Abstract
Introduction
In East Asia, hepatocellular carcinoma (HCC) is one of the most commonly diagnosed cancer types. Long noncoding RNA (lncRNA) prostate androgen-regulated transcript 1 (PART1) was reported to play crucial roles in regulating cancer progression. However, roles and mechanisms of action of PART1 in hepatocellular carcinoma (HCC) still remain unknown.
Methods
Quantitative real-time polymerase chain reaction (RT-qPCR) method was used to detect the PART1 expression level in HCC cells. Cell proliferation, colony formation, and transwell invasion assays were performed to investigate the biological roles of PART1 on HCC cell behaviors. Bioinformatic analysis methods were performed to analyze connections of microRNA-590-3p (miR-590-3p) with PART1 or high mobility group box 2 (HMGB2) in HCC. Moreover, expression levels of PART1, miR-590-3p, and HMGB2 in HCC tissues and normal tissues were analyzed at ENCORI.
Results
PART1 expression was found to be significantly upregulated in HCC tissues and cells. Functionally, silencing of PART1 significantly suppressed HCC cell proliferation, colony formation and invasion in vitro, while forcing PART1 exerts opposite biological effects. Mechanically, miR-590-3p/HMGB2 axis was downstream target of PART1, and silencing of miR-590-3p or forcing of HMGB2 could rescue the stimulation effects of PART1 overexpression on HCC cell behaviors.
Discussion
Our results provided evidence that PART1 serves as oncogenic lncRNA through sponging miR-590-3p to upregulate HMGB2 expression in HCC.
Keywords: PART1, miR-590-3p, HMGB2, ceRNA, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the main subtype of liver cancer with a poor prognosis.1,2 The infection of hepatitis B virus (HBV) is the main cause for the development of HCC.2 The long-term overall survival of HCC remains undesired even with the recent appearance of tumor targeted therapeutic agents.3
Long noncoding RNA (lncRNA) is a type of noncoding RNA with the length of over 200 nucleotides.4 The importance of lncRNA in regulating multiple cancer progressions including HCC has been recognized.5–7 Hence, investigating acting mechanisms of lncRNAs in HCC could provide novel therapeutic targets for HCC.
PART1, located at chromosome 5q12 60,487,713–60,547,657, is a lncRNA that is firstly identified to be overexpressed in prostate cancer.8 Liu et al showed that high PART1 level was closely correlated with prognosis of laryngeal squamous cell carcinoma cancer patients and could be used as prognostic biomarkers and therapeutic targets.9 Hu et al revealed PART1 was overexpressed in bladder cancer, and silencing of PART1 inhibits cancer cell growth through promoting apoptosis.10 Recently, Zhu et al investigated the functions of PART1 in non-small-cell lung cancer and found PART1 could promote cancer development via regulating Janus kinase and signal transducer and activator of transcription proteins pathway.11 A recent study indicated that PART1 could be used as a predictor for overall survival and recurrence-free survival of HCC patients.12 However, biological roles and underlying mechanisms of PART1 in HCC remain poorly investigated.
High mobility group box 2 (HMGB2) is found highly expressed in cancers and functions as an oncogene.13–15 For instance, HMGB2 could regulate gastric cancer progression under the regulation of lncRNA and miRNA.13,14 Also, there are several targets for HMGB2 including LDHB and FBP1 have been found in cancers.15
This work was conducted to investigate the roles of PART1 in HCC by analyzing how it influences cancer hallmarks. We also investigate miRNA between PART1 and HMGB2 to understand the mechanisms of PART1 in HCC.
Materials and Methods
Cell Culture
HCC cells (SMMC-7721 and Huh7) and normal hepatic cell line LO2 were purchased from Cell Bank of Chinese Academy Sciences. Cells were cultured with DMEM supplemented with 10% FBS (Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a 37°C humidified incubator contains 5% CO2.
Cell Transfection
Small interfering RNA against PART1 (si-PART1, 5′-GCAAAUGAAAGCUACCAAU-3′), against HMGB2 (si-HMGB2, 5ʹ-AAGACCAUGUCUGCAAAGGAG-3ʹ), negative control (si-NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-590-3p mimic (5ʹ-UAAUUUUAUGUAUAAGCUAGU-3ʹ), and matched negative control (mi-NC, 5ʹ-AUUAUGUUACGGAUUAUUAUA-3ʹ) were purchased from GeneChem (Shanghai, China). pcDNA3.1 contains sequence of PART1 (pPART1) was obtained from GenePharm. Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.) was used for cell transfection.
Cell Proliferation Assay
Cells were seeded into 96-well plate at density of 2⨰103 cells/well and incubated for 0, 24, 48, and 72 h. Cell counting kit-8 (CCK-8) reagent (Beyotime, Haimen, Jiangsu, China) was added to the plate and further incubated for four hours. Optical density was observed at the wavelength of 450 nm using a microplate reader.
Colony Formation Assay
Six hundred cells were incubated in a six-well plate and allow to grow for 14 days. Colonies were fixed with methanol, stained with crystal violet, and counted under a microscope. Colony numbers were calculated using formula: colonies from five independent fields/5.
Transwell Invasion Assay
We seeded 5⨰104 cells in serum-free DMEM on upper chamber of Matrigel coated insert. The lower chamber was filled with DMEM containing FBS.After 24 h incubation, invaded cells were fixed with paraformaldehyde, stained with crystal violet, and washed with PBS. Invaded cell numbers were counted under an optical microscope.
Quantitative Real-time Polymerase Chain Reaction (RT-qPCR)
TRIzol reagent (Beyotime) was used to isolate RNA sample from cultured cells. Then, RNA was reverse transcribed into complementary DNA using PrimeScript RT kit (Takara, Dalian, Liaoning, China). RT-qPCR was conducted at ABI 7500 system (Foster City, CA, USA) using SYBR Green (Takara). Relative gene expression level was calculated with 2−ΔΔCt method. Primers used were: PART1: 5ʹ‐AAGGCCGTGTCAGAACTCAA‐3ʹ (forward) and 5ʹ‐GTTTTCCATCTCAGCCTGGA‐3ʹ (reverse); HMGB2: 5ʹ‐GGGGAAGAAAAAGGACCCCA‐3ʹ (forward) and 5ʹ‐GCTGACTGCTCAGACCACAT‐3ʹ (reverse); GAPDH: 5ʹ-AACGTGTCAGTGGTGGACCTG-3ʹ (forward); and 5ʹ-AGTGGGTGTCGCTGTTGAAGT-3ʹ (reverse); miR-590-3p: 5ʹ-GCGCTAATTTTATGTATAA-3ʹ (forward); and 5ʹ-GTGCAGGGTCCGAGGT-3ʹ (reverse); U6 snRNA: 5ʹ‐CTCGCTTCGGCAGCACA‐3ʹ (forward) and 5ʹ‐AACGCTTCACGAATTTGCGT‐3ʹ (reverse).
Immunoblotting Assay
Total proteins from cells were extracted using RIPA lysis buffer (Beyotime) and quantified with BCA kit (Beyotime). Twenty-five micrograms of protein sample was separated at 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis and transferred to PVDF membrane. Then the membrane was incubated with primary antibodies (anti-HMGB2: ab124670, anti-GAPDH: ab181602, Abcam, Cambridge, MA, USA). PVDF membrane was further incubated with horseradish peroxidase-conjugated secondary antibodies (ab6721, Abcam). Band signal was detected with BeyoECL kit (Beyotime).
Targets Prediction Using Bioinformatic Tool
miRNA target for PART1 was analyzed at LncBase V 2.0. Target gene for miRNA was analyzed at TargetScan. miR-590-3p and HMGB2 were selected for further analysis using these two bioinformatic tools.
Luciferase Reporter Assay
pmiR-Report luciferase vector contains wild-type (wt) sequence of PART1 or HMGB2 was named as wt-PART1/HMGB2. Mutant luciferase vectors were constructed using site-direct mutagenesis kit (Takara) and named as mt-PART1/HMGB2. Cells were transfected with luciferase reporter vectors and miRNAs using Lipofectamine 2000. After 48 h transfection, relative luciferase activity was measured using Dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
Expression Level of PART1, miR-590-3p, and HMGB2 in HCC Tissues Was Detected at ENCORI
Expression levels of PART1, miR-590-3p, and HMGB2 in HCC tissues and normal tissues were analyzed at ENCORI website.
Silencing of PART1 Hinders HCC Progression
BALB/c nude mice were used to detect the roles of PART1 on HCC progression in vivo. Study protocol was approved by the Ethics Committee of Affiliated Hospital of Youjiang Medical University for Nationalities and performed in accordance with National Guidelines for Experimental Animal Welfare issued by the Ministry of Science and Technology, China. Cells transfected with PART1 knockdown (sh-PART1 (5ʹ-ACCTCATGCAGCTTGACTGTGATTCATCAAGAGTGAATCACAGTCAAGCTGCATTT-3ʹ) or sh-NC (5ʹ-ACCTCGTACGATTGCGCGTCTTAATTCAAGAGATTAAGACGCGCAATCGTACTT-3ʹ)) or stable PART1 overexpression were injected into the flank of nude mice. Tumor width and length were measured every seven days to calculate tumor volume. After four weeks, the mice were killed to measure tumor weight. Furthermore, RNA samples were extracted from these tissues to measure the relative gene expression level.
Statistical Analysis
GraphPad Prism 8.0 (GraphPad Inc., San Diego, CA, USA) was used to analyze data obtained from three independent experiments. Differences were calculated using one-way ANOVA and Tukey's post hoc test or Student’s t-test. Data were presented as mean ± SD. P -value < 0.05 was an indicator for significant difference.
Results
PART1 Expression was Elevated in HCC
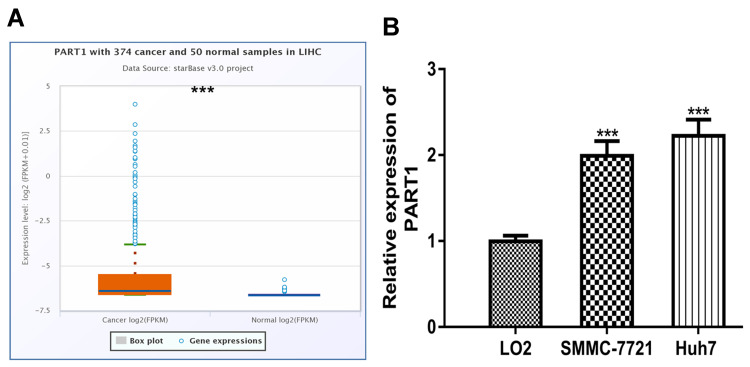
ENCORI analysis showed PART1 was significantly elevated in HCC tissues compared with normal tissues (Figure 1A). Moreover, RT-qPCR analysis result demonstrated PART1 was upregulated in HCC cells in comparison with normal cells (Figure 1B).
Figure 1.
Expression of PART1 in HCC. (A) Expression of PART1 in HCC tissues and normal tissues. (B) Expression of PART1 in HCC cell lines and normal cell line. * P<0.001. PART1: prostate androgen-regulated transcript 1.
Abbreviation: HCC, hepatocellular carcinoma.
Knockdown of PART1 Suppresses HCC Cell Proliferation, Colony Formation, and Migration
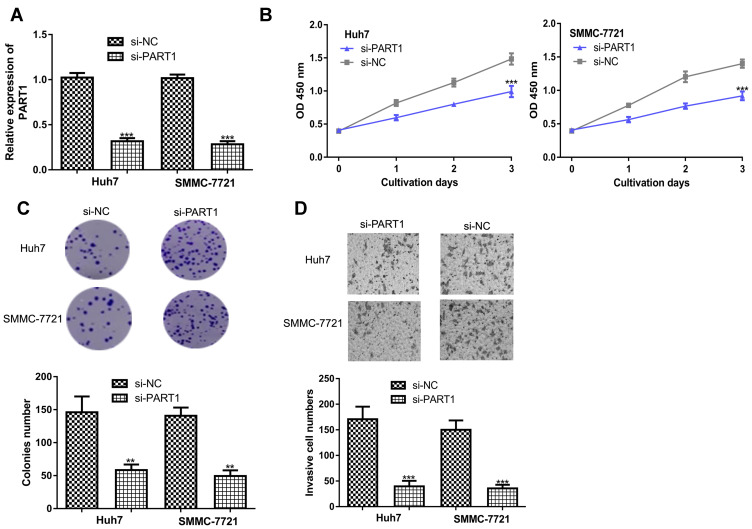
Introduction of si-PART1 significantly decreased PART1 expression in HCC cells (Figure 2A). CCK-8 assay revealed knockdown of PART1 inhibits HCC cell proliferation rate (Figure 2B). Colony formation assay confirmed the results of CCK-8 assay (Figure 2C). Moreover, decreased invasion ability was observed in HCC cells with si-PART1 transfection (Figure 2D).
Figure 2.
PART1 knockdown inhibits HCC cell proliferation, colony formation and invasion. (A) Transfection efficacy of si-PART1 and si-NC in HCC cells. (B) CCK-8 assay revealed HCC cell proliferation was suppressed by PART1 knockdown. (C) Colony numbers were reduced by PART1 knockdown in HCC cells. (D) Transwell invasion assay found that PART1 knockdown suppressed invasion of HCC cells. *P<0.001, **P<0.01.
Abbreviations:PART1, prostate androgen-regulated transcript 1; HCC, hepatocellular carcinoma; si-PART1, small interfering RNA targeting PART1; si-NC, negative control siRNA; CCK-8, cell counting kit-8.
Overexpression of PART1 Promotes HCC Cell Proliferation, Colony Formation, and Migration
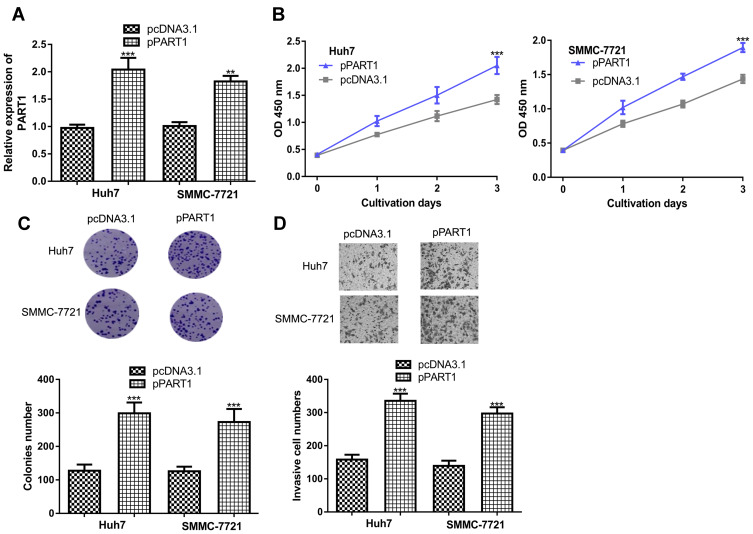
In addition, gain-of-function experiments were conducted to investigate the roles of PART1. Transfection of pPART1 significantly increased the levels of PART1 in HCC cells (Figure 3A). CCK-8 and colony formation assays confirmed PART1 overexpression promotes HCC cell growth (Figure 3B and C). Moreover, invasive cell numbers were significantly increased after PART1 overexpression (Figure 3D).
Figure 3.
PART1 overexpression promotes HCC cell proliferation, colony formation and invasion. (A) Transfection efficacy of pPART1 and pcDNA3.1 in HCC cells. (B) CCK-8 assay revealed HCC cell proliferation was stimulated by PART1 overexpression. (C) Colony numbers were increased by PART1 overexpression in HCC cells. (D) Transwell invasion assay found that PART1 overexpression promoted invasion of HCC cells. *P<0.001, **P<0.01.
Abbreviations:PART1, prostate androgen-regulated transcript 1; HCC, hepatocellular carcinoma; CCK-8, cell counting kit-8.
PART1 Regulates miR-590-3p and HMGB2 in HCC
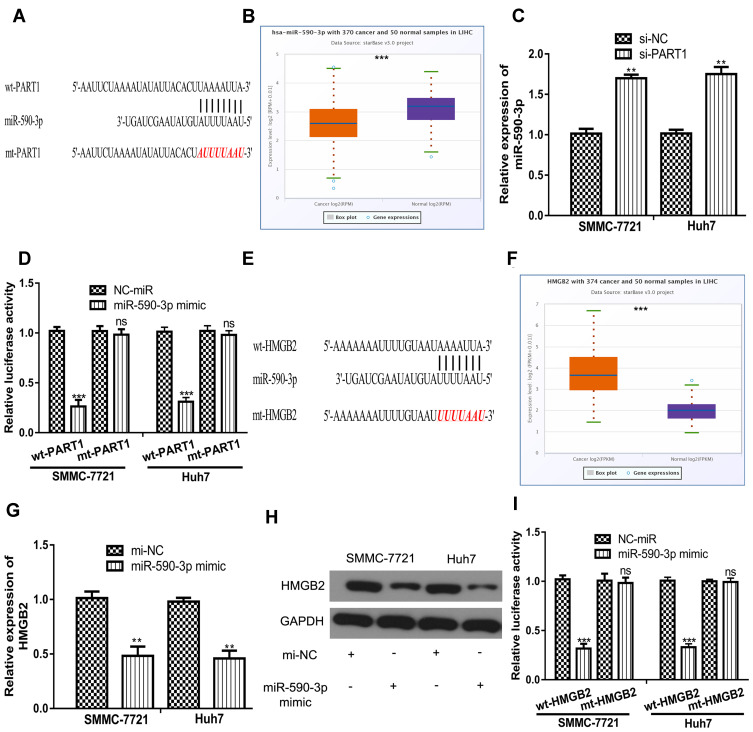
Growing evidence indicated PART1 serves as competing endogenous RNA (ceRNA) to sequester miRNA and regulate target gene expression.10,11 miR-590-3p was revealed to be the potential target for PART1 (Figure 4A). miR-590-3p level was found to be decreased in tumor tissues compared with normal tissues (Figure 4B). Moreover, the knockdown of PART1 significantly increased miR-590-3p levels in HCC cells (Figure 4C). Importantly, forcing miR-590-3p expression reduced relative luciferase activity of HCC cells transfected with wt-PART1 but not mt-PART1 (Figure 4D). Moreover, miR-590-3p was found to bind with HMGB2 through TargetScan analysis (Figure 4E). HMGB2 level was found to be elevated in HCC tissues compared with normal tissues (Figure 4F). Interestingly, transfection of miR-590-3p mimic decreased HMGB2 mRNA levels in HCC cells (Figure 4G). Western blot confirmed that forcing miR-590-3p expression decreased HMGB2 protein levels in HCC cells (Figure 4H). As expected, forcing miR-590-3p expression inhibited relative luciferase activity of HCC cells transfected with wt-HMGB2 (Figure 4I).
Figure 4.
PART1 regulates the levels of miR-590-3p and HMGB2 in HCC cells. (A) Interaction between PART1 and miR-590-3p. (B) Expression of miR-590-3p in HCC tissues and normal tissues. (C) Knockdown of PART1 increased the levels of miR-590-3p in HCC cells. (D) miR-590-3p overexpression inhibits the relative luciferase activity of cells transfected with wt-PART1. (E) Interaction between HMGB2 and miR-590-3p. (F) Expression of HMGB2 in HCC tissues and normal tissues. (G) miR-590-3p overexpression decreased the mRNA levels of HMGB2 in HCC cells. (H) miR-590-3p overexpression decreased the protein levels of HMGB2 in HCC cells. (I) miR-590-3p overexpression inhibits the relative luciferase activity of cells transfected with wt-HMGB2. *P<0.001, **P<0.01.
Abbreviations: ns, not significant; PART1, prostate androgen-regulated transcript 1; HCC, hepatocellular carcinoma; miR-590-3p, microRNA-590-3p; mi-NC, negative control miRNA; HMGB2, high mobility group box 2; wt, wild-type; mt, mutant.
miR-590-3p and HMGB2 Mediates the Oncogenic Role of PART1 in HCC
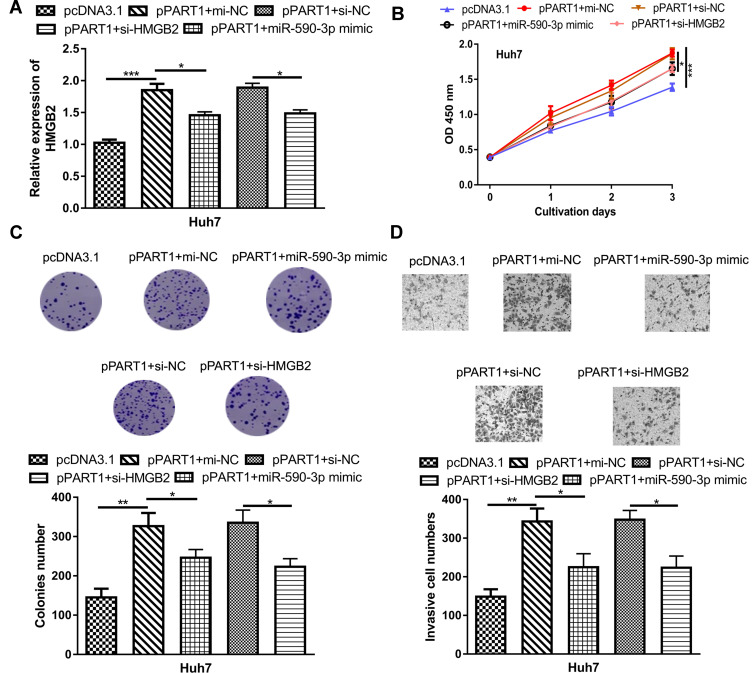
To further explore the roles of miR-590-3p and HMGB2 in HCC, rescue experiments were conducted. RT-qPCR showed overexpression of miR-590-3p or knockdown of HMGB2 partially reversed the effects of pPART1 on HMGB2 expression (Figure 5A). CCK-8 assay, colony formation assay, and transwell invasion assay indicated that the stimulation effects of pPART1 on HCC cells could be abrogated by miR-590-3p overexpression or HMGB2 knockdown (Figure 5B–D).
Figure 5.
miR-590-3p overexpression and HMGB2 overexpression partially reverses PART1 overexpression-induced effects on HCC cells. (A) HMGB2 expression in HCC cell transfected with pPART1, pPART1+miR-590-3p mimic, or pPART1+si-HMGB2. (B) CCK-8 assay, (C) colony formation assay, and (D) transwell invasion assay were performed to detect the proliferation, and invasion of HCC cells in each group. *P<0.05, **P<0.01, ***P<0.001,.
Abbreviations:PART1, prostate androgen-regulated transcript 1; HCC, hepatocellular carcinoma; miR-590-3p, microRNA-590-3p; HMGB2, high mobility group box 2; si-HMGB2, small interfering RNA targeting HMGB2; CCK-8, cell counting kit-8.
Roles of PART1 on HCC Tumor Growth in vivo
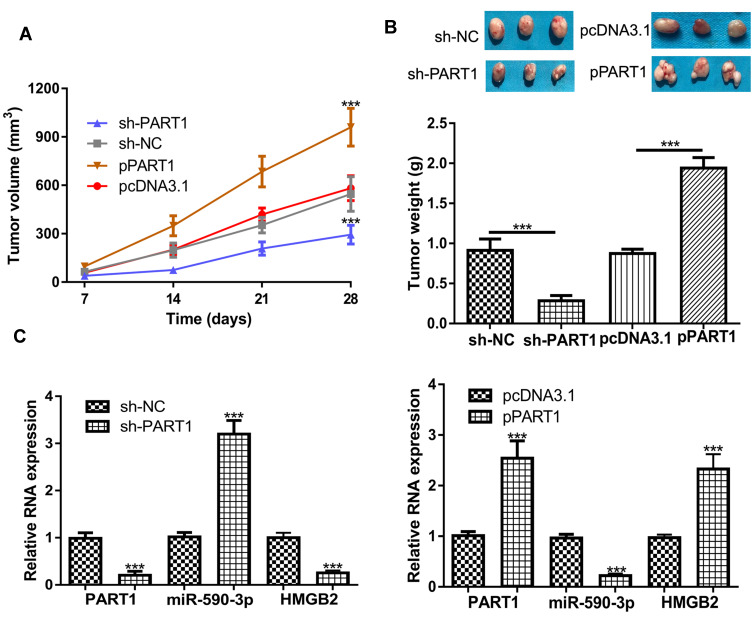
To analyze the roles of PART1 in vivo, cells with PART1 knockdown or overexpression were used to establish xenograft model. The results in Figure 6A and B showed that both tumor volume and tumor weight were decreased by sh-PART1 compared with sh-NC. On the contrary, tumor volume and weight were increased after HMGB2 overexpression (Figure 6A and B). Furthermore, we showed the expression of PART1 and HMGB2 was significantly decreased by PART1 knockdown, while increased by PART1 overexpression (Figure 6C). Moreover, the expression of miR-590-3p was dramatically increased by PART1 knockdown and decreased by PART1 overexpression (Figure 6C).
Figure 6.
Effects of PART1 on HCC tumor growth in vivo. (A) Tumor volume after PART1 knockdown or overexpression was calculated every week. (B) Mice were killed and tumor weight was examined after PART1 knockdown or overexpression. (C) The expressions of PART1, miR-590-3p and HMGB2 were detected after PART1 knockdown or overexpression. *P<0.001.
Abbreviations:PART1, prostate androgen-regulated transcript 1; HCC, hepatocellular carcinoma; miR-590-3p, microRNA-590-3p; HMGB2, high mobility group box 2; sh-HMGB2, short hairpin RNA targeting HMGB2; sh-NC, negative control shRNA.
Discussion
Growing evidence indicated lncRNAs played crucial roles in regulating HCC progression.16,17 lncRNA activated by transforming growth factor beta was revealed as a predictor for poorer overall survival of HCC patients, and regulated HCC cell autophagy via activating Yes-associated protein and autophagy-related protein five expression.16 Moreover, a recent study showed small nucleolar RNA host gene 7 (SNHG7) increased expression in HCC tissues compared with those in normal tissues.17 Mechanism studies showed SNHG7 could promote HCC tumorigenesis via sponging miR-122-5p to upregulate ribosomal protein L4 expression.16
In this study, our RT-qPCR data and ENCORI analysis data showed PART1 was highly expressed in HCC cells and tissues. Moreover, silencing of PART1 could inhibit HCC cell growth and invasion. Importantly, forcing of PART1 could promote HCC cell growth and invasion. In vivo analyses indicated that silencing of PART1 could suppress tumor growth, while forcing of PART1 exerts opposite effects on tumor growth. These results collectively may suggest the oncogenic role of PART1 in HCC, a role that was reported in previous studies.10,11 A very recent study conducted by Zhou et al indicated PART1 could boost HCC progression via the miR-149-5p/MAP2K1 pair in vitro.18 Our work can be regarded as another proof for the oncogenic role of PART1 in HCC. However, there is a difference between our current work and this work, which is that we provided in vivo analysis results to support the oncogenic role of PART1.
It has been widely recognized lncRNA exerts biological roles via regulating miRNA and target gene. We found miR-590-3p, downregulated expression in HCC, was a putative target for PART1. We confirmed the direct interaction of PART1 and miR-590-3p through luciferase activity reporter assay. miR-590-3p was reported to function as either tumor promoter or suppressor gene in cancers.19,20 Considering the distinct role of miR-590-3p in cancers, hence the roles of miR-590-3p in HCC was explored. We validated the interaction of miR-590-3p and HMGB2. HMGB2 was reported to function as an oncogene in cancers.12–14 In our work, we also found HMGB2 expression was elevated in HCC tissues and cells. Importantly, miR-590-3p overexpression or HMGB2 knockdown partially reversed PART1 overexpression on HCC cell proliferation, colony formation and invasion. The limitation of this work is that we did not explore the expression levels of this newly identified ceRNA triplet in HCC clinical samples. In addition, the clinical significance of these genes in HCC was not investigated. Hence, further experiments are still needed to validate the importance of this PART1/miR-590-3p/HMGB2 axis in HCC.
Conclusion
To conclude, our work provided evidence that PART1 was elevated expression in HCC. In vitro and in vivo assays revealed PART1 could promote HCC tumor progression by regulating miR-590-3p/HMGB2. Our work provided novel evidence regarding mechanisms underlying HCC pathogenesis.
Funding Statement
This work was supported by Science and Technology Project (AD17129025).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Waidmann O. Recent developments with immunotherapy for hepatocellular carcinoma. Expert Opin Biol Ther. 2018;18(8):905–910. doi: 10.1080/14712598.2018.1499722 [DOI] [PubMed] [Google Scholar]
- 4.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36(2):423–434. doi: 10.1159/000430109 [DOI] [PubMed] [Google Scholar]
- 7.Wang B, Tang D, Zhang Z, Wang Z. Identification of aberrantly expressed lncRNA and the associated TF-mRNA network in hepatocellular carcinoma. J Cell Biochem. 2019. doi: 10.1002/jcb.29384. [DOI] [PubMed] [Google Scholar]
- 8.Lin B, White JT, Ferguson C, et al. PART‐1: a novel human prostate‐specific, androgen‐regulated gene that maps to chromosome 5q12. Cancer Res. 2000;60(4):858‐863. [PubMed] [Google Scholar]
- 9.Liu Y, Ye F. Construction and integrated analysis of crosstalking ceRNAs networks in laryngeal squamous cell carcinoma. Peer J. 2019;7:e7380. doi: 10.7717/peerj.7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu X, Feng H, Huang H, et al. Downregulated long noncoding RNA PART1 inhibits proliferation and promotes apoptosis in bladder cancer. Technol Cancer Res Treat. 2019;18:1533033819846638. doi: 10.1177/1533033819846638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu D, Yu Y, Wang W, et al. Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med. 2019;8(13):6064–6081. doi: 10.1002/cam4.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye J, Zhang J, Lv Y, et al. Integrated analysis of a competing endogenous RNA network reveals key long noncoding RNAs as potential prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120(8):13810–13825. doi: 10.1002/jcb.28655 [DOI] [PubMed] [Google Scholar]
- 13.Cui G, Cai F, Ding Z, Gao L. HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum Pathol. 2019;84:133–141. doi: 10.1016/j.humpath.2018.09.017 [DOI] [PubMed] [Google Scholar]
- 14.Xu CH, Xiao LM, Liu Y, et al. The lncRNA HOXA11-AS promotes glioma cell growth and metastasis by targeting miR-130a-5p/HMGB2. Eur Rev Med Pharmacol Sci. 2019;23(1):241–252. doi: 10.26355/eurrev_201901_16770 [DOI] [PubMed] [Google Scholar]
- 15.Fu D, Li J, Wei J, et al. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal. 2018;16(1):8. doi: 10.1186/s12964-018-0219-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang CZ, Yan GX, Dong DS, Xin H, Liu ZY. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol. 2019;25(35):5310–5322. doi: 10.3748/wjg.v25.i35.5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Sun L, Wang L, Yao B, Mo H, Yang W. LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed Pharmacother. 2019;118:109386. doi: 10.1016/j.biopha.2019.109386 [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Wang P, Tu M, Huang Y, Xiong F, Wu Y. Long non-coding RNA PART1 promotes proliferation, migration and invasion of hepatocellular carcinoma cells via miR-149-5p/MAP2K1 axis. Cancer Manag Res. 2020;12:3771–3782. doi: 10.2147/CMAR.S246311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng ZY, Xu XH, Cen DZ, Luo CY, Wu SB. miR-590-3p promotes colon cancer cell proliferation via Wnt/β-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur Rev Med Pharmacol Sci. 2017;21(21):4844–4852. [PubMed] [Google Scholar]
- 20.Zu C, Liu S, Cao W, et al. MiR-590-3p suppresses epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma by inhibiting SIP1 expression. Oncotarget. 2017;8(21):34698–34708. doi: 10.18632/oncotarget.16150 [DOI] [PMC free article] [PubMed] [Google Scholar]