Abstract
Abstract
Bacteria belonging to Rhodococcus genus represent ideal candidates for microbial biotechnology applications because of their metabolic versatility, ability to degrade a wide range of organic compounds, and resistance to various stress conditions, such as metal toxicity, desiccation, and high concentration of organic solvents. Rhodococcus spp. strains have also peculiar biosynthetic activities that contribute to their strong persistence in harsh and contaminated environments and provide them a competitive advantage over other microorganisms. This review is focused on the metabolic features of Rhodococcus genus and their potential use in biotechnology strategies for the production of compounds with environmental, industrial, and medical relevance such as biosurfactants, bioflocculants, carotenoids, triacylglycerols, polyhydroxyalkanoate, siderophores, antimicrobials, and metal-based nanostructures. These biosynthetic capacities can also be exploited to obtain high value-added products from low-cost substrates (industrial wastes and contaminants), offering the possibility to efficiently recover valuable resources and providing possible waste disposal solutions. Rhodococcus spp. strains have also recently been pointed out as a source of novel bioactive molecules highlighting the need to extend the knowledge on biosynthetic capacities of members of this genus and their potential utilization in the framework of bioeconomy.
Key points
• Rhodococcus possesses promising biosynthetic and bioconversion capacities.
• Rhodococcus bioconversion capacities can provide waste disposal solutions.
• Rhodococcus bioproducts have environmental, industrial, and medical relevance.
Keywords: Rhodococcus, Antimicrobials, Bioflocculants, Biosynthesis, Bioconversion, Biosurfactants, Carotenoids, Lipids, Metal-based nanostructures, Siderophores
Introduction
Rhodococcus genus belongs to the Actinobacteria phylum and comprises genetically and physiologically diverse bacteria, which are distributed in various water, soil, and marine habitats, also including harsh ecological niches such as arctic, desert, and heavily contaminated sites (Cappelletti et al. 2019a, 2019b). The wide distribution of Rhodococcus spp. strains is due to their extraordinary metabolic versatility, which is comparable to that described only in a few other bacterial genera, and to their unique environmental persistence and robustness (LeBlanc et al. 2008; Cappelletti et al. 2016).
Bacterial strains belonging to Rhodococcus genus have been largely studied for their application in bioremediation, biotransformation, and biocatalysis due to their capacity to biodegrade a wide range of organic compounds, including toxic and recalcitrant molecules such as chlorinated aliphatic and aromatic hydrocarbons, N- and S-heterocyclic compounds, and synthetic polymers (e.g., polyethylene) (Martínková et al. 2009; Cappelletti et al. 2015, 2017, 2018; Ciavarelli et al. 2012; Krivoruchko et al. 2019). During the last decade, it has become obvious that various species of this genus have also interesting capabilities regarding the biosynthesis of lipids and other valuable compounds. These biosynthetic capacities are often associated to the ability of Rhodococcus to resist to different environmental stresses (LeBlanc et al. 2008; Orro et al. 2015; Cappelletti et al. 2016) and allow these bacteria to cope with the presence of hydrophobic substrates in the growth medium, limited nitrogen sources, oxidative stressors, among others. The most extensively reported compounds that are biosynthesized by Rhodococcus spp. strains are glycolipid biosurfactants, carotenoids, triacylglycerols, and polyhydroxyalkanoates (PHAs) (Krivoruchko et al. 2019). Recently, novel siderophores and new antibiotics have been reported to be produced by Rhodococcus spp. strains as possible competition strategies. Further, several Rhodococcus spp. strains have recently been described to be able to produce metal-based nanostructures as the result of metal and metalloid [metal(loid)] bioconversion into their elemental state (Presentato et al. 2016, 2018a, 2018b; Firrincieli et al. 2019).
Compared to the many papers summarizing the biotechnological potential of Rhodococcus cells in association with their biodegradative enzymes for bioremediation and biocatalysis (Martínková et al. 2009; Kim et al. 2018; Krivoruchko et al. 2019; Jiao et al. 2020), this review gives an overview on the relevant biosynthetic capacities of bacteria belonging to this genus. These anabolic functions are at the basis of Rhodococcus-mediated bioproduction of commercially valuable compounds and bioactive molecules, as well as bioconversions of organic and inorganic contaminants/wastes into high value-added products.
Biosurfactants
Biosurfactants are a heterogeneous group of amphiphilic surface-active compounds that are produced by various microorganisms including bacteria, yeast, and fungi and find wide applications in several industries as demulsifiers; wetting, foaming, and spreading agents; functional food ingredients; and soaps/detergents (Santos et al. 2016). Biological compounds with surfactant activities can generally be classified into low-molecular-weight (glycolipids, lipopeptides, and phospholipids) and high-molecular-weight (polysaccharides, proteins, lipoproteins, or polymeric compounds) substances, the first being considered actual biosurfactant and the second generally described as bioemulsifiers (Franzetti et al. 2010). They possess several advantages as compared to the chemical surfactants including biodegradability, lower critical micelle concentrations (CMC), reduced toxicity, higher stability, and the possibility to be produced by renewable raw material (Banat et al. 2010).
Several Rhodococcus spp. strains produce biosurfactants in response to the presence of water-insoluble substrates (e.g., hydrocarbons) (Whyte et al. 1999; Cappelletti et al. 2019a), and in some cases also on water-soluble substances (e.g., ethanol and glucose) (Table 1) (Pirog et al. 2004; Ciapina et al. 2006). The formation of biosurfactants is believed to improve the utilization (bioavailability) of these water-insoluble compounds as growth substrates by facilitating their entry into cells (Lang and Philp 1998; Yakimov et al. 1999; Philp et al. 2002; Peng et al. 2007; Cappelletti et al. 2019b).
Table 1.
Biosynthetic activities of the most representative Rhodococcus spp. strains and growth substrate or waste residue used for the production of each valuable compound category
| Product category | Strain/s | Substrate and/or growth conditionsa | Features of the product and/or biosynthetic process | Reference |
|---|---|---|---|---|
| Biosurfactants | R. erythropolis DSM 43215 | Cells growing on C14–C15 n-alkanes or kerosene | Production of extracellular trehalose-monomycolates and trehalose-dimycolates | Kim et al. 1990 |
| Rhodococcus H13-A | Cells growing on n-alkanes or fatty alcohols | Production of glycolipid which is able to solubilize polyaromatic hydrocarbons | Page et al. 1999 | |
| R. opacus 1CP | Cells growing on C10, C12, C14, C16 n-alkanes | Production of trehalose dinocardiomycolates with double bonds | Niescher et al. 2006 | |
| R. erythropolis ATCC 4277 | Cells growing on glycerol or n-hexadecane | Production of extracellular biosurfactant on glycerol, and cell-bound biosurfactant on n-hexadecane | Ciapina et al. 2006 | |
| R. erythropolis strain EK-1 | Cells growing on hydrocarbons (C16 n-alkane and paraffin), ethanol or glucose | Production of cell-bound non-ionic trehalolipids on hydrocarbons, production of bioemulsifier on soluble substrates | Pirog et al. 2008 | |
| R. erythropolis SD-74 | Cells growing on n-hexadecane | Production of succynoil trehalolipids | Tokumoto et al. 2009 | |
| R. erythropolis 3C-9 | Cells growing on n-hexadecane | Production of extracellular trehalose lipids and free fatty acids | Peng et al. 2007 | |
| R. erythropolis 16 LM.USTHB | Cells growing on sunflower frying oil | Production of extracellular glycolipids | Sadouk et al. 2008 | |
| Rhodococcus sp. BS32 | Cells growing on rapeseed oil | Production of extracellular biosurfactants | Ruggeri et al. 2009 | |
| R. erythropolis sp. P6-4P | Cells growing on fish waste compost | Production of biosurfactant that is mainly composed of fatty acids | Kazemi et al. 2009 | |
| Bioflocculants | R. erythropolis S-1 | Cells growing on sorbitol, mannitol, ethanol, glucose, or fructose | Production of the peptidic bioflocculant named NOC-1, it is one of the best performing bioflocculant described up to date | Kurane et al. 1994 |
| R. erythropolis ATCC 10543 | Cells growing on pre-treated sludge and livestock wastewater | Production of a polysaccharidic bioflocculant | Peng et al. 2014 | |
| R. erythropolis | Cells growing on potato starch wastewater | Production of a polysaccharidic bioflocculant | Guo et al. 2018 | |
| Carotenoids | R.luteus, R. coprhilus, R. lentifragmentus, R. maris | Cells growing on Sauton agar medium | Production of β-carotene | Ochiyama et al. 1989 |
| R. equi, R. rubroperctinctus, R. aichiensis, R. sputi, R. chubuensis, R. obuensis, R. bornchialis, R. roseus, R. rhodochrous, R. rhodnii, R. terrae | Cells growing on defined medium l-asparagine and glycerol as main nitrogen and carbon sources, respectively | Production of γ-carotene-like compound | Ochiyama et al. 1989 | |
| R. erythropolis IBBPo1 | Cells growing on n-alkane | Production of lycopene at higher level | Stancu et al. 2015 | |
| R. rhodochrous RNMS1 | ND | Production of γ-carotene derivatives | Takaichi et al. 1990 | |
| R. erythropolis AN12 | Cells growing on rich medium (i.e., nutrient broth-based medium) | Production of γ-carotene derivatives | Tao et al. 2004 | |
| Rhodococcus sp. CIP | Cells growing on rich medium | Production of OH-chlorobactene glucoside hexadecanoate and related rare carotenoids | Osawa et al. 2011 | |
| R. opacus PD630 | Glycerol and ammonium acetate as main carbon and nitrogen sources, respectively | Production of uncharacterized carotenoids | Thanapimmetha et al. 2017 | |
| R. pyridinivorans NT2 | Cells growing on 2,4-dinitrotoluene | Production of a carotenoid mixture composed by 4-keto- γ-carotene and γ -carotene, lycopene, and β-carotene | Kundu et al. 2015 | |
| Triacylglycerols (TAGs) | R. opacus PD630 | Cells growing on phenyldecane | Accumulation of TAGs containing phenyldecanoic acid residues | Alvarez et al. 2002 |
| Cells growing on n-alkane | Accumulation of fatty acids with the same carbon skeleton of the alkane used for growth | Alvarez 2003 | ||
| Cells growing on citrate, succinate, propionate, valerate, saturated, and mono-unsaturated fatty acids with C15 and C17 chain length | Accumulation of TAGs with increased fraction of odd-numbered fatty acids | Alvarez et al. 1997 | ||
| Cells growing on C15–C18 n-alkanes, phenylacetic acid, phenyldecane, gluconic acid, acetic acid, propionic acid, fructose, and olive oil | First work reporting the ability of PD630 strain to produce and accumulate TAGs using different carbon sources | Alvarez et al. 1996 | ||
| Cells growing on sugar beet molasses, and sucrose | Production of large amounts of TAGs was demonstrated using a high-cell-density cultivation in a 500-l pilot-plant scale | Voss and Steinbüchel 2001 | ||
| R. opacus MITXM-61 (i.e., the spontaneous mutant of R. opacus PD630 carrying exogenous genes of xylA and xylB from Streptomyces padanus) | Cells growing on xylose/glucose, or corn stover hydrolysates | Robust growth and TAG biosynthesis on high concentrations of xylose and simultaneous utilization of xylose and glucose in the corn stover hydrolysate | Kurosawa et al. 2013, 2014 | |
| R. opacus PD630 pEC-K18mob2::bglABCTF | Cells growing on glucose and cellobiose | Acquisition of cellobiose utilization for growth and TAG accumulation | Hetzler and Steinbüchel 2013 | |
| R. opacus MITGM-173 (i.e., adaptively evolved R. opacus PD630) | Cells growing on glycerol/glucose/xylose | Higher glycerol utilization as compared to the parental strain | Kurosawa et al. 2015b | |
| R. erythropolis strains DSMZ 43060, 17 and DM1–21; R. fascians strains F7, S1.17b, 123 and D188–5; R. opacus strains PD630 and MR22; R. jostii strains RHA1 and 602; R. equi ATCC 6939; R. opacus PD630 pTip-QC2 /glpK1D1F7 | Cells growing on glycerol | Rhodococcus strains differed in terms of number of days required for TAGs production; R. equi produced lower amounts of TAGs as compared to the other species; R. opacus PD630 improved TAG production when expressing glpK1D1 from R. fascians | Herrero et al. 2016 | |
| Rhodococcus sp. 602 | Cells growing on gluconate, benzoate, n-hexadecane, naphthalene, or naphthyl-1-dodecanoate | TAGs with fatty acids of different chain length were accumulated depending on the growth substrate, also under resting cell condition | Silva et al. 2010 | |
| R. jostii LGK (R. jostii RHA1 bearing plasmid pTAC-lgk) | Cells growing on glucose, levoglucosan | RHA1 expressing lgk gene from Lipomyces starkeyi YZ-215 acquired the capacity to utilize levoglucosan for growth and TAG accumulation | Xiong et al. 2016a | |
| R. jostii RHA3 (R. jostii RHA1 bearing both araBAD and araFGH) | Cells growing on arabinose | RHA1 expressing araBAD and araFGH from E. coli acquired the capacity to utilize l-arabinose for growth and TAG accumulation | Xiong et al. 2016b | |
| R. opacus strains PD630 and MR22, R. wratislaviensis V, R. jostii RHA1, R. erythropolis DSMZ 43060, R. fascians F7, R. equi ATCC 6939, R. jostii RHA1 pJAM2/ltp1 | Cells growing on olive mill wastes | R. opacus, R. wratislaviensis, and R. jostii were more efficient at producing cell biomass and lipids than R. fascians, R. erythropolis, and R. equi. The overexpression of ltp1 (a gene encoding a fatty acid importer) in R. jostii RHA1 promoted an increase of 3.4-fold in lipids production | Herrero et al. 2018 | |
| Rhodococcus sp. YHY01 | Cells growing on oil palm biomass and barley straw lignin | Utilization of various aromatic compounds derived from lignocellulosic biomasses for growth and TAG accumulation | Bhatia et al. 2017, 2019 | |
| Polyhydroxyalkanoate (PHAs) | R. ruber NCIMB 40126 | Cells growing on acetate, lactate, succinate, fructose, glucose, and molasses | Accumulation of PHA with 3 HV and 3HB monomers. The relative amount of each monomer changed depending on the substrate, but HV was generally the predominant | Haywood et al. (1991) |
| R. ruber NCIMB 40126 | Cells growing on 4-hydroxybutyrate and 1,4-butanediol | Accumulation of PHA mainly composed by 3 HV and 3HB but also incorporating 4HB | Haywood et al. (1991) | |
| R. ruber NCIMB 40126 | Cells growing on 5-chlorovalerate | Accumulation of PHA mainly composed by 3 HV and 3HB but also incorporating 5HB | Haywood et al. (1991) | |
| R. ruber NCIMB 40126 | Cells growing on hexanoate or 2-hexenoate | Production of PHA containing 3HB, 3 HV, and 3-hydroxyhexanoate (3HHx) monomer units | Haywood et al. (1991) | |
| R. ruber NCIMB 40126 | Cells growing on valeric acid and 2-pentenoic acid | Accumulation of almost pure poly(3-hydroxyvalerate) | Haywood et al. (1991) | |
| R. ruber NCIMB 40126 | Cells growing on glucose and valerate | Accumulation of the copolymer poly(3HB-co-3 HV) as main PHA | Alvarez et al. 1997b | |
| R. rhodochrous ATCC 19070 | Cells growing on acetate, lactate, fructose, glucose, and sucrose | Accumulation of PHA with 3 HV and 3HB monomers. The relative amount of each monomer changed depending on the substrate, but HV was generally the predominant | Haywood et al. (1991) | |
| R. erythropolis DSMZ 43060, R.fascians D188–5, R. opacus MR22 | Cells growing on glucose, gluconate, and valerate | Accumulation of small amounts of homopolymer PHB | Alvarez et al. 1997b | |
| R. aetherivorans IAR | Cells growing on toluene | Accumulation of the copolymer poly(3HB-co-3 HV) as main PHA | Hori et al. 2009 | |
| Polyunsaturated fatty acids (PUFA) | R. aetherivorans BCP1 | Cells growing on cyclopentane carboxylic acid, cyclohexane carboxylic acid, and glucose | Production of linoleic acid increased on CPCA as compared to the other C sources | Presentato et al. 2018a |
| Antimicrobials | R. jostii K01-B0171 | Cells growing on rich medium with mannitol, glucose, yeast extract, ammonium succinate as carbon and nitrogen sources | Production of lariantin A and B | Inokoshi et al. (2012) |
| R. erythropolis JCM 6824 | Cell growing on succinate, sucrose, and casamino acids | Production of aurachin RE | Kitigawa et al. (2018) | |
| Rhodococcus sp. Acta 2259 | Cells growing in rich medium | Production of four aurachins | Nachtigall et al. 2010 | |
| Rhodococcus sp. Mer-N1033 | Cells growing in rich medium | Production of rhodopeptins | Chiba et al. 1999 | |
| R. fascians 307CO | Co-culturing with Streptomyces padanus | Production of rhodostreptomycin A and B | Kurosawa et al. 2008 | |
| R. erythropolis JCM 2895 | Soft-agar assay with rich medium medium | Production of a bacteriocin-like molecule | Kitigawa et al. 2018 | |
| R. equi and R. erythropolis strains, Rhodococcus enclensis NIO-1009 | Genome-based analyses | Production of humimycin A and B | Chu et al. 2016 | |
| Siderophores | R. erythropolis IGTS8 | Cells growing in defined salt media under iron limitation conditions | Production of heterobactin A and B | Carrano et al. 2001 |
| R. erythropolis PR4 | Cells growing on glucose, under iron-depleted conditions | Production of heterobactin A, heterobactins S1 and S2 | Bosello et al. 2013 | |
| R. jostii RHA1 | Cells growing on glucose under iron-depleted conditions | Production of rhodochelin | Bosello et al. 2011 | |
| R. rhodochrous OFS | Cells growing on hexadecane in iron-deficient minimal medium | Production of rhodobactin | Dhungana et al. 2007 | |
| R. erythropolis S43 | Cells growing on glucose, under iron-depleted conditions | Production of siderophore biding trivalent arsenic, AsO33- [also referred to as As(III)] | Retamal-Morales et al. 2018b | |
| Metal(loid) nanomaterials | Rhodococcus sp. strain | Resting cells and HAuCl4 as precursor | Production of intracellular crystalline AuNPs (ca. 12 nm) | Ahmad et al. 2003 |
| Rhodococcus sp. NCIM2891 | Cells growing using sodium acetate or cell-free extracts, AgNO3 as precursor | Production of intracellular crystalline AgNPs (ca. 10 nm) | Otari et al. 2012 | |
| R. pyridinivorans NT2 | Cells growing in rich medium or cell-free extracts; ZnSO4·H2O as precursor | Production of extracellular spherical and hexagonal crystalline ZnO NPs (ca.100 nm) | Kundu et al. 2014 | |
| R. aetherivorans BCP1 | Cells growing in rich medium; TeO32− as precursor | Production of intracellular TeNRs (from 100 to 500 nm) | Presentato et al. 2016 | |
| Resting cells; TeO32− as precursor | Production of intracellular crystalline TeNPs and TeNRs (from 200 to 700 nm) | Presentato et al. 2018b | ||
| Cells growing in rich medium; SeO32− as precursor | Production of SeNPs and SeNRs (from 50 to 600 nm) | Presentato et al. 2018a | ||
| R. erythropolis ATCC 4277 | Cells growing in rich medium or in a stirred tank reactor; sulfate mineral coal tailings as precursor | Production of crystalline Fe2O3 NPs (ca. 50 or 100 nm) | Maas et al. 2019a, 2019b |
aIn Cx, x indicates the number of carbons in the alkane chain
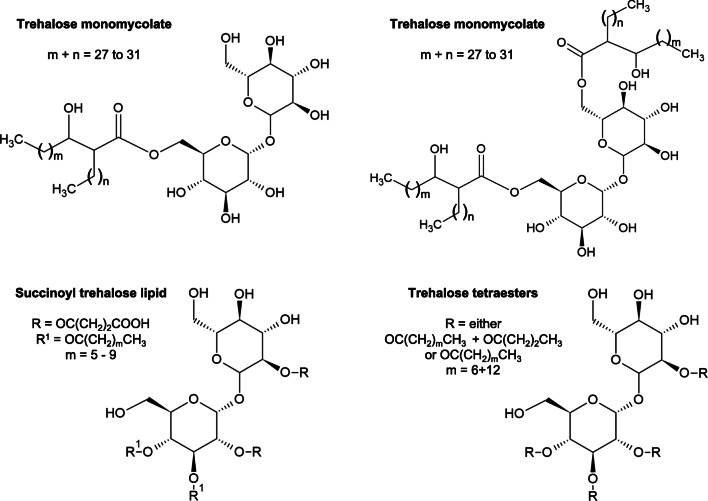
Rhodococci typically produce trehalose-based glycolipid biosurfactants, which possess various hydrophobic moieties and chemical compositions (Kuyukina and Ivshina 2010) (Fig. 1). Trehalolipids (TP) are a class of glycolipid biosurfactants with interesting physicochemical and biological properties that have been studied for i) environmental applications as emulsifiers in bioremediation (e.g. oil-spill treatment), ii) microbial-enhanced oil recovery, and iii) cosmetic and food industries (Christofi and Ivshina 2002; Pacheco et al. 2010). Besides, these compounds can be used for medical application for their immune-stimulating, antitumor, and antiviral properties (Ortiz et al. 2008; Kuyukina et al. 2015).
Fig. 1.
Chemical structures of the main biosurfactants produced by Rhodococcus spp. strains (modified from Franzetti et al. 2010; Kuyukina and Ivshina 2010)
TP are generally produced by rhodococci as either extracellular or cell wall-associated compounds during the growth on hydrocarbons (Lang and Philp 1998; Yakimov et al. 1999). Nitrogen limitation was also shown to favor the formation of anionic trehalose tetraesters using these growth substrates (Ristau and Wagner 1983; Kim et al. 1990). In those Rhodococcus spp. strains where the biosurfactants are excreted from the cell, the emulsification of the hydrocarbons with aqueous culture medium results in a very large surface area of contact between the cell and these compounds. Nevertheless, TP are more frequently retained by the Rhodococcus cell and localized on the outer cell surface (Rapp et al. 1979), increasing the hydrophobicity of the cell surface that facilitates the attachment and subsequent uptake of the hydrophobic compounds to be used as growth substrate (Kretschmer and Wagner 1983; Bredholt et al. 1998; Lang and Philp 1998; Cappelletti et al. 2019a). These biosurfactants, which bind to the cells, have more limited industrial applications because of problems with their recovery. The mechanism involved in the production and retention of biosurfactants by Rhodococcus cells appeared to be related to the type of substrate and the cell growth phase (Bredholt et al. 2002). Particularly, a marine Rhodococcus strain grown in the presence of sunflower oil, produced trehalose-based biosurfactants that were bound to cells during the first growth phase, while a strong increase of extracellular trehalolipid level was observed after the onset of stationary growth phase, reaching a maximum level of ~75% of the total trehalolipids present in the liquid medium (White et al. 2013). Further, the formation of anionic trehalose ester in R. opacus 1CP was associated with both biomass growth and n-alkane consumption, whereas the trehalose lipid production by R. erythropolis DSM43215 seemed to be uncoupled from growth and occurred in the stationary phase or under resting cells condition (Kim et al. 1990). An additional aspect regards the substrate used for the biosurfactant production, as R. erythropolis ATCC 4277 produced extracellular glycolipids upon growth on glycerol as sole carbon source, while partially cell-bound biosurfactants were obtained when n-hexadecane was added as only carbon and energy source (Ciapina et al. 2006). The presence of hydrocarbons (liquid paraffin and hexadecane) favored the generation of cell-bound trehalose-based glycolipids in R. erythropolis EK-1 cells. Conversely, strain EK-1 produced extracellular bioemulsifiers when the cells were grown on soluble substrates (ethanol and glucose) (Pirog et al. 2008; Franzetti et al. 2010). These considerations highlight the wide diversity in biosurfactant production mechanisms among different Rhodococcus strains and the possibility to drive the production of specific surfactant molecules by selecting the growth substrate. In this regard, supplying R. erythropolis SD-74 cultures with different n-alkanes as carbon source led to the biosynthesis of succinoyl trehalolipids featuring acyl groups of the same carbon chain length as the growth substrate (Tokumoto et al. 2009), suggesting the possibility to direct the synthesis of specific biosurfactants by providing alkanes with specific chemical structure (Lang and Philp 1998; Inaba et al. 2013; Cappelletti et al. 2019a). This property is incredibly useful for the commercial production, as most biosurfactant producers do not synthesize specifically defined derivatives. On the other hand, different Rhodococcus spp. strains produced diverse biosurfactants using the same hydrocarbon as a carbon source. For instance, Rhodococcus sp. SD-74 generated extracellular succinoyl trehalose lipids when cultivated on n-hexadecane (Tokumoto et al. 2009), while the same substrate in R. erythropolis 3C-9 favored the synthesis and release of two types of biosurfactants, i.e., trehalose lipids and free fatty acids—the latter being rarely reported in Rhodococcus spp., with the exception of R. ruber (Kuyukina et al. 2001; Peng et al. 2007). Other active compounds with surfactant activity produced by Rhodococcus were extracellular polysaccharides (EPS), which, in the case of R. rhodochrous S-2, enhanced its tolerance to the aromatic fraction (AF) of crude oil and facilitated the growth of indigenous bacteria resulting in the promotion of AF degradation in seawater-based medium (Iwabuchi et al. 2002).
Biosurfactants produced by some Rhodococcus species were reported to be more efficient in reducing the surface and interfacial tensions between aqueous and oil phases and to have lower critical micelle concentrations (CMCs) than many synthetic surfactants (Lang and Philp 1998). For instance, the biosurfactant produced by Rhodococcus sp. strain H13-A was up to 35-fold more effective than the synthetic Tween 80 counterpart in solubilizing polyaromatic hydrocarbons from a complex mixture into an aqueous solution (Page et al. 1999). Furthermore, in several cases, the biosurfactants from Rhodococcus were also found to have extremely low toxicity, sometimes lower as compared to that of biosurfactants isolated from other bacterial genera. Indeed, biosurfactants produced by and extracted from R. ruber AC 235 were 100–1000 times less toxic than synthetic commercial surfactants (Inipol EAP22, Corexit 9597, Finasol OSR-5) and 13 times less toxic than rhamnolipids from Pseudomonas aeruginosa (Kuyukina et al. 2001).
To reduce the cost of producing biosurfactant with rhodococci, specific approaches, such as the exploitation of low-cost substrates, were used. Raw material accounts for almost 30% of the overall cost of a microbial surfactant production and the investigation on potential usage of low-cost raw materials (e.g., industrial and/or municipal wastes) is necessary to develop economically sustainable processes (George and Jayachandran 2013). In this context, R. erythropolis 16 LM.USTHB showed the capacity to convert residual sunflower frying oil, a cheap renewable substrate, into extracellular glycolipids, which effectively lowered the surface tension of the crude broth (Sadouk et al. 2008). Ruggeri et al. (2009) isolated Rhodococcus sp. BS32, a strain able to produce extracellular biosurfactants growing on rapeseed oil. Fish waste compost was an effective source of nutrient-rich organic matter for the growth of R. erythropolis sp. P6-4P and the production of biosurfactants (Kazemi et al. 2009).
Despite promising features of the biosurfactants produced by rhodococci as compared to the well-understood biochemical and molecular bases of rhamnolipids synthesized by Pseudomonas strains (George and Jayachandran 2013), less information is available for the genetic and biochemistry of TL generation in Rhodococcus. It seems that the hydrophilic (the trehalose moiety) and hydrophobic (the mycolic acid moiety) portions of the trehalolipid molecules are synthesized independently and are subsequently esterified (Kretschmer and Wagner 1983; Kuyukina and Ivshina 2010). During biosynthesis, trehalose is first esterified with mycolic acids synthesized in the cytoplasm to form the trehalose monomycolate, which is believed to be the precursor of the di- or trimycolates produced at the plasma membrane (Lang and Philp 1998). Moreover, an enoyl-acyl carrier protein reductase, InhA, was indicated to be part of the fatty acid synthase-II system (FASII) involved in the process of the elongation of medium-sized fatty acids leading to long-chained hydrophobic moieties of trehalose mycolates in Rhodococcus spp. (Asselineau et al. 2002). The trehalose-6-phosphate synthase, OtsA, was reported to be the key enzyme involved in the synthesis of the sugar residue of the trehalose lipids biosynthesis (Tischler et al. 2013). The presence of an alternative trehalose biosynthesis pathway mediated by maltooligosyltrehalose synthase (TreY) and maltooligosyltrehalose trehalohydrolase (TreZ) enzymes was predicted in the biosurfactant producer R. erythropolis B7g (Retamal-Morales et al. 2018a). This pathway for trehalose biosynthesis uses oligo/polymalto-dextrins/glycogen as substrate (Tropis et al. 2005).
Bioflocculants
Flocculants are used as additives that induce aggregation or agglomeration of colloidal and other particles to form large particles (flocs) which settle allowing the clarification of the system in sedimentation and clarification processes (Lee et al. 2014). In this context, flocculants have been extensively applied for removing turbidity, suspended and dissolved solids, colors and dyes, and chemical oxygen demand (COD) in tap- and wastewater treatment processes. Compared with traditional inorganic and organic synthetic flocculants such as polyacrylamides, bioflocculants are more readily biodegradable, as well as less toxic to humans and environments (Salehizadeh et al. 2018).
Bioflocculants produced by Rhodococcus spp. strains received a large amount of attention due to their strong flocculating activities (Peng et al. 2014; Jiang et al. 2019). They were reported to mainly consist of polypeptide and lipid assemblies, especially mycolate-containing glycolipids (Finnerty 1992), which should allow the bacteria to more easily access the growth substrate. Specifically, bioflocculants seemed to be produced by those Rhodococcus spp. strains that do not synthesize biosurfactants during the utilization of long-chain alkanes (Bouchez-Naïtali et al. 2001). One of the first studies on this topic indicated that the production of an extracellular flocculant by R. erythropolis S-1 cells was strongly dependent on the type of substrate added. Although glucose, fructose, sorbitol, and mannitol were all efficiently used for bioflocculant production by S-1 strain, ethanol resulted to be the best performing carbon source for this purpose (Kurane et al. 1994), whereas organic acids and phenol did not induce any bioflocculant generation. The bioflocculant produced by S-1 resulted to be usable on a wide range of suspended solids, such as acid and alkaline soils, as well as India ink (Kurane et al. 1994). The same Rhodococcus strain could also use n-pentadecane as a carbon source, yet in this culture medium, the flocculant was found to be located on the cell surface, influencing the cell appearance during S-1 growth. Indeed, cells forming fibrous flocs that floated at the surface were observed upon S-l cultivation in the medium containing n-pentadecane, whereas when glucose was used as the carbon source, the cells were dispersed (Takeda et al. 1991). Characterization analyses performed on the bioflocculant produced by S-1 revealed that its main component was a protein (Kurane et al. 1994; Takeda et al. 1991). This bioflocculant was named NOC-1 and it has been used in both environmental pollution control and industrial wastewater treatment. R. erythropolis S-1 is currently recognized as one of the best flocculating bacteria and was used as a reference strain in studies on microbial flocculant production (Jiang et al. 2019).
Several studies investigated the possibility to reduce the cost of bioflocculant development, by using economical culture media for bacterial growth (Table 1). For instance, using pre-treated sludge and livestock wastewater as growth substrate, R. erythropolis ATCC 10543 produced a polysaccharidic bioflocculant featuring a strong flocculating potential on the same wastewaters within a wide pH range (2–12) (Peng et al. 2014). A recent study reported on the use of potato starch wastewater by another R. erythropolis strain to produce, parallelly to its growth, a bioflocculant, which showed a polysaccharide nature. The effectiveness of the bioflocculant was harnessed by treating the potato starch wastewater at neutral pH and analyzing the residual organics (COD and BOD), ammonium, phosphorus, turbidity, and chroma. This bioflocculant could be successfully applied to the potato starch wastewater treatment in a sequencing batch reactor (SBR), indicating that the compound was active in a real application scenario (Guo et al. 2018).
Carotenoids
Carotenoids are pigments ranging in color from yellow to orange and deep red that are soluble in lipids. Chemically, they are usually tetraterpenoids composed of a 40-carbon atom polyene skeleton, which is either acyclic or terminated by one or two cyclic end groups. In nature, carotenoids protect cell membranes against damage by light, oxidation, and free radicals (Thanapimmetha et al. 2017); further, several carotenoids have been proved to play an important role in the prevention of human diseases and maintaining good health, due to antioxidant and anticancer properties (Rao and Rao 2007). Moreover, in humans and animals, α-carotene, β-carotene, and β-cryptoxanthin are precursors of vitamin A; thus, carotenoids are widely used as food additives, as well as pharmaceuticals, poultry, and cosmetic products (Saini and Keum 2017). Nowadays, there is an increasing demand for “natural” carotenoids to replace the chemically synthesized ones due to the impelling scientific interest towards green technologies and the public awareness about the possible toxicity of synthetic food additives. The microbial production of carotenoids is considered as a promising alternative, as carotenoid-rich microbial biomass can be grown in bioreactors using inexpensive substrates and under controllable culture conditions to maximize production (Agarwal and Rao 2000; Thanapimmetha et al. 2017).
Many Rhodococcus spp. strains can produce different types of carotenoid pigments (Table 1) (Takaichi et al. 1990; Tao et al. 2004), which are located either intracellularly (e.g., in lipid droplets or in the vicinity of the plasma membrane) or around the hydrophobic Rhodococcus cell wall. As they are non-photosynthetic bacteria, the role of carotenoids was generally associated with cell protection from various oxidative damages, as demonstrated by testing cell response to H2O2 treatment and to methylene blue-sensitized photo-oxidation (Osawa et al. 2011; Bequer Urbano et al. 2014). Further, the increase of carotenoid accumulation in these bacteria or the modification of carotenoid profile was associated with the type of growth and the presence of organic solvents. Indeed, carotenoids were suggested to have an antioxidant role in Rhodococcus cells grown as biofilm, whose development could induce an oxidative stress in individual cells within the community due to an increase of reactive oxygen species (ROS) levels (Zheng et al. 2013; Kundu et al. 2015). Rhodococcus erythropolis IBBPo1 cells exposed for 1 or 24 h to 1% alkanes instead showed a modification of the carotenoid profile with an increased level of lycopene (the intermediate in the biosynthesis of γ-carotene and β-carotene) as compared to the control, likely as a response to these organic solvents (Stancu 2015). Lastly, carotenoid formation in a Rhodococcus strain isolated from Azolla symbiotic cavities was reported to be induced by cells’ exposition to the light, which led to a sevenfold higher accumulation of a yellowish-orange pigment than dark-incubated cells (Cohen et al. 2004).
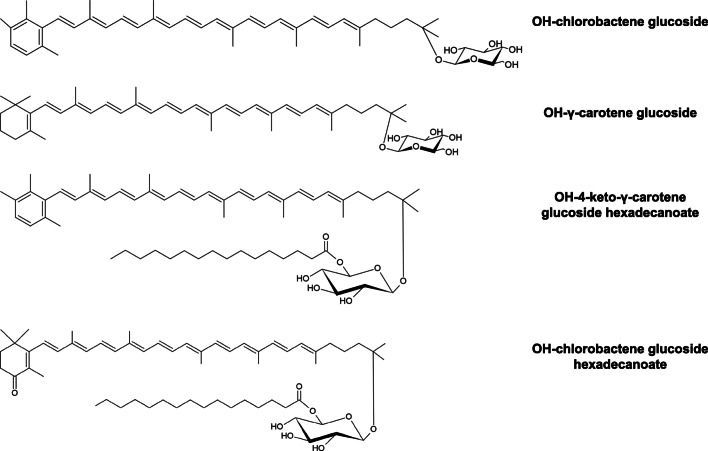
The first study on the ability of Rhodococcus strains to produce carotenoids was performed by Ichiyama et al. (1989), who cultivated 16 Rhodococcus strains, each belonging to a different species, using glycerol as the main carbon source, at pH 7 and at 37 or 28 °C (Ichiyama et al. 1988, 1989). Based on thin-layer chromatography, the strains produced 11 different carotenoid-type pigments, whose nature was only partly identified. Nevertheless, Rhodococcus species were divided into three groups depending on the type of pigment synthesized, i.e., β-carotene, a γ-carotene-like substance, or neither of these carotenes; some species also produced derivatives of β- and γ-carotene, such as myxoxanthophyll-like, zeaxanthin-like, and β-citraurin-like carotenoids. In this work, the biosynthesis of β-carotene was suggested to originate from a conversion of γ-carotene. The presence of the enzyme catalyzing the transformation of γ-carotene into β-carotene distinguished the species under analysis (Ichiyama et al. 1989). Among these, R. maris and R. ruber were subsequently reported as β-carotene (bicyclic carotenoid) producers without being further characterized (Ichiyama et al. 1989). On the other hand, R. rhodochrous RNMS1 and R. erythropolis AN12 were described to produce only monocyclic carotenoids such as γ-carotene derivatives (Takaichi et al. 1990; Tao et al. 2004). This aspect was related in the AN12 strain with the peculiar activity of the lycopene β-monocyclase encoded by the crtLm gene, which produced almost exclusively γ-carotene from lycopene. In relation to this, Rhodococcus is one of the few bacterial genera, which are able to produce only monocyclic carotenoids such as γ-carotene derivatives. More frequently, monocyclic carotenoids are part of the mixture formed during the synthesis of bicyclic carotenoids (e.g., β-carotene). The asymmetrically acting lycopene β-cyclase from AN12 strain is therefore of great biotechnological significance to obtain asymmetric carotenoids of commercial interest, which are generally very difficult to produce chemically (Tao et al. 2004). An additional work by Tao and Cheng (2004) further described the remaining of the carotenoid synthesis genes (crt genes) in AN12. Among these genes, crtO encodes a β-carotene ketolase, whose heterologous expression in an Escherichia coli strain accumulating β-carotene resulted in the production of canthaxanthin (β,β-carotene-4,4′-dione) (Tao and Cheng 2004), which is an orange-to-red keto-carotenoid exhibiting strong antioxidant activities and the ability to reduce and protect against UV light-induced tumor/types of cancers (Rao and Rao 2007). Additionally, members of genus Rhodococcus are among the few selected microbes producing carotenoids with conjugated keto functions (Ichiyama et al. 1989). In particular, keto-carotenoids are characterized by high polarity which facilitates their absorption and distribution once ingested and positively influences their antioxidant activity in membranes (Borroni et al. 2017). Recently, during a screening for antioxidative carotenoids of bacterial origin, a novel aromatic carotenoid (OH-chlorobactene glucoside hexadecanoate) and rare carotenoids (OH-chlorobactene glucoside, OH-carotene glucoside, and OH-4-keto-carotene glucoside hexadecanoate) were found to be produced by the orange-pigmented Rhodococcus sp. CIP isolated from a soil sample (Fig. 2). The produced carotenoids showed potent antioxidative activities, as indicated by analyzing the 1O2 quenching abilities of the compounds. The biosynthetic pathway for these promising carotenoids showed the γ-carotene as the main metabolic intermediate, which is further modified by the activities of sequential enzymes, such as a desaturase (CrtU), a hydroxylase (CrtC), and a glucosyltransferase (CruC), an acyltransferase (CruD), or a β-carotene ketolase (CtrO) (Osawa et al. 2011).
Fig. 2.
Chemical structures of the main carotenoids produced by Rhodococcus sp. 1CP
In the view of setting the bases for a possible industrial process, the carotenoid content of the biomass, the culture productivities, and biomass yields were evaluated in batch and fed-batch cultures of R. opacus PD630 using glycerol and ammonium acetate as cheap carbon and nitrogen sources, respectively. These experiments indicated that carotenoid production was dependent on the biomass growth, while no effect was observed upon modifications of the growth medium pH (Thanapimmetha et al. 2017). Under similar growth conditions, adding glucose as a co-substrate in a glycerol-based culture medium and controlling the nitrogen supply improved the production of biomass, lipid, and carotenoids. Particularly, a repeated fed-batch operation with controlled feeding of carbon- and nitrogen-substrates increased the productivity of carotenoids in PD630 up to 3.5-fold, and a 5-fold increase in the final lipid concentration (Suwaleerat et al. 2017). In addition to the use of glycerol as cheap substrate, organic contaminants are promising carbon sources for the sustainable production of carotenoids using Rhodococcus spp. strains. In this respect, a few studies have reported the capacity of Rhodococcus spp. strains to accumulate carotenoids growing on hydrocarbon and nitroaromatic compounds (Table 1) (Stancu 2015; Kundu et al. 2015).
Intracellular accumulation of storage compounds
Rhodococcus spp. strains are able to synthesize and accumulate storage lipids in the form of polyhydroxyalkanoates (PHAs) (Anderson and Dawes 1990), triacylglycerols (TAGs) (Alvarez et al. 1996), or wax esters (WEs) (Lanfranconi and Alvarez 2017; Alvarez and Steinbüchel 2019) as an adaptation response to specific growth conditions and nutrients.
In addition to the accumulation of lipid-based molecule, Rhodococcus spp. can also produce inclusions enriched in polyphosphate (polyP) and/or carbohydrates (i.e., glycogen and trehalose) (Hernández et al. 2008). In this respect, the microbial polyP production has been described as a promising approach to remove or recover phosphate from wastewaters (Wang et al. 2018). PolyP granules are formed as a response to unfavorable environmental conditions, such as osmotic and oxidative stresses, low environmental phosphate load, desiccation, and heat shock. Indeed, oligotrophic growth conditions determined the accumulation in R. erythropolis N9T-4 of polyP granules, which were named oligobodies due to their analogies with acidocalcisomes of eukaryotic cells (Yoshida et al. 2017; Presentato et al. 2018a). Similar polyP granules were intracellularly generated by R. aetherivorans BCP1 cells upon utilization of cyclohexane carboxylic acid (CHCA, a naphthenic acid molecule) as the sole carbon and energy source (Fig. 3). Recently, the intracellular accumulation of polyP was proposed as an energy sink to replace ATP, which may have a regulatory effect towards specific enzymatic activities (Hernández et al. 2008). On the other hand, the accumulation of glycogen and trehalose in Rhodococcus was considered a carbon and energy storage strategy occurring in response to either N-limiting conditions of growth or hyperosmotic stress (Hernández et al. 2008).
Fig. 3.
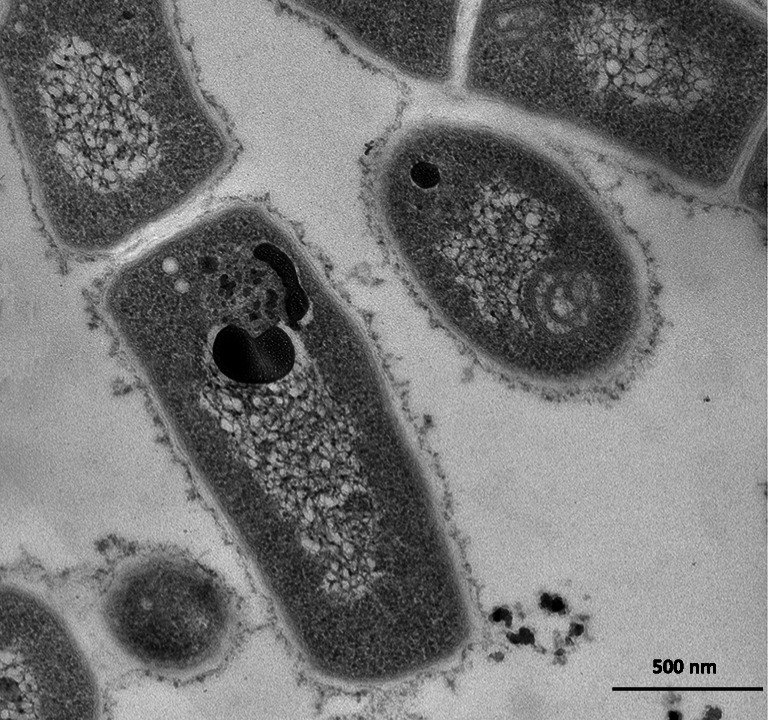
Transmission electron microscopy (TEM) image of Rhodococcus aetherivorans BCP1 cells grown on naphthenic acids. The electron-dense intracellular body represents a possible polyP granule (from Presentato et al. 2018a)
Triacylglycerols
The ability to accumulate triacylglycerols (TAGs, glycerol esters with long-chain length fatty acids) and wax esters (WE, esters of primary long-chain fatty acids and primary long-chain fatty alcohols) has been reported for only some bacterial genera including Streptomyces, Rhodococcus, Mycobacterium, and Nocardia (Alvarez et al. 2019). Bacterial TAGs were proposed to function as reserve compounds for carbon and energy, although other roles have been also discussed in relation to the regulation of cellular membrane fluidity and with the sink for reducing equivalents (Alvarez and Steinbuchel 2002). These lipids have relevant applications in the production of food additives, cosmetics, lubricants, oleochemicals, candles, and biofuels (Alvarez and Steinbüchel 2002). Alternative sources for TAG production are derived from the agriculture, although the advantages of using microorganisms over agricultural sources are numerous and mainly relating to the high variability of the fatty acid composition produced, as well as the better accessibility of microorganisms to genetic and metabolic engineering.
Among the TAG accumulating genera, Rhodococcus is one of the most promising since bacterial strains of this genus can accumulate significant amounts of lipids (above 20% of the cell dry weight), being for this reason referred to as “oleaginous” (Table 1). In particular, R. opacus and R. jostii strains are considered oleaginous models as they produce large amounts of TAGs (up to 87% of the cellular dry weight in R. opacus PD630) (Fig. 4) (Alvarez et al. 1996; Alvarez and Steinbüchel 2002; Alvarez 2016), which are visible inside the cells as insoluble inclusions surrounded by a thin membrane (Alvarez et al. 1996; Alvarez 2003). They also showed the ability to accumulate unusual acyl moieties in TAGs, such as phenyldecanoic acid (Alvarez et al. 2002).
Fig. 4.
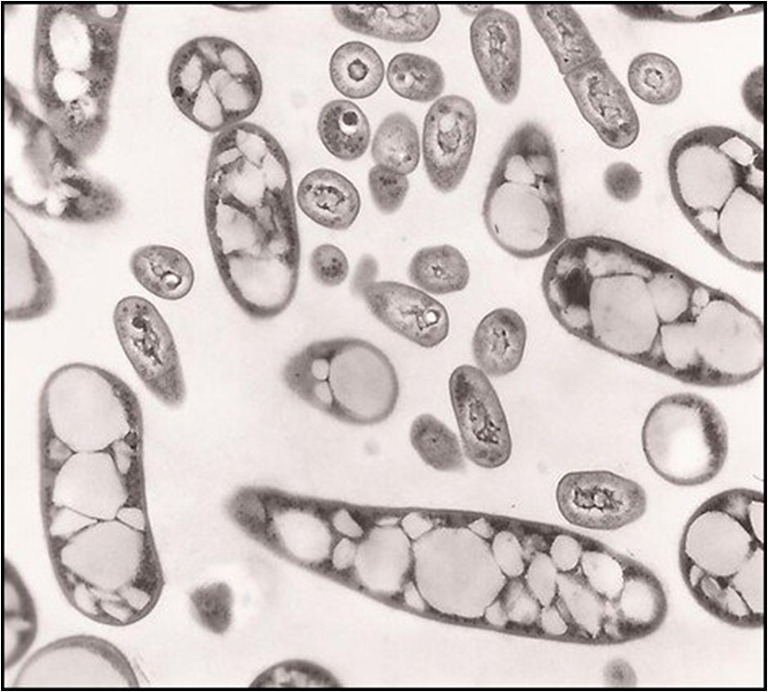
Transmission electron microscopy (TEM) image of Rhodococcus opacus PD630 cells containing TAG granules (from Alvarez et al. 2013)
As compared to R. opacus and R. jostii, R. fascians, R. erythropolis, and R. equi generally produced lower amounts of TAGs during cultivation on sugars, organic acids, or hydrocarbons (Table 1) (Alvarez et al. 1997; Alvarez 2003; Herrero et al. 2018). The different biotransformation performance is due to genetic and physiological differences among Rhodococcus spp. strains. The high lipid production performance of R. opacus, R. jostii, and R. wratilaviensis was attributed to their large genomes and high number of genes coding for transporters and enzymes involved in the lipid metabolism (Cappelletti et al. 2019a). On the other hand, R. fascians and R. erythropolis were the most efficient species in TAG accumulation from glycerol. Despite the smaller genome, these Rhodococcus spp. possessed the genes glpFK1D1, which are involved in glycerol degradation and are absent in R. opacus and R. jostii (Herrero et al. 2016).
In addition to the species/strain itself, other factors influence the amount, composition, and structure of lipids in Rhodococcus such as the carbon source used, the time of cultivation, and the amount of carbon and nitrogen present in the culture medium (Alvarez et al. 1997; Wältermann et al. 2005). Different Rhodococcus spp. strains have shown the ability to produce and accumulate TAG using different substrates including defined carbons sources like sugars and organic acids (Alvarez et al. 1996, 1997, 2000; Silva et al. 2010), single organic contaminants (e.g., aliphatic and aromatic hydrocarbons) (Alvarez et al. 1997; Silva et al. 2010) but also complex carbon sources present in agro-industrial wastes, such as sugar beet molasses, orange, olive mill wastes, whey, oil palm biomass and corn stover (Voss and Steinbuchel 2001; Kurosawa et al. 2014; Bhatia et al. 2017; Herrero et al. 2018).
The growth substrate supplied to the culture medium influenced the amount and chemical nature of the accumulated TAGs. For instance, in R. opacus PD630 culture, the supply of growth substrates which are intermediates of the TCA cycle (i.e., citrate and succinate) or which are delivered to the TCA cycle, i.e., acetate, and odd-numbered organic acids (i.e., propionate or valerate), induced an increase of the fraction of odd-numbered fatty acids (both saturated and mono-unsaturated, with chain length of C15 and C17) in TAG compared to the growth on fructose or gluconate (Alvarez et al. 1997). Conversely, when the cells were cultivated on n-alkanes, the accumulated fatty acids were related to the carbon skeleton of the respective alkane used for growth. For instance, when R. opacus PD630 cells were cultivated on hexadecane as the sole carbon source, palmitic acid (C16:0) was the predominant fatty acid occurring in cells, whereas during PD630 growth on pentadecane, cells accumulated only odd-numbered fatty acids directly related to the carbon chain of the specific alkane and to its β-oxidation derivatives (Alvarez 2003). These results suggested that β-oxidation pathway does not only represent a catabolic route in oleaginous Rhodococcus spp. strains, but it is also a source of fatty acids. Further, the alkanes that are incorporated into cellular lipids are not completely degraded to acetyl-CoA (Alvarez 2003; Cappelletti et al. 2019b). These studies highlighted the possibility to drive lipid storage to specific branched-chain and odd-numbered fatty acids in R. opacus by only changing the growth substrate, making this species a valuable TAG producer for next-generation biofuels (Tsitko et al. 1999).
The limitation of the nitrogen source in the presence of an excess of carbon in the culture medium was reported to be the main trigger for TAG accumulation, causing an imbalance between C and N in cells, which redirects the carbon metabolism to lipogenesis (Alvarez et al. 2000). Early studies described the improved capacity of R. opacus PD630 to accumulate TAGs when the cells were growing under nitrogen limiting conditions (ca. 20 times less nitrogen than control) and in the presence of high amounts of gluconate (Alvarez et al. 2000). These results indicated that, in contrast to what was observed in many bacteria, PD630 cells do not block lipid metabolism under growth-limiting conditions and generate acyl-residues from the available carbon source leading to TAG production. Yet, TAGs were mobilized and used as carbon and energy source when R. opacus PD630 cells were incubated under carbon starvation conditions in the presence of a nitrogen source (Alvarez et al. 2000; Alvarez and Steinbüchel 2002; Alvarez 2019). Finally, TAG biosynthesis was dependent on the time of bacterial cultivation and, therefore, the growth phase. Indeed, R. opacus PD630 demonstrated shifts in the composition of TAGs during the progression from exponential to stationary phase under unbalanced growth conditions (Alvarez et al. 2000).
Due to the capability of Rhodococcus spp. strains to convert lignin-derived aromatic compounds (e.g., 4-hydroxybenzoate, benzoate, phenol, vanillate, guaiacol, and trans-p-coumaric acid, p-coumaric acids, cresol, and 2,6-dimethoxyphenol) into TAGs, members of this genus are promising microbial hosts for lignocellulosic biomass conversion into biofuels (Kosa and Ragauskas 2012; Bhatia et al. 2019). Diverse genetic, physiological, and biochemical studies along with more recent “omic” works provided indications on the metabolic pathways and regulatory mechanisms involved in the biosynthesis and accumulation of TAGs from different carbon sources, including lignocellulose biomasses (Chen et al. 2014; Dávila Costa et al. 2015). On the basis of these studies, genetic and metabolic engineering strategies were proposed to develop sustainable processes of biofuel production using Rhodococcus spp. strains, mostly R. opacus PD630 and R. jostii RHA1 (Castro et al. 2016; Anthony et al. 2019). In this context, R. opacus PD630 and R. jostii RHA1 cells expressing specific heterologous genes, e.g., xylA and xylB from Streptomyces lividans TK23, and bglABC operon from Thermobifida fusca, were shown to acquire the capacity to degrade cellulose, arabinose, and xylose from lignocellulosic biomass and to simultaneously produce lipids (Table 1) (Hetzler et al. 2013; Xiong et al. 2012, 2016a, 2016b; Hernández et al. 2015; Kurosawa et al. 2013). Through Adaptive Laboratory Evolution (hereafter: ALE) procedures, a series of R. opacus PD630 strains were generated, which were able to better utilize aromatics and produce lipids, to consume multiple carbon sources simultaneously (i.e., glycerol, glucose, and xylose in R. opacus MITGM-173, Table 1), and to better tolerate inhibitors (e.g., phenolic compounds), which are usually present in lignocellulosic hydrolysates (Kurosawa et al. 2015a, 2015b; Yoneda et al. 2016; Henson et al. 2018). Genome-based manipulation strategies based on CRISPR-Cas9 and recombineering, were successfully applied in Rhodococcus spp. strains, opening new frontiers for genetic and metabolic engineering of members of this genus aimed at optimizing biofuel production from lignocellulose bioconversion (DeLorenzo et al. 2018; Liang et al. 2020). In order to implement metabolic engineering strategies for waste bioconversion into valuable compounds, a genome-scale model was also developed in R. jostii RHA1 to describe and predict the accumulation rate of three types of carbon storage compounds (i.e., glycogen, polyhydroxyalkanoate, and triacyloglycerols) using different carbon sources (glucose or acetate) and under growth conditions typically occurring in activated sludge bioreactor systems for wastewater recovery (Tajparast and Frigon 2015, 2018).
Polyhydroxyalkanoates
Polyhydroxyalkanoates (PHAs) are polyesters of various hydroxycarboxylic acids, which are produced by a variety of bacterial species generally under nutrient-restricting conditions in the presence of carbon in excess (Anderson and Dawes 1990). They are accumulated by bacteria as intracellular hydrophobic inclusions as carbon and energy storage or as electron sinks of redundant reducing power (Muhammadi et al. 2014). Accumulated PHAs are typically degraded by intracellular depolymerases and metabolized as carbon and energy sources as soon as the supply of the limiting nutrient is restored (Steinbüchel et al. 1992). Based on the type of monomer(s) constituent, PHAs can be homopolymers (e.g., polyhydroxybutyrate [PHB]), copolymers (e.g., poly(3-hydroxybutyrate-co-3-hydroxyvalerate [poly(3HB-co-3 HV) or PHBV]), or terpolymers (e.g., poly(3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate) [P-3HB-co-3 HV-co-3HHx]) (Haywood et al. 1991). These polymers show good biodegradability, insolubility in water, non-toxicity, thermoplasticity, and/or elastomericity, which support their suitability for applications in the packaging industry, medicine, pharmacy, agriculture, food, and chemical industries (Anderson and Dawes 1990). Indeed, PHA is considered the green polymers of the future, since they are expected to gradually substitute conventional plastics (e.g., polypropylene (PP) and low-density polyethylene (LDPE)) with similar physicochemical, thermal, and mechanical properties (Kourmentza et al. 2017). Since the chemical synthesis of PHA is not feasible, microorganisms represent the only source for these polymers.
Members of Rhodococcus genus are able to synthesize PHAs in variable amounts and composition depending on the bacterial strain and the carbon source (Haywood et al. 1991; Pieper and Steinbüchel 1992; Alvarez et al. 1997a,b; Alvarez and Steinbüchel 2002; Alvarez 2003) (Table 1, Fig. 5). One of the most extensively studied strains is R. ruber NCIMB 40126 which was found to be able to accumulate a peculiar PHA containing a copolymer of 3-hydroxyvalerate (3 HV) and 3-hydroxybutyrate (3HB) monomer units (i.e., poly(3HB-co-3 HV) or PHBV) upon cultivation under N-limiting conditions in the presence of different substrates such as acetate, lactate, succinate, fructose, glucose, and molasses (Table 1). In particular, this strain, as well as other members of genus Rhodococcus (e.g., R. aetherivorans IAR), showed the peculiar capacity to synthesize the commercially important PHBV from single carbon sources which were characterized by chemical structure unrelated with that of the copolymer (Hori et al. 2009). The relative amount of each monomer changed depending on the carbon source, although HV was always predominant over HB (HV content range 60–91%) (Haywood et al. 1991). Several Rhodococcus strains, including NCIMB 40126 and R. aetherivorans IAR, NCIMB 40126 was also capable of incorporating 4HB and 5 HV monomer units into PHAs when provided with substrates which are precursors of these monomers (i.e., 4-hydroxybutyrate and 5-chlorovalerate, respectively) (Haywood et al. 1991). Further, this R. ruber strain accumulated almost pure poly(3-hydroxyvalerate) when supplied with valeric acid and 2-pentenoic acid, while when hexanoate or 2-hexenoate were used as substrates, a PHA containing 3HB, 3 HV, and 3-hydroxyhexanoate (3HHx) monomer units was produced (Haywood et al. 1991) (Fig. 5). In a different study performed by Alvarez et al. (1997b), R. ruber NCIMB 40126 showed higher accumulation of PHA than TAGs on valerate and glucose, whereas it seemed to accumulate only TAGs on hexadecane (Table 1) (Alvarez 1997b). R. fascians D 188–5, R. erythropolis DSMZ 43060, and R. opacus MR22 accumulated homopolymers of polyhydroxybutyrate (PHB) in addition to minor amounts of diacylglycerols and wax esters from various carbon sources, although the main carbon storage was TAG (Alvarez 1997b). R. opacus PD630 has been described to accumulate significant amounts of TAGs but not PHA from various carbon sources (Alvarez et al. 1996).
Fig. 5.
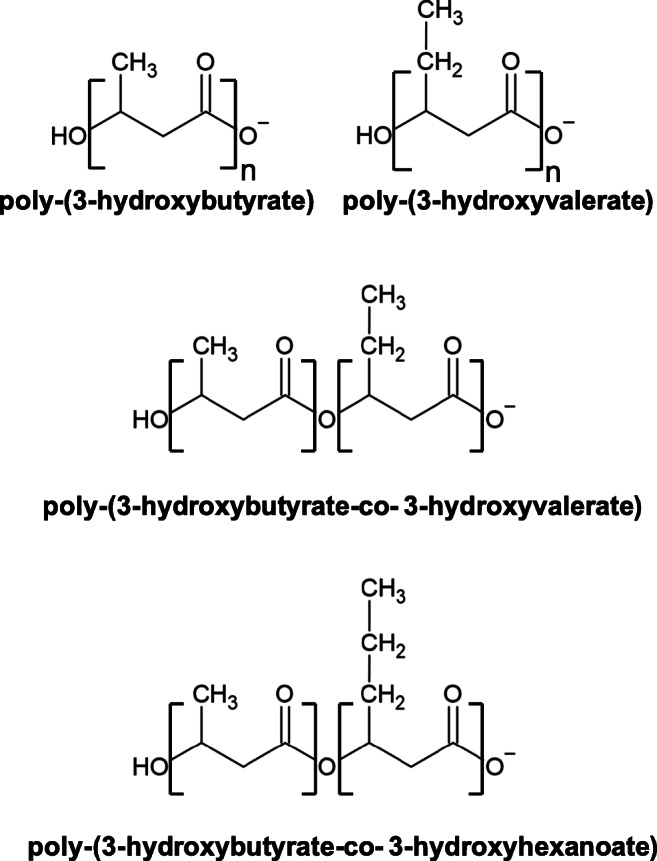
Chemical structures of the main polyhydroxyalkanoates (PHAs) produced by Rhodococcus spp. strains
The possibility to combine biodegradation of toxic and contaminant molecules with PHA production was investigated to achieve a cost reduction of the biodegradable plastic production process, along with the effective utilization of wastes or toxic compounds. In this context, R. aetherivorans IAR was able to produce PHBV from toluene, which is the volatile organic compound (VOC) most abundantly emitted in the environment of some countries (e.g., Japan) (Hori et al. 2009). R. aetherivorans BCP1 accumulated electron-transparent intracellular inclusions composed by neutral lipids during the growth on naphthenic acid models (Presentato et al. 2018a). Although the chemical nature of these inclusion bodies is still undefined, recent experiments showed that, based on the different NAs used for growth, BCP1 cells accumulated either PHB or poly(3HB-co-3 HV) (Cappelletti, unpublished results).
Notably, IAR strain could also accumulate TAG along with PHA on toluene and acetate (Hori et al. 2009), being both PHBV and TAGs simultaneously synthesized and accumulated before the nitrogen source was exhausted. Subsequently, the accumulation of both the storage compounds continued, whereas only the TAG was synthesized even after the carbon source depletion, although at a low rate. This lipid production profile was different from that of R. ruber NCIMB 40126 cells growing on glucose, during which only the accumulation of PHBV started during the exponential growth phase (Alvarez et al. 2000). When the complete consumption of the nitrogen source occurred, PHBV content reached a maximum in NCIMB 40126 cells and the biosynthesis and accumulation of TAGs started. These results demonstrated that the accumulation of TAGs continued after carbon source exhaustion in both the strains, whereas, after substrate depletion, PHBV was mobilized and started to be degraded. With the aim of optimizing the bioprocess, this time course study gave indications on the culture and harvesting conditions that resulted in both high cellular content of these storage compounds and PHAs and TAGs of desired composition (Alvarez et al. 2000). The utilization of specific metabolic inhibitors led to the production of either PHA or TAG, defining possible selective biosynthesis strategies (Alvarez et al. 2000). These experiments also demonstrated that the biosynthetic route of PHA and fatty acids in R. ruber NCIMB 40126 compete for the common precursors, acetyl-CoA and propionyl-CoA, during cell growth under storage conditions (Alvarez et al. 2000).
PHA synthases represent the key enzymes of PHA biosynthesis, which catalyze the stereoselective conversion of (R)-3-hydroxyacyl-CoA substrates to PHA with the concomitant release of CoA (Hernández et al. 2008). The phaC gene from R. ruber is the only gene encoding for a PHA synthase identified and cloned from a member of Rhodococcus (Pieper and Steinbüchel 1992). This PhaC enzyme is a short-chain length class I PHA synthase, which comprises only one type of subunit that utilize CoA thioesters of 3-hydroxy fatty acids with 3 to 5 carbon atoms (Rehm 2003). Downstream of the R. ruber phaC, a gene coding for a putative PHA depolymerase (PhaZ) was identified, which is predicted to be the key enzyme for PHA mobilization. In the model strain R. jostii RHA1, three different chromosomal loci including the phaC and phaZ were recognized. Despite being similar to that described in R. ruber, this pha gene organization is different from that of other Gram-negative bacterial strains accumulating short-chain length PHA, which also typically include other genes involved in the PHA biosynthetic pathway such as phaA and phaB (Rehm and Steinbüchel 1999).
Polyunsaturated fatty acids
Polyunsaturated fatty acids (PUFAs) are fatty acids containing more than two double bonds. Important PUFAs include omega-3 and omega-6 (ω-3 FA and ω-6 FA) fatty acids that are named based on the position of the first double bond from the methyl-end in the fatty acid chain. They are critical nutrients for human health, as they modulate brain development and cognition, as well as many diseases, such as cardiovascular disease, cancers, and diabetes (Lee et al. 2016). Furthermore, specific PUFAs are precursors of molecules regulating inflammatory and immune responses (Calder 2013). Humans must acquire PUFAs via foods or nutritional supplements because they do not contain delta-12 desaturases and convert molecules to PUFAs very slowly. Because of the nutraceutical and pharmacological importance, biotechnology methods have been used to clone and introduce numerous desaturases into diverse organisms to yield PUFAs with critical significance in diet. Many groups have investigated the use of marine algae as a commercial source (Patel et al. 2019); however, bioengineered bacterial strains may result in superior producers, due to the large biomass accumulation and high growth rate. In this context, R. opacus was proposed as a host strain of polyunsaturated fatty acid synthase genes (pfa genes from marine bacterial strain Shewanella baltica) expression for PUFA production (Blakie 2015), because of the ability of this species to accumulate high amount of fatty acids.
PUFA biosynthetic process has been mainly described in marine bacteria (Nichols 2003), but recently, Rhodococcus erythropolis and R. aetherivorans strains showed the ability to accumulate PUFAs, likely as a response mechanism associated with specific stress conditions, such as the exposition to a high salt concentration, extremely low temperatures, toxic metals, and the growth on toxic organic compounds (e.g., naphthenic acids) (de Carvalho 2012; de Carvalho et al. 2014; Presentato et al. 2018a). In particular, R. aetherivorans BCP1 cells accumulated significant amounts (around 7% of the total cell fatty acid content) of linoleic acid (omega-6, 9cis,12cis-C18:2) during the growth on the NA, cyclopentane carboxylic acid (CPCA). The molecular mechanisms supporting PUFA accumulation in Rhodococcus spp. strains have not been elucidated yet. Recent RT-qPCR experiments on RNA extracted from R. aetherivorans BCP1 cells grown on naphthenic acids did not clearly show the upregulation of any specific gene among those predicted to encode desaturases and polyketide synthases (Cappelletti, unpublished results).
Antimicrobials
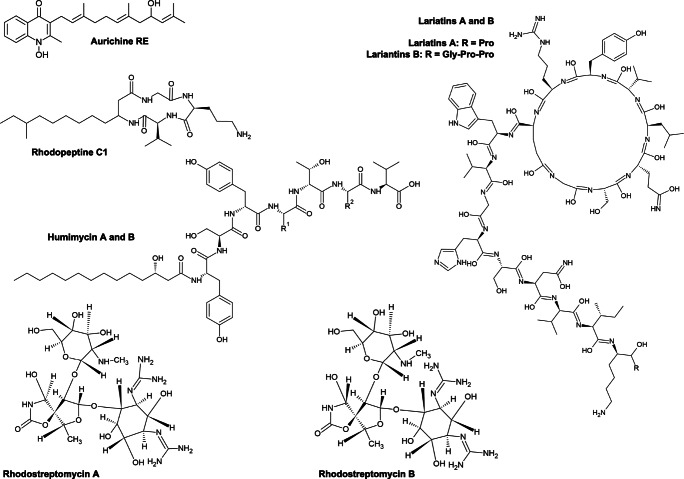
Antibiotics revolutionized the treatment and prevention of numerous types of infections and deadly diseases. As a side effect, many pathogenic bacteria have developed antibiotic resistance, which causes problems in the treatment and threatens modern healthcare. Increasing efforts are in progress to find new antibiotics or antimicrobial strategies to fight these strains. However, it is frequent to rediscover compounds that are already known, and this aspect presently slows down substantial developments in novel drug discovery. More than half of the presently known antibiotics have been isolated from Streptomyces spp. (Nepal and Wang 2019). Despite belonging to the same phylum (Actinobacteria), the knowledge on the production of antimicrobial compounds by Rhodococcus spp. strains is only a very recent matter of study (Kitigawa et al. 2018; Elsayed et al. 2017). Over the last 15 years, a few studies have reported this genus to be a good source of novel antibiotics (Table 1). To date, the lariantin peptide antibiotics, the polyketide aurachin RE, the rhodopeptins, and the humimycins are the natural and important products with antimicrobial activity that have been isolated from Rhodococcus strains (Fig. 6).
Fig. 6.
Chemical structures of the main antibiotics found to be produced up to date by Rhodococcus spp. strains
One of the first studies on antimicrobial production by Rhodococcus reported the strain R. jostii K01-B0171 to be able to produce lariantin A and B, which are antimycobacterial cyclic peptides consisting of 18 and 20 l-amino acid residues, respectively. They are defined as “lasso” peptides because of their structure having a macrolactam ring between the α-amino group of amino-terminal Gly1 and the γ-carboxyl group of Glu8 and a carboxy-terminal tail of the peptide looped back and threaded through the macrolactam ring (Fig. 1). These two peptides selectively inhibited the growth of Mycobacterium smegmatis and Mycobacterium tuberculosis, probably via targeting mycobacterial cell wall synthesis (Iwatsuki et al. 2006, 2007). The main biosynthetic gene cluster of the lariatins in R. jostii K01-B0171 was described to consist of five genes, larABCDE, in which larA encodes the precursor peptide which is post-translationally modified by LarB and LarD to produce lariatin. The larE gene encodes a possible ABC transporter involved in the mature lariatin export (Inokoshi et al. 2012).
By screening 80 Rhodococcus strains from several bacterial culture collections, Kitigawa et al. (2018) showed that 18 strains of five different Rhodococcus species had antimicrobial activities. Among these, a high number of strains belonging to R. erythropolis species were found to produce antibiotics. These R. erythropolis strains were divided into three groups depending on the antibiotic production (Kitigawa et al. 2018). In particular, one of the antibiotic-producing strains, R. erythropolis JCM 6824, was described to produce a new quinoline antibiotic aurachin RE which exhibited a strong activity against a broad range of Gram-positive bacteria. From a chemical point of view, aurachin RE has a structure very similar to aurachin C, which derives from the Gram-negative bacterium Stigmatella aurantiaca Sga15 (Fig. 7). However, compared to aurachin C, aurachin RE exhibited a wide and strong antimicrobial spectrum against both high- and low-GC Gram-positive bacteria (Kitagawa and Tamura 2008). Additional four aurachin molecules were isolated from Rhodococcus sp. Acta 2259 (Nachtigall et al. 2010). These compounds showed inhibitory activities against the growth of numerous Gram-positive bacteria, such as Staphylococcus epidermidis DSM 20044, Bacillus subtilis DSM 347, and Propionibacterium acnes DSM 1897, but not against Gram-negative bacteria (Nachtigall et al. 2010). Conversely, the cyclic tetrapeptide rhodopeptins that were first isolated from Rhodococcus sp. Mer-N1033 exhibited antifungal activity against Candida albicans and Cryptococcus neoformis, but they showed no antibacterial activity (Chiba et al. 1999).
Fig. 7.
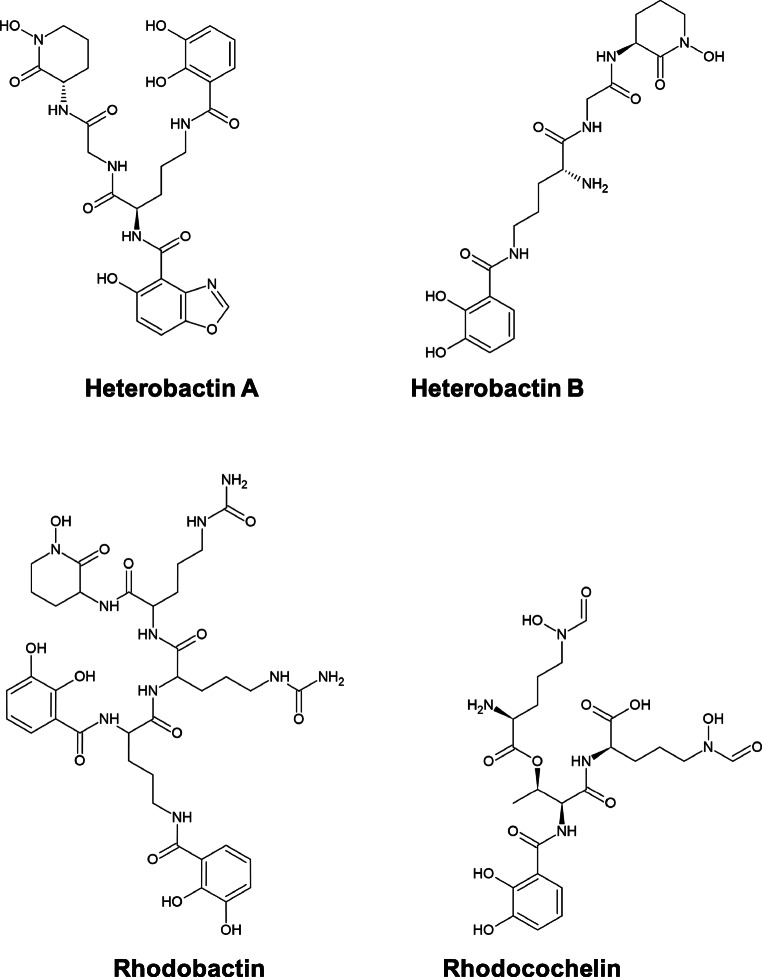
Structures of the main siderophores produced by Rhodococcus spp. strains (modified from Bosello et al. 2012)
The two antibiotics rhodostreptomycin A and B were instead isolated from culture broths of R. fascians 307CO cells after being subject to competitive co-culture experiments with the antibiotic producer Streptomyces padanus. These antibiotics were found to be biosynthesized following horizontal gene transfer of a large DNA segment derived from the Streptomyces strain. From a chemical point of view, rhodostreptomycins were identified as two isomers of a new class of aminoglycosides, which greatly differed in the structure from the Streptomyces-produced actinomycins (Kurosawa et al. 2008). Rhodostreptomycins exhibited good antibiotic activities against an extensive range of Gram-negative and Gram-positive bacteria, such as Streptomyces padanus, Escherichia coli, Staphylococcus aureus, Bacillus subtilis, and Helicobacter pylori. Rhodostreptomycin B was found to be more potent than rhodostreptomycin A, suggesting that their stereochemistry difference influenced the biological activity (Kurosawa et al. 2008) (Fig. 7).
Recently, the first bacteriocin-like molecule and its structural gene (rap) were obtained from Rhodococcus erythropolis JCM 2895. A 5.4-kb circular plasmid harbors rapA and rapB genes encoding for RapA and RapB proteins. RapA is a small, heat-stable, water-soluble protein showing antimicrobial activity against other R. erythropolis strains, while RapB is an immunity protein against RapA that may be located on the cell membrane (Kitigawa et al. 2018). Bacteriocins are usually peptides synthesized by ribosomes and have a narrow spectrum of activity; they are produced to inhibit the growth of competitor organisms in their environment to outcompete for nutrients (Gillor et al. 2008).
Differently to the other antibiotic compounds, the structure of humimycins was initially predicted through a bioinformatic approach (named syn-BNP, standing for synthetic-bioinformatic natural product), which scanned hundreds of bacterial genomes residing in the human body for clusters of genes that were likely to produce non-ribosomal peptides that form the basis of many antibiotics (Fig. 7) (Chu et al. 2016). Gene clusters encoding non-ribosomal peptide synthases producing humimycin A and B were detected from genomes of Rhodococcus equi and R. erythropolis strains. The genome of Rhodococcus enclensis NIO-1009 also showed a biosynthetic gene cluster coding for humimycin B (Ceniceros et al. 2016). The humimycin antibiotics are lipid II flippase inhibitors which hamper bacterial cell wall biosynthesis. They were found to be particularly effective against pathogenic and multidrug-resistant Staphylococcus and Streptococcus strains (Chu et al. 2016).
Siderophores
Siderophores are low-molecular-weight compounds, which are released by some bacteria to chelate extracellular iron and transport it inside the cell. The production and release of siderophores allows bacteria to accumulate an intracellular pool of iron and to satisfy the nutritional iron requirement also under metal-limiting conditions (Kraemer 2004). Siderophores were also reported to bind other essential metals such as zinc, manganese, molybdenum, and vanadium for their acquisition, as well as to interact with heavy metals to prevent their cellular entry (Johnstone and Nolan 2015). Further siderophore functions include the protection against oxidative stress and the enhancement of antibiotic targeting and delivery (Johnstone and Nolan 2015). Based on the chemical structure, siderophores are classified as phenolate, hydroxamate, catecholate, (hydroxy-)carboxylate, and mixed types (Miethke and Marahiel 2007), and their biosynthesis occurs via different mechanisms (Miethke and Marahiel 2007). Areas of siderophore application include agriculture, medicine, pharmacology, bioremediation, biodegradation, and food industry. For instance, siderophores can be used to enhance plant growth due to their uptake by rhizobia, while in bioremediation strategies, siderophores can mobilize heavy metals and radionuclides (De Serrano 2017).
Rhodococcus spp. strains were described to be able to produce chemically diverse siderophores, such as rhodochelin, rhodobactin, heterobactin A, rhequichelin, and rhequibactin (Table 1 and Fig. 7). Carrano et al. (2001) were among the first groups to isolate a new class of siderophores, named heterobactins, from R. erythropolis IGTS8 culture. Among the three heterobactins that were produced by strain IGTS8, the two more abundant were named heterobactin A and B. From a structural point of view, heterobactins were catecholate-hydroxamate mixed-type siderophores, which contained both hydroxamate and catecholate donor groups. Heterobactins were also isolated from R. erythropolis PR4 by Bosello et al. (2013), who, through a bioinformatic analysis of the bacterial genome, identified the gene cluster responsible for heterobactin A’s biosynthesis. The biosynthesis of this siderophore was described to involve the activity of the modular enzymes nonribosomal peptide synthetases (NRPSs), in which each module adds a specific monomer to the peptide backbone. Similarly, a genomic study conducted on the other four Rhodococcus strains showed the presence of a high number of gene clusters encoding for NRPSs compared to other Actinobacteria (Doroghazi and Metcalf 2013).
Analogously to heterobactin, rhodochelin and rhodobactin belong to the hydroxamate–catecholate mixed type family and were isolated from Rhodococcus jostii RHA1 and Rhodococcus rhodochrous OFS, respectively (Dhungana et al. 2007; Bosello et al. 2011). While the gene clusters which determine rhodobactin synthesis were not identified, the gene clusters involved in the synthesis of rhodochelin were described in RHA1. This biosynthetic process involved the functional cross-talk between three distantly located NRPS gene clusters (Bosello et al. 2011), which were identified in most of Rhodococcus spp. strains (Bosello et al. 2011; Bosello et al. 2012).
Additional works assessed the capacity of siderophores produced by R. erythropolis S43 to bind trivalent arsenic, or arsenite [As(III)]. Interestingly, the arsenic-binding activity of the siderophore-like compounds from S43 was higher than the iron-chelating one. Although the capacity of the siderophore to bind and sequester toxic metals could not be clearly associated with arsenic resistance/tolerance mechanisms, the study provided the basis for a first analysis of different actinobacterial strains, including R. erythropolis S43, as a source of arsenic-binding compounds. When overproduced or in association with plants, this type of molecules could be used to decontaminate soil or water, as well as for other potential biotechnological applications (Retamal-Morales et al. 2018b).
Metal(loid)-based nanostructures
Nanotechnology is defined as the world of “very small material” and it is based on the manipulation of matter at either molecular or atomic level (Horikoshi and Serpone 2013). The prefix nano is related to materials that feature at least one dimension in the nanorange (1–100 nm), where physical–chemical properties (i.e., high surface-to-volume ratio, large surface energy, and high spatial confinement) become enhanced as compared to their bulk counterparts (Cao 2004). Metal or metalloid (metal(loid)) nanostructures (NSs) have widespread applications in biomedicine/biotechnologies, energy production, environmental engineering, material science, and optoelectronics (Cao 2004; Horikoshi and Serpone 2013). Different physical–chemical methods efficiently produce high-quality nanomaterials (NMs) of various compositions and morphologies (including nanoparticles (NPs), nanocrystals (NCs), nanorods (NRs), nanowires (NWs), and nanotubes (NTs)) (Rao et al. 2004); however, these approaches mostly rely on dangerous operational conditions (e.g., high temperature and pressure), as well as the use of toxic and harsh chemicals (Zhang et al. 2006; CDC 2014). Moreover, there are still important challenges to be faced in this industry, among which the most urgent are represented by the generation of NSs featuring homogeneous size (i.e., monodispersity) and shape, as well as thermodynamic stability (Piacenza et al. 2018). The natural ability of microorganisms to cope with metal(loid) compounds, simultaneously generating NMs, has taken the form of an innovative and eco-friendly bioprocess.
For the most part, bacterial cells cope and interact with large amounts of metal(loid) compounds by exploiting biosorption, bioaccumulation, and biotransformation processes, which are used for either detoxification or energy purposes (Li et al. 2011; Pantidos and Horsfall 2014). Bacteria belonging to the Rhodococcus genus tolerate the presence of various metal(loid)s, in many cases, due to their biotic conversion of metal(loid)s into a less toxic form. In this context, Rhodococcus spp. have been shown to bioconvert several metal(loid) precursors including gold (Au), silver (Ag), zinc oxide (ZnO), tellurium (Te), selenium (Se), and arsenic (As) synthesizing NMs, which are observed as either intra- or extracellular products, as a function of the strain and growth conditions (Fig. 8) (Ahmad et al. 2003; Otari et al. 2012; Subbaiya et al. 2014; Kundu et al. 2014; Presentato et al. 2016, 2018b, 2018c). The first study reporting rhodococci for metal(loid) NS production highlighted the ability of a Rhodococcus sp. strain to synthesize relatively monodisperse spherical AuNPs having ca. 12 nm as the average diameter and a good crystalline structure, as compared to chemogenic AuNMs (Ahmad et al. 2003). Similarly, Rhodococcus sp. NCIM2891 proved to be an efficient biocatalyst for the generation of intracellular, small (ca. 10 nm), spherical, and crystalline AgNPs (Otari et al. 2012), which were also described both as an antimicrobial, to be used in commercialized textiles and wound dressings, or an antitumor agent (Subbaiya et al. 2014). Iron oxide (Fe2O3) NPs were biosynthesized by R. erythropolis ATCC 427 cultures during biomining processes (Maas et al. 2019a, 2019b). In this context, parameters of the designed implants, as the stirring rate and oxygen flow rate, appeared to determine the size of the produced Fe2O3 NPs, allowing to obtain fairly monodisperse populations (50 or 100 nm), as well as α-Fe2O3 and β-Fe2O3 crystalline structures (Maas et al. 2019a, 2019b).
Fig. 8.
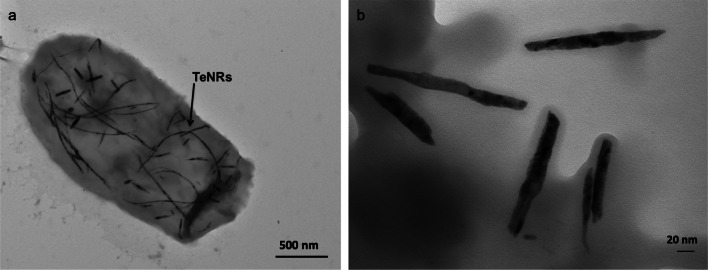
Tellurium-based nanorods (TeNRs) intracellularly produced by Rhodococcus aetherivorans BCP1 and visible inside the cell (a) or in the cell-free extract (after cell sonication) (b)
The role of rhodococci in the microbial nanotechnology field was deeper analyzed by Kundu and co-workers (2014), who focused on biosynthesis of extracellular ZnO NPs by R. pyridinivorans NT2. The produced ZnO NPs showed a quasi-spherical and hexagonal morphology, a large polydispersity (average size 100–120 nm), and a low tendency to aggregate. Nevertheless, these biogenic nanoproducts were as pure and crystalline as those commercially available, showing good absorbance and fluorescence properties (Kundu et al. 2014). Biogenic ZnONPs were also used as a coating for textile cotton surfaces, displaying good UV-blocking, self-cleaning, and antimicrobial properties (Kundu et al. 2014). Finally, these NPs resulted to be efficacious against colon carcinoma cells (HT-29), without showing cytotoxicity against normal peripheral blood mononuclear cells (Kundu et al. 2014).
To date, R. aetherivorans BCP1 represents one of the most versatile and well-known strain belonging to this genus for NM biosynthesis. This fundamental technological trait relies, for the most part, on the ability of this bacterium to handle very high concentrations of the toxic oxyanions TeO32− and SeO32−, biotically converting them in their elemental forms (Te0 and Se0) generating Te or Se NSs, such as NRs or NPs (Figs. 8 and 9). Moreover, by playing with the physiological state of BCP1 cells, as well as the amount of precursor concentration supplied to the culture medium, this strain could improve the average length of the intracellularly produced TeNRs, which featured a regular single-crystalline structure (Fig. 8) (Presentato et al. 2018c). These TeNRs also possessed an electrical conductivity comparable to chemogenic TeNMs, further supporting the technological value of Te-based nanoproducts synthesized by BCP1 cells (Presentato et al. 2018c). Curiously, exposing the same strain to SeO32−, the production of SeNPs and NRs was associated with the cell membrane (Fig. 9) (Presentato et al. 2018b). Another distinctive case is represented by BCP1 incubated with increasing concentrations of arsenate [AsO43-, also referred to as As(V)] in the presence of glucose as the sole carbon and energy source. Here, some preliminary TEM analyses showed the presence of electron-dense hexagonal nanoplates (possibly associated to metal(loid) biosorption) that deserve further analysis (Firrincieli et al. 2019).
Fig. 9.
Selenium-based nanorods (NRs) and nanoparticles (NPs) produced by Rhodococcus aetherivorans BCP1 and visible on the cell membrane (a) or in the cell-free extract (b)
The physical–chemical characterization of the metal(loid) NMs produced by Rhodococcus spp. also highlighted the absence of NS aggregation phenomena under diverse conditions, indicating their high thermodynamic stability and allowing to avoid post-production treatments of these biogenic NMs to ensure their high quality and applicability. This crucial feature seemed to rely on the presence of organic molecules derived from bacterial cells, which may simultaneously act as both electrostatic and steric stabilizers for the NMs in suspension (Piacenza et al. 2018). This aspect was deeply investigated for the ZnO NPs produced by R. pyridinivorans NT2 (Kundu et al. 2014), as well as biogenic Se or Te NMs synthesized by R. aetherivorans BCP1 (Presentato et al. 2016, 2018b, 2018c). Proteins and peptides were found to be important components of the biogenic NM extracts recovered from both rhodococci strains, as tyrosine (Kundu et al. 2014) or tryptophan residues (Presentato et al. 2016, 2018b, 2018c) were detected within the organic material recovered from NT2 or BCP1 cells, respectively. Additionally, intra and/or extracellular heterocyclic metabolites (in NT2 strain; Kundu et al. 2014) and intracellular amphiphilic biomolecules (in BCP1 strain; Presentato et al. 2016, 2018b, 2018c) remained associated with the NMs despite the cells were subjected to several washing steps.
Although rhodococci have been seen so far as good biocatalysts for the synthesis of metal(loid) NMs, yet the biochemical mechanism(s) behind their formation is still poorly understood. Some insights into these bioprocesses have been provided, including indications about biosorption phenomena deriving from electrostatic interactions between the charged metal(loid)s and enzyme functional groups located at the bacterial cell membrane level (Manimaran and Kannabiran 2017). Moreover, NAD(P)H-dependent reductases or secreted reductases were found to be responsible for the direct reduction of metal(loid) ions in different Rhodococcus strains (Manivasagan et al. 2016), including Rhodococcus sp. NCIM2891(Otari et al. 2012), R. pyridinivorans NT2 (Kundu et al. 2014), and R. erythropolis ATCC 427 (Maas et al., 2019). Painter-type reactions involving thiol-containing molecules (e.g., glutathiones and mycothiols) were suggested to mediate the intracellular or membrane-bound bioreduction of metalloid oxyanions, such as SeO32− and TeO32−, by R. aetherivorans BCP1 (Presentato et al. 2016, 2018b, 2018c). In all cases, the bioreduction of metal(loid) precursor results in a highly localized concentration of elemental atoms, which would aggregate with each other to counteract their thermodynamic instability, and eventually assemble forming defined NSs (Ahmad et al. 2003; Adil et al. 2015). This phenomenon seems to be determined by both the thermodynamics of the metal(loid) element itself and the biomolecules in close proximity of the cluster forming elemental atoms (Piacenza et al. 2018). Indeed, the nature and the strength of the interaction occurring between proteins and/or specific enzymes with Au or Zn atoms appeared to control the size, shape, and crystallinity of the Au and ZnO NPs produced by Rhodococcus spp. (Ahmad et al. 2003) and R. pyridinivorans NT2 (Kundu et al. 2014). On the other hand, upon increasing the level of bioconversion of TeO32− within BCP1 cells, Te atoms collapse forming the so-called Te nucleation seeds, whose concentration is directly proportional to the oxyanion bioreduction extent. Once a critical concentration of Te seeds is reached in the intracellular environment, they aggregate firstly forming amorphous TeNPs, which, due to their relative thermodynamic instability, tend to dissolve and deposit along one preferential axis forming TeNRs, whose growth is likely mediated by the amphiphilic biomolecules present within the biogenic TeNM extracts (Presentato et al. 2018c).
Conclusions and perspectives
Members of the Rhodococcus genus have been extensively described for their broad biodegradation activities and unique environmental persistence and robustness, mostly in association with their application in bioremediation strategies. During the last 20 years, the metabolic versatility of Rhodococcus spp. strains have been extended to their biosynthetic capabilities, which can find applications in industrial, environmental, and medical fields, including biofuel generation, metal biorecovery, and novel drug discovery. These biosynthetic pathways are often associated with cell response to challenging conditions of growth, e.g., starvation, hydrophobicity of the substrate and include (i) the production of biosurfactants or bioflocculants that facilitate the contact and metabolism of substrate compounds including organic contaminants, (ii) the generation of carotenoids to counteract oxidative stress, (iii) the accumulation of storage compounds (i.e., TAGs and PHAs), which is enhanced under nitrogen/carbon unbalanced growth conditions, (iv) the production of metal-based nanomaterials to detoxify toxic metal(loids), and (v) the biosynthesis of antimicrobials to outcompete other microorganisms. Some of the compounds produced by Rhodococcus (i.e., biosurfactants, bioflocculants, carotenoids) were demonstrated to have potential application to real industrial settings, due to their higher performance as compared to those produced by other organisms along with their lower toxicity as compared to the chemically synthesized counterpart. Additionally, the production of valuable compounds by Rhodococcus such as TAGs and PHAs could be associated with bioconversion of renewable feedstocks and low-cost wastes making it possible to exploit the metabolic versatility of members of this genus in sustainable and ecofriendly processes. In particular, much attention was paid to the study of lignocellulosic biomass conversion into TAGs by R. opacus PD630, which was proved to enhance to some extent through mutagenesis and genetic modification approaches.
Some challenges are well known and still ongoing for Rhodococcus strains used in industrial settings that are associated with the relatively slow growth, complex cell cycle, and low metabolic activity under non-optimal environmental conditions (Krivoruchko et al. 2019). Nevertheless, the application range of Rhodococcus biosynthetic processes has good chances to strongly increase in the near future thanks to the wide metabolic versatility and peculiar cell persistence under stress conditions of this bacterial genus. In this context, further research is required to expand our knowledge on regulatory mechanisms of Rhodococcus biosynthetic capacities in order to improve the production yields and efficiency but also to extend these studies to a larger number of members of this genus. Indeed Rhodococcus spp. are known to be extraordinarily diversified in terms of genomic contents, metabolic pathways, and evolutionary adaptations, even when considering strains of the same species (Cappelletti et al. 2019b). In this respect, different Rhodococcus spp. strains showed the ability to produce distinct compounds (also depending on the available substrates and the culture conditions) and comparative genomics studies highlighted the biosynthetic potentials of these bacteria that are still understudied (Ceniceros et al. 2017) which might predict new perspectives in terms of bioactive molecules identification and novel natural product discovery. Despite some knowledge is available on the major metabolic pathways involved in a few valuable compound bioproduction (e.g., TAG synthesis and accumulation in R. opacus PD630), our present knowledge on the genetic traits along with the metabolic pathways and regulation mechanisms of most of the biosynthetic activities in Rhodococcus spp. strains, is fragmentary (for biosurfactants and carotenoids), absent (for bioflocculants, antibiotics, metal-based nanostructures) or limited (for TAGs and PHAs) to only those strains, which are considered model strains of this genus (i.e., R. opacus PD630 and R. jostii RHA1) for their genetic tractability. In particular, more extensive multi-omic studies and metabolic engineering strategies of Rhodococcus strains are needed to delve into the major gene clusters involved in the anabolic pathways of interest and to identify specific genetic traits to be targeted for synthetic biology approaches. In this regard, novel genetic toolkits based on CRISPR/Cas9 system and recombineering have been recently applied for metabolic engineering of model strains of this genus (De Lorenzo et al. 2018; Liang et al. 2020), but they are mostly at the proof-of-concept stage and need to be further implemented to be used for the development of Rhodococcus-based microbial cell factories for efficient biotechnology applications. The extension of metabolic and genetic analyses of non-model Rhodococcus spp. strains and the advancements of synthetic biology tools for their genome-scale engineering will provide new opportunities for the use of an increasingly large number of strains of this genus in targeted bioconversion and biodegradation processes.
Author contributions
MC conceived the study and developed the whole manuscript by writing and integrating the different text parts. EP wrote the paragraph about the metal-based nanostructure; AP contributed to this paragraph and prepared the associated figs. AP also contributed to the first draft of the paragraph about TAG and PHA accumulation. AF contributed to the manuscript editing and to preparing the figures showing chemical structures. RJT and DZ revised the manuscript and provided useful suggestions. All authors read and approved the manuscript.
Funding
Open access funding provided by Alma Mater Studiorum - Università di Bologna within the CRUI-CARE Agreement. This study was funded by RFO grants and G.&V. Rossi funding (University of Bologna).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alvarez HM (2016) Triacylglycerol and wax ester-accumulating machinery in prokaryotes. Biochimie 120:28–39. 10.1016/j.biochi.2015.08.016 [DOI] [PubMed]
- Adil SF, Assal ME, Khan M, Al-Warthan A, Siddiqui MR, Liz-Marzán LM. Biogenic synthesis of metallic nanoparticles and prospects toward green chemistry. Dalton Trans. 2015;44:9709–9717. doi: 10.1039/c4dt03222e. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao AV. Carotenoids and chronic diseases. Drug Metabol Drug Interact. 2000;17:189–210. doi: 10.1515/DMDI.2000.17.1-4.189. [DOI] [PubMed] [Google Scholar]
- Ahmad A, Senapati S, Khan MI, Kumar R, Ramani R, Srinivas V, Sastry M. Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete, Rhodococcus species. Nanotechnology. 2003;14:824–828. doi: 10.1088/0957-4484/14/7/323. [DOI] [Google Scholar]
- Alvarez HM. Relationship between β-oxidation pathway and the hydrocarbon-degrading profile in actinomycetes bacteria. Int Biodeterior Biodegradation. 2003;52:35–42. doi: 10.1016/S0964-8305(02)00120-8. [DOI] [Google Scholar]
- Alvarez HM, Steinbüchel A. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol. 2002;60:367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- Alvarez HM, Steinbüchel A. Biology of triacylglycerol accumulation by Rhodococcus. In: Alvarez HM, editor. Biology of Rhodococcus. 2. Switzerland: Springer Nature; 2019. pp. 299–332. [Google Scholar]
- Alvarez HM, Mayer F, Fabritius D, Steinbüchel A. Formation of intracytoplasmic lipid inclusions by Rhodococcus opacus strain PD630. Arch Microbiol. 1996;165:377–386. doi: 10.1007/s002030050341. [DOI] [PubMed] [Google Scholar]
- Alvarez HM, Kalscheuer R, Steinbüchel A. Accumulation of storage lipids in species of Rhodococcus and Nocardia and effect of inhibitors and polyethylene glycol. Fett-Lipid. 1997;99:239–246. doi: 10.1002/lipi.19970990704. [DOI] [Google Scholar]
- Alvarez HM, Kalscheuer R, Steinbüchel A. Accumulation and mobilization of storage lipids by Rhodococcus opacus PD630 and Rhodococcus ruber NCIMB 40126. Appl Microbiol Biotechnol. 2000;54:218–223. doi: 10.1007/s002530000395. [DOI] [PubMed] [Google Scholar]
- Alvarez HM, Luftmann H, Silva RA, Cesari AC, Viale A, Wältermann M, Steinbüchel A. Identification of phenyldecanoic acid as a constituent of triacylglycerols and wax ester produced by Rhodococcus opacus PD630. Microbiology. 2002;148:1407–1412. doi: 10.1099/00221287-148-5-1407. [DOI] [PubMed] [Google Scholar]
- Alvarez HM, Silva RA, Herrero M, Hernández MA, Villalba MS. Metabolism of triacylglycerols in Rhodococcus species: insights from physiology and molecular genetics. J Mol Biochem. 2013;2:2119–2130. doi: 10.1007/s00253-012-4360-1. [DOI] [Google Scholar]
- Alvarez HM, Herrero OM, Silva RA, Hernández MA, Lanfranconi MP, Villalba MS. Insights into the metabolism of oleaginous Rhodococcus spp. Appl Environ Microbiol. 2019;85:8075–8012. doi: 10.1128/AEM.00498-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/MMBR.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony WE, Carr RR, DeLorenzo DM, Campbell TP, Shang Z, Foston M, Moon TS, Dantas G. Development of Rhodococcus opacus as a chassis for lignin valorization and bioproduction of high-value compounds. Biotechnol Biofuels. 2019;12:192. doi: 10.1186/s13068-019-1535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselineau C, Asselineau J, Lanéelle G, Lanéelle MA. The biosynthesis of mycolic acids by Mycobacteria: current and alternative hypotheses. Prog Lipid Res. 2002;41:501–523. doi: 10.1016/S0163-7827(02)00008-5. [DOI] [PubMed] [Google Scholar]
- Banat IM, Franzetti A, Gandolfi I, Bestetti G, Martinotti MG, Fracchia L, Smyth TJ, Marchant R. Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol. 2010;87:427–444. doi: 10.1007/s00253-010-2589-0. [DOI] [PubMed] [Google Scholar]
- Bhatia SK, Kim J, Song HS, Kim HJ, Jeon JM, Sathiyanarayanan G, Yoon JJ, Park K, Kim YG, Yang YH. Microbial biodiesel production from oil palm biomass hydrolysate using marine Rhodococcus sp. YHY01. Bioresour Technol. 2017;233:99–109. doi: 10.1016/j.biortech.2017.02.061. [DOI] [PubMed] [Google Scholar]
- Bhatia SK, Gurav R, Choi TR, Han YH, Park YL, Park JY, Jung HR, Yang SY, Song HS, Kim SH, Choi KY, Yang YH. Bioconversion of barley straw lignin into biodiesel using Rhodococcus sp. YHY01. Bioresour Technol. 2019;289:121704. doi: 10.1016/j.biortech.2019.121704. [DOI] [PubMed] [Google Scholar]
- Blakie B (2015) Production of omega-3 fatty acids in Rhodococcus opacus PD630. Dissertation, University of Otago, New Zealand
- Borroni V, Gonzalez MT, Carelli AA. Bioproduction of carotenoid compounds using two-phase olive mill waste as the substrate. Process Biochem. 2017;54:128–134. doi: 10.1016/j.procbio.2017.01.003. [DOI] [Google Scholar]
- Bosello M, Robbel L, Linne U, Xie X, Marahiel MA. Biosynthesis of the siderophore rhodochelin requires the coordinated expression of three independent gene clusters in Rhodococcus jostii RHA1. J Am Chem Soc. 2011;133:4587–4595. doi: 10.1021/ja1109453. [DOI] [PubMed] [Google Scholar]
- Bosello M, Mielcarek A, Giessen TW, Marahiel MA. An enzymatic pathway for the biosynthesis of the formylhydroxyornithine required for rhodochelin iron coordination. Biochemistry. 2012;51:3059–3066. doi: 10.1021/bi201837f. [DOI] [PubMed] [Google Scholar]
- Bosello M, Zeyadi M, Kraas FI, Linne U, Xie X, Marahiel MA. Structural characterization of the heterobactin siderophores from Rhodococcus erythropolis PR4 and elucidation of their biosynthetic machinery. J Nat Prod. 2013;76:2282–2290. doi: 10.1021/np4006579. [DOI] [PubMed] [Google Scholar]
- Bouchez-Naïtali M, Blanchet D, Bardin V, Vandecasteele JP. Evidence for interfacial uptake in hexadecane degradation by Rhodococcus equi: the importance of cell flocculation. Microbiology. 2001;147:2537–2543. doi: 10.1099/00221287-147-9-2537. [DOI] [PubMed] [Google Scholar]
- Bredholt H, Josefsen K, Vatland A, Bruheim P, Eimhjellen K. Emulsification of crude oil by an alkane-oxidizing Rhodococcus species isolated from seawater. Can J Microbiol. 1998;44:330–340. doi: 10.1139/w98-005. [DOI] [Google Scholar]
- Bredholt H, Bruheim P, Potocky M, Eimhjellen K. Hydrophobicity development, alkane oxidation, and crude-oil emulsification in a Rhodococcus species. Can J Microbiol. 2002;48:295–304. doi: 10.1139/w02-024. [DOI] [PubMed] [Google Scholar]
- Calder PC. Omega-3 polyunsaturated fatty acids and inflammatory processes: nutrition or pharmacology? Br J Clin Pharmacol. 2013;75:645–662. doi: 10.1111/j.1365-2125-2012.04374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. Introduction. In: Cao G, editor. Nanostructures and nanomaterials, synthesis, properties and applications. 1. London: Imperial College Press; 2004. pp. 1–17. [Google Scholar]
- Cappelletti M, Presentato A, Milazzo G, Turner RJ, Fedi S, Frascari D, Zannoni D. Growth of Rhodococcus sp. strain BCP1 on gaseous n-alkanes: new metabolic insights and transcriptional analysis of two soluble di-iron monooxygenase genes. Front Microbiol. 2015;6:393. doi: 10.3389/fmicb.2015.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti M, Fedi S, Zampolli J, Di Canito A, D'Ursi P, Orro A, Viti C, Milanesi L, Zannoni D, Di Gennaro P. Phenotype microarray analysis may unravel genetic determinants of the stress response by Rhodococcus aetherivorans BCP1 and Rhodococcus opacus R7. Res Microbiol. 2016;167:766–773. doi: 10.1016/j.resmic.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Cappelletti M, Frascari D, Pinelli D, Mezzettia F, Fedi S, Zannoni D. Aerobic cometabolism of 1,1,2,2-TeCA by a propane-growing microbial consortium (C2): diversity of alkane monooxygenase genes and design of an on-site bioremediation process. Int Biodeterior Biodegr. 2017;119:649–660. doi: 10.1016/j.ibiod.2016.09.019. [DOI] [Google Scholar]
- Cappelletti M, Pinelli D, Fedi S, Mezzetti F, Zannoni D. Aerobic co-metabolism of 1,1,2,2-tetrachloroethane by Rhodococcus aetherivorans TPA grown on propane: kinetic study and bioreactor configuration analysis. J Chem Technol Biotechnol. 2018;93:155–165. doi: 10.1002/jctb.5335. [DOI] [Google Scholar]
- Cappelletti M, Fedi S, Zannoni D. Degradation of alkanes in Rhodococcus. In: Alvarez HM, editor. Biology of Rhodococcus. 2. Switzerland: Springer; 2019. pp. 137–171. [Google Scholar]
- Cappelletti M, Zampolli J, Di Gennaro P, Zannoni D. Genomics of Rhodococcus. In: Alvarez HM, editor. Biology of Rhodococcus. 2. Switzerland: Springer; 2019. pp. 23–60. [Google Scholar]
- Carrano CJ, Jordan M, Drechsel H, Schmid DG, Winkelmann G. Heterobactins: a new class of siderophores from Rhodococcus erythropolis IGTS8 containing both hydroxamate and catecholate donor groups. Biometals. 2001;14:119–125. doi: 10.1023/A:1016633529461. [DOI] [PubMed] [Google Scholar]
- Castro AR, Rocha I, Alves MM, Pereira MA (2016) Rhodococcus opacus B4: a promising bacterium for production of biofuels and biobased chemicals. AMB Express 6:35. 10.1186/s13568-016-0207-y [DOI] [PMC free article] [PubMed]
- Ceniceros A, Dijkhuizen L, Petrusma M, Medema MH. Genome-based exploration of the specialized metabolic capacities of the genus Rhodococcus. BMC Genomics. 2017;18:593. doi: 10.1186/s12864-017-3966-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention National Institute for Occupational Safety and Health (CDC) (2014) Current strategies for engineering controls in nanomaterial production and downstream handling processes. https://www.cdc.gov/niosh/docs/2014-102/pdfs/2014-102.pdf
- Chen Y, Ding Y, Yang L, Yu J, Liu G, Wang X, Zhang S, Yu D, Song L, Zhang H, Zhang C, Huo L, Huo C, Wang Y, Du Y, Zhang H, Zhang P, Na H, Xu S, Zhu Y, Xie Z, He T, Zhang Y, Wang G, Fan Z, Yang F, Liu H, Wang X, Zhang X, Zhang MQ, Li Y, Steinbüchel A, Fujimoto T, Cichello S, Yu J, Liu P. Integrated omics study delineates the dynamics of lipid droplets in Rhodococcus opacus PD630. Nucleic Acids Res. 2014;42:1052–1064. doi: 10.1093/nar/gkt932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Agematu H, Kaneto R, Terasawa T, Sakai K, Dobashi K, Yoshioka T. Rhodopeptins (Mer-N1033), novel cyclic tetrapeptides with antifungal activity from Rhodococcus sp. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot. 1999;52:695–699. doi: 10.7164/antibiotics.52.695. [DOI] [PubMed] [Google Scholar]
- Christofi N, Ivshina IB. Microbial surfactants and their use in field studies of soil remediation. J Appl Microbiol. 2002;93:915–929. doi: 10.1046/j.1365-2672.2002.01774.x. [DOI] [PubMed] [Google Scholar]
- Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, Reddy BV, Charlop-Powers Z, Zebroski HA, Gallardo-Macias R, Jaskowski M, Satish S, Park S, Perlin DS, Freundlich JS, Brady SF. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat Chem Biol. 2016;12:1004–1006. doi: 10.1038/nchembio.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapina EM, Melo WC, Santa Anna LM, Santos AS, Freire DM, Pereira N., Jr Biosurfactant production by Rhodococcus erythropolis grown on glycerol as sole carbon source. Appl Biochem Biotechnol. 2006;31:880–886. doi: 10.1385/ABAB:131:1:880. [DOI] [PubMed] [Google Scholar]
- Ciavarelli R, Cappelletti M, Fedi S, Pinelli D, Frascari D. Chloroform aerobic cometabolism by butane-growing Rhodococcus aetherovorans BCP1 in continuous-flow biofilm reactors. Bioprocess Biosyst Eng. 2012;35:667–681. doi: 10.1007/s00449-011-0647-3. [DOI] [PubMed] [Google Scholar]
- Cohen M, Meziane T, Yamasaki H. A photocarotenogenic Rhodococcus sp. isolated from the symbiotic fern Azolla. Endocytobiosis Cell Res. 2004;15:350–355. [Google Scholar]
- Dávila Costa JS, Herrero OM, Alvarez HM, Leichert L. Label-free and redox proteomic analyses of the triacylglycerol-accumulating Rhodococcus jostii RHA1. Microbiology. 2015;161:593–610. doi: 10.1099/mic.0.000028. [DOI] [PubMed] [Google Scholar]
- de Carvalho CCCR. Adaptation of Rhodococcus erythropolis cells for growth and bioremediation under extreme conditions. Res Microbiol. 2012;163:125–136. doi: 10.1016/j.resmic.2011.11.003. [DOI] [PubMed] [Google Scholar]
- de Carvalho CCCR, Marques MPC, Hachicho N, Heipieper HJ. Rapid adaptation of Rhodococcus erythropolis cells to salt stress by synthesizing polyunsaturated fatty acids. Appl Microbiol Biotechnol. 2014;98:5599–5606. doi: 10.1007/s00253-014-5549-2. [DOI] [PubMed] [Google Scholar]
- De Serrano LO. Biotechnology of siderophores in high-impact scientific fields. Biomol Concepts. 2017;8:683–611. doi: 10.1515/bmc-2017-0016. [DOI] [PubMed] [Google Scholar]
- DeLorenzo DM, Rottinghaus AG, Henson WR, Moon TS. Molecular toolkit for gene expression control and genome modification in Rhodococcus opacus PD630. ACS Synth Biol. 2018;7:727–738. doi: 10.1021/acssynbio.7b00416. [DOI] [PubMed] [Google Scholar]
- Dhungana S, Michalczyk R, Boukhalfa H, Lack JG, Koppisch AT, Fairlee JM, Johnson MT, Ruggiero CE, John SG, Cox MM, Browder CC, Forsythe JH, Vanderberg LA, Neu MP, Hersman LE. Purification and characterization of rhodobactin: a mixed ligand siderophore from Rhodococcus rhodochrous strain OFS. Biometals. 2007;20:853. doi: 10.1007/s10534-006-9079-y. [DOI] [PubMed] [Google Scholar]
- Doroghazi JR, Metcalf WW. Comparative genomics of Actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics. 2013;14:611. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed Y, Refaat J, Abdelmohsen UR, Fouad MA. The genus Rhodococcus as a source of novel bioactive substances: a review. J Pharma Phytochem. 2017;6:83–92. [Google Scholar]
- Finnerty WR. The biology and genetics of the genus Rhodococcus. Annu Rev Microbiol. 1992;46:193–218. doi: 10.1146/annurev.mi.46.100192.001205. [DOI] [PubMed] [Google Scholar]
- Firrincieli A, Prepentato A, Favoino G, Marabottini R, Allevato E, Stazi SR, Scarascia Mugnozza G, Harfouche A, Petruccioli M, Turner RJ, Zannoni D, Cappelletti M. Identification of resistance genes and response to arsenic in Rhodococcus aetherivorans BCP1. Front Microbiol. 2019;10:888. doi: 10.3389/fmicb.2019.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzetti A, Gandolfi I, Bestetti G, Smyth TJP, Banat IM. Production and applications of trehalose lipid biosurfactants. Eur J Lipid Sci Technol. 2010;112:617–627. doi: 10.1002/ejlt.200900162. [DOI] [Google Scholar]
- George S, Jayachandran K. Production and characterization of rhamnolipid biosurfactant from waste frying coconut oil using a novel Pseudomonas aeruginosa D. J Appl Microbiol. 2013;114:373–383. doi: 10.1111/jam.12069. [DOI] [PubMed] [Google Scholar]
- Gillor O, Etzion A, Riley MA. The dual role of bacteriocins as anti- and probiotics. Appl Microbiol Biotechnol. 2008;81:591–606. doi: 10.1007/s00253-008-1726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Liu J, Yang Y, Zhou Y, Jiang S, Chen C. Fermentation and kinetics characteristics of a bioflocculant from potato starch wastewater and its application. Sci Rep. 2018;8:3631. doi: 10.1038/s41598-018-21796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood GW, Anderson AJ, Williams DR, Dawes EA, Ewing DF. Accumulation of a poly(hydroxyalkanoate) copolymer containing primarily 3-hydroxyvalerate from simple carbohydrate substrates by Rhodococcus sp. NCIMB 40126. Int J Biol Macromol. 1991;13:83–88. doi: 10.1016/0141-8130(91)90053-W. [DOI] [PubMed] [Google Scholar]
- Henson WR, Campbell T, DeLorenzo DM, Gao Y, Berla B, Kim SJ, Foston M, Moon TS, Dantas G. Multi-omic elucidation of aromatic catabolism in adaptively evolved Rhodococcus opacus. Metab Eng. 2018;49:69–83. doi: 10.1016/j.ymben.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Hernández MA, Mohn WW, Martínez E, Rost E, Alvarez AF, Alvarez HM. Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics. 2008;12:600. doi: 10.1186/1471-2164-9-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández MA, Comba S, Arabolaza A, Gramajo H, Alvarez HM. Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus spp. strains. Appl Microbiol Biotechnol. 2015;99:2191–2207. doi: 10.1007/s00253-014-6002-2. [DOI] [PubMed] [Google Scholar]
- Herrero OM, Moncalián G, Alvarez HM. Physiological and genetic differences amongst Rhodococcus species for using glycerol as a source for growth and triacylglycerol production. Microbiology. 2016;162:384–397. doi: 10.1099/mic.0.000232. [DOI] [PubMed] [Google Scholar]
- Herrero OM, Villalba MS, Lanfranconi MP, Alvarez HM. Rhodococcus bacteria as a promising source of oils from olive mill wastes. World J Microbiol Biotechnol. 2018;34:114. doi: 10.1007/s11274-018-2499-3. [DOI] [PubMed] [Google Scholar]
- Hetzler S, Steinbüchel A. Establishment of cellobiose utilization for lipid production in Rhodococcus opacus PD630. Appl Environ Microbiol. 2013;79:3122–3125. doi: 10.1128/AEM.03678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzler S, Bröker D, Steinbüchel A. Saccharification of cellulose by recombinant Rhodococcus opacus PD630 strains. Appl Environ Microbiol. 2013;79:5159–5166. doi: 10.1128/AEM.01214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Abe M, Unno H. Production of triacylglycerol and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by the toluene-degrading bacterium Rhodococcus aetherivorans IAR1. J Biosci Bioeng. 2009;108:319–324. doi: 10.1016/j.jbiosc.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Horikoshi S, Serpone N. General introduction to nanoparticles. In: Horikoshi S, Serpone N, editors. Microwaves in nanoparticle synthesis: fundamentals and applications. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2013. pp. 1–24. [Google Scholar]
- Ichiyama S, Shimokata K, Tsukamura M. Relationship between mycobacterial species and their carotenoid pigments. Microbiol Immunol. 1988;32:473–479. doi: 10.1111/j.1348-0421.1988.tb01407.x. [DOI] [PubMed] [Google Scholar]
- Ichiyama S, Shimokata K, Tsukamura M. Carotenoid pigments of genus Rhodococcus. Microbiol Immunol. 1989;33:503–508. doi: 10.1111/j.1348-0421.1989.tb01999.x. [DOI] [PubMed] [Google Scholar]
- Inaba T, Tokumoto Y, Miyazaki Y, Inoue N, Maseda H, Nakajima-Kambe T, Uchiyama H, Nomura N. Analysis of genes for succinoyl trehalose lipid production and increasing production in Rhodococcus sp. strain SD-74. Appl Environ Microbiol. 2013;79:7082–7090. doi: 10.1128/AEM.01664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokoshi J, Matsuhama M, Miyake M, Ikeda H, Tomoda H (2012) Molecular cloning of the gene cluster for lariatin biosynthesis of Rhodococcus jostii K01-B0171. Appl Microbiol Biotechnol 95 (2):451–460 [DOI] [PubMed]
- Iwabuchi N, Sunairi M, Urai M, Itoh C, Anzai H, Nakajima M, Harayama S. Extracellular polysaccharides of Rhodococcus rhodochrous S-2 stimulate the degradation of aromatic components in crude oil by indigenous marine bacteria. Appl Environ Microbiol. 2002;68:2337–2343. doi: 10.1128/AEM.68.5.2337-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki M, Tomoda H, Uchida R, Gouda H, Hirono S, Omura S. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J Am Chem Soc. 2006;128:7487. doi: 10.1021/ja056780z. [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Uchida R, Takakusagi Y, Matsumoto A, Jiang CL, Takahashi Y, Arai M, Kobayashi S, Matsumoto M, Inokoshi J, Tomoda H, Omura S. Lariatins, novel anti-mycobacterial peptides with a lasso structure, produced by Rhodococcus jostii K01-B0171. J Antibiot. 2007;60:357–363. doi: 10.1038/ja.2007.48. [DOI] [PubMed] [Google Scholar]
- Jiang J, Liu L, Nie W, Chen Y, Wang Z. Screening of a high bioflocculant-producing bacterial strain from an intensive fishpond and comparison of the bioflocculation effects with Rhodococcus erythropolis. Aquac Res. 2019;50:1047–1056. doi: 10.1111/are.13977. [DOI] [Google Scholar]
- Jiao S, Li F, Yu H, Shen Z. Advances in acrylamide bioproduction catalyzed with Rhodococcus cells harboring nitrile hydratase. Appl Microbiol Biotechnol. 2020;104:1001–1012. doi: 10.1007/s00253-019-10284-5. [DOI] [PubMed] [Google Scholar]
- Johnstone TC, Nolan EM. Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi K, Zhang B, Lye LM (2009) Production of biosurfactant by Rhodococcus erythropolis sp. cultivated in a novel fish waste compost extract substrate. Conference: CSCE Annual Conference At: London, Canada
- Kim JS, Powalla M, Lang S, Wagner F, Lunsdorf H, Wray V. Microbial glycolipid production under nitrogen limitation and resting cell conditions. J Biotechnol. 1990;13:257–266. doi: 10.1016/0168-1656(90)90074-L. [DOI] [PubMed] [Google Scholar]
- Kim D, Choi KY, Yoo M, Zylstra GJ, Kim E. Biotechnological potential of Rhodococcus biodegradative pathways. J Microbiol Biotechnol. 2018;28:1037–1051. doi: 10.4014/jmb.1712.12017. [DOI] [PubMed] [Google Scholar]
- Kitagawa W, Tamura T. Three types of antibiotics produced from Rhodococcus erythropolis strains. Microbes Environ. 2008;23:167–171. doi: 10.1264/jsme2.23.167. [DOI] [PubMed] [Google Scholar]
- Kitigawa W, Mitsuhashi S, Hata M, Tamura T. Identification of a novel bacteriocin-like protein and structural gene from Rhodococcus erythropolis JCM 2895, using suppression-subtractive hybridization. J Antibiot. 2018;71:872–879. doi: 10.1038/s4129-018-0078-3. [DOI] [PubMed] [Google Scholar]
- Kosa M, Ragauskas AJ. Bioconversion of lignin model compounds with oleaginous rhodococci. Appl Microbiol Biotechnol. 2012;93:891–900. doi: 10.1007/s00253-011-3743-z. [DOI] [PubMed] [Google Scholar]
- Kourmentza C, Placido J, Venetsaneas N, Burniol-Figols A, Varrone C, Gavala HN, Reis MAM. Recent advances and challenges towards sustainable polyhydroxyalkanoate (PHA) production. Bioengineering. 2017;4:55. doi: 10.3390/bioengineering4020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM. Iron oxide dissolution and solubility in the presence of siderophores. Aquat Sci. 2004;66:3–18. doi: 10.1007/s00027-003-0690-5. [DOI] [Google Scholar]
- Kretschmer A, Wagner F. Characterization of biosynthetic intermediates of trehalose dicorynomycolates from Rhodococcus erythropolis grown on n-alkanes. Appl Environ Microbiol. 1983;44:864–870. doi: 10.1016/0005-2760(83)90053-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivoruchko A, Kuyukina M, Ivshina I. Advanced Rhodococcus biocatalysts for environmental biotechnologies. Catalysts. 2019;9:236–236. doi: 10.3390/catal9030236. [DOI] [Google Scholar]
- Kundu D, Hazra C, Chatterjee A, Chaudhari A, Mishra S. Extracellular biosynthesis of zinc oxide nanoparticles using Rhodococcus pyridinivorans NT2: multifunctional textile finishing, biosafety evaluation and in vitro drug delivery in colon carcinoma. J Photochem Photobiol B. 2014;140:194–204. doi: 10.1016/j.jphotobiol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Kundu D, Hazra C, Chaudhari A. Biodegradation of 2,4-dinitrotoluene with Rhodococcus pyridinivorans NT2: characteristics, kinetic modeling, physiological responses and metabolic pathway. RCS Adv. 2015;5:38818–38829. doi: 10.1039/C5RA02450A. [DOI] [Google Scholar]
- Kurane R, Hatamochi K, Kakuno T, Kiyohara M, Hirano M, Taniguchi Y. Production of a bioflocculant by Rhodococcus erythropolis S-1 grown on alcohols. Biosci Biotechnol Biochem. 1994;58:428–429. doi: 10.1271/bbb.58.428. [DOI] [Google Scholar]
- Kurosawa K, Ghiviriga I, Sambandan TG, Lessard PA, Barbara JE, Rha C, Sinskey AJ. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J Am Chem Soc. 2008;30:1126–1127. doi: 10.1021/ja077821p. [DOI] [PubMed] [Google Scholar]
- Kurosawa K, Wewetzer SJ, Sinskey AJ. Engineering xylose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Biotechnol Biofuels. 2013;6:134. doi: 10.1186/1754-6834-6-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa K, Wewetzer SJ, Sinskey AJ. Triacylglycerol production from corn Stover using a xylose-fermenting Rhodococcus opacus strain for lignocellulosic biofuels. J Microbial Biochem Technol. 2014;6:254–259. doi: 10.4172/1948-5948.1000153. [DOI] [Google Scholar]
- Kurosawa K, Laser J, Sinskey AJ. Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels. 2015;8:76. doi: 10.1186/s13068-015-0258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa K, Radek A, Plassmeier JK, Sinskey AJ. Improved glycerol utilization by a triacylglycerol-producing Rhodococcus opacus strain for renewable fuels. Biotechnol Biofuels. 2015;8:31. doi: 10.1186/s13068-015-0209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyukina MS, Ivshina IB. Rhodococcus biosurfactants: biosynthesis, properties, and potential applications. In: Alvarez HM, editor. Biology of Rhodococcus. 1. Berlin: Springer-Verlag; 2010. pp. 292–313. [Google Scholar]
- Kuyukina MS, Ivshina IB, Philp JC, Christofi N, Dunbar SA, Ritchkova MI. Recovery of Rhodococcus biosurfactants using methyl tertiary-butyl ether extraction. J Microbiol Methods. 2001;46:149–156. doi: 10.1016/S0167-7012(01)00259-7. [DOI] [PubMed] [Google Scholar]
- Kuyukina MS, Ivshina IB, Baeva TA, Kochina OA, Gein SV, Chereshnev VA. Trehalolipid biosurfactants from nonpathogenic Rhodococcus actinobacteria with diverse immunomodulatory activities. New Biotechnol. 2015;32:559–568. doi: 10.1016/j.nbt.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Lanfranconi MP, Alvarez HM. Rewiring neutral lipids production for the de novo synthesis of wax esters in Rhodococcus opacus PD630. J Biotechnol. 2017;260:67–73. doi: 10.1016/j.jbiotec.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Lang S, Philp JC. Surface-active lipids in rhodococci. Antonie Van Leeuwenhoek. 1998;74:59–70. doi: 10.1023/A:1001799711799. [DOI] [PubMed] [Google Scholar]
- LeBlanc JC, Gonçalves ER, Mohn WW. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl Environ Microbiol. 2008;74:2627–2636. doi: 10.1128/AEM.02711-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Robinson J, Chong MF. A review on application of flocculants in wastewater treatment. Process Saf Environ. 2014;92:489–508. doi: 10.1016/j.psep.2014.04.010. [DOI] [Google Scholar]
- Lee JM, Lee H, Kang S, Park WJ. Fatty acid desaturases, polyunsaturated fatty acid regulation, and biotechnological advances. Nutrients. 2016;4:8. doi: 10.3390/nu8010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xu H, Chen ZS, Chen G. Biosynthesis of nanoparticles by microorganisms and their applications. J Nanomater. 2011;2011:270974. doi: 10.1155/2011/270974. [DOI] [Google Scholar]
- Liang Y, Jiao S, Wang M, Yu H, Shen Z. A CRISPR/Cas9-based genome editing system for Rhodococcus ruber TH. Metab Eng. 2020;57:13–22. doi: 10.1016/j.ymben.2019.10.003. [DOI] [PubMed] [Google Scholar]
- Maas D, Valerio A, Lourenco LA, de Oliveira D, Hotza D. Biosynthesis of iron oxide nanoparticles from mineral coal tailings in a stirred tank reactor. Hydrometallurgy. 2019;184:199–205. doi: 10.1016/j.hydromet.2019.01.010. [DOI] [Google Scholar]
- Maas D, de Medeiros MM, Cesa Rovaris B, Bernardin AM, de Oliveira D, Hotza D. Biomining of iron-containing nanoparticles from coal tailings. Appl Microbiol Biotechnol. 2019;103:7231–7240. doi: 10.1007/s00253-019-10001-2. [DOI] [PubMed] [Google Scholar]
- Manimaran M, Kannabiran K. Actinomycetes-mediated biogenic synthesis of metal and metal oxide nanoparticles: progress and challenges. Lett Appl Microbiol. 2017;64:401–408. doi: 10.1111/lam.12730. [DOI] [PubMed] [Google Scholar]
- Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Actinobacteria mediated synthesis of nanoparticles and their biological properties: a review. Crit Rev Microbiol. 2016;42:209–221. doi: 10.3109/104084X.2014.917069. [DOI] [PubMed] [Google Scholar]
- Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V. Biodegradation potential of the genus Rhodococcus. Environ Int. 2009;35:162–177. doi: 10.1016/j.envint.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammadi S, Afzal M, Hameed S. Bacterial polyhydroxyalkanoates-eco-friendly next generation plastic: production, biocompatibility, biodegradation, physical properties and applications. Green Chem Lett Rev. 2014;8:356–377. doi: 10.1080/17518253.2015.1109715. [DOI] [Google Scholar]
- Nachtigall J, Schneider K, Nicholson G, Goodfellow M, Zinecker H, Imhoff JF, Süssmuth RD, Fiedler HP. Two new aurachins from Rhodococcus sp. Acta 2259. J Antibiot. 2010;63:567–569. doi: 10.1038/ja.2010.79. [DOI] [PubMed] [Google Scholar]
- Nepal KK, Wang G. Streptomycetes: surrogate hosts for the genetic manipulation of biosynthetic gene clusters and production of natural products. Biotechnol Adv. 2019;37:1–20. doi: 10.1016/j.biotechadv.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DS. Prokaryotes and the input of polyunsaturated fatty acids to the marine food web. FEMS Microbiol Lett. 2003;219:1–7. doi: 10.3390/molecules23102583. [DOI] [PubMed] [Google Scholar]
- Niescher S, Wray V, Lang S, Kaschabek SR, Schlömann M. Identification and structural characterisation of novel trehalose dinocardiomycolates from n-alkane-grown Rhodococcus opacus 1CP. Appl Microbiol Biotechnol. 2005;70:605–611. doi: 10.1007/s00253-005-0113-8. [DOI] [PubMed] [Google Scholar]
- Orro A, Cappelletti M, D’Ursi P, Milanesi L, Di Canito A, Zampolli J, Collina E, Decorosi F, Viti C, Fedi S, Presentato A, Zannoni D, Di Gennaro P. Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: genetic determinants and metabolic abilities with environmental relevance. PLoS One. 2015;10:e0139467. doi: 10.1371/journal.pone.0139467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A, Teruel JA, Espuny MJ, Marqués A, Manresa A, Aranda FJ. Interactions of a Rhodococcus sp. biosurfactant trehalose lipid with phosphatidylethanolamine membranes. Biochim Biophys Acta. 2008;1778:2806–2813. doi: 10.1016/j.bbamem.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Osawa A, Kasahara A, Masttuoka S, Gassel S, Sandmann G, Shindo K. Isolation of a novel carotenoid, OH-chlorobactene glucoside hexadecanoate, and related rare carotenoids from Rhodococcus sp. cip and their antioxidative activities. Biosci Biotechnol Biochem. 2011;75:2142–2147. doi: 10.1271/bbb.110441. [DOI] [PubMed] [Google Scholar]
- Otari SV, Patil RM, Nadaf NH, Ghosh SJ, Pawar SH (2012) Green biosynthesis of silver nanoparticles from an actinobacteria Rhodococcus sp. mater Lett 72:92-94. 10.1016/j.matlet.2011.12.109
- Pacheco GJ, Ciapina EM, Gomes Ede B, Junior NP. Biosurfactant production by Rhodococcus erythropolis and its application to oil removal. Braz J Microbiol. 2010;41:685–693. doi: 10.1590/S1517-83822010000300019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page CA, Bonner JS, Kanga SA, Mills MA, Autenrieth RL. Biosurfactant solubilization of PAHs. Environ Eng Sci. 1999;16:465–474. doi: 10.1089/ees.1999.16.465. [DOI] [Google Scholar]
- Pantidos N, Horsfall LE. Biological synthesis of metallic nanoparticles by bacteria, fungi and plants. J Nanomed Nanotechnol. 2014;5:5. doi: 10.4172/2157-7439.1000233. [DOI] [Google Scholar]
- Patel A, Matsakas L, Hrůzová K, Rova U, Christakopoulos P. Biosynthesis of nutraceutical fatty acids by the oleaginous marine microalgae Phaeodactylum tricornutum utilizing hydrolysates from organosolv-pretreated birch and spruce biomass. Mar Drugs. 2019;17:119. doi: 10.3390/md17020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F, Liu Z, Wang L, Shao Z. An oil-degrading bacterium: Rhodococcus erythropolis strain 3C-9 and its biosurfactants. J Appl Microbiol. 2007;102:1603–1611. doi: 10.1111/j.1365-2672.2006.03267.x. [DOI] [PubMed] [Google Scholar]
- Peng L, Yang C, Zeng G, Wang L, Dai C, Long Z, Liu H, Zhong Y. Characterization and application of bioflocculant prepared by Rhodococcus erythropolis using sludge and livestock wastewater as cheap culture media. Appl Microbiol Biotechnol. 2014;98:6847–6858. doi: 10.1007/s00253-014-5725-4. [DOI] [PubMed] [Google Scholar]
- Philp JC, Kuyukina MS, Ivshina IB, Dunbar SA, Christofi N, Lang S, Wray V. Alkanotrophic Rhodococcus ruber as a biosurfactant producer. Appl Microbiol Biotechnol. 2002;59:318–324. doi: 10.1007/s00253-002-1018-4. [DOI] [PubMed] [Google Scholar]
- Piacenza E, Presentato A, Turner RJ. Stability of biogenic metal(loid) nanomaterials related to the colloidal stabilization theory of chemical nanostructures. Crit Rev Biotechnol. 2018;25:1–20. doi: 10.1080/07388551.2018.1440525. [DOI] [PubMed] [Google Scholar]
- Pieper U, Steinbüchel A. Identification, cloning and sequence analysis of the poly(3-hydroxyalkanoic acid) synthase gene of the gram-positive bacterium Rhodococcus ruber. FEMS Microbiol Lett. 1992;75:73–79. doi: 10.1111/j.1574-6968.1992.tb05396.x. [DOI] [PubMed] [Google Scholar]
- Pirog TP, Shevchuk TA, Voloshina IN, Karpenko EV. Production of surfactants by Rhodococcus erythropolis strain EK-1, grown on hydrophilic and hydrophobic substrates. Appl Biochem Microbiol. 2004;40:470–475. doi: 10.1023/B:ABIM.0000040670.33787.5f. [DOI] [PubMed] [Google Scholar]
- Pirog TP, Korzh YV, Shevchuk TA, Tarasenko DA. Peculiarities of C2 metabolism and intensification of the synthesis of surface-active substances in Rhodococcus erythropolis EK-1 grown in ethanol. Microbiology. 2008;77:665–673. doi: 10.1134/S0026261708060039. [DOI] [PubMed] [Google Scholar]
- Presentato A, Piacenza E, Anikovskiy M, Cappelletti M, Zannoni D, Turner RJ (2016) Rhodococcus aetherivorans BCP1 as cell factory for the production of intracellular tellurium nanorods under aerobic conditions. Microb Cell Factories 15:204. 10.1186/s12934-016-0602-8 [DOI] [PMC free article] [PubMed]
- Presentato A, Cappelletti M, Sansone A, Ferreri C, Piacenza E, Demeter MA, Crognale S, Petruccioli M, Milazzo G, Fedi S, Steinbüchel A, Turner RJ, Zannoni D. Aerobic growth of Rhodococcus aetherivorans BCP1 using selected naphthenic acids as the sole carbon and energy sources. Front Microbiol. 2018;9:672. doi: 10.3389/fmicb.2018.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presentato A, Piacenza E, Anikovskiy M, Cappelletti M, Zannoni D, Turner RJ. Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. New Biotechnol. 2018;41:1–8. doi: 10.1016/j.nbt.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Presentato A, Piacenza E, Darbandi A, Anikovskiy M, Cappelletti M, Zannoni D, Turner RJ. Assembly, growth and conductive properties of tellurium nanorods produced by Rhodococcus aetherivorans BCP1. Sci Rep. 2018;8:3923. doi: 10.1038/s41598-018-22320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AV, Rao LG. Carotenoids and human health. Pharmacol Res. 2007;55:207–216. doi: 10.1016/j.phrs.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Rao CN, Muller A, Cheetham AK. The chemistry of nanomaterials: synthesis, properties and applications. Wiley-VCH Verlag GmbH: Weinheim; 2004. pp. 1–11. [Google Scholar]
- Rapp P, Bock H, Wray V, Wagner F. Formation, isolation and characterization of trehalose dimycolates from Rhodococcus erythropolis grown on n-alkanes. J Gen Microbiol. 1979;115:491–503. doi: 10.1099/00221287-115-2-491. [DOI] [Google Scholar]
- Rehm BH. Polyester synthases: natural catalysts for plastics. Biochem J. 2003;376:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BHA, Steinbüchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macrol. 1999;25:3–19. doi: 10.1016/S0141-8130(99)00010-0. [DOI] [PubMed] [Google Scholar]
- Retamal-Morales G, Heine T, Tischler JS, Erler B, Gröning JAD, Kaschabek SR, Schlömann M, Levicán G, Tischler D. Draft genome sequence of Rhodococcus erythropolis B7g, a biosurfactant producing actinobacterium. J Biotechnol. 2018;20:38–41. doi: 10.1016/j.biotec.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Retamal-Morales G, Mehnert M, Schwabe R, Tischler D, Zapata C, Chávez R, Schlömann M, Levicán G. Detection of arsenic-binding siderophores in arsenic-tolerating Actinobacteria by a modified CAS assay. Ecotoxicol Environ Saf. 2018;15(157):176–181. doi: 10.1016/j.ecoenv.2018.03.087. [DOI] [PubMed] [Google Scholar]
- Ristau E, Wagner F. Formation of novel anionic trehalose tetraesters from Rhodococcus erythropolis under growth-limiting conditions. Biotechnol Lett. 1983;5:95–100. doi: 10.1007/BF00132166. [DOI] [Google Scholar]
- Ruggeri C, Franzetti A, Bestetti G, Caredda P, La Colla P, Pintus M, Sergi S, Tamburini E. Isolation and characterization of surface active compounds producing bacteria from hydrocarbon-contaminated environments. Int Biodeterior Biodegrad. 2009;63:936–942. doi: 10.1016/j.biod.2009.05.003. [DOI] [Google Scholar]
- Sadouk Z, Hacene H, Tazerouti A. Biosurfactants production from low cost substrate and degradation of diesel oil by Rhodococcus strain. Oil Gas Sci Technol. 2008;63:747–753. doi: 10.2516/ogst:2008037. [DOI] [Google Scholar]
- Saini RK, Keum YS. Progress in microbial carotenoids production. Indian J Microbiol. 2017;57:129–130. doi: 10.1007/s12088-016-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehizadeh H, Yan N, Farnood R. Recent advances in polysaccharide bio-based flocculants. Biotechnol Adv. 2018;36:92–119. doi: 10.1016/j.biotechadv.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Santos DK, Rufino RD, Luna JM, Santos VA, Sarubbo LA. Biosurfactants: multifunctional biomolecules of the 21st century. Int J Mol Sci. 2016;17:401. doi: 10.3390/ijms17030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RA, Grossi V, Olivera N, Alvarez HM. Characterization of the indigenous Rhodococcus sp. 602, a strain able to accumulate triacylglycerides from naphthyl-compounds under nitrogen-starved conditions. Res Microbiol. 2010;161:198–207. doi: 10.1016/j.resmic.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Stancu MM. Response of Rhodococcus erythropolis strain IBBPo1 to toxic organic solvents. Braz J Microbiol. 2015;46:1009–1018. doi: 10.1590/S1517-838246420140462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992;103:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- Subbaiya R, Preetha L, Gayathril S, Swarnalatha WA, Selvam MM (2014) Synthesis and characterization of silver nanoparticles from Rhodococcus-2891 and its antitumor activity against lung cancer cell line (A549). In: International conference on science, engineering and management research (ICSEMR 2014). IEEE, Chennai
- Suwaleerat T, Thanapimmetha A, Saisriyoot M, Chisti Y, Srinophakun P. Enhanced production of carotenoids and lipids by Rhodococcus opacus PD630. J Chem Technol Biotechnol. 2017;93:2160–2169. doi: 10.1002/jctb.5554. [DOI] [PubMed] [Google Scholar]
- Tajparast M, Frigon D. Genome-scale metabolic model of Rhodococcus jostii RHA1 (iMT1174) to study the accumulation of storage compounds during nitrogen-limited condition. BMC Syst Biol. 2015;9:43. doi: 10.1186/s12918-015-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajparast M, Frigon D. Predicting the accumulation of storage compounds by Rhodococcus jostii RHA1 in the feast-famine growth cycles using genome-scale flux balance analysis. PLoS One. 2018;13:e0191835. doi: 10.1371/journal.pone.0191835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaichi S, Ishidsu JI, Seki T, Fukada S. Carotenoid pigments from Rhodococcus rhodochrous RNMS1: two monocyclic carotenoids, a carotenoid monoglycoside and carotenoid glycoside monoesters. Agric Biol Chem. 1990;54:1931–1937. doi: 10.1271/bbb1961.54.1931. [DOI] [Google Scholar]
- Takeda M, Kurane R, Nakamura I. Localization of a biopolymer produced by Rhodococcus erythropolis grown on n-pentadecane. Agric Biol Chem. 1991;55:2665–2666. doi: 10.1271/bbb.1961.55.2665. [DOI] [Google Scholar]
- Tao L, Cheng Q. Novel beta-carotene ketolases from non-photosynthetic bacteria for canthaxanthin synthesis. Mol Gen Genomics. 2004;272:530–537. doi: 10.1007/s00438-004-1038-8. [DOI] [PubMed] [Google Scholar]
- Tao L, Picataggio S, Rouviere PE, Cheng Q. Asymmetrically acting lycopene beta-cyclases (CrtLm) from non-photosynthetic bacteria. Mol Gen Genomics. 2004;271:180–188. doi: 10.1007/s00438-003-0969-1. [DOI] [PubMed] [Google Scholar]
- Thanapimmetha A, Suwaleerat T, Saisriyoot M, Chisti Y, Srinophakun P (2017) Production of carotenoids and lipids by Rhodococcus opacus PD630 in batch and fed-batch culture. Bioprocess Biosyst Eng 40:133–143. 10.1007/s00449-016-1681-y [DOI] [PubMed]
- Tischler D, Niescher S, Kaschabek SR, Schlömann M. Trehalose phosphate synthases OtsA1 and OtsA2 of Rhodococcus opacus 1CP. FEMS Microbiol Lett. 2013;342:113–122. doi: 10.1111/1574-6968. [DOI] [PubMed] [Google Scholar]
- Tokumoto Y, Nomura N, Uchiyama H, Imura T, Morita T, Fukuoka T, Kitamoto D (2009) Structural characterization and surface-active properties of succinoyl trehalose lipid produced by Rhodococcus sp. SD-74. J Oleo Sci 58:97–102. 10.5650/jos.58.97 [DOI] [PubMed]
- Tropis M, Meniche X, Wolf A, Gebhardt H, Strelkov S, Chami M, Schomburg D, Krämer R, Morbach S, Daffé M. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae. J Biol Chem. 2005;280:26573–26585. doi: 10.1074/jbc.M502104200. [DOI] [PubMed] [Google Scholar]
- Tsitko IV, Zaitsev GM, Lobanok AG, Salkinoja-Saloneni MS. Effect of aromatic compounds on cellular fatty acid composition of Rhodococcus opacus. Appl Environ Microbiol. 1999;65:853–855. doi: 10.1128/AEM.65.2.853-855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano SB, Di Capua C, Cortez N, Farías ME, Alvarez HM. Triacylglycerol accumulation and oxidative stress in Rhodococcus species: differential effects of pro-oxidants on lipid metabolism. Extremophiles. 2014;18:375–384. doi: 10.1007/s00792-013-0623-8. [DOI] [PubMed] [Google Scholar]
- Voss I, Steinbüchel A. High cell density cultivation of Rhodococcus opacus for lipid production at a pilot scale. Appl Microbiol Biotechnol. 2001;55:547–555. doi: 10.1007/s002530000576. [DOI] [PubMed] [Google Scholar]
- Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla H-J, Kalscheuer R, Stöveken T, von Landenberg P, Steinbüchel A. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang X, Hui K, Wei W, Zhang W, Miao A, Xiao L, Yang L. Highly effective polyphosphate synthesis, phosphate removal, and concentration using engineered environmental bacteria based on a simple solo medium-copy plasmid strategy. Environ Sci Technol. 2018;52:214–222. doi: 10.1021/acs.est.7b04532. [DOI] [PubMed] [Google Scholar]
- White DA, Hird LC, Ali ST (2013) Production and characterization of a trehalolipid biosurfactant produced by the novel marine bacterium Rhodococcus sp. strain PML026. J Appl Microbiol 115:744–755. 10.1111/jam.12287 [DOI] [PubMed]
- Whyte LG, Slagman SJ, Pietrantonio F, Bourbonniere L, Koval SF, Lawrence JR, Innis WE, Greer SW. Physiological adaptations involved in alkane assimilation at a low temperature by Rhodococcus sp. strain Q15. Appl Environ Microbiol. 1999;65:2961–2968. doi: 10.1128/AEM.65.7.2961-2968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Chen S. Engineering of a xylose metabolic pathway in Rhodococcus spp. strains. Appl Environ Microbiol. 2012;78:5483–5491. doi: 10.1128/AEM.08022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Lian J, Yu X, Garcia-Perez M, Chen S. Engineering levoglucosan metabolic pathway in Rhodococcus jostii RHA1 for lipid production. J Ind Microbiol Biotechnol. 2016;43:1551–1560. doi: 10.1007/s10295-016-1832-9. [DOI] [PubMed] [Google Scholar]
- Xiong X, Wang X, Chen S. Engineering of an L-arabinose metabolic pathway in Rhodococcus jostii RHA1 for biofuel production. J Ind Microbiol Biotechnol. 2016;43:1017–1025. doi: 10.1007/s10295-016-1778-y. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, Giuliano L, Bruni V, Scarfì S, Golyshin PN. Characterization of antarctic hydrocarbon-degrading bacteria capable of producing bioemulsifiers. New Microbiol. 1999;22:249–256. [PubMed] [Google Scholar]
- Yoneda A, Henson WR, Goldner NK, Park KJ, Forsberg KJ, Kim SJ, Pesesky MW, Foston M, Dantas G, Moon TS. Comparative transcriptomics elucidates adaptive phenol tolerance and utilization in lipid-accumulating Rhodococcus opacus PD630. Nucleic Acids Res. 2016;44:2240–2254. doi: 10.1093/nar/gkw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Yano T, Kedo K, Fujiyoshi T, Nagai R, Iwano M, Taguchi E, Nishida T, Takagi H. A unique intracellular compartment formed during the oligotrophic growth of Rhodococcus erythropolis N9T-4. Appl Microbiol Biotechnol. 2017;101:331–340. doi: 10.1007/s00253-016-7883-z. [DOI] [PubMed] [Google Scholar]
- Zhang B, Ye X, Dai W, Hou W, Zuo F, Xie Y. Biomolecule-assisted synthesis of single-crystalline selenium nanowires and nanoribbons via a novel flake-cracking mechanism. Nanotechnology. 2006;17:385–390. doi: 10.1088/0957-4484/17/2/007. [DOI] [Google Scholar]
- Zheng YT, Toyofuku M, Nomura N, Shigeto S. Correlation of carotenoid accumulation with aggregation and biofilm development in Rhodococcus sp. SD-74. Anal Chem. 2013;85:7295–7301. doi: 10.1021/ac401188f. [DOI] [PubMed] [Google Scholar]