Abstract
MicroRNAs (miRs) are powerful regulators of CNS development and diseases. Plasma and cerebrospinal fluid (CSF) miRs have recently been implicated as potential new sources for biomarker development. Previously we showed that miR-124–3p, an essential miR for neuronal identity, is highly abundant in neuronal exosomes and its expression decreases in spinal cord of ALS model SOD1G93A mice. In the current study, we found a disease associated reduction of miR-124–3p levels specifically in spinal neurons using in situ hybridization. By employing our recently developed exosome reporter mice in combination with sciatic nerve injections, we observed an increased association of miR-124–3p with spinal motor neuron-derived exosomes in SOD1G93A mice, even at the pre-symptomatic stage. Sciatic nerve injection delivered miR-124–3p is also more frequently localized outside of spinal motor neurons in SOD1G93A mice. Subsequent quantitative analysis of miR-124–3p levels in CSF exosomes from ALS patients found a significant correlation between CSF exosomal miR-124–3p levels and disease stage (indicated by the ALSFRS-R score) of (male) ALS patients. These results provide preliminary evidence to support the potential use of CSF exosomal miR-124–3p as a disease stage indicator in ALS.
Introduction
Amyotrophic Lateral Sclerosis (ALS) is a typical adult-onset neurodegenerative disease in which the majority of upper and lower motor neurons (UMNs and LMNs) are degenerated. Current understanding of pathogenic mechanisms of ALS has mostly been gained from studies of familial ALS that are caused by genetic mutations of individual genes, including sod1, tardbp, and c9orf72, etc. (1). Studies from human postmortem tissues and animal models, especially transgenic SOD1 mutant models indicate that MN degeneration is likely caused by a series of pathological events that involve both MNs and neighboring glial cells (2, 3). Altered glia to neuron pathways, such as induction of a pro-inflammatory environment, reduced glutamate uptake (4) and metabolic support (5), and abnormal maturation of newly formed oligodendrocytes (6), have all been implicated in the pathogenesis of ALS (1).
MicroRNAs (miRs) are powerful regulators of CNS development, especially miR-124–3p (the abundant and active isoform of miR-124) and miR-9 are critical determinants for neuronal identity (7, 8). Both positive (miR-124, −132, −133b, and −212) and negative (miR-34a, −181a, and −134) miR regulators of synaptic connectivity and plasticity have been identified (9). Of which, miR-218 is selectively expressed in MNs and its deficiency leads to neuromuscular junction defects and progressive MN loss (10). ALS associated genes tardbp (11) and fus (12) both promote miR biogenesis by either interacting with nuclear Drosha and cytoplasmic Dicer complex or by recruiting Drosha during the processing of a subset of pri- and pre-miRs. Their mutations in ALS may impair the biogenesis of a subset of miRs, as indicated by several miR profiling studies in ALS (13, 14). Dysregulated miRs have been implicated in the pathogenesis of ALS. Up-regulation of miR-155 has been found in the spinal cord of end-stage SOD1G93A mice and human ALS post-mortem samples to promote inflammation and its suppression significantly extends the survival of SOD1G93A mice (15, 16). Microglial NF-kB signaling is modulated by miR-125b which also contributes to elevated inflammation in SOD1G93A mice (17). In addition, muscle-specific miR-206 is increased in SOD1G93A mice to help compensate the axonal reinnervation of muscle (18).
Recent studies have begun to unveil that miRs can shuttle intercellularly as a genetic signal through secreted extracellular vesicles (EVs), especially exosomes (19). Exosomes are small vesicles (50–150nm in diameter) derived from intraluminal vesicles (ILVs) of endosomes in mammalian cells, which often contain a subset of miRs distinct from host cells (20). We have previously shown that neuronal miR-124–3p can be transferred into astrocytes through secreted neuronal exosomes and up-regulates glutamate transporter GLT1 expression in astrocytes (21). Although changes of miR levels have been investigated in various ALS models and human samples (plasma and CSF) (14, 22, 23), these studies often lack cellular resolution (due to bulk tissue), not follow the spatial dynamics of individual miR (intra- vs. extracellular), or have not correlated with disease severity of ALS patients. In the current study, we determined the intercellular localization of miR-124–3p, a miR abundant in neuronal exosomes (24), in the SOD1G93A mouse model of ALS. Our results also unveiled a strong positive correlation between exosomal miR-124–3p levels in CSF and disease severity of (male) human ALS patients.
Results
Intercellular localization of miR-124–3p in spinal cord of SOD1G93A mice
Our previous miR profiling of neuronal exosomes found that miR-124–3p is one of the most abundant miRs in neuronal exosomes (24). MiR-124–3p was also found as the top up-regulated miR in sporadic ALS CSF samples compared to control CSF samples. Interestingly, miR-124–3p is the only miR that is highly abundant in neuronal exosomes and is also changed in human ALS CSF samples, after comparing the highly abundant miRs in neuronal exosomes (24) and changed miRs in ALS CSF samples (23) (data not shown). This makes it an excellent candidate to examine its expression and intracellular/extracellular localization changes in the ALS mouse model. Expression of miR-124–3p is essential for neuronal differentiation (7). It specifically provides a permissive environment for the formation of spinal motor neurons by inducing expression of miR-375 in neural progenitor cells (25). Previously, we found that miR-124–3p levels are reduced in the spinal cord of end-stage SOD1G93A mice (21). To determine whether the overall transcription of miR-124 is altered in disease, we measured both mature miR-124–3p and the primary transcript of miR-124–3p (pri-miR-124) levels from lumbar cord of SOD1G93A and control (wild-type) mice at pre-symptomatic (P60) and mid-disease (P100–110) stages by TaqMan qPCR. All the original Ct values of miR-124–3p are within 28, significantly lower than typical background values (32–34). The expression levels of miR-124–3p in lumbar cord are comparable between SOD1G93A and control mice at P60 (Fig. 1A), while miR-124–3p levels are reduced 40% (p = 0.002) in SOD1G93A lumbar cord when the disease progresses to mid-stage (Fig. 1A). We also measured pri-miR-124 levels and found no significant change of pri-miR-124 in either pre-symptomatic or mid-stage (P100–110d) SOD1G93A mice as compared to wild-type littermates (Fig. 1B). These results confirm that there is a disease associated reduction of mature miR-124–3p in lumbar cord tissue of SOD1G93A mice and also suggest that the overall transcription of miR-124 before and during disease, indicated by the levels of primiR-124, remains unchanged despite the loss of mature miR-124–3p in the lumbar cord tissue of diseased SOD1 mice.
Figure 1. Mature miR-124–3p expression is decreased in SOD1G93A spinal motor neurons.
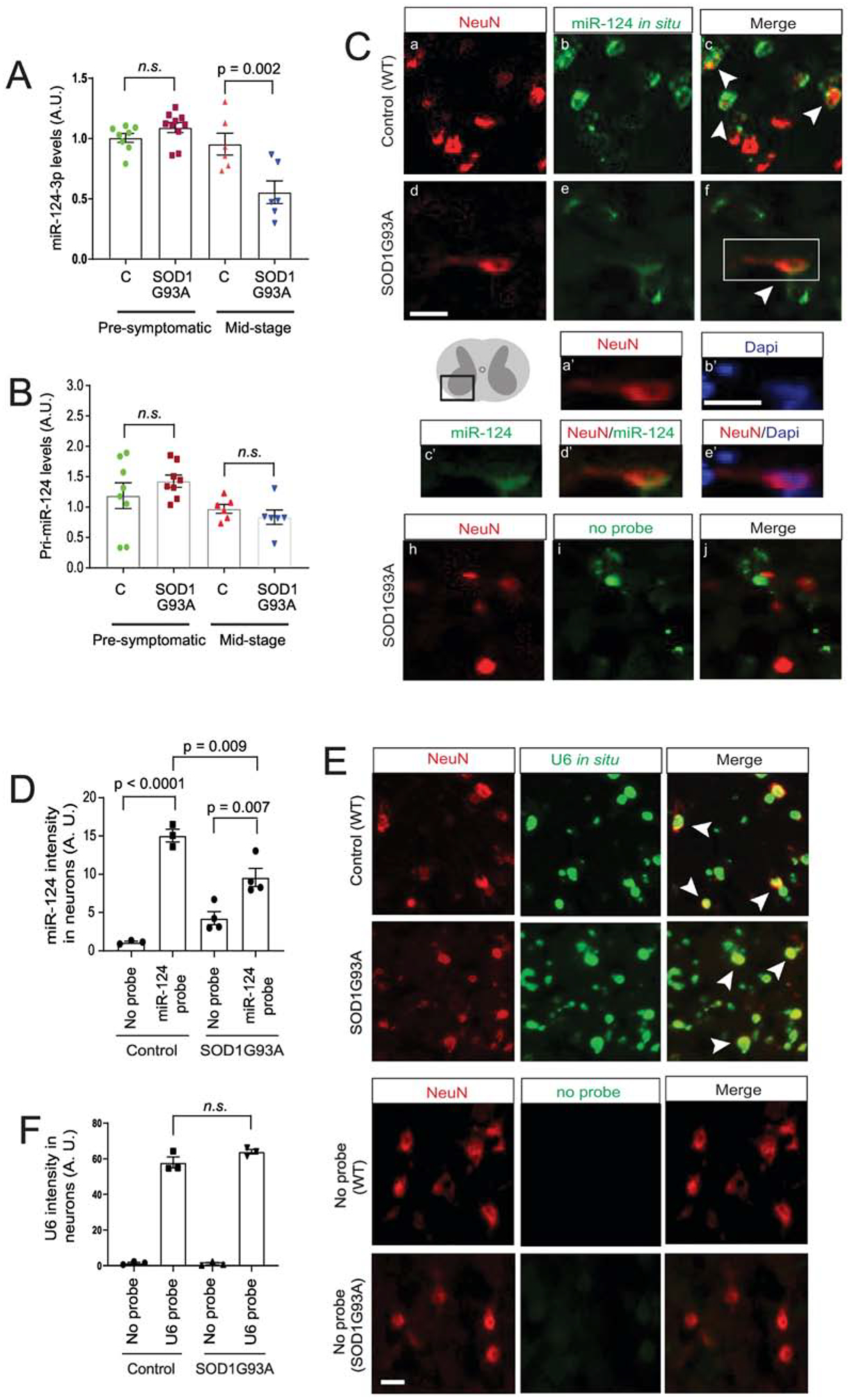
TaqMan probe-based qPCR detection of mature miR-124–3p (A) and pri-miR-124 expression (B) in lumbar cords isolated from SOD1G93A mice as compared to control wild-type littermates during pre-symptomatic (P60) and mid (P100–110)-stages of disease. N = 6–8 mice/group; p values were determined using the Student’s t-test; C: control wild type mice; miR-124–3p and pri-miR-124 is normalized by U6 snRNA or β-actin, respectively. (C) Representative images of miR-124–3p in situ hybridization and NeuN immunostaining from ventral lumbar cord of control littermates and diseased SOD1G93A mice (P100–110). Scale bar: 20 μm; white arrows: miR-124–3p in situ signals inside NeuN+ neurons. a’-e’, magnified view of NeuN, miR-124–33p, Dapi or merged signals from the box in f; scale bar: 20 μm; (D) Quantification of miR-124–3p (and no-probe control) in situ hybridization fluorescence signals inside NeuN+ neurons from ventral lumbar cord of control littermates and diseased SOD1G93A mice (P100–110). N = 3–4 mice with 15–20 neurons/mouse per group; p values were determined in One-way ANOVA analysis with post-hoc Tukey’s test; Representative images (E) and quantification (F) of U6 RNA in situ hybridization and NeuN immunostaining from ventral lumbar cord of control littermates and diseased SOD1G93A mice (P100–110). N = 3 mice with 15–20 neurons/mouse per group; n.s.; not significant; p values were determined from the Tukey post-hoc test after the one-way ANOVA test.
As the spinal cord tissue is composed of heterogeneous cell populations and miR-124–3p has been found in glial cells in vivo (26), we further determined specific miR-124–3p changes in spinal neurons by performing in situ hybridization of miR-124–3p in conjunction with immunostaining of a neuronal marker NeuN. We had attempted to combine ChAT (a motor neuron marker) immunostaining with miR-124–3p in situ, however, the acetylation and hybridization solutions used during the in situ hybridization procedure hindered a clear ChAT immunostaining signal. Alternatively, the NeuN immunostaining signals remain clear following the in situ hybridization procedure. Representative images from ventral horn of spinal cord (the diagram in Fig. 1C) of control (wild-type littermates) and SOD1G93A mice (P100) following miR-124–3p in situ and NeuN staining are shown in Fig. 1C a–f. Expression of miR-124–3p in neurons is indicated with white arrows. A no-probe control with NeuN staining was also performed in both control and SOD1G93A spinal cord sections (Fig. 1C, h–j, only SOD1G93A sample showed) to determine the background fluorescence signal. Wild-type littermates with no-probe control show a minimal background signal in NeuN+ cells but signals increased > 10-fold with the miR-124–3p probe (Fig. 1D). In diseased SOD1G93A spinal cord tissue, a modestly increased background signal in the no-probe condition was observed, likely due to increased background fluorescence of tissues. Although miR-124–3p signals with the miR-124–3p probe in NeuN+ cells are still significantly increased compared to the no-probe control (p = 0.007, Fig. 1D), miR-124–3p signals in NeuN+ cells in diseased SOD1G93A mice are 35% less than in control mice (p = 0.009). These data clearly indicate that the loss of miR-124–3p occurred in diseased neurons. To determine whether the observed loss of miR-124–3p is specific in diseased neurons, we next performed in- situ hybridization of the small nucleolar U6 RNA in lumbar cord of control and diseased (P100) SOD1G93A mice. In contrast to the decreased miR-124–3p signals in diseased neurons, we observed comparable expression levels of U6 RNA in NeuN+ cells of lumbar cord tissue from both control wild-type littermates and diseased SOD1G93A mice (white arrows, Fig. 1E, and quantification in Fig. 1F), indicating that the observed decrease of miR-124–3p in neurons is unlikely due to a global RNA metabolism change or a catastrophic apoptotic status in diseased (motor) neurons.
Although miR biogenesis typically involves several processing steps (27), the transcription of miR genes is often the primary factor determining the expression levels of mature miRs. Therefore, it is intriguing that the mature miR-124–3p (Fig. 1A, D) but not its overall transcription (indicated by the unchanged pri-miR-124, Fig. 1B) is decreased in neurons in the lumbar cord of diseased SOD1G93A mice. As we previously showed that neuronal miR-124–3p can be packed into exosomes and secreted extracellularly (21, 24), we ask the question: whether miR-124–3p secretion from spinal (motor) neurons is altered in the spinal cord of SOD1G93A mice? To test this possibility, we delivered a mixture of Cy5-miR-124–3p and Fluro-Gold (FG) dye (1.5 and 0.5μL, respectively) into spinal motor neurons through retrograde transport of axons following injections into peripheral sciatic nerves (see experimental diagram in Fig. 2A) at the pre-symptomatic stage (P60). This approach selectively delivers Cy5-miR-124–3p into spinal motor neurons but not other neurons or glial cells at the ventral horn, thus allowing in situ examination of Cy5-miR-124–3p secretion from spinal motor neurons. We chose sciatic nerves over other peripheral nerves because of their larger size for convenient injection. In addition, sciatic nerves are derived from MNs at lumbar segments 2–4, which are enlarged and thus facilitate examination of labeled spinal motor neurons and secretion of miR-124–3p. In control wild-type mice, peripherally delivered FG/Cy5-miR-124–3p (2μL) clearly labels spinal motor neurons (Fig. 2Ba) with Cy5-miR-124–3p being mostly intracellular in FG+ spinal motor neurons (Fig. 2Bc, c’, yellow arrows). Peripherally delivered FG/Cy5-miR-124–3p (same quantity as in control injections) similarly labeled a subset of spinal motor neurons (Fig. 2Bd) in the lumbar cord of SOD1G93A mice (P60, pre-symptomatic stage). Interestingly, while there is still a significant number of Cy5-miR-124–3p puncta inside FG+ spinal motor neurons (Fig. 2Be, f, f’, yellow arrows) in the lumbar cord of SOD1G93A mice, substantial extracellular Cy5-miR-124–3p puncta were also observed around FG+ spinal motor neurons (Fig. 2Bf, f’, white arrows).
Figure 2. Increased extracellular localization of miR-124–3p secreted from lumbar cord motor neurons in diseased SOD1G93A mice.
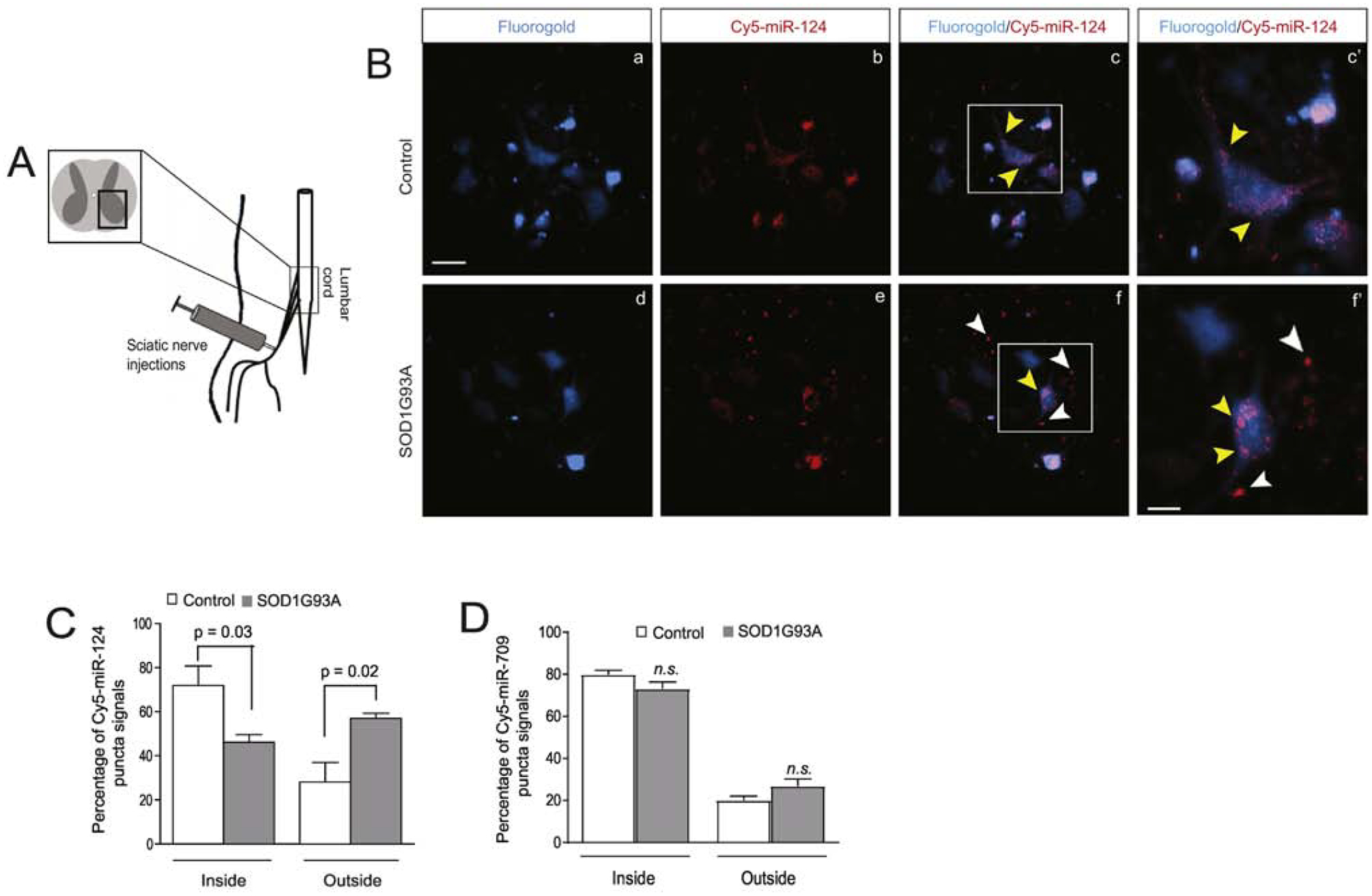
(A) Diagram of peripheral sciatic nerve injections of FluoroGold (FG)/Cy5-miR-124–3p to control and SOD1G93A mice. (B) Retrogradely transported FG/Cy5-miR-124–3p labels lumbar cord motor neurons in both control littermates and SOD1G93A mice following peripheral injection of sciatic nerves. c’ and f’: magnified view of highlighted areas in c and f; white arrows indicate extracellularly localized Cy5-miR-124–3p and yellow arrows indicate intracellularly localized Cy5-miR-124–3p. Scale bar: 40 μm (unmagnified images); 20 μm (magnified images); Quantification of Cy5-miR-124–3p (C) and Cy5-miR-709 (D) inside and outside of FG+ motor neurons in both control and SOD1G93A mice. N = 21 neurons/group, p values were determined using the Student’s t-test.
Based on FG labeling of spinal motor neurons, we quantified the number of Cy5-miR-124–3p puncta inside and outside of FG+ spinal motor neurons and subsequently calculated the percentage of intra- and extracellularly localized Cy5-miR-124–3p. As shown in Fig. 2C, we found that the percentage of intracellularly localized Cy5-miR-124–3p is 25% less (p = 0.03) in FG+ spinal motor neurons of SOD1G93A mice than in control wild-type mice. In contrast, the percentage of extracellularly localized Cy5-miR-124–3p increases 35% (p = 0.02) in the lumbar cord of SOD1G93A mice as compared to control mice (Fig. 2C), indicating that a significant portion of Cy5-miR-124–3p is secreted extracellularly from FG+ spinal motor neurons. Although FG-labeled spinal motor neurons appear intact and are morphologically comparable in both control and SOD1G93A at pre-symptomatic stage (P60), as spinal motor neurons are susceptible to degeneration in the spinal cord of SOD1G93A mice, it is still possible that Cy5-miR-124–3p is apoptotically and passively released to the extracellular space from degenerating spinal motor neurons in SOD1G93A mice during the two-week window following FG/Cy5-miR-124–3p injections. To rule out this possibility, a mix of FG and Cy5-miR-709, a miR that is abundantly expressed in liver but lowly expressed in the CNS (28), was similarly delivered, using the same peripheral injection approach, to lumbar cord motor neurons of control and SOD1G93A mice (P60). Unlike Cy5-miR-124–3p, the intra- and extracellular percentage of peripherally delivered Cy5-miR-709 is not altered (Fig. 2D) in spinal cord of SOD1G93A mice compared to the intra- and extracellular distribution of Cy5-miR-709 in control wild-type lumbar cord, indicating that the increase of extracellularly localized Cy5-miR-124–3p in SOD1G93A lumbar cord appears specific and selective, and is unlikely to result from random or accidental events, such as the loss of cellular integrity of Cy5-miR-124–3p-containing spinal motor neurons.
Increased association of extracellular miR-124–3p with spinal motor neuron-derived exosomes in spinal cord of SOD1G93A mice
Emerging evidence has begun to demonstrate that miRs can be packed into exosomes for extracellular secretion (19). The presence of vesicular membrane also helps shield miRs from degradation in the interstitial environment. Our previous neuronal exosomal miR profiling found that miR-124–3p is highly abundant in neuronal exosomes (24). We therefore hypothesized that the observed increase of extracellularly localized miR-124–3p is due to alterations in neuronal exosome secretion or exosomal association with miR-124–3p in spinal motor neurons of SOD1G93A mice. We previously generated Cre-dependent ILV/exosome reporter (CD63-GFPf/f) mice by inserting a loxP floxed stop codon upstream of human CD63 tagged with copGFP (a GFP variant) and demonstrated the secretion of neuron-derived exosomes in situ in the motor cortex (24) using immuno-EM. We have previously validated that the induced CD63-GFP faithfully mimics endogenous CD63 to label secreted extracellular vesicles using immuno-EM of CD63 and engineered GFP tag, confocal imaging, and immunoblotting in vitro and in vivo (24). This mouse tool allows us to examine the in situ distribution and localization of ILVs/secreted exosomes, especially from spinal cord neurons, in spinal cord sections.
To visualize induced CD63-GFP+ ILVs/exosomes from spinal motor neurons, we first injected a mixture of AAV9-CaMKII-Cre and FG dye into sciatic nerves of CD63-GFPf/+ mice, as shown in the diagram of Fig 3A, to induce expression of CD63-GFP in FG+ spinal motor neurons (Fig. 3A, a). The selective delivery of FG/AAV9-CaMKII-Cre into spinal motor neurons through sciatic nerves also helps determine the source of secreted CD63-GFP+ exosomes from FG+ spinal motor neurons. Peripherally delivered Cre efficiently and selectively induces CD63-GFP expression inside FG+ spinal motor neurons (Fig. 3B, b–d). Meanwhile, we also observed CD63-GFP+FG− puncta scattered around FG+ spinal motor neurons (Fig. 3B, e–g). Based on the density and size of CD63-GFP+FG+ and CD63-GFP+FG− puncta from spinal cord of CD63-GFPf/+ mice, we found that CD63-GFP+FG+ puncta, generally inside FG+ spinal motor neurons, are within a size range of 0.8–1.6 μm in diameter (Fig. 3C), -typical of ILV-containing endosomes, while the size of CD63-GFP+FG− puncta is significantly smaller (0.5–0.7 μm) in diameter (Fig. 3C). The lack of FG labeling and distinctly smaller size of CD63-GFP+FG− puncta suggest that they are extracellularly localized and secreted exosomes (or other EVs) from AAV9-CaMKII-Cre-transduced FG+ spinal motor neurons in CD63-GFPf/+ mice. We further examined CD63 localization by immuno-EM of CD63-GFP in spinal cord sections of CaMKII-CreER+CD63-GFPf/+ mice to overcome the detection limit of confocal microscopy on smaller sized CD63-GFP+ puncta. Extracellular (near synapse) CD63-GFP+ immunogold (IG) labeling particles (5 nm) were found (red arrows, Fig. 3D, b), consistent with confocal microscopy observations. The dendrites (D.) and axon terminals (A.) are indicated by clear post-synaptic density (PSD, white arrows) and synaptic vesicles (SV, yellow arrows), respectively, in the immuno-EM images.
Figure 3. Increased association between miR-124–3p and motor neuron-derived exosomes in lumbar cord of SOD1G93A mice.
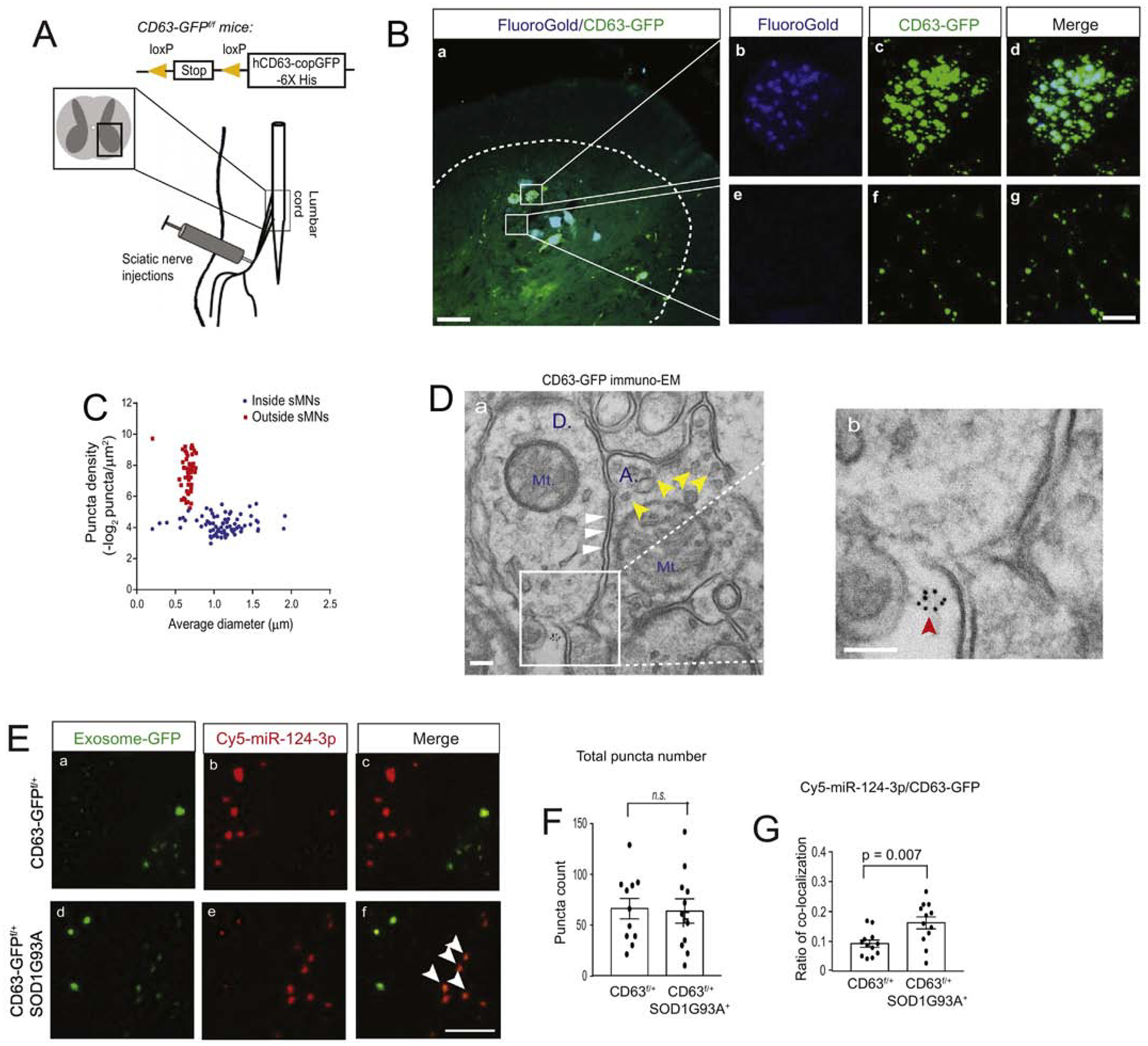
(A) Diagram of peripheral sciatic nerve injections of AAV9-CaMKII-Cre/FG or AAV9-CaMKII-Cre/FG/Cy5-miR-124–3p to CD63-GFPf/+ and CD63-GFPf/+SOD1G93A mice. (B) Representative images of AAV9-CaMKII-Cre-induced CD63-GFP expression and FG labeling of motor neurons in the lumbar cord of CD63-GFPf/+ mice; the dashed line shows the grey matter of lumbar cord; Scale bar: 50μm (a) and 10μm (b-g); magnified images to show intracellular and extracellular CD63-GFP labeling; (C) Quantification of the density and size of CD63-GFP+ puncta in AAV9-CaMKII-Cre and FG injected lumbar cord. Extracellular density was calculated by the total number of extracellular puncta divided by the total extracellular area of CD63-GFP+ puncta. Intracellular density was calculated by the total number of intracellular puncta divided by the individual FG+ spinal motor neurons area. Both extra- and intracellular density was –log2 converted; N = 30–40 images/4 mice/group; (D) Representative immuno-EM of CD63-GFP in the spinal cord of CaMKII-CreER+CD63f/+ mice. Immunogold labeling of CD63-GFP was performed on the CaMKII-CreER+CD63f/+ mouse spinal cord. Red arrow: extracellularly localized CD63 puncta; white arrows: post-synaptic density area; yellow arrows: synaptic vesicles; Mt: mitochondria; A: axonal terminal; D: dendritic spine; Scale bar: 100 nm. (E) Representative confocal images of CD63-GFP+ exosomes and Cy5-miR-124–3p following the injection of a mixture of FG/AAV9-CaMKII-Cre/Cy5-miR-124–3p into the sciatic nerves of CD63-GFPf/+ and CD63-GFPf/+SOD1G93A mice at P60. White arrows: co-localized CD63-GFP+ exosomes and Cy5-miR-124–3p; Scale bar: 5 μm; Quantification of the total number of extracellular CD63-GFP+ exosomes (F) and the ratio (G) of co-localized Cy5-miR-124–3p puncta with CD63-GFP+ exosomes; N = 12 images/4 mice/group, p value was determined using the two-tailed Student’s t-test.
To examine whether the secretion of spinal motor neuron-derived exosomes and their association with miR-124–3p is altered in lumbar cord of SOD1G93A mice, we peripherally injected a mixture of FG, AAV9-CaMKII-Cre, and Cy5-miR-124–3p (0.5, 1.5, and 1.5μL, respectively) to sciatic nerves of either CD63-GFPf/+ littermates or CD63-GFPf/+SOD1G93A mice at the pre-symptomatic stage (P60) to selectively label lumbar cord MN-derived exosomes, as shown in Fig. 3A, and to illustrate extracellularly localized Cy5-miR-124–3p. Representative extracellularly localized spinal motor neuron-derived CD63-GFP+ exosomes and Cy5-miR-124–3p in the spinal cord of both control and SOD1G93A mice are shown in Fig. 3E. It is clearly noted that there is an increased co-localization between CD63-GFP+ exosomes and extracellular Cy5-miR-124–3p puncta in lumbar cord of SOD1G93A mice (white arrows, Fig. 3E, f,). Indeed, quantification showed a 60% increase (p = 0.007) in the number of extracellular Cy5-miR-124–3p puncta that are co-localized with extracellular CD63-GFP+ exosomes in the SOD1G93A lumbar cord relative to control (Fig. 3G). However, the total number of extracellularly localized CD63-GFP+ exosome puncta appears unchanged in lumbar cord of control and SOD1G93A mice (Fig. 3F). These results suggest that while the overall secretion of exosomes from spinal motor neurons at the pre-symptomatic stage does not appear to be affected, the association between Cy5-miR-124–3p and spinal motor neuron-derived exosomes is significantly increased. As miRs are sorted and packed into exosomes, it is likely that there is an increased packaging of Cy5-miR-124–3p into spinal motor neuron-derived exosomes for extracellular release in the SOD1G93A condition.
Exosome-associated miR-124–3p levels in human CSF positively correlate with disease severity in ALS patients
Encouraged by the observation that miR-124–3p is increasingly associated with spinal motor neuron-derived exosomes and extracellularly localized in SOD1G93A mice, we decided to test whether secreted miR-124–3p levels in the CSF are correlated with disease severity. As the total volume of mouse CSF is very limited (2–5 μL), it can be technically highly challenging to isolate exosome-associated miRs from mouse CSF. We therefore decided to measure exosome-associated miR-124–3p levels in human CSF samples and to determine whether it correlates with disease severity of ALS patients. Human CSF samples from ALS patients (n = 14), disease controls (DCs, n = 9), and healthy controls (HCs, n = 9) were obtained from the Northeast ALS Consortium (NEALS) Biofluid Repository. All sample information is summarized in Table 1. The disease severity of ALS patients is indicated by the ALSFRS-R score, as previously described (29).
Table 1:
Clinical information of human subjects whose CFS samples used in this study. Sample collection was done within one month of ALSFRS-R score screening.
| Patient Code | Gender | Age at sample collection | Height | Weight | ALSFRS-R score at screening |
|---|---|---|---|---|---|
| Health Control (HC) | |||||
| HC #1 | F | 54.88 | NA | NA | |
| HC #2 | F | 49.77 | 159.5 | 65.5 | |
| HC #3 | F | 64.31 | 156.2 | 64.5 | |
| HC #4 | F | 55.90 | NA | 108.4 | |
| HC #5 | M | 75.72 | 182.8 | 88.9 | |
| HC #6 | M | 31.52 | 177.8 | 73.7 | |
| HC #7 | M | 46.65 | 190 | 107.5 | |
| HC #8 | M | 50.47 | 177.9 | 80.6 | |
| HC #9 | M | 56.14 | 193 | 97.7 | |
| Disease Control (DC) | |||||
| DC #1 | F | 59.29 | 152.4 | 46.3 | |
| DC #2 | F | 63.70 | 155 | 58.2 | |
| DC #3 | F | 29.75 | NA | 76.4 | |
| DC #4 | M | 56.80 | 177.8 | 69.9 | |
| DC #5 | M | 46.82 | 176.2 | 82.1 | |
| DC #6 | M | 56.80 | 177.8 | 69.9 | |
| DC #7 | M | 62.84 | 175.3 | 99.3 | |
| DC #8 | M | 50.12 | 170.2 | 93.2 | |
| DC #9 | M | 48.30 | 195.58 | 214.4 | |
| ALS cases (ALS) | |||||
| ALS #1 | F | 58.71 | 157.5 | 95.1 | 42 |
| ALS #2 | F | 68.17 | 158.75 | 50.5 | 36 |
| ALS #3 | F | 55.31 | 165.1 | 57 | 33 |
| ALS #4 | F | 62.23 | 137.2 | 64.0 | 45 |
| ALS #5 | F | 66.13 | 161.0 | 66.2 | 44 |
| ALS #6 | M | 53.56 | 181.6 | 76.5 | 44 |
| ALS #7 | M | 43.62 | 185.42 | 111.8 | 38 |
| ALS #8 | M | 39.79 | 184.73 | 88.4 | 42 |
| ALS #9 | M | 58.81 | 177.8 | 91.5 | 40 |
| ALS #10 | M | 72.93 | 182.6 | 100.5 | 43 |
| ALS #11 | M | 54.91 | 180.5 | 81.1 | 41 |
| ALS #12 | M | 50.58 | 170.6 | 68.4 | 36 |
| ALS #13 | M | 57.80 | 180.3 | 81.6 | 45 |
| ALS #14 | M | 50.58 | 177.8 | 82.5 | 41 |
We first analyzed the size and quantity of extracellular vesicles from representative human CSF samples after centrifugal concentration with Centricon filters (30 KDa cut-off) using the qNano particle analyzer (30, 31) and found that EV size is primarily within 50–150 nm range with the mean peak value 82 (representative HC) or 110 (representative ALS) nm (Fig. 4A), typical of in vitro prepared exosomes but not larger size microvesicles. The mean number of EVs from representative HC and ALS CSF samples are 8.8 × 108 particles/mL (HC) and 11 × 108 particles/mL (ALS), respectively. We then precipitated all exosomes from individual CSF samples (2 mL/subject) and determined miR-124–3p levels from total isolated exosomal RNA using qPCR. The total exosome isolation reagent has been previously validated to effectively isolate EVs from biofluid including CSF (32, 33). Expression of CD81, a well-validated exosome marker, on isolated CSF exosomes (with mean protein concentration: 2.1 μg/μL) was also clearly detected by immunoblot (Fig. 4A). We also tested the possibility of detecting miR-124–3p in the exosome-free fraction of CSF and found no detectable miR-124–3p (data not shown), consistent with the notion that CSF typically has a high RNase environment in which the naked RNA is generally degraded. The overall distribution of miR-124–3p levels among ALS patients, DCs, and HCs, indicated by its Ct values, were mostly within the range of 28–34 and the mean value (indicated by the dashed lines) from ALS patients (32.7 ± 0.3), DCs (31.7 ± 0.2), and HCs (32.2 ± 0.2) appears not significantly different (Fig. 4B). Interestingly, we observed a clear trend with increased miR-124–3p (lower Ct values) in CSF exosomes in male ALS patients, though not statistically significant (p= 0.09, Fig. 4C). In addition, the exosomal miR-124–3p levels in CSF of male ALS patients are well correlated with the ALSFRS-R score (R2=0.73, P = 0.002, Fig. 4D) of these patients. Male ALS patients in a more advanced stage, indicated by a lower ALSFRS-R score, tend to have much higher miR-124–3p levels, indicated with lower Ct values (Fig. 4D). This correlation between miR-124–3p levels and disease severity was not found in female ALS patients, likely due to the smaller sample size (n = 5) of female ALS patients. Meanwhile, we found no correlation between miR-124–3p levels in CSF exosomes and weight/height in both male and female ALS patients, DCs, and HCs (Fig. 4E–F). The levels of miR-124–3p in CSF exosomes are also not affected by the age of both male and female human subjects (Fig. 4G–H).
Figure 4. Positive correlation of CSF miR-124–3p abundance and disease severity in male ALS patients.
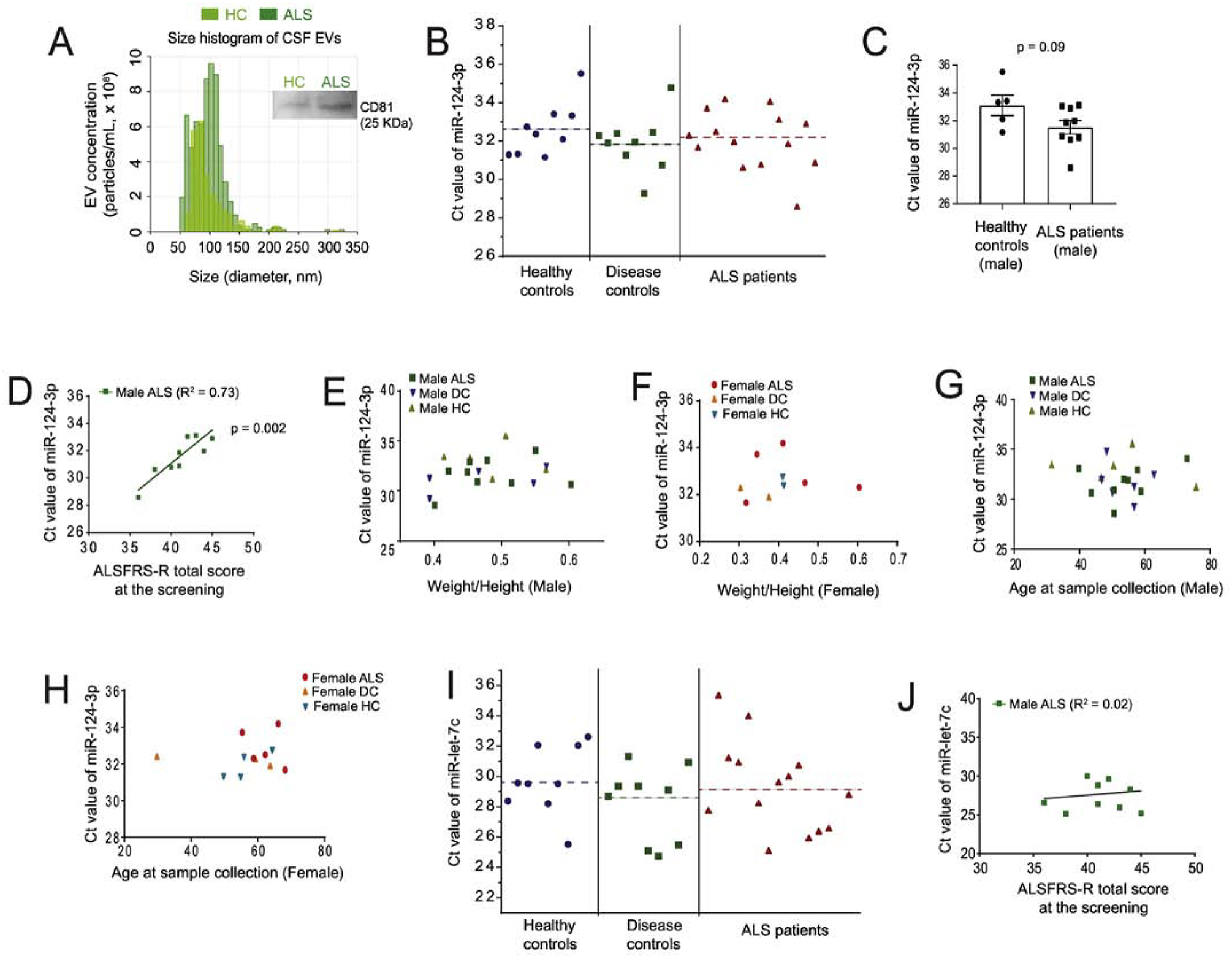
(A) Representative size distribution histogram of exosomes from human CSF samples (HC and ALS) determined by the qNano particle analyzer. CSF was concentrated 6 x using Centricon (30 KDa cut-off) filters by centrifugation; A representative CD81 immunoblot from the same HC and ALS samples was shown on the side; (B) Ct values of miR-124–3p measured in three sample groups: healthy controls (HC), disease controls (DC), and ALS patients. Dashed lines indicate the mean Ct value for each group; (C) Ct values of miR-124–3p between male HC (n = 5) and male ALS patients (n = 9); p value determined from two-tailed unpaired t-test. (D) Correlation between Ct values of miR-124–3p in male ALS patients and their disease stage determined by the ALSR-total score at screening. Pearson r = 0.85; p = 0.002 determined from the liner regression student’s t-test; Plot between Ct values of miR-124–3p and the weight/height ratio of male (E) and female (F) HC, DC, and ALS patients at sample collection. Plot between Ct values of miR-124–3p with the age of male (G) and (H) female HC, DC, and ALS patients at sample collection. (I) Ct values of miR-let-7c measured in HC, DC, and ALS patients. Dashed lines indicate the mean Ct value for each group; (J) Plot between Ct values of miR-let-7c in male ALS patients and their disease severity determined by the ALSFRS-R total score at screening. The number of samples used, and the rest of data info are compiled in Table 1. N = 9 HC, 9 DC, and 14 ALS (9 male and 5 female). Sample collection was done within one month of ALSFRS-R score screening. p value determined from the liner regression student’s t-test;
To further determine whether the correlation between miR-124–3p and male ALS patients is specific, we also measured the levels of let-7c, another miR that is highly abundant in neuronal exosomes based on our previous neuronal exosomal miR profiling results (24), from CSF exosomes of all human subjects. Similar to miR-124–3p, let-7c levels in the CSF exosomes of ALS patients (mean: 29.0 ± 0.1) are not significantly different from that of DCs (mean: 28.8 ± 0.3) and HCs (mean: 29.6±0.4) (Fig. 4I). However, we found no correlation between let-7c levels and the ALSFRS-R score of either male or female ALS patients (Fig. 4J). Thus, the highly positive correlation between miR-124 levels (lower Ct values) in CSF exosomes and the disease severity (lower ALSFRS-R score) of male ALS patients appears non-accidental and specific.
Discussion
In this study, we characterized altered intercellular localization of miR-124–3p and elucidated the role of spinal motor neuron-derived exosomes in the extracellular secretion of miR-124–3p in spinal cord of SOD1G93A mice. We further presented preliminary evidence to support the potential of using exosomal miR-124–3p in CSF as an indicator for monitoring disease severity in ALS.
MiR-124–3p is one of the most abundant neuronal miRs in the CNS, which is essential for neuronal differentiation during early embryogenesis by antagonizing the anti-neural REST/SCP1 pathway (7). The formation of spinal motor neurons is also facilitated by miR-124–3p-mediated silencing of REST during early development (25). Although miR-124 levels were not found to be different in the CSF of SOD1G93A rats as compared to control (14), our previous (21) and current studies found that miR-124–3p levels are significantly decreased in spinal cord tissue, especially in spinal (motor) neurons. Interestingly, the overall transcription of miR-124–3p, indicated by the pri-miR-124 levels, appears unaltered in spinal cord of SOD1G93A mice. Given that miR-124–3p transcription in glial cells is generally much lower than that in neurons (24), the sustained levels of pri-miR-124 in spinal cord of SOD1G93A mice support the idea that the overall transcription of miR-124 in neurons is unchanged. Alternatively, the increased association between miR-124–3p and spinal motor neuron-derived exosomes in SOD1G93A condition suggests that overexpression of SOD1G93A in spinal motor neurons may alter the sorting and packaging of miR-124–3p into exosomes for secretion and subsequently leads to increased extracellular localization of miR-124–3p. As axon transport deficits especially the slowing of axon transport have been observed in SOD1G93A mice as early as P36 (34, 35), our injected Cy5-miR-124–3p or AAV9-CaMKII-Cre with FG dye is likely to be less retrograde transported into spinal motor neurons (for Figs. 2 and 3 respectively) and consequently our results may underestimate the increased extracellular exosome labeling and extracellular miR-124–3p localization in SOD1G93A condition, which further supports our observation that miR-124–3p and spinal motor neuron-derived exosomes are increasingly localized extracellularly in SOD1G93A condition. Whether and how exosomally secreted miR-124–3p is involved in ALS pathogenesis is unknown, but it is possible that some of exosomally secreted miR-124–3p are internalized into neighboring astroglia to mediate pathological neuron to astroglial communication, as we recently showed with astroglial reporter mice and in vivo delivery of miR-124–3p (21, 24).
Searching for and validating biomarkers for the diagnosis and monitoring of disease progression in ALS has been one of the central goals in ALS study and is also clinically important. Previous efforts towards identifying biomarkers have been associated with excitotoxicity, oxidative stress, inflammation, neurodegeneration, and metabolic dysfunction in disease (36, 37). Recently, exosomal cargos have emerged as a new category of potential biomarkers for ALS (38). Several proteins that are implicated in the pathogenesis of ALS, including mutant SOD1 (39), TDP43 oligomers (40), FUS (41), and dipeptide repeat protein (DRP) derived from the non-ATG translation of c9orf72 hexanucleotide repeat expansions (42) have been identified in secreted exosomes from cultures. TDP43 C-terminal fragments have also been detected in the CSF of ALS patients (43). In addition to the protein cargo, CNS cell-derived exosomes also contain a unique subset of miRs (24, 44), becoming an additional source for biomarker development in ALS. In particular, the vesicular protection of miRs in exosomes/EVs in CSF potentially helps generate more consistent and reliable results from CSF miR profiling.
Our results showed that the association of miR-124–3p and spinal motor neuron-derived exosomes and extracellular localization of miR-124–3p has already increased even at the pre-symptomatic stage (P60) before a large number of spinal motor neurons undergo degeneration, indicating that the extracellular change in miR-124–3p is likely an early pathological event in the SOD1G93A model. Since we showed that the overall miR-124–3p levels in spinal cord of control and SOD1G93A mice are comparable at the pre-symptomatic stage (21), the observed localization change of miR-124–3p in the SOD1G93A condition is likely to be independent from the overall miR-124–3p expression level. As the disease progresses, miR-124–3p is likely to be further released from degenerating spinal motor neurons, resulting in additional increase in CSF. These results provided interesting evidence that miR-124–3p in CSF exosomes may have potential as a new prognosis biomarker in ALS. In particular, the early increase of extracellularly localized miR-124–3p at the pre-symptomatic stage (P60) suggests that CSF miR-124–3p has the potential to indicate early disease progression before widespread degeneration of spinal motor neuron occurs. Although the current study examined the correlation between a neuronal exosomal miR (miR-124–3p) and disease severity, as glial cells are also known to secrete miR-containing exosomes (45), whether glial exosome-contained miRs are correlated with disease severity remains to be explored in the future.
Although our study found no significant difference in miR-124–3p levels in CSF exosomes between DCs, HCs, and ALS patients, a recent study found that miR-124–3p is the top up-regulated miR in sporadic ALS CSF samples compared to control CSF samples using a deep sequencing approach (23). These differences are likely caused by different experimental approaches (qPCR vs. sequencing) and their sensitivity or sample sizes/disease stages. Nevertheless, both studies support the potential of using CSF exosomal miR-124–3p as a possible indicator for disease stage/progression. Meanwhile, it is intriguing but also puzzling why we only observed a clear correlation between CSF exosomal miR-124–3p levels and disease severity in male (but not female) patients. It is likely that the sample size of female patients is too small. It is also possible that this correlation may reflect a pathogenic process that is tightly correlated with particular genetic mutations (such as SOD1G93A). Indeed, male ALS patients tend to have earlier onset and female ALS patients have a slower disease progression (46). Therefore, it is likely that the CSF miR-124–3p changes occur earlier and are more significant in male ALS patients than in female ALS patients, making their correlation with disease severity more evident. It is worth noting that the ALSFRS-R scores in this study are in a less severe range (35–45), also likely indicating relatively early disease stages from which the CSF samples were collected. The intrinsic differences of miRNA expression profiles during development and in diseases (47, 48) may also contribute to a gender-specific correlation between miR-124–3p and disease severity in ALS. Future studies to examine the miR-124–3p dynamics in other ALS mutant conditions and also test the correlation between CSF exosomal miR-124–3p and disease severity from sporadic or familial ALS patients carrying different mutations will be important to fully evaluate the clinical potential of CSF exosomal miR-124–3p in the indication of disease progression of ALS.
Methods
Animals
The CD63-GFPf/f knock-in mice were generated by homologue recombination, as we previously described (24). Briefly, the human CD63-copGFP-6xHis cassette was subcloned onto the Rosa-CAG targeting vector, downstream of the CAG promoter and upstream of the 3’ arm, to generate the final CD63-copGFP-6xHis targeting vector. The targeting vectors were linearized and transfected into the ES cell line to induce homologue recombination. Chimeric mice were bred with C57BL/6J mice to obtain germline transmission. B6SJL-Tg (SOD1*G93A)1Gur/J mice were purchased from Jax Laboratories (# 002726). Both male and female mice were used in all experiments. All mice were maintained on a 12h light/dark cycle with food and water ad libitum. Care and treatment of animals in all procedures strictly followed the NIH Guide for the Care and Use of Laboratory Animals and the Guidelines for the Use of Animals in Neuroscience Research and the Tufts University IACUC.
Sciatic Nerve Injection
Control wild-type littermates, SOD1G93A, CD63-GFPf/+, or CD63-GFPf/+SOD1G93A (P60) mice were deeply anaesthetized for surgery with ketamine (100 mg/kg) plus xylazine (10 mg/kg) in saline by intraperitoneal (IP) injection. The hindquarter of the nerve to be injected was shaved using electric clippers, and the skin was sterilized using betadine and sterile alcohol. A horizontal incision through the skin was made on the mouse hind limb. The sciatic nerve was located by opening the fascial plane between the gluteus maximus and the anterior head of the biceps femoris. The sciatic nerve was then lifted from the muscle bellies and propped up using a spatula, taking care not to damage the nerve. For control and SOD1G93A mice (P60), a mixture of FluoroGold (1 μL) and Cy5-miR-124–3p or Cy5-miR-709 (2 μL, 100μM) was injected into the sciatic nerves using a Hamilton syringe (10 μL). For CD63-GFPf/+ and CD63-GFPf/+SOD1G93A mice (P60), a mixture of AAV9-CAMKII-Cre virus (2 μL, from Penn Vector Core), FluoroGold (1 μL), and Cy5-miR-124–3p (2 μL, 100μM) was then injected into the sciatic nerves. Cy5-miR-124–3p or Cy5-miR-709 was synthesized by IDT DNA Technologies with a 2’ O-Methyl group on each nucleotide and phosphoorothioate bonds between the first and last three nucleotides.
Immuno-EM Microscopy
Immuno-EM was carried out in the Harvard EM facility. Adult mice (P60) were perfused with 4% paraformaldehyde (PFA) and 0.1% glutaraldehyde. The spinal cord was dissected out and post-fixed in 4% PFA for 2 hrs and then spinal cord slices (100 μm) were prepared using a vibratome. The slices were then quenched, permeabilized, and blocked in blocking buffer (3% bovine serum albumin, 5% normal donkey serum, and 0.1% Triton X-100) at 4°C. Anti-human CD63 (BD Pharmingen, #556019) antibody was then added and incubated overnight at 4°C. After wash, slices were incubated with Protein A-gold for 1 hour at 25°C. The images were taken using the JEOL 1200EX transmission electron microscope.
Tissue processing, confocal imaging, and quantification
Mice were deeply anesthetized with Ketamine (100mg/kg) + Xylazine (10mg/kg) in saline by intraperitoneal (IP) injection and perfused intracardially with 4% paraformaldehyde (PFA) in PBS. The lumbar spinal cords were dissected and kept in 4% PFA overnight at 4°C, then cryoprotected by immersion in 30% sucrose for 48h. Spinal cords were embedded and frozen in OCT-Compound Tissu-Tek® (Sakura, Tokyo Japan). Lumbar cord (L3-L5) sections (20 μm) were prepared with a cryostat (Thermo HM525) and mounted on glass SuperFrost+ slides (Fisher Scientific). The slides were rinsed three times in PBS before mounting. Confocal images were taken using the A1R confocal laser scanning microscope (15–20 μm Z stack with 0.5–1 μm step) magnified with 40X (numerical aperture 0.8) or 60X (numerical aperture 1.0) objectives (Nikon Instruments INC. Melville, NY). Low magnification images were taken using the Zeiss Axio imager with ApoTome. For the quantification of Cy5-miR signals, the far-red channel of the confocal image (63x) was thresholded to remove background signals. The puncta were then analyzed using the Analyze Particles tool in Image J to obtain particle counts. The intracellular and extracellular Cy5-miR signals were determined by manually examining the overlaying Cy5-miR signals with the FluoroGold channel. The percentage of intracellular and extracellular Cy5-miR signals were then calculated from the total Cy5-miR signals. For the quantification of CD63-GFP+ puncta in spinal cord, confocal images (40x) were split into individual channels and converted to 8-bit greyscale. Regions of Interest (ROI) were drawn around motor neurons based on the FluoroGold fluorescence. Images were first thresholded to remove the background. The CD63-GFP puncta were then analyzed using the Analyze Particles tool to obtain particle count and particle area inside and outside of the ROIs, which was then converted to radius and then diameter (μm). For the quantification of the co-localization between extracellular CD63-GFP+ and Cy5-miR-124–3p puncta, green (CD63-GFP) and far-red (Cy5-miR-124–3p) channels of confocal images (40x) were first thresholded and their co-localization was identified and quantified using the JacoP plugin for Image J.
In situ hybridization and immunohistochemistry
Locked-Nucleic-Acid (LNA) fluorescence in situ hybridization was performed as previously described (49) with modifications. Using RNase-free technique, mice were perfused with PBS, and freshly dissected lumbar cord tissue was embedded in OCT-Compond Tissu-Tek® (Sakura, Tokyo Japan). Lumbar cords were sectioned (10μm) on a cryostat (Thermo HM525) and then mounted on glass SuperFrost+ slides (Fisher Scientific). Slides were briefly fixed for 1 minute with freshly prepared 4% PFA. Slides were incubated in acetylation solution for 10 minutes, and then in hybridization solution at 60˚C for 4 hours. For hybridization, slides were hybridized with denatured DIG-labeled miR-124–3p or U6 RNA LNA probe (80 nM, Exiqon) overnight at 60˚C. Slides were then washed and incubated in blocking solution (1% BSA, 5% goat-serum, and 0.2% Triton-X 100 in PBS) for 1 hour. To amplify the LNA-probe signal, slides were incubated with HRP anti-DIG (1:1,000, 11207733910, Roche) for 1 hour, then incubated with biotin-tyramide (1:1,000, Sigma) for 1 hour, then incubated with DTAF-conjugated streptavidin (1:500, 016–010-084, Jackson ImmunoResearch) for 1 hour. The NeuN immunostaining (1:500, MAB377, Millipore) was performed following standard immunohistochemistry procedures. The slides were then mounted with ProLong Gold Anti-Fade mounting medium (Thermo Fisher Scientific).
Immunoblot
Prepared CSF exosome samples were briefly sonicated, and the total protein amount was determined by the Bradford protein assay. A total of 30 μg proteins were loaded on 4–15% gradient SDS-PAGE gels and the protein mixtures were separated by size using gel electrophoresis. Separated proteins were transferred onto a PVDF membrane (0.22 μm, Bio-Rad) at constant 2.5Amp for 10 minutes using the Bio-Rad Trans-Blot Turbo Transfer System. The membrane was blocked with 3% nonfat milk in TBST (Tris buffer saline with 0.1% Tween 20) then incubated with the appropriate primary antibody overnight at 4°C. On the following day, the membrane was incubated with anti-CD81 antibody (1:200, clone B-11, Santa Cruz) and subsequently with HRP-conjugated anti-mouse secondary antibody (1:5000, abcam) diluted in TBST for 1 hour at RT. Bands were visualized by ECL Plus chemiluminescent substrate (Thermo Fisher Scientific) at the Bio-Rad ChemiDoc MP imaging system.
RNA isolation and miR qPCR
Total RNA was extracted from lumbar cord tissue or from exosomes of CSF using TRiZol reagent, following the manufacturer’s instruction. The quantity and quality of isolated miRNA was determined using the Agilent BioAnalyser with the pico RNA kit according to the manufacturer’s instructions. The miRNA was converted to cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) with specific primers for each individual miR and control U6 small nuclear (sn) RNA. For pri-miR-124–3p, random primers were used for the reverse transcription. The relative quantity of individual miRs was measured by qPCR using SyberGreen. The miR-124–3p and pri-miR-124 is normalized by U6 snRNA or β-actin, respectively.
CSF EV isolation and qNano particle analysis
Tunable resistive pulse sensing (TRPS) (30, 31) by qNano instrument 60 (Izon Science, MA, USA) was used to measure the size distribution and quantity of particles in isolated exosome fractions, as described in the instrument. An aliquot of exosomes fraction or calibration particles included in the reagent kit were placed in the Nanopore (NP150, Izon Science). CSF samples were first concentrated 6 x using Centricon (30 KDa cut-off) filters by centrifugation. Samples were then measured at 44.9 mm stretch with a voltage of 0.76 V at 1-pressure levels of 14 mbar. Particles were detected in short pulses of the current (blockades). The calibration particles were measured directly after the experimental sample under identical conditions. The data was analyzed using the Izon software (version 3.2). The Total Exosome Isolation Reagent (#4484453, Thermo Fisher Scientific)-mediated precipitation approach was used to isolate EVs from CSF for qPCR and western blot. CSF samples (2 mL/subject) were centrifuged at 2,000g for 10 min at 4°C. The supernatants were mixed with equal volume (2 mL) of the Total Exosome Isolation Reagent (4484453, Thermo Fisher Scientific) overnight and then centrifuged at 10,000g for 1 hour at 4°C to pellet all precipitated exosomes.
Subjects
All CSF samples of ALS patients, neurological disease controls (DCs) and healthy controls (HCs) were obtained from Northeast Amyotrophic Lateral Sclerosis Consortium (NEALS) Biofluid Repository (Table 1). Samples were received frozen and stored at −80°C. All the cases were selected by the NEALS Biofluid Repository to have the best possible matching by age, gender, weight and height, etc (Table 1).
Experimental design and statistical analysis
All experiments and analysis were performed blindly (with different lab members) without knowing the sample identity in advance. Sample size and statistical approach used for each experiment are described in each method section and in figure legends. All analyses were performed using GraphPad Prism 7. For graphs with error bars, error bars are presented as SEM. No custom code was used in the analysis. For multiple groups (> 2), one-way ANOVA was used to analyze the variance, followed by a Tukey post-hoc test to compare multiple groups. For two-group comparison, two-tailed student t-test was used. Statistical significance was tested at a 95% (P<0.05) confidence level and p values are shown in each graph.
Highlights.
miR-124–3p expression levels are decreased in mid-staged SOD1G93A spinal (motor) neurons.
Increased extracellular localization of miR-124–3p in lumbar cord in diseased SOD1G93A mice.
Increased association between miR-124–3p and neuron-derived exosomes in SOD1G93A mice.
Positive correlation of CSF miR-124–3p abundance and disease severity in male ALS patients.
Acknowledgments
This work was supported by NIH grants DA042342, NS087391, RF1AG057882 (YY) and The Packard center for ALS research (YY). We thank Yang Tian in sample collection of SOD1G93A mice. We thank Rachel Jarvis for proofreading the manuscript. We thank NEALS Biofluid Repository for providing human CSF samples and clinical measurements of human subjects.
Abbreviations
- miRs
MicroRNAs
- ALS
Amyotrophic Lateral Sclerosis
- UMNs and LMNs
upper and lower motor neurons
- EVs
extracellular vesicles
- ILVs
intraluminal vesicles
- DCs
disease controls
- HCs
healthy controls
- ROI
regions of interest
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare no conflict of interest in this study.
References
- 1.Taylor JP, Brown RH Jr. and Cleveland DW (2016) Decoding ALS: from genes to mechanism. Nature, 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleveland DW and Rothstein JD (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci, 2, 806–819. [DOI] [PubMed] [Google Scholar]
- 3.Ilieva H, Polymenidou M and Cleveland DW (2009) Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol, 187, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothstein JD, Martin LJ and Kuncl RW (1992) Decreased glutamate transport by the brain and spinal cord in amyotrophic lateral sclerosis. N Engl.J Med, 326, 1464–1468. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, Liu Y, Tsingalia A, Jin L, Zhang PW et al. (2012) Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature, 487, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang SH, Fukaya M, Yang JK, Rothstein JD and Bergles DE (2010) NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron, 68, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaco C, Otto S, Han JJ and Mandel G (2006) Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A, 103, 2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Packer AN, Xing Y, Harper SQ, Jones L and Davidson BL (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci, 28, 14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNeill E and Van Vactor D (2012) MicroRNAs shape the neuronal landscape. Neuron, 75, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amin ND, Bai G, Klug JR, Bonanomi D, Pankratz MT, Gifford WD, Hinckley CA, Sternfeld MJ, Driscoll SP, Dominguez B et al. (2015) Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science, 350, 1525–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawahara Y and Mieda-Sato A (2012) TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A, 109, 3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E and Bozzoni I (2012) FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J, 31, 4502–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Santis R, Santini L, Colantoni A, Peruzzi G, de Turris V, Alfano V, Bozzoni I and Rosa A (2017) FUS Mutant Human Motoneurons Display Altered Transcriptome and microRNA Pathways with Implications for ALS Pathogenesis. Stem Cell Reports, 9, 1450–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoye ML, Koval ED, Wegener AJ, Hyman TS, Yang C, O’Brien DR, Miller RL, Cole T, Schoch KM, Shen T et al. (2017) MicroRNA Profiling Reveals Marker of Motor Neuron Disease in ALS Models. J Neurosci, 37, 5574–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koval ED, Shaner C, Zhang P, du Maine X, Fischer K, Tay J, Chau BN, Wu GF and Miller TM (2013) Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet, 22, 4127–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butovsky O, Jedrychowski MP, Cialic R, Krasemann S, Murugaiyan G, Fanek Z, Greco DJ, Wu PM, Doykan CE, Kiner O et al. (2015) Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol, 77, 75–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parisi C, Napoli G, Amadio S, Spalloni A, Apolloni S, Longone P and Volonte C (2016) MicroRNA-125b regulates microglia activation and motor neuron death in ALS. Cell Death Differ, 23, 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR and Olson EN (2009) MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science, 326, 1549–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Li S, Li L, Li M, Guo C, Yao J and Mi S (2015) Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics, 13, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombo M, Raposo G and Thery C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- 21.Morel L, Regan M, Higashimori H, Ng SK, Esau C, Vidensky S, Rothstein J and Yang Y (2013) Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem, 288, 7105–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinchetti P, Rizzuti M, Faravelli I and Corti S (2018) MicroRNA Metabolism and Dysregulation in Amyotrophic Lateral Sclerosis. Mol Neurobiol, 55, 2617–2630. [DOI] [PubMed] [Google Scholar]
- 23.Waller R, Wyles M, Heath PR, Kazoka M, Wollff H, Shaw PJ and Kirby J (2017) Small RNA Sequencing of Sporadic Amyotrophic Lateral Sclerosis Cerebrospinal Fluid Reveals Differentially Expressed miRNAs Related to Neural and Glial Activity. Front Neurosci, 11, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Men Y, Yelick J, Jin S, Tian Y, Chiang MSR, Higashimori H, Brown E, Jarvis R and Yang Y (2019) Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat Commun, 10, 4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhinge A, Namboori SC, Bithell A, Soldati C, Buckley NJ and Stanton LW (2016) MiR-375 is Essential for Human Spinal Motor Neuron Development and May Be Involved in Motor Neuron Degeneration. Stem Cells, 34, 124–134. [DOI] [PubMed] [Google Scholar]
- 26.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM and Weiner HL (2011) MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med, 17, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He L and Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet, 5, 522–531. [DOI] [PubMed] [Google Scholar]
- 28.Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G and Calin GA (2008) MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia, 22, 1095–1105. [DOI] [PubMed] [Google Scholar]
- 29.Cedarbaum JM, Stambler N, Malta E, Fuller C, Hilt D, Thurmond B and Nakanishi A (1999) The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci, 169, 13–21. [DOI] [PubMed] [Google Scholar]
- 30.Maas SL, Broekman ML and de Vrij J (2017) Tunable Resistive Pulse Sensing for the Characterization of Extracellular Vesicles. Methods Mol Biol, 1545, 21–33. [DOI] [PubMed] [Google Scholar]
- 31.Vogel R, Willmott G, Kozak D, Roberts GS, Anderson W, Groenewegen L, Glossop B, Barnett A, Turner A and Trau M (2011) Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal Chem, 83, 3499–3506. [DOI] [PubMed] [Google Scholar]
- 32.Soares Martins T, Catita J, Martins Rosa I, A.B.d.C.E.S. O and Henriques AG (2018) Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One, 13, e0198820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rider MA, Hurwitz SN and Meckes DG Jr. (2016) ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci Rep, 6, 23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilsland LG, Sahai E, Kelly G, Golding M, Greensmith L and Schiavo G (2010) Deficits in axonal transport precede ALS symptoms in vivo. Proc Natl Acad Sci U S A, 107, 20523–20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson TL and Cleveland DW (1999) Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci, 2, 50–56. [DOI] [PubMed] [Google Scholar]
- 36.Turner MR, Kiernan MC, Leigh PN and Talbot K (2009) Biomarkers in amyotrophic lateral sclerosis. The Lancet. Neurology, 8, 94–109. [DOI] [PubMed] [Google Scholar]
- 37.Chen X and Shang HF (2015) New developments and future opportunities in biomarkers for amyotrophic lateral sclerosis. Transl Neurodegener, 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanninen KM, Bister N, Koistinaho J and Malm T (2016) Exosomes as new diagnostic tools in CNS diseases. Biochim Biophys Acta, 1862, 403–410. [DOI] [PubMed] [Google Scholar]
- 39.Grad LI, Yerbury JJ, Turner BJ, Guest WC, Pokrishevsky E, O’Neill MA, Yanai A, Silverman JM, Zeineddine R, Corcoran L et al. (2014) Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci U S A, 111, 3620–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iguchi Y, Eid L, Parent M, Soucy G, Bareil C, Riku Y, Kawai K, Takagi S, Yoshida M, Katsuno M et al. (2016) Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain, 139, 3187–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamelgarn M, Chen J, Kuang L, Arenas A, Zhai J, Zhu H and Gal J (2016) Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim Biophys Acta, 1862, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westergard T, Jensen BK, Wen X, Cai J, Kropf E, Iacovitti L, Pasinelli P and Trotti D (2016) Cell-to-Cell Transmission of Dipeptide Repeat Proteins Linked to C9orf72-ALS/FTD. Cell Rep, 17, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinacker P, Hendrich C, Sperfeld AD, Jesse S, von Arnim CA, Lehnert S, Pabst A, Uttner I, Tumani H, Lee VM et al. (2008) TDP-43 in cerebrospinal fluid of patients with frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Arch Neurol, 65, 1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lafourcade C, Ramirez JP, Luarte A, Fernandez A and Wyneken U (2016) MiRNAs in Astrocyte-Derived Exosomes as Possible Mediators of Neuronal Plasticity. J Exp Neurosci, 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jovicic A and Gitler AD (2017) Distinct repertoires of microRNAs present in mouse astrocytes compared to astrocyte-secreted exosomes. PLoS One, 12, e0171418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCombe PA and Henderson RD (2010) Effects of gender in amyotrophic lateral sclerosis. Gend Med, 7, 557–570. [DOI] [PubMed] [Google Scholar]
- 47.Guo L, Liang T, Yu J and Zou Q (2016) A Comprehensive Analysis of miRNA/isomiR Expression with Gender Difference. PLoS One, 11, e0154955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui C, Yang W, Shi J, Zhou Y, Yang J, Cui Q and Zhou Y (2018) Identification and Analysis of Human Sex-biased MicroRNAs. Genomics Proteomics Bioinformatics, 16, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silahtaroglu AN (2010) LNA-FISH for detection of microRNAs in frozen sections. Methods Mol Biol, 659, 165–171. [DOI] [PubMed] [Google Scholar]


