Abstract
Methodological improvements in both single particle cryo-electron microscopy (cryo-EM) and hydrogen/deuterium exchange mass spectrometry (HDX-MS) mean that the two methods are being more frequently used together to tackle complex problems in structural biology. There are many benefits to this combination, including for the analysis of low-resolution density, for structural validation, in the analysis of individual proteins versus the same proteins in large complexes, studies of allostery, protein quality control during cryo-EM construct optimization, and in the study of protein movements/dynamics during function. As will be highlighted in this review, through careful considerations of potential sample and conformational heterogeneity, many joint studies have recently been demonstrated, and many future studies using this combination are anticipated.
Keywords: protein conformation, cryo-EM, HDX-MS, structural heterogeneity
Combine and conquer
Electron microscopy (EM) and mass spectrometry (MS) are two methods that have become increasing prevalent over the last 10–20 years, as more information can be obtained with less material and in much less time. More specifically, single particle cryo-EM and hydrogen/deuterium exchange MS (HDX-MS), each, in their own way, have become transformative in structural biology. The subject of this review is the combination and complementarity of these two methods, with slightly more emphasis on HDX-MS. The combination of the two techniques is not meant to supplant one or the other technique, but rather to support each other, i.e., what does one learn new about structure if there is both cryo-EM and HDX-MS data?
The more structural techniques one can combine, the more comprehensive a picture can be obtained. A short list of methods with medium to high resolving power that might be combined includes X-ray crystallography, NMR, EM (both negative stain EM and cryo-EM), small angle x-ray scattering (SAXS), HDX-MS, molecular modeling, molecular dynamics simulations, and crosslinking MS. Each of these methods has strengths and weakness; for example, cryo-EM, negative stain EM, and SAXS provide information about the overall shape of a molecule, while HDX-MS cannot conclude much of anything about overall shape, though it is very sensitive to conformational changes. Therefore, the combination of cryo-EM and HDX-MS becomes especially valuable, and sometimes essential, in the analysis of complicated and large complexes, for protein machines in motion, when membranes and membrane mimetics become part of structural analysis, and for resolved protein folding studies.
HDX-MS (reviewed in [1–4]) provides information about substitution of deuterium for backbone amide hydrogens. Most studies measure the amount and rate of deuteration, factors influenced by all aspects of protein structure and solvent accessibility. Often, studies compare the exchange between various conformational states (e.g., free vs. ligand-bound) in order to determine what parts of a structure change as a result of some variable (e.g., binding, PTMs, solvent conditions, pH, etc.). The resolution of the exchange data is generally determined by the short peptides that average 10–15 residues in length and are formed after exchange is quenched and proteolytically digested. Single-particle cryo-EM (reviewed in [5–7]) provides a 3-dimensional picture of macromolecules and complexes by averaging single particle images collected from transmission electron microscopy at cryogenic temperatures. Images of many thousands of particles trapped in random orientations within vitreous ice are sorted, classified, aligned, and averaged to compute a 3-dimensional shape reconstruction for model refinement. Reconstruction of a model often involves the integration of data from several different experimental approaches: structures of individual domains or proteins obtained from x-ray crystallography or NMR, the location of proximal sites from cross-linking MS, and molecular modeling (Box 1 and examples below). The final refined model(s), especially in the case of some large and complex protein machines, can be fit with crystal structures of individual components or, as in the case of some modern cryo-EM, determined directly without the need for crystal structure data [8].
Box 1 – Synergy of methods.
Combinations of structural methods can be very useful for efficiently assembling larger protein complexes and for validation of the structural juxtaposition of different components. The generation of high-resolution X-ray structures of individual domains and/or proteins are essential to build-up structures from medium resolution cryo-EM data (in the 4–7 Å range). This is especially important when mapping structures into areas of limited density.
It is increasingly common for protein cross-linking to be used to ensure the integrity of protein complexes during the cryo-EM process, and such procedures lead naturally into the synergistic use of cryo-EM and crosslinking MS. An excellent review [47] of this approach is highly recommended.
Finally, the addition of molecular modeling approaches can be very useful for obtaining structures of larger complexes from any combination of cryo-EM, HDX-MS and additional approaches. Homology modeling can be very useful when a structure of a related protein is known, and the homology model can be validated by HDX-MS both alone and in the larger complex. Molecular docking can also be used in combination with HDX-MS interface data and structural data to validate molecular interfaces.
Why combinations are increasing in last few years
A revolution has happened in the fields of both cryo-EM and HDX-MS in the last decade, as described in Figure 1. Due to technological advances, the quality of data and the ease of measurement (including for novices) has improved for both techniques, meaning the types/sizes of molecules that can be studied has also changed. In the case of cryo-EM, overall resolution has generally improved, and smaller and smaller proteins can now be studied. In contrast, for HDX-MS, larger and larger proteins, or even complex protein systems, can now be studied (Figure 1). This convergence has meant that things that used to be out of the range of HDX-MS and only accessible by cryo-EM are now accessible by both techniques, and vice versa; the two methods can now “meet in the middle”.
Figure 1.

HDX-MS goes larger while cryo-EM goes smaller. Improvements to both cryo-EM and HDX-MS have expanded the range of proteins and proteins systems that can be studied. The size and complexity of systems that HDX-MS can access, historically limited to small to medium sized entities (i.e. in the range 5 to 100 kDa), has increased over time (bottom). In contrast, the size of proteins that cryo-EM can access, historically limited to large protein complexes and systems (i.e. larger than ~500 kDa), has gotten smaller over time (top). The number near each structure is the molecular weight (in kilodaltons) of the structure shown. Left-to-right: ubiquitinconjugating enzyme UbcH5b (PDB ID: 2ESK), glycogen phosphorylase b (PDB ID: 1GPB), ClpX-ClpP complex (PDB ID: 6POS), Plasmodium falciparum 20S proteasome in complex with activators (PDB ID: 6MUV).
The main drivers for improvements in both methods have been hardware and software. In cryo-EM, direct electron-detection cameras [9] have vastly improved the possible resolution, while improved computational power and imaging processing software make particle picking, classification, averaging, and model building faster and better [10–12]. Automation in cryo-EM has also occurred, contributing, inter alia, to overall better data quality and allowing non-experts to obtain high quality data. Improvements in software and automation have also occurred in HDX-MS but perhaps the most important HDX-MS development has been better overall peak capacity. For HDX-MS, peak capacity during the chromatography and mass spectrometry steps has been historically limiting because in order to retain as much deuterium label as possible, chromatography must be performed at 0 °C in a short time. Such analysis conditions are not conducive to highly efficient separations, meaning that mixtures of hundreds of peptides could not generally be studied. Improvements, including ultra-performance liquid chromatography (UPLC) [13] and ion mobility spectrometry [14], have vastly improved HDX-MS peak capacity at the restrictive HDX quench conditions.
In all of this, however, is the important consideration of unique sequence (UqSeq) and multimeric protein assemblies (Figure 2A). As examples, HDX-MS of 500 kDa of protein is not especially challenging when that 500 kDa is composed of 10 identical copies of a 450 residue monomer (Figure 2A, Case 1); what the mass spectrometer “sees” is 10x of only 450 amino acids of UqSeq. In contrast, in a 10-member complex where each monomer has a unique sequence (Figure 2A, Case 2), 500 kDa of protein also enters the mass spectrometer but because the sequences of each monomer are not identical as in Case 1, the mass spectrometer sees 4,500 amino acids of UqSeq. Such high UqSeq analysis is much more complicated in terms of the LC separation and data analysis. Low UqSeq analyses by HDX-MS have been possible for many years, whereas what is new in recent years is high UqSeq studies.
Figure 2.
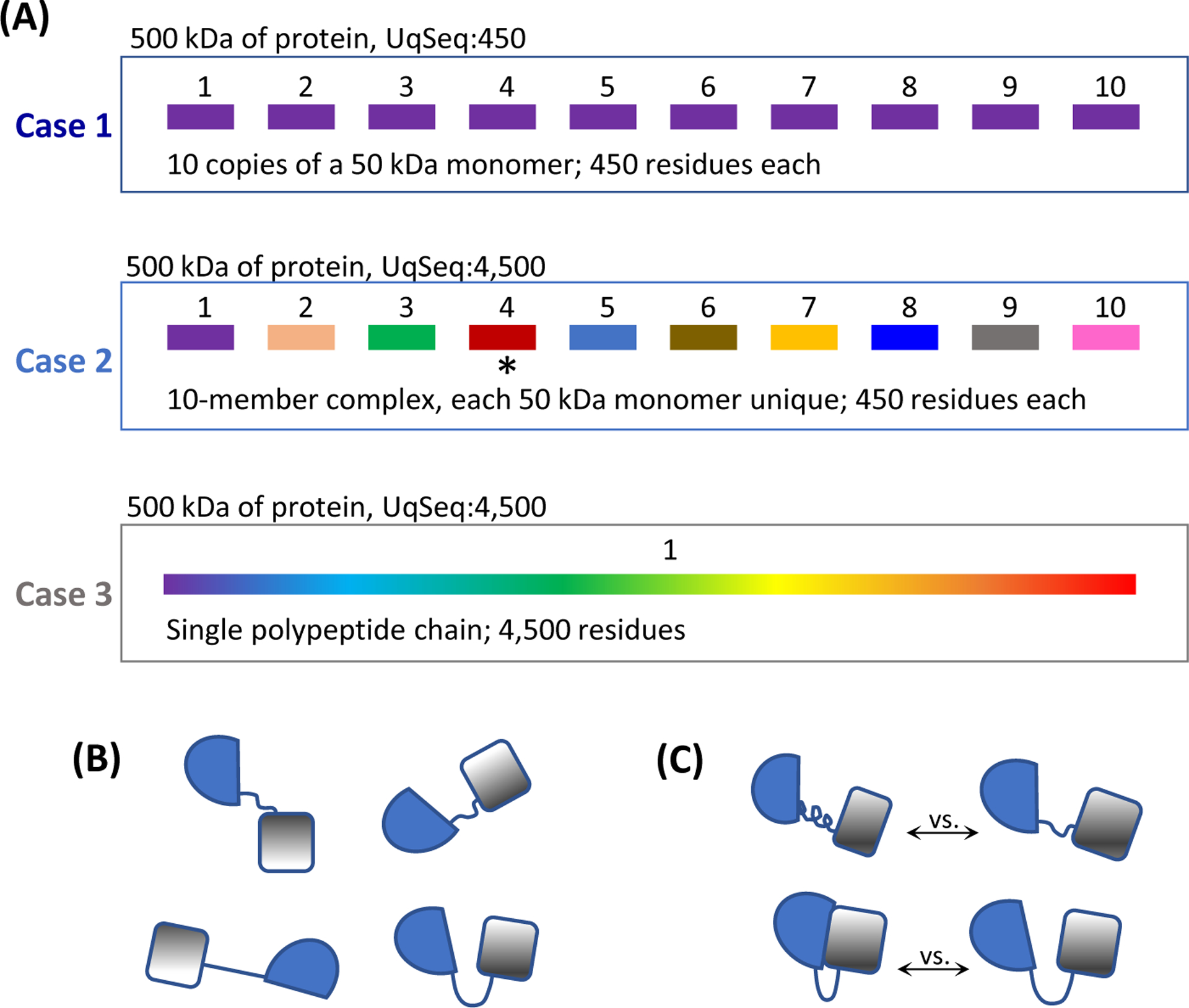
Considerations in HDX-MS and cryo-EM measurements. (A). The concept of unique sequence (UqSeq) in terms of what a mass spectrometer “sees”. Each of these three cases involves a total of 500 kDa of protein. In case 1 (top), there are 10 copies of the identical 450-residue monomer (50 kDa) so the mass spectrometer sees 10x of 450 residues because upon digestion, the same peptides will be produced from each of the ten monomers. In case 2 (middle), there are 10 monomers, each with different sequence; the mass spectrometer sees 1x of 4,500 residues upon digestion because different peptides will be produced from each different monomer. Case 3 (bottom) is similar to case 2, except there is one monomer of 4,500 residues. (B). Four different potential orientations of an example 2-domain protein. Each different arrangement could be observed and differentiated by cryo-EM but HDX-MS could likely not tell these apart as backbone amide exchange is the same in each domain orientation. (C). Two examples where HDX-MS could tell different domain orientations apart, including (top) rearrangement of the linker between domains to change structure/HDX of the linker and (bottom) creation of a new interface between domains that would likely reduce HDX at the interface.
Viruses are classic examples, studied by both cryo-EM and HDX-MS (e.g. [15–17]), of large structures with low UqSeq where many copies of the identical monomer exist. More recently, analysis of HDX-MS results in combination with cryo-EM structures of intact virus particles revealed conformational changes in dengue virus monomers versus whole virus particles at various temperatures relevant to the virus life cycle [18], and also probed turnip crinkle virus particle breathing [19]. While these systems are low UqSeq (for example, turnip crinkle virus particles are composed of 80 copies of the same 38 kDa monomer with identical UqSeq), the HDX-MS data reflect the ensemble average of all these monomers in the virus particle, and therefore HDX in distinct monomers, or even in classes of monomers (e.g. all those at an interface), cannot be determined. Both HDX-MS and cryo-EM have the issue of heterogeneity to contend with, and do so in different ways, as discussed below.
In the following sections, the convergence of cryo-EM and HDX-MS around basic questions will be discussed. The questions and types of analyses covered include: 1) is the conformation in solution as assessed by HDX-MS consistent with structural modeling and/or changes observed in the basic shape(s) from cryo-EM; 2) what are the movements/dynamics of proteins in solution versus the overall shape of the protein or protein complex; 3) is there conformational heterogeneity in solution or in multiple species interconverting over a given time-frame; and 4) how does structure change during protein folding? In many of these situations, the more UqSeq that can be followed by HDX-MS, the larger the complex that can be studied and perhaps even the identical protein preparation used for both cryo-EM and HDX-MS.
Are solution data consistent with structural observations and modeling?
One of the most straightforward combinations of cryo-EM and HDX-MS is to understand if what is seen in solution (by HDX-MS) is consistent with the structural determination/modeling (by EM), and vice versa. The combination of the two techniques can also be used to provide information on movements within the complex and to fill in “missing data”, or that which is currently unobtainable by the respective technique, as well as characterize how complexes are assembled and provide insights into their mechanisms. Some aspects of these combinations and their interplay are described next, with recent examples.
Structure validation.
Multiple studies have combined HDX-MS and EM (both negative stain EM and cryo-EM) to determine if the HDX-MS information for a protein was consistent with results from EM. Although more can generally be gained with cryo-EM, sometimes negative stain EM can prove valuable. One example is a study [20] of rubisco activase interacting with rubisco where conclusions about the positioning of rubisco activase on the rubisco multimer from negative stain EM supported HDX-MS data showing that the C-terminal region of the rubisco large subunit was moved by rubisco activase to expose areas near the C-terminus in tertiary structure. In a second example, Manthei et al. [21] measured HDX-MS protection of lecithin:cholesterol acyltransferase and high-density lipoproteins and found the results consistent with a negative stain EM model (and crosslinking MS data) of the complex, including the way in which crystallographic data were placed on the low-resolution negative stain model (see also Box 1). In a last example [22], negative stain EM and HDX-MS showed no changes in shape or deuteration, respectively, after furin-cleavage between the prodomain and growth factor region of growth differentiation factor 8 (GDF8), in the so-called latent form. However, EM heterogeneity and increased HDX in certain regions were seen in GDF8 after Tolloid cleavage of the prodomain that produces the so-called primed form, thereby supporting a model that involved conformational changes upon Tolloid cleavage but not upon furin cleavage.
Studies that combined HDX-MS and cryo-EM for structural validation are more numerous. As examples, both [23] and [24] used HDX-MS data to validate their cryo-EM structure. Further, Ye et al. [25] performed HDX-MS and concluded it showed good agreement with cryo-EM data, particularly in the placement of secondary structural elements from X-ray structures. Bardiaux et al. [26] used cryo-EM, NMR, HDX-MS, and molecular modeling to construct a structural model of the pilus in the type-2 secretion system or the type IV pilus. In their final model, the secondary structure and interfaces between monomers in the 30-monomer pilus assembly determined by HDX-MS was consistent with data from structural and modeling approaches; notably, HDX-MS and cryo-EM agreed in the positioning of a central helix in the center of the fiber. Again, combinations of methods – of which HDX-MS and cryo-EM are frequent major players – are often needed to validate structures, particularly of difficult systems (see also Box 1).
Quality control for altered forms.
HDX-MS can be used to check that changes made to protein(s) to help improve or make cryo-EM studies possible do not alter the protein structure in some critical way. Examples of modifications that could be used to improve cryo-EM suitability and quality include adding disulfide bonds, making mutants, adding tags for stabilization, or binding to antibodies (Fabs) to stabilize a particular protein. In an example by Zhang et al. [27], the quality of cryo-EM and SAXS both was improved by stabilizing insulin degrading enzyme (IDE) with Fab binding. HDX-MS demonstrated that Fab binding did not alter the conformation of IDE itself and therefore that Fab binding did not introduce non-biological artifacts into the structure. Upon binding to insulin, reduced HDX was observed in regions with higher B-factors by cryo-EM. These changes were consistent with a functional model for catalysis wherein substrate stabilizes the IDE catalytic domain.
Stoichiometry and domain movement.
Synergy between the two methods opens new information that one method cannot obtain on its own. While cryo-EM reveals domain orientation and stoichiometry, HDX-MS generally is not able to provide such information. In each of the previous examples, EM was essential for obtaining the stoichiometry of the overall complex, whereas HDX-MS provided no stoichiometry information. In the example of the 80 copies of turnip crinkle virus monomer [19], HDX-MS sees this as one protein, albeit a concentration 80-fold higher than if there were the same number of moles of a monomeric version. HDX-MS also cannot reveal gross movements that do not change the backbone amide hydrogen environment (such as domain movements as shown in Figure 2B), but it is sensitive to differences in structural forms where backbone amide hydrogen exchange could be perturbed, say in structural remodeling or the formation of a new interface (Figure 2C). Therefore, unlike cryo-EM, HDX-MS could prove useful in narrowing of small conformational changes to short regions that could be missed in cryo-EM, and could also provide data on flexibility and flexible regions.
Fuzzy EM density.
Though cryo-EM can reveal domain orientation, highly flexible regions may be difficult to deal with and can result in unresolved or low-resolution density in the final models. In these situations, HDX-MS can often provide some clues about these regions: are they low resolution because there are large domain movements but the overall smaller secondary structural elements are folded (meaning HDX is slow), or is the whole region exchanging fast implying that there is no organized secondary structure and/or a highly flexible tertiary fold, or is there too much conformational heterogeneity (see section below) to produce a high quality density map?
For example, Cash et al. [28] observed that regions with low-resolution density in cryo-EM appeared to have stable structures by HDX-MS and the regions could therefore be mapped to helices of certain regions of the protein missing from the rest of the model. In another example (there are many), Twomey et al. [29] found no density for the UT3 domain of Ufd1, but HDXMS data for the domain indicated a folded structure, and while density was seen for the UT6 domain, the resolution was too low for modeling and the loops connecting various domains were not visible. The HDX-MS data indicated that the loops were generally highly solvent exposed, supporting the idea that the domains were connected by flexible linkers and providing an explanation for why the EM density may have been of low resolution. Changes in the HDX-MS of loops and unstructured regions, generally areas of low cryo-EM density, could be related to protection or formation of structure. Such an application is central to HDX-MS studies of protein folding, as described in a section below.
Missing states.
A recent HDX-MS study [30] of the multi-drug transporter P-glycoprotein gave results that appeared to contradict the cryo-EM structure. During transport, this membrane-embedded transporter binds to two ATP molecules, the nucleotide binding domains dimerize in a head-to-tail arrangement (occluding the binding pocket from the intracellular environment), and ATP hydrolysis results in a post-transport outward-facing conformation which was observed by cryo-EM [31]. The HDX-MS, however, revealed much higher exchange in the extracellular loops than could be rationalized by the cryo-EM structure [30]. Rather, HDX-MS of an intermediate “occluded” structure, in which both the extracellular and intracellular portions of the protein were closed, suggested that the cryo-EM structure reflected this occluded structure rather than the outward-facing structure.
Whole and the sum of the parts.
By measuring HDX-MS for an individual protein in both isolation and in an assembled complex, and then interpreting the data for the complex assembly in light of the cryo-EM structure of that larger assembly, one can learn how the assembly is put together. An example is the recent structure of the ASB9-Cullin 5 E3 ligase, a complex of five proteins that binds the dimeric substrate, creatine kinase (CK). Structures of ASB9 with adapters, EloB/C, and of two pieces of Cul5 were available from x-ray crystallography. Cryo-EM was used to determine the structure of the CK-bound ASB9-EloB/C and of the full-length Cul5. HDX-MS was then used to determine how these two pieces assemble in the final ligase and how the ring-box protein is bound to the Cul5 [32]. Conformational changes might also occur during complex assembly. This concept is also well illustrated by two examples with the Nef protein of HIV-1, which interacts with subunits of AP-1 or AP-2 complexes at the membrane. In one study [33], HDX-MS monitored how deuteration of HIV-1 Nef alone compared to deuteration when Nef was part of larger complexes, including the C-terminal domain of AP-1 subunit μ1 and either tetherin or MHC-I. Flexibility differences were interpreted via a cryo-EM structure of the AP-1:Arf1:tetherin:Nef complex to understand cargo preferences for tetherin or MHC-I. The same group later studied SIV Nef in AP-2 complexes [34] where additional parts of Nef were seen protected from HDX in the entire assembled complex relative to Nef alone. In the larger assembled complex, reduced exchange was found within a particular helix of the AP-2 β2 subunit in the presence of both simian tetherin and SIV Nef. These data suggested refolding of this region of AP-2 in the larger complex, a hypothesis also supported by mutagenesis studies in the same report. Consistent with both the HDX-MS and mutagenesis, and first seen in their cryo-EM structure, a helix in the β2 subunit of the AP-2 complex becomes remodeled in the presence of binding partners tetherin or HIV Nef.
Allostery.
Allosteric effects can be revealed by analysis of both HDX-MS and the cryo-EM structure, but not necessarily with just the cryo-EM structure alone. In a study of the transcription initiation factor σS [35], σS was analyzed by HDX-MS when alone and then when bound to its activator protein Crl. HDX data for most parts of σS alone were consistent with a cryo-EM structure (a complex of σ2-RNA polymerase holoenzyme, Crl, and nucleic acid scaffold) except for one part (σ3.1) which was rapidly deuterated in σS alone but appeared well folded in the larger complex analyzed by cryo-EM. When free σS was bound to the activator Crl, there was protection from HDX at both the σS:Crl interface defined by cryo-EM, and at regions far from the interface seen in cryo-EM structure. These results led to the hypothesis of binding-induced allosteric structural changes that were tied to the function and activation mechanism. Long-range allostery was also revealed in the study by Cash et al. [28]. HDX-MS data were found consistent with previous crystallography and cryo-EM results for the protein P-Rex1, both when P-Rex1 was alone and when in a complex with G-proteins (P-Rex1-Gβγ complex). During complex formation, changes in HDX were found distant from the Gβγ-binding site revealed by the P-Rex1-Gβγ cryo-EM structure.
Movement and dynamics in function.
The suggestion by Richard Feynman in studying biological systems to “just look at the thing” [36] is addressed by cryo-EM; however, Jeremy Knowles’ observation that “studying the photograph of a racehorse cannot tell you how fast it can run” [37] is also a valid point and remains an issue. A well-known solution to the “racehorse problem” is that frames of a movie are obtained so there is both a picture and an indication of how fast it could run. The combination of cryo-EM and HDX-MS can provide some motion picture information (although perhaps not the entire film) on a molecular level, as pointed out by [25] and illustrated with their study and several others (e.g. Refs. [29, 38, 39]). To access the mechanism, or what the protein(s) is/are doing during function, HDX-MS and cryo-EM can be used to study structural changes in each of perhaps multiple functional forms. For example, Faull et al. [38] illustrate this point in their analyses of CSN, a 331 kDa regulator complex, when bound to the CRL2 E3 ligase. HDX-MS was used to understand stepwise activation and to watch changes to exchange during assembly. Then, they used PLIMSTEX [40], a type of HDX which can measure intramolecular affinity specific to particular subunits, a difficult task, to follow how particular subunits may interact. The HDX-MS results were compared to and found to be consistent with a mechanism hinted at by the assembled complex seen in cryo-EM. Taken with other biochemical data, HDX-MS and cryo-EM helped build a hypothesis of how the assembly of the complex contributes to its function.
Heterogeneity
What has been alluded to in some of the previous applications and examples, but not yet addressed, is heterogeneity. Proteins exist as populations of molecules and any population may present heterogeneity, perhaps introducing a major obstacle to analysis for both cryo-EM and HDX-MS. Some considerations regarding heterogeneity are shown in Box 2, and a few of the many examples of how heterogeneity plays a role in cryo-EM and HDX-MS are discussed in the following sections.
Box 2 – Issues in heterogeneity.
While cryo-EM is a static picture, a freeze-frame at the moment the sample was frozen, multiple conformations can exist at the moment of freezing and this can be captured; particle selection and sorting of the images helps classify what existed in the snapshot. HDX-MS is a solution technique which typically captures a time-course of labeling over at least 3–4 orders of time magnitude; proteins are able to change conformation in solution during labeling and this flux can sometimes be detected. The information provided by both methods about conformational heterogeneity is important. The way in which data are taken and the population of molecules being sampled are both unique for cryo-EM and HDXMS, influencing the results and interpretations. When interpreted together, many different issues and questions concerning conformational heterogeneity, can be addressed.
Understanding when conformational heterogeneity is real and when it is an experimental artifact is not straightforward, nor is interpreting the data when it reports overlapping results for coexisting multiple conformations. How many different structural forms coexist and how can this be known? Have the “right” particles been picked, is the classification optimized? Has particle selection favored one conformation and, appropriately or inappropriately, discarded data for another? If samples are not handled properly in HDX-MS, artifactual or “false” isotope pattern signatures for multiple populations [48] can be introduced and may be confused with real population distributions. Can overlapping populations in HDX-MS be deconvoluted? Might it be better to force a single state dominate the population and thereby reduce the heterogeneity? If so, how could this be done, would it be a functional state?
Compositional heterogeneity can also confound both cryo-EM and HDX-MS analyses. Issues such as heterogeneous post-translational modifications and sub-stoichiometric binding of some components of a complex can be misinterpreted. Quantitative mass spectrometry can be extremely helpful in resolving both of these issues, and it is recommended prior to all structural analyses. The quality and homogeneity of samples is a major bottleneck in cryo-EM analyses. Optimization of sample preparation including large-scale buffer screening, DSF and multi-angle light scattering coupled to size exclusion chromatography (SEC-MALS) can greatly facilitate successful analyses both by cryo-EM and by HDX-MS. While HDX-MS is not ideal as a high-throughput screening tool, once conditions are sufficiently narrowed by other techniques, HDXMS can become very helpful. In fact, screening of some sample conditions by HDX-MS prior to structure determination is becoming widely utilized by x-ray crystallographers and hopefully will also be useful for cryo-EM.
Compositional heterogeneity
Heterogeneity in primary structure (sequence), tertiary structure, and quaternary assembly can all present issues for cryo-EM and HDX-MS, but in different and unique ways. Quaternary assembly is one issue that must be carefully considered, especially for HDX-MS data interpretation; luckily, cryo-EM is ideally suited to reveal domain orientation and stoichiometry, thereby aiding HDX-MS. In a highly symmetrical virus particle, for example, all monomeric units may have equal surrounding environments meaning equal deuteration of each monomer. Other homo- and hetero-oligomeric assemblies can present differently. Figure 3 provides several specific examples of mixed multimers. In Figure 3A, two dimer arrangements of a heterotetramer (α2β2) are shown, either (case 1) α2β2β2α2 or case 2 (α2β2α2β2). The environments around each α and β subunit in these simple examples are different and would certainly result in different HDX-MS data at the peptide-level, and perhaps even unique cryo-EM structures. Because parts of proteins that are in different environments would incorporate deuterium differently (e.g., in two identical-sequence monomers with different binding interfaces), multiple populations would be apparent in the mass spectra (Figure 4A) at both the intact protein and peptide-level.
Figure 3.
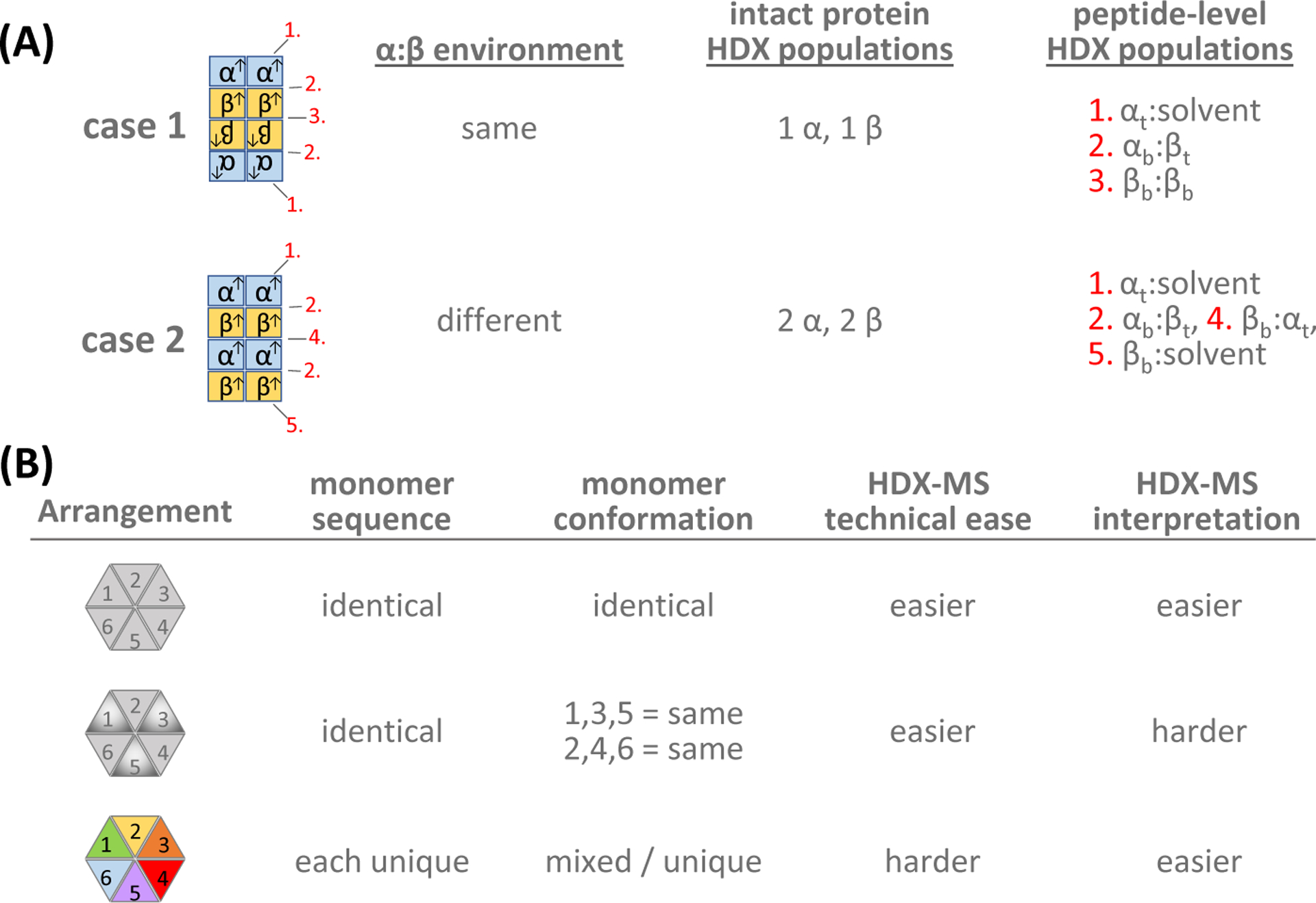
Examples of types of hetero-oligomers and their properties in HDX-MS. (A). Two cases of the dimerization of a heterotetramer (α2β2) where the arrangement is toe-to-toe (case 1, α2β2β2α2) or head-to-toe (case 2, α2β2α2β2). The environment of each α and β monomer is not the same in case 1 vs. case 2. All α monomers would have the same deuteration in case 1, as would all β monomers, where as in case 2 α and β monomers exposed to solvent would exchange differently from those protected on either side by another monomer . Peptide-level exchange experiments could distinguish between parts of α or β at several different monomer:monomer interfaces (2,3,4) or monomer:solvent interfaces (1,5) at the top (as drawn here) of each monomer (i.e., αt or βt) or bottom of each monomer (i.e., αb or βb). (B). Three potential arrangements of hexameric ATPases where (top) all monomers have identical sequence and conformation, (middle) where all monomers have identical sequence but 3 sample one conformation and 3 sample another, or (bottom) where all monomers have different sequences and perhaps each monomer has a different conformation. Perhaps counterintuitively, because technical hurdles of UqSeq complexity can likely be overcome, cases with unique sequences of each monomer (bottom) may lead to the best final conclusions because the interpretation is specific to each monomer.
Figure 4.

Examples of theoretical mass spectra during peptide-level HDX-MS of heterogeneous samples. (A). The spectra themselves can reveal how many populations co-exist at any one labeling time point. Signatures for 1, 2, 3, or even >3 co-existing populations are obvious in well-resolved isotope clusters; isotope clusters are not always well-resolved in the m/z dimension as shown in this theoretical data and can present as wide isotope distributions (bottom example >3), which do not always indicate how many populations may co-exist. Additionally, not all peptides from a protein may show more than one population, revealing which parts of a protein are conformationally heterogeneous in solution. Experimental artifacts can also present “false” populations [48]. (B). Example spectra frequently encountered during protein folding experiments. The folding time is shown next to each example spectrum. In classic pulsed labeling [46], the most unfolded species (example fold, green) is least protected and incorporates the most deuterium label during a short pulse of D2O, while the most folded species (example fold, purple) is most protected from deuteration. Many 2-state folders present only two species (purple and green) whose relative intensity changes during progression of the folding reaction; multi-state folders can present signatures for folding intermediate(s) (example fold, blue).
A 2019 paper by Brunle et al. [41] described a study of a molybdenum storage protein where cryo-EM and HDX-MS were applied to understand the quaternary assembly of a heterododecamer where a dimer of two (αβ)3 hexamers was arranged upside down on each other (β3α3α3β3). There were three α subunits at the dimer interface and three β subunits not at the interface. Both α and β were unique sequences of ~275 residues each, for a total of 550 UqSeq. A 3.2 Å resolution cryo-EM structure helped explain the quaternary structure and stoichiometry, while HDX-MS of various states revealed that there were functional changes in conformation only at the ATP-binding sites of the β subunits, and no changes at all in the α subunits. The results contributed to the model of this large assembly in which the β subunit was mostly responsible for pumping molybdenum whereas α subunits were passive and held the complex together.
Conformational heterogeneity
Further, studies of multimeric ATPases not only provide some excellent examples of the combination of cryo-EM and HDX-MS, they also illustrate homo- versus hetero-oligomeric considerations (Figure 3B), and the conformational heterogeneity that can exist depending on functional state. In the recent ATPase studies, there are observations of homo-oligomers (Saccharomyces cerevisiae Hsp104 which has six subunits with identical sequence [25], S. cerevisiae Cdc48 with six identical-sequence subunits [29]) and hetero-oligomers (S. cerevisiae Rpt1–6 in the19S base which has six different-sequence subunits [42], Bos taurus TriC with eight different-sequence subunits [39]). These papers demonstrate trapping or enrichment of select conformational states with binding to small molecules as a way of reducing the conformational heterogeneity in both cryo-EM and HDX-MS. This strategy is not always possible, and sometimes it is not known for all types of proteins that multiple functional and conformational states exist or how to trap them (see also Box 2). In the S .cerevisiae Hsp104 study [25], assembled hexamer was compared to monomer, showing some key HDX differences of particular peptides involved when the hexamer is assembled and when the ATPase is functioning. Distinct HDX-MS signals for two populations (see example in Figure 4A) supported the cryo-EM of Hsp104 +ADP or +AMPPNP, where one monomer was in one conformation and the other five monomers in another conformation. While the signals for each individual monomer were not resolvable (the sequence is identical for all monomers, see Figure 2A, case 1, and Figure 3B, top) the peak intensity ratio 1:5 was clear: one monomer being deuterated in one way and the other five in another way. In the presence of ATP, or ATPγS, this 1:5 distribution disappeared, consistent with the protein becoming symmetrical and flat as shown by the cryo-EM, and the population of molecules all synchronizing to one conformation. While the study of S. cerevisiae Cdc48 [29] measured differences in HDX-MS deuteration levels for cdc48 +ADP-BeF4 or +ADP, there is likely much more to the story because spectral heterogeneity information (i.e. population distributions) was not yet extracted from the dataset (see reprocessing recommendations below). Additionally, Balchin et al. [39] reported cryo-EM and HDX-MS of actin in the presence of TriC (hetero-oligomer, eight subunits) and GroEL (homo-oligomer of two 7-subunit rings). As illustrated in Figure 3B, HDX-MS of something like GroEL ([43] for example) is technically easier compared to HDX-MS of TriC due to the UqSeq differences (548 UqSeq for GroEL; 4,374 UqSeq for TRiC). However, TRiC information is more rich because the unique behavior of individual subunits of TriC (Figure 4 of Ref. [39]) could be identified by HDX-MS and correlated to the cryo-EM structure, something not possible in HDX-MS of GroEL or any other homo-oligomeric assembly.
Mass spectra have a lot to say.
When studying protein complexes with multiple monomeric units that might be doing different things conformationally, one must look at the spectra and the isotope patterns (Figure 4A). It is not enough to simply look at the amount of deuterium that was incorporated and ignore how the spectra look because the shape of each spectrum provides population information. An exciting example of careful spectral analysis was the HDX-MS study of the viral envelope protein conformations in the Dengue virus [44]. The envelope proteins were observed in two distinct (either pentameric or hexameric) positions by cryo-EM; however, the absence of two distinct HDX-MS profiles for the viral envelope proteins led researchers to further experiments to understand the apparent “spectral averaging” (meaning distinct higher-mass, more deuterated envelopes appeared merged or averaged with distinct lower-mass, less deuterated envelopes) of the two viral envelope protein positions. Sharma et al. [44] showed that antibodies and/or divalent cations caused the proteins to be fixed in one orientation or another, helping to explain the differing immune system reactions to various serotypes of Dengue. Such detailed analyses of mass spectra are time-consuming and become more difficult for large protein systems with a lot of UqSeq. There are perhaps older published datasets where reprocessing with emphasis on spectra and populations could be revealing, and newer datasets where extracting population information would be valuable but is not done because it is at present very time consuming.
Protein folding
Finally, the use of HDX in protein folding and unfolding, combined with cryo-EM, should be mentioned. These newer studies are not simply protein folding from a denatured state (e.g. [45]), but rather folding/unfolding in the presence of chaperones or molecular machines. The geometry of various proteins involved in unfolding (e.g., rubisco activase [20] as described above) can be determined with EM and the unfolding of the substrate followed in detail by HDX-MS. Another exciting combination takes advantage of low-resolution density and pulsed-labeling HDX-MS [46] wherein a protein is allowed to fold for a given time, then pulsed with a short exposure to deuterium to label only those position that are not folded (Figure 4B). Ordered regions versus not ordered regions in cryo-EM can be compared to protection from exchange. In Balchin et al. [39], this strategy was used to monitor actin folding in GroEL and TRiC. Cryo-EM structures (Figure 3 of Ref. [39]) showed shape profile changes that localized the substrate. Then, protection from HDX was monitored in the same state to identify what was folded in the “blobs” seen in the EM. For TRiC, this strategy identified where the unfolded species was in the chaperonin, what structure existed, and then what protection was afforded to the subunits of TRiC as a result of substrate interactions. Additionally, Twomey et al. [29] trapped an unfolded substrate in the Cdc48 complex and protection from exchange in the loading cofactor Ufd1:Npl4 could be observed by HDX-MS; this was consistent with the positioning of the unfolded ubiquitin chain in both the cofactor and the ATPase rings.
Concluding remarks
The combination of cryo-EM with HDX-MS is being used more and more frequently. Now that even large systems in excess of 5,000–10,000 UqSeq are possible by HDX-MS, the applications of the combination would seem to only grow in coming years. Processing, software, and automation (see Outstanding Questions) can only improve the number of applications and the speed with which studies could be performed. The connection between the two methods is now made mostly manually, and this is a bottleneck in terms of interpretation. While the end result of cryo-EM is a beautiful 3-dimmensional model, it is much harder to digest pages upon pages of HDX-MS information. There must be better ways to display and therefore interpret data. The way in which HDX-MS data are displayed could also be improved, even to the point that it accompanies the structural data in the PDB. Sample heterogeneity will continue to be an issue, and it must be considered in all types of experiments that combine these techniques. HDX-MS has more to offer in this regard than is currently being exploited. Population distributions are evident right in the data and better ways to deal with this information in timely ways must be found so its potential can be maximized. Taken all together, the more these two methods can be connected, the more they will together elucidate exciting biology.
Outstanding Questions.
As datasets become larger and larger, it is harder and harder to show HDX-MS data succinctly. The final result of cryo-EM is a 3D model, while the final result of HDX-MS is much harder to appreciate in a single picture. Could HDX-MS be summarized in equally easy-to-understand ways? Can new methods to display data be generated, along with software methods to easily implement these ideas?
Can HDX-MS data be included in databases alongside cryo-EM (or X-ray, NMR) structures? Why not have it all together?
Sometimes one acquires data on a complex with intention of just looking at one protein, but the data for the other proteins is there and could be mined for information at a later date, or in the context of other data about the other units present. Can the HDX-MS processing go faster? While current processing speed is an improvement over 5–10 years ago, large systems demand even better and faster analysis while maintaining robustness.
How does one deal with heterogeneity? Should one try to enrich a single form or instead just analyze the whole population and try to sort it out that way? Can individual conformations even be forced for all proteins?
Can more information be squeezed out of the data? In HDX-MS, what do the isotope patterns look like, especially in cases where there are clearly hetero-oligomers and multiple populations in the cryo-EM? Can better algorithms and software be made to speed up spectral mining?
Highlights.
Cryo-electron microscopy (cryo-EM) and hydrogen/deuterium exchange-mass spectrometry (HDX-MS) each provide unique information that, when combined, can validate each other
Technology improvements now allow large, complex systems to studied with HDX-MS, as well as provide more rapid, higher-resolution analysis by cryo-EM
Cryo-EM yields static pictures while HDX-MS provides dynamics and flexibility information, particularly for regions invisible to cryo-EM
Comparing HDX-MS in isolated proteins to that in large complexes shows how intermolecular interactions may affect dynamics, as well as reveals and localizes allostery
Cryo-EM structures can be used in the interpretation of HDX-MS data, especially when crystal structures do not exist
Exciting new possibilities include detailed functional studies of large molecular machines and studies of protein folding
Acknowledgements
We gratefully acknowledge financial support from the National Institutes of Health R01-CA233978 (JRE), R01-AI043957 (JRE), S10-OD016234 (EAK), and R01-HL127041 (EAK).
Glossary
- B-factors
a factor that describes the spread of electron density as caused by thermal motion, also known as the Debye-Waller factor or temperature-factor.
- Crosslinking MS
A technique where a reactive chemical entity (the crosslinker) covalently bonds to a target molecule, most often a protein, and a mass spectrometer is used to determine where the covalent addition occurred by means of a molecular weight increase in identified proteolysis products. Mass measurements before and after covalent addition can reveal which regions were within the right physical distance in three-dimensional space to allow the crosslinking reaction to occur, thus providing distance constraints for molecular model building.
- Hydrogen/deuterium exchange (HDX)
isotope exchange of deuterium for hydrogen at labile positions in proteins. Exchange at the backbone amide nitrogens is most often followed by any method sensitive to the differences between isotopes (e.g., NMR, MS, density)
- Mass spectrometry (MS)
measurement of the mass/charge ratio of molecules in the gaseous state through their response to electromagnetic fields or flight time in vacuum conditions. Proteins and peptides in solution are often charged and introduced into the gas-phase through electrospray ionization before introduction to the vacuum region of the instrument.
- Molecular dynamics
computer simulation of the physical movements of atoms and molecules, calculated using molecular mechanics force fields which describe the forces acting upon atoms.
- Molecular modeling
describing and studying molecules with models, most often computational in modern times but historically even including with clay or wood. Many types of data and known physical properties (such and typical bond length, angle, etc.) may contribute to the generation of an accurate model.
- Negative stain EM
a type of transmission electron microscopy where electron-rich heavy-metal salts (e.g. uranyl acetate) are adsorbed to biological molecules in order to increase the contrast in the electron beam. Images are of lower resolution than those of cryo-electron microscopy.
- Peak capacity
In separations, the number of species that can be separated with a resolution of unity (i.e., well resolved from one another) within a given time interval. Peak capacity can be calculated from the widths of chromatographic peaks in a chromatogram. The sharper the peaks, the higher the peak capacity.
- Protein–Ligand Interactions by Mass Spectrometry, Titration and hydrogen/deuterium EXchange (PLIMSTEX)
a method used to measure in inter- and intramolecular dissociation constants of protein-ligands complexes by their differential labeling in D2O at different concentrations of ligand.
- Resolving power
the ability to separate unique entities as separate from one another. In microscopy, including cryo-EM, resolving power describes the ability to separate or distinguish two or more small or adjacent images as being separate. Low resolving power produces images where details cannot be distinguished.
- Single particle cryo-electron microscopy (cryo-EM)
a method for constructing a 3-dimensional structure from thousands of transmission electron images of randomly oriented individual (single) particles frozen in vitreous ice, using alignment, averaging, and computational reconstruction.
- Small angle x-ray scattering (SAXS)
a low-resolution method to study the overall shape and structure of biological macromolecules by monitoring the scattering of x-rays at small angles (typically 0.1 – 10° from the incident x-ray beam trajectory).
- Unique sequence (UqSeq)
the amino acid sequence that is unique to a given protein, oligomer, or higher-order protein complex. A 200 amino acid dimer of two identical 100-residue proteins (homodimer) has 100 amino acids of UqSeq while a 200-residue dimer of two distinct 100-residue proteins (heterodimer) has 200 amino acids of UqSeq.
- Ultra-performance liquid chromatography (UPLC)
a separation method that uses small particles (sub 2 micron) and high-pressure pumps to increase the efficiency of separation in liquid chromatography.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trabjerg E et al. (2018) Conformational analysis of complex protein states by hydrogen/deuterium exchange mass spectrometry (HDX-MS): Challenges and emerging solutions. TrAC Trends in Analytical Chemistry 106, 125–138. [Google Scholar]
- 2.Engen JR and Wales TE (2015) Analytical Aspects of Hydrogen Exchange Mass Spectrometry. Annu Rev Anal Chem (Palo Alto Calif) 8, 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng J et al. (2019) Protein dynamics and conformational changes explored by hydrogen/deuterium exchange mass spectrometry. Curr Opin Struct Biol 58, 305–313. [DOI] [PubMed] [Google Scholar]
- 4.Weis DD (2016) Hydrogen Exchange Mass Spectrometry of Proteins: Fundamentals, Methods, and Applications, John Wiley & Sons, Ltd. [Google Scholar]
- 5.Danev R et al. (2019) Cryo-Electron Microscopy Methodology: Current Aspects and Future Directions. Trends Biochem Sci 44 (10), 837–848. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y (2018) Single-particle cryo-EM-How did it get here and where will it go. Science 361 (6405), 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Y et al. (2015) A primer to single-particle cryo-electron microscopy. Cell 161 (3), 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzik MA Jr. et al. (2017) Achieving better-than-3-A resolution by single-particle cryo-EM at 200 keV. Nat Methods 14 (11), 1075–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMullan G et al. (2016) Direct Electron Detectors. Methods Enzymol 579, 1–17. [DOI] [PubMed] [Google Scholar]
- 10.Scheres SH (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180 (3), 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murshudov GN (2016) Refinement of Atomic Structures Against cryo-EM Maps. Methods Enzymol 579, 277–305. [DOI] [PubMed] [Google Scholar]
- 12.DiMaio F and Chiu W (2016) Tools for Model Building and Optimization into Near-Atomic Resolution Electron Cryo-Microscopy Density Maps. Methods Enzymol 579, 255–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wales TE et al. (2008) High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem 80 (17), 6815–6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iacob RE et al. (2008) Ion mobility adds an additional dimension to mass spectrometric analysis of solution-phase hydrogen/deuterium exchange. Rapid Commun Mass Spectrom 22 (18), 2898–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L and Smith DL (2005) Capsid structure and dynamics of a human rhinovirus probed by hydrogen exchange mass spectrometry. Protein Sci 14 (6), 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L et al. (2001) Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci 10 (6), 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuma R et al. (2001) Hydrogen-deuterium exchange as a probe of folding and assembly in viral capsids. J Mol Biol 306 (3), 389–396. [DOI] [PubMed] [Google Scholar]
- 18.Lim XX et al. (2017) Epitope and Paratope Mapping Reveals Temperature-Dependent Alterations in the Dengue-Antibody Interface. Structure 25 (9), 1391–1402. [DOI] [PubMed] [Google Scholar]
- 19.Ramesh R et al. (2019) Uncovering metastability and disassembly hotspots in whole viral particles. Prog Biophys Mol Biol 143, 5–12. [DOI] [PubMed] [Google Scholar]
- 20.Bhat JY et al. (2017) Mechanism of Enzyme Repair by the AAA(+) Chaperone Rubisco Activase. Mol Cell 67 (5), 744–756. [DOI] [PubMed] [Google Scholar]
- 21.Manthei KA et al. (2020) Structural analysis of lecithin:cholesterol acyltransferase bound to high density lipoprotein particles. Commun Biol 3 (1), 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le VQ et al. (2018) Tolloid cleavage activates latent GDF8 by priming the pro-complex for dissociation. EMBO J 37 (3), 384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y et al. (2020) FACT caught in the act of manipulating the nucleosome. Nature 577 (7790), 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maity K et al. (2019) Cryo-EM structure of OSCA1.2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc Natl Acad Sci U S A 116 (28), 14309–14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye X et al. (2019) Hydrogen exchange reveals Hsp104 architecture, structural dynamics, and energetics in physiological solution. Proc Natl Acad Sci U S A 116 (15), 7333–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardiaux B et al. (2019) Dynamics of a type 2 secretion system pseudopilus unraveled by complementary approaches. J Biomol NMR 73 (6–7), 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z et al. (2018) Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme. Elife 7, e33572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cash JN et al. (2019) Cryo-electron microscopy structure and analysis of the P-Rex1-Gbetagamma signaling scaffold. Sci Adv 5 (10), eaax8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Twomey EC et al. (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 365 (6452), eaax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopcho N et al. (2019) Dynamics of ABC Transporter P-glycoprotein in Three Conformational States. Sci Rep 9 (1), 15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y and Chen J (2018) Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359 (6378), 915–919. [DOI] [PubMed] [Google Scholar]
- 32.Lumpkin RL et al. (2020) Structure and dynamics of the ASB9 CUL-RING E3 Ligase. Nat Commun, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris KL et al. (2018) HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation. Cell 174 (3), 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buffalo CZ et al. (2019) Structural Basis for Tetherin Antagonism as a Barrier to Zoonotic Lentiviral Transmission. Cell Host Microbe 26 (3), 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J et al. (2019) Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor. Elife 8, e50928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feynman RP (1999) There’s Plenty of Room at the Bottom. In Feynman and Computation: Exploring the Limits of Computers (Hey AJG ed), pp. 63–76, Perseus Books. [Google Scholar]
- 37.Cornish-Bowden A (1995) Fundamentals of Enzyme Kinetics, 2nd edn, Portland Press. [Google Scholar]
- 38.Faull SV et al. (2019) Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat Commun 10 (1), 3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balchin D et al. (2018) Pathway of Actin Folding Directed by the Eukaryotic Chaperonin TRiC. Cell 174 (6), 1507–1521. [DOI] [PubMed] [Google Scholar]
- 40.Zhu MM et al. (2003) Quantification of protein-ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. J Am Chem Soc 125 (18), 5252–5253. [DOI] [PubMed] [Google Scholar]
- 41.Brunle S et al. (2019) Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism. Proc Natl Acad Sci U S A 116 (52), 26497–26504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y et al. (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351 (6275), aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q et al. (2013) Nucleotide-induced conformational changes of tetradecameric GroEL mapped by H/D exchange monitored by FT-ICR mass spectrometry. Sci Rep 3, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma KK et al. (2019) Infectivity of Dengue Virus Serotypes 1 and 2 Is Correlated with E-Protein Intrinsic Dynamics but Not to Envelope Conformations. Structure 27 (4), 618–630. [DOI] [PubMed] [Google Scholar]
- 45.Yang H and Smith DL (1997) Kinetics of cytochrome c folding examined by hydrogen exchange and mass spectrometry. Biochemistry 36 (48), 14992–9. [DOI] [PubMed] [Google Scholar]
- 46.Deng Y et al. (1999) Comparison of continuous and pulsed labeling amide hydrogen exchange/mass spectrometry for studies of protein dynamics. J Am Soc Mass Spectrom 10 (8), 675–684. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt C and Urlaub H (2017) Combining cryo-electron microscopy (cryo-EM) and cross-linking mass spectrometry (CX-MS) for structural elucidation of large protein assemblies. Curr Opin Struct Biol 46, 157–168. [DOI] [PubMed] [Google Scholar]
- 48.Fang J et al. (2011) False EX1 signatures caused by sample carryover during HX MS analyses. Int J Mass Spectrom 302 (1–3), 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]


