Abstract
Histone post-translational modifications (PTMs) have emerged as exciting mechanisms of biological regulation, impacting pathways related to cancer, immunity, brain function, and more. Over the past decade alone, several histone PTMs have been discovered, including acylation, lipidation, monoaminylation, and glycation, many of which appear to have crucial roles in nucleosome stability and transcriptional regulation. In this review, we discuss novel histone PTMs identified within the past 10 years, with an extended focus on enzymatic versus nonenzymatic mechanisms underlying modification and adduction. Furthermore, we consider how these novel histone PTMs might fit within the framework of a so-called ‘histone code’, emphasizing the physiological relevance of these PTMs in metabolism, development, and disease states.
A Growing List of Histone Modifications
The collection of histone PTMs (see Glossary) expands each year, acquiring new chemical moieties capable of covalent attachment to various amino acids on histone proteins. While a comprehensive catalog of known histone PTMs was published in 2015 [1], the discovery of novel histone modifications has been outpacing our ability to investigate and associate them with biological purpose. As the foundation of chromatin, histones spool genomic loci by winding ~146 base pairs of DNA around histone octamers. These complexes form tightly wound nucleosomes that mediate accessibility to allow for the recruitment of regulatory machinery to DNA, thereby influencing transcription [2]. Evolutionarily conserved N- and C-terminal tails protrude from each core histone, containing numerous sites that are amenable to modification. While much chromatin research has focused on select N- and C-terminal residues, modifications additionally occur on histone fold domains to impact cellular programs [3].
Certain PTMs are capable of altering electrostatic interactions between positively charged histone residues and the negatively charged DNA backbone to allow for (un)winding of these transcriptional units. Moreover, histone PTMs can prevent or encourage effector protein binding and structurally safeguard from inadvertent adduction, together resulting in distinct molecular consequences. Yet, causal functions for newly discovered histone PTMs, along with the sheer number of combinatorial possibilities, have been difficult to tease apart. Despite the challenges of these tasks, current technologies and our understanding of histone interactions have come a long way, when 30 years spanned the identification of histone acetylation [4,5] to its connection to active transcription [6] (Figure 1; reviewed in [7]). In comparison, in 2019 alone, three novel histone PTMs were characterized with proposed regulatory and physiological roles by independent groups (see later).
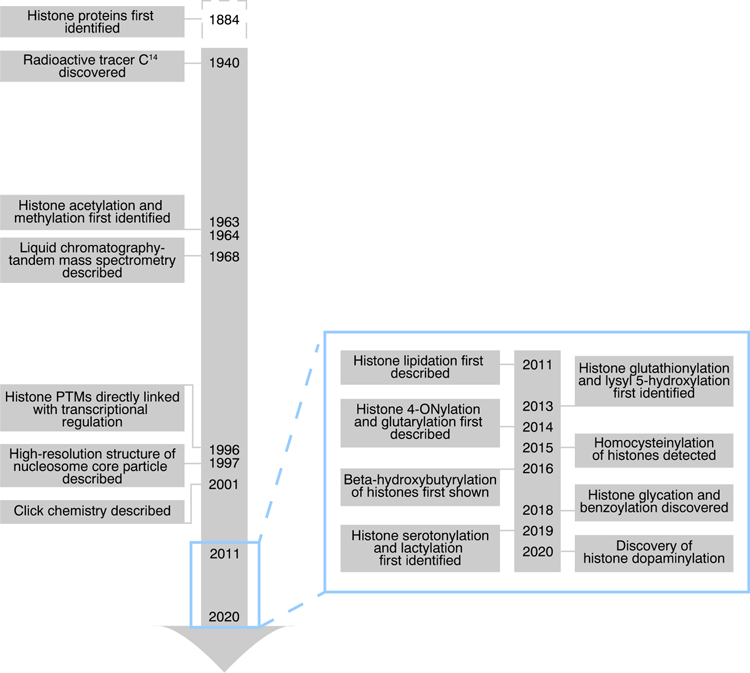
Figure 1. Selected Milestones in Histone Post-Translational Modification (PTM) Discovery.
Timeline showing a brief history of representative technological advancements that paved the way for histone PTM discovery during the 20th century. Left: In 1884, histone proteins were first identified by A. Kossel using acid extraction on avian red blood cells [96]. In 1940, M. Kamen and S. Ruben synthesized the radioactive isotope carbon-14 (C14) [97]. In 1963–1964, histone lysine acetylation and methylation were first identified using C14 labeling and acid extractions [4,5]. In 1968, V, Tal’Roze et al. described electron ionization, and were the first to report the development of liquid chromatography-tandem mass spectrometry (LC-MS/MS) [98]. In 1996, J. Brownell et al. reported direct evidence that histone acetyltransferase Gcn5 acetylates chromatin to activate gene expression [6] and, in the same year, J. Taunton et al. cloned human histone deacetylase I, previously associated with transcriptional changes [99]. In 1997, K. Luger et al. showed how histone proteins interacted with 146 base pairs of DNA by determining the crystal structure of the nucleosome core particle at 2.8- Å resolution [100]. In 2001, K. Sharpless and colleagues at Scripps Research Institute fully described click chemistry, a term he coined in 1998 [21]. Sky-blue inset: timeline of novel histone PTM discovery over the past decade (2011–mid-2020).
In this review, we discuss histone PTMs that have been identified within the past decade, specifically histone acylations, monoaminylations, lipidation, hydroxylation, and other nonenzymatic PTMs (Figure 2, Key Figure and Table 1), as well as their potential relevance to biological processes and disease. We also include extended discussions on both enzymatic and nonenzymatic mechanisms. Moreover, we consider how these PTMs might fit within the framework of the so-called ‘histone code hypothesis’.
Figure 2.
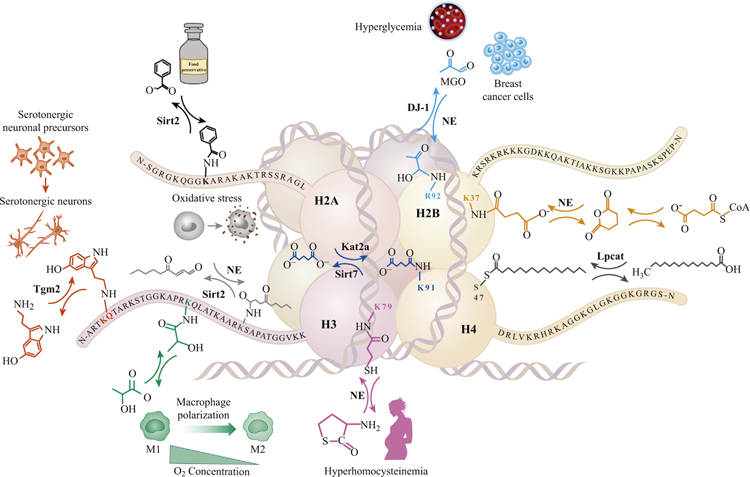
H2A is modified at lysine (K) 9 by benzoylation, removed by Sirtuin 2 (Sirt2) and influenced by the food preservative sodium benzoate (black). H2B receives nonenzymatic (NE) adduction by (i) succinyl-coA on K37, which undergoes intramolecular catalysis to form a highly reactive electrophilic anhydride (right, orange); and (ii) methylglyoxal (MGO) glycation, prevalent in hyperglycemia and breast cancer tumor cells, on arginine (R) 92 and is removed by the deglyase DJ-1 (sky-blue). H3 is (i) serotonylated by transglutaminase 2 (Tgm2) at glutamine 5, neighboring a methylated lysine 4 (H3 K4me3Q5ser) and is altered by differentiation of serotonergic neuronal precursors (far left, vermilion); (ii) lactylated at H3 K18 under hypoxic conditions and macrophage response to pathogens (blue-green); (iii) adducted by the peroxidized lipid electrophile 4-oxo-2-noneal, 4-ONylating H3 K27 under oxidative stress and can be removed by Sirt2 (left, light gray); and (iv) adducted on the globular site H3 K79 by homocysteine thyolactone, which is increased under conditions of hyperhomocysteinemia (e.g., pregnant women at risk of offspring neural tube defects; red-purple). H4 is (i) lipidated at serine 47 by the enzyme lysophosphatidylcholine acyltransferase 1 (Lpcat1), catalyzing addition of a 16-carbon fatty palmitic acid (right, dark gray); and (ii) glutarylated at H4 K91 by the histone glutaryltransferase Kat2a and removed by Sirtuin 7 (Sirt7, dark blue).
Key Figure.
Schematic Representation of Select Novel Histone Post-Translational Modifications (PTMs) Influenced by Normal Physiological and Disease States to Impact Chromatin
Table 1.
Summary of Novel Histone PTMs (2011 to mid-2020)
| Histone PTM | Reaction | Donor precursor | Writer | Eraser | Function | Physiological relevance | Refs |
|---|---|---|---|---|---|---|---|
| Glutarylation | Acylation | Glutarate | Kat2a, intramolecular catalysis | Sirt7 | Nucleosome destabilization, permissive transcription | Glutaric acidemia | [15,19,22] |
| Lactylation | Acylation | Lactate | p300 | Permissive transcription | Macrophage response, hypoxia | [23] | |
| Benzoylation | Acylation | Benzoate | Sirt2 | Permissive transcription | Sodium benzoate treatment | [27] | |
| S-palmitoylation | S-acylation | Palmitic acid | Cell signaling | [33] | |||
| O-palmitoylation | O-acylation | Palmitic acid | Lpcat1 | Reduced transcription | Cell signaling | [36] | |
| Serotonylation | Transamidation | Serotonin | Tgm2 | Permissive transcription | Neuronal differentiation | [39] | |
| Dopaminylation | Transamidation | Dopamine | Tgm2 | Altered transcription | Drug-seeking behaviors | [47] | |
| 5-Hydroxylysine | Hydroxylation | 2-Oxoglutarate | Jmjd6 | Testes, development | [50] | ||
| Glycation | Maillard | Methylglyoxal, monosaccharides | Nonenzymatic | DJ-1 | Altered nucleosome stability | Breast cancer, hyperglycemia | [59,60] |
| 4-Oxononanoylation | Ketoamide adduction | 4-Oxo-2-nonenal | Nonenzymatic | Sirt2 | Nucleosome destabilization | Lipid peroxidation | [63,64] |
| Acrolein adduct | Michael addition | Acrolein | Nonenzymatic | Nucleosome destabilization | Cigarette smoke, lipid peroxidation | [65,66] | |
| S-glutathionylation | Disulfide formation | Glutathione | Nonenzymatic | Nucleosome destabilization | Aging | [69] | |
| Homocysteinylation | Thiolation | Homocysteine thyolactone | Nonenzymatic | Reduced transcription | Hyperhomocysteinemia | [72,73] |
Histone Acylation
Histone PTMs derived from acylation reactions have dominated histone PTM discovery. An alphabet of acyl modifications, including acetylation, butyrylation, and crotonylation, share many characteristics: (i) analogous chemical structures stemming from carboxylic acid derivatives containing a carbonyl and alkyl group with differing hydrocarbon chain properties; (ii) linkage to lysine ε-amines on N-terminal histone tails; (iii) binding of mutual protein machinery; and (iv) an intricate connection with cellular metabolism. For example, acetyl-coA is the primary source for histone lysine acetylation (Kac), where dose-dependent increases in mitochondrial glucose metabolism increase ATP-citrate lyase (ACLY) cleavage of citrate to acetyl-coA and correlate with increased histone Kac levels [8]. Moreover, recent studies demonstrated that acetyl-coA levels also stem from the oxidation of fatty acids, including octanoate (used in food flavoring and perfumes), where supplementation dose dependently increases histone acetylation in a glucose- and ACLY-independent manner, relying on the enzyme acetyl-CoA synthetase short chain family member 2 (ACSS2) [9]. In neurons, ACSS2 also generates acetyl-coA from alcohol metabolism in liver, likewise influencing histone Kac abundance [10]. Thus, mammalian cells have evolved multiple nutrient-sensing pathways for histone Kac regulation, suggesting its importance in many biological processes.
Similarly, nonacetyl histone acylations depend on nutrient-sensing pathways for modification. Short-chain fatty acids (SCFA) produce labile thioester donor groups, such as propionyl-coA and crotonyl-coA, with characteristic short hydrophobic tails [11,12]. When these short acyl groups modify lysine residues, their charges become neutralized, reducing histone–DNA electrostatic interactions, leading to more open and accessible chromatin, similar to effects of histone Kac. Additionally, the extent of this neutralization depends on the hydrophobicity of the acyl group: for example, greater transcriptional activation is linked to crotonylation (planar four-carbon chain) versus acetylation (two-carbon chain) [13]. Acidic SCFA-derived acyl donors instead add negative charges to lysines via their carboxylate groups, suggesting that acidic acylation alters transcription by repelling DNA and/or impacting chromatin stability [14–16].
Importantly, the enzymes that classically read, write, and erase histone Kac also affect and/or bind to other acylation PTMs; for example, the histone acetyltransferase (HAT) p300/CBP writes histone acetylation, butyrylation, propionylation, and crotonylation, with differential p300 reactivity relying on acyl-chain length [11,17]. Therefore, increases in certain acyl donors may bind and, in turn, deplete available HATs, concurrently diminishing the transfer of less-abundant donors. This competition is reinforced by limited lysines, the residue of choice for acylation, owing to the nucleophilic reactivity of ε-amines when deprotonated, occurring at high pH with enzymatic assistance [18]. However, whether this seesaw of acylation PTMs produces functionally redundant or distinct effects remains less understood. In this section, we first summarize recently identified histone acylation PTMs (Table 1), and discuss new proposed biological roles for these modifications (Box 1).
Box 1. Advances in Histone Acylation Biology.
As tools to investigate histone acylation have become more abundant and readily available, the involvement of histone PTMs in the regulation of biological processes is becoming clearer. Advances in gut microbiota research indicate that histone acylations depend on microbial carbohydrate fermentation for SCFA production, linking these unique tissues, which are constantly impacted by diet, to chromatin regulation. For example, high-sucrose and high-fat diets (containing few carbohydrates) reduce histone acetylation levels in colon, liver, and white adipose tissues in mice [77]. Similarly, microbiota depletion impairs histone crotonylation in the colon, while it is conversely elevated by both crotonate and butyrate supplementation [78], with the effect of butyrate likely caused by its additional function as a histone deacetylase (HDAC) inhibitor. Thus, evidence that diet improves symptoms in cancers, irritable bowel syndrome, and Kabuki disorder, among others [79–81] may be partially explained by microbial SCFA production and regulation of histone PTM stoichiometry. Furthermore, the recently characterized acyl-PTM, histone lysine β-hydroxybutyrylation, stems from the ketone β-hydroxybutyrate, another known HDAC inhibitor, and is upregulated by fasting and ketogenic diets [82]. Conversely, these data suggest that pathological nutritional states lead to aberrant histone acylation states.
In addition to diet, histone acylations are influenced by other physiological events. For example, stress impacts all tissues in the body; in a mouse model of social defeat that mimics stress-affective disorders, histone crotonylation levels in the mouse prefrontal cortex were found to be reduced in ‘defeated’ mice, functionally inhibiting neuronal plasticity [83]. In a separate study, acetate from liver alcohol metabolism was shown to cross the critical placental barrier separating maternal and fetal circulation, serving as a substrate for histone acetylation in fetal brain that may contribute to fetal alcohol syndrome [10]. Furthermore, with the established connection between H3 lysine 27 acetylation (H3K27ac) and active enhancers [84], this PTM has been heavily studied as a regulator of gene expression programs that define cellular identity. Used especially as a marker of super-enhancers, H3K27ac density may alter higher-order nucleosome compaction to boost transcription. Other histone lysine acylation sites have also recently been described to mark active enhancers [85,86], painting a more complex picture not only of enhancer regulation, but also histone acylation in general.
Histone Glutarylation
Protein lysine glutarylation (Kglu) was first discovered in 2014, whereby Kglu enrichment was highest in mitochondria compared with other organelles [15]. The acidic Kglu side chain is structurally similar to malonylated and succinylated lysines with carboxylate groups and increasing carbon chain lengths, a characteristic important for the mechanism of acyl PTMs, as discussed later [19]. Furthermore, the donor source for these PTMs differs (e.g., glutaryl-coA, from tryptophan and lysine metabolism versus succinyl-coA, a Krebs cycle intermediate [20]), which suggests an important regulatory crosstalk mechanism between cellular energy status and acyl-PTM state.
Click chemistry methods have proven valuable and are utilized heavily throughout work discussed in this review [21]. To identify endogenously glutarylated proteins, click chemistry-assisted fluorescence cell imaging was used, where the precursor glutarate was modified by installing a terminal alkyne with high reaction specificity for an azide reporter molecule. Under normal cellular conditions, the modified glutarate can be metabolized to glutaryl-coA, where transfer of this donor group metabolically labels proteins endogenously subjected to glutarylation [15]. Subsequently, the reporter is added and ‘clicked’ onto the alkyne to produce a fluorescent signal [21]. Using this strategy, most glutarylated proteins were observed in the nucleus; liquid chromatography with tandem mass spectrometry (LC-MS/MS) identified 27 histone Kglu sites in HeLa cells, distributed across all core histones, with six Kglu sites unique to glutarylation [15]. Glutarylated histone H4 lysine 91 (H4K91glu) displays high modification stoichiometry (Box 2) and is positioned on the histone globular fold domain, where its negative charge weakens nucleosome assembly and DNA winding considerably (Figure 2, dark blue) [20]. Saccharomyces cerevisiae strains generated to mimic and maintain the negative charge of H4K91glu by mutating the lysine to glutamic acid (H4K91E) proliferate slowly and have ineffective DNA repair processes compared with control strains, supporting nucleosome destabilization by H4K91E/glu during these processes [22]. Genomically, H4K91glu is enriched at transcriptional start sites (TSS) and correlates with permissive transcription, consistent with observed DNA unwinding. Characterization of H4K91glu regulatory effectors pinpointed the nuclear protein, Sirtuin 7 (Sirt7), as a deglutarylase, where depletion of Sirt7 readily increases H4K91glu levels. Moreover, the known histone acetyl/succinyltransferase KAT2A was described as a potential histone glutaryltransferase, where specificity of its transferase activity may be contingent on other proteins present, such as components of the α-ketoadipate dehydrogenase complex, which were proposed to interact with KAT2A to promote H4K91glu levels [22]. With glutarylated nonhistone proteins implicated in the metabolic disorder glutaric acidemia [15], regulation of histone glutarylation may itself be crucial for these physiological processes.
Box 2. Modification Stoichiometry: An Indication of Functionality?
PTM/modification stoichiometry refers to the fraction of histones modified by a select PTM at a specific amino acid residue (Equation I) [87]:
| [1] |
Histone PTM sites that are highly abundant have high modification stoichiometry, while low-abundance PTM sites have low modification stoichiometry. Due to greater prevalence and detectability, high stoichiometric PTMs are assumed to have crucial roles in biology, with the capability to regulate large transcriptional networks. However, low stoichiometric PTMs may have the capability to respond more robustly to cellular signals (i.e., avoid ceiling effects and/or saturation). Therefore, whether PTM stoichiometry correlates with PTM importance and/or function remains unclear, although recent studies examining novel histone PTMs may provide some answers.
Early histone PTM discovery was accomplished with a less sensitive toolkit than what is available today, and specific histone acetylation sites went undetermined until 2006 [88]. Notably, the stoichiometries of histone acetylation sites vary from ~55% (H3 lysine 23) to 0.3% (H3 lysine 27) in Drosophila melanogaster cells [89], suggesting that abundances for PTMs of the same type differ across histone residues. Today, a handful of methods are used to detect lower abundant PTMs (b0.01% occupancy), including affinity enrichment strategies, and isotopic labeling, although there are still limitations to these approaches (see [87,90] for more detailed reviews of these methods). For example, several histone PTMs discussed in this review were previously undetected until the application of antibody-based affinity enrichment, where antibodies are designed to recognize a specific PTM moiety with either sequence-independent (e.g., pan anti-Kla) or sequence-dependent (e.g., anti-H3Q5ser) sites. These antibody-based strategies allow enough enrichment for HPLC-MS/MS detection of the desired PTM for comparison between experimental groups, but neglect alternative PTMs at the same residues, obstructing both stoichiometric and absolute measurements [90].
Despite lacking quantification, recent studies suggest that low-abundance histone PTMs are functionally significant. For example, increasing histone KHcy disrupts levels of acetylation and methylation that, together, likely produce a distinct transcriptional program [72]. Moreover, the serotonylated H3Q5ser sits adjacent to H3K4me3, a well-studied mark linked with permissive transcription [39]. Despite H3Q5ser being low in abundance, its regulation of H3K4me3 via the combinatorial H3K4me3Q5ser may serve as a mechanism to magnify its influence. Another study showing that histone crotonylation associates with greater transcriptional activation compared with histone acetylation suggests that the relationship between PTM occupancy and downstream consequences is not a one-size-fits-all [13]. The stability of the PTM is another important consideration, because dynamic PTMs may be critically relevant but more difficult to capture. There may also be differences in PTM abundance across cell types, age, sex, or cellular compartments, or in responsiveness to environmental cues. Another crucial consideration involves the majority of stoichiometric assessments occurring in vitro, which is informative but cannot completely mimic the in vivo condition. Together, attaining an accurate representation of histone PTM stoichiometry is difficult for any PTM, regardless of abundance. Technological advancements in MS sensitivity or in strategies that would enrich specific histone residues in a PTM-independent manner would facilitate precise stoichiometric measurements, and would likely result in further histone PTM discovery in the future.
Histone Lactylation
Histone lysine lactylation (Kla) joined the metabolite-derived histone acylation club in 2019 [23]. Traditionally thought of as a waste product derived from anaerobic respiration and exercise, lactate has gained attention as an important energy source in brain, skeletal muscles, and stem cells [24]. It is also a primary fuel under hypoxic conditions and is implicated in macrophage responses to bacterial infections and tumor cell metabolism via the Warburg effect, where cells favor energy from glycolysis versus the efficient oxidative phosphorylation [25]. Similar to its precursor, histone Kla is associated with glycolysis. Taking advantage of these systems that increase subcellular lactate levels, glucose supplementation preferably increases histone Kla, but not Kac. Moreover, histone Kla has distinct dynamics, increasing in abundance over 24 h postbacterial challenge, suggesting distinct regulation of histone lactylation versus acetylation in M1 macrophages despite similar glycolytic origins [23].
Identified in HeLa cells and mouse bone-marrow derived macrophages by high-performance LC (HPLC)-MS/MS, lactylated H3 lysine 18 (H3K18la) was assessed by chromatin immunoprecipitation coupled to sequencing (ChIP-sequencing) and RNA-sequencing in M1 macrophages compared with H3K18ac to deduce lactylation-specific properties (Figure 2, blue-green) [23]. Both histone PTMs were enriched around TSSs and, surprisingly, ~68% of genes marked by increased H3K18la levels during M1 macrophage polarization were not associated with increased H3K18ac levels, suggesting lactylation-specific regulation of this process. Interestingly, functional annotation analysis of those unique H3K18la-enriched genes implicated biological processes that are not inflammatory in nature or indicative of M1 macrophages, but instead enriched more in M2-like pathways, such as wound healing [23]. Given that M2-like phenotypes are linked with tumor-associated macrophages that produce immunosuppression and angiogenesis within tumor microenvironments [26], macrophage histone lactylation may have additional roles in tumor progression. Lastly, using a recombinant transcription assay, H3 lactylation was shown to directly activate transcription, and, under these in vitro conditions, was transferred to histones via p300 [23].
Histone Benzoylation
The first histone PTM described with an aromatic acyl group, histone lysine benzoylation (Kbz), was discovered in 2018 [27]. Histone Kbz stems from the donor benzoyl-coA, an endogenous intermediate formed by host microbes, and the exogenous compound sodium benzoate, which is commonly used as a food preservative [28]. Histone Kbz was discovered by HPLC-MS/MS with a large mass shift of ~104 Da. Given its weight (at least double that of other SCFA-derived PTMs) and aromatic ring, Kbz has strong hydrophobic properties and likely occupies considerable physical space, suggesting that its presence enhances chromatin accessibility. Evolutionarily, histone pan-benzoylation has been detected in Drosophila, mouse, and human cell lines [27], supporting a potentially conserved function for this PTM. Interestingly, benzoyl-coA and global histone Kbz levels were found to be increased in vitro by a concentration of sodium benzoate supplementation that is less than the 0.1% allowed in foods by the US Food and Drug Administration [27].
To identify enzymes that preferentially regulate histone Kbz, a screen of several histone deacetylases (HDACs) determined that the NAD-dependent Sirt2 protein serves as a debenzoylase, with a Michaelis constant that is 8.8-fold higher for benzoylated versus acetylated histone peptides (Figure 2, black) [27]. ChIP-sequencing of histone pan-Kbz demonstrated genomic enrichment of pathways, including phospholipase D signaling and glycerophospholipid metabolism, that are not observed by genomic enrichment of histone pan-Kac. These data suggest that histone Kbz has functions distinct from acetylation, although both PTMs correlate strongly with permissive gene expression. How the distinct effects of Kbz are mediated remain unknown, although its large structure and hydrophobicity serve as prime candidates, as suggested by in silico modeling that hints at enhancement of Kbz binding to Sirt2 via hydrophobic interactions [27]. Further work examining the role of histone benzoylation and its downstream consequences in vivo will be fascinating, especially considering its uncommon structure and the pervasiveness of this molecule in the food industry.
Histone Fatty-Acylation
Histone acyl-PTMs are not limited to proteins or metabolites, but also include lipids. Protein modification by lipids, including fatty acids, phospholipids, and steroids, occurs broadly throughout eukaryotic cells, increasing both protein hydrophobicity and membrane affinity [29]. To date, two types of histone lipidation state have been identified: S-palmitoylation/S-acylation and O-palmitoylation, both usually describing the addition of the 16-carbon palmitic acid [30]. Here, we discuss histone PTMs generated from acylation reactions with long-chain fatty acids [31].
S-acylation is the best-characterized protein lipidation state to date. Cysteine residues have sulfur-containing thiol side chains that attack electrophilic species, including molecules with carboxylic acid substituents, such as palmitic acid [32]. The resulting thioester bond between cysteine and palmitate is energetic and labile, allowing for reversibility. O-palmitoylation is less studied, with nucleophilic attack by a serine hydroxyl group forming an oxygen ester bond. Although hydroxyls are less nucleophilic compared with thiols, the resulting bond with palmitate is more stable and may result in distinct properties [30].
Click chemistry was again used to assess lipidated histones, where fatty acid analogs were ‘clicked’ onto reporter molecules linked to a cleavable biotin moiety. Using this strategy, metabolically labeled lipidated proteins were captured via biotin–streptavidin interactions and identified by MS, with lipidation of H3 variants being observed [33]. All H3 variants share a cysteine residue at position 110 that is ripe for fatty acylation, but when mutated to an alanine abrogates lipidation levels [33], suggesting that this residue is a primary site for fatty acylation. While the functional impact of H3 cysteine 110 lipidation was not examined in this study, based upon prior studies where mutagenesis of cysteine 110 to glutamic acid (C110E) was found to disrupt histone tetramer formation [34], addition of a long-chain lipid to this residue might similarly destabilize the nucleosome [35]. Furthermore, H3.1 and testes variant H3.1t have a cysteine residue at position 96 on the nucleosome surface [35], potentially doubling the capacity for S-acylation compared with other H3 variants, suggesting that these variants have differential functions when lipidated.
In another study, lipidation was detected on H4, where the presence of zero cysteines and two serines in the H4 sequence suggests that lipidation occurs via O-palmitoylation [36]. Mutagenesis of the serine at position 47 reduces H4 palmitoylation levels and substantially decreases RNA synthesis, suggesting transcriptional repression (Figure 2, dark gray). Furthermore, the enzyme Lpcat1 was proposed as the H4 palmitoylation writer in mouse lung epithelial cells [36]. Further work will be required to better understand the precise implications of site-specific histone lipidations, although the potential for scaffolding interactions, signal transduction, and altered protein stability exists, as described for other lipidation events. Moreover, given the existence of lysine lipidation in cytoplasmic proteins [37,38], other histone lipid-PTMs may occur endogenously.
Histone Monoaminylation
In 2019, histone serotonylation was described as a novel PTM [39] (Table 1). Serotonin (5-HT, 5-hydroxytryptamine) is commonly known as a brain neurotransmitter. Disruptions in its signaling are implicated in numerous neuropsychiatric disorders, including a spectrum of autism, depression, and anxiety-related syndromes. Serotonin is a biogenic monoamine and, for many years, was shown to modify cytosolic proteins through the enzymatic activities of the tissue transglutaminase 2 (Tgm2) protein, a process referred to as ‘monoaminylation’ [40]. Tgm2 can target peptide-associated glutamines for transamidation, in which the active cysteine at position 227 of Tgm2 binds glutamine δ-carbons, forming a thioester bond, followed by monoamine attack at the glutamine γ-carboxamide to generate monoaminylated proteins [41]. In particular, serotonylation of cytosolic substrates has been described to activate cytoskeletal proteins and small GTPases to physiologically impact platelets and myocytes, suggesting that protein monoaminylation dictates important regulatory outcomes in vivo [42,43].
Previous observations of extravesicular 5-HT (as well as other monoamines) in the nucleus in brain [44,45], along with known Tgm2 transamidation activities against histone proteins (e.g., glutamine-lysine crosslinking events), encouraged interrogations of histone monoaminylation [46]. Indeed, click chemistry using a labeled 5-HT analog provided initial evidence for Tgm2-dependent serotonylation in cells, occurring specifically on histone H3 [39]. Subsequent in vitro Tgm2 enzymatic assays, along with in vivo LC-MS/MS, identified H3Q5 as a dominant site of serotonylation, a PTM that exists both in isolation (H3Q5ser) and in combination with neighboring lysine 4 methylation, a transcriptionally permissive mark when tri-methylated (H3K4me3Q5ser). H3K4me3Q5ser is detected across multiple tissues, with modest enrichment in serotonergic colon and brain, and is evolutionarily conserved [39].
Using a combination of human pluripotent stem cell-derived 5-HTergic neurons and rat 5-HT neuronal precursors (RN46A-B14), dramatic upsurges of H3K4me3Q5ser were observed upon cellular differentiation, correlating with H3K4me3Q5ser increases occurring in mouse fetal brain from early to late gestation [39]. Comparing enrichment profiles for H3K4me3Q5ser versus H3K4me3 alone across differentiation, this PTM was observed to display several unique alterations in its genomic distribution, which positively correlated with differentially expressed genes, suggesting that the combinatorial PTM associates with permissive transcription (Figure 2, vermilion). Transduction of mutant histone variant H3.3 (a variant that incorporates into chromatin post mitotically) containing Q5 mutated to an alanine (Q5A) into RN46A-B14 cells reduced serotonylation levels, resulting in slower growth of neuronal processes and a significant reversal of gene expression programs important for differentiation. These data support histone serotonylation as an important factor for serotonergic neuronal differentiation. Furthermore, H3K4me3Q5ser potentiates interactions with the transcription factor complex TFIID compared with H3K4me3 alone, which may contribute to its permissive regulation [39]. Future work carving out roles for histone serotonylation in the various biological processes where 5-HT signaling is altered, including stress, infection, and antidepressant usage, will be critical to understanding its biological relevance in vivo.
In 2020, another histone monoaminylation mark was added to the PTM catalog: dopaminylation [47]. Dopamine (3-hydroxytyramine) is a neurotransmitter metabolized from tyrosine and containing a catechol moiety. This biogenic monoamine is heavily implicated in learning and memory, reward, addiction, and motor function [48].
Using a combination of LC-MS/MS and enzymatic assays, we showed that, similar to histone serotonylation, histone dopaminylation is transamidated by Tgm2 onto H3Q5, both alone (H3Q5dop) and in combination with K4 tri-methylation (H3K4me3Q5dop), and associates with transcriptional regulation in brain [47]. In the ventral tegmental area (VTA), a dopamine-synthesizing brain region involved in behavioral reward including responses to drugs of abuse, H3Q5dop levels were reduced in both rat and human postmortem brain tissues after chronic cocaine use [47]. In rat VTA, the directionality of H3Q5dop dysregulation was dependent on the timing of drug intake and withdrawal, where H3Q5dop levels were increased a month after cocaine access ended in rats.
To better understand the functional impact of increased H3Q5dop during abstinence, dominant negative viral vectors were delivered into rat VTA to reduce H3Q5dop levels following cocaine exposures. RNA-sequencing on virally infected tissues revealed that attenuated H3Q5dop levels reversed the transcriptional patterning normally induced during cocaine withdrawal. Moreover, reducing H3Q5dop blocked cocaine-seeking behaviors in rats, along with aberrant drug cue-induced release of dopamine into nucleus accumbens, a key brain reward structure that receives dopaminergic innervation from the VTA [47]. These data suggest that dopamine additionally has critical neurotransmission-independent roles in mediating aberrant gene expression associated with the precipitation of drug relapse vulnerability.
Whether histone dopaminylation enhances the canonical effects of H3K4me3 similar to H3K4me3Q5ser remains to be seen. Moreover, given that serotonylation and dopaminylation modify the same glutamine questions whether these PTMs are restricted to specific brain regions or exist in competition within common brain structures. Additionally, other monoamines also likely modify histones, thus compelling their discovery alongside the necessary readers and erasers of these PTM states.
Histone Hydroxylation
The first histone hydroxylation PTM was identified during a large-scale screen in 2011, but without subsequent investigations, biological roles for tyrosine hydroxylation remain unknown [49]. A more recent study further described hydroxylation of histone lysine carbon chains, despite the lack of MS detection in previous work [50] (Table 1). Histone 5-hydroxylysine modifications, catalyzed by the known lysyl hydroxymethylase and arginine demethylase Jumonji domain-containing 6 (Jmjd6) in mouse embryos, were detected by amino acid composition analysis, allowing for the detection of even minute histone 5-hydroxylysine levels (<0.1% of histone lysine PTMs) [51]. Interestingly, synthetic peptides containing 5-hydroxymethylated lysines are resistant to enzymatic deposition of acetyl and methyl groups, and vice versa, signifying that the presence of histone lysyl 5-hydroxymethylation may function to reduce acetylation and methylation levels [51]. Therefore, while the effects of histone 5-hydroxylation remain unknown, its regulation of other PTMs suggests, at least indirectly, that it promotes downstream consequences on transcription. However, further work is required to fully understand the physiological and transcriptional implications of this competitive relationship, including whether histone dehydroxylases exist that might influence these interactions.
Nonenzymatic Histone PTMs
While we classically consider all histone PTMs to have writers, recent studies have demonstrated that this assumption is incorrect. In fact, advances in nonenzymatic mechanisms of adduction suggest that histones are vulnerable to numerous small, reactive molecules, including metabolites and toxins within the nuclear environment. As we previously alluded to, N-terminal histone tails are rich in lysines and arginines, which have potent nucleophilic side chains capable of aberrant linkages to a host of exogenous and transiently electrophilic species. Given that these nonenzymatic PTMs are generally lowly abundant and can be generated by metabolism of exogenous compounds or during sample processing (such as for some methionine oxidation [52]), more work is needed to distinguish between artificial by-products with unintended effects versus conserved biological pathways. Importantly, some nonenzymatic histone adductions appear to have designated erasers, including HDACs, where the reversibility of these nonenzymatic histone PTMs, despite their low concentrations, implies functionality. Here, we briefly discuss select nonenzymatic histone PTMs described over the past decade (Table 1) (for an elegant review of additional nonenzymatic histone PTMs, see [53]).
Nonenzymatic Histone Acylation
The complexity of histone PTMs deepens because acylation reactions are now known to occur, under certain circumstances, without enzymes, creating more questions than answers with regard to the prospect of a deliberate histone code. Although many acyl-PTMs can react nonenzymatically to histones in vitro, acidic acyl-PTMs are most likely to occur without enzymatic assistance. Indeed, acidic acyl-coA donors are particularly reactive, outcompeting acetyl-coA and other donors in protein PTM deposition [54].
These acidic acyl donors are distinguished by charge and a terminal carboxylic acid, which can undergo intramolecular general base catalysis to produce five to six-carbon cyclic anhydride intermediates, rendering instability to these metabolites that accelerates histone acylation (Figure 2, orange) [55]. Succinyl-coA and glutaryl-coA serve as such donors, but not malonyl-coA (with three carbons, it cannot undergo intramolecular catalysis), supporting the idea that differential acylation reactivity is due to anhydride formation and not simply charge. With these parameters, 3-hydroxy-3-methylglutaryl-coA, 3-methylglutaconyl-coA, and 3-methylglutaryl-coA have been identified as highly reactive species that are capable of lysine PTMs with potential to influence chromatin regulation [19].
Currently, however, additional studies are lacking with respect to these PTMs, with particular scarcity in the implementation of in vivo systems for downstream assessments of biological function. it is likely that nonenzymatic histone acylation would produce similar results as enzymatically mediated acylation with respect to transcriptional and nucleosome regulation, although this question has thus far been difficult to address. Separation of enzymatic-dependent versus -independent histone acylation states remains challenging, especially given the abundance of HATs in the nuclear environment. However, one recent study described a preference for nonenzymatic histone acetylation at select lysine residues versus those targeted by HATs [56], suggesting that this pathway is biologically meaningful and not simply the consequence of metabolic by-products.
Histone Glycation
Advanced glycated end products (AGEs) are biomarkers for diabetes, atherosclerosis, and neurodegenerative disorders, and are prevalent in tumors [57]. Nonenzymatic glycation involves covalent binding of a reducing sugar or reactive carbonyl species with nucleophilic lysines and arginines that rearrange to form AGEs. AGEs detrimentally increase oxidative stress (glycoxidation) and crosslink with other proteins that inflict cellular damage and toxicity [58]. Histones are vulnerable to glycation, where accumulation of glycated-PTMs may be particularly harmful owing to the long half-lives of histones [59].
Two labs recently demonstrated that the glycolytic by-product methylglyoxal (MGO) facilitates histone glycation, with greater reactivity against arginine residues [59,60]. MGO treatments drive DNA–histone and histone–histone crosslinking in vitro that can disrupt both nucleosome assembly and chromatin accessibility, contrasting with prior work suggesting that H2A glycation increases nucleosome stability [61], potentially due to site specificity of this modification. With regards to transcriptional regulation, no changes in total abundance of RNA transcripts following MGO treatment were observed in one study [60], although in another study, DNA accessibility was reduced following MGO treatment [59]. Physiologically, high histone glycation levels have been observed in hyperglycemic and breast cancer models, and are reduced by the previously identified [62] deglycase enzyme DJ-1 via hydrolysis of the aminocarbinol MGO-arginine intermediate (Figure 2, sky-blue) [60]. Notably, histone glycation reduces the abundance of other PTMs, such as ubiquitination and acetylation, where HDAC inhibition before MGO treatment prevents glycation, suggesting a protective mechanism through occupancy of the site by acetylation [59]. Thus, despite glycation readily forming nonenzymatic adducts under pathophysiological conditions, targeting DJ-1 for deglycation may be useful for disorders with widespread AGEs, although further work examining the causality of histone glycation in disease states and its transcriptional impact is needed.
Histones and Oxidative Stress
Numerous fields have implicated reactive oxygen species (ROS) generation, such as free radicals or peroxides, in disease pathophysiology, many of which can originate from normal metabolism or exogenous toxins. For example, lipids are peroxidized to produce reactive electrophilic aldehydes, including 4-oxo-2-nonenal (4-ONE), that readily bind proteins. Histone PTM by 4-ONE impacts all core histones in vitro, occurring at histone tail lysines via ketoamide adduction or histidines via Michael addition, where H3 and H4 4-ONylation inhibit nucleosome assembly [63] and Sirt2 was interestingly suggested to remove this 4-ONylated mark (Figure 2, light gray) [64]. Another reactive aldehyde, referred to as acrolein, commonly found in cigarette smoke, also adducts to histones and dysregulates nucleosome assembly. However, acrolein PTM uniquely reacts with non-DNA-bound free H3, impacting nuclear import and incorporation, leading to depleted nuclear H3 levels [65,66].
Glutathione is the most abundant thiol in cells, with numerous antioxidant effects and protein PTM capacity. Cysteine S-glutathionylation PTM levels rapidly increase during oxidative stress, where glutathione forms transient disulfide bonds with protein cysteine residues to facilitate cellular redox pathways and/or buffer cysteines from ROS adduction [67]. Therefore, dynamic regulation of glutathione levels may serve as a biological switch, underlying its importance to cellular homeostasis [68]. Of histone proteins, only H3 variants have cysteine residues, sharing a cysteine at position 110 that reduces nucleosome stability when S-glutathionylated [69]. H3.1 and H3.1t have an additional cysteine at position 96 [33], doubling their potential to sense cellular redox status via S-glutathionylation [69]. Thus, histones may sense cellular redox states and respond via chromatin changes, with mechanistic implications for aging and numerous diseases.
Histone Homocysteinylation
Homocysteine is a nonproteogenic amino acid derived from methionine metabolism. Excess homocysteine levels (hyperhomocysteinemia) correlate with risk for atherosclerosis, early pregnancy terminations, and neural tube defects, as well as serum protein modification by its metabolite homocysteine thyolactone (HTL) [70]. The cyclic thioester HTL can react with lysine ε-amines, producing protein homocysteinylation and enzymatic impairment [71]. A recent study described histone lysine homocysteinylation (KHcy) in vitro, where increased H3 KHcy alters the abundance of H3 methylation and acetylation [72], suggesting that homocysteinylation competes with, or regulates, other PTMs and their downstream effects.
Another study investigated how hyperhomocysteinemia contributes to offspring neural tube defects. Chicken embryos that exhibit neural tube defects following HTL treatment display high levels of H3 lysine 79 homocysteinylation (K79Hcy) in brain tissues. Using ChIP-sequencing on embryonic brains, H3K79Hcy was found to be enriched at loci related to neurodevelopment, with corresponding decreases in the expression of those genes [73], providing a potential function for histone homocysteinylation (Figure 2, red-purple). How histone KHcy imparts these effects, by readers and/or structural changes, remains to be uncovered. Moreover, further work investigating histone KHcy involvement in basal and disease states would be valuable.
Novel PTMs and the Histone Code
What do these novel PTMs mean in the context of the ‘histone code hypothesis’? It is possible that different PTM combinations reflect cellular metabolic states requiring distinct transcriptional programs. For example, in developing sperm, histone acylation patterning influences transcription and histone retention depending on combinations of H4 lysine 5 and lysine 8 acetylations with butyrylation, resulting in differential interactions with the bromodomain testis-associated HAT [74]. This study implies that, despite the similarity of their side chains (one versus three carbons) and both correlating with permissive transcription [74], histone acetylation and butyrylation are functionally distinct. This may hold true for other structurally similar histone PTMs, where perhaps binding of unique reader proteins is important for differential downstream effects, including the extent of transcriptional activation. Moreover, that readers with even slightly different binding and/or signaling capabilities can influence the signaling of another PTM reader, or together produce a unique cellular cascade, suggests an extra level of regulation for the histone code.
In another study, increasing histone KHcy suppressed Kac and methylation levels, correlating with previous observations of excess homocysteine and reduced methyl donors in Alzheimer’s disease [72,75]. Correspondingly, another proposed function of histone acetylation is safeguarding lysine residues from nonenzymatic adduction of toxins (i.e., physically occupying the lysine binding site), thereby barring PTMs such as glycation and their downstream effects [59].
Characterization of H3K4me3Q5ser suggests that some PTMs function most prominently to reinforce principal PTMs; for example, Q5ser may stabilize H3K4me3, thereby potentiating its permissive status [39]. Such reinforcement may be tissue or circumstance specific, perhaps explaining the existence of several apparently redundant PTMs throughout eukaryotic systems. Additionally, the combinatorial H3K4me3Q5ser PTM may function distinctly from H3Q5ser alone, where deconvoluting this combinatorial modification, or considering Q5ser in conjunction with additional PTMs (e.g., neighboring phosphorylated threonines), might also be informative [39]. Given the recent nature of these discoveries, few studies have examined in detail the consequences of PTMs discussed throughout this review, let alone their activities in combination with additional PTMs. Assessing the possibility that these new histone modifications add to an increasingly complicated, multivalent code will necessitate future top- and middle-down MS-based methodologies, among others, to fully address this controversial question [76].
Concluding Remarks
As studies continue to link histone PTMs with transcriptional regulation, biology, and disease, important questions remain (see Outstanding Questions). Causal studies are needed to verify these relationships, since most, if not all, of these PTMs similarly occur on cytosolic and nonhistone nuclear proteins. Moreover, site-specific characterizations of histone PTMs and their downstream effects will be informative. Fortunately, these tasks are now easier with innovative techniques recently developed that permit site- and locus-specific manipulations of select histone PTMs (Box 3). Together, the histone PTMs discussed in this review represent an expanding catalog of chemical modifications capable of impacting cellular programs and physiology, underlining the contributions of chromatin regulation to disease pathophysiology (Figure 2). If the past decade is any indication, future work in this field will most certainly lead to the identification of additional novel histone PTMs, where pinpointing their precise biological functions and enzymatic regulators may aid in future drug and biomarker discovery efforts.
Outstanding Questions.
Given the structural similarity of several histone acyl PTMs, how are the specific cellular consequences of unique histone acylations elicited?
What are the biological effects of histone lipidation?
How is the deacylase enzyme Sirt2 specified for removal of short-chain versus long-chain fatty acid-derived histone PTMs?
What other biogenic monoamines can act as chromatin modifiers?
What is the function of histone lysine and tyrosine hydroxylation?
Do nonenzymatic histone acylations have biological purposes, or are they simply metabolic by-products gone wrong?
Do histones sense cellular redox states via a balance between nonenzymatic addition of ROS and/or reactive nitrogen species versus glutathione?
How does emerging evidence of nonenzymatic histone PTMs fit within the context of a deliberate ‘histone code?’
Given the specific dynamics of individual histone PTMs (cell type specific, sex dependent, developmentally regulated, cell cycle dependent, altered by gene × environment interactions, etc.), let alone in conjunction with other PTMs, how can we rectify the sheer number of histone PTM combinatorial possibilities within the framework of a so-called ‘histone code hypothesis’?
Do histone PTMs causally influence transcriptional or other DNA-related signaling programs to impact biology and disease, or is it all correlative?
Can we target enzymatic regulators of novel histone PTMs to aid in future drug or biomarker discovery efforts?
With the advancement of technology allowing detection of several low abundance histone PTMs this decade alone, will there be development of even more sensitive techniques leading to additional histone PTM discovery over the next decade?
Box 3. Epigenome Editing to Assess Causality of Histone Modifications.
Researchers have recently begun using ‘epigenomic editing’ approaches using zinc finger proteins (ZFPs), transcription activator-like effectors (TALEs), and, more recently, CRISPR/Cas9 nuclease systems. With these technologies, DNA-binding effector domains can be guided to specified DNA sequences, allowing for assessments of individual histone PTMs in vivo (reviewed in [91]). For example, in a rodent model of stress and drug abuse, ZFPs and TALEs were engineered to bring the p65 domain of NFkB to the locus of the FosB gene, a gene previously associated with the pathophysiology of addictive and depressive-like behaviors [92]. Guidance of this p65 domain to the FosB locus in mouse brain tissue increased histone acetylation across the FosB promoter by recruitment of the histone acetyltransferase CREB, activating FosB expression and enhancing behavior associated with stress and cocaine addiction [92]. In another study utilizing CRISPR/Cas9 technology, a synthetic HDAC was produced by fusing the full-length human HDAC3 with a dCas9 (deactivated endonuclease activity) and targeted to specific loci by guide RNAs (gRNA) [93]. This synthetic HDAC-dCas9 successfully catalyzed deacetylation of H3K27 near the Mecp2 promoter and correspondingly reduced Mecp2 expression [93], providing a potential approach to specifically manipulate expression of this gene, which is crucial in the origin of Rett Syndrome.
Another exciting method recently developed involves trans-splicing of histones using ultrafast split-inteins that, in two steps, can de novo generate semisynthetic chromatin fused with site-specific PTMs [94]. For example, to test the specific role of H2B lysine 120 ubiquitination (H2BK120ub) on canonical enhancement of H3 lysine 79 dimethylation (H3K79me2), David et al. generated constitutive expression of a folded N-terminal intein fragment fused to the C-terminal of histone H2B in human embryonic kidney cells [95]. Introduction of the cognate C-terminal intein fragment fused to a ubiquitin moiety on H2BK120 resulted in high-affinity binding of the two fragments, followed by self-excision of the inteins, leaving behind H2B with the engineered K120ub PTM that, as predicted, stimulated H3K79me2 levels, suggesting the involvement of these semisynthetic histones in functional crosstalk with endogenous histone PTMs. Given that specific histone PTMs are notoriously difficult to manipulate, especially ubiquitination, which, with its large size (~8.5 kDa), has no appropriate amino acid mimic, chromatin tagging using ultrafast trans-splicing split-inteins may provide a powerful tool to determine causality of histone PTMs [95]. Moreover, as the complexity of the ‘histone code’ deepens, using these techniques to further investigate causality of combinatorial PTMs will be necessary.
Highlights.
Histone post-translational modifications (PTMs) regulate several biological processes, including transcription and nucleosome assembly, that can contribute to functional consequences in disease, physiology, and behavior.
Over the past decade, new histone PTMs (monoaminylation, lipidation, hydroxylation, etc.) have been identified using technological advances that allow the detection of low-abundance PTMs; yet, our understanding of these PTMs alone or in combination, such as in the context of a ‘histone code’, remains poor.
New studies reveal a class of nonenzymatic histone PTMs derived from covalent binding of highly reactive species, refuting the common notion that histone PTMs require writers, readers, and erasers for biological significance.
Despite numerous studies highlighting the importance of histone PTMs in various biological pathways, their causal roles remain undefined and will require innovative techniques to manipulate them in a cell type-, locus-, and site-specific manner.
Acknowledgments
We apologize to those researchers whose work was not cited due to lack of space. This work was supported by the National Institute of Health DP1 DA042078, R21 DA044767, R01 HD097088, and R01 MH116900 (to I.M.). We thank C. Walters for useful discussion on this manuscript.
Glossary
- Amino acid composition analysis
biochemical technique used to determine the identity of proteins, peptides, macromolecules, and so on, by hydrolyzing the sample to its amino acid constituents and separating by chromatography, allowing the quantitation of the amino acid content
- Biogenic monoamines
class of molecules structurally categorized as containing one amino group connected to an aromatic ring by a two-carbon chain; includes neurotransmitters, such as serotonin, dopamine, norepinephrine, and histamine
- Click chemistry
describes the irreversible, highly specific, and thermodynamic reactions that conjugate a select biomolecule with a reporter or probe of choice in water with minimal by-products [e.g., copper (I)-catalyzed azide-alkyne cycloaddition]
- Core histones
DNA-binding proteins that together form the nucleosome, including the canonical histones H2A, H2B, H3, and H4, and their variants
- Electrophile
a chemical species that accepts an electron pair to form new covalent bonds with a nucleophile
- Erasers
enzymes that catalyze removal of histone PTMs
- Histone code hypothesis
postulates that transcription is influenced by the presence and combination of histone PTMs
- Histone post-translational modifications (PTMs)
chemical moieties covalently bound to histone proteins that can alter interactions with DNA
- Intramolecular general base catalysis
reaction occurring at neutral pH where an unprotonated carboxylate attacks the carbonyl group within the same molecule, cyclizing to produce five–six carbon reactive anhydrides that exhibit potent electrophilicity
- Liquid chromatography with tandem mass spectrometry (LC-MS/MS)
technique that separates a compound mixture by its chemical properties using high-performance liquid chromatography (HPLC) columns, followed by ionization and further separation based on mass:charge ratios. Next, select ions of similar mass: charge ratios are split into smaller daughter ions in a second ionization event and separated based on mass: charge ratios, measured by an ion detector that transmits this information to a computer for analysis
- Long-chain fatty acids
as subset of fatty acids containing ~12–20 carbon atoms, characteristically exhibiting water insolubility, pronounced lipophilicity, and incorporation into micelles and lipid bilayers (not observed for short chain fatty acids); examples include palmitic acid and myristic acid
- Michael addition
chemical reaction describing the addition of nucleophilic enolates of carbonyl compounds, including ketones and aldehydes, to α,β-unsaturated carbonyl compounds via 1,4-addition to produce 1,5- dicarbonyl compounds
- Modification stoichiometry
fraction of a select PTM at a specific site compared with the total quantity of that site; also known as PTM site occupancy or fractional occupancy
- Nucleophile
chemical species that donates an electron pair to form new covalent bonds with an electrophile
- Readers
protein effectors with domains that recognize and bind histone PTMs to mediate downstream cellular effects
- Short-chain fatty acids (SCFAs)
a subset of fatty acids containing fewer than six carbon atoms, produced by gut microbiota; examples include butyrate, propionate, crotonate, and acetate
- Thioester
molecules with the chemical group C-S-CO-C, which serve as reactive electrophiles
- Writers
enzymes that Catalyze addition of histone PTMs
References
- 1.Zhao Y et al. (2015) Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. Biol 7, a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg RD et al. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 [DOI] [PubMed] [Google Scholar]
- 3.Mersfelder EL et al. (2006) The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res 34, 2653–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips DM (1963) The presence of acetyl groups of histones. Biochem. J 87, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allfry VG et al. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell JE et al. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 [DOI] [PubMed] [Google Scholar]
- 7.Struhl K (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12, 599–606 [DOI] [PubMed] [Google Scholar]
- 8.Wellen KE et al. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science (80-.) 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonnell E et al. (2016) Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep 17, 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mews P et al. (2019) Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabari BR et al. (2015) Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y et al. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y et al. (2016) Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 62, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z et al. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan M et al. (2014) Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19, 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabari BR et al. (2017) Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol 18, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaczmarska Z et al. (2017) Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol 13, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith BC et al. (2009) Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta 1789, 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner GR et al. (2017) A class of reactive Acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab 25, 823–837 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trefely S et al. (2020) Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Mol. Metab Published online February 14, 2020. 10.1016/j.molmet.2020.01.005 [DOI] [PMC free article] [PubMed]
- 21.Kolb HC et al. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed 40, 2004–2021 [DOI] [PubMed] [Google Scholar]
- 22.Bao X et al. (2019) Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Mol. Cell 76, 660–675 e9 [DOI] [PubMed] [Google Scholar]
- 23.Zhang D et al. (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks GA (2018) The science and translation of lactate shuttle theory. Cell Metab 27, 757–785 [DOI] [PubMed] [Google Scholar]
- 25.Liberti MV et al. (2016) The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci 41, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benoit M et al. (2008) Macrophage polarization in bacterial infections. J. Immunol 181, 3733–3739 [DOI] [PubMed] [Google Scholar]
- 27.Huang H et al. (2018) Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun 9, 3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piper JD, et al. Benzoate and sorbate salts: a systematic review of the potential hazards of these invaluable preservatives and the expanding spectrum of clinical uses for sodium benzoate. Compr. Rev. Food Sci. Food Saf 16, 868–880 [DOI] [PubMed] [Google Scholar]
- 29.Jiang H et al. (2018) Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev 118, 919–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chamberlain LH et al. (2015) The physiology of protein S-acylation. Physiol. Rev 95, 341–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schönfeld P et al. (2016) Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J. Lipid Res 57, 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bischoff R et al. (2012) Amino acids: chemistry, functionality and selected non-enzymatic post-translational modifications. J. Proteome 75, 2275–2296 [DOI] [PubMed] [Google Scholar]
- 33.Wilson JP et al. (2011) Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell. Proteomics 10 M110.001198–M110.001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banks DD et al. (2004) Folding mechanism of the (H3–H4) 2 histone tetramer of the core nucleosome. Protein Sci 13, 1304–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hake SB et al. (2006) Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. U. S. A 103, 6428–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou C et al. (2011) Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J. Biol. Chem 286, 28019–28025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H et al. (2016) Lysine fatty acylation promotes lysosomal targeting of TNF-α. Sci. Rep 6, 24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H et al. (2013) SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrelly LA et al. (2019) Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walther DJ et al. (2011) Novel roles for biogenic monoamines: from monoamines in transglutaminase-mediated post-translational protein modification to monoaminylation de-regulation diseases. FEBS J 278, 4740–4755 [DOI] [PubMed] [Google Scholar]
- 41.Stamnaes J et al. (2008) The propensity for deamidation and transamidation of peptides by transglutaminase 2 is dependent on substrate affinity and reaction conditions. Biochim. Biophys. Acta Protein Proteomics 1784, 1804–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walther DJ et al. (2003) Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell 115, 851–862 [DOI] [PubMed] [Google Scholar]
- 43.Watts SW et al. (2009) Serotonylation of vascular proteins important to contraction. PLoS ONE 4, e5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young AB et al. (1971) Nuclear localization of histamine in neonatal rat brain. Science 173, 247–249 [DOI] [PubMed] [Google Scholar]
- 45.Colgan LA et al. (2009) Activity-dependent vesicular monoamine transporter-mediated depletion of the nucleus supports somatic release by serotonin neurons. J. Neurosci 29, 15878–15887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballestar E et al. (1996) Core histones are glutaminyl substrates for tissue transglutaminase. J. Biol. Chem 271, 18817–18824 [DOI] [PubMed] [Google Scholar]
- 47.Lepack AE et al. (2020) Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nutt DJ et al. (2015) The dopamine theory of addiction: 40 years of highs and lows. Nat. Rev. Neurosci 16, 305–312 [DOI] [PubMed] [Google Scholar]
- 49.Tan M et al. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webby CJ et al. (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science (80-.) 325, 90–93 [DOI] [PubMed] [Google Scholar]
- 51.Unoki M et al. (2013) Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem 288, 6053–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghesquière B et al. (2014) Proteomics methods to study methionine oxidation. Mass Spectrom. Rev 33, 147–156 [DOI] [PubMed] [Google Scholar]
- 53.Zheng Q et al. (2019) (De)Toxifying the epigenetic code. Chem. Res. Toxicol 32, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simithy J et al. (2017) Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun 8, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bender ML (1960) Mechanisms of catalysis of nucleophilic reactions of carboxylic acid derivatives. Chem. Rev 60, 53–113 [Google Scholar]
- 56.Kuo Y-M et al. (2013) Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS ONE 8, e54896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hellwig M et al. (2014) Baking, ageing, diabetes: a short history of the Maillard reaction. Angew. Chem. Int. Ed. Eng 53, 10316–10329 [DOI] [PubMed] [Google Scholar]
- 58.Vlassara H (2005) Advanced glycation in health and disease: role of the modern environment. Ann. N. Y. Acad. Sci 1043, 452–460 [DOI] [PubMed] [Google Scholar]
- 59.Zheng Q et al. (2019) Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun 10, 1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galligan JJ et al. (2018) Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. U. S. A 115, 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mir AR et al. (2014) Methylglyoxal mediated conformational changes in histone H2A-generation of carboxyethylated advanced glycation end products. Int. J. Biol. Macromol 69, 260–266 [DOI] [PubMed] [Google Scholar]
- 62.Richarme G et al. (2015) Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem 290, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galligan JJ et al. (2014) Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J. Am. Chem. Soc 136, 11864–11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin J et al. (2016) SIRT2 Reverses 4-oxononanoyl lysine modification on histones. J. Am. Chem. Soc 138, 12304–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen D et al. (2013) Cigarette smoke component acrolein modulates chromatin assembly by inhibiting histone acetylation. J. Biol. Chem 288, 21678–21687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang L et al. (2016) Mechanisms underlying acroleinmediated inhibition of chromatin assembly. Mol. Cell. Biol 36, 2995–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grek CL et al. (2013) Causes and consequences of cysteine s-glutathionylation. J. Biol. Chem 288, 26497–26504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong Y, et al. S-Glutathionylation: from molecular mechanisms to health outcomes. Antioxid. Redox Signal 15, 233–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.García-Giménez JL et al. (2013) Histone h3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid. Redox Signal 19, 1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marczak L et al. (2011) Analysis of site-specific N-homocysteinylation of human serum albumin in vitro and in vivo using MALDI-ToF and LC-MS/MS mass spectrometry. J. Proteome 74, 967–974 [DOI] [PubMed] [Google Scholar]
- 71.Jakubowski H (1999) Protein homocysteinylation: possible mechanism underlying pathological consequences of elevated homocysteine levels. FASEB J 13, 2277–2283 [PubMed] [Google Scholar]
- 72.Xu L et al. (2015) Crosstalk of homocysteinylation, methylation and acetylation on histone H3. Analyst 140, 3057–3063 [DOI] [PubMed] [Google Scholar]
- 73.Zhang Q et al. (2018) Elevated H3K79 homocysteinylation causes abnormal gene expression during neural development and subsequent neural tube defects. Nat. Commun 9, 3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goudarzi A et al. (2016) Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell 62, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuo J-M et al. (2011) Is hyperhomocysteinemia an Alzheimer’s disease (AD) risk factor, an AD marker, or neither? Trends Pharmacol. Sci 32, 562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rando OJ (2012) Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev 22, 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krautkramer KA et al. (2016) Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fellows R et al. (2018) Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paul B et al. (2015) Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 7, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bultman SJ (2017) Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol. Nutr. Food Res 61, 1500902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamin JS et al. (2017) A ketogenic diet rescues hippocampal memory defects in a mouse model of Kabuki syndrome. Proc. Natl. Acad. Sci 114, 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Z et al. (2016) Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation. Mol. Cell 62, 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y et al. (2019) Chromodomain Y-like protein-mediated histone crotonylation regulates stress-induced depressive behaviors. Biol. Psychiatry 85, 635–649 [DOI] [PubMed] [Google Scholar]
- 84.Creyghton MP et al. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A 107, 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pradeepa MM et al. (2016) Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet 48, 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taylor GCA et al. (2013) H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res 23, 2053–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prus G et al. (2019) Analysis and interpretation of protein post-translational modification site stoichiometry. Trends Biochem. Sci 44, 943–960 [DOI] [PubMed] [Google Scholar]
- 88.Kim SC et al. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 89.Feller C et al. (2015) Global and specific responses of the histone acetylome to systematic perturbation. Mol. Cell 57, 559–571 [DOI] [PubMed] [Google Scholar]
- 90.Huang H et al. (2015) Quantitative proteomic analysis of histone modifications. Chem. Rev 115, 2376–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pulecio J et al. (2017) CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell 21, 431–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heller EA et al. (2014) Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors TL-17. Nat. Neurosci 17, 1720–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwon DY et al. (2017) Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat. Commun 8, 15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shah NH et al. (2014) Inteins: Nature’s gift to protein chemists. Chem. Sci 5, 446–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.David Y et al. (2015) Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem 7, 394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kossel A (1884) Ueber einen peptonartigen bestandtheil des zellkerns. Z Physiol. Chem 8, 511–515 [Google Scholar]
- 97.Ruben S et al. (1941) Long-lived radioactive carbon: C14. Phys. Rev 59, 349–354 [Google Scholar]
- 98.Tal’roze GV et al. (1968) Capillary system for the introduction of liquid mixtures into an analytical mass spectrometer. Russ. J. Phys. Chem 42, 1658–1664 [Google Scholar]
- 99.Taunton J et al. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science (80-.) 272, 408–411 [DOI] [PubMed] [Google Scholar]
- 100.Luger K et al. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]