Abstract
Burkitt lymphoma (BL) is a malignant tumor in children. Although BL is generally curable, early relapse and refractoriness may occur. Some molecular indicators have been recently suggested for BL diagnosis, but large heterogeneity still exists. This study aimed at providing clinical molecular targets and methods that may help improve diagnosis and treatment of childhood BL. Only children patients were included in the study, and targeted gene sequencing was conducted to identify tumor specific mutations. The mRNA and protein level expression of potential target genes were measured by real-time PCR and immunohistochemistry. The relationship between BL specific gene mutation and differential expression with clinical features was analyzed. The results showed that i) detailed analysis of c-MYC/BCL2/BCL6 gene loci alteration and gene expression would help in accurate diagnosis and treatment determination of childhood BL; ii) loss-of-function mutations in SOCS1 or CIITA gene might be used as malignant markers for BL diagnosis and prognosis; iii) specific mutations of CD79A, MYD88, KLF2, DNMT3A and NFKBIE genes often concurrently existed in BL and showed association with benign clinical outcomes; iv) the high expression of MYC, TCF3 and loss-of-function ID3 genes in tumor may be potential therapeutic targets and could be used for treatment monitoring; and v) four MYC-translocation negative cases were re-defined as high-grade B-cell lymphoma-not otherwise specified (HGBL-NOS) but showed similar clinical outcomes and molecular features to other BL cases in the study, suggesting more studies needed to explore the molecular mechanisms and clinical significance of this provisional tumor entity.
Abbreviations: BL, Burkitt lymphoma; HGBL-NOS, high-grade B-cell lymphoma-not otherwise specified; NHL, Non-Hodgkin lymphoma; WHO, World Health Organization; BCR, B cell receptor; TGS, targeted gene sequencing; T, tumor; N, normal sample; IHC, immunohistochemistry; FFPE, formalin-fixed paraffin-embedded; NGS, next-generation sequencing; qRT-PCR, quantitative reverse transcription-PCR; LN, normal lymph node; HRM, high resolution melting; FISH, fluorescence in situ hybridization; LDH, lactate dehydrogenase; UA, uric acid; bHLH, basic helix-loop-helix; TLR, Toll-like receptor
Keywords: Childhood Burkitt lymphoma, Clinical molecular target, Targeted gene sequencing, Gene mutation, Gene translocation, Differential gene expression
Highlights
-
•
Detailed analysis of c-MYC/BCL2/BCL6 gene alteration and expression may help in accurate diagnosis and treatment;
-
•
The MYC-translocation negative cases (HGBL-NOS) showed similar clinical outcomes and molecular features to other cases;
-
•
Loss-of-function mutations of SOCS1 or CIITA gene could be used as malignant markers for diagnosis and prognosis;
-
•
Concurrent mutations in CD79A, MYD88, KLF2, DNMT3A and NFKBIE genes associated with benign clinical outcomes;
-
•
High expression of MYC, TCF3 and loss-of-function ID3 gene in tumor may be potential therapeutic targets.
Introduction
Non-Hodgkin lymphoma (NHL) is the fourth most common malignant tumor in pediatric tumors. The rate of new cases of childhood NHL was 1.4 per 100,000 children per year in the United States [1] and about 620 children were diagnosed with NHL in 2014 [2]. In Shanghai (China), the incidence rate of childhood NHL is 14.1/8.4 (male/female) per 1,000,000 children per year in 2003–2005 [3]. In the United States, Burkitt lymphoma (BL) accounts for 40% in pediatric NHL [4], and the very similar incidence of BL was found in China [5]. So approximately 1000 children would be diagnosed with BL per year in China.
BL is an invasive disease originated from follicular germinal center cells and has the characteristics of short doubling times and strong invasiveness in tumor cells. High doses and short durations of rituximab, an anti-CD20 monoclonal antibody, which showed good efficacy in patients with various CD20-expressing lymphoid malignancies [6], combined with multi-drug chemotherapy have been applied to BL patients [7]. The survival rate of BL has been greatly improved, with the 5-year event-free survival rate (EFS) exceeded 80% recently [8]. However, high-intensity chemotherapy regimens have significant acute side effects. In addition, some patients with relapsed/refractory disease are highly resistant to chemotherapy and are difficult to cure. Therefore, the development of target drugs with low toxicity and minimal side effects, as well as the establishment of accurate diagnostic methods, are the key to improve the treatment and cure rate with BL.
By studies using gene-expression profiling, Dave et al. suggested that unlike most B-cell lymphomas, the formation of BL may not depend on the NF-κB signaling pathway, but have a unique pathogenic mechanism [9,10]. The translocation of MYC gene locus (8q24) is a characteristic marker of BL, with t(8;14) (q24;q32) in 70–80% of BL and t(2;8) (p12;q24) or t(8;22) (q24;q11) in 10–15% of BL [[11], [12], [13], [14]]. Recent studies have found that a subset of lymphomas resemble BL morphologically and show similar clinical course, which lack of MYC rearrangement but with 11q aberration [15,16]. Although the number of the cases reported is still limited, it has been considered as a new provisional entity designated Burkitt-like lymphoma with 11q aberration in recently revised World Health Organization (WHO) classification of lymphoid neoplasms [17]. In the updated WHO classification, the cases with BL-like morphology or intermediate between diffuse large B cell lymphoma (DLBCL) and BL, which lack a MYC and BCL2 and/or BCL6 rearrangement has been classified as high-grade B-cell lymphoma-not otherwise specified (HGBL-NOS), while all large B-cell lymphoma (LBCL) with MYC and BCL2 and/or BCL6 rearrangements has been included in HGBL, with MYC and BCL2 and/or BCL6 rearrangements [17]. However, the clinical practices for these BL related tumor entities have kept changing recently but still uncertain [18].
Recent research have reported more candidate genes and pathways been involved in BL tumorigenesis, such as the B cell receptor (BCR), PI3K-AKT signaling (TCF3, ID3, PTEN), apoptosis (TP53), cell cycle regulation (CCND3), epigenetic regulation (ARID1A, SMARCA4, KMT2D), and G protein-coupled receptor signaling (GNA13, RHOA, P2RY8) [10,[19], [20], [21], [22], [23]]. The synergy between the PI3K signaling pathway and the MYC gene has been suggested to play an important role in BL formation [10,24]. However, the clinical impact of those genes in BL is largely unknown. Thus focused analysis of the genetic variation and differential expression of these candidate genes may provide more molecular targets and methods to improve the diagnosis and treatment of BL. Moreover, due to the very low incidence of childhood BL, previous studies usually consisted of adults or mixed-age cases. So the genetic background of the reported cases was not uniform enough to identify the accurate molecular targets for childhood BL.
Therefore, this study conducted the targeted gene sequencing (TGS) to identify specific mutations of selected genes in only childhood BL cases and paired analyzed the mutations in tumor (T) versus normal tissue (N) DNA from the same patient. At the same time, the mRNA and protein level expression of genes related to MYC, PI3K and NF-κB signaling pathways were examined to search the differentially expressed genes in childhood BL. At last, correlation analysis was performed to identify the potential clinical impact of the above molecular events.
Materials and methods
Patients
Biopsies from 19 patients who diagnosed with BL and underwent lymphoma resection at Shanghai Children's Hospital from September 2015 to May 2018 were collected in compliance with local ethics regulation. This study was approved by the Shanghai Children's Hospital Ethics Committee, Shanghai, China. All patients provided their informed consent for this study. Pathological review was performed in each case, including tissue cell morphology analysis and immunohistochemistry (IHC) analysis of protein expression of BCL2, BCL6, CD3, CD10, CD20, EBV, Ki67, TdT, and so on (Table S1). Diagnostic identification was reviewed according to the most recent WHO classification requirements related to BL [17]. Detailed clinical features are shown in Table 1.
Table 1.
Summary of clinical features of the childhood Burkitt Lymphoma cases, immunohistochemistry analysis, c-MYC loci, and targeted gene sequencing performed in the study.
| Case number | Clinical features |
IHC detection of protein expression |
c-MYC trans | Diagnosis | TGS analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Age | Clinical stage | Prognosis | LDH | UA | Other symptoms | MYC | ID3 | TCF3 | CCND3 | TNFAIP3 | CCND2 | c-FLIP | CD44 | ||||
| BL-01 | M | 3y | II | Benign | N | N | +++ | + | − | ++ | − | + | ++ | − | Yes | BL | √ | |
| BL-02 | M | 11y | III | Benign | N | N | +++ | ++ | + | + | ++ | + | ++ | + | No | HGBL-NOS | √ | |
| BL-03 | M | 4y | III | Poor | N | N | R & R | ++ | +++ | − | ++ | + | + | +++ | − | Yes | BL | √ |
| BL-04 | F | 3y | III | Benign | >N | N | / | / | / | / | / | / | / | / | Yes | BL | √ | |
| BL-05 | F | 10y | III | Benign | N | N | +++ | + | − | + | + | + | ++ | +/− | No | HGBL-NOS | √ | |
| BL-06 | M | 4y | III | Benign | >N | >N | +++ | ++ | − | + | + | − | ++ | − | Yes | BL | / | |
| BL-07 | M | 5y | II | Benign | N | N | ++ | + | − | + | − | + | ++ | − | Yes* | BL | √ | |
| BL-08 | F | 7y | III | Benign | >N | N | +++ | ++ | − | ++ | + | + | +++ | − | No | HGBL-NOS | / | |
| BL-09 | M | 5y | III | Benign | >N | >N | TLS | +++ | +++ | − | + | + | + | +++ | − | Yes | BL | / |
| BL-10 | F | 1y | III | Benign | >N | N | +++ | ++ | + | + | + | + | ++ | − | Yes | BL | / | |
| BL-11 | M | 4y | III | Benign | >N | >N | +++ | ++ | − | + | ++ | − | +++ | − | Yes | BL | / | |
| BL-12 | M | 6y | III | Benign | >N | N | ++ | ++ | − | + | ++ | + | ++ | − | No | HGBL-NOS | / | |
| BL-13 | M | 6y | IV | Poor | N | N | R & M | +++ | +++ | + | + | − | + | +++ | − | Yes | BL | / |
| BL-14 | M | 9y | III | Benign | N | N | +++ | + | + | + | + | + | ++ | − | Yes | BL | √ | |
| BL-15 | M | 10y | III | Poor | >N | >N | R & M | +++ | + | − | + | − | + | +++ | − | Yes | BL | √ |
| BL-16 | M | 10y | III | Benign | N | >N | ++ | ++ | − | + | + | + | ++ | − | Yes | BL | √ | |
| BL-17 | M | 6y | II | Benign | N | N | +++ | + | − | + | + | + | ++ | − | Yes | BL | √ | |
| BL-18 | M | 2y | IV | Poor | >N | >N | Death | +++ | +++ | − | +++ | ++ | + | +++ | − | Yes | BL | √ |
| BL-19 | M | 2y | III | Benign | N | >N | +++ | + | − | + | − | + | ++ | − | Yes | BL | √ | |
Abbreviations: LDH, lactate dehydrogenase; UA, uric acid; IHC, immunohistochemistry; trans, translocation; TGS, targeted gene sequencing; M, male; F, female; y, years; N, normal value; >N, higher than normal value; R & R, relapse and refractoriness; TLS, tumor lysis syndrome; R & M, relapse and metastasis; −, no expression; +, weak positive expression; ++, moderate positive expression; +++, strong positive expression; /, data not available; *, untypical break-apart; BL, Burkitt lymphoma; HGBL-NOS, high-grade B-cell lymphoma-not otherwise specified; √, analysis performed.
Sample collection
Pathologically identified formalin-fixed paraffin-embedded (FFPE) lymphoma tissues were obtained. A total of 12 cases were obtained for next-generation sequencing (NGS) analysis, for which 6 with bone marrow smear and 6 with normal tissue were used as normal sample (N) for paired comparison against the tumor (T) from the same patient. Eighteen cases were analyzed by real-time quantitative reverse transcription-PCR (qRT-PCR) and IHC for selected gene expression, in which 20 normal childhood lymph node (LN) paraffin embedded tissues were used as normal control samples. See Table 1 for additional details.
Nucleic acid extraction
DNA and RNA were extracted by using the GeneRead™ DNA FFPE Kit (Cat. #180134, QIAGEN, Germany) and RecoverAll™ Total Nucleic Acid Isolation Kit (Cat. #AM1975, Thermo Fisher Scientific, USA) respectively. The quality and purity were measured by using Qubit 3.0 (Thermo Fisher Scientific, USA), and the samples were frozen at −80 °C until use.
Targeted sequencing analysis
A customer-designed DNA panel of 46 lymphoma-related genes was manufactured by QIAGEN (Germany) using QIAseq™ technology (Table S2). The main steps included library construction, template preparation and machine sequencing. Library construction was performed according to the operating instructions for the QIAseq™ Targeted DNA Panel. Template preparation and machine sequencing were conducted using the Ion Chef™ and Ion S5™ system (Thermo Fisher Scientific, USA) according to the manufacturer's instructions.
The sequences were aligned to the GRCh37 (hg19) reference genome in QIAGEN official website (https://www.qiagen.com). Then the VCF files were uploaded to IVA software (https://apps.ingenuity.com) and compared according to the software instructions. The paired analysis of NGS data was performed between N and T of each case and by choosing filter parameters, such as “read depth ≥50 (at least)” and “allele fraction ≥1% (at least)”. Genetic abnormalities evaluated in this study included missense mutation, frameshift mutation, nonsense mutation, insertion mutation, deletion mutation, splice site mutation and untranslated region mutation. The mutations identified were compared with known mutations in the COSMIC database, Ensembl database, and SNP database.
High resolution melting (HRM) analysis of gene mutations
Primers were designed to amplify mutated regions identified by NGS (Table S3). Real-time PCR was carried out in a 10 μL reaction mixture with 10 ng template DNA and SSO Evagreen Supermix with low ROX (Cat. #172-5201AP, BioRad, USA), according to the manufacturer's instructions in Rotor-Gene Q (QIAGEN, Germany). Three independent replicates were done to each experiment and at each time, the samples were tested in duplicates.
Quantitative reverse transcription-PCR (qRT-PCR) analysis of mRNA level expression
SuperScript® III First-Strand Synthesis SuperMix (Thermo Fisher Scientific, USA) was used for cDNA synthesis with DNA-free Kit DNase Treatment and Removal Reagents (Thermo Fisher Scientific, USA) for removing residual DNA. Quantitative PCR was carried out according to the manufacturer's instructions. Gene-specific primer sequences (Table S4) were synthesized by Sangon Biotech Company (Shanghai, China). PCR cycle conditions were: 95 °C for 30 s, followed by 45 cycles of 95 °C (5 s) and 60 °C (30 s). Melting curve was checked to confirm specific amplification. Three independent replicates were used to collect the overall data, and at each time, the sample reactions were performed in duplicates. The average value of the cycle threshold (Ct) was used. A no template cDNA control sample was used to monitor cross contamination and 18S rRNA was used as reference gene. The ratio of the target gene to the reference gene (ΔCt) was used to calculate the relative expression level of each target gene.
Immunohistochemistry (IHC) analysis of protein level expression
The IHC assays were carried out on FFPE tissue sections with streptavidin-biotin peroxidase method by using rabbit polyclonal antibodies according to the previous study [25]. The primary antibodies of target proteins were listed in Table S5. The expression of these proteins was scored using a semiquantitative system based on the intensity and percentage of staining, and marked as negative (−), weak positive (+), moderate positive (++) or strong positive (+++) [26]. Negative (−) and weak positive (+) were regarded as low expression; moderate positive (++) and strong positive (+++) were considered as high expression. Immunostaining results were independently evaluated by two pathologists who were blinded to the clinicopathological features. Appropriate positive and negative controls were included in each protein assay.
Fluorescence in situ hybridization (FISH) analysis of gene locus
c-MYC translocation was detected by using c-MYC (8q24) Gene Fragmentation Probe (Cat. #F.01054-01, AP Technologies, China), MYC Gene Fragmentation and Recombination Detection Kit (Cat. #JLB301023, Jinlu Bio, China) and Vysis IGH/MYC/CEP 8 Tri-Color DF FISH Probe Kit (Cat. #04N10-020, Abbott, USA) according to the manufacturer's protocol. The aberration of chromosome 11q was detected by using 11q23/CEN11 Gene Copy Number Test Kit (Cat. #JLB301066, Jinlu Bio, China) and 11q24.1/CEN11 Gene Copy Number Test Kit (Cat. #JLB301067, Jinlu Bio, China). The copy number variation and break-apart of BCL2 and BCL6 was detected by using Vysis BCL2 Break Apart FISH Probe Kit (Cat. #05N51-020, Abbott, USA) and Vysis BCL6 Break Apart FISH Probe Kit (Cat. #01N23-020, Abbott, USA) respectively. The results were observed using a Leica DM6000B fluorescence microscope under a 100× lens, and images were acquired by Cyto Vision software (Leica Biosystems, German).
Statistical methods
Data analysis was performed using SAS 9.4 software and SPSS 13.0 software. GraphPad Prism 7.0 software was used for mapping analysis. The expression of genes of interest in T and LN was compared using the Student's t-test. Correlation analysis of clinical features was performed using the Fisher's Exact test. Statistical significance was defined by P values of <0.05.
Results
Clinical features of the BL cases
This study included 19 cases of childhood BL. The median age of the cohort was 5.7 years, ranging from 1 to 11 years old, and the male to female ratio was 1:0.27. Three patients were diagnosed as clinical stage II and the other sixteen cases were clinical stage III/IV. Among them, 4 patients had poor prognosis, 9 cases had high lactate dehydrogenase (LDH) values, and 7 cases had high uric acid (UA) values (Table 1).
Genetic abnormalities in the BL cases
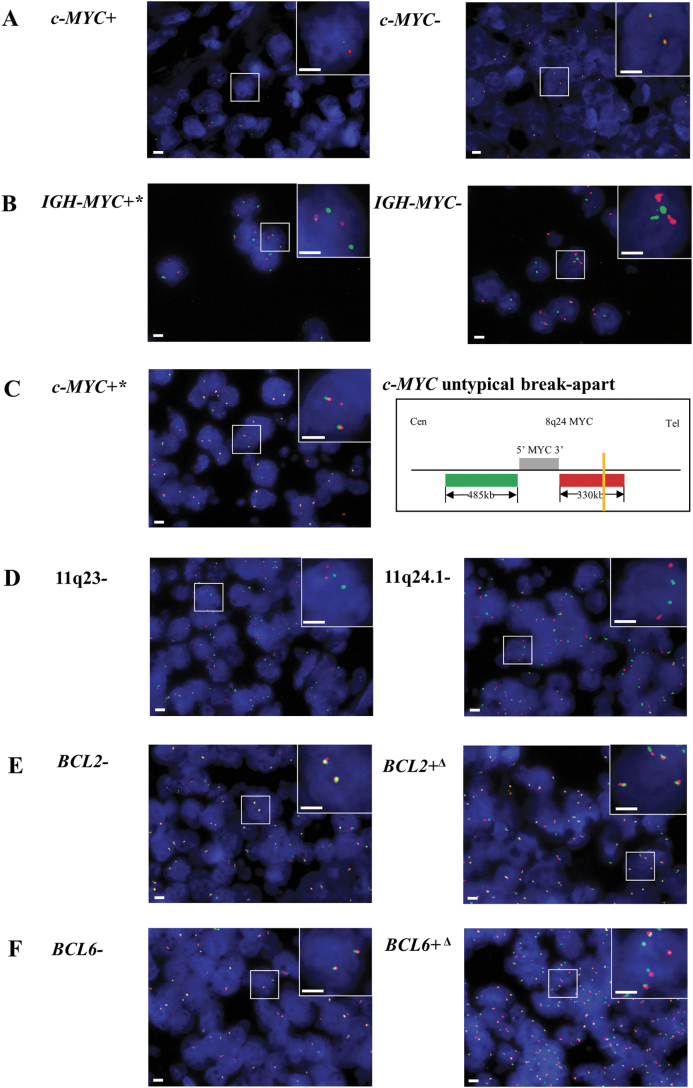
Detection of c-MYC gene locus (8q24) abnormality was performed by FISH test in all BL cases in the study. The c-MYC translocation was identified in 14 out of 19 cases by FISH using c-MYC break-apart probes (Table 1 and Fig. 1A). For the five c-MYC break-apart negative cases, FISH detection of IGH-MYC fusion, 11q aberration and the copy number variation and break-apart of BCL2 or BCL6 loci were further applied (Table S6 and Fig. 1B–F). One more case (BL-07) was identified as c-MYC translocation positive after using IGH-MYC fusion probe and additional MYC break-apart probe for FISH detection of MYC loci. In IGH-MYC FISH, 2 red signals and 3 green signals were observed in some cells of BL-07, with 1 pair of fused red-green signals plus one red and 2 green signals scattered (Table S6 and Fig. 1B). In additional MYC break-apart FISH analysis of this case, it showed 3 red signals and 2 green signals in some cells with 2 pairs of fused red-green signals plus a single red signal drifted aside (Table S6 and Fig. 1C). The results revealed that BL-07 harbored an “untypical” break-apart in the MYC locus (8q24) with a breakpoint at the 3′ downstream of MYC gene (Fig. 1C). Since none of 11q aberration, BCL2 break-apart or BCL6 break-apart was detected in the remaining four c-MYC translocation negative cases, the four cases were re-defined as high-grade B-cell lymphoma-not otherwise specified (HGBL-NOS) according to the 2016 revision of the WHO classification of lymphoid neoplasms [17]. However, there's no specific guideline for clinical practice available to HGBL-NOS till 2019. So the four HGBL-NOS cases were treated as same as BL and they showed no difference in treatment response or in any other clinical features when compared with the c-MYC translocation positive BL cases (Table 1).
Fig. 1.
Fluorescence in situ hybridization (FISH) analysis of c-MYC, BCL2 and BCL6 gene loci and chromosome 11q aberration in childhood Burkitt lymphoma cases using (A) c-MYC break-apart probes, (B) IGH-MYC probes, (C) additional c-MYC break-apart probes for untypical break-apart of c-MYC loci, (D) 11q aberration probes, (E) BCL2 break-apart probes and (F) BCL6 break-apart probes. Scale bar, 2 μm. c-MYC+, c-MYC break-apart positive; c-MYC-, c-MYC break-apart negative; IGH-MYC+*, IGH-MYC untypical fusion; IGH-MYC-, IGH-MYC fusion negative; c-MYC+*, c-MYC untypical break-apart; 11q23-, 11q23 copy number normal; 11q24.1-, 11q24.1 copy number normal; BCL2-, BCL2 break-apart negative; BCL2+Δ, BCL2 copy number anomaly; BCL6-, BCL6 break-apart negative; BCL6+Δ, BCL6 copy number anomaly.
The IHC results of BCL2 showed that 5/18 cases were positive (+), 3/18 cases were negative (−), and 10/18 cases hardly to give a definite conclusion (+/−). Among most cases, the expression of BCL2 was week or patchy if positive (+ or +/−) (Table S1). FISH examination of BCL2 or BCL6 loci aberration was also performed onto the five cases with BCL2 expression (+) (Table S6). None of BCL2 or BCL6 translocation was detected in any case. One case (BL-05) was identified with BCL2 and BCL6 gene loci copy number variation (3–4 copies), but this case showed no distinct clinical outcomes nor association with any other molecular features (Tables 1, S1 and S6).
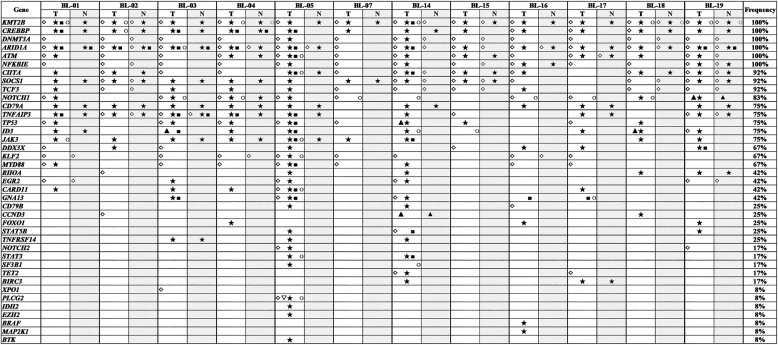
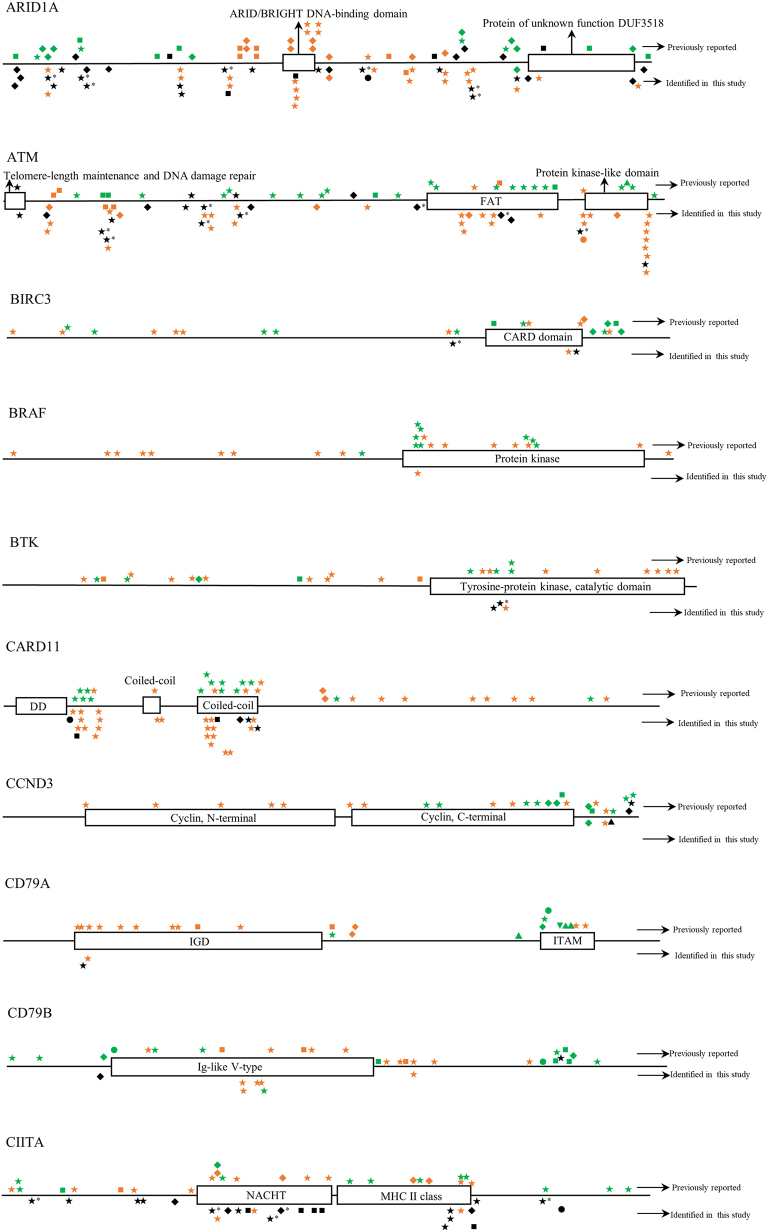
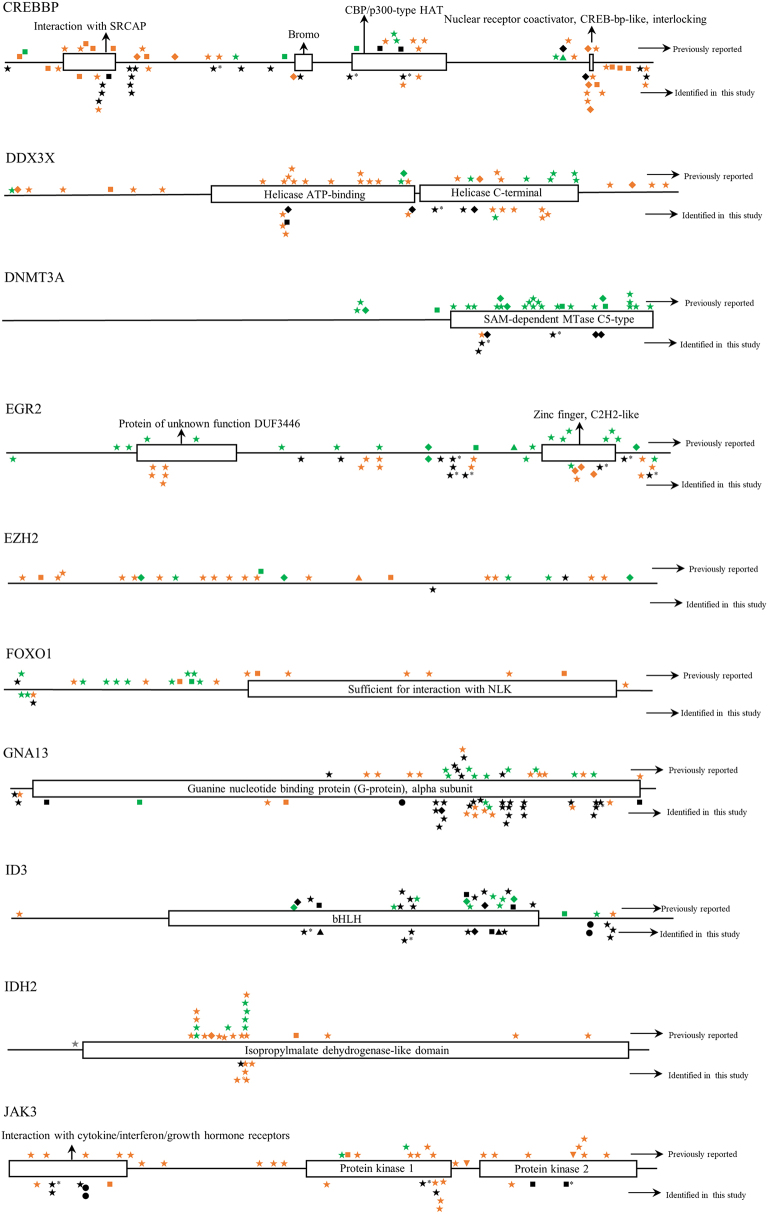
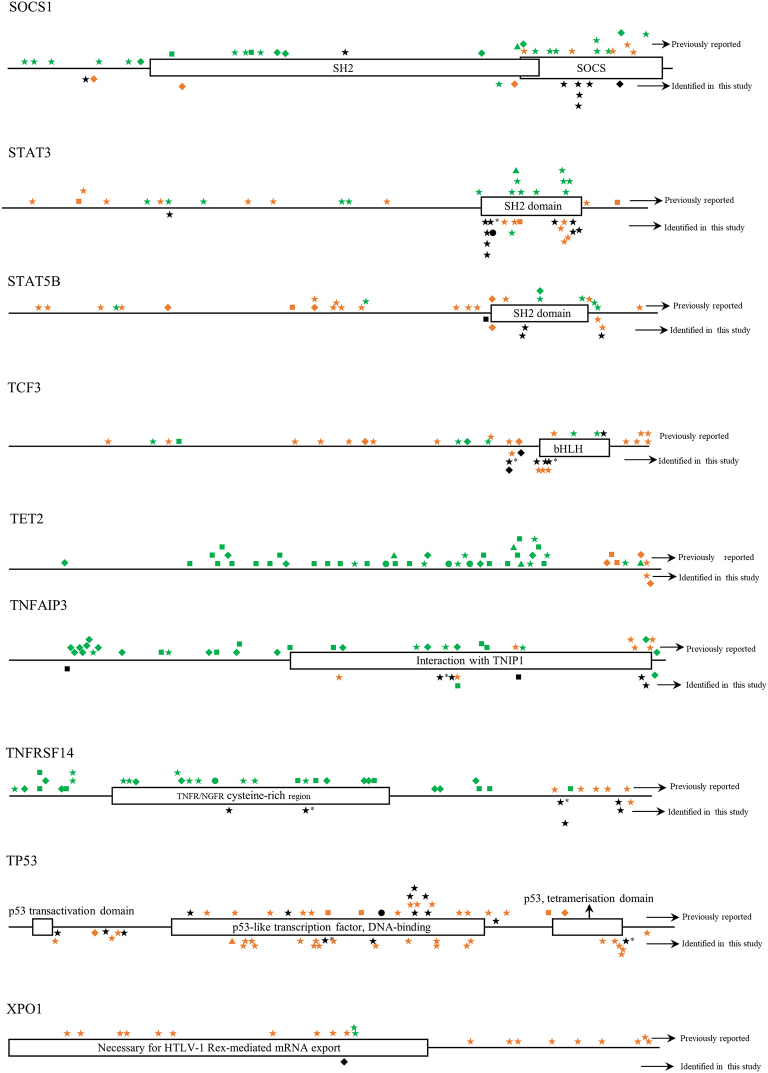
Twelve BL cases with paired tumor (T) and normal (N) DNA sample available were analyzed using a self-designed QIAseq™ DNA panel of 46 genes (Tables 1 and S2). The sequence depth across all samples spanned from 427 to 2385, and the mean depth was 1034. In total, 624 mutations in 39 different genes were detected. Case BL-05 had a maximum of 32 gene mutations, while BL-15 had a minimum of 13 gene mutations. The mean number of mutated genes detected per case was 20. The most frequently mutated genes were KMT2B (100%), CREBBP (100%), DNMT3A (100%), ARID1A (100%), ATM (100%), NFKBIE (100%), CIITA (92%), SOCS1 (92%) and TCF3 (92%) (Fig. 2). All mutations identified in the study were checked with COSMIC database, Ensembl database, and SNP database. Many novel mutations were identified for the first time in the study. But for BRAF, MAP2K1 and TET2 genes, all mutations detected at this time were known mutations which had been reported in one or more of the above databases [[27], [28], [29]]. The detailed distribution of mutations within each gene was shown in Fig. S1.
Fig. 2.
Summary of gene mutations detected by targeted gene sequencing in the study. T, tumor; N, normal sample; ◇, frameshift mutation; ▽, insertion mutation; ▲, deletion mutation; ★, missense mutation; ■, nonsense mutation; ○, splice site or untranslated region mutation.
Fig. S1.
The distribution of mutations identified in each gene in childhood Burkitt lymphoma cases of this study. Dark shapes represent the mutations detected in Burkitt lymphoma; green shapes represent the mutations detected in other lymphomas; orange shapes represent the mutations detected in diseases other than lymphoma. *, mutations belong to single nucleotide polymorphism; ◆, frameshift mutation; ▼, insertion mutation; ▲, deletion mutation; ★, missense mutation; ■, nonsense mutation; ●, splice site or untranslated region mutation; FAT, focal adhesion targeting; CARD, caspase recruitment domains; DD, death domain; IGD, Ig-like domain; ITAM, immuno-receptor tyrosine-based activation motif; NACHT, NAIP, CIITA, HET-E, and TP1; bHLH, basic helix-loop-helix; ID, linker domain; TIR, Toll-IL-1 receptor; LNR, Lin12/Notch related; SH2, Src homology domain; SOCS, suppressor of cytokine signaling.
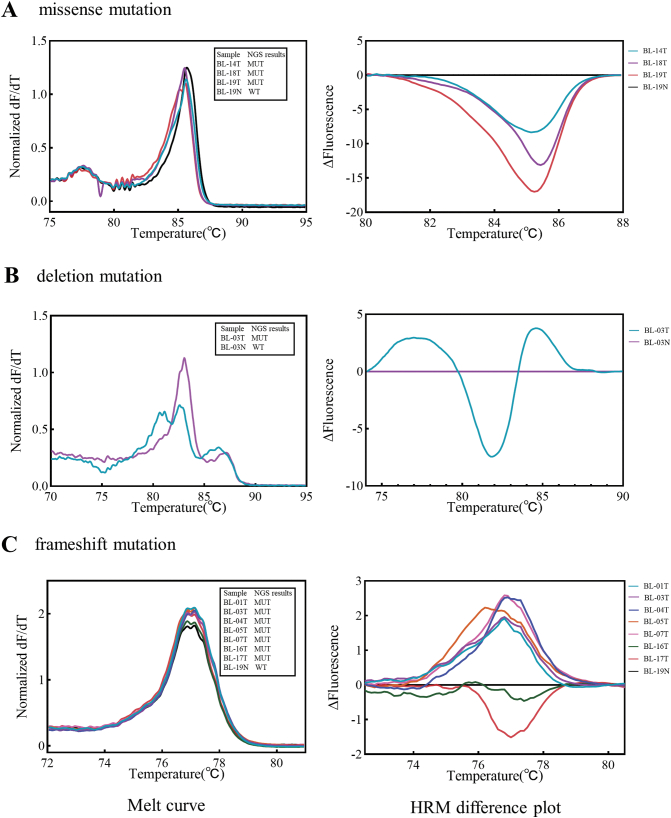
To verify the targeted sequencing results, real-time PCR with high resolution melting (HRM) was performed to check the mutations picked up by the sequencing analysis. An example for missense mutation, the ID3 (L64F) mutation in tumors of BL-14, BL-18 and BL-19 cases which detected by sequencing, were all identified by HRM (Fig. 3A). For deletion mutation, ID3 (R72_I77del) mutation that was identified in tumor of BL-03 by TGS analysis was also confirmed by using HRM (Fig. 3B). For an example of frameshift mutation, DNMT3A (K829fs*25) was detected in seven cases by TGS. Six of the mutations were confirmed by HRM analysis while only one showed similar melting curve as the normal sample (Fig. 3C). So, most of the mutations identified by TGS were confirmed by HRM. The mutation results identified by the two technical platforms were consistent.
Fig. 3.
Confirmation of mutations identified by targeted gene sequencing using high resolution melting assays. A, The example of missense mutation (ID3-L64F); B, The example of deletion mutation (ID3-R72_I77del); C, The example of frameshift mutation (DNMT3A-K829fs*25). T, tumor; N, normal sample; MUT, mutation; WT, wild-type; HRM, high resolution melting. All experiments were performed ≥ 3 times independently.
Differential gene expression in BL cases
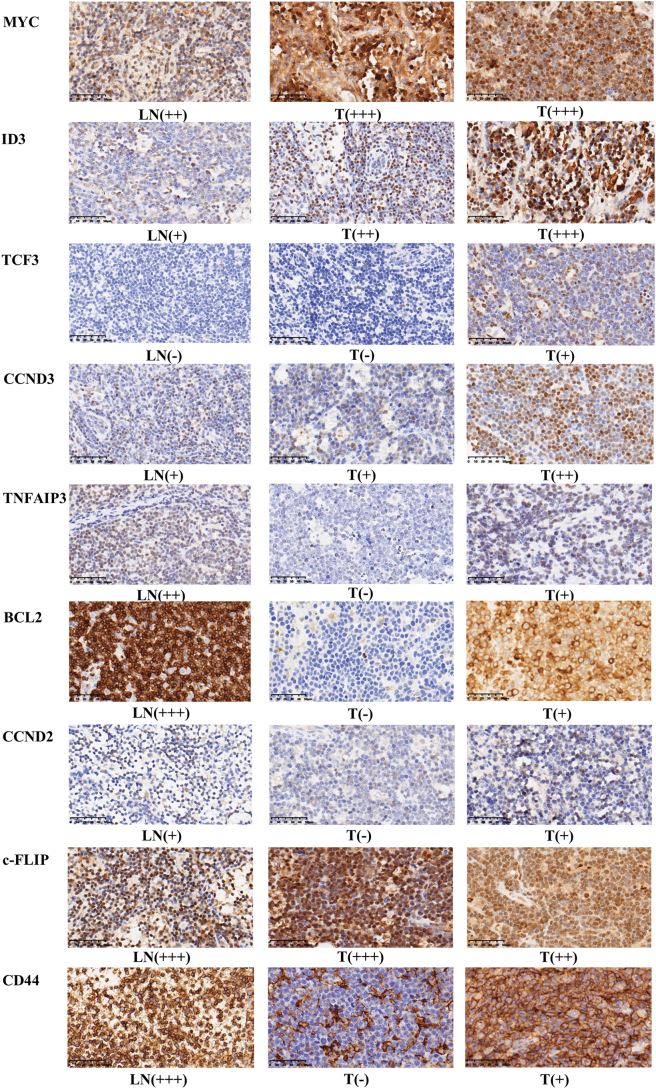
The expression of MYC, PI3K and NF-κB signaling pathway-related genes, including MYC, ID3, TCF3, CCND3, TNFAIP3, BCL2, CCND2, c-FLIP and CD44, were analyzed at both mRNA and protein levels in 18 cases (Tables 1, S1, Fig. 4, Fig. 5). Results demonstrated that MYC had higher expression in tumor than in normal lymph node. PI3K synergy-related genes, such as ID3 and TCF3, also showed higher expression in tumors. In contrast, genes related to the NF-κB pathway, such as TNFAIP3, BCL2, CCND2, c-FLIP and CD44, showed no difference or lower expression in tumor when compared to normal lymph node.
Fig. 4.
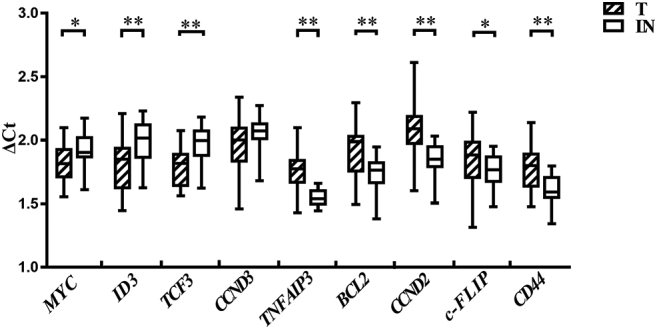
The mRNA expression of potential target genes in childhood Burkitt lymphoma. T, tumor; LN, normal lymph node; △Ct, the ratio of Ct value between target gene and reference gene; *, P ≤ 0.05; **, P ≤ 0.01. All experiments were performed ≥ 3 times independently.
Fig. 5.
The protein expression of potential target genes in childhood Burkitt lymphoma. Scale bar, 50 μm. LN, normal lymph node; T, tumor; −, no expression; +, weak positive expression; ++, moderate positive expression; +++, strong positive expression.
Correlation analysis of molecular features with clinical features
The molecular features including c-MYC translocation, differential gene expression and gene mutations found in the study were statistically analyzed with clinical features, such as age, clinical stage, LDH level, UA level and prognosis to identify the correlations between molecular marker and clinical outcomes.
Four BL cases carrying SOCS1 somatic mutation with frameshifts (A37fs*48, A164fs*41 and L196fs*76) were associated with higher UA level (P = 0.002) (Table 2). When mutations involved frameshifts or existed in functional domains, such as L84*, R160H and R188H, the cases were significantly associated with late clinical stages (P = 0.045) (Table 2). Nine cases carrying CIITA frameshift or nonsense mutations, such as V554fs*12, L833fs*57, T319fs*18, Q676* and K632*, were also correlated with late clinical stages (P = 0.045) (Table 2). Cases with ID3 gene mutation (C47_Y48del and R72_I77del) and high expression were associated with poorer prognosis than other cases (P = 0.055) (Table 3).
Table 2.
Malignant gene mutations identified in the study by correlation analysis.
| Gene | Type | Clinical features |
|||||
|---|---|---|---|---|---|---|---|
| UA |
P | Clinical stage |
P | ||||
| >N | N | III or IV | II | ||||
| SOCS1 | MUT | 4 | 0 | 0.002 | 7 | 0 | 0.045 |
| WT | 0 | 8 | 2 | 3 | |||
| CIITA | MUT | 4 | 6 | 0.515 | 9 | 1 | 0.045 |
| WT | 0 | 2 | 0 | 2 | |||
Abbreviations: UA, uric acid; N, normal value; >N, higher than normal value; MUT, mutation; WT, wild-type.
Table 3.
ID3 loss-of-function mutation and high expression in tumor associated with poor prognosis.
| Gene | Type | Prognosis |
P | |
|---|---|---|---|---|
| Benign | Poor | |||
| ID3 | MUT & H | 0 | 2 | 0.055 |
| Others | 8 | 1 | ||
Abbreviations: MUT, mutation; H, high expression.
In this study, 9 out of 12 BL cases had CD79A mutations. One of the mutations (A32G) was previously reported in ulcerative colitis-associated colorectal neoplasia [30], another was C30S, which was firstly identified in the study. Cases with such CD79A gene mutations showed normal LDH and/or UA levels (P = 0.045) (Table 4). In 8 of 12 BL cases, frameshift or missense mutations of MYD88 gene were detected, such as A13fs*38, R45fs*6, R153Gln and S222N. Cases with the above mutations in either CD79A or MYD88 were associated with normal LDH level (P = 0.045), normal LDH and/or UA levels (P = 0.015) and benign prognosis (P = 0.045) when compared to the cases with intact CD79A and MYD88 genes (Table 4). The above 8 cases with MYD88 mutations also harbored KLF2 mutations, which mainly were frameshifts (A182fs*108, G151fs*139 and G208fs*82). Thus, as similar as the cases with CD79A or MYD88 mutations, cases with CD79A or KLF2 mutations were also associated with normal LDH level (P = 0.045), normal LDH and/or UA levels (P = 0.015) and benign prognosis (P = 0.045) (Table 4). The tumor specific mutation (somatic mutation) of KLF2, MYD88, DNMT3A and NFKBIE genes were concurrently existed in some BL cases, which usually showed normal UA level (P = 0.033) (Table 5).
Table 4.
Benign gene mutations identified in this study by correlation analysis.
| Gene | Type | Clinical features |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LDH |
P | UA |
P | LDH & UA |
P | Prognosis |
P | ||||||
| >N | N | >N | N | >N | others | Benign | Poor | ||||||
| CD79A | MUT | 8 | 1 | 0.127 | 8 | 1 | 0.127 | 0 | 9 | 0.045 | 8 | 1 | 0.127 |
| WT | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 2 | |||||
| MYD88 | MUT | 7 | 1 | 0.236 | 7 | 1 | 0.067 | 0 | 8 | 0.091 | 7 | 1 | 0.236 |
| WT | 2 | 2 | 1 | 3 | 2 | 2 | 2 | 2 | |||||
| KLF2 | MUT | 7 | 1 | 0.236 | 7 | 1 | 0.067 | 0 | 8 | 0.091 | 7 | 1 | 0.236 |
| WT | 2 | 2 | 1 | 3 | 2 | 2 | 2 | 2 | |||||
| CD79A and/or MYD88 | MUT | 1 | 9 | 0.045 | 2 | 8 | 0.067 | 0 | 10 | 0.015 | 9 | 1 | 0.045 |
| WT | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 2 | |||||
| CD79A and/or KLF2 | MUT | 1 | 9 | 0.045 | 2 | 8 | 0.067 | 0 | 10 | 0.015 | 9 | 1 | 0.045 |
| WT | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 2 | |||||
Abbreviations: LDH, lactate dehydrogenase; UA, uric acid; N, normal value; >N, higher than normal value; MUT, mutation; WT, wild-type.
Table 5.
Genes with somatic mutations in c-MYC translocation positive Burkitt lymphoma cases associated with normal UA level.
| Gene | Type | UA |
P | |
|---|---|---|---|---|
| >N | N | |||
| KLF2 | MUT | 1 | 6 | 0.033 |
| WT | 3 | 0 | ||
| MYD88 | MUT | 1 | 6 | 0.033 |
| WT | 3 | 0 | ||
| DNMT3A | MUT | 1 | 6 | 0.033 |
| WT | 3 | 0 | ||
| NFKBIE | MUT | 1 | 6 | 0.033 |
| WT | 3 | 0 | ||
Abbreviations: UA, uric acid; N, normal value; >N, higher than normal value; MUT, mutation; WT, wild-type.
A detailed comparison of the fifteen MYC-translocation positive (BL) cases to the four MYC-translocation negative (HGBL-NOS) cases revealed that the two groups of patients showed very similar clinical outcomes and little difference in molecular features. Only the frameshift or functional domain mutation of RHOA gene (P = 0.045) and protein expression of CD44 (P = 0.039) seems associated with HGBL-NOS cases (Table 6).
Table 6.
RHOA functional mutation and CD44 expression associated with MYC translocation positive cases.
| Gene | Type |
MYC-translocation |
P | |
|---|---|---|---|---|
| Positive (BL) | Negative (HGBL-NOS) | |||
| RHOA | MUT | 1 | 2 | 0.045 |
| WT | 9 | 0 | ||
| CD44 | + & +/− | 0 | 2 | 0.039 |
| – | 14 | 2 | ||
Abbreviations: MUT, mutation; WT, wild-type; +, week positive expression; −, no expression; BL, Burkitt lymphoma; HGBL-NOS, high-grade B-cell lymphoma-not otherwise specified.
Discussion
The 2016 version of the WHO classification of lymphoid neoplasms added three new tumor entities related to BL, the Burkitt-like lymphoma with 11q aberration, HGBL-NOS and HGBL with MYC and BCL2 and/or BCL6 rearrangements. They are defined largely according to the status of MYC and BCL2 or BCL6 translocations although there's no specific guidelines about which LBCL should have the related FISH analyses [17]. Since the series of childhood BL cases in the study were registered from 2015 till 2018, the updated criteria suggested in the 2016 version of WHO classification were not fully applied. Plus a controversial issue not fully resolved in BL diagnosis is whether true BL without MYC translocations really exists. Therefore, this research carefully reviewed all the cases about their diagnosis according to the updated WHO classification and NCCN Guidelines related to Burkitt lymphomas [18]. The results showed that 15 cases were normal BL while 4 cases were re-defined as HGBL-NOS because they had BL-like histopathology but lack a MYC, BCL2 and BCL6 rearrangement. Since there had been no guideline for clinical practices applied to HGBL-NOS till 2019, the four HGBL-NOS cases were treated as same as BL cases. However, these cases showed the similar clinical course and molecular features to the MYC-translocation positive BL cases. Although BCL2 gene translocation was not found in any cases in the study, the IHC results showed that 5/18 cases (28%) with BCL2 expression, which consistent with a previous study reported that the BCL2 expression found in about 33% of BLs [31]. Among most cases in the series, the expression of BCL2 was week or patchy if positive, which consistent with the NCCN Guidelines for Pediatric Aggressive Mature B-Cell Lymphomas. Since the guidelines for BL-related clinical practice have been constantly changing recently, our results suggested that detailed detection of c-MYC/BCL2/BCL6 gene loci alterations together with the protein expression would help in accurate diagnosis and treatment determination for childhood BLs.
In this study, a self-designed lymphoma NGS panel was used to analyze a series of childhood BL cases. This panel included recurrently mutated genes reported in lymphomas with impact on treatment decision, diagnosis or prognosis, or with potential clinical impact [32]. Recently, several genes were found to be mutated recurrently in BL, including CREBBP (12%), ARID1A (14%), TCF3 (37%), NOTCH1 (8%), TP53 (26%) and ID3 (34%) [20,33]. All of these genes were included in the panel, and even higher mutation frequencies were observed in this study. For example, mutations of CREBBP and ARID1A were observed in all cases (100%), and there were high mutation rates for TCF3 (92%), NOTCH1 (83%), TP53 (75%) and ID3 (75%). While previous studies focused on adolescents and adults, this study has more important implications for the pathological development of BL in childhood cases. Consistent with a previous study [20], many silencing events, such as nonsense mutations and frameshift mutations, constituted a substantial proportion of CREBBP and ARID1A mutations in the study, suggesting that these genetic alterations may result in loss of function of these genes during BL development. The above genes with high frequency of mutations may be potential drug targets for precision treatment of BL. For examples, cyclophosphamide, fludarabine and mycophenolate mofetil could be used for BL cases with TCF3 mutations or TP53 mutations [34], while histone deacetylase inhibitors (HDACi) [35,36] might be helpful for cases with CREBBP or ARID1A mutations.
Besides genetic abnormalities, the expression of genes relevant to BL pathogenesis was detected in this study at the mRNA and protein levels. MYC was shown to be highly expressed in BL, which is consistent with the results of a previous study using microarrays [37]. Elevated MYC level may be a direct consequence of genomic aberrations involving the MYC locus [38]. Thus MYC function as an oncogene to trigger cell proliferation and tumorigenesis [39]. In recent years, several MYC-targeted gene inhibitors have been evaluated in clinical trials, such as Alisertib and Romidepsin, which regulate MYC expression, and rituximab, which targets MYC translocation [40].
In this study, ID3 gene was found to be more highly expressed in BL than in normal lymph node tissue, which is consistent with the results of a previous study using microarrays [9]. This may due to increased MYC signaling in BL for ID3 gene is a direct transcriptional target of MYC [10,19,20]. In B cells, ID3 can inhibit cell cycle progression and cell proliferation through its basic helix-loop-helix (bHLH) domain, which interacts with other bHLH proteins, such as TCF3, and inhibits their function [[41], [42], [43], [44]]. This study identified that two cases with ID3 high expression and small deletions in the bHLH domain had poorer prognosis than other cases. Deletion mutation in bHLH domain may cause ID3 loss of function. High expression of loss-of-function ID3 was associated with poor prognosis, suggesting that ID3 is an important target gene during BL tumorigenesis. The specific mutations of ID3 identified in the study by NGS, such as L64F and R72_I77del, were confirmed by using HRM, which proved the reliability of the mutations detected by targeted sequencing, and also provided an individual mutation detection method which might be used during treatment monitoring. TCF3 is highly expressed in B-cell development [45,46]. But TCF3 could be inhibited by ID3 through the interaction between bHLH domains. In this study, TCF3 were more highly expressed in tumor than in LN, indicating that TCF3 expression was not inhibited by ID3. This may due to ID3 proteins have loss-of-function mutations in its bHLH domain. Therefore, the results suggested that the loss-of-function mutation of ID3 and up-regulation of TCF3 expression could be used as molecular markers for prognosis and treatment monitoring of childhood BL.
Correlation analysis showed that the BL cases with either CD79A mutation or MYD88 mutation usually had normal LDH and/or UA levels and significantly better prognosis than the cases with intact CD79A and MYD88 genes. The same results were identified in CD79A and KLF2 genes. These findings suggested that the specific mutations identified in this study in CD79A, MYD88 and KLF2 genes were benign markers for childhood BL, which is different from the roles of these genes in other B cell lymphomas [[47], [48], [49]]. Moreover, the cases with MYD88 mutations also had KLF2 mutations, suggesting that the 2 gene mutations were always concurrently existed in the BL cases. Therefore, we checked the details of these gene mutations. For CD79A, mutations found so far in other types of lymphomas affect the immune-receptor tyrosine-based activation motif (ITAM), which is phosphorylated after antigen-induced BCR-aggregation and is crucial for the initiation of antigen-dependent downstream signaling of the NF-κB pathway [47,[50], [51], [52], [53]]. In contrast, mutations found in this study were centered in the Ig-like domain (IGD), which is different from mutation sites identified in previous studies. The mutations in IGD may affect CD79A binding to membrane immunoglobulin, which may consequently attenuate the BCR induced NF-κB activation and be benefit to tumor treatment. MYD88, which mediates the Toll-like receptor (TLR) signaling pathway, consists of a death domain (DD) at the N terminus, an intermediate linker domain (ID), and a Toll-IL-1 receptor (TIR) domain at the C terminus [54]. It was reported that mutations of MYD88, especially L265P in the TIR domain, can activate the NF-κB signaling pathway [48]. However, frameshift mutations of MYD88 in this study were located in the DD domain at the N terminus, which differed from the C-terminal mutations found previously in other B cell lymphomas, such as L265P. These mutations in BL cases would cause MYD88 protein truncation and therefore loss its function of mediating the TLR signaling induced NF-κB activation. KLF2 has recently been shown to be a negative regulator of inflammation and NF-κB activity [[55], [56], [57], [58]]. Functional assays in a previous study showed that frameshift, nonsense or missense mutations inactivate the ability of KLF2 to suppress NF-κB activation by TLR or BCR signalings [49]. The frameshift mutations and nonsense mutations found in this study could truncate the KLF2 protein and impair its inhibition of the NF-κB pathway. However, all the 8 cases harboring KLF2 loss-of-function mutations also carried loss-of-function MYD88. So, it was supposed that the activity of the NF-κB signaling pathway in these cases was not high. And this was proved by the expression analysis results of NF-κB target genes like TNFAIP3, BCL2, CCND2, c-FLIP and CD44, which were inhibited in BL. In addition, among c-MYC translocation cases, those with somatic mutations in KLF2, MYD88, DNMT3A or NFKBIE genes often show normal UA values, while the cases with intact such genes had higher UA level. In summary, the NF-κB signaling pathway related gene mutations found in the study did not lead to NF-κB activation, but specific mutations of genes such as CD79A, MYD88, KLF2, DNMT3A and NFKBIE showed association with some benign clinical outcomes. Thus, these genes may be worthy to check in future to see if they could be clinically used as benign markers for BL prognosis and the mechanisms involved.
More importantly, this study identified some genes that could be used as malignant markers for childhood BL diagnosis and prognosis, such as SOCS1 and CIITA. Four BL cases with frameshift or nonsense somatic mutation of SOCS1 gene had significant higher UA values than other cases. Seven cases with frameshift or functional domain mutations of SOCS1 gene were associated with late clinical stages. A previous study showed that classical Hodgkin lymphoma (cHL) patients harboring tumor cells with major mutations (insertion or deletion mutations and/or truncating mutations) in SOCS1 had a significantly shorter overall survival (OS) than those with minor mutations (without alteration of the length of the encoded protein) [59]. This may due to SOCS1 is a tumor suppressor [60]. Its SOCS box could mediate ubiquitination, which is significant for the process of malignancy [61]. In this study, frameshift or nonsense mutations could cause the truncation of the SOCS box and impair its function as a tumor suppressor, thus contributing to the poor clinical results of the cases with such SOCS1 mutations. Moreover, nine cases with loss-of-function (frameshift or nonsense) mutations of CIITA gene had correlation with late clinical stages. Previous studies suggested that loss-of-function mutations in CIITA would lead to lack of MHC class II protein expression in tumor cells and reduce cell immunogenicity [62]. Therefore, cases with loss-of-function mutations in SOCS1 and CIITA genes were expected to show poor clinical outcomes and could be used for prognosis and as drug targets.
Conclusions
In summary, this study identified some important molecular markers with potential clinical impact on childhood BL. Detailed analysis of c-MYC/BCL2/BCL6 gene loci alteration and gene expression would help in accurate diagnosis and treatment determination. Four MYC-translocation negative BL cases were re-defined as HGBL-NOS in the study. But they showed very similar clinical outcomes and molecular features to MYC-translocation positive BL cases. Loss-of-function mutations in SOCS1 and CIITA genes may be used as malignant markers for childhood BL diagnosis and prognosis, while specific mutations in genes such as CD79A, MYD88, KLF2, DNMT3A and NFKBIE may be as benign markers for childhood BL prognosis. High expression of MYC, TCF3 and loss-of-function ID3 genes in tumor provided important therapeutic targets and could be used for treatment monitoring. Moreover, drugs targeting TCF3, TP53, CREBBP, ARID1A, SOCS1, c-MYC and BCL2 genes may also provide new strategies for childhood BL treatment. However, since limited number of cases was available to this study due to the low incidence of childhood BL, the above results need to be further verified by studies with more samples in future.
The following are the supplementary data related to this article.
Table S1 The histological description and antibodies application for the diagnosis of childhood Burkitt lymphoma.
Table S2 List of the target genes on the self-designed sequencing panel.
Table S3 Primers for high resolution melting analysis.
Table S4 Primers for real-time quantitative reverse transcription-PCR analysis.
Table S5 Primary antibodies for immunohistochemistry analysis.
Table S6 Results of fluorescence in situ hybridization (FISH) analysis of c-MYC, BCL2 and BCL6 gene loci and chromosome 11q aberration of the childhood Burkitt lymphoma cases.
CRediT authorship contribution statement
Jing Zhang: Formal analysis, Investigation, Writing - Original Draft, Visualization; Leijun Meng: Resources, Formal analysis, Investigation; Weiyun Jiang: Formal analysis, Investigation, Methodology; Hong Zhang: Resources; Aiwu Zhou: Formal analysis; Naiyan Zeng: Conceptualization, Methodology, Supervision, Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This study was supported by the translational research funding from Yu Kang Biotechnology Co., Ltd in Jiaxing, Zhejiang, China, and a grant (No. 81372234) from the National Natural Science Foundation of China.
Ethics approval statement
This study was approved by the Shanghai Children's Hospital Ethics Committee, Shanghai, China (2016R043-E04).
Data availability statement
The sequence data that support the findings of this study have been deposited in NCBI Sequence Read Archive (SRA) with the accession codes “SUB7208508” and “SUB7195810”. All data generated or analyzed during this study are included either in this article or in the supplementary material.
References
- 1.National Cancer Institute. https://seer.cancer.gov/. Accessed 15 October 2019.
- 2.Ward E., DeSantis C., Robbins A., Kohler B., Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014;64(2):83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 3.Bao P.P., Zheng Y., Gu K., Wang C.F., Wu C.X., Jin F. Trends in childhood cancer incidence and mortality in urban Shanghai, 1973–2005. Pediatr. Blood Cancer. 2010;54(7):1009–1013. doi: 10.1002/pbc.22383. [DOI] [PubMed] [Google Scholar]
- 4.American Cancer Society https://www.cancer.org/ (Accessed 15 October 2019)
- 5.Shi Y., Sun Y., Liu T. Chinese norms for the diagnosis and treatment of malignant lymphoma (2015 edition) Chinese Journal of Oncology. 2015;37(02):148–158. [PubMed] [Google Scholar]
- 6.Salles G., Barrett M., Foa R., Maurer J., O'Brien S., Valente N. Rituximab in B-cell hematologic malignancies: a review of 20 years of clinical experience. Adv. Ther. 2017;34(10):2232–2273. doi: 10.1007/s12325-017-0612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M., Jin L., Yang J., Duan Y.L., Huang S., Zhou C.J. Clinical and prognostic analysis of 186 children with Burkitt's lymphoma. Zhonghua Er Ke Za Zhi. 2018;56(8):605–610. doi: 10.3760/cma.j.issn.0578-1310.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Worch J., Rohde M., Burkhardt B. Mature B-cell lymphoma and leukemia in children and adolescents-review of standard chemotherapy regimen and perspectives. Pediatr. Hematol. Oncol. 2013;30(6):465–483. doi: 10.3109/08880018.2013.783891. [DOI] [PubMed] [Google Scholar]
- 9.Dave S.S., Fu K., Wright G.W., Lam L.T., Kluin P., Boerma E.J. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 2006;354(23):2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz R., Young R.M., Ceribelli M., Jhavar S., Xiao W., Zhang M. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molyneux E.M., Rochford R., Griffin B., Newton R., Jackson G., Menon G. Burkitt's lymphoma. Lancet. 2012;379(9822):1234–1244. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand S., Berger R., Philip T., Bernheim A., Bryon P.A., Bertoglio J. Variant translocation in a nonendemic case of Burkitt's lymphoma: t (8;22) in an Epstein-Barr virus negative tumour and in a derived cell line. Eur. J. Cancer. 1981;17(5):577–584. doi: 10.1016/0014-2964(81)90060-8. [DOI] [PubMed] [Google Scholar]
- 13.Bernheim A., Berger R., Lenoir G. Cytogenetic studies on African Burkitt's lymphoma cell lines: t(8;14), t(2;8) and t(8;22) translocations. Cancer Genet. Cytogenet. 1981;3(4):307–315. doi: 10.1016/0165-4608(81)90039-x. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser-McCaw B., Epstein A.L., Kaplan H.S., Hecht F. Chromosome 14 translocation in African and North American Burkitt's lymphoma. Int. J. Cancer. 1977;19(4):482–486. doi: 10.1002/ijc.2910190408. [DOI] [PubMed] [Google Scholar]
- 15.Salaverria I., Martin-Guerrero I., Wagener R., Kreuz M., Kohler C.W., Richter J. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014;123(8):1187–1198. doi: 10.1182/blood-2013-06-507996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreiro J.F., Morscio J., Dierickx D., Marcelis L., Verhoef G., Vandenberghe P. Post-transplant molecularly defined Burkitt lymphomas are frequently MYC-negative and characterized by the 11q-gain/loss pattern. Haematologica. 2015;100(7):e275–e279. doi: 10.3324/haematol.2015.124305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swerdlow S.H., Campo E., Pileri S.A., Harris N.L., Stein H., Siebert R. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NCCN Clinical Practice Guidelines in Oncology-Pediatric Aggressive Mature B-Cell Lymphomas (2020 Version 2)[DB/OL]. http://www.nccn.org. [DOI] [PubMed]
- 19.Richter J., Schlesner M., Hoffmann S., Kreuz M., Leich E., Burkhardt B. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 2012;44(12):1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 20.Love C., Sun Z., Jima D., Li G., Zhang J., Miles R. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abate F., Ambrosio M.R., Mundo L., Laginestra M.A., Fuligni F., Rossi M. Distinct viral and mutational spectrum of endemic Burkitt lymphoma. PLoS Pathog. 2015;11(10) doi: 10.1371/journal.ppat.1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaymaz Y., Oduor C.I., Yu H., Otieno J.A., Ong'echa J.M., Moormann A.M. Comprehensive transcriptome and mutational profiling of endemic Burkitt lymphoma reveals EBV type-specific differences. Mol. Cancer Res. 2017;15(5):563–576. doi: 10.1158/1541-7786.MCR-16-0305-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouska A., Bi C., Lone W., Zhang W., Kedwaii A., Heavican T. Adult high-grade B-cell lymphoma with Burkitt lymphoma signature: genomic features and potential therapeutic targets. Blood. 2017;130(16):1819–1831. doi: 10.1182/blood-2017-02-767335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander S., Calado D.P., Srinivasan L., Kochert K., Zhang B., Rosolowski M. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell. 2012;22(2):167–179. doi: 10.1016/j.ccr.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hans C.P., Weisenburger D.D., Greiner T.C., Gascoyne R.D., Delabie J., Ott G. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 26.Fan W., Fan S.S., Feng J., Xiao D., Fan S., Luo J. Elevated expression of HSP10 protein inhibits apoptosis and associates with poor prognosis of astrocytoma. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0185563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.COSMIC. https://cancer.sanger.ac.uk/cosmic. Accessed 26 May 2019.
- 28.Ensembl. http://grch37.ensembl.org/index.html. Accessed 26 May 2019.
- 29.dbSNP. https://www.ncbi.nlm.nih.gov/snp/. Accessed 26 May 2019.
- 30.Chakrabarty S., Varghese V.K., Sahu P., Jayaram P., Shivakumar B.M., Pai C.G. Targeted sequencing-based analyses of candidate gene variants in ulcerative colitis-associated colorectal neoplasia. Br. J. Cancer. 2017;117(1):136–143. doi: 10.1038/bjc.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida M., Ichikawa A., Miyoshi H., Kiyasu J., Kimura Y., Niino D. Low incidence of MYC/BCL2 double-hit in Burkitt lymphoma. Pathol. Int. 2015;65(9):486–489. doi: 10.1111/pin.12333. [DOI] [PubMed] [Google Scholar]
- 32.Rosenquist R., Rosenwald A., Du M.Q., Gaidano G., Groenen P., Wotherspoon A. Clinical impact of recurrently mutated genes on lymphoma diagnostics: state-of-the-art and beyond. Haematologica. 2016;101(9):1002–1009. doi: 10.3324/haematol.2015.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forero-Castro M., Robledo C., Lumbreras E., Benito R., Hernandez-Sanchez J.M., Hernandez-Sanchez M. The presence of genomic imbalances is associated with poor outcome in patients with Burkitt lymphoma treated with dose-intensive chemotherapy including rituximab. Br. J. Haematol. 2016;172(3):428–438. doi: 10.1111/bjh.13849. [DOI] [PubMed] [Google Scholar]
- 34.My Cancer Genome. https://www.mycancergenome.org/. Accessed 10 June 2019.
- 35.Andersen C.L., Asmar F., Klausen T., Hasselbalch H., Gronbaek K. Somatic mutations of the CREBBP and EP300 genes affect response to histone deacetylase inhibition in malignant DLBCL clones. Leukemia Research Reports. 2012;2(1):1–3. doi: 10.1016/j.lrr.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukumoto T., Park P.H., Wu S., Fatkhutdinov N., Karakashev S., Nacarelli T. Repurposing pan-HDAC inhibitors for ARID1A-mutated ovarian cancer. Cell Rep. 2018;22(13):3393–3400. doi: 10.1016/j.celrep.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S., Day N.S., Miles R.R., Perkins S.L., Lim M.S., Ayello J. Comparative genomic expression signatures of signal transduction pathways and targets in paediatric Burkitt lymphoma: a Children's Oncology Group report. Br. J. Haematol. 2017;177(4):601–611. doi: 10.1111/bjh.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klapproth K., Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br. J. Haematol. 2010;149(4):484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 39.Dang C.V., O'Donnell K.A., Zeller K.I., Nguyen T., Osthus R.C., Li F. The c-Myc target gene network. Semin. Cancer Biol. 2006;16(4):253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 40.ClinicalTrials.gov. https://clinicaltrials.gov/ct2/home. Accessed 30 May 2019.
- 41.Greenough A., Dave S.S. New clues to the molecular pathogenesis of Burkitt lymphoma revealed through next-generation sequencing. Curr. Opin. Hematol. 2014;21(4):326–332. doi: 10.1097/MOH.0000000000000059. [DOI] [PubMed] [Google Scholar]
- 42.Benezra R, Davis RL, Lassar A, Tapscott S, Thayer M, Lockshon D, et al. Id: a negative regulator of helix-loop-helix DNA binding proteins. control of terminal myogenic differentiation. Annals of the New York Academy of Sciences. 1990;599:1–11. [DOI] [PubMed]
- 43.Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 44.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6(11):1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 45.Hsu L.Y., Lauring J., Liang H.E., Greenbaum S., Cado D., Zhuang Y. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19(1):105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- 46.Beck K., Peak M.M., Ota T., Nemazee D., Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 2009;206(10):2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis R.E., Ngo V.N., Lenz G., Tolar P., Young R.M., Romesser P.B. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi D. Role of MYD88 in lymphoplasmacytic lymphoma diagnosis and pathogenesis. Hematology American Society of Hematology Education Program. 2014;2014(1):113–118. doi: 10.1182/asheducation-2014.1.113. [DOI] [PubMed] [Google Scholar]
- 49.Clipson A., Wang M., de Leval L., Ashton-Key M., Wotherspoon A., Vassiliou G. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177–1185. doi: 10.1038/leu.2014.330. [DOI] [PubMed] [Google Scholar]
- 50.Awasthi Aradhana, Rolland Delphine C.M., Elmacken Mona, Reggio Christopher, Ayello Janet, van de Ven Carmella. A comparative global phosphoproteomics analysis of obinutuzumab (GA101) versus rituximab (RTX) against RTX sensitive and resistant Burkitt lymphoma (BL) demonstrates differential phosphorylation of signaling pathways proteins. Blood. 2014;124(21):5481. doi: 10.18632/oncotarget.23040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aradhana Awasthi, Delphine C.M. Rolland, Janet Ayello, Carmella van de Ven, Venkatesha Basrur, Kevin Conlon, et al. A comparative global phosphoproteomics analysis of obinutuzumab (GA101) versus rituximab (RTX) against RTX sensitive and resistant Burkitt lymphoma (BL) demonstrates differential phosphorylation of signaling pathway proteins after treatment. Oncotarget. 2017;8(69):113895–909. [DOI] [PMC free article] [PubMed]
- 52.Dwivedi P., Muench D.E., Wagner M., Azam M., Grimes H.L., Greis K.D. Time resolved quantitative phospho-tyrosine analysis reveals Bruton's Tyrosine kinase mediated signaling downstream of the mutated granulocyte-colony stimulating factor receptors. Leukemia. 2019;33(1):75–87. doi: 10.1038/s41375-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dwivedi P., Muench D.E., Wagner M., Azam M., Grimes H.L., Greis K.D. Phospho serine and threonine analysis of normal and mutated granulocyte colony stimulating factor receptors. Sci Data. 2019;6(1):21. doi: 10.1038/s41597-019-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner N., Núñez G. MyD88: a critical adaptor protein in innate immunity signal transduction. J. Immunol. 2013;190(1):3–4. doi: 10.4049/jimmunol.1203103. [DOI] [PubMed] [Google Scholar]
- 55.Mahabeleshwar G.H., Kawanami D., Sharma N., Takami Y., Zhou G., Shi H. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity. 2011;34(5):715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heo K.S., Chang E., Takei Y., Le N.T., Woo C.H., Sullivan M.A. Phosphorylation of protein inhibitor of activated STAT1 (PIAS1) by MAPK-activated protein kinase-2 inhibits endothelial inflammation via increasing both PIAS1 transrepression and SUMO E3 ligase activity. Arterioscler. Thromb. Vasc. Biol. 2013;33(2):321–329. doi: 10.1161/ATVBAHA.112.300619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nayak L., Goduni L., Takami Y., Sharma N., Kapil P., Jain M.K. Kruppel-like factor 2 is a transcriptional regulator of chronic and acute inflammation. Am. J. Pathol. 2013;182(5):1696–1704. doi: 10.1016/j.ajpath.2013.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zahlten J., Steinicke R., Opitz B., Eitel J., N'Guessan P.D., Vinzing M. TLR2- and nucleotide-binding oligomerization domain 2-dependent Kruppel-like factor 2 expression downregulates NF-kappa B-related gene expression. J. Immunol. 2010;185(1):597–604. doi: 10.4049/jimmunol.0901798. [DOI] [PubMed] [Google Scholar]
- 59.Mellert K., Martin M., Lennerz J.K., Ludeke M., Staiger A.M., Kreuz M. The impact of SOCS1 mutations in diffuse large B-cell lymphoma. Br. J. Haematol. 2019;187(5):627–637. doi: 10.1111/bjh.16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science (New York, N.Y.) 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe D., Ezoe S., Fujimoto M., Kimura A., Saito Y., Nagai H. Suppressor of cytokine signalling-1 gene silencing in acute myeloid leukaemia and human haematopoietic cell lines. Br. J. Haematol. 2004;126(5):726–735. doi: 10.1111/j.1365-2141.2004.05107.x. [DOI] [PubMed] [Google Scholar]
- 62.Steidl C., Shah S.P., Woolcock B.W., Rui L., Kawahara M., Farinha P. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471(7338):377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The histological description and antibodies application for the diagnosis of childhood Burkitt lymphoma.
Table S2 List of the target genes on the self-designed sequencing panel.
Table S3 Primers for high resolution melting analysis.
Table S4 Primers for real-time quantitative reverse transcription-PCR analysis.
Table S5 Primary antibodies for immunohistochemistry analysis.
Table S6 Results of fluorescence in situ hybridization (FISH) analysis of c-MYC, BCL2 and BCL6 gene loci and chromosome 11q aberration of the childhood Burkitt lymphoma cases.
Data Availability Statement
The sequence data that support the findings of this study have been deposited in NCBI Sequence Read Archive (SRA) with the accession codes “SUB7208508” and “SUB7195810”. All data generated or analyzed during this study are included either in this article or in the supplementary material.