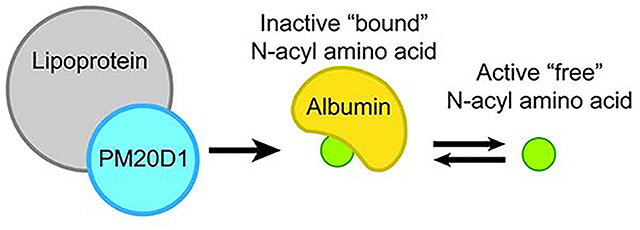
SUMMARY
N-acyl amino acids are a family of cold-inducible circulating lipids that stimulate thermogenesis. Their biosynthesis is mediated by a secreted enzyme called PM20D1. The extracellular mechanisms that regulate PM20D1 or N-acyl amino acid activity in the complex environment of blood plasma remains unknown. Using quantitative proteomics, here we show that PM20D1 circulates in tight association with both low- and high-density lipoproteins. Lipoprotein particles are powerful co-activators of PM20D1 activity in vitro and N-acyl amino acid biosynthesis in vivo. We also identify serum albumin as a physiologic N-acyl amino acid carrier, which spatially segregates N-acyl amino acids away from their sites of production, confers resistance to hydrolytic degradation, and establishes an equilibrium between thermogenic “free” versus inactive “bound” fractions. These data establish lipoprotein particles as principal extracellular sites of N-acyl amino acid biosynthesis and identify a lipoprotein-albumin network that regulates the activity of a circulating thermo- genic lipid family.
Graphical Abstract
eTOC BLURB
N-acyl amino acids are a family of lipid hormones that stimulate thermogenesis. Kim et al. uncover lipoprotein particles as sites for N-acyl amino acid biosynthesis and serum albumin as a physiologic N-acyl amino acid carrier. These highly abundant plasma proteins regulate the metabolism and bioactivity of a circulating lipid family.
INTRODUCTION
Circulating metabolites are powerful regulators of intercellular communication and organismal energy homeostasis. While many classical examples include endocrine hormones such as steroids, biogenic amines, and the thyroid hormones, many new signaling molecules are still being uncovered (Cao et al., 2008; Grevengoed et al., 2019; Leiria et al., 2019; Liu et al., 2013; Lynes et al., 2017; Roberts et al., 2014; Yore et al., 2014). Recently, we have reported that certain N-acyl amino acids, including N-acyl-phenylalanines and N-acyl-leucines, are circulating lipid metabolites that regulate energy metabolism (Lin et al., 2018; Long et al., 2016, 2018). These “thermogenic” N-acyl amino acids are physiologically elevated in the blood following chronic cold exposure and robustly stimulate UCP1-independent mitochondrial uncoupling (Keipert et al., 2017; Lin et al., 2018) and whole-body energy expenditure (Long et al., 2016).
The biosynthesis of N-acyl amino acids is mediated by PM20D1, an extracellular N-acyl amino acid synthase/hydrolase (Long et al., 2016, 2018). PM20D1 is one of five members of the mammalian M20 peptidase family (Kim et al., 2019) and is endogenously secreted from several tissues including brown fat, liver, kidney, and intestine (Schaum et al., 2018). Genetic elevation of circulating PM20D1 drives the biosynthesis of a broad panel of plasma N-acyl amino acids including N-oleoyl phenylalanine and N-oleoyl leucine and leads to a hypermetabolic phenotype (Long et al., 2016). Conversely, PM20D1-knockout mice exhibit glucose intolerance and insulin resistance (Long et al., 2018). In humans, the PM20D1 gene has been to both obesity and Alzheimer’s disease (Benson et al., 2019; Bycroft et al., 2018; Sanchez-Mut et al., 2018; Sudlow et al., 2015). More recently, a second intracellular N-acyl amino acid synthase/hydrolase has also been reported (Kim et al., 2020). Taken together, these data provide powerful enzymological and genetic evidence that PM20D1 and N-acyl amino acids are critical regulators of energy metabolism in both rodents and humans.
While the purified PM20D1 polypeptide is sufficient to catalyze N-acyl amino acid synthesis and hydrolysis in vitro (Long et al., 2016), how the PM20D1/N-acyl amino acid pathway is regulated in a complex physiologic environment such as blood remains unknown. Using a quantitative proteomic approach, here we show that PM20D1 circulates in tight association with both low- and high-density lipoprotein particles. Lipoprotein particles are co-activators of PM20D1 activity in vitro and N-acyl amino acid biosynthesis and in vivo. We also demonstrate that albumin is a physiologic N-acyl amino acid carrier, which spatially segregates N-acyl amino acids away from their site of biosynthesis, confers resistance to hydrolytic degradation, and acts as a buffer between biologically active “free” versus inactive “bound” N-acyl amino acids. These data establish lipoproteins as biosynthesis sites and serum albumin as a plasma carrier for a family of circulating lipid metabolites.
RESULTS
Proteomic identification of PM20D1 interacting proteins
To identify proteins that interact with PM20D1, primary brown adipose tissue (BAT) and inguinal white adipose tissue (iWAT) adipocytes were transduced with adenoviruses expressing a C-terminally flag-tagged PM20D1 construct or LacZ control. On day 6 of differentiation, at a time when the adipocytes were robustly expressing and secreting PM20D1-flag, conditioned media was harvested. A mild anti-FLAG immunoaffinity purification (IP) protocol was then used to capture PM20D1-flag and any PM20D1 interacting proteins. Silver staining of the immunoaffinity purified elution from either LacZ- or PM20D1-flag transduced cells revealed enrichment of a ~60kDa band corresponding to PM20D1-flag as well as several other bands (Fig. 1a and b). Most notably, several very high molecular weight proteins (>250kDa) and a low molecular weight protein (~25kDa) were reproducibly identified in anti-flag immunoaffinity purifications from both BAT and iWAT cells.
Fig. 1. PM20D1 interacts with lipoproteins in conditioned media of adipocytes.
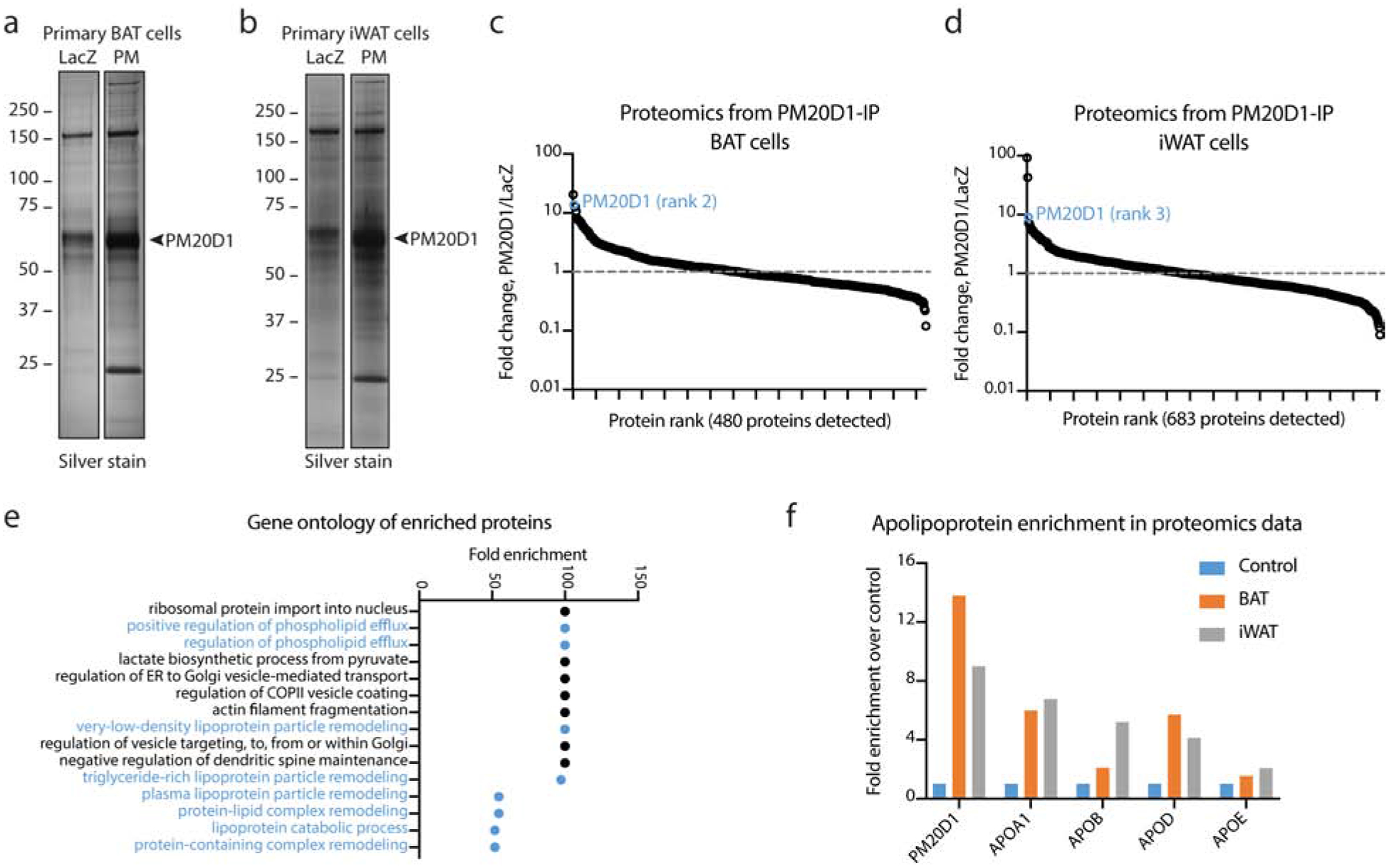
(a, b) Silver stain of PM20D1-flag immunoaffinity purified samples from primary brown (BAT, a) or primary inguinal (iWAT, b) adipocytes transduced with adenovirus expressing PM20D1-flag (right lanes) or LacZ control (left lanes). Cells were transduced on day 2 and conditioned media was harvested on day 6.
(c, d) Fold enrichment across the entire quantitative shotgun proteomics dataset of proteins in PM20D1 versus LacZ immunoaffinity purifications from BAT (c) or iWAT (d) cells. PM20D1 is highlighted in blue.
(e) Gene ontology of enriched biological processes from PM20D1 interacting proteins from the combined BAT or iWAT datasets. PM20D1 interacting proteins were defined as those with >2-fold change and at least two detected peptides. Only those processes with fold enrichment >50 and raw P-value < 0.001 are shown.
(f) Fold enrichment of the indicated apolipoproteins from PM20D1 immunoaffinity purifications in BAT (orange) or iWAT (grey) samples. See also Table S1 and S2.
To determine the identity of these proteins, LacZ and PM20D1 samples were labeled with tandem mass tags for quantitation, pooled, and analyzed by shotgun proteomics (Tables S1 and S2). In total, 480 and 683 proteins with at least one peptide were detected in the BAT and iWAT samples, respectively (Fig. 1c and d). As expected, PM20D1 was abundantly detected (207 and 171 peptides in BAT and iWAT samples, respectively) and highly enriched in PM20D1-IP versus LacZ-IP (14- and 9-fold, respectively). Using a cut-off of at least 2-fold enrichment in the PM20D1 samples and requiring at least 2 peptides per protein to avoid ambiguous protein assignments, a total of 45 and 55 proteins were identified to be enriched in the PM20D1-IP in BAT and iWAT, respectively. These proteins were pooled together and subjected to Gene Ontology Enrichment Analysis. Remarkably, 8 out of 15 biological processes (53%) with at least 50-fold enrichment were classified as related to lipoprotein metabolism (Fig. 1f). Confirming this global analysis, manual inspection of the proteomics dataset revealed that several apolipoproteins were enriched in PM20D1-IP samples from both BAT and iWAT cells (Fig. 1g). These enriched apolipoproteins include APOA1 and APOB, the major proteinaceous components of high- and low-density lipoproteins, respectively. These data therefore demonstrate that PM20D1 robustly interacts with lipoproteins in conditioned media of both brown and iWAT adipocytes.
PM20D1 is exclusively localized to lipoproteins in mice and human plasma
Lipoproteins are a heterogeneous collection of circulating particles that function to traffic triglyceride and cholesterol between tissues. In addition to apolipoproteins, >50 additional lipoprotein-associated polypeptides have also been identified. Surprisingly, PM20D1 has not been previously identified as a lipoprotein-associated factor. We therefore sought to validate whether PM20D1 interacts with lipoproteins in vivo, and if so, on which lipoprotein fractions. Because we have not yet identified an anti-PM20D1 antibody of sufficient sensitivity and selectivity for measurement of endogenous PM20D1 levels, instead we used two orthogonal approaches as surrogate measurements: (1) anti-Flag western blotting for overexpressed epitope-tagged PM20D1 and (2) N-acyl amino acid enzyme activity assays on plasma.
First, mice were transduced with epitope-tagged PM20D1-flag using adeno-associated virus (AAV serotype 8). We had previously demonstrated that this virus largely infects the liver and results in the robust secretion of PM20D1-flag into the blood (Long et al., 2016). As a control, mice were transduced with AAV-GFP. After a one-week transduction period, plasma was harvested from AAV-PM20D1 and AAV-GFP mice (n=5/group). First, we measured total N-acyl amino acids in both treatment groups by targeted liquid chromatography-mass spectrometry (LC-MS). As expected (Long et al., 2016), several N-acyl amino acids were dramatically increased by PM20D1 overexpression, including N-oleoyl-leucine (5.8-fold) and N-oleoyl-phenylalanine (10.3-fold) (Fig. 2a). Next, plasma from each treatment group was pooled and fractionated by fast protein liquid chromatography (FPLC) using a size-exclusion column (SEC) to separate the lipoprotein fractions away from albumin and other highly abundant proteins in blood. Under this protocol, the vast majority of proteins, including albumin were found in fraction 18–19 as indicated by the high protein concentration in those fractions (orange curves, Fig. 2b, c). APOB-containing low-density lipoproteins were predominantly found in the earliest fractions (fractions 9–16) and APOA1-containing high-density lipoproteins eluted in fraction 17 (Fig. 2b, c). PM20D1-flag was most abundantly detected in fractions 12–16 and coeluting with APOB, though some was also found in the APOA1+ fraction 17 (Fig. 2b). PM20D1-flag was absent in fractions 17 and 18 where the majority of plasma proteins eluted (Fig. 2b), establishing that PM20D1 is entirely associated to lipoproteins. Importantly, the PM20D1-flag band was specific because no corresponding band was found in any AAV-GFP fractions (Fig. 2c). By quantitation of total anti-flag band intensities, 91% was localized to an APOB+ fraction and the remaining 9% on an APOA1+ fraction (Fig. 2b).
Fig. 2. PM20D1 endogenously localizes to lipoproteins in vivo.
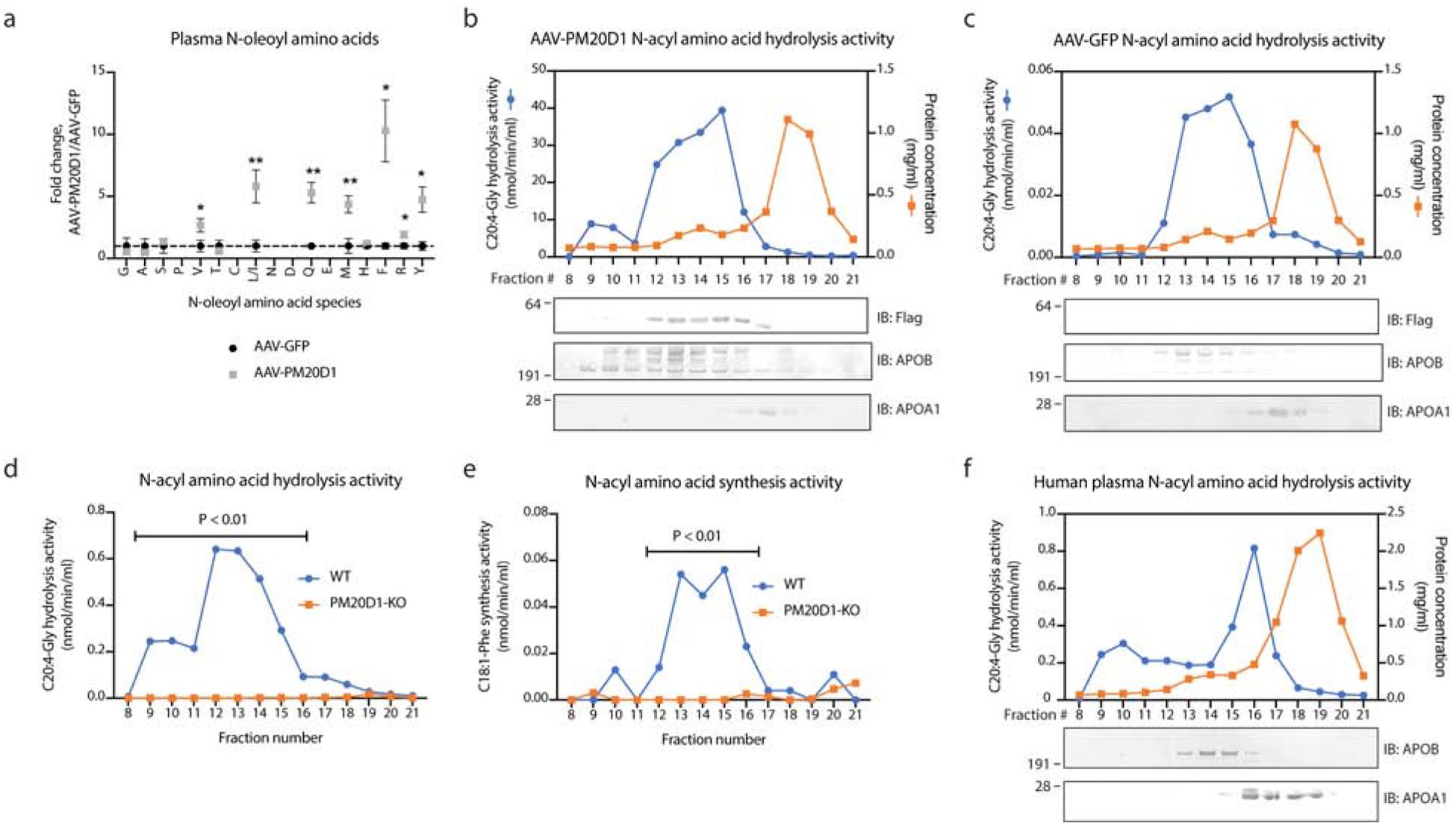
(a) Total plasma N-acyl amino acid levels from AAV-GFP (black) or AAV-PM20D1 (gray) transduced mice. Plasma was harvested after a one-week transduction period using a dose of 10e10 GC/mouse (intravenously).
(b,c) C20:4-Gly hydrolysis activity (blue traces), protein concentrations (orange traces), and Western blots (bottom panels) of fractionated mouse plasma one week after transduction by AAV-PM20D1-flag (a) or AAV-GFP (b).
(d,e) C20:4-Gly hydrolysis (c) and C18:1-Phe synthesis (d) activity of fractionated mouse plasma from either WT (blue trace) or PM20D1-KO (orange trace) mice.
(f) C20:4-Gly hydrolysis activity (blue traces), protein concentrations (orange traces), and Western blots (bottom panels) of fractionated pooled human plasma.
For (b-e), plasma from five mice per group was pooled together and separated by fast protein liquid chromatography-size exclusion (FPLC-SEC). For (a), (d), and (e), Student’s two-tailed t-test for AAV-GFP versus AAV-PM20D1 or WT versus PM20D1-KO across the indicated fractions was used to determine statistical significance. Data are shown as means ± SEM.
To confirm the lipoprotein association of PM20D1 using a second approach, a highly selective and previously validated mass spectrometry-based PM20D1 enzyme activity assay was used (Long et al., 2016, 2018). Fractionated plasma samples from AAV-PM20D1 and AAVGFP mice were incubated with a prototypical N-acyl amino acid substrate N-arachidonoyl glycine (C20:4-Gly) and hydrolysis to arachidonic acid was monitored by LC-MS (Fig. 2b, c). In AAV-PM20D1 samples, the C20:4-Gly hydrolysis activity largely co-localized in the same fractions in which the PM20D1-flag protein was detected (Fig. 2b). In AAV-GFP control mice, a C20:4-Gly hydrolysis activity was also robustly detected in the lipoprotein fractions (Fig. 2c), with the predominant difference versus AAV-PM20D1 samples being only in the magnitude of activity (average of 11.9 and 0.2 nmol/min/ml across all the fractions for AAV-PM20D1 and AAV-GFP, respectively, P = 0.008). To critically test whether endogenous PM20D1 is responsible for the activity observed in AAV-GFP control mice, we isolated plasma from a cohort of PM20D1-WT and PM20D1-KO animals under basal conditions without any viral transduction. Plasma was again fractionated using the same FPLC-SEC protocol as described above. In PM20D1-WT mice, an N-acyl amino acid hydrolysis activity was again detected in fractions 9–15, with peak activity in fractions 12 and 13 (Fig. 2d). All of the N-acyl amino acid hydrolysis activity was abolished in PM20D1-KO plasma (Fig. 2d). Parallel experiments monitoring the conversion of oleic acid to N-oleoyl-phenylalanine also demonstrate that PM20D1-containing lipoproteins can catalyze N-acyl amino acid biosynthesis (Fig. 2e). Taken together, these data demonstrate that overexpression of PM20D1 drives localization of PM20D1 protein exclusively to lipoprotein fractions. Moreover, PM20D1 is also endogenously trafficked on lipoproteins.
In humans, the PM20D1 gene also encodes a classically secreted enzyme that catalyzes bidirectional N-acyl amino acid synthesis/hydrolysis (Benson et al., 2019; Long et al., 2016). Considering species differences in lipoprotein composition between rodents and humans, as well as the lack of a previously reported N-acyl amino acid hydrolysis activity in human plasma, it has remained unclear whether a PM20D1 activity is detectable in human plasma and whether such an activity would localize to lipoproteins. To address this question, commercially available pooled human plasma was fractionated by FPLC-SEC. The fractionated human plasma was then analyzed by N-acyl amino acid hydrolysis activity and Western blotting. As shown in Fig. 2g, human plasma N-acyl amino acid hydrolysis activity was also localized to earlier lipoprotein fractions with peak activity observed in fraction 16 corresponding to an APOA1+ high density lipoprotein fraction, though activity was detectable in all lipoprotein fractions. This N-acyl amino acid hydrolysis activity was once again clearly separated away from the majority of proteins which eluted in fraction 18 and 19. The human N-acyl amino acid hydrolysis activity was approximately 10-fold higher than that observed in mice (Fig. 2g). We therefore conclude that human plasma N-acyl amino acid hydrolysis activity is also associated to lipoproteins.
Lipoproteins co-activate PM20D1 and drive N-acyl amino acid biosynthesis
We next evaluated the possibility that lipoproteins might be an important in vivo regulator of PM20D1 activity. We generated recombinant, purified mouse PM20D1 with a C-terminal flag tag. We also purified low- and high-density lipoproteins (LDL and HDL, respectively) from PM20D1-KO mice by FPLC (see methods). Importantly, these isolated lipoproteins are devoid of any endogenous PM20D1 which would confound downstream enzyme activity measurements. PM20D1-deficient lipoproteins or PBS control were then incubated with recombinant PM20D1 overnight and in vitro enzyme activity assays were performed the next day. As expected, essentially no background activity was detected in control samples or PM20D1-KO lipoprotein only samples when recombinant PM20D1 was omitted (Fig. 3a). Recombinant PM20D1 (0.1 μg per reaction) yielded an enzyme activity of 130 nmol/min/mg hydrolysis activity for the conversion of C20:4-Gly to arachidonic acid. Co-incubation of recombinant PM20D1 with LDL resulted in a +154% increase in enzyme activity to 330 nmol/min/mg (Fig. 3a). Similar experiments with HDL also resulted in an increased activity, though of a lower magnitude than LDL (+49% increase versus PM20D1 alone, Fig. 3a). These results demonstrate that lipoprotein particles powerfully co-activate PM20D1 enzyme activity in vitro.
Fig. 3. Lipoproteins are PM20D1 co-activators in vitro and in vivo.
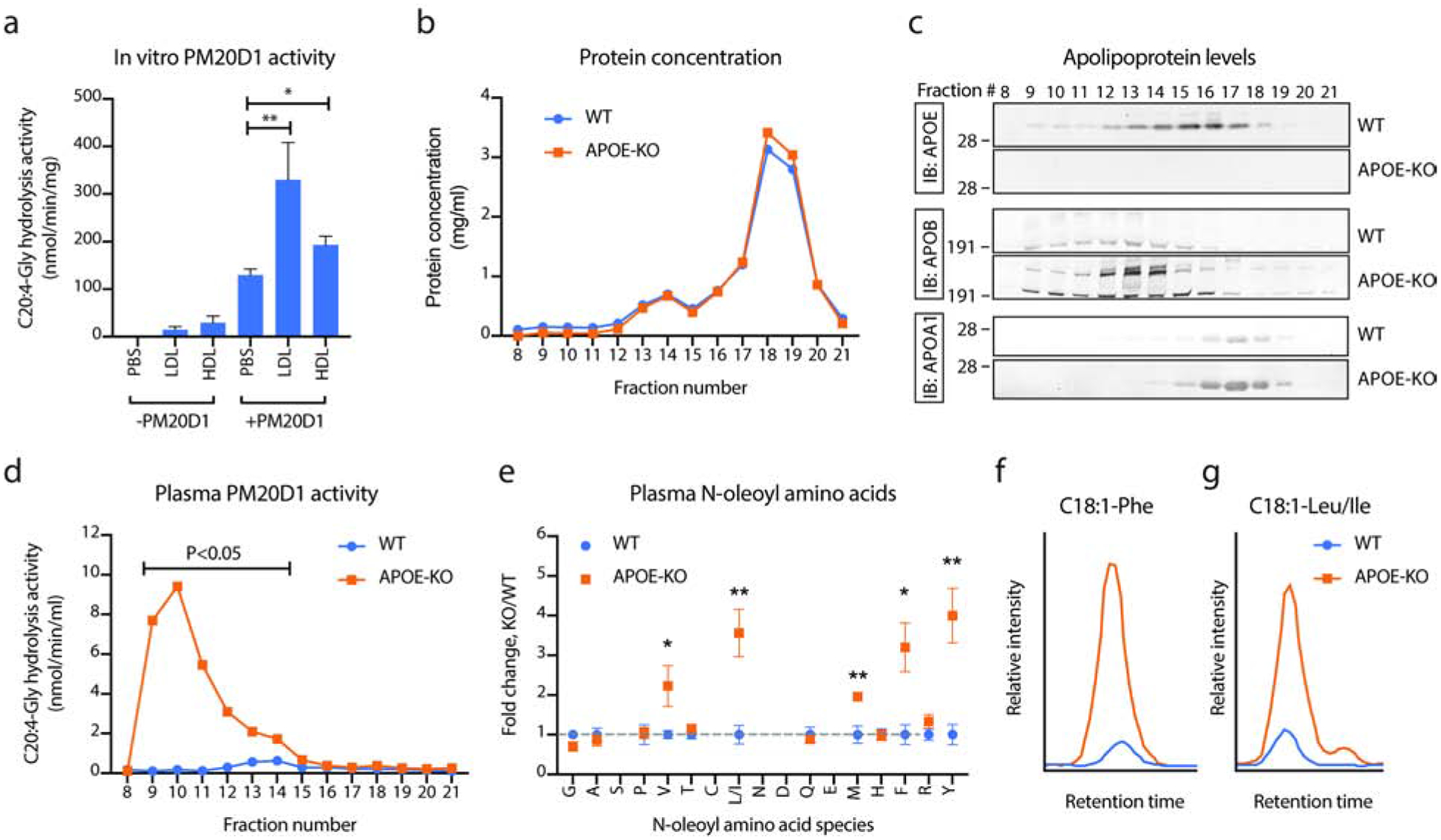
(a) C20:4-Gly hydrolysis activity of mammalian recombinant, purified mouse PM20D1 (0.1 ug/reaction) alone or following incubation with APOB+ or APOA1+ lipoproteins (LDL and HDL, respectively) isolated from PM20D1-KO mice. N=5/group.
(b-d) Protein concentrations (b), apolipoprotein levels (c), and PM20D1 activity as measured by C20:4-Gly hydrolysis activity (d) of fractionated mouse plasma from either WT (blue trace) or APOE-KO (orange trace) mice.
(e) Total plasma N-acyl amino acid levels from WT (blue) or APOE-KO (orange) mice. N=5/group.
(f,g) Representative LC-MS traces of C18:1-Phe (f) and C18:1-Leu/Ile (g) from WT (blue) or APOE-KO (orange) mice.
For (a), LDL or HDL was isolated from plasma of five pooled PM20D1-KO mice. For (b-d), plasma from five mice per group was pooled together and separated by fast protein liquid chromatography-size exclusion (FPLC-SEC). For (e), N=5/group.
For (a), (d), and (e), Student’s two-tailed t-test was used for the indicated comparison or for WT versus APOE-KO mice. * P < 0.05, ** P < 0.01. Data are shown as means ± SEM. See also Table S3 and S4.
To determine whether lipoprotein particles could also augment PM20D1 activity in vivo, we turned to the classical APOE-knockout mouse model. APOE is a ligand for several lipoprotein receptors and APOE-KO mice consequently exhibit dramatic accumulation of lipoproteins in the blood. While APOE-KO mice are classically used as a model for atherosclerosis (Plump et al., 1992), these animals also exhibit a robust hypermetabolic phenotype characterized by resistance to diet-induced obesity and increased whole body energy expenditure (Chiba et al., 2003; Gao et al., 2007; Zhi et al., 2006). However, the precise molecular pathways underlying the metabolic phenotype in APOE-KO mice has remained unknown. Pooled WT and APOE-KO plasma was again fractionated by FPLC-SEC. As expected, the majority of the proteins including albumin eluted in fractions 18–19, with lipoproteins eluting in earlier fractions (Fig. 3b). Western blotting of the FPLC fractions from APOE-KO mice confirmed complete loss of APOE protein and concomitant elevations in APOB and APOA1 (Fig. 3c). Remarkably, PM20D1 activity was dramatically elevated in lipoprotein fractions from APOE-KO mice versus WT mice (Fig. 3d), with the largest increases in enzyme activity observed over fractions 9 to 14. To dissect whether this dramatic increase in enzyme activity was due to increased PM20D1 activity alone or some combination of increased PM20D1 protein levels and activity, we pooled the intermediate FPLC fractions 10–12 from WT or APOEKO mice and performed shotgun proteomics. By spectral counting, we observed an 8-fold increase in PM20D1 protein levels in APOE-KO versus WT lipoprotein fractions (Supplemental Tables 3 and 4). By N-acyl amino acid hydrolysis activity, we observed a 30-fold difference in activity across these same fractions (6.0 versus 0.2 nmol/min/ml in APOE-KO versus WT samples). Taken together, we conclude that APOE-KO mice have dramatically increased lipoprotein-associated N-acyl amino acid hydrolysis activities which are due to both increased PM20D1 protein levels and also increased PM20D1 activity.
To determine if the observed increased PM20D1 activity would translate into altered circulating N-acyl amino acids, a panel of N-oleoyl and N-arachidonoyl amino acids from WT or APOE-KO mice was measured by targeted LC-MS (Fig. 3e). Consistent with the increased PM20D1 activity (Fig. 3d), APOE-KO mice also exhibited elevations in several N-acyl amino acids, including a 3.2-fold and 3.6-fold increases in N-oleoyl-phenylalanine and N-oleoylleucine, respectively (Fig. 3e–g). Remarkably, the pattern of N-acyl amino acid accumulation in APOE-KO mice in terms of both N-acyl amino acid species and magnitude was nearly identical to that observed in the AAV-PM20D1 mice (Fig. 3e versus Fig. 2a). Plasma free fatty acid levels have been previously reported to be similar between APOE-KO and WT mice, suggesting that changes to free fatty acids are not likely to be a driver of these N-acyl amino acid changes (Grubb et al., 2014). These data establish that APOE-KO mice exhibit elevation of N-acyl amino acids in blood. We conclude that lipoproteins are powerful co-activators of PM20D1 activity in vitro and N-acyl amino acid biosynthesis in vivo. These observations also suggest that elevation of thermogenic N-acyl amino acids might contribute to the hypermetabolic phenotypes observed in APOE-KO mice.
Serum albumin is a physiologic plasma N-acyl amino acid carrier and confers hydrolytic stability
Because of the lipoprotein localization for circulating PM20D1, we next considered the possibility that N-acyl amino acids might themselves also be constituents of lipoprotein particles. Alternatively, N-acyl amino acids may be localized to a site distinct from their biosynthetic origin. To differentiate between these possibilities, N-acyl amino acids from FPLC fractionated plasma of WT mice were analyzed by targeted LC-MS. To increase the signal to noise for detecting N-acyl amino acids across these diluted fractions, consecutively pooled FPLC fractions were analyzed. As shown in Fig. 4a, N-acyl amino acids were not found in the same FPLC fractions as PM20D1 or as other lipoproteins, demonstrating that these lipids are not constituents of lipoprotein particles. Instead, N-acyl amino acids were primarily detected in fraction 18–19, the same FPLC fractions where the majority of plasma proteins including albumin elute (Fig. 2b and Fig. 3b).
Fig. 4. N-acyl amino acids are trafficked by albumin in the circulation.
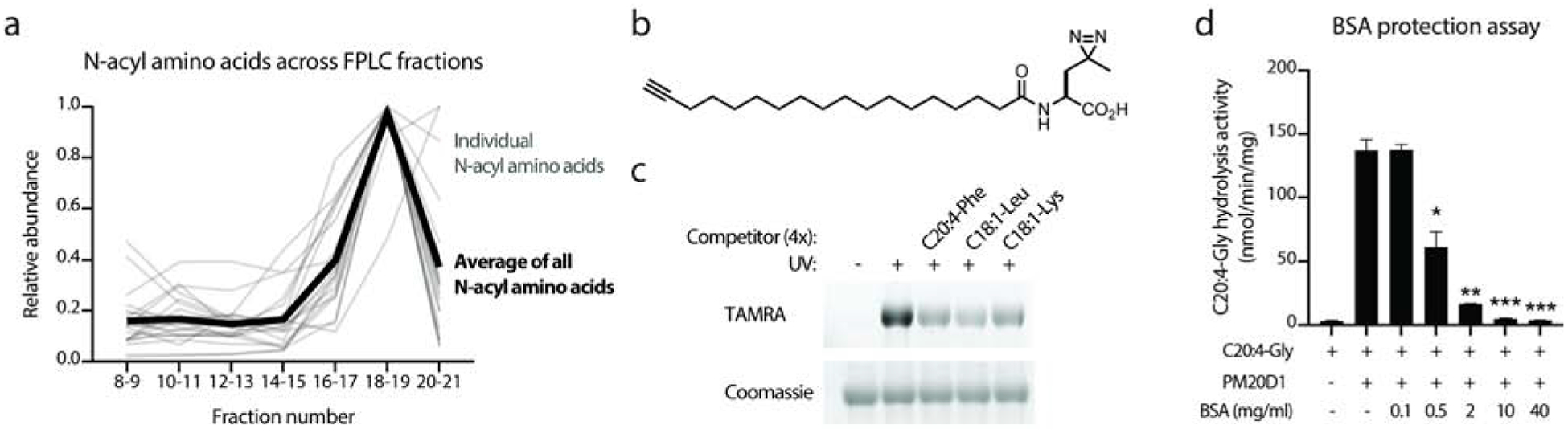
(a) Quantitation of N-acyl amino acid levels across the indicated size exclusion chromatography fractions.
(b) Chemical structure of an N-acyl amino acid photocrosslinking analog.
(c) TAMRA fluorescence (top) and Coomassie staining (bottom) of N-acyl amino acid photocrosslinker (50 μM) labeling of bovine serum albumin (1 mg/ml) in the presence of absence of the indicated “cold” competitor (200 μM).
(d) Conversion of C20:4-Gly to arachidonic acid by recombinant PM20D1 in the absence or presence of increasing concentrations of fatty acid free BSA.
For (a), plasma from five wild-type mice was pooled together and separated by fast protein liquid chromatography-size exclusion (FPLC-SEC). Consecutive fractions were pooled and analyzed by LC-MS (see Methods). For (d), N=3/group, * P < 0.05, ** P < 0.01, *** P < 0.001 versus hydrolysis activity in the absence of BSA. Data are shown as means ± SEM.
Beyond its most well-known characteristic as the most abundant individual protein in plasma (~40 mg/ml), albumin is also recognized as a key plasma carrier for many circulating hormones including steroids (Hammond, 2016) and thyroid hormones (Petitpas et al., 2003). The observation of N-acyl amino acids co-eluting in fractions 18–19 also suggested that plasma albumin may also function as a carrier for circulating N-acyl amino acids. A photocrosslinking analog of N-acyl amino acids was therefore used to probe the direct biochemical interaction of N-acyl amino acids with serum albumin (Long et al., 2016). This “photoprobe” contains a diazirine moiety which enables UV-dependent crosslinking and an alkyne handle for downstream click chemistry detection and visualization (Fig. 4b). Importantly, this photoprobe has been previously validated to also exhibit mitochondrial uncoupling in cells, demonstrating that the introduction of these chemical modifications does not alter the bioactivity of this compound (Long et al., 2016). Recombinant bovine serum albumin (BSA) was incubated in the presence of photoprobe (50 μM) or with photoprobe and four-fold molar excess of N-acyl amino acid competitors (200 μM). These samples were UV irradiated to crosslink photoprobe to albumin and copper-mediated click chemistry with TAMRA-azide was used to install a fluorophore on to any crosslinked photoprobe. As a negative control for UV-dependent crosslinking, photoprobe was incubated with albumin in the presence of ambient light. All samples were visualized by in-gel fluorescence for TAMRA. A robust, UV-dependent labeling with N-acyl amino acid photoprobe was observed, establishing that this N-acyl amino acid photocrosslinker can directly interact with albumin (Fig. 4c). Importantly, the photocrosslinking signal was competed in the excess presence of one of three “cold” N-acyl amino acids including N-arachidonoyl-phenylalanine, N-oleoyl-leucine, or N-oleoyl-lysine (Fig. 4c). Taken together, these data demonstrate that N-acyl amino acids directly bind albumin.
Lastly, we reasoned that N-acyl amino acid binding to albumin may provide a steric “protective” mechanism to confer resistance to PM20D1-mediated hydrolysis. To directly test this possibility, in vitro hydrolysis activity assays were performed with recombinant PM20D1 in the presence or absence of BSA. Remarkably, co-incubation of physiologic concentrations of BSA (40 mg/ml) with C20:4-Gly provided complete resistance to hydrolysis by PM20D1 (Fig. 4d). A dose course of BSA revealed an ED50 of 0.5 mg/ml for this protective effect (Fig. 4d). These data demonstrate that albumin-bound N-acyl amino acids are resistant to hydrolytic degradation. We speculate that this protective mechanism, along with the spatial segregation of N-acyl amino acids away from their sites of biosynthesis, may explain how PM20D1 can catalyze an otherwise thermodynamically unfavorable “synthase” reaction.
A protein binding equilibrium regulates biologically active “free” versus “bound” N-acyl amino acids
For other lipophilic hormones, protein binding separates biologically active “free” versus biologically inactive “bound” populations (Ekins, 1990; Goldman et al., 2017; Hammond, 2016). To determine whether protein binding to N-acyl amino acids might serve a similar function, we first sought to determine the fraction of N-acyl amino acids that was free versus associated to protein (“bound”). Towards this end, total plasma from five pooled mice was fractionated using a 3 kDa molecular weight cut off filter and N-acyl amino acids in both the flow through and the retained fraction were analyzed by LC-MS/MS (Fig. 5a). On average, 96.5% of total plasma N-acyl amino acids were bound to protein (Fig. 5b). This figure is of a similar magnitude to that previously observed for circulating testosterone (98% bound), estrogen (98% bound), and thyroxine (99.9% bound) (Ekins, 1990; Goldman et al., 2017; Hammond, 2016). These data therefore demonstrate that N-acyl amino acids are in equilibrium between free and protein-bound states in the plasma.
Fig. 5. Characterization of biologically active “free” N-acyl amino acids versus biologically inactive “bound” N-acyl amino acids.
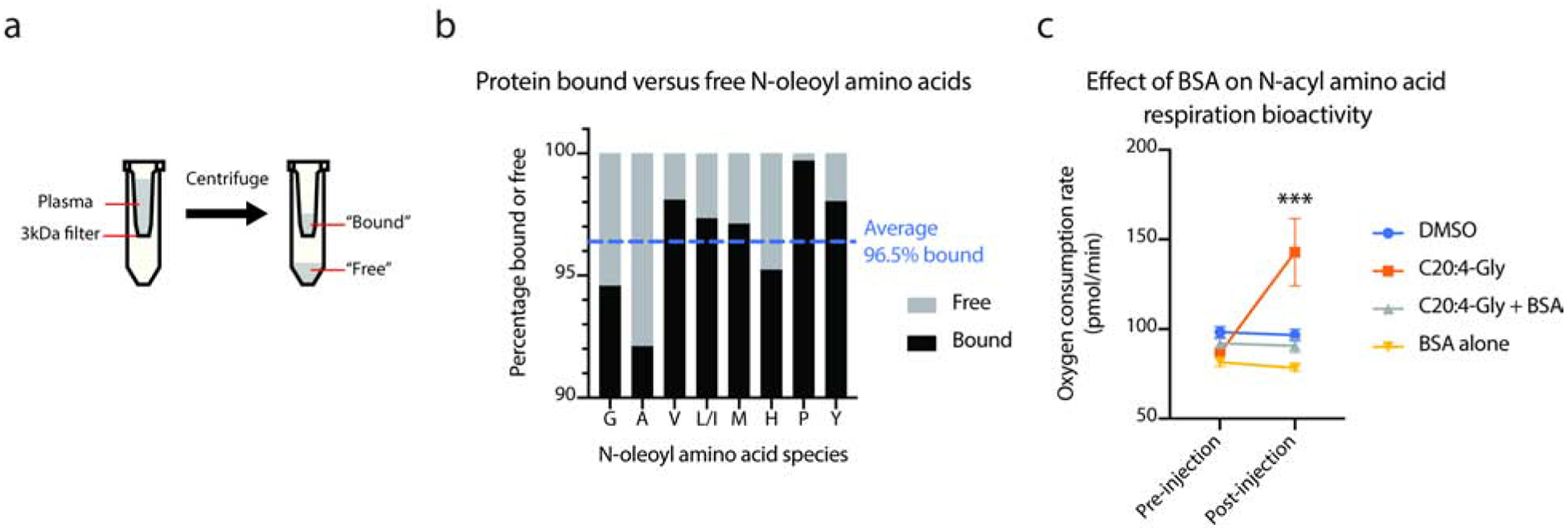
(a) Schematic of the biochemical strategy for separation of protein-bound versus free N-acyl amino acids from plasma.
(b) Fraction of detected N-acyl amino acids in the free fraction versus protein-bound fraction from pooled plasma.
(c) Bioactivity of “free” C20:4-Gly versus “bound” BSA-C20:4-Gly in stimulating cellular respiration in C2C12 cells.
For (c), all experiments were performed in the presence of oligomycin to selectively measure uncoupled respiration. *** P < 0.001 versus DMSO. N=4 wells/condition. Data are shown as means ± SEM.
We next sought to determine if albumin-bound N-acyl amino acids and free N-acyl amino acids represented a biologically inactive and biologically active state, respectively. As a bioactivity assay, we used a respiration assay on murine C2C12 cells in which the cellular oxygen consumption rate is measured as a function of compound treatment. As expected, treatment of cells with the N-acyl amino acid C20:4-Gly robustly stimulated cellular respiration by +65% over pre-treatment levels, whereas treatment with DMSO control did not stimulate respiration (Fig. 5c). Pre-incubation of a C20:4-Gly with BSA (40 mg/ml) completely abolished the respiratory bioactivity of this N-acyl amino acid (Fig. 5c). Importantly, BSA alone did not have any effects on cellular respiration (Fig. 5c). We therefore conclude that “free” N-acyl amino acids represent the biologically active form of these circulating molecules, and that circulating N-acyl amino acid bioactivity is determined by plasma protein binding in a similar manner to that of other lipophilic hormones.
DISCUSSION
N-acyl amino acids and their biosynthetic enzyme, PM20D1, are a circulating biochemical pathway that controls mitochondrial respiration with potential disease relevance to human obesity and neurodegenerative diseases (Benson et al., 2019; Long et al., 2016; Sanchez-Mut et al., 2018, 2020). Here we establish a unifying framework for understanding the functional organization and regulation of this pathway in blood plasma. We demonstrate that PM20D1 is an exclusively lipoprotein-associated enzyme. Lipoprotein association powerfully co-activates PM20D1 activity in vitro and in vivo. We also identify serum albumin as a physiologic N-acyl amino acid carrier, which spatially segregates these bioactive lipids away from their site of biosynthesis, confers hydrolysis resistance, and functions to buffer biologically active “free” versus biologically inactive “bound” N-acyl amino acid forms. These studies establish that the surface of PM20D1-containing lipoproteins represents a major site for N-acyl amino acid biosynthesis. Furthermore, N-acyl amino acids share plasma transport characteristics with other lipophilic hormones including thyroid hormone and sex steroids (Fig. 6). Whether PM20D1 and/or N-acyl amino acids might reciprocally regulate atherogenesis remains an open question.
Fig. 6. Working model for plasma protein regulation of PM20D1 and N-acyl amino acids.
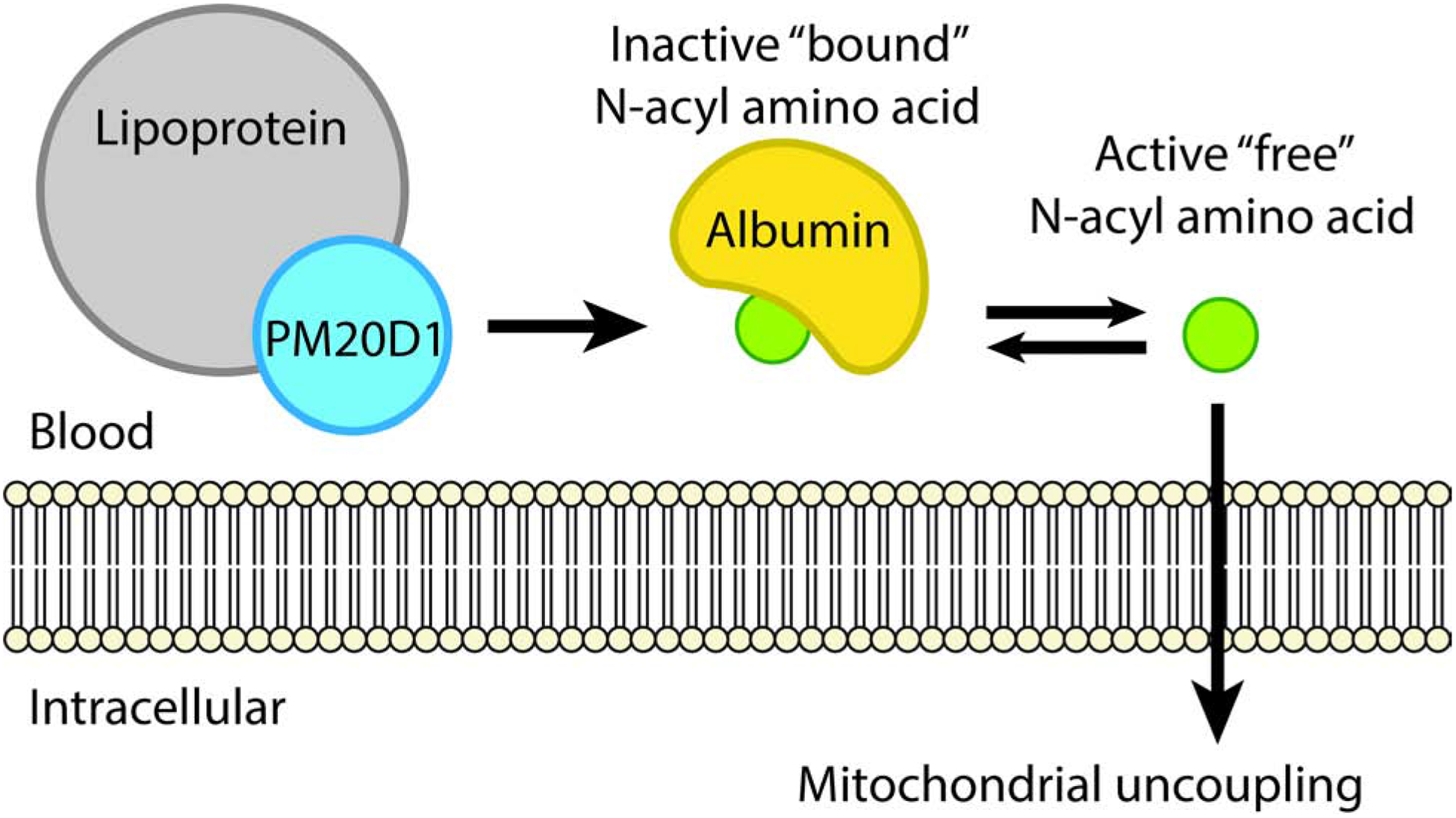
Lipoprotein association functions to co-activate PM20D1 activity, which then synthesizes N-acyl amino acids. These lipids then circulate in an inactive albumin-bound as well as a biologically active free form. Only free N-acyl amino acids can stimulate cellular respiration.
As a genetic test of this model, we demonstrate that elevation of lipoproteins in the APOE-KO mouse was sufficient to fully drive all downstream aspects of the PM20D1/N-acyl amino acid pathway, including augmentation of circulating PM20D1 activity and increased plasma N-acyl amino acid levels. In future studies, the physiological relevance of PM20D1 activation by lipoproteins beyond these genetic models still remains to be determined. Besides its classical use as an atherosclerosis model, APOE-KO mice also exhibit a paradoxical leanness, increased energy expenditure, and protection from diet-induced obesity (Chiba et al., 2003; Gao et al., 2007; Zhi et al., 2006). This phenotype is consistent with what has previously been shown with systemic overexpression of PM20D1 in mouse models (Long et al., 2016). Interestingly, APOE-KO mice also exhibit robust protection from Alzheimer’s disease at both biochemical and functional levels (Bales et al., 1997, 1999; Ganor et al., 2018), a second phenotype consistent with overexpression of PM20D1 in the brain (Sanchez-Mut et al., 2018). The activation of the PM20D1/N-acyl amino acid pathway may therefore be a contributor to the protection from metabolic and neurological diseases observed in APOE-KO mice.
The identification of serum albumin as a physiologic N-acyl amino acid carrier provides key insights into the metabolism, trafficking, and bioactivity of these thermogenic lipids. Albumin binding to N-acyl amino acids confers hydrolysis resistance as well as spatial segregation from their sites of biosynthesis. These two observations provide a plausible biochemical mechanism for how PM20D1 can catalyze an otherwise thermodynamically unfavorable condensation reaction. Serum albumin binding to N-acyl amino acid binding also establishes that N-acyl amino acids share a common carrier mechanism with other lipophilic hormones including thyroid hormone and sex steroids. All of these lipid hormones exhibit significant (>95%) protein binding in the plasma. Furthermore, protein binding determines biologically active “free” versus biologically inactive “bound” forms. As with the other lipophilic hormones, this free-to-bound equilibrium likely represents a key regulatory node for the bioactivity of N-acyl amino acids. Beyond albumin, that thyroid hormone and sex steroids also have other dedicated high affinity plasma carriers (e.g., TBG and SHBG, respectively). We hypothesize that additional analogous plasma carriers might also regulate the transport and/or availability of biologically active “free” N-acyl amino acids. The future identification of these potential plasma carriers may enable manipulation of the free-to-bound ratio in plasma and downstream N-acyl amino acid uncoupling bioactivity.
Lastly, our studies identify a potential opportunity for pharmacological and therapeutic manipulation for PM20D1. PM20D1 activity can be co-activated by lipoprotein association, an observation similar to lipoprotein-mediated allosteric modulation for other key circulating enzymes including LPL, LCAT, and secreted PLA2s (Burke and Dennis, 2009; Cooke et al., 2018; Goldberg et al., 1990). When available, compounds that mimic lipoprotein co-activation, as has been already described for APOC2 mimetics (Amar et al., 2015) or small molecule LCAT activators (Manthei et al., 2018), may be useful for increasing PM20D1 activity and circulating N-acyl amino acids for the treatment of age-associated metabolic or neurological diseases.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jonathan Z. Long (jzlong@stanford.edu).
Materials Availability.
This study did not generate new unique reagents.
Data and Code Availability.
This study did not generate/analyze any datasets/code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and primary cultures.
HEK293T (female) and C2C12 (female) cells were obtained from ATCC (CRL-3216 and CRL-1772, respectively) and cultured in DMEM with L-glutamine, 4.5 g/l glucose and sodium pyruvate (Corning 10013CV) supplemented with 10% FBS (Corning 35010CV). For primary iWAT and BAT cells from male mice, the stromal-vascular fraction of inguinal (iWAT) pad from 4–12 week old mice was dissected, minced, and digested for 45 min at 37°C in PBS containing 10 mM CaCl2, 2.4 U/ml dispase II (Roche), and 1.5 U/ml collagenase D (Roche). The stromal-vascular fraction of brown fat (BAT) pads from newborn (P1–P14) pups was dissected, minced, and digested for 45 min at 37°C in PBS containing 1.3 mM CaCl2, 120 mM NaCl, 5 mM KCl, 5 mM glucose, 100 mM HEPES, 4% BSA, and 1.5 mg/ml collagenase B (Roche). Digested tissue was diluted with adipocyte culture media (DMEM/F-12, GlutaMAX supplement, Life Technologies, with 10% FBS and pen/strep) and centrifuged (600 × g, 10 min). The pellet was resuspended in 10 ml adipocyte culture media, strained through a 40 μm filter, and plated. Differentiation was induced by adipogenic cocktail containing 5 μg/ml insulin (Sigma), 5 μM dexamethasone (Sigma), 250 μM isobutylmethylxanthine (Sigma), and 1 μM rosiglitazone (Cayman) for 2 days. Two days after induction, cells were maintained in adipocyte culture media containing 5 μg/ml insulin and 1 μM rosiglitazone. Cells were incubated at 37°C, 5% CO2 for growth.
General animal information.
Animal experiments were performed according to procedures approved by the Stanford University IACUC. Mice were maintained in 12 h light-dark cycles at 22°C and fed a standard irradiated rodent chow diet. All experiments on wild-type mice were performed with male C57BL/6J mice purchased from Jackson Laboratories (stock number 000664). APOE-KO mice were purchased from Jackson Laboratories (stock number 002052). Global Pm20d1 knockout mice have been previously described (Long et al., 2018) and are available from Jackson Laboratories (stock number 032193, MGI:6201144). For AAV injections, 7-week old male mice (C57BL/6J) were injected via tail vein at a dose of 10e10 GC/mouse diluted in saline in a total volume of 100 μl/mouse. All blood was collected into lithium heparin tubes (BD) via submandibular bleed and immediately spun (5,000 rpm, 10 min, 4°C) to isolate the plasma.
METHOD DETAILS
Materials.
N-arachidonoyl glycine was purchased from Cayman. N-acyl amino acid photocrosslinking probe was synthesized as previously described (Long et al., 2016). Phenylalanine (BP391100) and oleic acid (AC270290050) were purchased from Fisher. BSA (protease free, fatty acid free, essentially globulin free) was purchased from Sigma (A7030). Pooled human plasma (IPLA-N) was purchased from Innovative Research. The following antibodies were used: anti-Flag M2 (Sigma F3165), anti-APOB (Abcam ab20737), anti-APOA1 (ab7614). The following plasmids were used: mouse AAV-PM20D1-flag (Addgene 132682), mouse PM20D1-flag (Addgene 84566). Adeno-associated viruses were produced at the Penn Vector Core. AAV-GFP virus was purchased from Penn Vector Core (AAV8.CB7.CI.eGFP.WPRE.rBG).
Immunoaffinity purification of PM20D1 from conditioned media.
Primary iWAT or BAT cells in 10cm plates were transduced with PM20D1-flag or LacZ control adenovirus on day 2 of differentiation. On day 6, the media was aspirated and refreshed with fresh complete media. 24 h later, total conditioned media was harvested and centrifuged (1000 × g) to remove debris. The conditioned media with protease inhibitors added was then incubated with anti-FLAG M2 magnetic beads (M8823, 50 μl per sample) overnight. The following day, the beads were washed three times with PBS (0.5 ml each) and proteins were eluted by 3xFLAG peptide (0.1 mg/ml, 100 μl in PBS).
Proteomics of PM20D1-interacting proteins.
On bead digestion was carried out as follows: supernatants were resuspended 1:1 with 1M urea, 50 mM Tris, pH 8.0 and digested it with trypsin (1 mg) at 37°C under vortex shaking overnight. The following morning, digested peptides were reduced with 5 mM TCEP, followed by alkylation with 10 mM Iodoacetamide, quenching alkylation with 5 mM DTT and finally quenching the digestion process with TFA. Acidified digested peptide were desalted over C18 StageTip following protocol described before (Ong et al., 2002). Briefly, the tips were prepared placing a small disc of Empore material 3M in an ordinary pipette tip, preparing a single tip for each sample. Tips were cleaned and secured with Methanol, activated with 50% acetonitrile, 0.1% FA, equilibrated with 0.1% FA. Acidified digested sample was added to the column and finally washed twice with 0.1% TFA solution. Liquid was passed through the pipette tip with a centrifugation. Peptides were then eluted with 80% acetonitrile, 0.1% FA buffer thrice and dried in a speedvac. Dried desalted peptides samples were reconstituted with 200 mM EPPS buffer, pH 8.0 and labelled with 10-plex tandem mass tag (TMT) reagent. TMT labeling reactions were performed for 1 hours at room temperature. Modification of tyrosine residue with TMT was reversed by the addition of 5% hydroxyl amine for 15 minutes and the reaction was quenched with 0.5% FA. Samples were combined, further desalted over stage-tip, finally eluted into an Autosampler Inserts (Thermo Scientific), dried in a speedvac and reconstituted with 5% Acetonitrile-5% FA for MS analysis. Labelled peptide sample from previous step was analyzed with an LC-MS3 data collection strategy (McAlister et al., 2014) on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) equipped with a Thermo Easy-nLC 1200 for online sample handling and peptide separations. Resuspended peptide from previous step was loaded onto a 100 μm inner diameter fused-silica micro capillary with a needle tip pulled to an internal diameter less than 5 μm. The column was packed in-house to a length of 35 cm with a C18 reverse phase resin (GP118 resin 1.8 μm, 120 Å, Sepax Technologies). The peptides were separated using a 180 min linear gradient from 5% to 42% buffer B (90% ACN + 0.1% formic acid) equilibrated with buffer A (5% ACN + 0.1% formic acid) at a flow rate of 550 nL/min across the column. The scan sequence for the Fusion Orbitrap began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 350 – 1350 m/z scan range, AGC target 9 × 103, maximum injection time 80 ms, dynamic exclusion of 90 seconds). The “Top10” precursors was selected for MS2 analysis, which consisted of CID (quadrupole isolation set at 0.7 Da and ion trap analysis, AGC 8 × 103, Collision Energy 35%, maximum injection time 150 ms). The top ten precursors from each MS2 scan were selected for MS3 analysis (synchronous precursor selection), in which precursors were fragmented by HCD prior to Orbitrap analysis (Collision Energy 55%, max AGC 1 × 105, maximum injection time 120 ms, resolution 50,000). A compendium of in-house software tools were used to for .RAW file processing and controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides as previously described. MS/MS spectra were searched against a Uniprot Mouse database (downloaded March, 2014) with both the forward and reverse sequences. Database search criteria are as follows: tryptic with two missed cleavages, a precursor mass tolerance of 50 ppm, fragment ion mass tolerance of 1.0 Da, static alkylation of cysteine (57.02146 Da), static TMT labeling of lysine residues and N-termini of peptides (229.162932 Da), and variable oxidation of methionine (15.99491 Da). TMT reporter ion intensities were measured using a 0.003 Da window around the theoretical m/z for each reporter ion in the MS3 scan. Peptide spectral matches with poor quality MS3 spectra were excluded from quantitation (<100 summed signal-to-noise across all channels and <0.5 precursor isolation specificity).
FPLC-SEC fractionation of plasma.
Plasma was fractionated by fast protein liquid chromatography (FPLC) using a Superose 6 increase 10/300 GL (size range 5K - 5 MDa) column (GE Healthcare Life Sciences, 29091596) on a GE AKTA Pure FPLC system. A total of 250 μl of plasma was eluted in cold PBS down the column and separated into 40 × 1 mL fractions.
Purification of murine recombinant PM20D1.
HEK293T cells were infected with retrovirus expressing mouse PM20D1–6xHis-Flag in the presence of polybrene (8 μg/ml). After two days, cells were selected with hygromycin (150 μg/ml, Sigma Aldrich). The stable HEK294T cells were then grown in complete media. At confluence, the media (~500 ml) was changed, harvested 24 h later, and concentrated ~10-fold in 30 kDa MWCO filters (EMD Millipore) according to the manufacturer’s instructions. The concentrated media was centrifuged to remove debris (600 × g, 10 min, 4°C) and the supernatant containing PM20D1-flag was decanted into a new tube. PM20D1-flag was immunoaffinity purified overnight at 4°C from the concentrated media using magnetic Flag-M2 beads (Sigma Aldrich). The beads were collected, washed three times in PBS, eluted with 3xFlag peptide (0.1 μg/ml in PBS, Sigma Aldrich), aliquoted, and stored at −80°C.
In vitro PM20D1 enzyme activity assays.
In vitro hydrolysis and synthesis reactions of N-acyl amino acids was measured by incubating recombinant PM20D1 (0.5 ug) with N-acyl amino acids (100 μM) or amino acid and fatty acid (1 mM each) in PBS for 1 hour at 37°C. To measure enzymatic activity across FPLC fractionated plasma, 100 μl of each fraction was incubated with N-acyl amino acids (100 μM) or amino acid and fatty acid (1 mM each) in PBS for 1 hour at 37°C. All reactions were terminated by quenching with 400 μl 2:1 chloroform:methanol. Samples were then vortexed thoroughly, centrifuged at 1,000 rpm for 10 minutes, and the organic layer was transferred to a mass spec vial for analysis by LC-MS.
UV crosslinking using N-acyl amino acid photoprobe.
All experiments were performed in a 96-well plate. N-acyl amino acid photoprobe (50 μM) was incubated with fatty acid free BSA (1 mg/ml) in PBS for 1 h at room temperature. For competition experiments, “cold” N-acyl amino acid competitors (200 μM, 4x excess) were added to fatty acid free BSA (1 mg/ml) for 30 min at room temperature prior to the addition of N-acyl amino acid photoprobe (50 μM). Samples were placed on ice and UV-irradiated (10 min, UV Stratalinker 2400). Control samples were left on ice under ambient light. After UV irradiation, click chemistry was performed as follows: To 50 μl mixture at 1 mg/ml was added 3 μl TBTA (stock solution: 1.7 mM in 4:1 v/v DMSO:t-BuOH), 1 ul CuSO4 (stock solution: 50 mM in water), 1 μl TCEP (freshly prepared, stock solution: 50 mM), and 1 μl TAMRA-N3 (stock solution: 1.25 mM in DMSO). Reactions were incubated at room temperature for 1h, and then quenched with 4x SDS loading buffer (17 μl). In-gel TAMRA fluorescence was visualized on a Typhoon FLA 9500 biomolecular imager (GE Healthcare Life Sciences).
Determination of free versus bound N-acyl amino acids in plasma.
100 μl of plasma was filtered using 3kDa molecular weight cut-off filters (Amicon UFC9003) for 1 h at 15,000 rpm (4°C). 100 μl ultrapure water was then added to either the eluant or the top unfiltered layer. These layers were transferred to glass vials. Next, 300 μl 2:1 v/v acetonitrile:methanol was added. The vial was vortexed, centrifuged at 15,000 rpm for 10 min, and then the supernatant was transferred for mass spectrometry analysis.
Cellular respiration measurements.
Oligomycin was purchased from EMD Millipore, and FCCP and rotenone were purchased from Sigma. C2C12 cells were seeded at 30,000 cells/well, respectively in an XFp cell culture microplate (Seahorse Bioscience) and analyzed the following day. On the day of analysis, the cells were washed once with Seahorse respiration buffer (8.3 g/l DMEM, 1.8 g/l NaCl, 1 mM pyruvate, 20 mM glucose, pen/strep), placed in 0.5 ml Seahorse respiration buffer, and incubated in a CO2-free incubator for 1 hr. Port injection solutions were prepared as follows (final concentrations in assay in parentheses): 10 μM oligomycin (1 μM final), 500 μM C20:4-Gly (50 μM final), and 30 μM rotenone (3 μM final). The Seahorse program was run as follows: basal measurement, 3 cycles; inject port A (oligomycin), 3 cycles; inject port B (compounds), 3 cycles; inject port C (rotenone), 3 cycles. Each cycle consisted of mix 4 min, wait 0 min, and measure 2 min. For data expressed as “pre” and “post,” the average of the three measurements following oligomycin injection were averaged for the “pre” state and the average of the three measurements following N-acyl amino acid injection were averaged for the “post” state. Experiments using BSA included fatty acid free BSA in the port injection solution (e.g., 500 μM C20:4-Gly with 40 mg/ml BSA).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical comparisons were performed using Student’s t-test by Excel or Prism. Statistical details of each experiment can be found in the figures and figure legends. No explicit power analysis was used to determine same size. Sample sizes were determined based on previous literature for biochemical or animal studies. Unless indicated otherwise, all experiments were performed once, with N corresponding to biological replicates. Outliers were not removed from analyses. The experimentalist was not blinded to sample or treatment conditions.
Supplementary Material
Table S1. Quantitative proteomics of PM20D1-flag immunoaffinity purification from conditioned media of iWAT cells. Related to Figure 1.
Table S2. Quantitative proteomics of PM20D1-flag immunoaffinity purification from conditioned media of BAT cells. Related to Figure. 1.
Table S3. Shotgun proteomics of wild-type mouse plasma FPLC fractions 10–12 (pooled). Related to Figure 3.
Table S4. Shotgun proteomics of APOE-KO FPLC fractions 10–12 (pooled). Related to Figure 3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal Anti-FLAG M2 antibody | Sigma-Aldrich | F1804 |
| Anti-Apolipoprotein A I antibody | Abcam | ab7614 |
| Anti-Apolipoprotein B antibody | Abcam | ab20737 |
| Anti-beta Tubulin antibody | Abcam | ab6046 |
| Bacterial and Virus Strains | ||
| GFP adeno-associated virus | Penn Vector Core | AAV8.CB7.CI.eGF P.WPRE.rB |
| PM20D1-flag adeno-associated virus | Long et al., Cell 2016 (PMID 27374330) | Addgene 132682 |
| PM20D1-flag adenovirus | Long et al., Cell 2016 (PMID 27374330) | N/A |
| LacZ adenovirus | Long et al., Cell 2016 (PMID 27374330) | N/A |
| Biological Samples | ||
| Pooled Human Plasma Apheresis Derived | Innovative Research | IPLA-N |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TBTA, Tris[(1-benzyl-1 H-1,2,3-triazol-4-yl)methyl]amine | Fisher | H66485–03 |
| TCEP HCl (tris(2-carboxyethyl)phosphine hydrochloride) | Fisher | J60316–06 |
| TAMRA, Tetramethylrhodamine Azide | Fisher | T10182 |
| Oligomycin | EMD Millipore | 1404-19-9s |
| FCCP, 2-[2-[4-(trifluoromethoxy)phenyl]hydrazinylidene]-propanedinitrile | Sigma | 370-86-5 |
| Rotenone | Sigma | 83-79-4 |
| Hygromycin | Sigma | 31282-04-9 |
| Polybrene, Hexadimethrine bromide | Sigma | 107689 |
| Dispase II | Roche | 17-105-041 |
| Collagenase D from Clostridium histolyticum | Roche | 50-100-3282 |
| Insulin from bovine pancreas | Sigma | I5500 |
| Dexamethasone | Sigma | D4902 |
| Isobutylmethylxanthine | Sigma | I7018 |
| Rosiglitazone | Cayman | 71740 |
| Bovine Serum Albumin; heat shock fraction, protease free, fatty acid free, essentially globulin free | Sigma | A7030 |
| C20:4-Gly | Cayman | 90051 |
| C20:4-Phe | Lin et al., J. Med. Chem. 2018 (PMID 29533650) | N/A |
| C18:1-Leu | Lin et al., J. Med. Chem. 2018 (PMID 29533650) | N/A |
| C18:1-Lys | Lin et al., J. Med. Chem. 2018 (PMID 29533650) | N/A |
| N-acyl amino acid photoprobe | Long et al., Cell 2016 (PMID 27374330) | N/A |
| Phenylalanine | Fisher | BP391100 |
| Oleic Acid | Fisher | AC270290050 |
| 3x FLAG Peptide | Sigma | F4799 |
| Critical Commercial Assays | ||
| Seahorse XFp FluxPak | Seahorse Bioscience | 103022–100 |
| Experimental Models: Cell Lines | ||
| HEK 293T (Homo-sapiens) | ATCC | CRL-3216 |
| C2C12 (M. musculus) | ATCC | CRL-1772 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J (M. musculus) | Jackson Laboratory | 000664 |
| PM20D1-KO, C57BL/6J-Pm20d 1 em1Brsp/J (M. musculus) | Jackson Laboratory | 032193 |
| APOE-KO, B6.129P2-Apoetm1Unc/J (M. musculus) | Jackson Laboratory | 002052 |
| Recombinant DNA | ||
| PM20D1-flag transfected construct (M. musculus) | Long et al., Cell 2016 (PMID 27374330) | Addgene 84566 |
| Software and Algorithms | ||
| Graphpad Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| 3 kDa Molecular Weight Cutoff Filter | Amicon | UFC9003 |
| 30 kDa Molecular Weight Cutoff Filter | Amicon | UFC803024 |
| Anti-flag M2 magnetic beads | Sigma | M8823 |
| Uniprot Mouse database | Uniprot | https://www.uniprot.org/ |
SIGNIFICANCE.
Circulating metabolites in blood constitute fundamental axes of chemical intercellular communication that regulate diverse aspects of mammalian physiology. N-acyl amino acids are one such family of amino acid-fatty acid conjugated lipid hormones. These molecules endogenously circulate in mouse and human plasma and are elevated following chronic exposure to cold environments. A subset of N-acyl amino acids with medium-chain fatty acid tails and neutral amino acid head groups can directly stimulate cellular respiration, thereby linking environmental cold with an increased physiologic thermogenic response. However, the mechanisms that regulate circulating N-acyl amino acid function in blood remains unknown. Here, we show that major blood plasma components, including lipoproteins and serum albumin, are involved in regulating N-acyl amino acid metabolism and bioactivity. First, we establish that the extracellular N-acyl amino acid biosynthetic enzyme PM20D1 does not circulate as a free enzyme, but is rather entirely localized to high- and low-density lipoproteins. This association co-activates PM20D1 activity and drives N-acyl amino acid biosynthesis. Second, we show that N-acyl amino acids are tightly associated to albumin, which spatially segregates N-acyl amino acids away from their site of biosynthesis, confers resistance to hydrolytic degradation, and acts as a buffer between biologically active “free” versus inactive “bound” N-acyl amino acids. These data establish that N-acyl amino acid biosynthesis occurs on the surface of PM20D1-containing lipoproteins and that N-acyl amino acids share plasma transport characteristics with other lipophilic hormones including thyroid hormone and sex steroids. Taken together, we demonstrate the presence of a plasma protein network that controls the metabolism and bioactivity of a circulating lipid family.
HIGHLIGHTS.
The N-acyl amino acid biosynthetic enzyme PM20D1 is localized to lipoproteins.
Lipoproteins co-activate PM20D1 activity and N-acyl amino acid biosynthesis.
Serum albumin is a physiologic plasma carrier for N-acyl amino acids.
Albumin determines an equilibrium between active versus inactive N-acyl amino acids.
ACKNOWLEDGEMENTS
We thank members of the Long, Svensson, and Spiegelman labs for helpful discussions. This work was supported by the US National Institutes of Health (DK105203 and DK124265 to JZL), by the Stanford ChEM-H Institute (DF and CRF), and by the Stanford Diabetes Research Center (P30DK116074).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amar MJA, Sakurai T, Sakurai-Ikuta A, Sviridov D, Freeman L, Ahsan L, and Remaley AT (2015). A novel apolipoprotein C-II mimetic peptide that activates lipoprotein lipase and decreases serum triglycerides in apolipoprotein E-knockout mice. J. Pharmacol. Exp. Ther [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, et al. (1997). Lack of apolipoprotein E dramatically reduces amyloid β-peptide deposition. Nat. Genet [DOI] [PubMed] [Google Scholar]
- Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, Delong CA, Piccardo P, Petegnief V, et al. (1999). Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson KK, Hu W, Weller AH, Bennett AH, Chen ER, Khetarpal SA, Yoshino S, Bone WP, Wang L, Rabinowitz JD, et al. (2019). Natural human genetic variation determines basal and inducible expression of PM20D1, an obesity-associated gene. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, and Dennis EA (2009). Phospholipase A 2 structure/function, mechanism, and signaling. J. Lipid Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, and Hotamisligil GS (2008). Identification of a Lipokine, a Lipid Hormone Linking Adipose Tissue to Systemic Metabolism. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Nakazawa T, Yui K, Kaneko E, and Shimokado K (2003). VLDL induces adipocyte differentiation in ApoE-dependent manner. Arterioscler. Thromb. Vasc. Biol [DOI] [PubMed] [Google Scholar]
- Cooke AL, Morris J, Melchior JT, Street SE, Gray Jerome W, Huang R, Herr AB, Smith LE, Segrest JP, Remaley AT, et al. (2018). A thumbwheel mechanism for APOA1 activation of LCAT activity in HDL. J. Lipid Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins R (1990). Measurement of free hormones in blood. Endocr. Rev [DOI] [PubMed] [Google Scholar]
- Ganor RS, Harats D, Schiby G, Rosenblatt K, Lubitz I, Shaish A, and Salomon O (2018). Elderly apolipoprotein E−/− mice with advanced atherosclerotic lesions in the aorta do not develop Alzheimer’s disease-like pathologies. Mol. Med. Rep [DOI] [PubMed] [Google Scholar]
- Gao J, Katagiri H, Ishigaki Y, Yamada T, Ogihara T, Imai J, Uno K, Hasegawa Y, Kanzaki M, Yamamoto TT, et al. (2007). Involvement of apolipoprotein E in excess fat accumulation and insulin resistance. Diabetes [DOI] [PubMed] [Google Scholar]
- Goldberg IJ, Scheraldi CA, Yacoub LK, Saxena U, and Bisgaier CL (1990). Lipoprotein apoC-II activation of lipoprotein lipase. Modulation by apolipoprotein A-IV. J. Biol. Chem [PubMed] [Google Scholar]
- Goldman AL, Bhasin S, Wu FCW, Krishna M, Matsumoto AM, and Jasuja R (2017). A reappraisal of testosterone’s binding in circulation: Physiological and clinical implications. Endocr. Rev [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed TJ, Trammell SAJ, McKinney MK, Petersen N, Cardone RL, Svenningsen JS, Ogasawara D, Nexøe-Larsen CC, Knop FK, Schwartz TW, et al. (2019). N -acyl taurines are endogenous lipid messengers that improve glucose homeostasis. Proc. Natl. Acad. Sci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb SC, Bult CJ, and Bogue MA (2014). Mouse Phenome Database. Nucleic Acids Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GL (2016). Plasma steroid-binding proteins: Primary gatekeepers of steroid hormone action. J. Endocrinol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keipert S, Kutschke M, Ost M, Schwarzmayr T, van Schothorst EM, Lamp D, Brachthäuser L, Hamp I, Mazibuko SE, Hartwig S, et al. (2017). Long-Term Cold Adaptation Does Not Require FGF21 or UCP1. Cell Metab [DOI] [PubMed] [Google Scholar]
- Kim JT, Li VL, Terrell SM, Fischer CR, and Long JZ (2019). Family-wide Annotation of Enzymatic Pathways by Parallel In Vivo Metabolomics. Cell Chem. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JT, Terrell SM, Li VL, Wei W, Fischer CR, and Long JZ (2020). Cooperative Enzymatic Control of N-acyl Amino Acids by PM20D1 and FAAH. Elife 9, e55211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiria LO, Wang CH, Lynes MD, Yang K, Shamsi F, Sato M, Sugimoto S, Chen EY, Bussberg V, Narain NR, et al. (2019). 12-Lipoxygenase Regulates Cold Adaptation and Glucose Metabolism by Producing the Omega-3 Lipid 12-HEPE from Brown Fat. Cell Metab [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Long JZ, Roche AM, Svensson KJ, Dou FY, Chang MR, Strutzenberg T, Ruiz C, Cameron MD, Novick SJ, et al. (2018). Discovery of Hydrolysis-Resistant Isoindoline N-Acyl Amino Acid Analogues that Stimulate Mitochondrial Respiration. J. Med. Chem 61, 3224–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. (2013). A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Bateman LA, Lin H, Kamenecka T, Lokurkar IA, Lou J, Rao RR, Chang MR, Jedrychowski MP, et al. (2016). The Secreted Enzyme PM20D1 Regulates Lipidated Amino Acid Uncouplers of Mitochondria. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Roche AM, Berdan CA, Louie SM, Roberts AJ, Svensson KJ, Dou FY, Bateman LA, Mina AI, Deng Z, et al. (2018). Ablation of PM20D1 reveals N -acyl amino acid control of metabolism and nociception. Proc. Natl. Acad. Sci 115, 201803389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynes MD, Leiria LO, Lundh M, Bartelt A, Shamsi F, Huang TL, Takahashi H, Hirshman MF, Schlein C, Lee A, et al. (2017). The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nat. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthei KA, Yang SM, Baljinnyam B, Chang L, Glukhova A, Yuan W, Freeman LA, Maloney DJ, Schwendeman A, Remaley AT, et al. (2018). Molecular basis for activation of lecithin: Cholesterol acyltransferase by a compound that increases HDL cholesterol. Elife [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, and Mann M (2002). Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics [DOI] [PubMed] [Google Scholar]
- Petitpas I, Petersen CE, Ha CE, Bhattacharya AA, Zunszain PA, Ghuman J, Bhagavan NV, and Curry S (2003). Structural basis of albumin-thyroxine interactions and familial dysalbuminemic hyperthyroxinemia. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump AS, Smith JD, Hayek T, Aalto-Setälä K, Walsh A, Verstuyft JG, Rubin EM, and Breslow JL (1992). Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell 71, 343–353. [DOI] [PubMed] [Google Scholar]
- Roberts LD, Bostrom P, O’Sullivan JF, Schinzel RT, Lewis GD, Dejam A, Lee YK, Palma MJ, Calhoun S, Georgiadi A, et al. (2014). beta-Aminoisobutyric Acid Induces Browning of White Fat and Hepatic beta-Oxidation and Is Inversely Correlated with Cardiometabolic Risk Factors. Cell Metab 19, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Heyn H, Silva BA, Dixsaut L, Garcia-Esparcia P, Vidal E, Sayols S, Glauser L, Monteagudo-Sánchez A, Perez-Tur J, et al. (2018). PM20D1 is a quantitative trait locus associated with Alzheimer’s disease. Nat. Med [DOI] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Glauser L, Monk D, and Gräff J (2020). Comprehensive analysis of PM20D1 QTL in Alzheimer’s disease. Clin. Epigenetics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, et al. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. (2015). UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, et al. (2014). Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi HH, Reardon CA, and Mazzone T (2006). Endogenous ApoE expression modulates adipocyte triglyceride content and turnover. Diabetes [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Quantitative proteomics of PM20D1-flag immunoaffinity purification from conditioned media of iWAT cells. Related to Figure 1.
Table S2. Quantitative proteomics of PM20D1-flag immunoaffinity purification from conditioned media of BAT cells. Related to Figure. 1.
Table S3. Shotgun proteomics of wild-type mouse plasma FPLC fractions 10–12 (pooled). Related to Figure 3.
Table S4. Shotgun proteomics of APOE-KO FPLC fractions 10–12 (pooled). Related to Figure 3.
Data Availability Statement
This study did not generate/analyze any datasets/code.


