Summary
Tumor heterogeneity and cisplatin resistance are major causes of tumor relapse and poor survival. Here, we show that in lung cancer, interaction between paxillin (PXN) and integrin β4 (ITGB4), components of the focal adhesion (FA) complex, contributes to cisplatin resistance. Knocking down PXN and ITGB4 attenuated cell growth and improved cisplatin sensitivity, both in 2D and 3D cultures. PXN and ITGB4 independently regulated expression of several genes. In addition, they also regulated expression of common genes including USP1 and VDAC1, which are required for maintaining genomic stability and mitochondrial function, respectively. Mathematical modeling suggested that bistability could lead to stochastic phenotypic switching between cisplatin-sensitive and resistant states in these cells. Consistently, purified subpopulations of sensitive and resistant cells re-created the mixed parental population when cultured separately. Altogether, these data point to an unexpected role of the FA complex in cisplatin resistance and highlight a novel non-genetic mechanism.
Subject Areas: Cell Biology, Mathematical Biosciences, Cancer
Graphical Abstract

Highlights
-
•
In lung cancer, interaction between PXN and ITGB4 contributes to cisplatin resistance.
-
•
Knocking down PXN and ITGB4 attenuated cell growth and improved cisplatin sensitivity.
-
•
PXN and ITGB4 regulate expression of USP1 and VDAC1, respectively.
-
•
The data underscore a novel non-genetic mechanism for chemoresistance in lung cancer.
Cell Biology; Mathematical Biosciences; Cancer
Introduction
Lung cancer is the most frequently diagnosed cancer and a leading cause of cancer-related death worldwide (Bray et al., 2018). Approximately 85% patients have a group of histological subtypes collectively known as non-small cell lung cancer (NSCLC), of which lung adenocarcinoma (LUAD) and lung squamous cell carcinoma are the most common subtypes, followed by large cell carcinoma (Salgia, 2016; Herbst et al., 2018). LUAD accounts for ∼40% of all lung cancers. These subtypes differ in their cell of origin and display different patterns of genomic alterations that influence their response to cytotoxic chemotherapies. Approximately 40% to 50% patients with NSCLC will be diagnosed with advanced or metastatic disease and are not candidates for curative therapy. Recent advances have transformed lung cancer care for a percentage of patients, with EGFR mutations, receiving first-line tyrosine kinase inhibitors (Tan et al., 2017). However, immunotherapy alone or in combination with platinum-based chemotherapy remains the recommended first-line treatment option for the remainder of these patients.
Cisplatin is a widely prescribed platinum-based compound that is effective against a wide spectrum of solid neoplasms, including testicular, bladder, ovarian, colorectal, lung, and head and neck cancers (Galluzzi et al, 2012, 2014). Cisplatin treatment is generally associated with high rates of clinical responses. However, in the vast majority of cases, malignant cells exposed to cisplatin activate a multipronged adaptive response that renders them less susceptible to the anti-proliferative and cytotoxic effects of the drug, and eventually resume proliferation. Nonetheless, the exact mechanism(s) underlying the emergence of drug resistance remains poorly understood, and a bewildering plethora of targets have been implicated in different cancer types (Sarin et al., 2017; Xiao et al., 2017; Hu et al., 2016; Deben et al., 2018). Furthermore, although changes in administration schedules, choice of methods, and frequency of toxicity monitoring have all contributed to incremental improvements, chemoresistance limits the clinical utility of cisplatin (Fennell et al., 2016).
The focal adhesion (FA) complex is a large macromolecular assembly through which mechanical force and regulatory signals are transmitted between the extracellular matrix and an interacting cell (Chen et al., 2003). Paxillin (PXN), integrins, and focal adhesion kinase (FAK) are among the major components of this complex. In LUAD, expression of PXN is correlated with tumor progression and metastasis (Song et al., 2010; Mackinnon et al., 2011). Furthermore, phosphorylation of PXN activates the MAPK/ERK pathway, increasing Bcl-2 expression and cisplatin resistance (Wu et al., 2014). Furthermore, certain PXN mutants, through their interactions with Bcl-2 and dynamin-related protein 1 (DRP1), also regulate cisplatin resistance in human lung cancer cells (Kawada et al., 2013).
Integrins are transmembrane receptors that facilitate cell-extracellular matrix adhesion; they form a critical link between the extracellular matrix and the cell interior by interacting with the FA via PXN. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals that regulate cell cycle, organization of the intracellular cytoskeleton, and movement of new receptors to the cell membrane (Giancotti and Ruoslahti 1999; Maziveyi and Alahari, 2017). Integrins are obligate heterodimers of one α and one β subunit. In mammals, there are 24 α and 9 β subunits (Alberts et al., 2014). Among the various β subunits, β1 is ubiquitously expressed in most cell types and can dimerize with multiple α subunits, forming receptors for extracellular matrix proteins. On the other hand, integrin β4 (ITGB4) is reported to be quite selective and heterodimerizes only with the α6 subunit and binds to laminin (Mainiero et al., 1997). ITGB4 is also unique because of its >1,000-amino acid-long cytoplasmic domain compared with the ∼50-amino acid-long domain of other β forms (Su et al., 2008). In NSCLC, the receptor tyrosine kinase MET interacts with ITGA6/B4, and this interaction is required for hepatocyte growth factor (HGF)-dependent tumor invasion (Trusolino et al., 2001). In addition, the presence of the ITGA6/B4 heterodimer in tumor-derived exosomes facilitates the creation of a microenvironment that promotes lung cancer (Hoshino et al., 2015). Together, these observations underscore the importance of the FA complex in NSCLC pathophysiology. However, how the interactions of the individual components of the FA complex may contribute to cisplatin resistance remains poorly understood.
In this article, we demonstrate that direct interaction between paxillin (PXN) and integrin β4 (ITGB4), two key components of the FA complex, contributes to cisplatin resistance in LUAD. Thus, knocking down PXN and ITGB4 in cisplatin-resistant NSCLC cells significantly attenuated cell growth and improved cisplatin sensitivity, and the double knockdown showed an additive effect both in 2D and 3D spheroid cultures. Mechanistically, PXN or ITGB4 independently regulated expression of several unique genes, and together regulate expression of USP1 and VDAC1 that are required for maintaining genomic stability and mitochondrial function, respectively. Mathematical modeling suggested that bistability could lead to stochastic phenotypic switching between cisplatin-sensitive and resistant states in these cells. Consistently, purified subpopulations of sensitive and resistant cells re-created the mixed parental population when cultured separately. Taken together, our data highlight a novel role for the FA complex in cisplatin resistance in LUAD and uncover a non-genetic mechanism underlying chemoresistance in the disease.
Results
Upregulation of Both PXN and ITGB4 but Not PXN Alone Correlates with Cisplatin Resistance
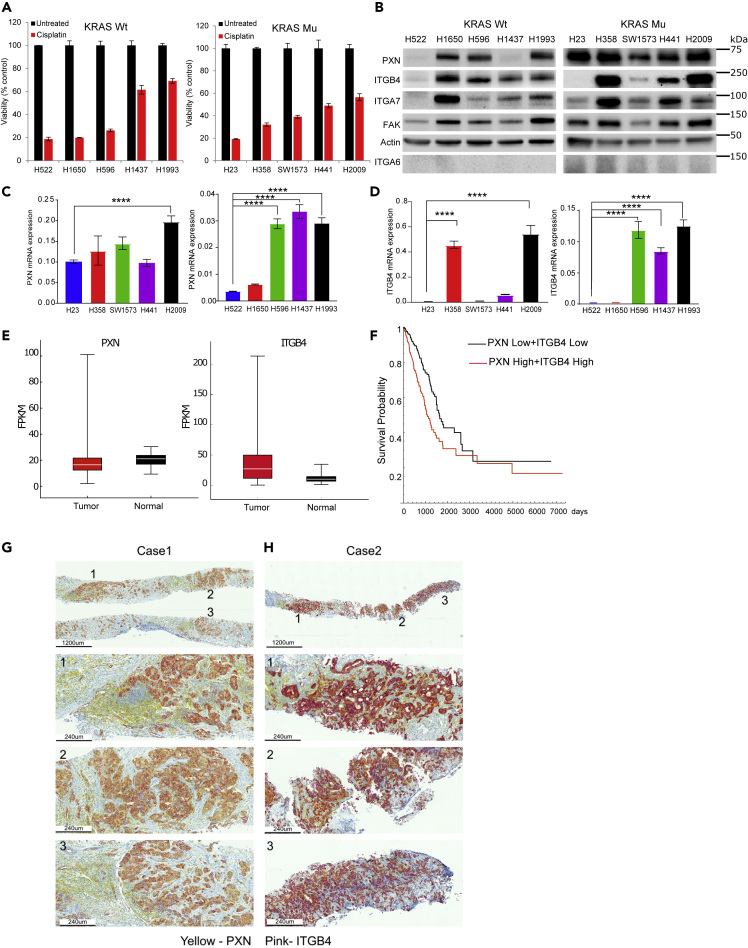
A gene set enrichment (GSE) analysis of the RNA sequencing (RNA-seq) data from the Molecular Signatures Database v6.2 (MSigDB), a collection of annotated gene sets (Broad Institute), identified a set of 11 genes that were upregulated in cisplatin-resistant cells compared with sensitive cells (Table S1). Four of these 11 genes, namely, PXN, ITGB4, ITGA7, and Rac Family Small GTPase 1, are constituents of the FA complex activation, formation, and downstream signaling pathways. Therefore, to verify the correlation of these genes with cisplatin resistance, we randomly selected five LUAD cell lines that harbored wild-type (WT) KRAS and five that carried a mutant (MT) version of KRAS, treated them with 10 μM cisplatin for 72 h, and determined cell viability using the CCK-8 viability assay. Among the KRAS WT cell lines, H1437 and H1993 were significantly more resistant, whereas among the KRAS MT cell lines, H441 and H2009 were relatively more resistant than the other three mutant cell lines (Figure 1A).
Figure 1.
PXN and ITGB4 Are Upregulated in Lung Cancer and in Cisplatin-Resistant LUAD Cell Lines
(A) Five LUAD cell lines with wild-type (WT) KRAS and five cell lines with mutant (MT) KRAS were treated with 10 μM cisplatin for 72 h, and cell viability was determined using Cell Counting Kit-8 assay.
(B) Immunoblot showing that the most cisplatin-resistant cell lines (H1993 and H2009) have high expression of both PXN and ITGB4.
(C and D) qPCR results demonstrating high mRNA expression of (C) PXN and (D) ITGB4 in cisplatin-resistant cell lines.
(E) Gene expression profiles of patients with LUAD were extracted from TCGA database, and expressions of PXN and ITGB4 were higher compared with normal tissue.
(F) Kaplan-Meier curves indicate significant survival difference (p = 0.000089) between the two groups (PXN low + ITGB4 low and PXN high + ITGB4 high).
(G) Immunohistochemistry staining of tissue from Case 1 (needle biopsy) showing ubiquitous expression of PXN (yellow) and weak expression of ITGB4 (pink).
(H) Staining of Case 2 (needle biopsy) showing ubiquitous expression of PXN (yellow) and high expression of ITGB4 (pink).
Data are represented as mean ± SD (∗∗∗∗p < 0.0001 one-way ANOVA).
An immunoblotting analysis of the cell lysates revealed that PXN and FAK were expressed in all the cell lines tested regardless of whether they were cisplatin sensitive or resistant (Figure 1B). The expression of ITGB4 and ITGA7 was variable, whereas RAC1 and ITGA6 were undetectable among the cell lines. ITGB4 expression was high in the two cisplatin-resistant cell lines, H2009 and H1993. Finally, a qPCR analysis using total RNA from the cell lines showed that the mRNAs for both PXN and ITGB4 were also upregulated in the H2009 and H1993 cell lines (Figures 1C and 1D). In contrast, very low expression of ITGA6 was observed in these cell lines compared with SW480 that was used as a positive control (Figure S1).
Co-expression of PXN and ITGB4 Correlates with Poorer Prognosis in Patients with LUAD
As LUAD cell lines overexpressing PXN were sensitive to cisplatin treatment when they did not express ITGB4, we hypothesized that there is a correlation between PXN and ITGB4 expression and cisplatin resistance. To test this hypothesis, gene expression profiles of 498 patients diagnosed with “lung adenocarcinoma” were extracted from The Cancer Genome Atlas (TCGA) database and analyzed for expression of ITGB4 and PXN (Figure 1E). The gene-level expression values FPKM (fragments per kilobase of exon model per million reads mapped) were analyzed according to the Cox proportional hazards model: Survival ∼ Sex + Stage + Age + PXN ∗ ITGB4. Stage and PXN ∗ ITGB4 showed significant contribution to the model. We separated the patients into two groups by the median value of the PXN∗ITGB4 product (Table S2). The Kaplan-Meier curves showed clear survival difference between the two groups, and the log rank test showed significant differences between the two curves (p = 0.000089) (Figure 1F). In addition, we performed immunohistochemistry (IHC) on needle biopsy specimens obtained from two de-identified cases from City of Hope. PXN staining was detected using yellow, and ITGB4, with pink color. Thus, the coexpression of the two molecules would generate an orange or bright red color based on the expression level. Indeed, we observed significant coexpression of ITGB4 and PXN in various regions of the tumor (Figures 1G and 1H). The case represented in Figure 1G had weak expression of ITGB4, and coexpression generated an orange color. The second case (Figure 1H) that had high expression of ITGB4 generated a reddish pattern, implying coexpression. We further extended the IHC staining to three more cases where staining of the whole tumor section was done. We observed variability in the expression of PXN and ITGB4 among cases and within the sample as well. PXN was expressed in most regions of the tumor, but ITGB4 expression was sporadic, and the level of ITGB4 expression was also variable among cases. Case 3 had high expression of ITGB4, Case 4 had intermediate expression, and Case 5 had weak expression (Figures S2A–S2C). The immunostaining also suggests that PXN expression was not only limited to the tumors but also could be observed in the surrounding lymphoid tissue. The PXN staining in normal lung tissue was weak compared with tumor tissue, reconfirming the upregulation of PXN in adenocarcinoma. The coexpression of PXN and ITGB4 (red or orange color) was limited to some regions of the tumor, emphasizing heterogeneity in their expression patterns (Figures S2A–S2C). This suggested that cisplatin treatment appears to favor the selection of a subclonal population of tumor cells having coexpression of ITGB4 and PXN. Therefore, next, we interrogated the role of ITGB4 in cisplatin resistance in those cell lines that had high expression of both ITGB4 and PXN.
Knocking down ITGB4 Attenuates Proliferation and Migration of Cisplatin-Resistant Cells
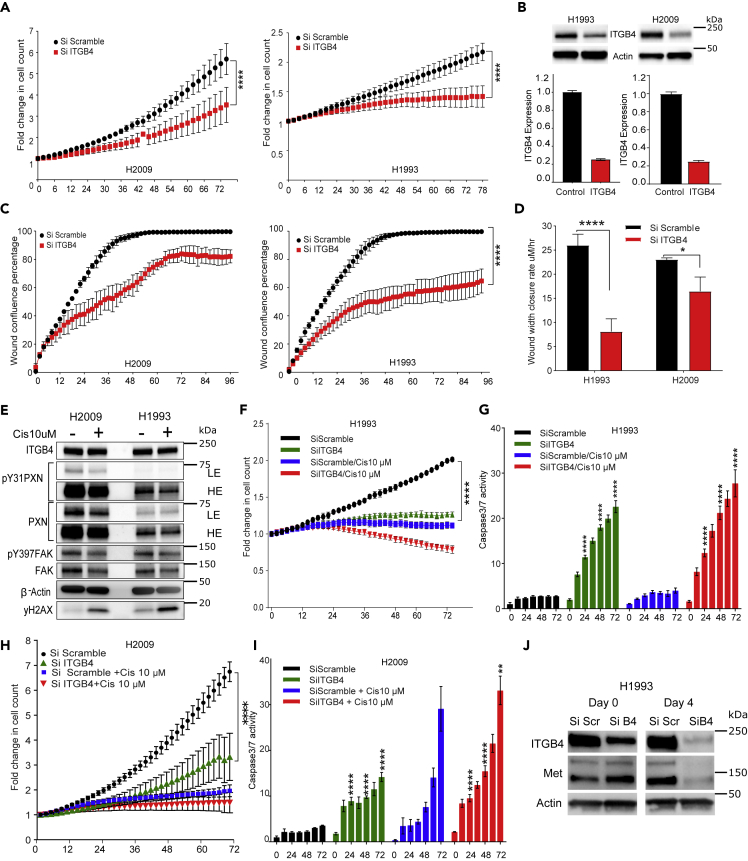
We infected H1993, H2009, H358, and A549 cells with IncuCyte NucLight lentivirus particles for generating stable cell lines expressing nucleus-restricted red fluorescence protein, mKate2 (Figure S3A). These stable cells were transiently transfected with a control (scrambled) and ITGB4-specific siRNA, and the effect of silencing its expression on cell proliferation was determined using the IncuCyte Live Cell Imaging System (Essen BioScience, Ann Arbor, MI). We tested three small interfering RNA (siRNA) constructs from OriGene in the commonly employed NSCLC cell lines H358 and A549. siRNA C was found to be most effective in knocking down ITGB4 and inhibiting proliferation (Figures S3B–S3D). Consistent with these findings, knocking down ITGB4 using siRNA C in H1993 and H2009 cells significantly attenuated their proliferation (Figure 2A). The knockdown was confirmed with immunoblotting analysis using total cell lysates and qPCR analysis with RNA extracted from transfected cells (Figure 2B). Henceforth, we used the siRNA C construct for the remainder of the experiments.
Figure 2.
Knocking Down ITGB4 Attenuates Proliferation and Migration and Increases Sensitivity of Cisplatin-Resistant Cells
H2009 and H1993 stable cell lines expressing mKate2 were transfected with control (Si Scramble) or ITGB4-specific (Si ITGB4) siRNA.
(A and B) (A) ITGB4 knockdown cells (red) had a significantly reduced proliferation rate than control cells (black). (∗∗∗∗p < 0.0001 two-way ANOVA). (B) Immunoblotting and qPCR data confirming the knockdown.
(C and D) (C) A scratch wound assay demonstrating the effect of knocking down ITGB4. ITGB4 knockdown (red) significantly halted the migration and did not close the wound completely after 96 h in both resistant cell lines. (∗∗∗∗p < 0.0001 two-way ANOVA). (D) Rate at which the wound closed was also significantly decreased in ITGB4 knockdown cells (red). (∗∗∗∗p < 0.0001 and ∗p < 0.0156 two-way ANOVA).
(E) Cisplatin (10 μM) treatment for 72 h reduced expression of phosphorylated PXN, PXN, and total FAK, but not ITGB4. (LE = low exposure, HE = high exposure).
(F) ITGB4 knockdown in H1993 cells inhibited proliferation and addition of cisplatin had a cytotoxic effect (∗∗∗∗p < 0.0001 two-way ANOVA).
(G) ITGB4 knockdown in H1993 cells increased caspase-3/7 activity. Treating ITGB4 knockdown cells with cisplatin had an added effect of inducing caspase activity, but drug treatment alone did not have a cytotoxic effect (∗∗∗∗p < 0.0001 two-way ANOVA).
(H) ITGB4 knockdown in H2009 cells inhibited proliferation by 72 h and addition of cisplatin had a cytostatic effect (∗∗∗∗p < 0.0001 two-way ANOVA).
(I) ITGB4 knockdown in H2009 cells did not induce caspase activity. Cisplatin treatment to ITGB4 knockdown cells for 72 h had an additive effect to induce caspase activity (∗∗∗∗p < 0.0001 two-way ANOVA).
(J) Immunoblot showing that MET protein expression in H1993 cells was reduced 4 days after knocking down ITGB4. Data are represented as mean ± SD.
Next, we calculated the effect of ITGB4 knockdown on doubling time of H2009 and H1993. In the H2009 cells, we observed that by 50 h post transfection, control (scramble siRNA) cells showed a 4-fold increase in cell count, indicating the doubling time to be approximately 25 h. However, in ITGB4 knocked down H2009 cells, the doubling time increased to 40 h. Similarly, in H1993, knocking down ITGB4 led to only a 0.2-fold increase in cell count by 72 h, leading to an estimated doubling time of approximately 120 h compared with 72 h for control cells (Figure S4A). We also knocked down ITGA7 in H2009 cell lines to validate its role in cell proliferation but did not observe any significant change (Figure S4B). In addition, we also determined expression of other integrin β forms to rescue the effect of ITGB4 knockdown and found an increase in the mRNA levels of ITGB1, ITGB2, and ITGB3 (Figure S4C). We further tested the inhibitory effect of ITGB3 inhibitors in the ITGB4 knocked down cells but found no significant difference in their proliferation (Figure S4D).
To discern the effect of knocking down ITGB4 on cell migration, H1993 and H2009 cells were transfected with the control (scrambled) or ITGB4-specific siRNA in 6-well plates; 12 h post transfection, the cells were trypsinized and reseeded at high density in a 96-well plate. Once wells were confluent, a scratch wound was generated using the WoundMaker tool (Essen Bioscience), and wound healing was observed in real time using IncuCyte (Figure S4E, Videos S1 and S2). Both cell lines transfected with the scramble siRNA achieved 100% confluency of the wound within 48 h, whereas cells in which ITGB4 was knocked down reached ∼60% and ∼80% confluency, respectively, and neither cell line reached 100% confluency even after 96 h (Figure 2C). We next estimated the rate of wound closure. The rate was determined by dividing the initial wound width by the time taken to close the wound. In the case of the H1993 cells, the wound closure rate was ∼25 μm/h, and on knocking down ITGB4, the rate significantly dropped to <10 μm/h. Similarly, in the case of the H2009 cells, the wound width closure rate dropped from 22 μm/h to about 18 μm/h (Figure 2D). Together, these results confirmed the significance of ITGB4 in modulating cell proliferation and migration in cisplatin-resistant LUAD cell lines.
Knocking down ITGB4 Increases Sensitivity to Cisplatin Treatment
We treated H1993 and H2009 cells with cisplatin for 3 days and observed reduction in the expression of PXN and FAK within 48 h of cisplatin treatment, whereas the ITGB4 expression remained unaltered (Figure S4F). We observed a reduction in the expression of total FAK and PXN as well as phospho PXN, but not in ITGB4 expression (Figure 2E). Next, we transiently transfected H1993 and H2009 cells with ITGB4 siRNA and 24 h post-transfection treated them with 10 μM cisplatin and then determined cell proliferation and caspase activity (Figure S5A, Videos S3, S4, S5, and S6). In H1993, cisplatin treatment inhibited cell proliferation significantly, and the fold change in cell count was ∼1, indicating a cytostatic effect. However, in ITGB4 knockdown cells, the fold change in response to cisplatin dipped to below 1, alluding to a cytotoxic effect (Figure 2F). Furthermore, live caspase-3/7 activity analyzed in the same cells showed a 5- to 6-fold increase in activity compared with control by 24 h, and by 72 h, caspase activity increased to 20-fold versus control (Figures S5B–S5E). Addition of cisplatin to the ITGB4 knockdown cells enhanced cisplatin sensitivity, but cisplatin treatment alone did not induce caspase activity, indicating that whereas the reduction in cell proliferation by cisplatin was cytostatic, knocking down ITGB4 with cisplatin present induced cytotoxicity (Figure 2G). In KRAS MT H2009 cells, a similar trend was observed, but cell proliferation did not drop below 1, and the induction of caspase activity by ITGB4 knockdown was weak. Knocking down ITGB4 induced caspase activity within 24 h of seeding, whereas cisplatin treatment alone induced apoptosis in H2009 cells by 72 h and the combination increased sensitivity to cisplatin (Figures 2H and 2I). ITGB4 knockdown in another KRAS WT cell line H1650 also inhibited cell proliferation and induced cell death. However, cell death was not due to caspase activation but appeared to be due to cell bursting, which is related to anoikis (Figures S5F and S5G, Videos S7 and S8). Furthermore, ITGB4 knockdown also sensitized H358 and A549 cells to 2.5 μM cisplatin (Figures S6A and S6B). Together, these data underscore the role of ITGB4 in imparting cisplatin resistance in these two LUAD cell lines irrespective of their KRAS status.
It has been shown that the cytoplasmic domain of ITGB4 acts as a scaffold for various kinases involved in activating the MAPK, PI3K, or Akt pathways (Trusolino et al., 2001). MET Proto-Oncogene, Receptor Tyrosine Kinase (MET) is also one of the interacting partners of ITGB4 and is responsible for HGF-dependent phosphorylation and activation of ITGB4 (Trusolino et al., 2001). Of the two cell lines used in the present study, H1993 is known to harbor MET amplification. Thus, we asked if ITGB4 knockdown disrupts MET activity. Indeed, we observed a decrease in MET expression in ITGB4 knocked down cells at the protein level, but there was no significant change in the mRNA expression, suggesting that ITGB4 may be involved in MET protein stability in these cells (Figures 2J and S6C). However, in H2009 cells that have no MET amplification, no detectable decrease in MET expression was observed (Figure S6C). These data suggested that the cells dependent on MET signaling (H1993) may be more sensitive to ITGB4 knockdown, whereas cells independent of MET signaling (H2009) may be insensitive.
Knocking down Both PXN and ITGB4 has a Combined Effect on Attenuating Cisplatin Resistance
As cisplatin treatment caused changes in expression of FAK and PXN but not ITGB4 (Figure 2E), and all three proteins are required for FA complex formation and activation, we asked if PXN knockdown alone can induce a similar phenotype or PXN and ITGB4 double knockdown can cooperate to change cell proliferation and survival rates. In H1993 cells, PXN knockdown inhibited cell proliferation by 15% after 96 h compared with 50% inhibition by ITGB4 knockdown and 70% inhibition caused by double knockdown (Figure 3A). Treating H1993 single and double knockdown cells with cisplatin induced cytotoxic effect; a drop in the fold change of cell count by 40% and 60%, respectively, was observed compared with control (scramble siRNA) cells treated with cisplatin. However, treating PXN knocked down cells with cisplatin had a weaker effect (Figure 3B). We further tested the effect of knocking down PXN on apoptosis and found that PXN knockdown did not activate caspase-3/7 but increased caspase activity when combined with ITGB4 knockdown. Furthermore, knocking down ITGB4 alone or together with PXN made the cells more prone to cisplatin toxicity by inducing apoptosis (Figure S5C). In H2009 cells, double knockdown inhibited cell proliferation significantly, indicating a cytostatic effect compared with scramble knockdown (Figure 3C). Unlike the cytotoxic effect observed in H1993 cells, we observed a cytostatic effect when these double knockdown cells were treated with cisplatin (Figure 3D). Again, double knockdown or ITGB4 single knockdown induced apoptosis significantly within 24 h of seeding compared with PXN or scramble knockdown. Addition of cisplatin further sensitized these cells as seen by an increase in apoptosis at 72 h (Figure S5E). Immunoblotting analysis confirmed knockdown of both ITGB4 and PXN (Figure 3E). In addition, we observed that PXN knockdown induced expression of the tumor suppressor p27 with simultaneous reduction in phosphorylation of the retinoblastoma protein Rb1, suggesting cell cycle inhibition. Indeed, a cell cycle analysis after knocking down PXN increased G1 population by 20% indicating slowing of G1-S transition, whereas knocking down ITGB4 resulted in a decrease of S phase population and accumulation in G1 as well as G2 populations. ITGB4 and PXN double knockdown increased further the population of both G1 and G2, indicating that cell cycle is interrupted at both G1-S and G2-M phases (Figure 3F).
Figure 3.
Knocking Down Both PXN and ITGB4 has a Synergistic Effect on Attenuating Cisplatin Resistance in 2D and 3D Cultures
(A) Double knockdown of both PXN and ITGB4 (red) in H1993 cells had a synergistic effect on inhibiting proliferation compared with single knockdown of either gene (green/blue) and control (black). (∗∗∗∗p < 0.0001 two-way ANOVA).
(B) Adding 10 μM cisplatin to the double knockdown cells (red) had an even greater effect on inhibiting proliferation compared with single knockdown of either gene (green/blue) and control (black) treated with cisplatin. (∗∗∗∗p < 0.0001 two-way ANOVA).
(C) Double knockdown of both PXN and ITGB4 (red) in H2009 cells also had a synergistic effect on inhibition of proliferation compared with single knockdown of either gene (green/blue) and control (black). (∗∗∗∗p < 0.0001 two-way ANOVA).
(D) With double knockdown of PXN and ITGB4 in combination with cisplatin (red), proliferation was greatly inhibited compared with single knockdown of either gene (green/blue) and control (black) treated with cisplatin. (∗∗∗∗p < 0.0001 two-way ANOVA).
(E) Immunoblotting confirmed siRNA-mediated knockdown of PXN and ITGB4. PXN knockdown alone increased expression of p27 and decreased levels of phospho-Rb (S807/811), indicating cell-cycle arrest.
(F) Cell cycle analysis revealed that knocking down PXN induced G1-S arrest, whereas knocking down ITGB4 arrested cells in G2-M. Double knockdown of PXN and ITGB4 arrested cells in G1-S and G2-M.
(G) H2009 cells expressing mKate2 were transfected with siRNA, seeded in a 96-well ultra-low attachment plate (5,000 cells/well), and allowed to form a compact spheroid overnight. Images acquired by the IncuCyte Live Cell Imaging System showed that spheroids with a single knockdown start disintegrating by day 3 and even earlier for double knockdown spheroids (Day 1).
(H) Immunoblotting confirmed that the PXN and ITGB4 siRNA-mediated knockdown was still effective after 72 h in 3D culture.
(I and J) To quantitate spheroid viability, (I) red fluorescence area and (J) mean intensity were measured. Both parameters showed that double knockdown had a synergistic effect on attenuating spheroid viability (∗∗∗∗p < 0.0001 two-way ANOVA). Data are represented as mean ± SD.
Knocking down PXN or ITGB4 Disrupts Spheroid Growth
To mimic conditions closer to in vivo, we studied the effect of knocking down PXN and ITGB4 in 3D spheroid cultures. H2009 cells engineered to express RFP (mKate2) were first transfected with scrambled or PXN/ITGB4-specific siRNA and then seeded in an ultra-low attachment round-bottom plate to generate spheroids. Within 4 h of seeding, we observed spheroid formation in wells seeded with cells that were transfected with the control siRNA, but not in the cells transfected with ITGB4 and PXN siRNA (Figure S7A, Videos S9, S10, S11, and S12). The double knockdown cells took much longer to form spheroids. Once spheroids formed, they were monitored for 5 days using the IncuCyte Live Cell Imaging System (Figure 3G). By immunoblotting analysis, we confirmed that spheroids continued to have diminished levels of ITGB4 and PXN by 72 h post-siRNA transfection (Figure 3H).
Spheroid growth and survival were quantitated by monitoring red fluorescence intensity and area in real time. We observed a reduction in spheroid growth in double knockdown cells by >40% compared with that seen in the spheroids formed by cells transfected with the scrambled siRNA (Figure 3I). In addition, spheroids formed from double knockdown cells showed a decline in red fluorescence intensity, and by day 4, the intensity was only ∼30% of the control (Figure 3J). These data suggest that the cell-to-cell interaction, which is essential for the spheroid formation, was disrupted by ITGB4 and PXN knockdown, which resulted in the observed decrease in spheroid integrity. In addition, we treated the single and double knockdown spheroids with cisplatin and observed that double knockdown spheroids were the most sensitive, followed by spheroids with only ITGB4 knocked down. However, PXN knockdown spheroids behaved similarly to the control group (Figures S7B and S7C). To confirm that the reduction in spheroid size was due to cell death, we assayed them for caspase-3/7 activity using the Essen BioScience assay. We observed 20-fold higher caspase-3/7 activity with double knockdown compared with scrambled siRNA by day 5 (Figure S7D). We also analyzed caspase activity in control spheroids and double knockdown spheroids treated with cisplatin and found that the cell death in double knockdown spheroids exhibited an additive effect (Figure S7E). Considered together, the data from the 2D and 3D culture experiments revealed that the interaction between PXN and ITGB4 is important for modulating cisplatin resistance in these LUAD cell lines.
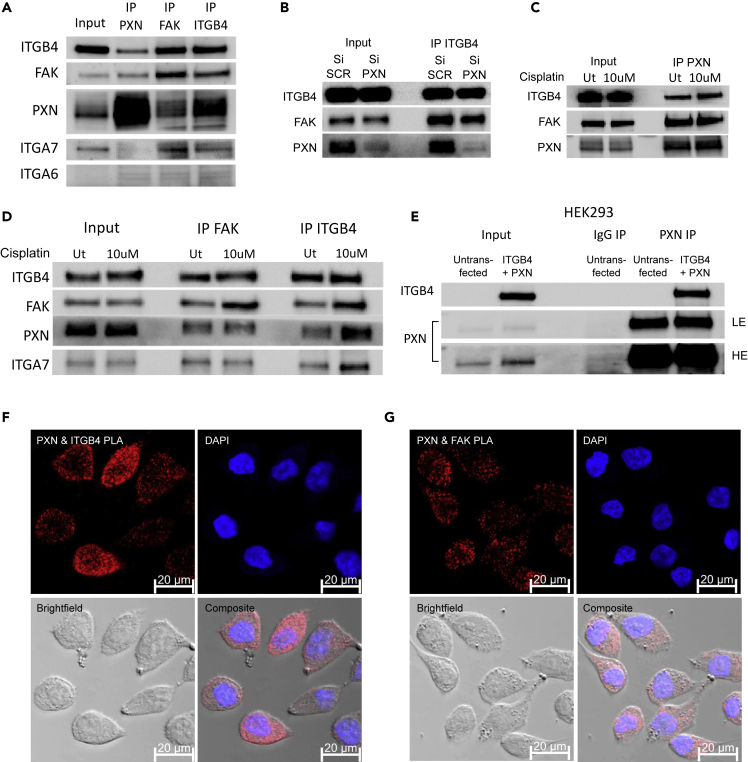
Biochemical Data Demonstrate Direct Interaction between PXN and ITGB4
From the foregoing, it follows that PXN and ITGB4 interact with each other, and this interaction is important in cisplatin resistance. ITGB4, PXN, and FAK are known to prime the formation of the FA complex. To determine these interactions, we performed co-immunoprecipitation (co-IP) experiments with an ITGB4 antibody as well as with FAK and PXN antibodies in separate experiments. Immunoblotting with ITGA6 antibody served as a positive control as it is known to interact with ITGB4 (Tözeren et al., 1994). Much to our surprise, we were unable to detect the ITGA6 protein in the pulldown, but were able to detect ITGA7 leading to the possibility of ITGB4 heterodimer with ITGA7 in the H2009 cells (Figure 4A). Furthermore, the immunoprecipitation data indicated a stronger interaction between FAK and ITGB4 compared with PXN and ITGB4, suggesting that FAK interaction with ITGB4 is PXN-dependent, or vice versa. To test this possibility, we knocked down PXN and repeated immunoprecipitation with the ITGB4 antibody. This experiment demonstrated that indeed, FAK interacts with ITGB4 irrespective of PXN (Figure 4B). We next performed immunoprecipitations using lysates from cells treated with cisplatin for 48 h to determine if cisplatin induced any changes in the interactions among ITGB4, FAK, and PXN (Figures 4C and 4D). By immunoprecipitating ITGB4, FAK, as well as PXN, we observed no changes in the interactions among FAK, PXN, and ITGB4 in the presence of cisplatin, which again indicated the involvement of the FA complex in imparting cisplatin resistance. We overexpressed ITGB4 and PXN in HEK293 cells and repeated the co-IP experiment using the PXN antibody and were able to detect ITGB4 using the ITGB4-specific antibody confirming the results from the endogenous IP (Figure 4E). Considered together, the co-IP data suggest that the interactions among PXN, ITGB4, and FAK are responsible for cisplatin resistance and that disrupting this interaction could make these cells susceptible to cisplatin.
Figure 4.
ITGB4/PXN/FAK Interact in the Focal Adhesion Complex
(A) Co-immunoprecipitation (co-IP) of H2009 whole-cell lysate with an ITGB4 antibody showing that FAK, PXN, and ITGA7 interact with ITGB4. Reverse co-IP with FAK and PXN antibody showing that ITGB4 binds to FAK and PXN.
(B) Knocking down PXN did not affect the interaction between FAK and PXN.
(C) PXN IP on H2009 cells after 48 h of cisplatin treatment (10 μM) did not affect interaction with ITGB4 and FAK.
(D) FAK IP and ITGB4 IP on H2009 cells after 48 h of cisplatin treatment (10 μM) did not affect interaction with PXN.
(E) We expressed truncated ITGB4 and PXN in HEK293 cells and then performed PXN co-IP (IgG used as control). The immunoblot confirms the interaction (LE = low exposure, HE = high exposure).
(F and G) Proximity ligation assay (PLA) demonstrated the colocalization of (F) PXN/ITGB4 and (G) PXN/FAK by the quantification of punctate red fluorescence, which indicates that the two molecules are in close proximity (between 30 and 40 nm).
To validate the interaction of ITGB4 and PXN in situ, we performed proximity ligation assay (PLA). We procured the Duo Link In Situ Detection Kit from Sigma and performed the experiment on the H2009 cells as per the manufacturer's protocol to detect the interaction of ITGB4 (rabbit antibody) and PXN (mouse antibody). The PXN and FAK interaction served as positive control, and for the negative control, we used individual antibodies for one specific target ITGB4, PXN, or FAK (Figure S8). The interaction of two molecules is detected by the red fluorescence emitted. The data validated the interaction of ITGB4 and PXN and suggested that the interaction was variable among cells. Thus, whereas some cells showed high fluorescence, the signal was weak in some cells. This is consistent with variable expression of ITGB4 observed in cell lines and patient needle biopsies IHC data. In contrast, the interaction between FAK and PXN was uniform across all the cells (Figures 4F and 4G).
RNA-Seq Analysis Suggests that Multiple Pathways Converge in Cisplatin Resistance
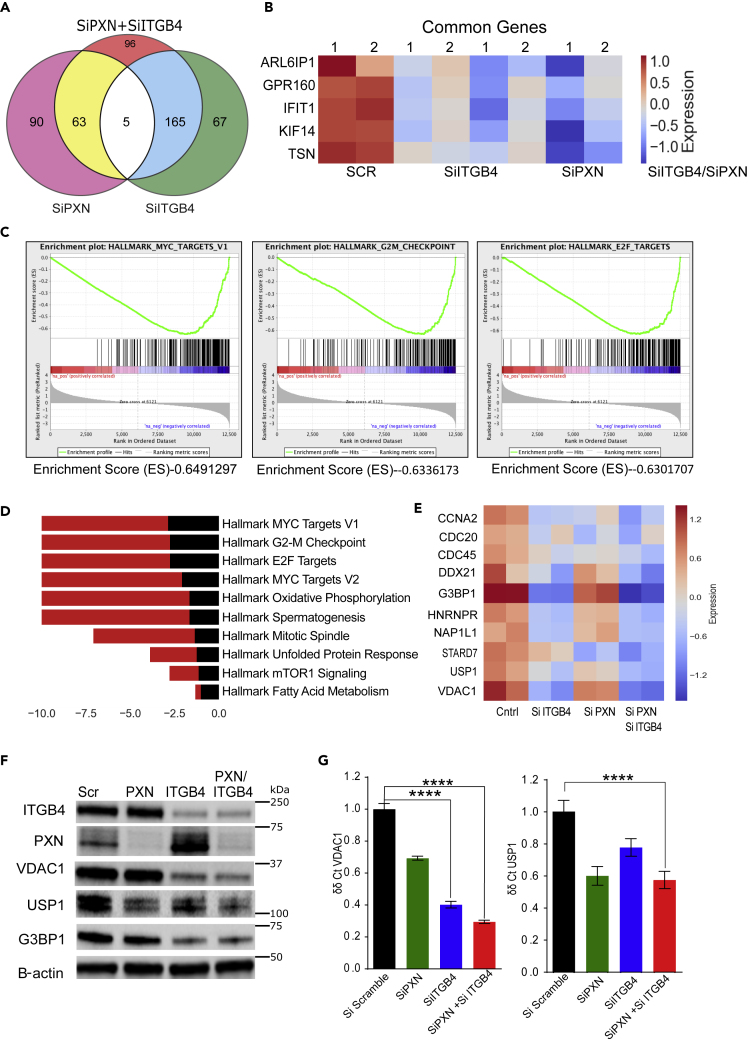
To identify the effect of ITGB4/PXN knockdown on signaling, we determined the changes in global gene expression patterns using RNA-seq. RNA was extracted from both single and double knockdown cells 48 h post-siRNA transfection, and total RNA-seq was performed as described in the Methods. A total of 30 million reads were analyzed for each condition. We found that 237 genes were downregulated when ITGB4 was knocked down and 158 genes were downregulated upon knocking down PXN. In the case of the double knockdown, we identified 329 genes that were downregulated (Figure 5A). Interestingly, five genes, ARL6IP1, GPR160, IFIT1, KIF14, and TSN, were common to both single and double knockdown samples (Figure 5B). Of these, ARL6IP1 is known to have an anti-apoptotic role and KIF14 is known to control cell division, cell cycle progression, and apoptosis. Comparative GSE analysis of the RNA-seq data between control and double knockdown cells showed significant enrichment of genes regulated by MYC, genes associated with G2-M checkpoint, and genes regulated by the E2-F transcription factors required for cell cycle progression (Figures 5C and 5D). Myc is known to induce tumorigenesis by regulating the expression of genes required to suppress tumor senescence, activate tumor stemness, modify the tumor microenvironment, induce angiogenesis, and suppress immune activation. In murine models, Myc knockout is enough to contain and eradicate KRAS-driven lung cancer (Soucek et al., 2013). Therefore, we determined the effect of ITGB4 and PXN knockdown in regulating MYC targets. The top 10 genes contributing significantly to downregulation of the MYC pathways were interrogated (Figure 5E). Differential expression analysis of RNA-seq yielded 96 unique genes that were downregulated only in double knockdown cells. DAVID analysis performed on these 96 genes indicated that they are involved in the regulation of transcription, G2/M transition, cell adhesion, and transforming growth factor (TGF)-β signaling (Table S3). DAVID analysis of the 206 genes that were downregulated in double knockdown as well as PXN or ITGB4 single knockdown are listed Table S4.
Figure 5.
Signaling Pathways of PXN and ITGB4 Converge in Mediating Cisplatin Resistance
(A) Total RNA was extracted from single and double knockdown cells 48 h after siRNA transfection. Total RNA-seq revealed the number of genes downregulated with single knockdown of PXN and ITGB4 and when both genes are knocked down simultaneously.
(B) Total RNA-seq after single and double knockdown of PXN and ITGB4 revealed that five common genes were downregulated compared with control cells (SCR).
(C) Major pathways affected by double knockdown of PXN and ITGB4 were MYC targets, G2-M checkpoint, and E2-F targets.
(D) Bar diagram showing the major Hallmark pathways affected by the double knockdown. The top 10 pathways were arranged in descending order of their enrichment scor23e.
(E) Heatmap representation of top 10 genes that belong to hallmark MYC target V1 pathways were analyzed after single or double knockdown. The heatmap is representative of two experimental repeats.
(F) Immunoblotting confirmed decreased expression of top three genes (G3BP1, USP1, and VDAC1) that are downregulated in MYC pathway after knockdown of PXN and/or ITGB4.
(G) qPCR results with single and double knockdown of PXN and ITGB4 confirmed downregulation of VDAC1 and USP1 at the mRNA level (∗∗∗∗p < 0.0001 two-way ANOVA). Data are represented as mean ± SD.
ITGB4 and PXN May Regulate Genomic Stability and Mitochondrial Function via USP1 and VDAC1
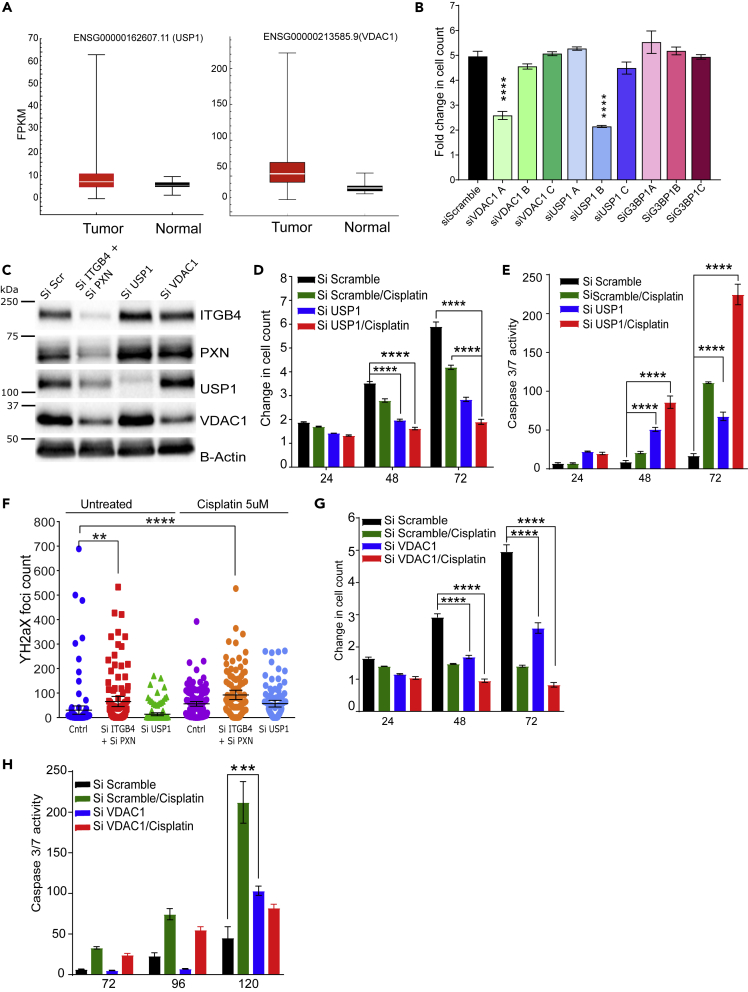
The Hallmark pathways represent specific well-defined biological processes and exhibit coherent expression. G3BP1, USP1, and VDAC1 were the top three downregulated genes that contribute to the MYC1 Targets V1 Hallmark pathway. To validate the RNA-seq data, we performed western blot and qPCR experiments. These results confirmed that knocking down either PXN or ITGB4 resulted in the decreased expression of all three genes, but in double knockdown, the decrease in expression is seen at both protein (Figure 5F) and RNA levels (Figure 5G). Furthermore, we analyzed the expression of these genes in lung TCGA dataset to determine the expression of USP1 and VDAC1 in tumor and normal samples. Both USP1 and VDAC1 mRNAs showed increased expression in tumor compared with normal tissue, and a Cox regression analysis on VDAC1 expression identified VDAC1 as an independent factor with significant prognostic value for worse overall survival (p < 0.0001) (Grills et al., 2011) (Figure 6A).
Figure 6.
Knocking Down MYC Pathway Genes USP1 and VDAC1 Attenuates Cisplatin Resistance and USP1 is Involved in Genomic Stability
(A) Gene expression profiles of patients with LUAD were extracted from TCGA database, and expressions of USP1 and VDAC1 were higher compared with normal tissue.
(B) siRNA constructs A, B, or C against top three downregulated genes (VDAC1, USP1, and G3BP1) were tested to determine the construct with maximum effect on inhibiting proliferation. (∗∗∗∗p < 0.0001 two-way ANOVA).
(C) Double knockdown of PXN and ITGB4 (Si ITGB4+Si PXN) decreased expression of USP1 and VDAC1. However, knocking down USP1 (Si USP1) or VDAC1 (Si VDAC1) did not affect expression of ITGB4 or PXN.
(D) In H2009 cells, knocking down USP1 (blue) attenuated proliferation (∗∗∗∗p < 0.0001 two-way ANOVA).
(E) In H2009 cells, knocking down USP1 (blue) induced apoptosis and sensitized cells to a lower dose of cisplatin (2 μM) (red) (∗∗∗∗p < 0.0001 two-way ANOVA).
(F) After knocking down PXN/ITGB4 and USP1, γH2AX foci were detected via immunofluorescence, imaged with confocal microscopy, and counted with QuPath image analysis software (∗∗p = 0.004, ∗∗∗∗p < 0.0001).
(G) Knocking down VDAC1 (blue) inhibited cell proliferation by 50% within 72 h, and cisplatin (red) addition has an additive effect to VDAC1 knockdown in inhibiting proliferation (∗∗∗∗p < 0.0001 two-way ANOVA).
(H) Knocking down VDAC1 (blue) could induce apoptosis only at a later time point (∗∗∗p = 0.0001 two-way ANOVA).
Data are represented as mean ± SD.
We next asked whether knocking down the USP1, G3BP1, or VDAC1 genes would recapitulate the phenotype of the ITGB4/PXN double knockdown. To this end, we tested three siRNA constructs for each gene USP1, VDAC1, and G3BP1 (OriGene). The optimal siRNA construct was selected by transfecting H2009 cells with 10 nM of each siRNA and screening for the phenotypes of reduced proliferation and induction of caspase activity (Figures 6B and S9A). For USP1 and VDAC1, siRNA sequences B and A were selected for all downstream experiments, respectively. The efficiency of these siRNAs in inhibiting gene expression was confirmed using immunoblotting. In addition, we also checked the possibility of decreased expression of ITGB4 and PXN upon knocking down USP1 or VDAC1 but did not observe any change (Figure 6C).
USP1 knockdown inhibited cell proliferation by more than 2-fold within 72 h of seeding and showed a simultaneous increase in caspase activity. Furthermore, addition of a lower dose of cisplatin (2.5 μM) to USP1 knockdown H2009 cells decreased their proliferation by 3-fold (Figure 6D) and increased apoptosis by 200-fold within 72 h (Figure 6E). We also tested the sensitization of H2009 and H1993 cells to the USP1-selective inhibitor ML323 (Liang et al., 2014). We treated cells with ML323 (5 and 10 μM) for 3 days but did not see any significant change in the cell proliferation rates (Figure S9B). These data indicate that H2009 and H1993 cell survival and proliferation is dependent on USP1 protein level, but not the interaction between USP1 and UAF, as ML323 is known to disrupt the USP1-UAF interaction, and therefore, may not decrease USP1 protein level. Hence, we could not reproduce the phenotype of USP1 knockdown using ML323. Furthermore, USP1 is a deubiquitinase known to increase DNA repair activity in cisplatin-treated cells (García-Santisteban et al., 2013). Therefore, we sought to measure changes in the γH2AX foci as a measure of DNA damage in the double knockdown (ITGB4/PXN) or USP1 knockdown cells. We observed that double knockdown cells have significantly higher γH2AX foci counts compared with control or USP1 knockdown. Addition of cisplatin in conjunction with USP1 knockdown further increased the extent of DNA damage (Figure 6F).
On the other hand, VDAC1 is an ion channel pump located in the mitochondrial and plasma membranes. Knocking down VDAC1 led to a decrease in cell proliferation by 50% within 72 h, but this did not contribute to apoptosis induction even in the presence of cisplatin. However, at a later time point (120 h), we observed a 2-fold increase in apoptosis (Figures 6G and 6H).
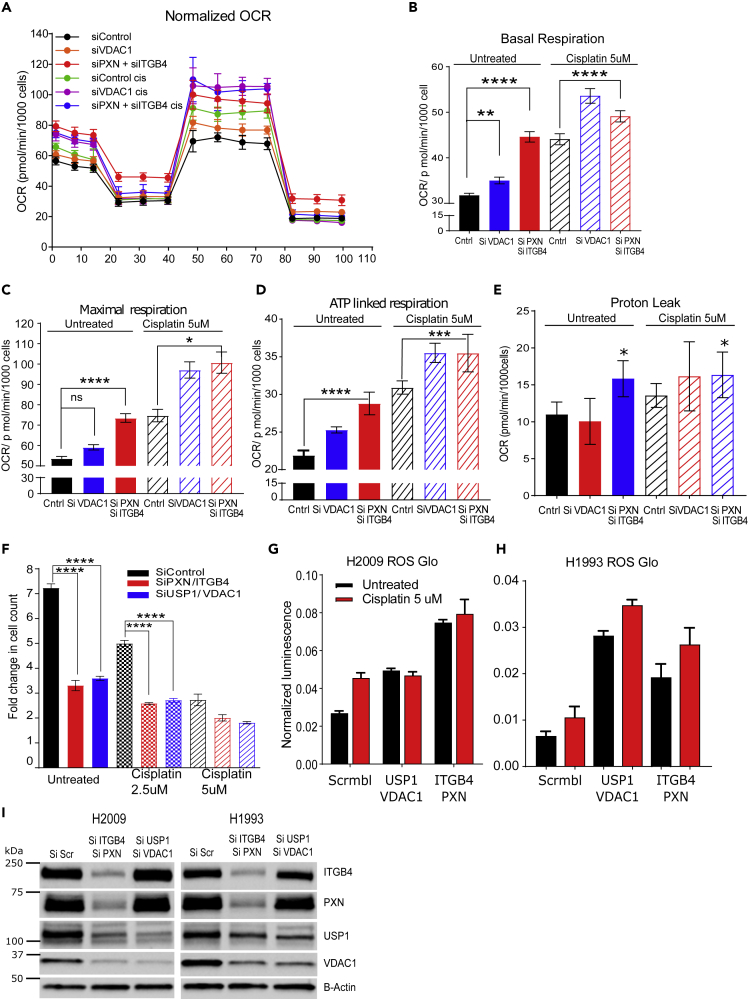
To discern the role of VDAC1 in mitochondrial function, we knocked down VDAC1 as well as PXN and ITGB4 in H2009 cells and analyzed mitochondrial respiration in the absence and presence of cisplatin using the Seahorse XF Analyzer (Agilent, Santa Clara, CA). Knocking down VDAC1 or ITGB4/PXN increased the oxygen consumption rate (OCR), which is measured by the basal as well as maximal mitochondrial OCR compared with the control, and these rates were further increased in the presence of cisplatin (Figures 7A–7C). Thus, it is possible that the increase in oxygen consumption can lead to mitochondrial stress and generation of reactive oxygen species (ROS), which in turn can inhibit cell viability.
Figure 7.
Both PXN and ITGB4 are Involved in Mitochondrial Function
(A) Using the Seahorse XF Analyzer, cellular mitochondrial activity was measured with knockdown of PXN/ITGB4 or VDAC1 and in the absence or presence of cisplatin.
(B and C) Knocking down PXN/ITGB4 or VDAC1 increased basal (B) and maximal (C) mitochondrial oxygen consumption rate per 1,000 cells (∗p = 0.021, ∗∗p = 0.0029, ∗∗∗∗p < 0.0001 one-way ANOVA).
(D and E) ATP-linked respiration (D) and proton leak (∗∗∗p = 0.0006, ∗∗∗∗p < 0.0001) (E) also showed an increase in the same knockdown cells. (∗p = 0.01 one-way ANOVA).
(F) Double knockdown of PXN/ITGB4 (red) and USP1/VDAC1 (blue) inhibited proliferation to a similar extent in the absence or presence of cisplatin (∗∗∗∗p < 0.0001 two-way ANOVA).
(G and H) Reactive oxygen species (ROS) production in (G) H2009 and (H) H1993 cells was measured using the ROS-Glo H2O2 Assay.
(I) H2009 and H1993 cells with PXN/ITGB4 or USP1/VDAC1 double knockdown was confirmed by immunoblot to be used for ROS production assay. Data are represented as mean ± SD.
Oxidative phosphorylation is an efficient but slow process of ATP production when compared with the glycolytic pathway, and is preferentially used by tumor cells. Pyruvate generated during glycolysis is oxidized by mitochondria for oxidative phosphorylation, which leads to mitochondrial ATP production, a required process for ATP-linked respiration (Divakaruni et al., 2014). Treating cells with oligomycin, which inhibits the electron transport chain and blocks mitochondrial ATP generation, can inhibit ATP-linked respiration. An increase in ATP-linked respiration is indicative of the availability of more substrates like pyruvate to drive oxidative phosphorylation. On the other hand, it also suggests an increase in ATP demand of the cell. Therefore, we calculated ATP-linked respiration and observed an increase in the linked respiration in double knockdown cells, which increased further upon addition of cisplatin. Control (scramble siRNA treated) cells in the presence of cisplatin showed increased ATP-linked respiration, which is indicative of a stress-induced increase in ATP demand. The same increase in ATP demand was also seen in these cells upon VDAC1 knockdown or double knockdown (Figure 7D).
Under ideal conditions, mitochondria maintain a proton motive potential for ATP generation. To maintain the proton motive force, mitochondria increase their oxygen consumption rate as measured by increases in basal and maximal respiration. We observed an increase in the proton leak in the case of ITGB4 and PXN double knockdown that may explain the increase in respiration to compensate for the loss in membrane potential (Figure 7E). Furthermore, respiratory capacity is measured by considering the ratio of maximal respiration compared with basal respiration. However, we did not observe a discernable difference across the samples (Figure S9C). Taken together, these data suggest that USP1 and VDAC1 are downstream of PXN and ITGB4, and thus, knocking down USP1 and VDAC1 should recapitulate the phenotype of PXN and ITGB4 knockdown.
Double Knockdown of USP1 and VDAC1 Recapitulates the Phenotype Caused by Silencing Both ITGB4 and PXN
H2009 cells were transfected with a combination of ITGB4 and PXN siRNAs or USP1 and VDAC1 siRNAs, and changes in cell proliferation rates were compared. Proliferation was reduced by 50% after knocking down either of these combinations. The addition of cisplatin to either knockdown pair had an additive effect in inhibiting cell proliferation, but there was no significant difference observed between either gene combinations (Figure 7F).
Irradiation is known to increase mitochondrial respiration, causing an increase in generation of mitochondrial ROS, which eventually induces apoptosis (Yamamori et al., 2012). Therefore, we asked if increased mitochondrial respiration leads to increased ROS generation in double knockdown cells. We observed an increase in the generation of ROS by knocking down either ITGB4/PXN or USP1/VDAC1 combination compared with the control (scramble siRNA) (Figures 7G and 7H). Addition of cisplatin to control cells increased ROS production but had no significant effect on the double knockdown cells, and no change in PXN and ITGB4 expression in USP1 and VDAC1 double knockdown cells was observed, indicating that the regulation of USP1 and VDAC1 by PXN and ITGB4 is unidirectional (Figure 7I).
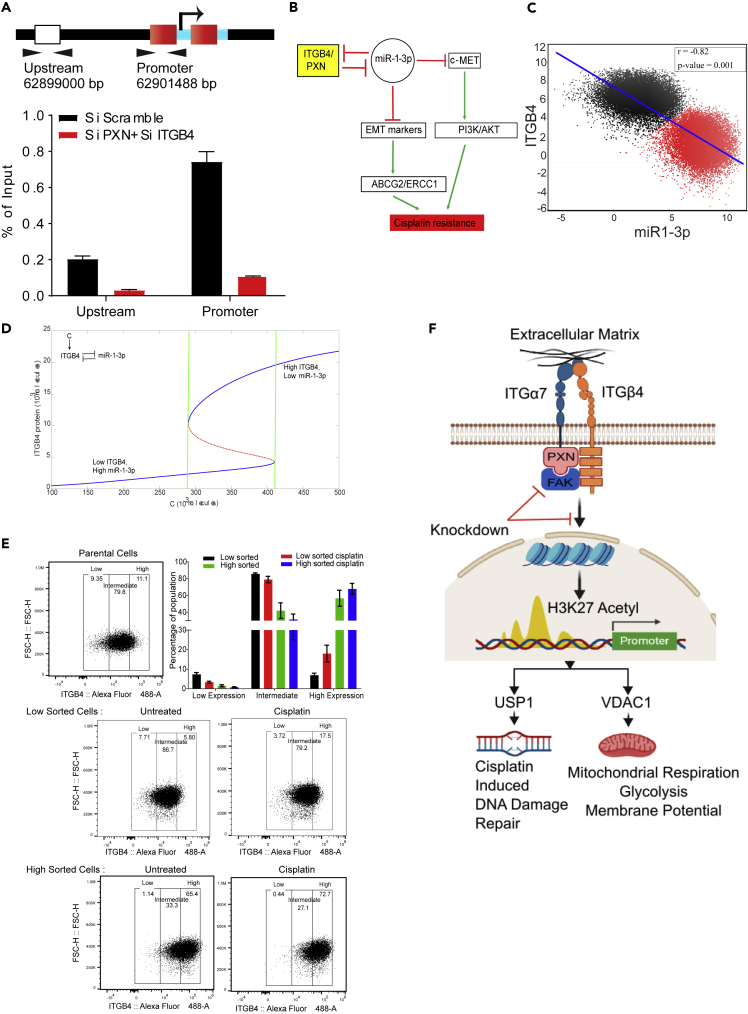
ITGB4 and PXN Regulate USP1 Transcription Epigenetically
Finally, as the double knockdown cells silenced USP1 expression, alluding to possible transcriptional regulation, we analyzed the upstream sequence of the USP1 promoter region using UCSC Genome Browser and identified two potential acetylated histone H3K27-binding sites. To determine the functional significance of these sites, we knocked down both PXN and ITGB4, and after 72 h, performed chromatin immunoprecipitation using an anti-acetylated histone H3K27 antibody (Diagenode, Denville, NJ). In the double knockdown samples, we observed reduced binding of acetylated H3K27 at the promoter, suggesting that USP1 expression is induced by hyper-acetylation (Figure 8A). Of note, the data also suggest that ITGB4 and PXN involvement is not limited to migration and invasion; they may also be involved in controlling the epigenetic landscape of lung cancer. However, additional studies are needed at the global level along with analyses of other histone acetylation states.
Figure 8.
Mathematical Modeling Suggests that Cisplatin Resistance Can Be Stochastic and Reversible
(A) Chromatin immunoprecipitation was performed with an acetylated H3K27 antibody 72 h after siRNA-mediated knockdown of PXN/ITGB4. With the knockdown, H3K27 acetylation at the promoter region of USP1 was greatly reduced compared with that of an upstream region, indicating the roles of ITGB4 and PXN in USP1 transcriptional activation.
(B) A double-negative feedback loop between ITGB4 and miR-1-3p leads to bistability.
(C) To test this mathematical model, RACIPE algorithm generated an ensemble (n = 100,000) with varying parameter sets then plotted to represent robust dynamical patterns. Results showed that ITGB4 and miR-1-3p exhibit bimodality: two distinct subpopulations of cells that are negatively correlated, reinforcing the previously described negative feedback loop.
(D) A mathematical model stimulating the dynamics of ITGB4 and miR-1-3p showed cisplatin resistance to be a reversible state. High ITGB4 and low miR-1-3p render cells to be resistant, whereas low ITGB4 and high miR-1-3p represents a more sensitive state.
(E) H2009 cells were stained with ITGB4 antibody conjugated to Alexa Fluor 488 and sorted based on gates set to high and low ~10% of ITGB4-expressing population using the FACSAria Fusion instrument. Sorted cells were subsequently cultured for 48 h and then treated with 1 μM cisplatin for 48 h. Then, using the Attune NxT Flow Cytometer, equal numbers of cells were stained again and analyzed to determine shifts in population between untreated and treated cells. Low sorted cells treated with cisplatin had a greater cell population that shifted toward higher ITGB4 expression. High sorted cells treated with cisplatin did not undergo significant changes in population compared with untreated.
(F) Schematic depicting the interaction between ITGB4 and PXN regulating downstream proteins USP1 and VDAC1 at the transcriptional level to coordinate cisplatin resistance. Data are represented as mean ± SD.
Mathematical Modeling Indicates Bistability
Analysis of RNA-seq data revealed that several microRNAs were also differentially regulated in response to single or double knockdown (Table S5). In particular, mir-1-3p was upregulated both in ITGB4 single knockdown and in the ITGB4/PXN double knockdown by 3.1-fold and 2.54-fold, respectively. Furthermore, it was reported that overexpressing ITGB4 downregulates miR-1-3p (Gerson et al., 2012). Together, these observations suggested that there exists a double-negative feedback loop between ITGB4 and miR-1-3p. Integrating these observations, we developed a mathematical model to simulate the dynamics of ITGB4/miR-1-3p feedback loop (Figure 8B). This model showed the existence of two stable states (phenotypes): one (sensitive) state represented by (low ITGB4, high miR-1-3p) and the other (resistant) state represented by (high ITGB4, low miR-1-3p) (Figure 8C). Moreover, the model suggested that it is possible for cells to switch their state under the influence of biological noise or stochasticity; thus, a sensitive cell can behave as resistant, and vice versa (Figure 8D).
To discern whether this co-existence of two states is a robust feature that can be expected from the given network topology, we implemented the algorithm RACIPE (Random Circuit Perturbation), which generates an ensemble of mathematical models with varying parameter sets (Huang et al., 2017). The results from this ensemble (n = 100,000) were then plotted together to identify robust dynamical features of the underlying regulatory network; each mathematical model can represent one cell, and this ensemble represents a cell population with varying levels of genetic/phenotypic heterogeneity. RACIPE results for the ITGB4/miR-1-3p feedback loop showed that both ITGB4 and miR-1-3p exhibit bimodality, i.e., two subpopulations (Figure 8C). These two subpopulations were also distinctly observed in a scatterplot of ITGB4/miR-1-3p showing negative correlation (r = - 0.82, p < 0.001), reinforcing our previous results indicating the existence of two states—high ITGB4, low miR-1-3p and low ITGB4, high miR-1-3p. All in all, these results indicate that ITGB4/miR-1-3p feedback loop enables phenotypic plasticity (Figure S9D).
To test the predictions made by the model, we treated H2009 cells with cisplatin for 4 days. On day 5, we subjected them to fluorescence-activated cell sorting (FACS) analysis using an ITGB4 antibody. We observed a 15% decrease in ITGB4 low-expressing cells compared with untreated cells, suggesting that the H2009 cells are inherently a mixed population with variable expression of ITGB4. In the presence of stress, the cells either increase the expression of ITGB4 or the cells having high ITGB4 get selected (Figure S9E). In addition, we gated and purified 10% of median low ITGB4-expressing cells (low sorted) and 10% of median high ITGB4 (high sorted) subpopulations from H2009 cells by FACS sorting and cultured them separately. We seeded the sorted cells in six independent wells and allowed them to proliferate. After 2 days, three of six wells were treated with a sub-lethal dose of cisplatin (1μM) for 2 days. The cells were then counted, and an equal number of cells were stained with ITGB4 antibody and propidium iodide and analyzed by Attune NxT Flow Cytometer. Interestingly, we observed that the ITGB4-low cells recreated ITGB4-intermediate and ITGB4-high population, whereas ITGB4-high sorted cells kept on proliferating and increasing the population of cells having highITGB4. The high sorted cells created an intermediate population of 30%, whereas the low sorted created a population of 80% similar to the parental cells suggesting that cells having intermediate or high ITGB4 expression were preferentially selected. Finally, we treated these cells with cisplatin and observed a shift in population for the low and intermediate gated cells toward the higher gate, suggesting that (cisplatin) stress favored the population with high expression of ITGB4 (Figure 8E).
Discussion
A Non-genetic Mechanism Drives Cisplatin Resistance in Lung Cancer
It is generally held that the antineoplastic effects of cisplatin are due to its ability to generate unrepairable DNA lesions resulting in either a permanent proliferative arrest (cellular senescence) or cell death due to apoptosis. The drug enters cells via multiple pathways and forms multiple DNA-platinum adducts, which results in dramatic epigenetic and/or genetic alternations. Such changes have been reported to occur in almost every mechanism supporting cell survival, including cell growth-promoting pathways, apoptosis, developmental pathways, DNA damage repair, and endocytosis (Shen et al., 2012; Rocha et al., 2018). Therefore, at a single-cell level, the genetic underpinning involved in cisplatin resistance is obvious. Drug resistance is also observed to be strongly influenced by intratumoral heterogeneity and changes in the microenvironment (Álvarez-Arenas et al., 2019), and heterogeneity is thought to arise from genetic mutations, and analogous to Darwinian evolution selection of tumor cells is believed to result from the adaptation to the microenvironment (Gerlinger and Swanton, 2010).
Contrary to the prevailing wisdom, however, it is now increasingly evident that non-genetic mechanisms may also play an important role as information transfer can occur horizontally via a Lamarckian mode of evolution (Álvarez-Arenas et al., 2019). Thus, a population of isogenic cells in the same environment can exhibit single-cell-level stochastic fluctuations in gene expression. Such fluctuations, known as gene expression noise or transcriptional noise, can result in isogenic cells “making” entirely different decisions with regard to their phenotype, hence their ability to adapt themselves to the same environmental perturbation (Balázsi et al., 2011; Farquhar et al., 2019; Engl, 2019). Transcriptional noise can arise from the intrinsic randomness of underlying biochemical reactions or processes extrinsic to the gene (Swain et al., 2002). Regardless, two main characteristics of gene expression noise are amplitude and memory. Amplitude, often measured by the coefficient of variation, defines how far cells deviate from the average. Memory describes the amount of time for which cells remain deviant once they depart from the average (Acar et al., 2005; Charlebois et al., 2011). Thus, it follows that the effect noise produces is likely reversible, underscoring its importance in phenotypic switching.
The present data suggest that cisplatin resistance can arise stochastically in response to drug treatment. Our data suggest that a double-negative feedback loop between ITGB4 and miR-1-3p results in bistability, facilitating a reversible phenotypic switch between cisplatin-sensitive and resistant states. Additional studies that are currently underway in our laboratory will help confirm this feedback circuit. Nonetheless, a recent study on oxaliplatin chemotherapy in pancreatic ductal adenocarcinoma (Kumar et al., 2019), where the authors studied a coarse-grained stochastic model to quantify phenotypic heterogeneity in a population of cancer cells, also mirrors the present findings, suggesting the phenomenon may be applicable across cancer types. Furthermore, a random population of cisplatin-resistant LUAD cells is heterogeneous and comprises individuals that either express high or low levels of ITGB4. These cells, therefore, are either more resistant or less resistant to cisplatin, respectively. However, when purified to homogeneity (>99%) and plated separately, the purified population re-created the heterogeneity. Taken together, these observations not only corroborate the bistability predicted by the model but also highlight the role of phenotypic switching in generating population heterogeneity in cancer.
An Unsuspected Role for the Focal Adhesion Complex in Cisplatin Resistance
One of our significant findings reported in this article is the critical role played by the major components of the FA complex. More specifically, as far as we are aware, the interaction between ITGB4 and PXN has not been suspected, much less investigated, in cisplatin resistance.
Human PXN is a 68-kDa (591 amino acids) protein (Salgia et al., 1995), and its upregulation is a recognized contributor to cisplatin resistance in lung cancer (Wu et al., 2014). The N terminus contains a proline-rich region that anchors SH3-containing proteins and five leucine-rich LD domains (LD1–LD5) with a consensus sequence LDXLLXXL (Turner, 1998, 2000; Kanteti et al., 2016). The LD2-LD4 region includes sequences for the recruitment of signaling and structural molecules, such as FAK, vinculin, and Crk. This region has also been reported to interact with integrin α, more specifically, integrin α4 (ITGA4) (Liu et al., 1999; Liu and Ginsberg, 2000). The C-terminal region is also involved in the anchoring of PXN to the plasma membrane and its targeting to FAs. It contains four cysteine-histidine-enriched LIM domains that form zinc fingers, suggesting that PXN could bind DNA and act as a transcription factor. Consistently, PXN is reported to locate to the nucleus, behavior that is regulated by its phosphorylation state (Dong et al., 2009; Ma and Hammes, 2018). In LUAD, expression of PXN is correlated with tumor progression and metastasis (Song et al., 2010; Mackinnon et al., 2011). Furthermore, phosphorylation of PXN activates the ERK pathway, increasing Bcl-2 expression and cisplatin resistance (Wu et al., 2014). Finally, specific PXN mutants, through their interactions with Bcl-2 and DRP1, also regulate cisplatin resistance in human lung cancer cells (Kawada et al., 2013).
In addition to the LD domains and the LIM domains, PXN also contains an SH3 domain-binding site and SH2 domain-binding sites (Salgia et al., 1995). Together, these motifs serve as docking sites for cytoskeletal proteins, tyrosine kinases, serine/threonine kinases, GTPase-activating proteins, and a host of other adaptor proteins that recruit additional enzymes into complex with PXN. Thus, PXN appears to serve as a docking protein to recruit signaling molecules to the FA complex and thereby coordinate downstream signaling (Schaller, 2001). It is now well recognized that cellular protein interaction networks are organized as scale-free networks and hence are remarkably resilient to perturbations (Barabasi and Albert, 1999; Barabasi, 2009). Thus, whereas disabling minor nodes does not significantly affect the continuity, and hence the functionality of the network, attacking the critical nodes occupying hub positions typically constituted by intrinsically disordered proteins can incapacitate the entire network (Schwartz et al., 2002). Therefore, PXN appears to constitute a critical hub and its malfunction appears to account for the failure of the cisplatin-resistant cells to tolerate the drug. Consistent with this argument, a significant portion of the N terminus of PXN is reported to be intrinsically disordered (Neerathilingam et al., 2016). Additional studies elucidating the structural biology of PXN and the PXN/ITGB4/FAK complex that are currently underway in our laboratories should provide a deeper understanding of the role of these proteins in cisplatin resistance.
Yet another phenomenon that contributes to cisplatin resistance in NSCLC is epithelial-to-mesenchymal transition (EMT) (Fischer et al., 2015; Zheng et al., 2015). Consistently, we observed that the Smad and TGF-β pathways that are known to be associated with EMT are affected by knocking down ITGB4 and PXN, suggesting that there is cross talk between the molecules that regulate EMT and signaling via the FA complex. Therefore, combining therapies that target the molecules involved in these two processes may prove more effective in treating these patients.
It is well established the FA complex serves as a conduit through which mechanical force and regulatory signals are transmitted between the extracellular matrix and an interacting cell (Chen et al., 2003). Furthermore, the role of PXN in cisplatin resistance has also been recognized (Wu et al., 2014). However, as far as we are aware, the involvement of ITGB4 and the interaction between these molecules with FAK in modulating cisplatin resistance has not been reported. Similarly, the interaction between ITGB4 and integrin alpha 7 has not been observed either. As summarized in Figure 8F, ITGB4 and PXN regulate transcription of USP1 and VDAC1 that play critical roles in tumor proliferation and sensitivity to cisplatin. The double knockdown of either ITGB4/PXN or USP1/VDAC1 combination increases mitochondrial oxygen consumption rate, leading to increased ROS generation resulting ultimately in increased DNA damage. Furthermore, the induction of DNA damage was cumulative when cisplatin was added to these cells. To repair this extensive DNA damage, cells need to activate DNA repair mechanisms like USP1, which were abrogated by ITGB4 and PXN double knockdown rendering the cells more sensitive to cisplatin. Taken together, our results suggest that PXN and ITGB4 are not only involved in tumor invasion and metastasis but also required for the regulation of the transcription of various genes involved in maintaining genomic stability and mitochondrial function (Figure 8F). These findings support a new role for the FA complex in cancer, particularly in LUAD.
Limitations of the Study
The study was conducted using siRNAs that silence gene expression transiently. To address this limitation, we transfected H2009 cells with two different concentrations of p-GFP-V-RS plasmid expressing short hairpin RNA against ITGB4. GFP-positive cells were followed after 48 h of transfection for 3 days. However, we did not discern any significant change in proliferation, whereas the control cells had 4-fold increase in cell growth. We also performed puromycin selection to generate a stable cell line, but the cells were growth-arrested as seen by their bigger, flattened adherent shape in contrast to a spindle shape of the cells transfected with the vehicle only (Figures S10A and S10B).
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ravi Salgia (rsalgia@coh.org).
Materials Availability
Cell lines and plasmids generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and Code Availability
The RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE155605 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155605).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
M.K.J. was supported by Ramanujan Fellowship (SB/S2/RJN-049/2018) provided by SERB, DST, Government of India. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number NIH/NCI 5P30CA033572-34, NIH 1U54CA209978-01A1, NIH R01CA218545, and NIH 1R01CA247471-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
Conceptualization, A.M., P.K., and R.S.; Methodology, A.M. and A. Nam; Formal Analysis, A.P., L.Y., A. Nathan., X.W., M.N., A.R.S., J.O., and M.K.J.; Investigation, A.M., A. Nam, S.S., and L.G.; Writing – Original Draft, A.M., A. Nam, and P.K.; Writing – Review & Editing, A.M., A. Nam., S.S., I.M., M.W.N., S.K.B., J.O., M.K.J., E.M., P.K., and R.S.; Visualization, A.M. and A. Nam; Supervision, P.K. and R.S.; Funding Acquisition, R.S.
Declaration of Interests
The authors declare that a patent application has been filed on their behalf.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101496.
Supplemental Information
References
- Acar M., Becskei A., van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- Alberts B., Johnson A.D., Lewis J., Morgan D., Raff M., Roberts K., Walter P. Sixth Edition. Garland Science Publishers; 2014. Molecular Biology of the Cell. [Google Scholar]
- Álvarez-Arenas A., Podolski-Renic A., Belmonte-Beitia J., Pesic M., Calvo G.F. Interplay of Darwinian selection, Lamarckian induction and microvesicle transfer on drug resistance in cancer. Sci. Rep. 2019;9:9332. doi: 10.1038/s41598-019-45863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi G., van Oudenaarden A., Collins J.J. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A.L. Scale-free networks: a decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Charlebois D.A., Abdennur N., Kaern M. Gene expression noise facilitates adaptation and drug resistance independently of mutation. Phys. Rev. Lett. 2011;107:218101. doi: 10.1103/PhysRevLett.107.218101. [DOI] [PubMed] [Google Scholar]
- Chen C.S., Alonso J.L., Ostuni E., Whitesides G.M., Ingber D.E. Cell shape provides global control of focal adhesion assembly. Biochem. Biophys. Res. Commun. 2003;307:355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- Deben C., Deschoolmeester V., De Waele J., Jacobs J., Van den Bossche J., Wouters A., Peeters M., Rolfo C., Smits E., Lardon F., Pauwels P. Hypoxia-Induced cisplatin resistance in non-small cell lung cancer cells is mediated by HIF-1α and mutant p53 and can be overcome by induction of oxidative stress. Cancers (Basel) 2018;10:E126. doi: 10.3390/cancers10040126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni A.S., Paradyse A., Ferrick D.A., Murphy A.N., Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309–354. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- Dong J.M., Lau L.S., Ng Y.W., Lim L., Manser E. Paxillin nuclear-cytoplasmic localization is regulated by phosphorylation of the LD4 motif: evidence that nuclear paxillin promotes cell proliferation. Biochem. J. 2009;418:173–184. doi: 10.1042/BJ20080170. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engl C. Noise in bacterial gene expression. Biochem. Soc. Trans. 2019;47:209–217. doi: 10.1042/BST20180500. [DOI] [PubMed] [Google Scholar]
- Farquhar K.S., Charlebois D.A., Szenk M., Cohen J., Nevozhay D., Balázsi G. Role of network-mediated stochasticity in mammalian drug resistance. Nat. Commun. 2019;10:2766. doi: 10.1038/s41467-019-10330-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell D.A., Summers Y., Cadranel J., Benepal T., Christoph D.C., Lal R., Das M., Maxwell F., Visseren-Grul C., Ferry D. Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat. Rev. 2016;44:42–50. doi: 10.1016/j.ctrv.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T., Choi H., El Rayes T., Ryu S., Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Senovilla L., Vitale I., Michels J., Martins I., Kepp O., Castedo M., Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Vitale I., Michels J., Brenner C., Szabadkai G., Harel-Bellan A., Castedo M., Kroemer G. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014;5:e1257. doi: 10.1038/cddis.2013.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Santisteban I., Peters G.J., Giovannetti E., Rodríguez J.A. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol. Cancer. 2013;12:91. doi: 10.1186/1476-4598-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br. J. Cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson K.D., Shearstone J.R., Maddula V.S., Seligmann B.E., Mercurio A.M. Integrin β4 regulates SPARC protein to promote invasion. J. Biol. Chem. 2012;287:9835–9844. doi: 10.1074/jbc.M111.317727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti F.G., Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- Grills C., Jithesh P.V., Blayney J., Zhang S.D., Fennell D.A. Gene expression meta-analysis identifies VDAC1 as a predictor of poor outcome in early stage non-small cell lung cancer. PLoS One. 2011;6:e14635. doi: 10.1371/journal.pone.0014635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Jin P., Liu W. Periostin contributes to cisplatin resistance in human non-small cell lung cancer A549 cells via activation of Stat3 and Akt and upregulation of survivin. Cell Physiol. Biochem. 2016;38:1199–1208. doi: 10.1159/000443068. [DOI] [PubMed] [Google Scholar]
- Huang B., Lu M., Jia D., Ben-Jacob E., Levine H., Onuchic J.N. Interrogating the topological robustness of gene regulatory circuits by randomization. PLoS Comput. Biol. 2017;13:e1005456. doi: 10.1371/journal.pcbi.1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanteti R., Batra S.K., Lennon F.E., Salgia R. FAK and paxillin, two potential targets in pancreatic cancer. Oncotarget. 2016;7:31586–31601. doi: 10.18632/oncotarget.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada I., Hasina R., Lennon F.E., Bindokas V.P., Usatyuk P., Tan Y.H., Krishnaswamy S., Arif Q., Carey G., Hseu R.D. Paxillin mutations affect focal adhesions and lead to altered mitochondrial dynamics: relevance to lung cancer. Cancer Biol. Ther. 2013;14:679–691. doi: 10.4161/cbt.25091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Cramer G.M., Dahaj S.A.Z., Sundaram B., Celli J.P., Kulkarni R.V. Stochastic modeling of phenotypic switching and chemoresistance in cancer cell populations. Sci. Rep. 2019;9:10845. doi: 10.1038/s41598-019-46926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Dexheimer T.S., Zhang P., Rosenthal A.S., Villamil M.A., You C., Zhang Q., Chen J., Ott C.A., Sun H. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Ginsberg M.H. Paxillin binding to a conserved sequence motif in the α4 integrin cytoplasmic domain. J. Biol. Chem. 2000;275:22736–22742. doi: 10.1074/jbc.M000388200. [DOI] [PubMed] [Google Scholar]
- Liu S., Thomas S.M., Woodside D.G., Rose D.M., Kiosses W.B., Pfaff M., Ginsberg M.H. Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature. 1999;402:676–681. doi: 10.1038/45264. [DOI] [PubMed] [Google Scholar]
- Ma X., Hammes S.R. Paxillin actions in the nucleus. Steroids. 2018;133:87–92. doi: 10.1016/j.steroids.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon A.C., Tretiakova M., Henderson L., Mehta R.G., Yan B.C., Joseph L., Krausz T., Husain A.N., Reid M.E., Salgia R. Paxillin expression and amplification in early lung lesions of high-risk patients, lung adenocarcinoma and metastatic disease. J. Clin. Pathol. 2011;64:16–24. doi: 10.1136/jcp.2010.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero F., Murgia C., Wary K.K., Curatola A.M., Pepe A., Blumemberg M., Westwick J.K., Der C.J., Giancotti F.G. The coupling of α6β4 integrin to Ras-MAP kinase pathways mediated by Shc controls keratinocyte proliferation. EMBO J. 1997;16:2365–2375. doi: 10.1093/emboj/16.9.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziveyi M., Alahari S.K. Cell matrix adhesions in cancer: the proteins that form the glue. Oncotarget. 2017;8:48471–48487. doi: 10.18632/oncotarget.17265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerathilingam M., Bairy S.G., Mysore S. Deciphering mode of action of functionally important regions in the intrinsically disordered paxillin (residues 1-313) using its interaction with FAT (focal adhesion targeting domain of focal adhesion kinase) PLoS One. 2016;11:e0150153. doi: 10.1371/journal.pone.0150153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha C.R.R., Silva M.M., Quinet A., Cabral-Neto J.B., Menck C.F.M. DNA repair pathways and cisplatin resistance: an intimate relationship. Clinics (Sao Paulo) 2018;73:e478s. doi: 10.6061/clinics/2018/e478s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgia R. Mutation testing for directing upfront targeted therapy and post-progression combination therapy strategies in lung adenocarcinoma. Expert Rev. Mol. Diagn. 2016;16:737–749. doi: 10.1080/14737159.2016.1181545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgia R., Li J.L., Lo S.H., Brunkhorst B., Kansas G.S., Sobhany E.S., Sun Y., Pisick E., Hallek M., Ernst T. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL. J. Biol. Chem. 1995;270:5039–5047. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- Sarin N., Engel F., Kalayda G.V., Mannewitz M., Cinatl J., Jr., Rothweiler F., Michaelis M., Saafan H., Ritter C.A., Jaehde U., Frötschl R. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS One. 2017;12:e0181081. doi: 10.1371/journal.pone.0181081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M.D. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20:6459–6472. doi: 10.1038/sj.onc.1204786. [DOI] [PubMed] [Google Scholar]
- Schwartz N., Cohen R., Ben-Avraham D., Barabási A.L., Havlin S. Percolation in directed scale-free networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2002;66:015104. doi: 10.1103/PhysRevE.66.015104. [DOI] [PubMed] [Google Scholar]
- Shen D.W., Pouliot L.M., Hall M.D., Gottesman M.M. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012;64:706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Li M., Tretiakova M., Salgia R., Cagle P.T., Husain A.N. Expression patterns of PAX5, c-Met, and paxillin in neuroendocrine tumors of the lung. Arch. Pathol. Lab. Med. 2010;134:1702–1705. doi: 10.5858/2009-0664-oar1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L., Whitfield J.R., Sodir N.M., Massó-Vallés D., Serrano E., Karnezis A.N., Swigart L.B., Evan G.I. Inhibition of Myc family proteins eradicates Kras driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Lv X., Miao J. Integrin beta 4 in neural cells. Neuromolecular Med. 2008;10:316–321. doi: 10.1007/s12017-008-8042-1. [DOI] [PubMed] [Google Scholar]
- Swain P.S., Elowitz M.B., Siggia E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. U S A. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.C., Mirzapoiazova T., Won B.M., Zhu L., Srivastava M.K., Vokes E.E., Husain A.N., Batra S.K., Sharma S., Salgia R. Differential responsiveness of MET inhibition in non-small-cell lung cancer with altered CBL. Sci. Rep. 2017;7:9192. doi: 10.1038/s41598-017-09078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tözeren A., Kleinman H.K., Wu S., Mercurio A.M., Byers S.W. Integrin alpha 6 beta 4 mediates dynamic interactions with laminin. J. Cell Sci. 1994;107:3153–3163. doi: 10.1242/jcs.107.11.3153. [DOI] [PubMed] [Google Scholar]
- Trusolino L., Bertotti A., Comoglio P.M. A signaling adapter function for alpha6beta4 integrin in the control of HGF-dependent invasive growth. Cell. 2001;107:643–654. doi: 10.1016/s0092-8674(01)00567-0. [DOI] [PubMed] [Google Scholar]
- Turner C.E. Paxillin. Int. J. Biochem. Cell Biol. 1998;30:955–959. doi: 10.1016/s1357-2725(98)00062-4. [DOI] [PubMed] [Google Scholar]
- Turner C.E. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2000;2:E231–E236. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
- Wu D.W., Wu T.C., Wu J.Y., Cheng Y.W., Chen Y.C., Lee M.C., Chen C.Y., Lee H. Phosphorylation of paxillin confers cisplatin resistance in non-small cell lung cancer via activating ERK-mediated Bcl-2 expression. Oncogene. 2014;33:4385–4395. doi: 10.1038/onc.2013.389. [DOI] [PubMed] [Google Scholar]
- Xiao L., Lan X., Shi X., Zhao K., Wang D., Wang X., Li F., Huang H., Liu J. Cytoplasmic RAP1 mediates cisplatin resistance of non-small cell lung cancer. Cell Death Dis. 2017;8:e2803. doi: 10.1038/cddis.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori T., Yasui H., Yamazumi M., Wada Y., Nakamura Y., Nakamura H., InanamI O. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic. Biol. Med. 2012;53:260–270. doi: 10.1016/j.freeradbiomed.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE155605 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155605).