Abstract
Purpose
Hodgkin's disease (HD) is a type of cancer affecting men in the reproductive age with potential consequences on their fertility status. This study aims to analyze sperm parameters, alterations in proteomic profiles and validate selected protein biomarkers of spermatozoa in men with HD undergoing sperm banking before cancer therapy.
Materials and Methods
Semen analysis was carried out in healthy fertile donors (control, n=42), and patients diagnosed with HD (patients, n=38) before cancer therapy. We compared proteomic profiles of spermatozoa from donors (n=3) and patients (n=3) using LTQ-Orbitrap Elite hybrid MS system.
Results
A total of 1,169 proteins were identified by global proteomic in both groups. The ingenuity pathway analysis revealed that differentially expressed proteins involved in capacitation, acrosome reaction, binding of sperm to the zona pellucida, sperm motility, regulation of sperm DNA damage, and apoptosis were significantly downregulated in HD patients. Validation of proteins implicated in sperm fertility potential by Western Blot demonstrated that peroxiredoxin 2 (PRDX 2) was underexpressed (p=0.015), and transferrin (p=0.045) and SERPIN A5 (p=0.010) protein levels were overexpressed in spermatozoa of men with HD.
Conclusions
Findings of this study indicates that the key proteins involved in sperm fertility potential are significantly altered in men with HD, which provides substantial explanation for the observed low sperm quality in HD subjects prior to cancer therapy. Furthermore, our results suggest PRDX 2, transferrin and SERPIN A5 as possible candidate proteins for assessing sperm quality in HD patients prior to cancer therapy.
Keywords: Hodgkin disease, Infertility, Mass spectrometry, Proteomics, Spermatozoa, Transferrin
INTRODUCTION
Hodgkin's disease (HD) is one of the most prevalent cancers in men in the reproductive age. According to the American Society of Cancer [1], about 4,570 men are estimated to be diagnosed with HD and about 590 men are projected to die from HD. The five years survival rate in people diagnosed with this HD is about 86% [2] and hence, fertility preservation is crucial for men diagnosed with HD. Sperm cryopreservation is recommended to all male patients before cancer therapy [3,4].
Lower sperm quality, such as sperm concentration or motility has been reported in HD patients even before cancer therapy when compared to healthy individuals [5,6,7]. Though the reasons for this lower sperm quality is not fully understood, psychological factors or immune-mediated mechanisms affecting the spermatogenesis have been hypothesized to be the possible contributing causes [8,9]. Molecular changes at the subcellular level of spermatozoa that can explain the low sperm quality in men with HD are still unknown.
Proteomic analysis of spermatozoa is a high-throughput technique that elucidates the complex nature of the sperm [10,11]. Advances in mass spectroscopy and analysis of proteomic profile using bioinformatic tools have been used in the discovery of potential biomarkers in tissues and biological fluids [10,11,12,13,14,15]. Literature review reveals that the proteomic profile of spermatozoa in HD patients has not been investigated until now. Therefore, we sought to investigate the sperm proteomic profiling of HD subjects in comparison to healthy fertile volunteers in order to elucidate the reason(s) for low sperm quality in HD subjects at the time of sperm banking. The objectives of this study are: (1) to analyze and compare semen parameters of men with HD, who banked their specimen before cancer treatment, with fertile donors; (2) to characterize and compare the spermatozoa protein profile among cancer patients and healthy fertile donors; and (3) to validate proteins of interest implicated in fertility that are differently expressed in Hodgkin's cancer patients when compared to controls.
MATERIALS AND METHODS
1. Study subjects and ethics statement
This study was approved by the Institutional Review Board (IRB) of Cleveland Clinic. All the participants signed an informed written consent at the time of sample collection at the Andrology Center, Cleveland Clinic (No. #13-1554). Semen samples were obtained from 42 healthy male donors of proven fertility (control group) who had fathered a child in the past 2 years before enrollment in the study and 38 patients with HD before cancer therapy (cancer group). All the subjects included in the control group had normal semen parameters [16].
Men between the ages of 20 to 45 years diagnosed with HD were included in the study and these patients have not yet started their oncological treatment. We have excluded men with azoospermia and those under the supportive medication, steroids or drugs. In addition, patients with systemic reproductive tract inflammation, smoking, sexually transmitted disease, and leukocytospermic were excluded from the current study.
2. Semen analysis
Semen analysis was performed according to the World Health Organization (WHO) guidelines (2010) [16] in samples from both control (n=38) and cancer (n=33) groups. Semen samples were collected by masturbation after sexual abstinence of at least 48 hours. Samples were allowed to liquefy completely for 20 minutes at 37℃, and semen analysis was performed according to WHO guidelines [16] using the Microcell counting chamber (Vitrolife, San Diego, CA, USA) to evaluate sperm count, motility and round cells. For the samples from the control group, after completion of routine semen analysis, the remainder of the sample was centrifuged for 7 minutes at 1,000×g. The clear seminal plasma was aspirated from the samples and spermatozoa were stored at −80℃ for proteomic studies. Semen samples from cancer group were cryopreserved with Freezing Medium Test Yolk Buffer (TYB) using an established protocol [4].
3. Preparation of semen samples
Semen samples from the control group were thawed at room temperature and centrifuged at 3,000×g for 30 minutes to remove the remnants of seminal plasma and washed with phosphate buffer saline (PBS). Cryopreserved samples from the cancer group were thawed to room temperature and centrifuged at 3,000×g for 30 minutes to remove the TYB and the supernatant was discarded. The pellet was then washed with PBS and centrifuged at 3,000×g for 30 minutes. This step was repeated 3 times to remove the remains of TYB.
4. Protein extraction and quantification
Proteins were extracted with radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma-Aldrich, St. Louis, MO, USA) containing the proteinase inhibitor cocktail (Roche, Indianapolis, IN, USA). RIPA was added to the sperm pellet (approximately 100 µL for 100×106 sperm) and incubated overnight at 4℃ to allow the complete lysis of the spermatozoa. The supernatant was aspirated after centrifugation at 13,000×g for 20 minutes to a clean microcentrifuge tube. The protein concentration was determined using a commercially available kit, bicinchoninic acid kit (Thermo, Rockford, IL, USA), following the manufacturer instructions.
5. Preparation of semen samples for proteomics
Proteomic analysis was performed on randomly selected donor (n=3) and HD (n=3) samples. Further, normalization of the samples was done by pooling the samples to overcome the biological variation [17,18]. The samples were mixed with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) buffer and subjected to one-dimensional-PAGE in triplicate runs. After completion of electrophoresis, the gel was cut into 6 pieces and digested using 5 µL trypsin (10 ng/µL), 50 mM ammonium bicarbonate and incubated overnight at room temperature. The samples (cut pieces) were alkylated with iodoacetamine and reduced with dithiothreitol. The peptides from the digested gel were extracted in two aliquots of 30 µL acetonitrile (10%) with formic acid (5%). The two aliquots were pooled together and evaporated to <10 µL and then diluted with 1% acetic acid to make up a final volume to 30 µL.
6. Liquid chromatography-mass spectrometer analysis
Proteomic profiling of spermatozoa was carried out using a Finnigan LTQ linear ion trap mass spectrometer liquid chromatography (LC)-mass spectrometer analysis (MS)/MS system. The peptides were fractionated by injecting 5 µL volumes into high-performance liquid chromatography column (Phenomenex Jupiter C18 reversed-phase capillary chromatography column; Phenomenex, Torrence, CA, USA). Fractions containing the peptides were eluted in acetonitrile/0.1% formic acid at a flow rate of 0.25 µL/min and were introduced into the source of the mass spectrometer on-line. The micro electrospray ion source was operated at 2.5 kV. A full spectral scan was performed by utilizing the data dependent multitask ability of the instrument to determine peptide molecular weights and amino acid sequence of the peptides [19].
7. Database searching and protein identification
Tandem mass spectra generated by LC-MS/MS system were retrieved using Proteome Discoverer ver. 1.4.1.288. Mascot (ver. 2.3.02; Matrix Science, London, UK), Sequest (ver. 1.4.0.288; Thermo Fisher Scientific, San Jose, CA, USA) and X! Tandem (ver. CYCLONE 2010.12.01.1; The GPM, thegpm.org) search was performed on all the MS/MS raw files. The search was limited to the human reference sequences database (http://www.hprd.org/) assuming the digestion enzyme trypsin. The mass tolerance for parent ion was set to 10 parts per million (ppm) and for fragment ion with 1.0 Da. The search results were uploaded into the Scaffold (ver. 4.0.6.1; Proteome Software Inc., Portland, OR, USA) program as previously described [20]. Protein probabilities were assigned by the Protein Prophet (Systems Biology, Seattle, WA, USA) algorithm. Annotation of proteins was performed using Gene Ontology (GO) terms from National Center for Biotechnology Information.
8. Quantitative proteomics
The relative quantification of the proteins was performed by comparing the number of spectra, termed spectral counts in both cancer and control groups. To achieve false detection rate <1%, protein identification criteria was established at >99% probability as explained in our previous study [21]. The abundance of the proteins was determined by matching the spectra (spectral counts or SpCs), and classified as high (H), medium (M), low (L), or very low (VL). To overcome the sample-to-sample variation, normalization of spectral counts was done using the normalized spectral abundance factor (NSAF). In general, longer proteins have more peptide identifications than shorter proteins. NSAF ratio determines the actual expression of the protein in the samples. Proteins with ratio <1 and >1 are considered underexpressed and overexpressed, respectively. Different constraints for fold-change cutoffs and significance tests (p-value) based on the average SpC from 3 replicate runs were applied to obtain the differentially expressed proteins (DEPs) [20]. Appropriate filters were applied used to minimize the errors due to the presence of low abundance proteins. Abundance and the expression of DEPs are based on the following criteria: (i) VL - SpC range, 1.7–7; NSAF ratio (≥2.5 for upregulated and ≤0.4 for downregulated proteins); and p≤0.001, (ii) L - SpC range, 8–19; NSAF ratio (≥2.5 for upregulated and ≤0.4 for downregulated proteins); and p≤0.01, (iii) M - SpC range, 20–79; NSAF ratio (≥2.0 for upregulated and ≤0.5 for downregulated proteins); and p≤0.05, (iv) H - SpC, >80; NSAF ratio (≥1.5 for upregulated and ≤0.67 for downregulated proteins); and p≤0.05.
9. Bioinformatic analysis
DEPs identified in both the study groups were subjected to functional annotation and enrichment analysis using publicly available bioinformatic annotation tools and databases such as GO Term Finder, GO Term Mapper, UniProt, and Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.niaid.nih.gov). Protein-protein interaction was demonstrated using Ingenuity Pathways Analysis (IPA) based on the criteria: experimental evidence, neighborhood, gene fusion, occurrence, co-expression, existing databases, and text mining. Proprietary software package Metacore™ (GeneGo Inc., St. Joseph, MI, USA) was also used to identify the upstream regulators involved in the enriched pathways. Based on their function and role in fertility potential in male six DEPs were chosen for validation by Western blot (WB).
10. Protein validation by Western blotting
The DEPs involved in the reproductive functions and fertilization process, and top canonical pathways were selected for validation. The function of these proteins are well described in the literature. From the identified proteins, six DEPs were selected for validation. The DEPs were validated by WB using n=6 for the control group and n=6 for cancer group. A total of 20 µg of spermatozoa protein was mixed with equal volume of loading buffer (125 mMol Tris-HCl, pH 6.8, 2% SDS, 5% glycerol, 0.003% bromophenol blue, and 1% β-mercaptoethanol). The sample mixture was boiled for 10 minutes and kept on the ice for 5 minutes. 30 µL of each sample was loaded into each well of a 4% to 15% SDS-PAGE and electrophoresed for 2 hours at 90 V along with a set of molecular weight marker (Sigma Chemical Co., St. Louis, MO, USA). The resolved protein bands were then transferred onto polyvinylidene difluoride (PVDF) membranes at 18 V for 30 minutes using a transfer buffer of 25 mMol Tris base, 192 mMol glycine, and 20% methanol. PVDF membranes were blocked with Tris-buffered saline-Tween-20 (TBST) with 5% bovine serum albumin and used for immunodetection of sperm proteins. For each protein analysis, specific primary antibodies were incubated at 4℃ overnight (Table 1). Next, the membranes were washed four times with TBST for 10 minutes and incubated with the secondary antibodies at room temperature for 1 hour (Table 1). Membranes were washed four times again with TBST (10 minutes) and finally treated with enhanced chemiluminescence (ECL) reagent (GE Healthcare, Marlborough, MA, USA) for 5 minutes. ECL reacted blots were exposed to Chemi-Doc (ChemiDoc™ MP Imaging System; Bio-Rad, Hercules, CA, USA) to detect the chemiluminescence signals.
Table 1. Primary and secondary antibodies used in this study.
| Primary | Secondary | |||||
|---|---|---|---|---|---|---|
| Protein | Manufacturer | Source | Dilution | Antibody | Manufacturer | Dilution |
| ACE | Abcam ab85955 | Rabbit | 1:1,000 | Anti-Rabbit | Abcam ab97051 | 1:10,000 |
| PRDX 2 | Abcam ab71533 | Goat IgG | ||||
| Transferin | Abcam ab82411 | |||||
| CCT3 | Abcam ab225878 | 1:5,000 | ||||
| SERPINA5 | Abcam ab172060 | Mouse | 1:500 | Anti-Mouse | Abcam ab6728 | 1:10,000 |
| Rabbit IgG | ||||||
ACE: angiotensin converter enzyme, PRDX2: peroxiredoxin-2, CCT3: T-complex protein 1 subunit gamma, IgG: immunoglobulin G.
11. Total protein staining
The total amount of protein present in the membranes was identified using a Colloidal Gold Total Protein Stain (Bio-Rad). The protocol was performed according to manufacturer instructions. Briefly, the membranes were washed twice for 10 minutes in the distilled water and stained with total colloidal gold protein stain for 2 hours at room temperature by gentle shaking. Stained membranes were washed twice with distilled water for 10 minutes, and the densitometry image was captured using calorimetric mode on Chemi-Doc (ChemiDoc™ MP Imaging System; Bio-Rad).
12. Statistical analysis
Data analysis was performed using MedCalc Statistical Software (ver. 17.8; MedCalc Software, Ostend, Belgium). Mann–Whitney test was carried out to compare the semen parameters of control and cancer groups, and the results were considered significant with p<0.05. The same test was used to compare the expression levels of the proteins validated using WB technique in both the groups.
RESULTS
1. Semen parameters in men with Hodgkin's disease
Semen analysis revealed significant decrease (p<0.05) in sperm concentration, total count and total motile sperm count in cancer group when compared to control group (Table 2).
Table 2. Sperm parameter of men from control and cancer group.
| Group | Volume (mL) | Concentration (×106/mL) | Motility (%) | Total count (×106) | Total motile sperm (×106) |
|---|---|---|---|---|---|
| Control (n=42) | 3.63±1.71 | 70.72±32.22 | 58.43±12.38 | 255.17±156.54 | 147.51±91.04 |
| Patients (n=38) | 3.48±1.78 | 35.98±23.34 | 53.60±24.51 | 119.74±83.48 | 75.49±65.36 |
| p-value | 0.62 | <0.0001 | 0.44 | <0.0001 | 0.0004 |
Values are presented as mean±standard deviation. Mann–Whitney test was carried out to compare the semen parameters of control and cancer groups, and the results were considered significant with p<0.05.
2. Proteomic shotgun analysis revealed a total of 1049 different peptides common to both experimental groups
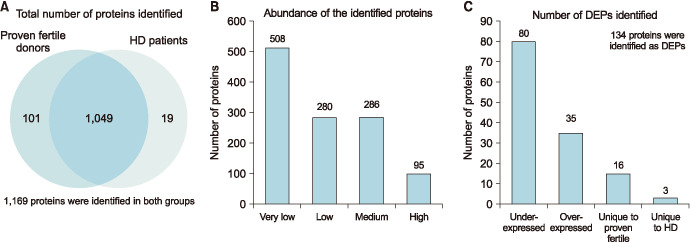
In this study we had used LC-MS/MS, a high throughput technique to obtain the proteome profile of spermatozoa from men diagnosed with HD before cancer therapy and compared this profile with the proteome profile of fertile donors. LC-MS/MS spectrometric analysis identified a total of 1,169 different peptides, out of which a total 1,049 proteins were common to both experimental groups, 101 were unique to the spermatozoa of proven fertile donors, and 19 were unique to the spermatozoa of patients with HD before cancer therapy (Fig. 1A). Relative abundance of the identified proteins revealed that 508 in very low abundance, 280 in low abundance, 286 in medium and 95 in high abundance (Fig. 1B). Among the identified proteins, 134 were detected as DEPs between the two experimental groups (Table 3). The sperm proteome of men with HD before cancer therapy had 35 overexpressed and 80 underexpressed DEPs compared to the sperm proteome of fertile donors (Fig. 1B). In the proteomic analysis of our study, we were also able to identify 16 DEPs unique to fertile donors and 3 DEPs unique to men with HD (Fig. 1C).
Fig. 1. (A) Total number proteins identified in the spermatozoa of proven fertile donors' group and patients with Hodgkin's disease (HD) before cancer therapy by liquid chromatography-mass spectrometer analysis (MS)/MS spectrometry. (B) Distribution of the identified proteins based on their relative abundance. (C) Differentially expressed proteins (DEPs) of experimental groups.
Table 3. Differently expressed proteins identified in sperm proteome of men with HD compared with donors.
| Protein | Accession No. | Average SpC | NSAF ratio | p-value | Expression | |
|---|---|---|---|---|---|---|
| Donors | Patients | Patients/Donors | ||||
| Tetratricopeptide repeat protein 25 | 13899233 | 12.7 | 0 | 0.00 | 0.00003 | Unique to donors |
| Mannose-1-phosphate guanyltransferase beta isoform 1 | 11761619 | 4.7 | 0 | 0.00 | 0.00000 | Unique to donors |
| 60S ribosomal protein l7a | 4506661 | 5.3 | 0 | 0.00 | 0.00000 | Unique to donors |
| Vitamin K epoxide reductase complex subunit 1-like protein 1 isoform 1 | 46309463 | 3.7 | 0 | 0.00 | 0.00000 | Unique to donors |
| Translocation protein SEC63 homolog | 6005872 | 2.0 | 0 | 0.00 | 0.00001 | Unique to donors |
| Dynein heavy chain 6, axonemal isoform X1 | 578802753 | 3.3 | 0 | 0.00 | 0.00001 | Unique to donors |
| Dynein intermediate chain 1, axonemal isoform 2 | 526479830 | 7.0 | 0 | 0.00 | 0.00003 | Unique to donors |
| Guanine nucleotide-binding protein subunit beta-2-like 1 | 5174447 | 2.0 | 0 | 0.00 | 0.00003 | Unique to donors |
| Mitochondrial import receptor subunit TOM22 homolog | 9910382 | 6.0 | 0 | 0.00 | 0.00009 | Unique to donors |
| Cation channel sperm-associated protein subunit beta precursor | 51339295 | 2.0 | 0 | 0.00 | 0.00009 | Unique to donors |
| Calcyphosin isoform b | 18104964 | 3.3 | 0 | 0.00 | 0.00018 | Unique to donors |
| Radial spoke head protein 6 homolog A | 13540559 | 3.3 | 0 | 0.00 | 0.00018 | Unique to donors |
| Glutamine--trna ligase isoform b | 441478305 | 3.7 | 0 | 0.00 | 0.00029 | Unique to donors |
| LETM1 domain-containing protein 1 isoform X7 | 578823569 | 2.3 | 0 | 0.00 | 0.00044 | Unique to donors |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | 111160296 | 7.0 | 0 | 0.00 | 0.00057 | Unique to donors |
| Nucleosome assembly protein 1-like 1 | 21327708 | 4.3 | 0 | 0.00 | 0.00059 | Unique to donors |
| Dynein intermediate chain 2, axonemal isoform X4 | 530412670 | 15.7 | 0.3 | 0.03 | 0.00012 | Underexpressed |
| Cytoplasmic dynein 1 heavy chain 1 | 33350932 | 11.3 | 0.7 | 0.04 | 0.00890 | Underexpressed |
| Mitochondrial fission 1 protein | 151108473 | 8.7 | 0.3 | 0.04 | 0.00162 | Underexpressed |
| V-type proton atpase subunit B, brain isoform | 19913428 | 18.7 | 0.7 | 0.05 | 0.00017 | Underexpressed |
| Alpha-soluble NSF attachment protein | 47933379 | 8.0 | 0.3 | 0.05 | 0.00232 | Underexpressed |
| Arachidonate 15-lipoxygenase B isoform d | 85067501 | 23.7 | 1.0 | 0.06 | 0.00013 | Underexpressed |
| 26S proteasome non-atpase regulatory subunit 4 | 5292161 | 9.0 | 0.7 | 0.09 | 0.00096 | Underexpressed |
| V-type proton atpase catalytic subunit A | 19913424 | 15.7 | 1.0 | 0.09 | 0.00404 | Underexpressed |
| Dipeptidyl peptidase 3 isoform 1 | 86792661 | 12.7 | 1.0 | 0.10 | 0.00073 | Underexpressed |
| Armadillo repeat-containing protein 4 isoform 1 | 585866234 | 18.7 | 1.7 | 0.12 | 0.00525 | Underexpressed |
| Thioredoxin domain-containing protein 3 | 148839372 | 18.3 | 2.3 | 0.13 | 0.00029 | Underexpressed |
| Long-chain-fatty-acid--coa ligase 3 | 42794754 | 9.7 | 1.0 | 0.13 | 0.00137 | Underexpressed |
| Exportin-1 isoform X1 | 530368070 | 8.3 | 2.0 | 0.13 | 0.00101 | Underexpressed |
| Protein ERGIC-53 precursor | 5031873 | 10.0 | 1.0 | 0.14 | 0.00368 | Underexpressed |
| Lipid phosphate phosphohydrolase 1 isoform 2 | 29171738 | 17.7 | 3.3 | 0.18 | 0.00241 | Underexpressed |
| Probable inactive serine protease 37 isoform 1 precursor | 285394164 | 9.0 | 1.3 | 0.18 | 0.00875 | Underexpressed |
| Surfeit locus protein 4 isoform 1 | 19557691 | 9.7 | 1.7 | 0.19 | 0.00200 | Underexpressed |
| Speriolin isoform 1 | 197276668 | 6.3 | 1.0 | 0.20 | 0.00041 | Underexpressed |
| Dynein heavy chain 17, axonemal | 256542310 | 88.0 | 9.0 | 0.20 | 0.00004 | Underexpressed |
| EF-hand calcium-binding domain-containing protein 1 isoform a | 13375787 | 11.3 | 2.0 | 0.20 | 0.00517 | Underexpressed |
| Cytosolic non-specific dipeptidase isoform X2 | 530414265 | 30.7 | 4.7 | 0.20 | 0.00006 | Underexpressed |
| Spectrin alpha chain, non-erythrocytic 1 isoform X1 | 578817797 | 8.0 | 1.0 | 0.21 | 0.00052 | Underexpressed |
| EF-hand calcium-binding domain-containing protein 14 | 7662160 | 6.7 | 1.0 | 0.22 | 0.00087 | Underexpressed |
| Dynein heavy chain 8, axonemal isoform X1 | 578811443 | 132.3 | 18.0 | 0.22 | 0.00009 | Underexpressed |
| Dynein heavy chain 7, axonemal | 151301127 | 18.0 | 2.3 | 0.22 | 0.00094 | Underexpressed |
| Heat shock protein 75, mitochondrial isoform 1 precursor | 155722983 | 8.3 | 1.7 | 0.23 | 0.00239 | Underexpressed |
| Carnitine O-palmitoyltransferase 1, muscle isoform isoform a | 4758050 | 11.3 | 3.0 | 0.23 | 0.00408 | Underexpressed |
| Actin-like protein 7A | 5729720 | 16.7 | 3.0 | 0.23 | 0.00013 | Underexpressed |
| Growth/differentiation factor 15 precursor | 153792495 | 9.0 | 1.7 | 0.23 | 0.00201 | Underexpressed |
| Adenylyl cyclase-associated protein 1 | 5453595 | 32.0 | 6.7 | 0.24 | 0.00071 | Underexpressed |
| Glutathione reductase, mitochondrial isoform 2 precursor | 305410789 | 18.7 | 3.3 | 0.24 | 0.00203 | Underexpressed |
| NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial isoform 1 precursor | 20149568 | 14.0 | 2.3 | 0.25 | 0.00445 | Underexpressed |
| L-amino-acid oxidase isoform 2 precursor | 384381475 | 76.0 | 16.0 | 0.25 | 0.00002 | Underexpressed |
| Purine nucleoside phosphorylase | 157168362 | 10.7 | 2.3 | 0.26 | 0.00259 | Underexpressed |
| Protein FAM209B isoform X2 | 578835992 | 8.0 | 2.0 | 0.28 | 0.00724 | Underexpressed |
| Heat shock protein 105 isoform 1 | 42544159 | 8.7 | 1.7 | 0.29 | 0.00781 | Underexpressed |
| Myoferlin isoform X2 | 530393412 | 39.3 | 6.0 | 0.29 | 0.00003 | Underexpressed |
| Protein-glutamine gamma-glutamyltransferase 4 | 156627577 | 232.3 | 35.7 | 0.29 | 0.00005 | Underexpressed |
| Myosin regulatory light chain 12B | 15809016 | 11.7 | 3.0 | 0.30 | 0.00106 | Underexpressed |
| Probable serine carboxypeptidase CPVL isoform X1 | 530384848 | 27.0 | 5.7 | 0.30 | 0.00204 | Underexpressed |
| Uncharacterized protein KIAA1683 isoform X1 | 530415216 | 23.3 | 6.3 | 0.32 | 0.02207 | Underexpressed |
| Protein FAM71B | 222418633 | 46.7 | 13.0 | 0.34 | 0.00178 | Underexpressed |
| 26S proteasome non-atpase regulatory subunit 6 isoform 2 | 7661914 | 18.7 | 4.7 | 0.34 | 0.00962 | Underexpressed |
| 26S proteasome non-atpase regulatory subunit 13 isoform 1 | 157502193 | 19.7 | 5.3 | 0.35 | 0.00841 | Underexpressed |
| Dnaj homolog subfamily B member 11 precursor | 7706495 | 15.0 | 3.7 | 0.35 | 0.00330 | Underexpressed |
| 26S proteasome non-atpase regulatory subunit 3 | 25777612 | 35.3 | 11.0 | 0.36 | 0.00033 | Underexpressed |
| Calpain small subunit 1 | 51599151 | 9.3 | 3.0 | 0.36 | 0.00997 | Underexpressed |
| Filamin-B isoform 2 | 105990514 | 25.7 | 5.7 | 0.37 | 0.00474 | Underexpressed |
| Disintegrin and metalloproteinase domain-containing protein 32 precursor | 148664238 | 6.0 | 1.7 | 0.37 | 0.00091 | Underexpressed |
| Casein kinase II subunit beta isoform 1 | 23503295 | 9.3 | 3.0 | 0.38 | 0.00436 | Underexpressed |
| Calcium-binding tyrosine phosphorylation-regulated protein isoform a | 24797108 | 63.3 | 23.7 | 0.38 | 0.00038 | Underexpressed |
| Glutamate carboxypeptidase 2 isoform 1 | 4758398 | 69.3 | 13.7 | 0.39 | 0.00100 | Underexpressed |
| Dipeptidyl peptidase 2 isoform X1 | 530426726 | 17.7 | 5.3 | 0.39 | 0.00112 | Underexpressed |
| Tissue alpha-L-fucosidase precursor | 119360348 | 19.0 | 5.3 | 0.39 | 0.00523 | Underexpressed |
| Acylamino-acid-releasing enzyme | 23510451 | 27.0 | 8.0 | 0.40 | 0.00434 | Underexpressed |
| T-complex protein 1 subunit zeta-2 isoform X1 | 578830267 | 36.7 | 13.3 | 0.42 | 0.00010 | Underexpressed |
| Cathepsin F precursor | 6042196 | 21.0 | 7.3 | 0.43 | 0.00109 | Underexpressed |
| Actin-related protein T2 | 29893808 | 45.7 | 15.7 | 0.44 | 0.00119 | Underexpressed |
| Transmembrane and coiled-coil domain-containing protein 2 | 56847610 | 23.3 | 9.0 | 0.45 | 0.00067 | Underexpressed |
| Peroxiredoxin-2 | 32189392 | 33.3 | 13.7 | 0.45 | 0.00393 | Underexpressed |
| Creatine kinase B-type | 21536286 | 51.0 | 17.7 | 0.46 | 0.00013 | Underexpressed |
| Beta-galactosidase-1-like protein isoform X1 | 530370954 | 35.3 | 12.3 | 0.46 | 0.00023 | Underexpressed |
| T-complex protein 1 subunit gamma isoform a | 63162572 | 128.7 | 50.3 | 0.46 | 0.00014 | Underexpressed |
| Retinal dehydrogenase 1 | 21361176 | 20.3 | 7.0 | 0.47 | 0.00457 | Underexpressed |
| T-complex protein 1 subunit theta isoform 1 | 48762932 | 77.7 | 28.0 | 0.48 | 0.00236 | Underexpressed |
| T-complex protein 1 subunit zeta isoform a | 4502643 | 71.7 | 28.7 | 0.49 | 0.00183 | Underexpressed |
| Glutamine synthetase isoform X1 | 578800828 | 23.3 | 8.3 | 0.50 | 0.00915 | Underexpressed |
| Isocitrate dehydrogenase [NADP] cytoplasmic | 538917681 | 59.0 | 21.7 | 0.50 | 0.00110 | Underexpressed |
| Elongation factor 1-delta isoform 1 | 304555581 | 32.3 | 13.0 | 0.50 | 0.00303 | Underexpressed |
| Hypoxia up-regulated protein 1 isoform X2 | 530397761 | 177.3 | 63.0 | 0.50 | 0.00124 | Underexpressed |
| T-complex protein 1 subunit beta isoform 1 | 5453603 | 120.7 | 50.3 | 0.50 | 0.00125 | Underexpressed |
| T-complex protein 1 subunit eta isoform a | 5453607 | 129.7 | 58.3 | 0.55 | 0.00129 | Underexpressed |
| Acrosin precursor | 148613878 | 255.7 | 136.0 | 0.58 | 0.00052 | Underexpressed |
| Ruvb-like 2 | 5730023 | 137.3 | 66.7 | 0.60 | 0.00609 | Underexpressed |
| T-complex protein 1 subunit alpha isoform a | 57863257 | 132.0 | 65.3 | 0.61 | 0.00065 | Underexpressed |
| T-complex protein 1 subunit delta isoform a | 38455427 | 108.3 | 54.3 | 0.61 | 0.00479 | Underexpressed |
| Sperm equatorial segment protein 1 precursor | 21717832 | 100.7 | 53.7 | 0.62 | 0.00076 | Underexpressed |
| Ruvb-like 1 | 4506753 | 99.7 | 51.3 | 0.65 | 0.00355 | Underexpressed |
| Endoplasmin precursor | 4507677 | 543.7 | 238.7 | 0.66 | 0.00276 | Underexpressed |
| Leucine-rich repeat-containing protein 37A2 precursor | 116325993 | 163.0 | 78.3 | 0.67 | 0.00214 | Underexpressed |
| Myeloperoxidase precursor | 4557759 | 69.3 | 81.0 | 1.53 | 0.01314 | Overexpressed |
| Nuclear pore membrane glycoprotein 210-like isoform 1 precursor | 117414168 | 126.7 | 120.7 | 1.56 | 0.00216 | Overexpressed |
| Nuclear pore complex protein Nup155 isoform 1 | 24430149 | 86.0 | 74.7 | 1.60 | 0.01024 | Overexpressed |
| Angiotensin-converting enzyme isoform 1 precursor | 4503273 | 141.3 | 142.3 | 1.65 | 0.00027 | Overexpressed |
| Lactotransferrin isoform 1 precursor | 54607120 | 702.3 | 829.3 | 1.67 | 0.00002 | Overexpressed |
| Clusterin preproprotein | 355594753 | 116.7 | 147.0 | 1.79 | 0.00055 | Overexpressed |
| Protein S100-A8 | 21614544 | 15.0 | 25.0 | 2.03 | 0.02068 | Overexpressed |
| Galectin-3-binding protein precursor | 5031863 | 17.3 | 26.0 | 2.23 | 0.01154 | Overexpressed |
| Plasma serine protease inhibitor preproprotein | 194018472 | 40.7 | 69.7 | 2.33 | 0.00003 | Overexpressed |
| Neutrophil gelatinase-associated lipocalin precursor | 38455402 | 9.3 | 20.0 | 2.39 | 0.00320 | Overexpressed |
| Erythrocyte band 7 integral membrane protein isoform a | 38016911 | 9.3 | 23.3 | 2.43 | 0.00067 | Overexpressed |
| Serotransferrin precursor | 4557871 | 11.3 | 21.3 | 2.47 | 0.04142 | Overexpressed |
| Myeloblastin precursor | 71361688 | 6.0 | 13.7 | 2.69 | 0.00081 | Overexpressed |
| Semenogelin-2 precursor | 4506885 | 261.3 | 606.0 | 2.83 | 0.00678 | Overexpressed |
| Sulfhydryl oxidase 1 isoform a precursor | 13325075 | 7.0 | 15.3 | 3.05 | 0.00162 | Overexpressed |
| Extracellular matrix protein 1 isoform 1 precursor | 221316614 | 7.3 | 16.0 | 3.06 | 0.00007 | Overexpressed |
| Epididymal secretory protein E3-alpha precursor | 11386189 | 4.7 | 12.7 | 3.25 | 0.00651 | Overexpressed |
| Alpha-1-antitrypsin precursor | 189163532 | 13.7 | 34.7 | 3.43 | 0.00003 | Overexpressed |
| Beta-2-microglobulin precursor | 4757826 | 4.0 | 11.7 | 3.57 | 0.00238 | Overexpressed |
| Prosaposin isoform a preproprotein | 11386147 | 20.0 | 42.7 | 3.71 | 0.00007 | Overexpressed |
| Prolactin-inducible protein precursor | 4505821 | 238.0 | 788.3 | 3.85 | 0.00122 | Overexpressed |
| Semenogelin-1 preproprotein | 4506883 | 94.0 | 393.7 | 5.08 | 0.00009 | Overexpressed |
| Lipoprotein lipase precursor | 4557727 | 4.3 | 17.0 | 5.55 | 0.00068 | Overexpressed |
| Fibronectin isoform 1 preproprotein | 47132557 | 112.7 | 505.0 | 6.44 | 0.00000 | Overexpressed |
| Zymogen granule protein 16 homolog B precursor | 94536866 | 5.7 | 29.7 | 6.65 | 0.00007 | Overexpressed |
| Integrin beta-2 isoform X1 | 578836536 | 2.3 | 15.0 | 8.92 | 0.00004 | Overexpressed |
| Integrin alpha-M isoform 1 precursor | 224831239 | 5.3 | 33.0 | 10.26 | 0.00078 | Overexpressed |
| Mucin-5B precursor | 301172750 | 22.0 | 214.3 | 12.40 | 0.00000 | Overexpressed |
| Mucin-6 precursor | 151301154 | 3.7 | 36.0 | 16.60 | 0.00050 | Overexpressed |
| Laminin subunit beta-2 isoform X1 | 530372442 | 1.3 | 17.3 | 22.70 | 0.00054 | Overexpressed |
| BPI fold-containing family B member 1 precursor | 40807482 | 4.7 | 95.3 | 27.49 | 0.00049 | Overexpressed |
| Bactericidal permeability-increasing protein precursor | 157276599 | 0.3 | 6.7 | 28.80 | 0.00085 | Overexpressed |
| Plasma protease C1 inhibitor precursor | 73858570 | 0.3 | 2.7 | 34.56 | 0.00067 | Overexpressed |
| Alpha-1-acid glycoprotein 1 precursor | 167857790 | 0.7 | 18.0 | 43.86 | 0.00001 | Overexpressed |
| Polymeric immunoglobulin receptor isoform X1 | 530366266 | 0.3 | 22.3 | 97.14 | 0.00486 | Overexpressed |
| Endothelial lipase precursor | 5174497 | 0.0 | 2.0 | - | 0.00000 | Unique to patients |
| Apolipoprotein A-IV precursor | 71773110 | 0.0 | 5.3 | - | 0.00022 | Unique to patients |
| Carcinoembryonic antigen-related cell adhesion molecule 8 precursor | 21314600 | 0.0 | 3.7 | - | 0.00099 | Unique to patients |
HD: Hodgkin's disease, SpC: spectral count, NSAF: spectral abundance factor.
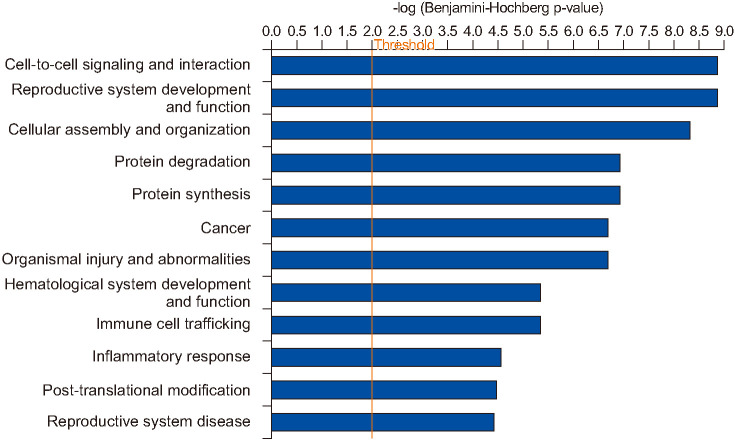
3. Ingenuity pathways Analysis revealed cell-to-cell signaling and interaction as top biofunction dysregulated in spermatozoa of men with Hodgkin's disease
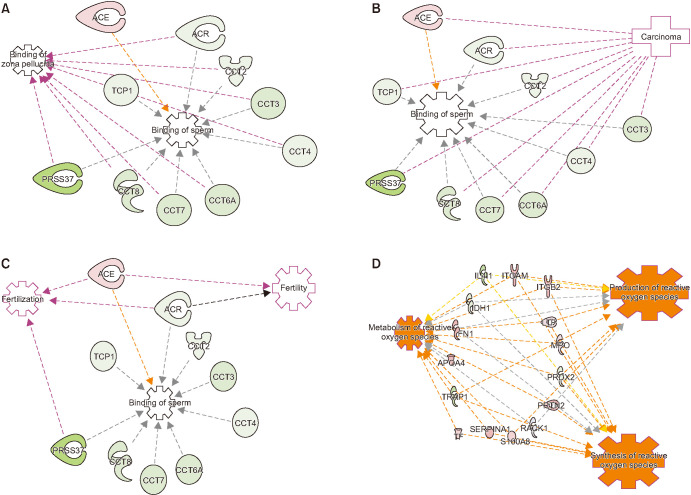
IPA analysis of the DEPs revealed that pathways associated with top diseases and biofunctions were dysregulated in HD group with a cut off equal to 4.44 and a p=0.01 (Fig. 2). The top three pathways identified in the diseases and biofunctions are cell-to-cell signaling and interaction, reproductive system development and function, and cellular assembly and organization (Fig. 2). Functional analysis of DEPs identified that majority of the sperm proteins were associated with sperm functions (Fig. 3). Binding of sperm and binding of zona pellucida processes were affected due to the aberrant expression of sperm proteins, such as some subunits of T-complex protein 1 (TCP1, CCT2, CCT3, CCT4, CCT6A, CCT7, and CCT8), acrosin (ACR), probable inactive serine protease 37 (PRSS37) and angiotensin-converting enzyme (ACE). Other DEPs have a connection between the binding to zona pellucida and binding to sperm, (Fig. 3A). Additionally, the DEPs (ACR, TCP1, CCT2, CCT3, CCT4, CCT6A, CCT7, and CCT8) were also involved in the carcinoma functions in spermatozoa of men with HD (Fig. 3B). IPA analysis also showed that the DEPs ACE and ACR were involved in several reproductive functions such as sperm binding, fertility and fertilization process of the spermatozoa (Fig. 3C). Furthermore, IPA analysis also showed that production of reactive oxygen species (ROS) with metabolism of ROS and synthesis of ROS were affected due to altered expression of several DEPs (IL4I1, ITGAM, ITGB2, LTF, MPO, PRDX2, PRTN3, RACK1, S100A8, SERPIN A1, TF, TRAP1, APOA4, FN1, and IDH1) (Fig. 3D).
Fig. 2. Diseases and biofunctions identified in the spermatozoa of proven fertile donors' group and patients with Hodgkin's disease before cancer therapy by Ingenuity Pathway Analysis with a cut off=4.44 and p=0.01.
Fig. 3. Ingenuity Pathway Analysis of the differently expressed protein in spermatozoa of proven fertile donors' group and patients with Hodgkin's disease before cancer therapy, with the connecting protein related to (A) binding of zona pellucida and binding of sperm; (B) biding of sperm and carcinoma; (C) binding of sperm, fertility and fertilization; and (D) metabolism, production and synthesis of reactive oxygen species. TCP1: T-complex protein 1 subunit alpha, CCT3: T-complex protein 1 subunit gamma, ACR: acrosin, PRSS37: probable inactive serine protease 37, ACE: angiotensin-converting enzyme, IL4I1: L-amino-acid oxidase, ITGAM: integrin alpha-M; IDH: isocitrate dehydrogenase, ITGB2: integrin subunit beta 2, LTF: lactotransferrin precursor, PRTN2: transcription regulatory protein 2, RACK: receptor for activated C kinase, S100A8: S100 calcium-binding protein A8, SERPIN: serine proteinase inhibitors, TF: transferrin, TRAP: heat shock protein 75 kDa mitochondrial percursor, APOA: apolipoprotein A, FN1: fibronectin, MPO: myeloperoxidase.
4. Western Blot validation of spermatozoa proteins in men with Hodgkin's disease
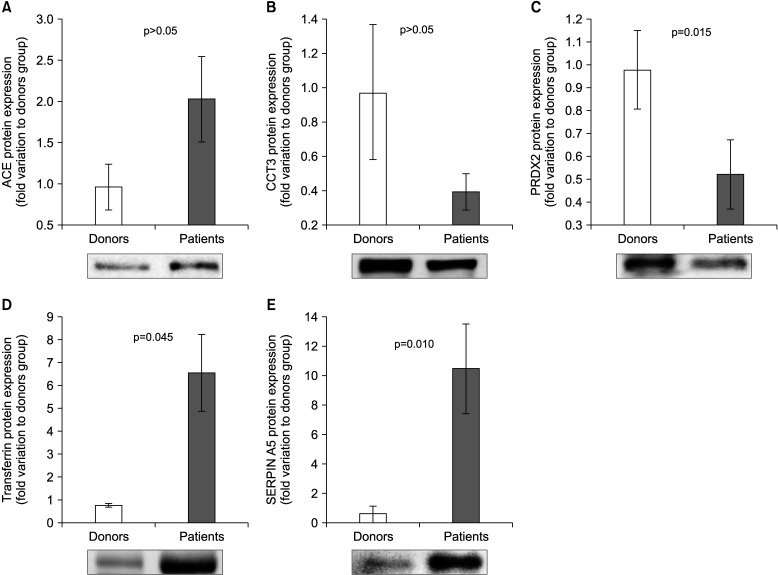
Based on selection criteria and the biological role, five proteins (ACE, PRDX2, CCT3, TF, and SERPINA5) were validated by WB. Our results showed that the expression of ACE was comparable in control and HD groups (Fig. 4A). The expression level of CCT3 was found to be similar in HD and control groups (Fig. 4C). Furthermore, our WB results revealed an underexpression (p=0.015) of PRDX 2 (Fig. 4D), while overexpression of TF (p=0.045) and SERPINA5 (p=0.010) in HD group when compared to fertile donor group.
Fig. 4. Protein expression levels of the differentially expressed proteins selected for validation by Western blot in spermatozoa of proven fertile donors' group and patients with Hodgkin Disease before cancer therapy. (A) Angiotensin converting enzyme (ACE); (B) T-complex protein 1 subunit gamma (CCT3); (C) Peroxiredoxin-2 (PRDX2); (D) Transferrin; (E) Plasma serine protease inhibitor (SERPIN A5). Results are expressed as mean±standard error of mean and in fold variation to donors' group.
DISCUSSION
Proteomic analysis using bioinformatic tools offers a comprehensive information regarding the protein distribution, and functional and molecular pathways associated with the peptides identified in spermatozoa [13]. Previous studies in the global proteomics analysis of spermatozoa resulted in the discovery of biomarkers for different pathologies [10,13,14,15,22]. In this study we used LC-MS/MS, a high throughput technique, to profile the spermatozoa proteins from men diagnosed with HD prior to cancer therapy and compared it with the proteome profile of fertile donors. The semen analysis was performed according to WHO guidelines (2010) [16], and the results indicated that the men from cancer group had lower sperm concentration, total sperm count and total motile count, while there was no significant difference in the motility. The quality of sperm in HD patients was lower when compared to fertile donors, however the sperm parameters values from patients were still within the WHO (2010) [16] reference values. The presence of lower sperm quality in men with cancer, particularly with HD obtained in our study is compliant with previous reports [5,6,7]. However, semen analysis alone can not predict whether the patients are fertile, subfertile or infertile. Semen parameters are used as a screening test to detect the contribution of male factor, but they fail to provide a full understanding of fertility potential [23,24]. As several factors (hyperactivation, capacitation, acrosome reaction, oxidative stress, sperm DNA fragmentation, etc.) in addition to semen quality contribute to the ability of spermatozoa to fertilize an oocyte, use of high-throughput techniques can allow a better understanding of the spermatozoal fertility potential at molecular level. In the present study, proteomic profiling of spermatozoa showed altered expression of key proteins associated with sperm function, which may contribute to the diminished fertility potential seen in some HD patients.
To minimize the variability existent in the complex nature of the spermatozoa [12], we had pooled the samples and run it in triplicate. LC-MS/MS spectrometry identified a total of 1,169 different peptides in the two experimental groups and about 1,049 were common to both experimental groups. The proteomic profile of spermatozoa is not new and similar studies have been performed in mature and immature ejaculated spermatozoa from fertile men [14], infertile men [11], and in spermatozoa of infertile men with bilateral varicocele [22]. However, this is the first report on the sperm proteins in men with HD before cancer therapy. To find an explanation for the lower sperm quality in these patients, we had chosen five proteins for validation. These proteins were selected based on their involvement in reproductive functions and fertilization process. Cell to cell signaling and interactions are crucial for the fertilization process [25,26], and the binding of sperm to zona pellucida is in fact crucial for the oocyte fertilization. Our analysis revealed that binding of zona pellucida and binding of sperm were dysregulated in spermatozoa of men with HD. ACE is a protein involved in fertility and our IPA analysis relates this protein to binding of sperm and zona pellucida, and interrelates the binding of sperm with fertilization process and fertility. ACE has been reported to be altered in infertile patients [13,27,28]. In our experiment, the proteomic analysis showed an overexpression in ACE, although the WB validation did not show any significant increase in expression. This protein has been described as a functional protein in sperm motility and capacitation [26,29]. Due to the localization of ACE in the peri-acrosomal section of the sperm head, and an agonist-induced function in acrosomal exocytosis, this protein consequently has a significant role in the fertility process [30]. Studies have reported a correlation between decrease in ACE with an increase in ROS and possible impairment of acrosome reaction [28,29]. PRDXs are central antioxidant enzymes and have a role in sperm function and male fertility [31]. PRDX2 is mostly found in the cytosol, eliminating ROS formed as a by-product of metabolism [32]. This protein was found to be underexpressed by the proteomic analysis and validated by WB. Since PRDX2 is an antioxidant enzyme, the underexpression of this protein can be related to low levels of oxidative stress. Transferrin which has been found to be overexpressed by proteomic analysis and validated by WB is also involved in sperm protection [33]. This protein decreases oxidative stress by reducing the availability of free iron [34].
The acrosome is crucial for the fertilization and it contains numerous hydrolytic enzymes, including ACR [35]. ACR is synthesized and stored in the sperm acrosome matrix in the form of proacrosin, an inactive form of ACR. During acrosome reaction, proacrosin transforms to intermediate and mature forms through autoactivation [36]. Previous studies have reported the importance of ACR in fertility and sperm ACR activity is significantly reduced in men with anti-sperm antibodies [37]. Another study reported that the ACR activity is a reflection of male fertility status [38]. ACR is a proteolytic enzyme capable of hydrolyzing the zona pellucida of oocyte, playing an important role in fertilization [39]. This is in line with our IPA analysis results which revealed that ACR was related to binding of sperm, zona pellucida and fertilization process. Our proteomics analysis demonstrated an under expression of ACR. The low expression of ACR in patients can be related with other protein identified by LC-MS/MS, the probable inactive serine protease 37 (PRSS37). PRSS37 is involved in acrosin activation [36], and abnormal activation of proacrosin/acrosin system has been reported in sperm with low levels PRSS37 [36]. Interestingly, our proteomic results showed an underexpression of PRSS37 (Table 3). SERPIN A5 was found to be overexpressed in spermatozoa of men with HD compared with proven fertile donors by LC-MS/MS spectroscopy with concordant results from validation. SERPIN A5 is localized in external plasma membrane and has many functions including inhibition of several serine proteases related to male reproductive tract [40,41,42]. This protein has a role in motility [40,41], fertilization process [42], and indirectly inhibits the degradation of semenogelin1 and 2 [43]. Our proteomic study results demonstrated the overexpression of semenogelin1 and 2 (Table 3), which is concordant with the overexpression of SERPIN A5 that inhibits the degradation of these proteins. Besides these functions, SERPIN A5 also inhibits acrosin and indirectly protects the male genital tract from the damaging effects of excess acrosin [42].
CCT3 is a subunit of T-complex protein 1, and a chaperone responsible for protein folding in the cells that requires the assistance of enzymes [44]. Low levels of this protein are related to low cell proliferation in cell counts, and induced cell apoptosis [45,46]. This fact can be indirectly responsible for the low sperm count in the HD group. Another fact to consider is the association between high levels of CCT3 and its negative correlation with survival in cancer patients [45,46]. The expression levels of CCT3 are low in sperm of HD patients, which is in compliance with the high survival rate for HD described by the American Cancer Society, [1].
CONCLUSIONS
This study aimed to explain reasons for poor semen quality seen in men diagnosed with HD prior to cancer therapy, as understanding the cellular and molecular mechanisms of cancer-associated poor semen quality may guide future research in fertility preservation. Our proteomic data showed an altered proteomic profile in spermatozoa of men with HD and our WB results validated the vital proteins involved in the fertility process. Validated proteins can serve as potential biomarkers to determine the sperm quality and fertility status in HD patients prior to cancer therapy.
ACKNOWLEDGEMENTS
Financial support for this study was provided by the American Center for Reproductive Medicine, Cleveland Clinic, OH, USA. Ana D. Martins was funded by FCT (SFRH/BD/108726/2015) and Fulbright (ID: E0585654). The authors would like to thank to: Dr. Belinda Willard (Director, Proteomics Core Laboratory, Lerner Research Institute, Cleveland Clinic) for her support in the proteomic analysis.
Footnotes
Conflict of Interest: The authors have nothing to disclose.
- Conceptualization: AA.
- Data curation: ADM, PNP
- Formal analysis: ADM, MKPS, SB.
- Investigation: AA.
- Methodology: ADM, PNP.
- Project administration: AA.
- Resources: ADM, MKPS.
- Software: PNP.
- Supervision: AA.
- Validation: ADM.
- Writing—original draft: ADM.
- Writing—review & editing: all authors.
Data Sharing Statement
The data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
References
- 1.American Cancer Society. Key statistics for hodgkin lymphoma [Internet] Atlanta: American Cancer Society; 2018. [cited 2018 Oct 23]. Available from: https://www.cancer.org/cancer/hodgkin-lymphoma/about/key-statistics.html. [Google Scholar]
- 2.Eichenauer DA, Becker I, Monsef I, Chadwick N, de Sanctis V, Federico M, et al. Secondary malignant neoplasms, progression-free survival and overall survival in patients treated for Hodgkin lymphoma: a systematic review and meta-analysis of randomized clinical trials. Haematologica. 2017;102:1748–1757. doi: 10.3324/haematol.2017.167478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal A, Ranganathan P, Kattal N, Pasqualotto F, Hallak J, Khayal S, et al. Fertility after cancer: a prospective review of assisted reproductive outcome with banked semen specimens. Fertil Steril. 2004;81:342–348. doi: 10.1016/j.fertnstert.2003.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S, Sharma R, Agarwal A. The process of sperm cryopreservation, thawing and washing techniques. In: MajzoubA, Agarwal A, editors. The complete guide to male fertility preservation. Cham: Springer; 2018. pp. 183–204. [Google Scholar]
- 5.van der Kaaij MA, Heutte N, van Echten-Arends J, Raemaekers JM, Carde P, Noordijk EM, et al. Sperm quality before treatment in patients with early stage Hodgkin's lymphoma enrolled in EORTC-GELA Lymphoma Group trials. Haematologica. 2009;94:1691–1697. doi: 10.3324/haematol.2009.009696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitoussi, Eghbali H, Tchen N, Berjon JP, Soubeyran P, Hoerni B. Semen analysis and cryoconservation before treatment in Hodgkin's disease. Ann Oncol. 2000;11:679–684. doi: 10.1023/a:1008353728560. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi H, Larson K, Sharma RK, Nelson DR, Evenson DP, Toma H, et al. DNA damage in patients with untreated cancer as measured by the sperm chromatin structure assay. Fertil Steril. 2001;75:469–475. doi: 10.1016/s0015-0282(00)01740-4. [DOI] [PubMed] [Google Scholar]
- 8.Chung K, Irani J, Knee G, Efymow B, Blasco L, Patrizio P. Sperm cryopreservation for male patients with cancer: an epidemiological analysis at the University of Pennsylvania. Eur J Obstet Gynecol Reprod Biol. 2004;113 Suppl 1:S7–S11. doi: 10.1016/j.ejogrb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 9.Barr RD, Clark DA, Booth JD. Dyspermia in men with localized Hodgkin's disease. A potentially reversible, immunemediated disorder. Med Hypotheses. 1993;40:165–168. doi: 10.1016/0306-9877(93)90205-5. [DOI] [PubMed] [Google Scholar]
- 10.Panner Selvam MK, Agarwal A. Update on the proteomics of male infertility: a systematic review. Arab J Urol. 2017;16:103–112. doi: 10.1016/j.aju.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samanta L, Sharma R, Cui Z, Agarwal A. Proteomic analysis reveals dysregulated cell signaling in ejaculated spermatozoa from infertile men. Asian J Androl. 2019;21:121–130. doi: 10.4103/aja.aja_56_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Mateo S, Martínez-Heredia J, Estanyol JM, Domínguez-Fandos D, Vidal-Taboada JM, Ballescà JL, et al. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics. 2007;7:4264–4277. doi: 10.1002/pmic.200700521. [DOI] [PubMed] [Google Scholar]
- 13.Sharma R, Agarwal A, Mohanty G, Hamada AJ, Gopalan B, Willard B, et al. Proteomic analysis of human spermatozoa proteins with oxidative stress. Reprod Biol Endocrinol. 2013;11:48. doi: 10.1186/1477-7827-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Z, Sharma R, Agarwal A. Proteomic analysis of mature and immature ejaculated spermatozoa from fertile men. Asian J Androl. 2016;18:735–746. doi: 10.4103/1008-682X.164924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thacker S, Yadav SP, Sharma RK, Kashou A, Willard B, Zhang D, et al. Evaluation of sperm proteins in infertile men: a proteomic approach. Fertil Steril. 2011;95:2745–2748. doi: 10.1016/j.fertnstert.2011.03.112. [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization. WHO laboratory manual for the examination and processing of human semen. Geneva: WHO; 2010. [Google Scholar]
- 17.Ayaz A, Agarwal A, Sharma R, Arafa M, Elbardisi H, Cui Z. Impact of precise modulation of reactive oxygen species levels on spermatozoa proteins in infertile men. Clin Proteomics. 2015;12:4. doi: 10.1186/1559-0275-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogle OA, Kumar K, Attardo-Parrinello C, Lewis SE, Estanyol JM, Ballescà JL, et al. Identification of protein changes in human spermatozoa throughout the cryopreservation process. Andrology. 2017;5:10–22. doi: 10.1111/andr.12279. [DOI] [PubMed] [Google Scholar]
- 19.Sharma R, Agarwal A, Mohanty G, Jesudasan R, Gopalan B, Willard B, et al. Functional proteomic analysis of seminal plasma proteins in men with various semen parameters. Reprod Biol Endocrinol. 2013;11:38. doi: 10.1186/1477-7827-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal A, Sharma R, Durairajanayagam D, Ayaz A, Cui Z, Willard B, et al. Major protein alterations in spermatozoa from infertile men with unilateral varicocele. Reprod Biol Endocrinol. 2015;13:8. doi: 10.1186/s12958-015-0007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteves SC, Agarwal A. Afterword to varicocele and male infertility: current concepts and future perspectives. Asian J Androl. 2016;18:319–322. doi: 10.4103/1008-682X.172820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal A, Sharma R, Durairajanayagam D, Cui Z, Ayaz A, Gupta S, et al. Spermatozoa protein alterations in infertile men with bilateral varicocele. Asian J Androl. 2016;18:43–53. doi: 10.4103/1008-682X.153848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiles KA, Schlegel PN. What do semen parameters mean? How to define a normal semen analysis. Andrology. 2015 doi: 10.4172/2167-0250.1000136. [DOI] [Google Scholar]
- 24.Wang C, Swerdloff RS. Limitations of semen analysis as a test of male fertility and anticipated needs from newer tests. Fertil Steril. 2014;102:1502–1507. doi: 10.1016/j.fertnstert.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okabe M. The cell biology of mammalian fertilization. Development. 2013;140:4471–4479. doi: 10.1242/dev.090613. [DOI] [PubMed] [Google Scholar]
- 26.Shibahara H, Kamata M, Hu J, Nakagawa H, Obara H, Kondoh N, et al. Activity of testis angiotensin converting enzyme (ACE) in ejaculated human spermatozoa. Int J Androl. 2001;24:295–299. doi: 10.1046/j.1365-2605.2001.00301.x. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal A, Durairajanayagam D, Halabi J, Peng J, Vazquez-Levin M. Proteomics, oxidative stress and male infertility. Reprod Biomed Online. 2014;29:32–58. doi: 10.1016/j.rbmo.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Ayaz A, Agarwal A, Sharma R, Kothandaraman N, Cakar Z, Sikka S. Proteomic analysis of sperm proteins in infertile men with high levels of reactive oxygen species. Andrologia. 2018;50:e13015. doi: 10.1111/and.13015. [DOI] [PubMed] [Google Scholar]
- 29.Zalata AA, Morsy HK, Badawy Ael-N, Elhanbly S, Mostafa T. ACE gene insertion/deletion polymorphism seminal associations in infertile men. J Urol. 2012;187:1776–1780. doi: 10.1016/j.juro.2011.12.076. [DOI] [PubMed] [Google Scholar]
- 30.Bromfield EG, McLaughlin EA, Aitken RJ, Nixon B. Heat Shock Protein member A2 forms a stable complex with angiotensin converting enzyme and protein disulfide isomerase A6 in human spermatozoa. Mol Hum Reprod. 2016;22:93–109. doi: 10.1093/molehr/gav073. [DOI] [PubMed] [Google Scholar]
- 31.Ryu DY, Kim KU, Kwon WS, Rahman MS, Khatun A, Pang MG. Peroxiredoxin activity is a major landmark of male fertility. Sci Rep. 2017;7:17174. doi: 10.1038/s41598-017-17488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang SW, Chae HZ, Seo MS, Kim K, Baines IC, Rhee SG. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J Biol Chem. 1998;273:6297–6302. doi: 10.1074/jbc.273.11.6297. [DOI] [PubMed] [Google Scholar]
- 33.van Tilburg MF, Sousa SD, Ferreira de Melo RB, Moreno FB, Monteiro-Moreira AC, Moreira RA, et al. Proteome of the rete testis fluid from tropically-adapted Morada Nova rams. Anim Reprod Sci. 2017;176:20–31. doi: 10.1016/j.anireprosci.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal A, Prabakaran SA. Mechanism, measurement, and prevention of oxidative stress in male reproductive physiology. Indian J Exp Biol. 2005;43:963–974. [PubMed] [Google Scholar]
- 35.Liu DY, Baker HW. Tests of human sperm function and fertilization in vitro. Fertil Steril. 1992;58:465–483. doi: 10.1016/s0015-0282(16)55247-9. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Shen C, Fan W, Chen Y, Zhang A, Feng Y, et al. Low levels of PRSS37 protein in sperm are associated with many cases of unexplained male infertility. Acta Biochim Biophys Sin (Shanghai) 2016;48:1058–1065. doi: 10.1093/abbs/gmw096. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Zhao E, Zhang C, Li X. The change of semen superoxide dismutase and acrosin activity in the sterility of male patients with positive antisperm antibody. Cell Biochem Biophys. 2015;73:451–453. doi: 10.1007/s12013-015-0663-z. [DOI] [PubMed] [Google Scholar]
- 38.Reichart M, Lederman H, Har-Even D, Kedem P, Bartoov B. Human sperm acrosin activity with relation to semen parameters and acrosomal ultrastructure. Andrologia. 1993;25:59–66. doi: 10.1111/j.1439-0272.1993.tb02683.x. [DOI] [PubMed] [Google Scholar]
- 39.Mao HT, Yang WX. Modes of acrosin functioning during fertilization. Gene. 2013;526:75–79. doi: 10.1016/j.gene.2013.05.058. [DOI] [PubMed] [Google Scholar]
- 40.Meijers JC, Kanters DH, Vlooswijk RA, van Erp HE, Hessing M, Bouma BN. Inactivation of human plasma kallikrein and factor XIa by protein C inhibitor. Biochemistry. 1988;27:4231–4237. doi: 10.1021/bi00412a005. [DOI] [PubMed] [Google Scholar]
- 41.España F, Gilabert J, Estellés A, Romeu A, Aznar J, Cabo A. Functionally active protein C inhibitor/plasminogen activator inhibitor-3 (PCI/PAI-3) is secreted in seminal vesicles, occurs at high concentrations in human seminal plasma and complexes with prostate-specific antigen. Thromb Res. 1991;64:309–320. doi: 10.1016/0049-3848(91)90002-e. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X, Geiger M, Ecke S, Bielek E, Donner P, Eberspächer U, et al. Inhibition of acrosin by protein C inhibitor and localization of protein C inhibitor to spermatozoa. Am J Physiol. 1994;267:C466–C472. doi: 10.1152/ajpcell.1994.267.2.C466. [DOI] [PubMed] [Google Scholar]
- 43.Kise H, Nishioka J, Kawamura J, Suzuki K. Characterization of semenogelin II and its molecular interaction with prostate-specific antigen and protein C inhibitor. Eur J Biochem. 1996;238:88–96. doi: 10.1111/j.1432-1033.1996.0088q.x. [DOI] [PubMed] [Google Scholar]
- 44.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 45.Cui X, Hu ZP, Li Z, Gao PJ, Zhu JY. Overexpression of chaperonin containing TCP1, subunit 3 predicts poor prognosis in hepatocellular carcinoma. World J Gastroenterol. 2015;21:8588–8604. doi: 10.3748/wjg.v21.i28.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Wang Y, Wei Y, Wu J, Zhang P, Shen S, et al. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett. 2016;372:101–109. doi: 10.1016/j.canlet.2015.12.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.