Summary
Brassinosteroids (BRs) regulate a variety of physiological processes in plants via extensive crosstalk with diverse biological signaling networks. Although BRs are known to reciprocally regulate circadian oscillation, the molecular mechanism underlying BR-mediated regulation of circadian clock remains unknown. Here, we demonstrate that the BR-activated transcription factor bri1-EMS-SUPPRESSOR 1 (BES1) integrates BR signaling into the circadian network in Arabidopsis. BES1 repressed expression of CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) at night by binding to their promoters, together with TOPLESS (TPL). The repression of CCA1 and LHY by BR treatment, which occurred during the night, was compromised in bes1-ko and tpl-8 mutants. Consistently, long-term treatment with BR shortened the circadian period, and BR-induced rhythmic shortening was impaired in bes1-ko and tpl-8 single mutants and in the cca1-1lhy-21 double mutant. Overall, BR signaling is conveyed to the circadian oscillator via the BES1/TPL-CCA1/LHY module, contributing to gating diurnal BR responses in plants.
Subject Areas: Chronobiology, Plant Biology
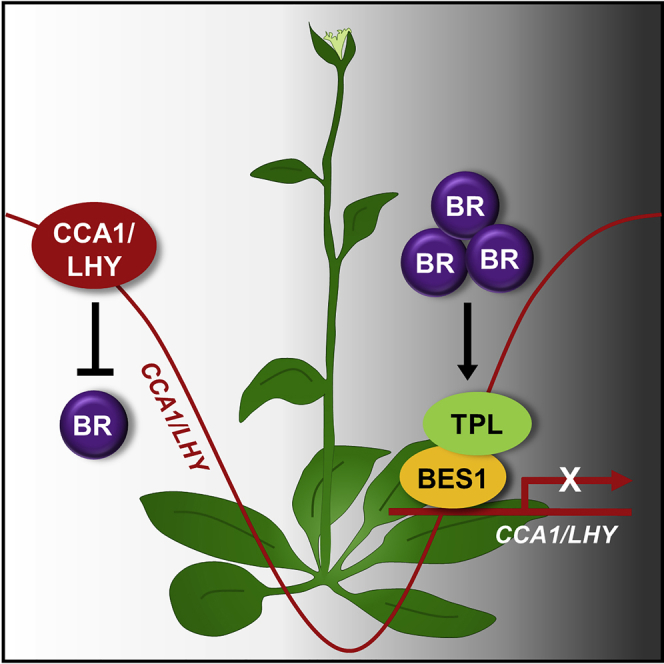
Graphical Abstract
Highlights
-
•
Circadian rhythm is shortened in the presence of phytohormone BR
-
•
BR signaling is relayed to circadian clock through BES1
-
•
BES1/TPL complex binds to CCA1/LHY promoter in the dark, repressing expression
-
•
The BES1/TPL-CCA1/LHY module shapes circadian gating of BR signaling
Chronobiology; Plant Biology
Introduction
Steroids are important for biological development in animals and plants. While animal steroid hormones are perceived by nuclear receptors (He et al., 2010; Sever and Glass, 2013), the plant steroid hormone brassinosteroid (BR) binds to the extracellular domain of the membrane-bound receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) (Kinoshita et al., 2005; Hothorn et al., 2011; Sun et al., 2013). The plasma-membrane-anchored co-receptor of BRI1, BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), activates intracellular signaling pathways comprising the serine/threonine protein kinase BRASSINOSTEROID-SIGNALING KINASE 1 (BSK1), the protein phosphatase bri1-SUPPRESSOR 1 (BSU1), the GLYCOGEN SYNTHASE KINASE 3 (GSK3)-like kinase BRASSINOSTEROID INSENSITIVE 2 (BIN2), PROTEIN PHOSPHATASE 2A (PP2A) phosphatases, and BRASSINAZOLE RESISTANT 1 (BZR1) family transcription factors (Ye et al., 2011; Wang et al., 2012).
In the absence of BRs, BIN2 inactivates BES1/BZR2 and BZR1 via phosphorylation (He et al., 2002; Wang et al., 2002; Yin et al., 2002). The phosphorylated BES1 and BZR1 not only retain their cytoplasmic localization but also exhibit reduced affinity for DNA (Gampala et al., 2007; Ryu et al., 2007, 2010). In the presence of BRs, BRI1 inactivates BIN2 and stimulates PP2A activity to catalyze the dephosphorylation of BES1 and BZR1 (Tang et al., 2011). Dephosphorylated BES1 and BZR1 then enter the nucleus, where they regulate the expression of BR-responsive genes by binding directly to their promoters (He et al., 2005; Yin et al., 2005; Sun et al., 2010).
BRs regulate a variety of developmental and physiological processes in plants including cell division, photomorphogenesis, reproductive organ development, stomata development, leaf senescence, and biotic and abiotic stress responses (Wang et al., 2012; Gruszka, 2013; Zhiponova et al., 2013; Wang et al., 2001; Planas-Riverola et al., 2019; Yu et al., 2018; Ackerman-Lavert and Savaldi-Goldstein, 2020). Consistently, the molecular crosstalk between the BR signaling pathway and other hormonal and developmental signaling pathways is starting to emerge.
In plants, the circadian clock maintains an endogenous rhythm with a period of ∼24 h (Covington et al., 2008; Mizuno and Yamashino, 2008; Hsu and Harmer, 2012). Extensive transcriptional feedback loops form the basic framework of the clock oscillator. Intricate connections among several transcriptional regulators, including CCA1, LHY, TIMING OF CAB EXPRESSION 1 (TOC1), PSEUDO-RESPONSE REGULATORs (PRRs), EARLY FLOWERING 3 (ELF3), ELF4, and LUX ARRYTHMO (LUX), are involved in the maintenance of circadian homeostasis (Alabadi et al., 2001; Nakamichi et al., 2010; Nusinow et al., 2011; Huang et al., 2012). The circadian oscillator diurnally regulates a variety of output pathways in plants, including primary and secondary metabolism, hormone biosynthesis and signaling, and responses to environmental challenges to induce relevant physiological changes at a specific time of the day (Dodd et al., 2005; Legnaioli et al., 2009; Singh and Mas, 2018). In addition, it is noteworthy that some circadian-controlled phytohormones reciprocally regulate core clock genes to ensure an accurate circadian gating of hormone metabolism and signaling (Covington and Harmer, 2007; Lee et al., 2016; Singh and Mas, 2018).
A bidirectional regulation has been suggested between the circadian clock and BR signaling. The majority of BR biosynthesis and signaling genes are clock-controlled (Bancos et al., 2006). Moreover, exogenous BR application also influences clock activity (Hanano et al., 2006). However, molecular factors and mechanisms underlying the link between BR signaling and the circadian clock remain unclear. In this study, we demonstrate that BES1 regulates CCA1 and LHY expression to relay BR signals to the clock oscillator. In the presence of BRs, BES1 binds to CCA1 and LHY promoters and represses their expression, especially at night. This BES1-CCA1/LHY module delicately regulates circadian oscillation to ensure acute activation of BR signaling at a specific time of day.
Results
BES1 Specifically Relays BR-Regulated Clock Activity
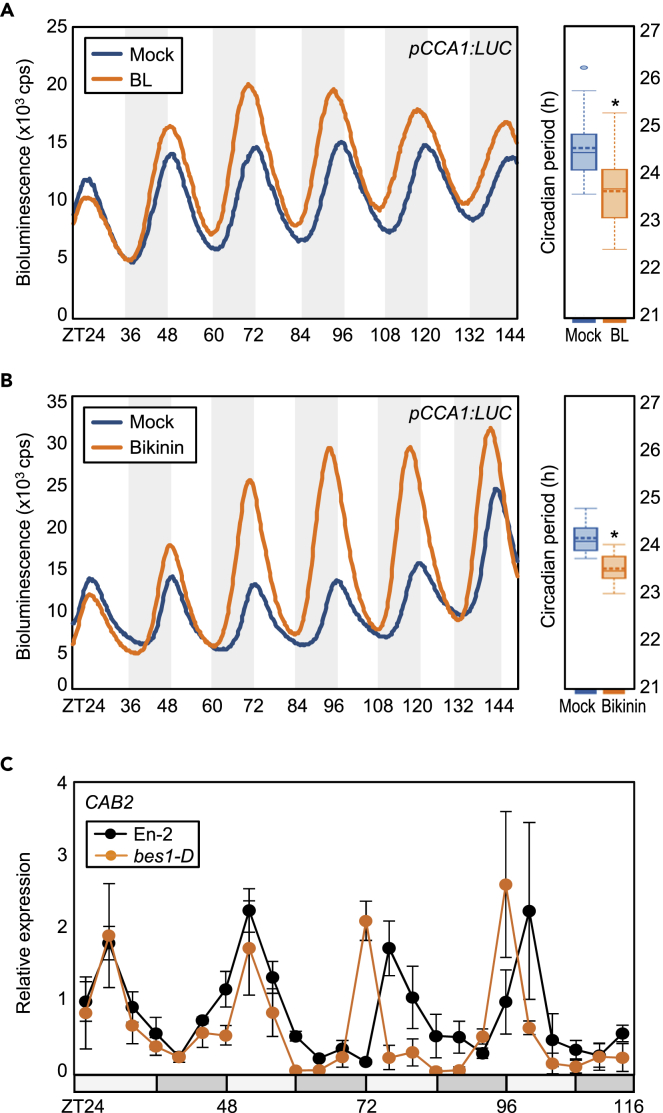
Although circadian clock activity exerts diurnal control over hormone signaling, it is also reciprocally regulated by hormone signaling (Covington and Harmer, 2007; Lee et al., 2016; Singh and Mas, 2018; Nohales and Kay, 2019). Plant hormones, specifically abscisic acid (ABA) and BRs, have a considerable impact on circadian oscillation (Hanano et al., 2006; Lee et al., 2016), although the underlying molecular mechanisms remain unknown. Therefore, we investigated the link between BR signaling and circadian networks. To test whether BRs influence clock oscillation in our growth conditions, we examined the effects of exogenous BR treatment on circadian oscillation by monitoring luciferase (LUC) activity in pCCA1:LUC transgenic plants. Exogenous treatment with epi-brassinolide (epi-BL) significantly shortened the circadian period (Figure 1A). Quantitative analysis revealed that mock-treated wild-type plants displayed circadian periods of approximately 24.6 h under free-running conditions, whereas epi-BL-treated wild-type plants showed a circadian period of approximately 23.7 h (Figure 1A).
Figure 1.
BES1 Is Responsible for BR-Dependent Circadian Control
(A) Effects of exogenous BR treatment on circadian period. pCCA1:LUC transgenic plants (Col-0 background) grown and entrained for 7 days on MS-solid medium with 3% sucrose under neutral day (ND) conditions were transferred to continuous dark (DD) conditions. During the free-running conditions, seedlings were incubated in MS-liquid medium supplemented with or without 100 μM epi-brassinolide (BL). Circadian period estimates of pCCA1:LUC activity in the presence or absence of 100 μM epi-BL was shown (right panel). Data represented as mean ± SEM were analyzed with using the Fast Fourier Transform Non-linear Least Squares (FFT-NLLS) suite of programs available in the Biodare2 software (n > 15). Significant differences between Mock and BL are indicated using an asterisk (∗p < 0.05; Student's t test).
(B) Effects of bikinin on circadian oscillation. Seven-day-old pCCA1:LUC transgenic plants grown under ND conditions were transferred to DD conditions. LUC activity was examined in the presence or absence of 40 μM bikinin (left panel). Circadian period estimates of pCCA1:LUC activity in the presence or absence of 40 μM bikinin was shown (right panel). Data represent mean ± SEM (n > 15) and were analyzed by FFT-NLLS. Significant differences between Mock and Bikinin are indicated using an asterisk (∗p < 0.05; Student's t test).
(C) Expression of CAB2 in bes1-D mutant. Seedlings grown under ND conditions for 2 weeks were transferred to continuous light (LL) conditions. Transcript levels of genes were determined by quantitative real-time RT-PCR (RT-qPCR). Gene expression values were normalized relative to the expression of EUKARYOTIC TRANSLATION INITIATION FACTOR 4a1 (eIF4a) and represented as n-fold relative to the value of the sample at ZT24. Data represent mean ± SEM.
To understand the molecular basis of BR-mediated regulation of the circadian clock, we interfered with BR signaling and examined circadian oscillation. Notably, a potent chemical inhibitor of GSK proteins, bikinin (Uehara et al., 2019), shortened the rhythmic period in pCCA1:LUC transgenic plants compared with mock treatment (Figure 1B), similar to the epi-BL treatment (Figure 1A). These observations suggest that the effect of BRs on circadian oscillation could be mediated by GSK kinases or by downstream components of the BR signaling pathway, such as BES1 and BZR1. Because circadian components are largely under transcriptional control, we focused on the potential role of BES1 and BZR1 in circadian oscillation.
We analyzed circadian oscillation in bes1-D and bzr1-1D mutants, which constitutively activate BES1 and BZR1, respectively (Wang et al., 2002, 2012; Li et al., 2018). Quantitative real-time RT-PCR (RT-qPCR) analysis of the CHLOROPHYLL A/B-BINDING PROTEIN 2 (CAB2) circadian marker gene revealed that bes1-D plants exhibited a shortened circadian period (Figure 1C), whereas bzr1-1D plants exhibited normal circadian oscillation (Figure S1), similar to wild-type plants under free-running conditions (Figure S1). However, bes1-ko single mutant did not display alterations in circadian oscillation, probably because the BR signaling was below the threshold required for circadian control under normal growth conditions (Figure S1). In support, the bri1-5 mutant also exhibited no alteration in circadian oscillation compared with wild-type under normal growth conditions (Figure S2). These results suggest that BES1 is responsible for controlling circadian oscillation especially in the presence of BRs.
BES1 Binds to CCA1 and LHY Promoters
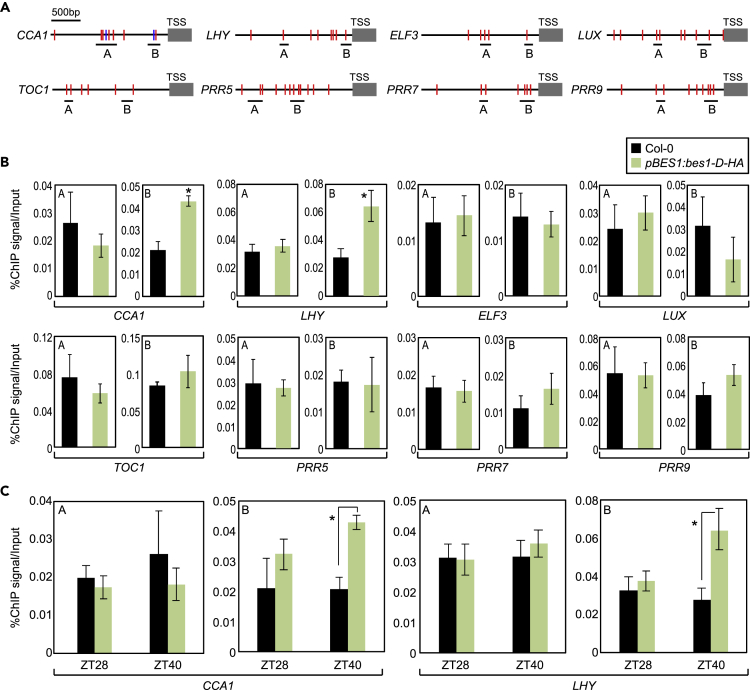
Next, we aimed to decipher how BES1 regulates circadian oscillation. To identify the target genes of BES1 in the circadian pathway, we analyzed cis-elements in the core clock gene promoters, which may be targeted by the atypical basic-helix-loop-helix (bHLH) DNA-binding motifs of BES1 (Nosaki et al., 2018). Promoter analysis revealed the presence of the conserved BR-response element (BRRE) (CGTG(T/C)G) and/or E-box (CAnnTG) motif in the promoter regions of several core clock genes (Li et al., 2018) (Figure 2A).
Figure 2.
BES1 Binds to CCA1 and LHY Promoters
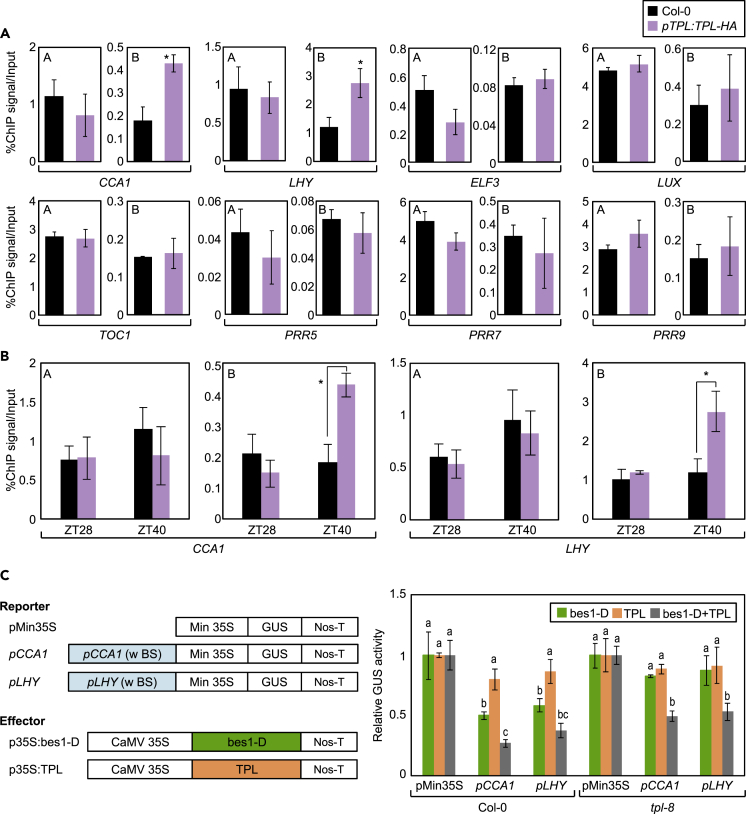
(A) Genomic structures of core clock genes. Underbars indicate the regions amplified by quantitative PCR (qPCR) following chromatin immunoprecipitation (ChIP). Red lines indicate E-boxes, and blue lines indicate BR-response elements (BRREs).
(B) Binding of BES1 to clock gene promoters at ZT40.
(C) Binding of BES1 to clock gene promoters. In (B) and (C), two-week-old seedlings entrained under ND cycles were subjected to LL conditions. Plants were harvested for ChIP analysis with anti-HA antibody. Data represent mean ± SEM. Significant differences between Col-0 and pBES1:bes1-D-HA plants are indicated using an asterisk (∗p < 0.05; Student's t test).
To determine whether BES1 binds to core clock gene promoters to control circadian oscillation, we employed pBES1:bes1-D-HA plants and performed chromatin immunoprecipitation (ChIP) analysis using an anti-HA antibody. Fragmented DNA was eluted from protein–DNA complexes and subjected to quantitative PCR (qPCR) analysis. DNA enrichment indicated that BES1 binds to CCA1 and LHY promoters (Figure 2B) but not to the promoters of other clock genes, including ELF3, LUX, TOC1, PRR5, PRR7, and PRR9 (Figures 2B and S3). In addition, coding regions of CCA1 and LHY loci were not targeted by BES1 (Figure S4), indicating that the promoter regions of CCA1 and LHY are the primary binding sites of BES1. In addition, BZR1 did not bind to any of the core clock gene promoters (Figure S5) and was independent of circadian regulation. Overall, our results indicate that BES1 is specifically associated with CCA1 and LHY genes.
To confirm the results, we performed transient β-glucuronidase (GUS) expression assays using Arabidopsis protoplasts. We designed two reporter constructs for each gene: one was a longer promoter sequence containing BES1-binding site (BS) (B regions in Figure 2A) and the other was a shorter promoter sequence without BES1-BS (Figure S6). The promoter sequences were transcriptionally fused to a cauliflower mosaic virus (CaMV) 35S minimal promoter. The reporter plasmids were cotransformed with effector plasmids, p35S:bes1-D and p35S:bzr1-1D, into Arabidopsis protoplasts. The cotransformation of p35S:bes1-D with pCCA1-reporter containing BS was found to repress the reporter gene expression (Figure S6). By contrast, cotransformation with pCCA1-reporter without BS did not repress the reporter gene expression (Figure S6). Furthermore, cotransfection of BS-containing reporter plasmids with the 35S:bzr1-1D construct negligibly influenced CCA1 and LHY expression (Figure S6), indicating that BES1 is specifically associated with the CCA1 and LHY loci and represses expression.
BR signaling components were predicted to be circadian-regulated (Figure S7). However, the BES1 gene did not exhibit rhythmic expression (Figure S7). Nonetheless, the stability of BES1 protein is known to be diurnally regulated by posttranslational modifications (Martinez et al., 2018). Notably, an active dephosphorylated form of BES1 is enriched during night time (Yang et al., 2017; Zhang et al., 2017; Martinez et al., 2018). Based on these observations, we speculated that BES1 regulation of the circadian clock may be relevant at night. To test this possibility, we examined the time of day when BES1 binds to CCA1 and LHY promoters. ChIP-qPCR analysis showed that BES1 bound CCA1 and LHY promoters primarily at zeitgeber time 40 (ZT40) (Figure 2C), consistent with the circadian accumulation of BES1 protein. These observations indicate that BES1 directly regulates the expression of CCA1 and LHY, especially during the night.
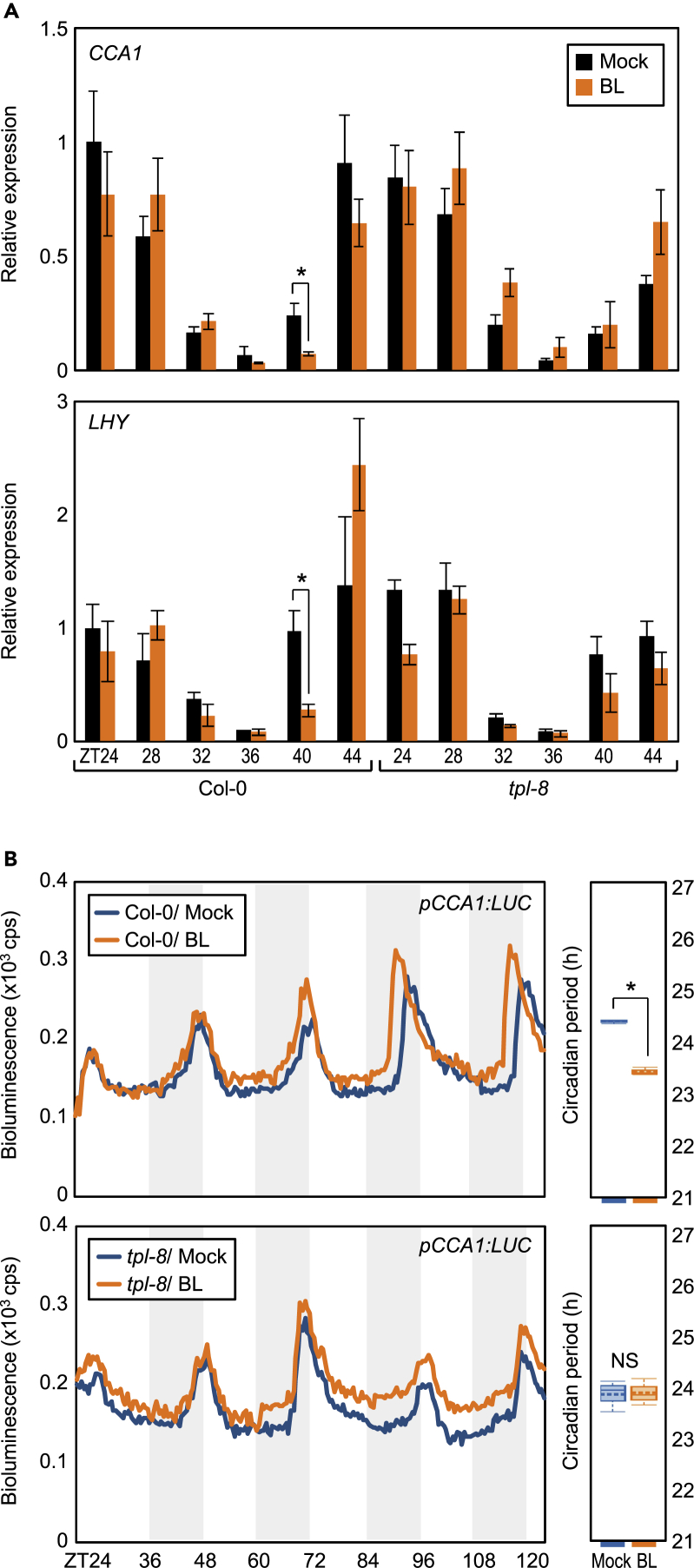
BR Shortens the Circadian Period via the BES1-CCA1/LHY Module
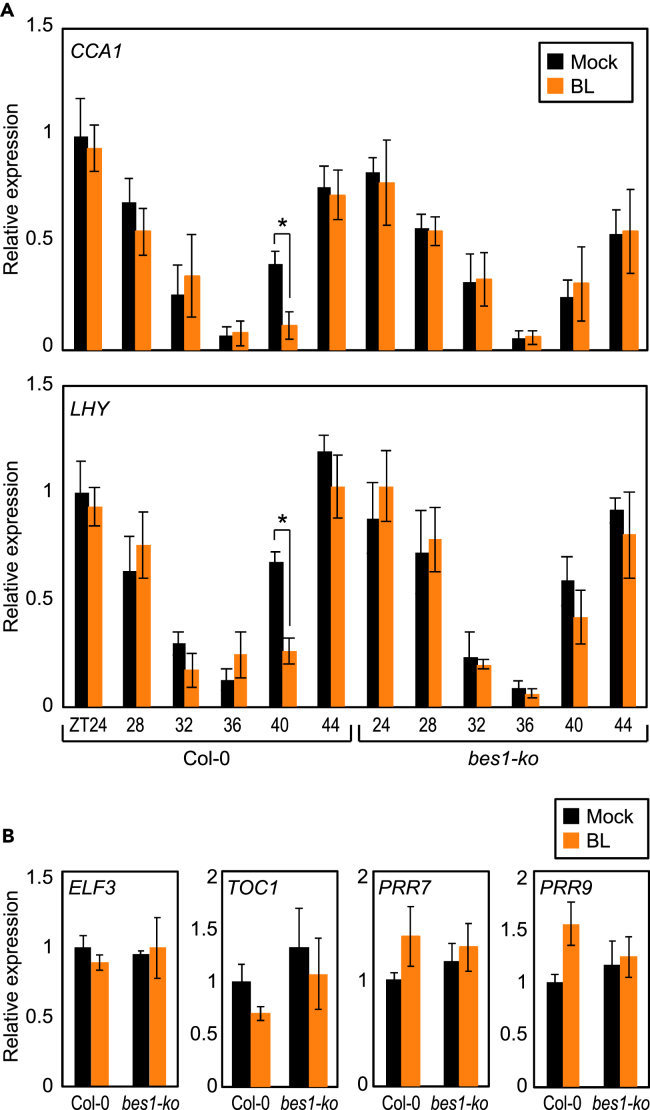
BES1 mediates the effects of BRs on the circadian clock by controlling the expression of CCA1 and LHY genes, in the presence of BR. To provide further support, we examined the transcript accumulation of CCA1 and LHY in wild-type seedlings subjected to a short-term (4 h) BR treatment at different times during the day. RT-qPCR analysis showed that BL suppressed CCA1 and LHY expression in wild type (Figure 3A), and this suppression occurred only from ZT40 to ZT44 (Figure 3A). Furthermore, this BR-dependent suppression of CCA1 and LHY was impaired in bes1-ko mutant plants (Figure 3A), whereas the expression of other clock genes was uninfluenced from ZT40 to ZT44 (Figure 3B). This result is consistent with the binding of BES1 to CCA1 and LHY promoters at night (Figure 2C), suggesting that the BES1-CCA1/LHY module is most likely responsible for the BR-regulated circadian rhythm.
Figure 3.
BES1 Represses CCA1 and LHY Expression
Two-week-old seedlings grown under ND conditions were transferred to MS-liquid medium supplemented with or without 1 μM epi-BL at the indicated ZT points and incubated for 4 h under LL conditions.
Transcript accumulation of CCA1 and LHY (A) and the other clock genes (B) was analyzed by RT-qPCR. Gene expression values were normalized relative to eIF4a expression and represented as n-fold relative to the value of the mock-treated wild-type sample. Data represent mean ± SEM (∗p < 0.05; Student's t test).
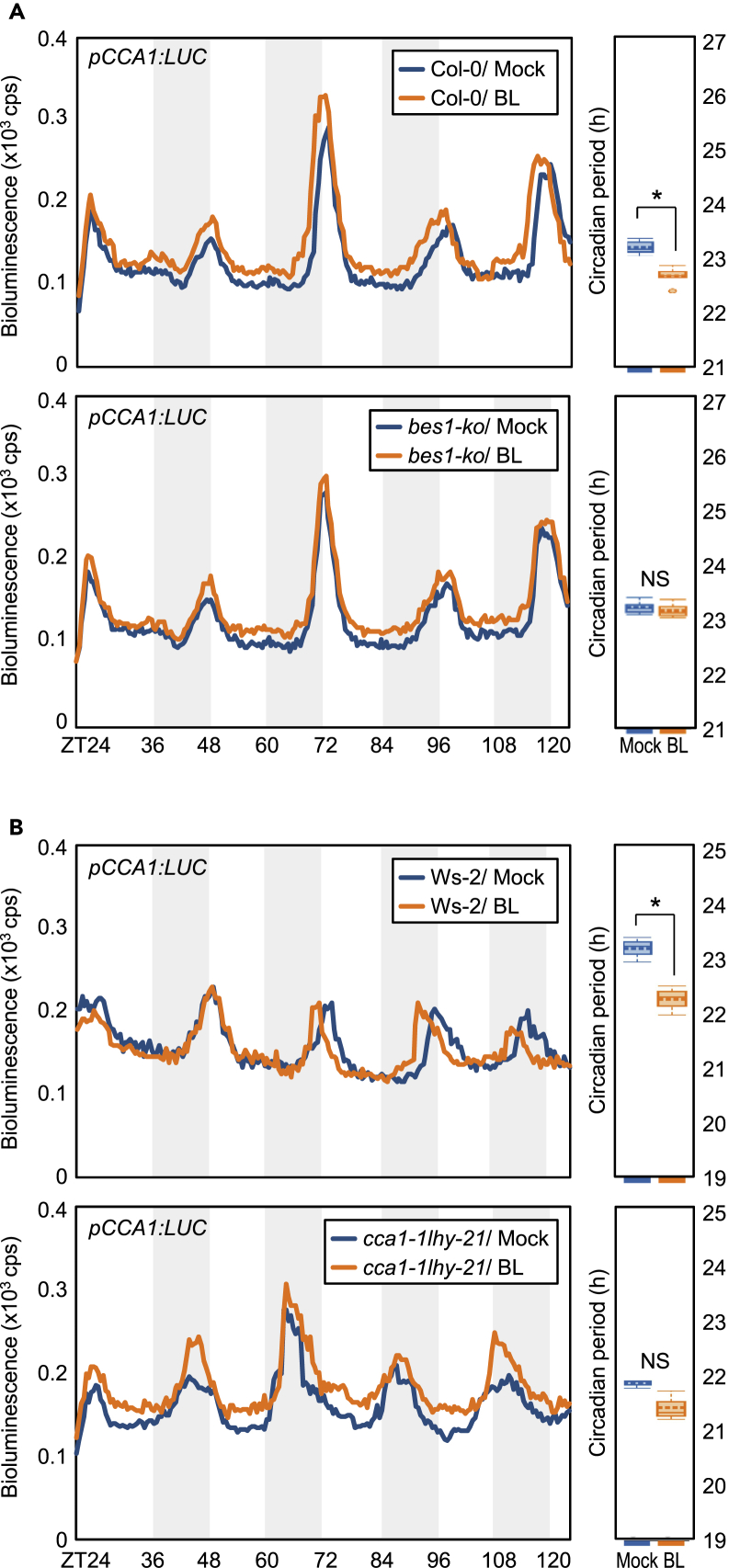
Furthermore, this result raised the possibility that BR-induced period shortening (Figure 1A) is also mediated by BES1 and CCA1/LHY. To test this possibility, we monitored the bioluminescence of protoplasts transiently expressing pCCA1:LUC. In wild-type protoplasts expressing pCCA1:LUC, LUC activity oscillation was similar to that observed in wild-type seedlings (Figures 1A and 4A), and long-term treatment with epi-BL reduced circadian period (Figures 4A and 4B). Notably, BR-dependent period shortening disappeared in bes1-ko single mutant (Figure 4A) and cca1-1lhy-21 double mutant protoplasts (Figure 4B). These results indicate that the BES1-CCA1/LHY module is important for reciprocal clock resetting in response to BR treatment.
Figure 4.
BR-Regulated Circadian Oscillation Is Compromised in bes1-ko and cca1-1lhy21 Mutants
(A and B) Bioluminescence circadian rhythms of bes1-ko (Col-0 background) (A) and cca1-1 lhy-21 (Ws-2 background) (B) mesophyll protoplasts transiently expressing the pCCA1:LUC construct in the presence or absence of 100 μM epi-BL. Mesophyll protoplasts of bes1-ko, cca1-1 lhy-21, and wild type were isolated from leaves of 3-week-old seedlings entrained MS-solid medium under ND cycles. Isolated protoplasts were transiently PEG-transfected with the pCCA1-LUC plasmid and then subjected to DD conditions to monitor bioluminescence traces. Data represent mean ± SEM (∗p < 0.05; Student's t test). Period estimates of pCCA1:LUC activity in the presence or absence of 100 μM epi-BL analyzed by FFT-NLLS in Biodare2 were shown (right panel). NS, not significant.
TPL Is Required for BES1-Dependent Repression of CCA1 and LHY
The BES1 protein acts either as a transcriptional activator or as a transcriptional repressor (Yin et al., 2002, 2005; Li et al., 2009; Espinosa-Ruiz et al., 2017). In this study, BES1 suppressed CCA1 and LHY expression, in the presence of BRs. We thus postulated that BES1 recruits additional trans-factors to repress gene expression. Previous studies showed that BES1 usually accompanies TPL and TPL-RELATED (TPR) proteins in processes requiring predominantly repressive activities (Ryu et al., 2014; Espinosa-Ruiz et al., 2017; Kim et al., 2019). Yeast-two-hybrid (Y2H) assays performed in this study supported the BES1-TPL interaction (Figure S8). However, none of the other clock components showed physical interaction with BES1 (Figure S8). Based on these observations, we speculated that the BES1-TPL transcriptional co-repressor complex primarily represses CCA1 and LHY expression to control circadian oscillation in response to BRs.
To support our claim, we examined the binding of TPL to CCA1 and LHY promoters using pTPL:TPL-HA transgenic plants. ChIP-qPCR analysis revealed that TPL directly associated with CCA1 and LHY loci (Figure 5A). The binding sites of TPL on CCA1 and LHY promoters overlapped with those of BES1 (Figures 5A and 2B, see also Figures S3, S4, S9, and S10). Furthermore, the binding of TPL to CCA1 and LHY promoters was observed specifically at night (Figure 5B), consistent with the timing of BES1 binding (Figure 2C).
Figure 5.
Repression of CCA1 and LHY by BES1 Is Dependent on TPL
(A) Binding of TPL to CCA1 and LHY promoters.
(B) Timing of TPL binding to CCA1 and LHY promoters at ZT40. In (A) and (B), 2-week-old plants grown under ND cycles were subjected to LL conditions and harvested at ZT28 and ZT40 for ChIP analysis with anti-HA antibody. Enrichment of putative TPL binding regions in clock gene promoters was analyzed by ChIP-qPCR. Data represent mean ± SEM (∗p < 0.05; Student's t test).
(C) Transient expression analysis using Arabidopsis protoplasts. Core elements containing BES1-binding sites (BS) of CCA1 and LHY promoters were cloned into the reporter plasmid. The effector and reporter constructs were transiently coexpressed in Col-0 and tpl-8 protoplasts. GUS activity in protoplasts was measured using a fluorometer. LUC gene expression was used to normalize GUS activity. Data represent mean ± SEM of biological triplicates. Different letters represent a significant difference at p < 0.05 (one-way ANOVA with Fisher's post hoc test).
We also performed transient coexpression analyses using Arabidopsis protoplasts. The core binding regions of BES1 in CCA1 and LHY promoters were cloned upstream of the β-glucuronidase (GUS) reporter gene. Recombinant reporter plasmids (pCCA1:GUS and pLHY:GUS) were cotransformed with effector plasmids (p35S:bes1-D and p35S:TPL) into Arabidopsis protoplasts isolated from wild-type and tpl-8 mutant leaves. In wild-type protoplasts, cotransfection of reporter plasmids with the bes1-D construct repressed CCA1 and LHY expression (Figure 5C). In addition, gene repression was synergistic when coexpressed with the TPL construct (Figure 5C). In tpl-8 mutant protoplasts, the repression of CCA1 and LHY by BES1 was impaired, but TPL complementation restored BES1-dependent repression of CCA1 and LHY (Figure 5C).
These results were supported by the short-term treatment of wild-type and tpl-8 mutant plants with epi-BL; although short-term epi-BL treatment reduced the expression of CCA1 and LHY genes from ZT40-44 in wild-type seedlings, this effect was compromised in tpl-8 mutant seedlings (Figure 6A), similar to that observed in bes1-ko mutant plants (Figure 3A). Furthermore, the effect of long-term treatment with BR on circadian oscillation was also mediated by TPL. BR-induced period shortening was impaired in tpl-8 protoplasts expressing pCCA1:LUC (Figure 6B), indicating that BES1 requires TPL to suppress CCA1 and LHY expression and thereby shorten the circadian period, in the presence of BR.
Figure 6.
TPL Mediates BL-Regulated Circadian Oscillation
(A) Impaired repression of CCA1 and LHY by BL in tpl-8. Two-week-old seedlings grown under ND conditions were transferred to MS-liquid medium supplemented with or without 1 μM epi-BL at the indicated ZT points and incubated for 4 h under LL conditions. Transcript accumulation was analyzed by RT-qPCR. Gene expression values were normalized relative to eIF4a expression and represented as n-fold relative to the value of the mock-treated wild-type sample. Data represent mean ± SEM (∗p < 0.05; Student's t test).
(B) Bioluminescence circadian expression of protoplasts isolated from Col-0 and tpl-8 leaves in the presence of 100 μM epi-BL. Mesophyll protoplasts of tpl-8 and wild type were isolated from leaves of 3-week-old seedlings entrained on MS-solid medium under ND cycles. Isolated protoplasts were transiently PEG-transfected with the pCCA1-LUC plasmid and then subjected to DD conditions to monitor bioluminescence traces. Data presented as mean ± SEM of biological triplicates were analyzed by FFT-NLLS. Statistical differences are indicated using an asterisk (∗p < 0.05; Student's t test).
Taken together, our results suggest that the BES1/TPL-CCA1/LHY module reciprocally regulates circadian oscillation. In the presence of BRs, BES1 is activated at night and binds to CCA1 and LHY promoters, together with TPL, to repress their expression. Consequently, BES1 delays the rising phase of CCA1 and LHY (Figure 7), which attenuate BR responses (Figure S11). This circadian phase resetting allows precise gating of BR signaling as well as plant growth and development at a specific time of day. In addition, the BES1/TPL-CCA1/LHY-module-dependent circadian control is likely relevant in tissues or cells with high concentrations of BRs.
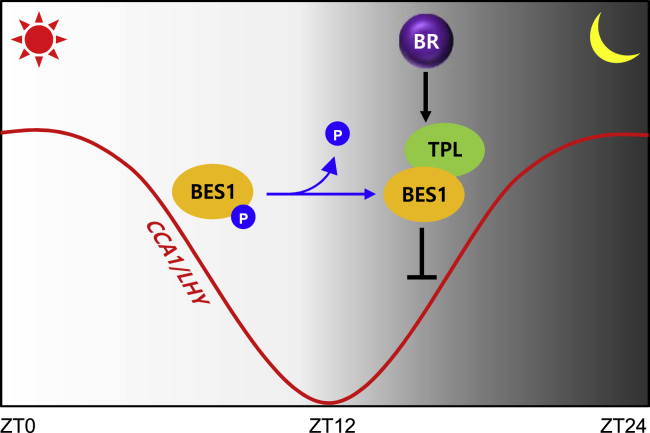
Figure 7.
Schematic Representation of CCA1 and LHY Repression by BES1 and TPL
BR-activated BES1 transcription factor binds to the promoters of morning-expressed clock genes CCA1 and LHY, especially at night. In the presence of BRs, BES1 recruits TPL to repress CCA1 and LHY expression, which interferes with BR signaling, at night and ensures acute gating of diurnal BR responses.
Discussion
Circadian Gating of Hormone Signaling
The circadian clock is a biological timekeeper that ensures an endogenous rhythm with a period of approximately 24 h. Correct matching of the endogenous rhythm with environmental cycles maximizes plant fitness and survival (Nozue et al., 2007; Nomoto et al., 2012; Lee et al., 2016). Approximately 50% of the Arabidopsis transcriptome is under the control of the circadian clock (Mizuno and Yamashino, 2008; Doherty and Kay, 2010); thus many biological processes are diurnally gated (Zhou et al., 2015; Lee et al., 2016; Nohales and Kay, 2019). For example, stress response and hormone-related pathways are under robust circadian regulation (Covington and Harmer, 2007; Grundy et al., 2015; Singh and Mas, 2018; Thines et al., 2019), and consequently many biological processes related to stress responses and hormone-mediated plant growth and development are gated at a specific time of day (Zhou et al., 2015; Lee et al., 2016; Nohales and Kay, 2019).
Accumulating evidence shows that circadian gating is shaped by bidirectional regulation. Not only the circadian clock regulate output pathways but also components of output pathways reciprocally regulate the clock oscillator (Lee et al., 2016). This bidirectional regulation readjusts circadian oscillation, thus allowing optimum coordination of output signaling with environmental cycles (Lee et al., 2016). For instance, bidirectional regulation has been demonstrated between circadian and ABA signaling. Plants are usually exposed to drought stress during the day, and the water-deficit crisis is generally at its peak in the afternoon (Legnaioli et al., 2009; Seo and Mas, 2015; Lee et al., 2016). Consistently, ABA metabolism and responses are gated primarily from midday to dusk. The majority of ABA signaling genes is diurnally activated and displays peak expression in the afternoon (Michael et al., 2008; Legnaioli et al., 2009; Lee et al., 2016). The CCA1 and Evening Complex transcriptional repressors are putative upstream regulators of stress signaling genes and allow their expression during the day (Legnaioli et al., 2009; Lee et al., 2016). In addition, ABA signaling reciprocally regulates circadian activity. The ABA-inducible MYB96 transcription factor activates the TOC1 gene by binding directly to its promoter (Lee et al., 2016). This reciprocal regulation of the circadian oscillator by output pathways most likely facilitates the readjustment of circadian oscillation in a sophisticated manner, ensuring near-perfect matching between diurnal physiological processes and environmental fluctuations to maximize plant performance under a given condition.
Bidirectional Regulation between BRs and the Circadian Clock
BR metabolism and signaling are intensively regulated by the circadian control. The BR biosynthesis gene CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD) exhibits rhythmic expression, with peak expression at dusk (Bancos et al., 2006). Furthermore, BR signaling components are circadian controlled and show diurnal changes in protein accumulation (Bancos et al., 2006; Martinez et al., 2018; Kim et al., 2019). For example, BES1 activity is shaped by diurnal light-dark cycles (Martinez et al., 2018). A couple of E3 ligases regulate the levels of phosphorylated and dephosphorylated BES1 proteins. During the light period, SINA OF ARABIDOPSIS THALIANAs (SINATs) accumulate to mediate the degradation of dephosphorylated BES1; thus a low ratio of dephosphorylated BES1 to phosphorylated BES1 (BES1/pBES1) is maintained to attenuate BR signaling (Yang et al., 2017). During the dark period, SINATs are degraded in a self-dependent manner, thus increasing the level of dephosphorylated BES1. In addition, pBES1 is degraded by the CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) protein, leading to a high ratio of BES1/pBES1 and activation of BR signaling (Kim et al., 2014; Yang et al., 2017).
We showed that BR signaling reciprocally influences circadian oscillation. BRs activated BES1, which subsequently repressed the expression of CCA1 and LHY genes during the night, together with TPL. Consistently, circadian oscillation in bes1-ko and tpl-8 single mutants and cca1-1lhy-21 double mutant was insensitive to BR treatment. These results indicate that the BES1-CCA1/LHY module plays a central role in relaying BR signaling to the circadian oscillator, establishing a bidirectional regulation between BR and circadian signaling.
BR regulation of the circadian clock likely strengthens BR responses at night. For instance, circadian gating of BR signaling is particularly important for cell elongation (Zhang et al., 2017). Consistently, the growth rate of hypocotyls in Arabidopsis is the highest at night (Nusinow et al., 2011; Soy et al., 2012; Pham et al., 2018). Reciprocal regulation of the circadian clock by BR-activated BES1 ensures circadian gating. BES1 suppresses CCA1 and LHY expression, which attenuate BR signaling, and delays the rising phase of the morning genes during the night, ensuring acute BR signaling at a specific time of day. Notably, this function is exaggerated by long-term exogenous BR treatment, which shortens the circadian period. Given that the circadian clock is globally linked to vast cellular networks, this signaling scheme is not limited to BR responses but is widely applicable to a variety of biological processes in plants.
Limitations of the Study
In this study, we demonstrated that the phytohormone BR regulates circadian clock through direct binding of BES1 to the promoters of CCA1 and LHY. We would like to note that the effects of BR on circadian rhythm is observed upon the treatment with high concentrations of epi-BL. We suspect that additional internal and external cues should be further considered to account for the physiological impact of BR in circadian clock.
In addition, we have demonstrated the bidirectional regulation between circadian clock and BR response, but the detailed molecular mechanism of how CCA1/LHY attenuate BR signaling should be elucidated in the future studies.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pil Joon Seo (pjseo1@snu.ac.kr).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The data used and analyzed during this study are available from the corresponding author on reasonable request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the Basic Science Research (NRF-2019R1A2C2006915) and Basic Research Laboratory (2020R1A4A2002901) programs provided by the National Research Foundation of Korea and by the Next-Generation BioGreen 21 Program (PJ01314501) provided by the Rural Development Administration.
Author Contributions
P.J.S. participated in the design of the study and wrote the article. H.G.L., J.H.W., Y.R.C., and K.L. performed the molecular experiments. H.G.L. and J.H.W. performed large-scale circadian time course experiments; all authors read and approved the final article.
Declaration of Interests
The authors declare no competing interests.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101528.
Supplemental Information
2
References
- Ackerman-Lavert M., Savaldi-Goldstein S. Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 2020;53:90–97. doi: 10.1016/j.pbi.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Bancos S., Szatmari A.M., Castle J., Kozma-Bognar L., Shibata K., Yokota T., Bishop G.J., Nagy F., Szekeres M. Diurnal regulation of the brassinosteroid-biosynthetic CPD gene in Arabidopsis. Plant Physiol. 2006;141:299–309. doi: 10.1104/pp.106.079145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Harmer S.L. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kevei E., Toth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Kay S.A. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Ruiz A., Martinez C., de Lucas M., Fabregas N., Bosch N., Cano-Delgado A.I., Prat S. TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development. 2017;144:1619–1628. doi: 10.1242/dev.143214. [DOI] [PubMed] [Google Scholar]
- Gampala S.S., Kim T.W., He J.X., Tang W., Deng Z., Bai M.Y., Guan S., Lalonde S., Sun Y., Gendron J.M. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy J., Stoker C., Carre I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 2015;6:648. doi: 10.3389/fpls.2015.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszka D. The brassinosteroid signaling pathway-new key players and interconnections with other signaling networks crucial for plant development and stress tolerance. Int. J. Mol. Sci. 2013;14:8740–8774. doi: 10.3390/ijms14058740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Domagalska M.A., Nagy F., Davis S.J. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells. 2006;11:1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- He J., Cheng Q., Xie W. Minireview: nuclear receptor-controlled steroid hormone synthesis and metabolism. Mol. Endocrinol. 2010;24:11–21. doi: 10.1210/me.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011;474:467–471. doi: 10.1038/nature10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.Y., Harmer S.L. Circadian phase has profound effects on differential expression analysis. PLoS One. 2012;7:e49853. doi: 10.1371/journal.pone.0049853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Perez-Garcia P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- Kim B., Jeong Y.J., Corvalan C., Fujioka S., Cho S., Park T., Choe S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014;77:737–747. doi: 10.1111/tpj.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Shim D., Moon S., Lee J., Bae W., Choi H., Kim K., Ryu H. Transcriptional network regulation of the brassinosteroid signaling pathway by the BES1-TPL-HDA19 co-repressor complex. Planta. 2019;250:1371–1377. doi: 10.1007/s00425-019-03233-z. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Cano-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Lee H.G., Mas P., Seo P.J. MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci. Rep. 2016;6:17754. doi: 10.1038/srep17754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T., Cuevas J., Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.-F., Lu J., Yu J.-W., Zhang C.-Q., He J.-X., Liu Q.-Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta. Gene Regul. Mech. 2018;1861:561–571. doi: 10.1016/j.bbagrm.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Martinez C., Espinosa-Ruiz A., de Lucas M., Bernardo-Garcia S., Franco-Zorrilla J.M., Prat S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018;37:e99552. doi: 10.15252/embj.201899552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49:481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales M.A., Kay S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2019;116:21893–21899. doi: 10.1073/pnas.1913532116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto Y., Kubozono S., Miyachi M., Yamashino T., Nakamichi N., Mizuno T. A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1965–1973. doi: 10.1093/pcp/pcs141. [DOI] [PubMed] [Google Scholar]
- Nosaki S., Miyakawa T., Xu Y., Nakamura A., Hirabayashi K., Asami T., Nakano T., Tanokura M. Structural basis for brassinosteroid response by BIL1/BZR1. Nat. Plants. 2018;4:771–776. doi: 10.1038/s41477-018-0255-1. [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farre E.M., Kay S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V.N., Kathare P.K., Huq E. Dynamic regulation of PIF5 by COP1-SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 2018;96:260–273. doi: 10.1111/tpj.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A., Gupta A., Betegon-Putze I., Bosch N., Ibanes M., Cano-Delgado A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development. 2019;146:dev151894. doi: 10.1242/dev.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Cho H., Bae W., Hwang I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014;5:4138. doi: 10.1038/ncomms5138. [DOI] [PubMed] [Google Scholar]
- Ryu H., Cho H., Kim K., Hwang I. Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells. 2010;29:283–290. doi: 10.1007/s10059-010-0035-x. [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Mas P. STRESSing the role of the plant circadian clock. Trends Plant Sci. 2015;20:230–237. doi: 10.1016/j.tplants.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Sever R., Glass C.K. Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 2013;5:a016709. doi: 10.1101/cshperspect.a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Mas P. A functional connection between the circadian clock and hormonal timing in Arabidopsis. Genes (Basel) 2018;9:567. doi: 10.3390/genes9120567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., Gonzalez-Schain N., Sentandreu M., Prat S., Quail P.H., Monte E. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 2012;71:390–401. doi: 10.1111/j.1365-313X.2012.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X.Y., Cao D.M., Tang W., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S., Oh E. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B., Chai J. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Yuan M., Wang R., Yang Y., Wang C., Oses-Prieto J.A., Kim T.W., Zhou H.W., Deng Z., Gampala S.S. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Parlan E.V., Fulton E.C. Circadian network interactions with jasmonate signaling and defense. Plants (Basel) 2019;8:252. doi: 10.3390/plants8080252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T.N., Mizutani Y., Kuwata K., Hirota T., Sato A., Mizoi J., Takao S., Matsuo H., Suzuki T., Ito S. Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U S A. 2019;116:11528–11536. doi: 10.1073/pnas.1903357116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., Wang X. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell. 2017;41:47–58 e4. doi: 10.1016/j.devcel.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Li L., Yin Y. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J. Integr. Plant Biol. 2011;53:455–468. doi: 10.1111/j.1744-7909.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yu M.-H., Zhao Z.-Z., He J.-X. Brassinosteroid signaling in plant–microbe interactions. Int. J. Mol. Sci. 2018;19:4091. doi: 10.3390/ijms19124091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Xu P., Wang W., Wang S., Caruana J.C., Yang H.Q., Lian H. Arabidopsis G-protein beta subunit AGB1 interacts with BES1 to regulate brassinosteroid signaling and cell elongation. Front. Plant Sci. 2017;8:2225. doi: 10.3389/fpls.2017.02225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhiponova M.K., Vanhoutte I., Boudolf V., Betti C., Dhondt S., Coppens F., Mylle E., Maes S., Gonzalez-Garcia M.P., Cano-Delgado A.I. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 2013;197:490–502. doi: 10.1111/nph.12036. [DOI] [PubMed] [Google Scholar]
- Zhou M., Wang W., Karapetyan S., Mwimba M., Marques J., Buchler N.E., Dong X. Redox rhythm reinforces the circadian clock to gate immune response. Nature. 2015;523:472–476. doi: 10.1038/nature14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2
Data Availability Statement
The data used and analyzed during this study are available from the corresponding author on reasonable request.