, Lowichik A, & Viskochil DH (2005). Fetus with interstitial del(5)(p13.1p14.2) diagnosed postnatally with Cornelia de Lange syndrome. Am J Med Genet A, 137A(3), 336–338. doi: 10.1002/ajmg.a.30856 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, … Cuppen E (2012). Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep, 1(6), 648–655. doi: 10.1016/j.celrep.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, … Jackson LG (2004). Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet, 36(6), 631–635. doi: 10.1038/ng1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kołodziejska KE, Dharmadhikari AV, … Bi W (2011). Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell, 146(6), 889–903. doi: 10.1016/j.cell.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MR, Flibotte S, Moerman DG, & Ahmed S (2011). DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science, 332(6028), 468–471. doi: 10.1126/science.1199022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, & Wilson RK (2012). Chromothripsis and human disease: piecing together the shattering process. Cell, 148(1–2), 29–32. doi: 10.1016/j.cell.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, & Cunningham F (2010). Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics, 26(16), 2069–2070. doi: 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, & Pellman D (2011). Cancer genomes evolve by pulverizing single chromosomes. Cell, 144(1), 9–10. doi: 10.1016/j.cell.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, … Larizza L (2006). X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet, 38(5), 528–530. doi: 10.1038/ng1779 [DOI] [PubMed] [Google Scholar]
- Nazaryan-Petersen L, Eisfeldt J, Pettersson M, Lundin J, Nilsson D, Wincent J, … Lindstrand A (2018). Replicative and non-replicative mechanisms in the formation of clustered CNVs are indicated by whole genome characterization. PLoS Genet, 14(11), e1007780. doi: 10.1371/journal.pgen.1007780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Pettersson M, Gustavsson P, Förster A, Hofmeister W, Wincent J, … Lindstrand A (2017). Whole-Genome Sequencing of Cytogenetically Balanced Chromosome Translocations Identifies Potentially Pathological Gene Disruptions and Highlights the Importance of Microhomology in the Mechanism of Formation. Hum Mutat, 38(2), 180–192. doi: 10.1002/humu.23146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehlivan D, Hullings M, Carvalho CM, Gonzaga-Jauregui CG, Loy E, Jackson LG, … Lupski JR (2012). NIPBL rearrangements in Cornelia de Lange syndrome: evidence for replicative mechanism and genotype-phenotype correlation. Genet Med, 14(3), 313–322. doi: 10.1038/gim.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F (2019). Chromoanagenesis: cataclysms behind complex chromosomal rearrangements. Mol Cytogenet, 12, 6. doi: 10.1186/s13039-019-0415-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, … Talkowski ME (2017). The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet, 49(1), 36–45. doi: 10.1038/ng.3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, & Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol, 29(1), 24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikhibrahim Z, Offermann A, & Perner S (2014). Words of wisdom: Re: Punctuated evolution of prostate cancer genomes. Eur Urol, 65(3), 666–667. doi: 10.1016/j.eururo.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Slamova Z, Nazaryan-Petersen L, Mehrjouy MM, Drabova J, Hancarova M, Marikova T, … Sedlacek Z (2018). Very short DNA segments can be detected and handled by the repair machinery during germline chromothriptic chromosome reassembly. Hum Mutat, 39(5), 709–716. doi: 10.1002/humu.23408 [DOI] [PubMed] [Google Scholar]
- Song X, Beck CR, Du R, Campbell IM, Coban-Akdemir Z, Gu S, … Lupski JR (2018). Predicting human genes susceptible to genomic instability associated with. Genome Res, 28(8), 1228–1242. doi: 10.1101/gr.229401.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, … Campbell PJ (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 144(1), 27–40. doi: 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, & Strachan T (2004). NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet, 36(6), 636–641. doi: 10.1038/ng1363 [DOI] [PubMed] [Google Scholar]
- Tubio JM, & Estivill X (2011). Cancer: When catastrophe strikes a cell. Nature, 470(7335), 476–477. doi: 10.1038/470476a [DOI] [PubMed] [Google Scholar]
- Yan J, Saifi GM, Wierzba TH, Withers M, Bien-Willner GA, Limon J, … Wierzba J (2006). Mutational and genotype-phenotype correlation analyses in 28 Polish patients with Cornelia de Lange syndrome. Am J Med Genet A, 140(14), 1531–1541. doi: 10.1002/ajmg.a.31305 [DOI] [PubMed] [Google Scholar]
- Yuan B, Neira J, Pehlivan D, Santiago-Sim T, Song X, Rosenfeld J, … Study D (2019). Clinical exome sequencing reveals locus heterogeneity and phenotypic variability of cohesinopathies. Genet Med, 21(3), 663–675. doi: 10.1038/s41436-018-0085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Pehlivan D, Karaca E, Patel N, Charng WL, Gambin T, … Lupski JR (2015). Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest, 125(2), 636–651. doi: 10.1172/JCI77435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda-Mendoza CJ, & Morton CC (2019). The Iceberg under Water: Unexplored Complexity of Chromoanagenesis in Congenital Disorders. Am J Hum Genet, 104(4), 565–577. doi: 10.1016/j.ajhg.2019.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Leibowitz ML, & Pellman D (2013). Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev, 27(23), 2513–2530. doi: 10.1101/gad.229559.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, … Pellman D (2015). Chromothripsis from DNA damage in micronuclei. Nature, 522(7555), 179–184. doi: 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, & Lupski JR (2009). Complex human chromosomal and genomic rearrangements. Trends Genet, 25(7), 298–307. doi: 10.1016/j.tig.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.