Abstract
Hepatic steatosis droplet quantification with histology biopsies has high clinical significance for risk stratification and management of patients with fatty liver diseases and in the decision to use donor livers for transplantation. However, pathology reviewing processes, when conducted manually, are subject to a high inter- and intra-reader variability, due to the overwhelmingly large number and significantly varying appearance of steatosis instances. Meanwhile, this process is challenging as there is a large number of overlapped steatosis droplets with either missing or weak boundaries. In this study, we propose a deep learning based region-boundary integrated network for precise steatosis quantification with whole slide liver histopathology images. The proposed model consists of two sequential steps: a region extraction and a boundary prediction module for foreground regions and steatosis boundary prediction, followed by an integrated prediction map generation. Missing steatosis boundaries are next recovered from the predicted map and assembled from adjacent image patches to generate results for the whole slide histopathology image. The resulting steatosis measures both at the pixel level and steatosis object level present strong correlation with pathologist annotations, radiology readouts and clinical data. In addition, the segregated steatosis object count is shown as a promising alternative measure to the traditional metrics at the pixel level. These results suggest a high potential of AI assisted technology to enhance liver disease decision support using whole slide images.
Keywords: histopathology analysis, deep learning, overlapped steatosis segmentation, integrated network, liver disease
Liver steatosis is a disease caused by an excessive accumulation of lipids in liver cells [1]. Clinically, it is important to accurately measure steatosis components, as steatosis quantification serves as a key step in the clinical decision to use donor livers for transplantation. It has been proved that liver transplant recipients with rich steatosis components tend to have a higher rate of primary graft dysfunction and/or renal failure in 80% of cases [2]. In spite of the recent advance in non-invasive diagnostics, histopathology review of steatosis components in liver tissue biopsies is an important factor for the assessment of fatty liver disease and other liver conditions. Staging of steatosis can provide guidance to clinicians with regard to diagnosis severity and the necessity for liver disease treatment [3]. In clinical practices, pathologists determine the degree of steatosis components by examining the Hematoxylin and Eosin (H&E) stained tissue slides. However, their estimations are prone to both large intra- and inter-observer variability due to unacceptable sampling bias and poor reproducibility [4]. With the advent of high throughput digital scanners, computer-based methods have been developed to automate tissue microscopy image processing in a large variety of analyses, ranging from histopathology object detection, segmentation, to classification [5–8] . Despite the active development in this field, the ability to extract clinically relevant phenotype information from the whole slide images remains limited [9]. Deep learning based methods have become popular in the computer vision domain, due to their state-of-the-art performance in a wide range of applications, including image classification [10, 11], object detection, and segmentation tasks [12]. Deep learning is a class of emerging machine learning methods that can computationally learn low level image features for computerized image analysis [13]. Deep learning works with artificial neural networks that are comprised of layers of nodes as analogous to perceptron neurons interconnected in human brains. In each layer, the deep learning model has a convolution layer that applies different image filters to convolve with input image for low level image feature extraction. The final layer compiles the weighted inputs to produce an output. Unlike the traditional machine learning methods, no manual feature engineering is required for deep learning models. As the model keeps exploring new image features and optimizing node connection weights during the training stage, it can achieve promising performance after the model is fully trained. Numerous image segmentation methods based on Convolutional Neural Networks (CNNs) have been proposed, including nuclei detection, segmentation, and gland segmentation, among others. For example, a CNN based model is trained for nucleus segmentation followed by a deformable shape model for touching nuclei separation [6]. In a similar study, CNN is applied to enhanced gray scale images for nuclei segmentation [14]. The segmented result is refined by morphological operators. In a gland segmentation study, CNN outperforms Support Vector Machine classifier that requires the handcrafted feature extraction for glandular structure segmentation in colon histology images [15]. Additionally, it has been demonstrated that fusing multiple channels with CNN models can lead to improved gland segmentation results [16, 17].
By comparison, Fully Convolutional Neural network (FCN) [18] is more efficient and accurate for the semantic segmentation scenarios, where fully connected layers are embedded in CNNs, enabling an end-to-end training and testing. With FCN as a building block, a novel Deep Contour-Aware Network (DCAN) with a unified multi-task learning framework is proposed [19]. Multi-level contextual features are explored based on an end-to-end FCN for accurate gland detection and segmentation. Additionally, a nucleus-boundary model is introduced to predict nuclei regions and their boundaries simultaneously by a FCN [20]. U-Net [12] is yet another popular model in the FCN family that employs a U-shape deep convolutional network designed for biomedical image segmentation problems with the state-of-the-art performance even when the amount of training data is limited. Additionally, Holistically-nested Neural Networks (HNN) have demonstrated their promising performances for object segmentation in the medical imaging domain [16, 17, 21]. Their main advantage is that the resulting performance can be continuously improved as the training data scale increases. Additionally, this model can capture the underlying structure complexity and appearance of overlapping objects through an automatic feature learning mechanism in the training stage. Meanwhile, multiple network aggregation for the enhanced performance has been proposed. For example, ENet based models are trained for nuclear region and boundary prediction [22]. This is followed by the third ENet to combine the output of region- and boundary-ENets.
In clinical practice, there is a lack of objective ways to quantify steatosis due to multiple challenges. First, it is challenging to detect and segment the steatosis components from whole slide liver histopathology images, as steatosis droplets are subject to large variation in shape, size and appearance in distinct tissue sections [23]. Additionally, a large number of steatosis droplets are found in clumps with missing or weak separating borders. While isolated steatosis droplets are mostly circular in shape, overlapped instances have irregular shapes. Numerous methods based on hand-crafted features have been proposed for histopathology structure segmentation, ranging from thresholding [24], watershed, deformable models, morphological operations to sophisticated methods such as graph based methods [25]. However, hand-crafted features are limited in representation power and subject to feature parameters. Given the large variations in structures of overlapped steatosis droplets, it is challenging to define robust features suitable for all cases. The resulting performance of traditional supervised learning methods, such as Support Vector Machine (SVM), Adaboost or Bayesian, can be significantly deteriorated, as they highly depend on these hand-crafted features. Due to the presence of overlapped steatosis droplets in large tissue areas, no prior image analysis method for overlapped steatosis droplet quantification is equipped with whole slide image analysis capability to improve the clinical decision support.
In this paper, we present a steatosis segmentation model to identify individual steatosis and delineate boundaries of overlapping steatosis instances in whole slide microscopy images of liver biopsies. Specifically, a region-based module is designed to segment the foreground steatosis droplet region from background pixels, while a boundary module is introduced to learn the perceptual boundary features for each overlapped steatosis region. Next, the region and boundary information are combined to train the third deep neural network responsible for dividing steatosis droplets in clumps. The proposed network architecture is named as DeEp LearnINg stEATosis sEgmentation (DELINEATE). We provide both patch-wise and whole slide steatosis prediction analysis. For each patch, the pixel-wise analysis strategy is applied. Although some partial steatosis components are found crossing adjacent patches in most cases, the border area in each patch lacks such contextual information. To address this problem, we use a spatial indexing based approach to identify partial steatosis droplet components from neighboring patches, and have them efficiently assembled by MaReIA [26], a tool we developed in our prior work.
We quantitatively assess the accuracy of our method and compare it with other state-of-the-art methods. Our method is systematically validated with whole slide liver histopathology images of 36 patients with Nonalcoholic Fatty Liver Disease (NAFLD) collected from Children’s Hospital of Atlanta and Emory University. The resulting DELINEATE steatosis measurements at both steatosis pixel level and isolated steatosis object level are strongly correlated with gold standard pathological review results, patient clinical data, and fat readout from MRI images of the same patient cohort. Statistical tests suggest that DELINEATE derived steatosis measures both at steatosis pixel and isolated steatosis object level are promising clinical indicators presenting statistically significant difference between (1) two diagnostic groups - Nonalcoholic Steatohepatitis (NASH) and Nonalcoholic Fatty Liver (NAFL), and (2) groups with and without lobular inflammation. In addition, steatosis object level measure is found as an informative alternative to the pixel level measure for differentiating histological steatosis grades.
Materials and Methods
Data pre-processing
Whole slide images, human annotations, radiology readouts and clinical data were obtained from the Children’s Hospital of Atlanta and Emory University. All liver tissue permanent sections were formalin-fixed and paraffin-embedded, and stained by H&E. Resulting images of permanent section slides were reviewed to exclude those with unacceptable tissue-processing artifacts. Portal tract areas with bile ducts and large vessels were manually excluded, leaving only hepatic lobules for quantitative analysis. Each whole slide image contains multiple tissue components. To reduce image size for analysis, we extracted complete tissue component images capturing minimum non-tissue areas by rotations at the highest image resolution level from original whole slide images [27]. The resulting whole tissue component images were still too large in size to feed into deep learning models for steatosis prediction. Therefore, each complete tissue component image was partitioned to non-overlapping image patches of size 512 × 512 pixels. This process results in 2,050 image patches that are divided into training and validation cohorts with a ratio of 80:20. All image patches were extracted at 20× objective magnification using OpenSlide [28]. As colors of H&E stained images depend on numerous factors related to the tissue preparation, staining, and scanning process [20], they can vary significantly. Thus, all images were normalized to a standard H&E calibration image by the stain color [29] before analysis. Whole slide images of human liver biopsies from 36 unique patients were analyzed, with an average image resolution of 30,000 × 20,000 by pixels.
DELINEATE architecture
As demonstrated in Fig. 1(A), the proposed DELINEATE model for steatosis segmentation has an end-to-end deep learning process in two stages. First, the foreground steatosis regions and their boundaries are identified and separated. Next, the resulting prediction outputs are combined in the integrative module for the clumped steatosis segmentation.
Figure 1.
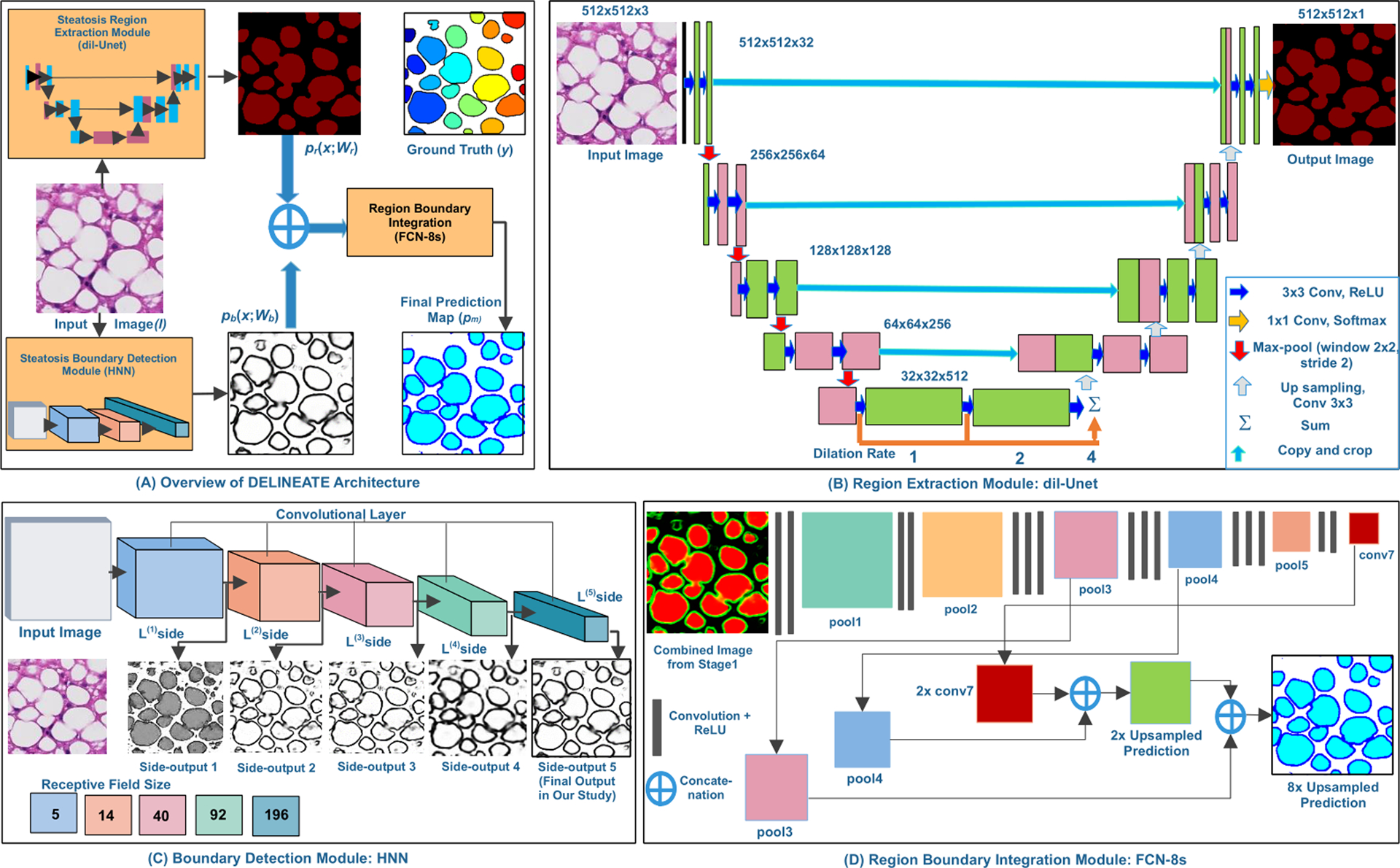
The DELINEATE Model. The DELINEATE model first identifies regions and boundaries of steatosis droplets individually (A). The resulting two output predictions are combined for generating an integrated prediction map where the clumped steatosis regions are separated. The region extraction module detects steatosis regions with a dil-Unet module (B). The steatosis boundary detection module is based on a Holistically-Nested Network (HNN) (C). The region-boundary integration network generates the final prediction output from the integrated region and boundary information (D).
The steatosis region extraction module has a modified U-Net [12] architecture consisting of four encoding and another four decoding layers, aiming at identification of the steatosis components from the background. Different levels of contextual feature maps are extracted by the encoding layers, while the decoding layers generate probability masks of steatosis regions. As a way to reduce spatial information loss, the high resolution feature maps from the encoder layers are connected to the corresponding decoder layers. Additionally, dilated convolution [30] is used to exponentially expand the receptive field, decrease space-invariance, and reduce detailed spatial information loss by max-pooling and strided convolution in the down-sampling path, contributing to a promising segmentation accuracy as demonstrated in Table 1. Three dilated convolution layers at a rate of 1, 2 and 4 are stacked at the bottleneck block where the feature maps have the lowest resolution. This architecture, named as dil-Unet, is illustrated in Fig. 1(B). Input images of size 512 × 512 are used for training and testing this model. All kernels used in each convolutional layer are initialized by the standard Xavier initialization [31] and the bias is initialized with zero.
Table 1:
Comprehensive performance comparison of steatosis segmentation methods.
| Models | Approach | Precision | Recall | F1-Score | Object wise Dice Index |
Object wise Hausdorff Distance |
|||
|---|---|---|---|---|---|---|---|---|---|
| Standard | FCN | 0.01 | 0.06 | 0.04 | 0.8338 | 3.8521 | |||
| Models | DeepLab V2 | 0.01 | 0.08 | 0.05 | 0.9083 | 5.3179 | |||
| dil-Unet + HNN + FCN-8s | 0.01 | 0.06 | 0.03 | 0.9492 | 3.4591 | ||||
| dil-Unet + HNN + FCN-4s | 0.01 | 0.06 | 0.03 | 0.9480 | 3.5753 | ||||
| Variations of | Unet + HNN + FCN-4s | 0.01 | 0.06 | 0.03 | 0.9489 | 3.4685 | |||
| Our Models | dil-Unet + HNN + dil-FCN | 0.01 | 0.06 | 0.03 | 0.9459 | 3.6658 | |||
| Unet + Unet + Unet | 0.04 | 0.07 | 0.05 | 0.9247 | 5.8289 | ||||
| Unet + Unet + FCN-8s | 0.03 | 0.06 | 0.04 | 0.9458 | 3.8773 | ||||
The steatosis region extraction network minimizes the softmax cross entropy loss Lr between the prediction map Pr and the target Yr :
| (1) |
Where K = 2 is the number of classes, yr is the binary indicator of the true label at pixel (i,j) in an image domain Ω, and Pr (i,j |c,wr, br) is the output of soft-max activation layer indicating the probability of the pixel (i,j) having label c. We use Adam optimizer [32] along with exponential learning rate decay to optimize the parameter set wr by back-propagation.
In addition to the region supporting information from dil-Unet, a complementary steatosis boundary detection module with Holistically-nested Neural Network (HNN) [33] derived from VGGNet is used to delineate the hidden boundaries of the overlapped steatosis droplets. The architecture of this module is depicted in Fig. 1(C). The module consists of five convolutional stages of distinct receptive field sizes and stride values (i.e. 1, 2, 4, 8, and 16), respectively. Additionally, it has M side-output layers serving as classifiers with weights w = (w(1),.… w(M)).
The steatosis boundary module is a combination of a HNN and a “weighted-fusion” layer trained in parallel during the training phase [21, 33]. The parameter set W is initialized by the pre-trained network [33] and updated with our training data (X, Yb). The training process minimizes the following loss function:
| (2) |
where lm is computed over all pixels of training image pair (X, Yb), and represents the image level loss function from side output m. Each side output lm is refined for minimization over iterations; is the output from the “weighted-fusion” layer with fusion weights {hi}. Sigmoid activation function σ (·) is used to compute the class probability of each pixel (i,j); Dist (·) is the cross-entropy loss with the fused predictions and the ground truth label maps. Resulting optimal parameters are found by the objective function minimization with stochastic gradient descent and back propagation in training. In the testing phase, the prediction is generated from side output layers and the weighted-fusion layer. The 5th side output is used to represent steatosis boundaries as it produces results with good contrast after careful visual inspections.
By experiments, neither region nor boundary information by its own is sufficient for accurate steatosis droplet segmentation. Additionally, neither direct combination nor simple concatenation of the two channels of outputs provides precise boundary information for overlapped steatosis droplets. As a result, a Fully Convolutional Network (FCN) [18] is used to integrate complementary information from steatosis region and boundary modules for final prediction.
Specifically, FCN-8s as illustrated in Fig. 1(D), with skip connections from pool3 and pool4 are used for better deep semantic information integration on the down-sampling path. The final output has three channels representing the probabilities of each pixel being background, boundary, or region class, respectively. The integrated network is trained with softmax cross entropy loss and Adam optimizer [32]. Dropout with probability of 0.3 is used to overcome the over-fitting problem.
Training and validation of data
Each whole tissue component image was partitioned to non-overlapping image patches for generating training and validation data. Each steatosis component boundary was annotated by domain experts and served as the ground truth for the steatosis boundary prediction module. The region labels derived from the gold standard boundaries were used for the steatosis region extraction module. Deep learning model training requires a large set of training data to avoid the over-fitting problem. For pathology image review, however, it is highly time-consuming to manually label all steatosis boundaries in a large image patch set by domain experts. Therefore, the limited human-annotated training data set for the steatosis region module was augmented by the horizontal flip, vertical flip, rotation in four degree angles, and re-scaling by 0.5 scales. Similarly, each image patch was rotated in 16 different angles and flipped at each angle to generate an augmented training set for the boundary module. A total of 1,471 patches of size 512 × 512 were used for this study where 735 were randomly selected for the region and boundary module training, with the remaining 736 for the integrative network training. Each cohort was split into 80:20 for training and validation image generation. The test set consists of 150 patches from a new set of tissues not seen in the training and validation sets. The region and boundary modules were trained separately. Their outputs were combined to train the integrated network in turn.
Post-processing
The FCN-8s integration network predicts steatosis region, boundary, and background classes. The resulting prediction map is binarized with a cutoff value of 0.5. Each connected component in the binary mask represents the internal region of a steatosis droplet. For overlapped steatosis droplet segregation, we further apply the high curvature point detection and an ellipse fitting quality assessment method in the post-processing [27]. Specifically, high curvature points on a steatosis contour are detected [34] and combined with adjacent high curvature points in aggregated point representations. High curvature points in all possible pairs are connected by straight lines for further division assessment. Each such candidate line partitions the overlapped steatosis region into two components. The partitioning quality is further assessed by fitting an ellipse for each component. For each divided component, we next compute the ratio of the intersection to union area of the resulting ellipse and the partitioned steatosis region. Of all possible candidate point pairs, we only connect the paired points with maximum ratios both greater than the fitting quality cutoff value of 0.7. Post-processing results are illustrated in Fig. S1.
Patch-wise segmentation assembly for whole tissue image analysis
Due to the limitation of GPU memory size, deep learning methods cannot process a single high-resolution whole slide histopathological image at once. Therefore, we divide each whole tissue image into 512 × 512 image patches for model training and prediction. As the FCN family algorithms do not provide accurate border area prediction [20], we use a generic MapReduce based Image Analysis framework (MaReIA) to avoid counting duplicates of steatosis droplets crossing patch borders [26]. The framework introduces an overlapping partitioning method that partitions whole tissue component images into patches with extended buffers for accurate segmentation, eliminating the boundary crossing object problem. The buffer zone is adjusted according to the histology structure size in such a way that it is large enough to completely contain histology objects of interest in the buffer zone. The block diagram in Fig. 5(B) illustrates individual steps of steatosis quantification with whole tissue component images. First, we extract the overlapping patches of size 512 × 512 from each whole tissue image. The resulting patches are processed by the steatosis segmentation pipeline. Next, all patches are merged with MaReIA that generates a steatosis prediction map for each whole tissue component. The resulting output is further polished by the post-processing step. One typical whole tissue steatosis segmentation result is demonstrated in Fig. 5(A). As the proposed solution to patch wise result aggregation is generic, it can be applied to a large number of whole slide image analysis research where aggregation of results from patches in whole slide images is important.
Figure 5.
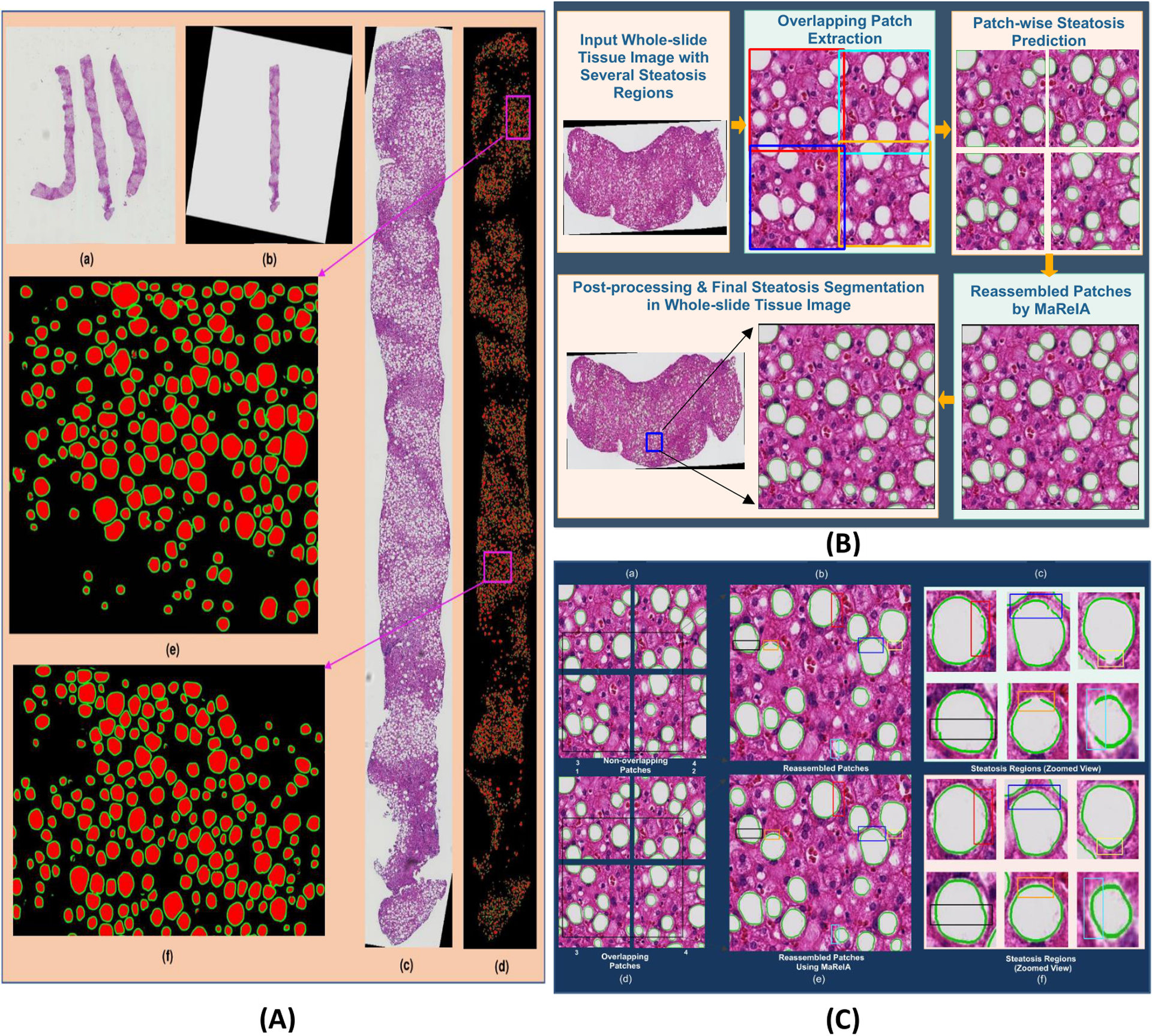
(A) Whole tissue steatosis prediction. (a) a low resolution whole slide liver image containing multiple tissue components; (b) one complete tissue component extracted at a low resolution; (c) the highest resolution tissue component extracted after rotation and interpolation; (d) steatosis regions and the boundary masks in the complete tissue component detected by DELINEATE model; (e-f): close-up views of two representative tissue regions in purple rectangles in (d). (B) Block diagram of steatosis quantification in whole slide liver tissue images. It consists of high resolution tissue component extraction, overlapped tissue region partitioning, steatosis segmentation by DELINEATE model, and patch-wise steatosis segmentation assembled by MaReIA. (C) Steatosis segmentation assembled by different methods. (a) typical four adjacent non-overlapping patches; (b) steatosis segmentation with simple concatenation; (c) close-up views of steatosis droplets with simple concatenation; (d) overlapping patches; (e) steatosis segmentation assembled by MaReIA; and (f) close-up views of assembled steatosis droplets by our proposed MaReIA.
Hardware and software specifications
The developed framework is implemented with the open-source deep learning library TensorFlow [35] and Keras [36]. The experiments is carried out on Tesla K80 and V100 GPUs with CUDA 9.1. Adam optimization algorithm [32] is used to train all three modules. The initial learning rate and learning rate decay are set as 0.0001 and 0.9 respectively for the steatosis region module, while the boundary detection module is trained with a learning rate of 0.001 and a weight decay of 0.0002. The parameters of boundary module and integration module are initialized by pre-trained VGG16 model [11]. In the training phase, the learning rate of the integration network FCN-8s is set as 1.00e−5.
Data availability
All source codes and annotation data related to this paper are available at GitHub [37]. We share image data in a public repository [38].
Results
Steatosis droplet segmentation using DELINEATE model
The overall DELINEATE framework is illustrated in Fig. 1. Our proposed DELINEATE model for steatosis segmentation is a region-boundary integrated network architecture and has an end-to-end deep learning process in two stages. First, the foreground steatosis droplet regions and their boundaries are identified separately. The resulting two output predictions are combined to create a final prediction map in the second stage where the clumped steatosis droplets are divided into separate components (Methods).
For steatosis region segmentation, we built the architecture based upon the FCN family model U-Net [12] by stacking dilated convolutional layers at the bottleneck of the U-Net model. The dilated convolution enhances the network performance with a wider receptive field without the down-sampling operation, resulting in more accurate segmentation than the standard U-Net model as demonstrated in Fig. 2(A). To better learn steatosis boundary features, we used Holistically-nested Neural Network (HNN) to capture low, middle and high level contour signatures from hierarchically embedded multi-scale edge fields [21, 33]. Instead of summing all side outputs by weights, we propose to retain the single side output from the fifth side as the steatosis boundary prediction result.
Figure 2.
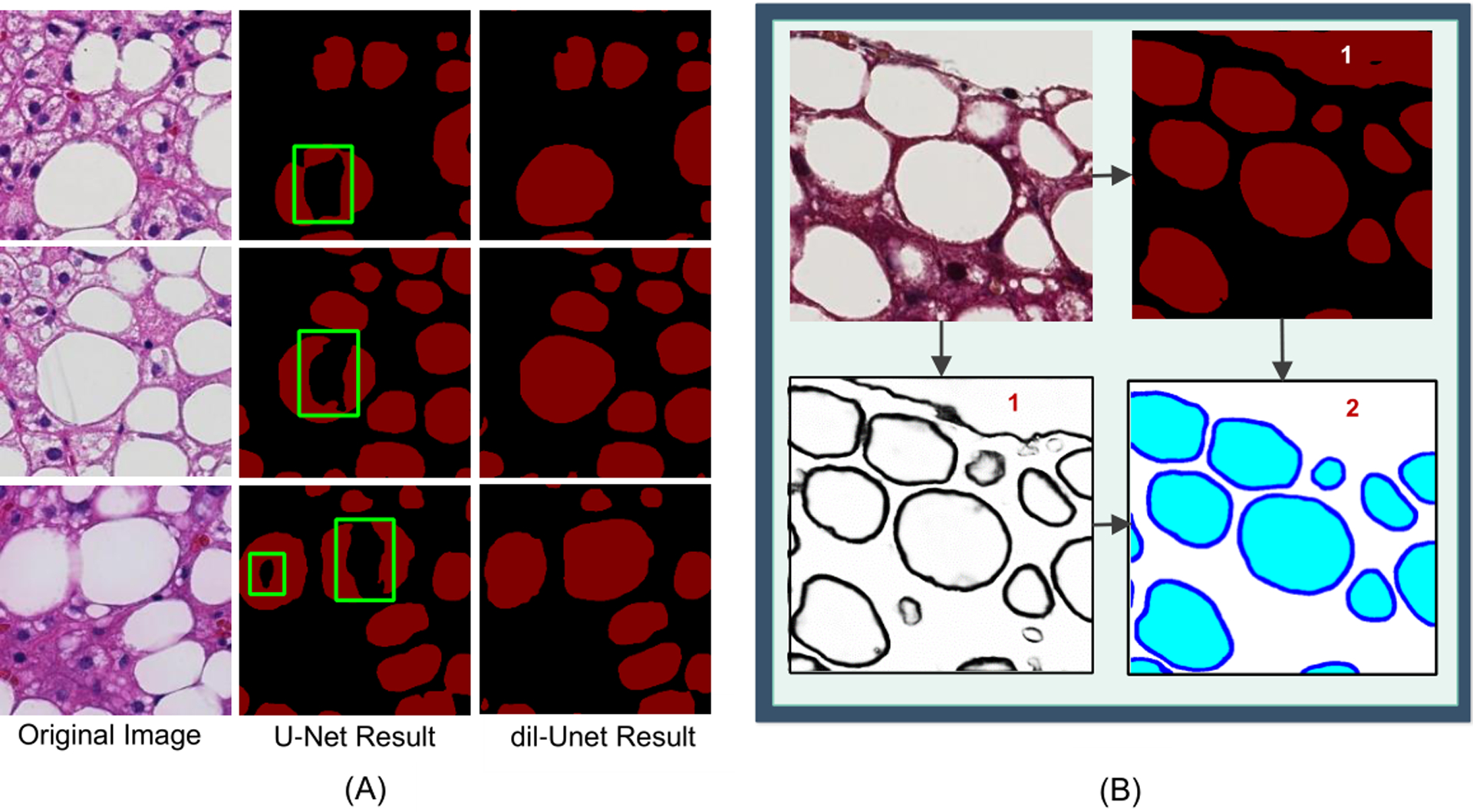
Comparison of segmentation results. Comparison of segmentation results between dil-Unet and the standard U-Net model (A). Left: original images; Middle: steatosis segmentation by U-Net model; Right: steatosis segmentation by the proposed dil-Unet model. By contrast, dil-Unet can recover steatosis regions with a substantially improved accuracy. Comparison of results from the DELINEATE model (B). Top-Left: input image; Top-Right: output from the region extraction module; Bottom-Left: output from the boundary detection module; and Bottom-Right: final output of the integration module. “1” labels the false positive steatosis region captured by the region prediction module, and “2” labels the corrected steatosis regions by the final integration module.
By detecting the boundary in an additional module, we can delineate the hidden boundaries of overlapped steatosis regions, and therefore, improve steatosis segmentation accuracy. Therefore, we further used a fully convolutional network [18] with skip connection to integrate the derived region and boundary information and produced the final prediction map of three classes: steatosis droplet region, steatosis boundary, and the background. Specifically, FCN with a transposed convolution layer having a stride 8 (FCN-8s) at the final layer was used to generate the resulting segmentation map. With extensive experiments, we demonstrate that such integrative network leveraging information from both the region and the boundary detection module will help remove large false positive regions (Fig. 2(B)).
Evaluation of DELINEATE segmentation accuracy
To evaluate the DELINEATE model accuracy, we applied our method to whole slide liver histopathology images of 36 children with Nonalcoholic Fatty Liver Disease (NAFLD) collected from Children’s Hospital of Atlanta and Emory University. The corresponding demographics, steatosis diagnostics, radiology measures, and clinical outcome of the patient cohort are summarized in Table S1. As each whole slide liver image may contain multiple tissue components, we retained each such tissue component in a separate image for analysis [27] (Method).
The segmentation accuracy of DELINEATE model was evaluated by five-fold cross-validation method. We randomly partitioned the dataset (Supplement S1: Dataset) into a training and a testing set by a ratio of 80:20. We trained the DELINEATE model with the training set and evaluated the accuracy of the model with testing. Steatosis quantification accuracy was measured both at the object level and the pixel level. The object-level measures include F1 score, Precision, Recall [39] and Hausdorff Distance. True Positive is counted when a segmented steatosis droplet shares more than 50% of its area with the ground truth. Otherwise, it is considered as a False Positive. Ground truth steatosis regions not segmented by DELINEATE are considered False Negative. To accurately assess overlapped steatosis segmentation results, we used object level Dice index [16, 19] for method evaluation at the pixel level. Without loss of generality, G is denoted as a ground truth set for steatosis instances and P as a set of machine segmented steatosis instances. For the i-th steatosis instance Gi in the ground truth set, we found the maximally overlapped segmented steatosis instance Pi in the same image and computed the Dice index D(Gi, Pi). Similarly, for the j-th segmented steatosis instance , we detected the maximally overlapped ground truth steatosis instance and computed the Dice index . The resulting object-level Dice score D(G, P ) is defined as follows:
| (3) |
where wi is the ratio of the number of pixels in the i-th ground truth instance to the sum of all pixels in all steatosis components in the ground truth image; is the ratio of the number of pixels in the i-th instance from deep learning model to the sum of all pixels in all steatosis components in the automated segmented image. NG and NP are the numbers of steatosis instances in the ground truth set and the corresponding machine segmented result set. As morphological features are important for steatosis droplet identification, we computed Hausdorff distance to evaluate shape similarity. We computed an object-level Hausdorff distance in the same way as for the object-level Dice score.
Table 1 summarizes the segmentation performance of our proposed DELINEATE model, and comparison results with other methods, including baseline FCN, DeepLab [40], and multiple variations of our proposed model. By contrast to other methods, DELINEATE dil-Unet+HED+FCN-8s achieves the best overall performance, as indicated by F1-score, Recall, object-wise Dice index and object-wise Hausdorff distance [41]. We represent DELINEATE model and its variations by connecting three modules with ‘+’ sign for easy interpretation. Compared to the other models, DELINEATE results in a higher object-wise dice index and a lower object-wise Hausdorff distance [41], indicating its superior performance. Note that DELINEATE model substantially outperforms state-of-the-art FCN and DeepLab models and achieves better performance on delineating overlapped steatosis droplets guided by joint region and boundary information. Neither FCN nor DeepLab can process touching steatosis droplets accurately with missing region-boundary integrative information, resulting in lower performance scores. The salient difference in performance across these methods is visually confirmed by Fig. 3 where each segmented steatosis region is illustrated in a unique color. It is noticeable that touching regions highlighted by black boxes are well separated by DELINEATE model, whereas they are incorrectly segmented by other methods for comparison. Green boxes are used to highlight challenging steatosis regions where only DELINEATE model can accurately recognize steatosis components. We further visualize patch-wise and instance-wise steatosis segmentation accuracy heat maps of one representative tissue component in Fig.S5 and Fig.S6 within Supplemental Information. These visual results demonstrate a high concordance between the DELINEATE segmentation results and annotations and confirm the efficacy of our proposed model.
Figure 3.
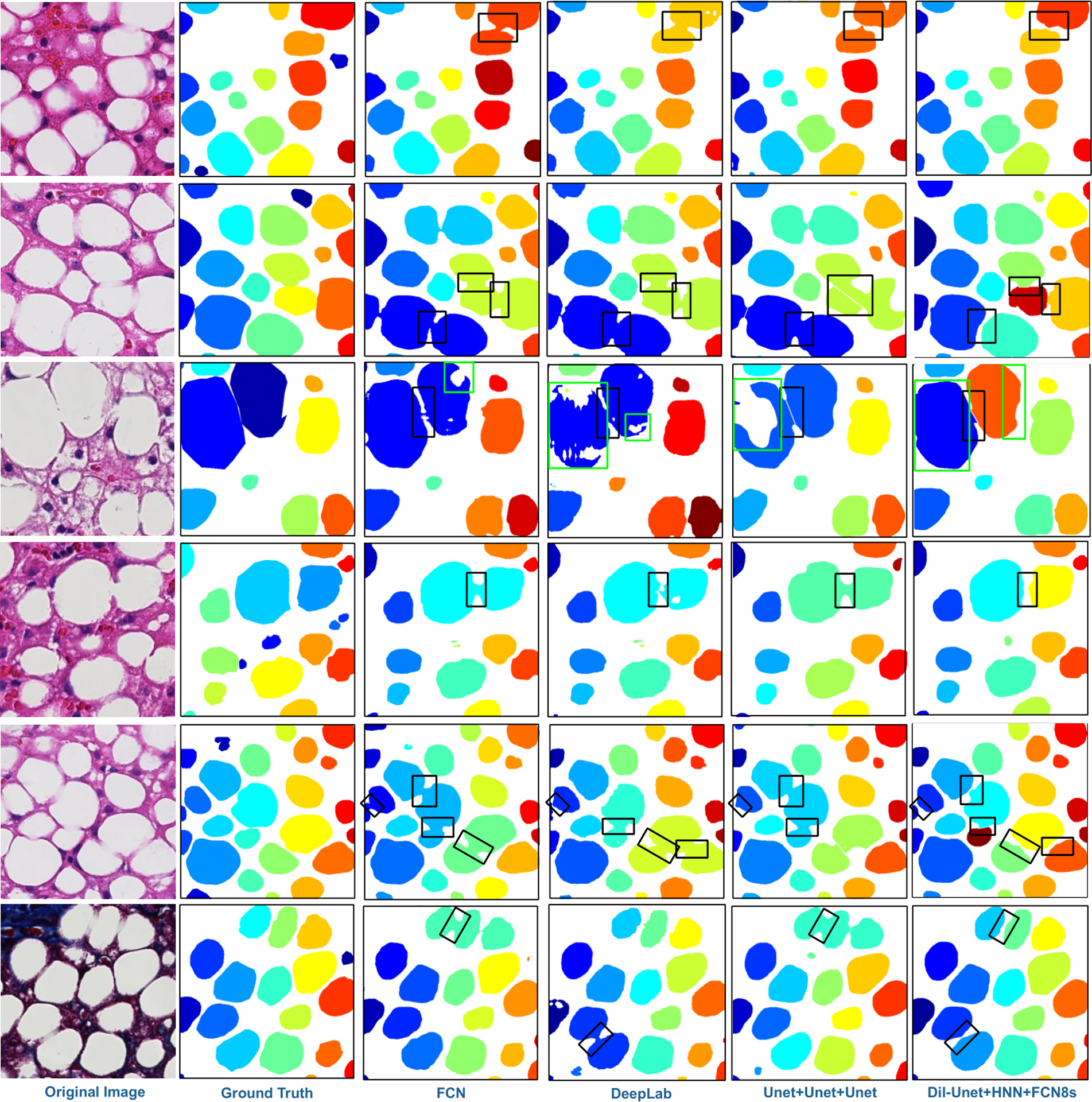
Visualization of segmented steatosis droplets in masks of distinct colors. From left to right column: original image, ground truth, results from FCN, DeepLab V2, U-Net+U-Net+U-Net (one variation of our proposed model), and dil-Unet+HNN+FCN-8s (proposed DELINEATE model), respectively. The clumped steatosis regions indicated by black boxes in all images are well separated by DELINEATE model but failed by other methods in the comparison study. Additionally, problematic regions in green boxes are only fully recovered by DELINEATE model.
DELINEATE correlation with pathological grading, radiology, and clinical data
The results produced by the DELINEATE model present strong correlations with liver tissue pathological grading, fat quantity from MRI data, and patient clinical information. The correlation analysis includes 36 children diagnosed with NAFLD. This cohort of patients underwent a liver biopsy at the Children’s Hospital of Atlanta between 2014 and 2016. NAFLD diagnosis was established by liver biopsy, and other etiologies were excluded by standard clinical and laboratory assessment. All liver biopsies were clinically diagnosed and each section was blindly reassessed by an expert pathologist at Emory University Hospital. Basic demographic characteristics were collected at the time of the biopsy. All parents or guardians signed an informed consent form and all children provided written assent in these studies, which were approved by the Emory University IRB board. Spearman’s correlation was used to analyze the correlation between two variables. Mann-Whitney test (for non-parametric data) was used to compare the difference between two groups. For comparisons across diverse steatosis groups, DELINEATE was logarithmically transformed before analysis. Analysis of Variance (ANOVA) was used to study the difference among four histological steatosis grading groups. This was followed by Tukey’s multiple comparison post-test. All statistical analyses were performed using R (version 3.4.2). Data were considered statistically significant for p-value < 0.05.
We present in Table 2 the Spearman correlation coefficients between steatosis measures from DELINEATE and all the following measures, including manual macrovesicular steatosis measure, manual total steatosis measure, fat readout from MRI images of the same patient cohort, and Positive Pixel Counting by Aperio [42], respectively. With DELINEATE, we computed both steatosis pixel percentage (DSP%) and isolated steatosis droplet count percentage (DSC%). Both measures were normalized by tissue sizes. Note that we report steatosis droplet count in percentage to match to steatosis measure representation in prior work on the assessment of steatosis measure [43] and from the Nonalcoholic Steatohepatitis Clinical Research Network [44]. For comparison, a third-party commercial software Aperio Positive Pixel Counting was used to measure steatosis pixel percentage (ASP%). Note that this software supports pixel-wise classification. However, it cannot group pixels to form individual steatosis. It can neither detect weak or missing boundaries for dividing steatosis droplets in clumps. DSP% and DSC% present strong correlations with all these histology and radiology measures that are manually confirmed. Note that DSC% from DELINEATE presents the strongest correlation with Total Steatosis and Macrovesicular Steatosis by histology review, while DSP% from DELINEATE has the best correlation with MRI fat quantification. Enhanced correlations between measures from DELINEATE and well-verified gold standard suggest accurate steatosis measures from histology images with our proposed DELINEATE method. In Fig. 4(A–C), we illustrate DSC% correlations with other histology and radiology measures, with overlaid linear regression lines in the scatter plots.
Table 2:
Correlation coefficients and p-values are presented for pairwise correlations using steatosis measures, results of a gold standard histology review, and manual fat readout from MRI images. Steatosis measures include DELINEATE Steatosis Pixel% (DSP%), DELINEATE Steatosis Count% (DSC%) and Aperio Steatosis Pixel% (ASP%), respectively.
| Correlation Measure | DSP % (p-value) | DSC %(p-value) | ASP % (p-value) |
|---|---|---|---|
| Macrovesicular steatosis% | 0.85(<0.001) | 0.90(<0.001) | 0.83(<0.001) |
| Total steatosis% | 0.85(<0.001) | 0.90(<0.001) | 0.84(<0.001) |
| MRI fat readout | 0.85(<0.001) | 0.82(<0.001) | 0.83(<0.001) |
| Aperio Pixel% | 0.94(<0.001) | 0.91(<0.001) | - |
Figure 4.
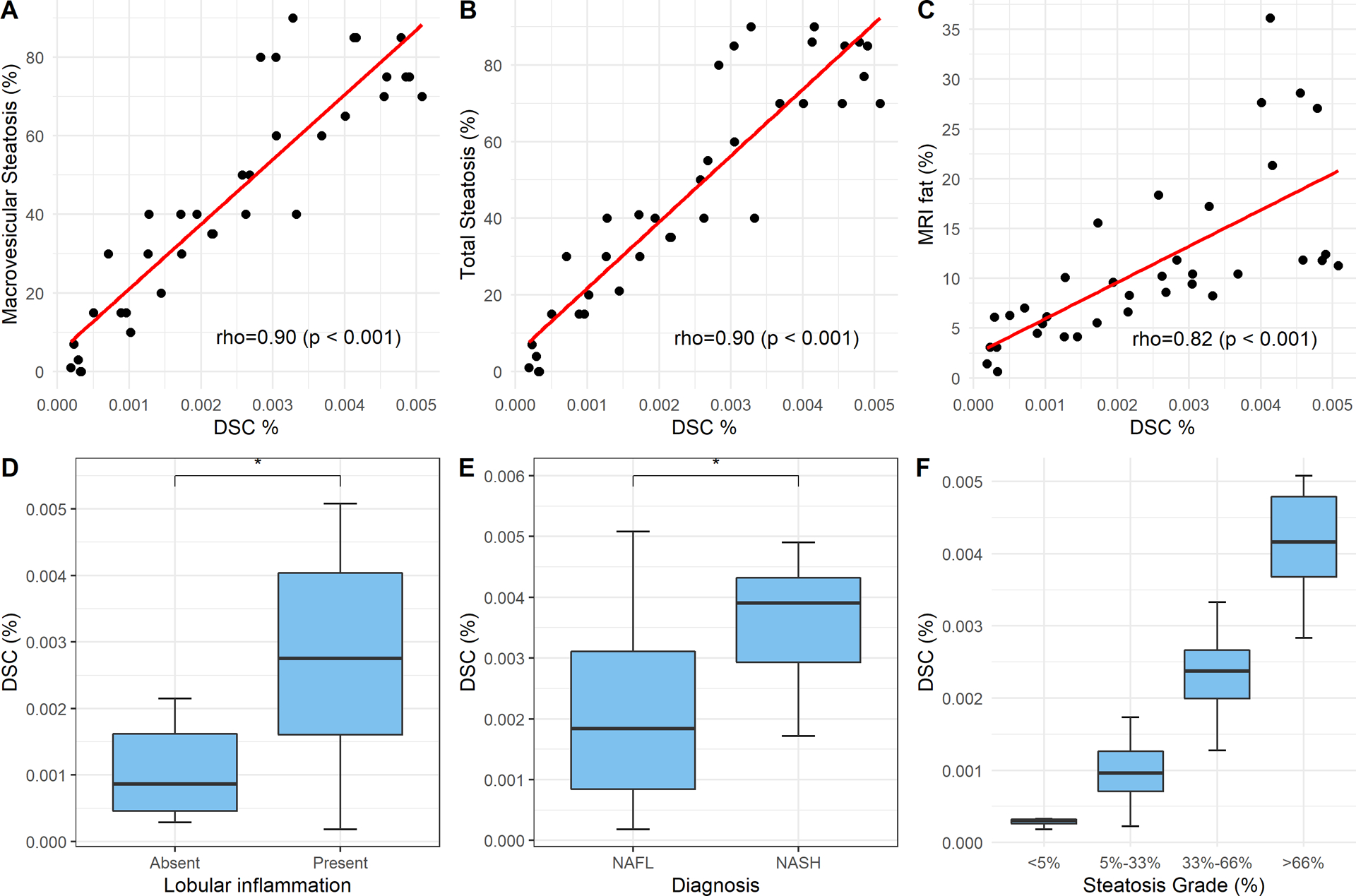
Pair-wise scatter plots with correlation coefficients (p-values) are illustrated for DELINEATE Steatosis Count% (DSC%) at individual droplet level and manual macrovesicular steatosis measures (A); manual total steatosis measures (B); manual fat readout from MRI images (C). We applied Mann-Whitney test to DSC% measures between Lobular Inflammation presence and absence (D); and between NAFL (i.e. Non-NASH) and NASH (E), respectively. We applied ANOVA to DSC% measurements of tissue samples among four manually graded histological steatosis percentage groups with p-value less than 5.25e−14 (F).
For group median and mean difference investigations, we applied Mann-Whitney test to steatosis measures between two diagnostic groups NAFL and NASH, and between groups with and without lobular inflammation, respectively. As this study cohort includes NASH cases, we can test the hypothesis that higher degrees of steatosis are associated with higher degrees of inflammation, which is in turn associated with more severe NASH cases. Three steatosis measures, ASP%, DSP% and DSC%, were used for group difference analysis. Specifically, box plots of these two analyses with DSC% are demonstrated in Fig.4(D–E). The mean, standard deviation and median of these measures and p-values of statistical tests are presented in Table 3. Notably, DELINEATE steatosis measures present statistically significant group difference in both analyses. Specifically, the steatosis object level measure DSC% produces the least p-value in lobular inflammation comparison, and both steatosis measures (DSC% and DSP%) yield a low p-value in diagnosis group comparison study.
Table 3:
Mean, median, and range of DELINEATE Steatosis Pixel% (DSP%), DELINEATE Steatosis Count% (DSC%), and Aperio Steatosis Pixel% (ASP%) are presented with the associated p-values of Mann-Whitney test.
| Steatosis Measure | Lobular Inflammation | Diagnosis | Overall | |||||
|---|---|---|---|---|---|---|---|---|
| Absent | Present | p-Value | NAFL | NASH | p-Value | |||
| DSP% | Mean(SD) | 5.24 (7.01) | 12.1 (8.20) | 0.036 | 8.75 (7.75) | 17.0 (7.64) | 0.010 | 10.6 (8.38) |
| Min, Median, Max | 0.538, 2.15, 21.5 | 0.300, 12.6, 27.4 | 0.300, 7.34, 26.2 | 2.20, 17.4, 27.4 | 0.300, 8.49, 27.4 | |||
| DSC% | Mean (SD) | 1.37e-3 (1.43e-3) | 2.72e-3 (1.52e-3) | 0.030 | 2.08e-3 (1.54e-3) | 3.62e-3 (1.11e-3) | 0.010 | 2.42e-3 (1.58e-3) |
| Min, Median, Max | 2.89e-4, 8.64e-4, 4.55e-3 | 1.84e-4, 2.76e-3, 5.08e-3 | 1.84e-4, 1.84e-3, 5.08e-3 | 1.72e-3, 3.9e-3, 4.9e-3 | 1.84e-4, 2.37e-3, 5.08e-3 | |||
| ASP% | Mean(SD) | 10.1 (8.04) | 16.5 (9.51) | 0.070 | 13.1 (8.72) | 22.2 (9.12) | 0.020 | 15.1 (9.49) |
| Min, Median, Max | 1.88, 9.57, 27.1 | 0.641, 18.1, 31.5 | 0.641, 11.9, 31.5 | 1.84, 24.2, 30.9 | 0.641, 13.5, 31.5 | |||
Additionally, we assessed the difference of steatosis measurements across four histological steatosis percentage grades by DSP%, DSC%, and ASP%, respectively. The resulting p-values of ANOVA and Tukey’s multiple comparison tests for ASP%, DSP%, and DSC% are presented in Table 4. Note that both DELINEATE steatosis measures demonstrate statistically significant difference across grades, with the proposed steatosis count measure DSC% presenting the least p-value. The box plot of this analysis with the use of DSC% is demonstrated in Fig. 4 (F). Table 5 contains the optimal threshold of DELINEATE Pixel, DELINEATE Count, and Aperio PPC for differentiation of patient groups by different steatosis grades assessed by an expert pathologist. Table 5 shows the area under the receiver operating curve (AUROC), sensitivity, specificity, and accuracy of these methods. Additionally, there was no significant difference for the AUROC across these methods in each stage.
Table 4:
ANOVA and Tukey’s multiple comparisons test with liver tissue steatosis measures across four manually annotated steatosis percentage groups with p-values adjusted by the Benjamini-Hochberg method. The adjusted p-value for DELINEATE Steatosis Pixel% (DSP%), DELINEATE Steatosis Count% (DSC%) and Aperio Steatosis Pixel% (ASP%) are shown in column 2, 3 and 4, respectively.
| Statistical Test | DSP% | DSC% | ASP% |
|---|---|---|---|
| ANOVA | 7.86e-09 | 5.25e-14 | 3.37e-09 |
| <5% vs 5%−33% | 0.005 | 0.25 | 0.69 |
| <5% vs 33%−66% | <0.001 | <0.001 | 0.002 |
| 5%−33% vs 33%−66% | <0.001 | <0.001 | 0.005 |
| <5% vs >66% | <0.001 | <0.001 | <0.001 |
| 5%−33% vs >66% | <0.001 | <0.001 | <0.001 |
| 33%−66% vs >66% | 0.01 | <0.001 | 0.001 |
Table 5:
Performance of DELINEATE Steatosis Pixel% (DSP%), DELINEATE Steatosis Count% (DSC%), and Aperio Steatosis Pixel%, (ASP%) for differentiation of patient groups of steatosis grades by pathologist assessment.
| Steatosis Measure | Steatosis Grade | |||
|---|---|---|---|---|
| 0 vs 1–3 | 0–1 vs 2–3 | 0–2 vs 3 | ||
| DSP% | Threshold | 1.26 | 7.34 | 11.91 |
| AUROC (95% CI) | 0.992 (0.971–1.00) | 0.977 (0.938–1.00) | 0.930 (0.851–1.00) | |
| Sensitivity | 96.80% | 91.30% | 92.30% | |
| Specificity | 100% | 100% | 82.60% | |
| Accuracy | 97% | 94% | 86% | |
| DSC% | Threshold | 0.0004145 | 0.0018375 | 0.002755 |
| AUROC (95% CI) | 0.977 (0.928–1.00) | 0.990 (0.970–1.00) | 0.983 (0.954–1.00) | |
| Sensitivity | 96.80% | 91.30% | 100% | |
| Specificity | 100% | 100% | 91.30% | |
| Accuracy | 97% | 94% | 94% | |
| ASP% | Threshold | 7.45 | 11.89 | 13.48 |
| AUROC (95% CI) | 0.922 (0.819–1.00) | 0.957 (0.883–1.00) | 0.946 (0.880–1.00) | |
| Sensitivity | 81.25% | 91.30% | 100% | |
| Specificity | 100% | 100% | 78.20% | |
| Accuracy | 83% | 94% | 86% | |
Segmentation improvement by enhanced deep learning network
DELINEATE model used a dilated version of the standard U-Net [12] model for enhanced steatosis segmentation. Dilated convolution operation can reduce the down-sampling operation and information loss, empowering the network with an exponential receptive expansion. As demonstrated in Fig. 2(A), certain steatosis regions bounded by green boxes in the middle column are missing from segmentation due to the down-sampling operations in the standard U-Net architecture. To compensate for such information loss, we incorporated dilated convolutional layers at the bottleneck block of U-Net model and managed to recover the missing steatosis regions with the dil-Unet architecture. The improved segmentation results illustrated in the right column in Fig. 2(A) well justifies the merit of the use of dilated convolutional layers.
We present in detail the way to combine the steatosis region and boundary prediction results with the third integration module in Methods Section. The top-right subfigure in Fig. 2(B) presents a typical image region where a large area (labeled by ‘1’) is falsely recognized as a steatosis component by the steatosis region module. This false positive (labeled by ‘2’) is removed from the final prediction result by the following integration module. The removal of falsely segmented regions by the third integration network further improves the overall performance of the DELINEATE model.
Whole tissue analysis and visualization
We further extended our analysis on steatosis component quantification to whole liver tissue images. A representative whole tissue image region at a low image resolution is presented in Fig. 5(A:(a)) where multiple tissue components are included. Each tissue component was extracted and rotated in a way such that the area of the resulting tissue bounding box was minimized, as illustrated in Fig. 5(A:(b)). With estimated tissue component location and rotation angle at a low image resolution, each tissue component was next extracted at the highest image resolution as depicted in Fig. 5(A:(c)). Each extracted whole tissue component at the highest resolution was further partitioned into multiple overlapping patches (512 × 512 pixels) with a buffer region of 16 pixel distance on each side by MaReIA [26] framework. Each patch was analyzed by the DELINEATE model for steatosis segmentation and the resulting steatosis prediction maps were assembled for each whole tissue component by the MaReIA framework. One representative steatosis segmentation result of a whole tissue component is presented in Fig. 5(A:(d)). Close-up views of two representative regions (purple boxes) from this tissue component are shown in Fig. 5(A :(e) and (f)), where recognized steatosis regions and boundaries are represented in red and green, respectively. These typical visual results demonstrate the efficacy of DELINEATE model for overlapped steatosis droplet segmentation.
The complete analysis procedure for each rotated whole tissue component image at the highest resolution is presented in Fig. 5(B). The overall steatosis prediction maps were assembled from patch-wise prediction maps with MaReIA. For comparisons, we extracted non-overlapping patches from each whole tissue component as illustrated in Fig. 5(C:(a)) and combined the adjacent patch prediction outputs with simple concatenation as depicted in Fig. 5(C:(b)). The end result in this way becomes much degraded as demonstrated in Fig. 5(C:(c)). Specifically, it is noticeable that steatosis components distributed to neighboring patches are not fully recovered. It is not rare to see broken, incomplete, or misaligned boundaries of steatosis droplets crossing patch borders. Some such examples are illustrated in Fig. 5(C:(c)) where problematic recovered boundaries are highlighted by rectangles. By contrast, the MaReIA framework can help assemble results in a much better way, as illustrated in Fig. 5(C:(f)).
Discussion
DELINEATE summary
Motivated by the strong feature representation capability and remarkably superior performance of the recently emerged deep neural networks, we have developed an end-to-end deep learning based model for overlapped steatosis droplet segmentation and quantification with liver whole slide histopathology images. The overall model consists of whole slide tissue extraction, color normalization, steatosis region prediction, boundary detection, integrated prediction map production, and post-processing. The developed DELINEATE model is in sharp contrast to prior work on liver steatosis quantification [9, 43, 45], as it does not require any manual feature engineering and presents promising accuracy, robustness and efficiency for delineating overlapped histology structures. Our study offers a new avenue for histology phenotype information extraction essential to clinical decision support. We provide a generic multi-layer framework that can be extended to analyses of a large set of histology structures with similar processing principles. To support analysis of extremely large-scale whole slide histopathology images, we have proposed a whole tissue component extraction method by estimating each tissue component location and rotation angle at multiple image resolution levels. For accurate measurement and better result visualization, we have also applied our assembling framework developed in our previous work to gracefully aggregate boundaries of steatosis droplets crossing neighboring image patches. We have systematically validated the efficacy of our approaches and correlated DELINEATE derived steatosis quantification measures with ground truth grading, pathologist measurements, radiology readouts, and clinical data, leading to multiple discoveries of high correlations.
The developed DELINEATE model is based on Deep Neural Network (DNN) that consists of two layers processing visual information with different focuses. The first layer has two parallel modules designed to extract multi-scaled image texture information within steatosis regions and on steatosis boundaries. The resulting visual hues from these two sources are further integrated by the third learning network that can make joint use of the perceptual information from steatosis internal regions and boundaries to delineate the missing or weak boundaries of overlapped steatosis droplets. In the post-processing step, we detect high curvature points on steatosis boundaries and assess ellipse fitting quality with identified steatosis contours to improve steatosis droplet recovery [27].
DELINEATE analysis consists of two phases, training and testing. Our DELINEATE model only needs to be trained once. After the model is trained, it can be used to analyze new cases for testing. DELINEATE model can generate patch-level prediction result in 0.26 second on average. The average number of image patches per whole-slide image is 1,600 in our dataset. Our current experiments are carried out on a machine with one CPU and one Graphical Processing Unit (GPU). We expect the execution time cost can be further significantly decreased to potentially less than a minute for each whole slide image by multiple CPUs and GPUs using parallel computing. For future clinical deployment, DELINEATE can be scheduled to run in the background and complete the analysis before a pathologist reviews tissue slides.
Clinical significance
Analysis of the steatosis extent is important for clinical care since systems that describe the severity of fatty liver disease rely on an accurate quantification of the degree of steatosis [1]. Hepatic steatosis is one of the most common incidental findings with radiology imaging, which leads to further patient evaluations in order to rule out liver diseases like NAFLD. Health implications of hepatic steatosis have been largely studied, and are associated with insulin resistance, dyslipidemia, and a chronic low-grade inflammation. Moreover, in transplantation, the graft survival is associated with the fat percentage of the graft, since hepatic steatosis increases the hepatocyte necrosis and impairs the regeneration. To date, liver biopsy remains an important information source for the assessment of fatty liver disease and other liver conditions. Steatosis droplets are accumulation of fat in liver tissues and present varying shapes even within a single tissue. Individual steatosis droplets are mostly circular in shape. However, the clumped steatosis regions have irregular shapes as the dividing boundaries for overlapped steatosis droplets are often weak or even missing. Additionally, tissue regions of interest are often selected from whole-slide images for clinical review by human pathologists. Such review bias and analysis obstacle on clumped steatosis regions limit quantification accuracy and impact the clinical decision. Our study has explored a new computational way to quantitatively measure overlapped steatosis components in liver biopsies for better clinical support of liver transplantation decision. The developed deep learning-based model DELINEATE can analyze the whole slide images in a definitive, robust, and consistent manner. As only annotated steatosis regions are marked as foreground in the training set, DELINEATE learns a large set of low level characteristics unique to the steatosis regions. Therefore, the fully trained model is able to selectively target the steatosis regions. Our proposed method DELINEATE can be a powerful addition to the current pathology analysis pipelines, as it can provide automated and precise analysis to complement standard pathological reviews.
Model optimization
We have systematically investigated multiple model variations at distinct stages and tested their performances with comprehensive comparison experiments. The first model U-Net+U-Net+U-Net consists of three U-Net modules for region extraction, boundary detection, and information integration. In addition, we have created the second model U-Net+U-Net+FCN-8s by replacing the U-Net module with FCN-8s at the integration layer. Neither of these two models produces consistently promising results evaluated by all metrics in Table 1. The third model variation is prompted by the observation that the boundary prediction results from U-Net module are incomplete in most cases. As the total number of boundary pixels is much less than that of pixels in steatosis droplet regions, it results in a numerical challenge for the network to converge in the training stage. Therefore, we further replace the boundary module in U-Net+UNet+FCN-8s with Holistically-nested Neural Network (HNN) [21, 33] for steatosis boundary prediction. The resulting network variation U-Net+HNN+FCN-8s consists of a U-Net module for steatosis region prediction, a HNN for boundary detection, and a FCN-8s integration network for region and boundary information synergy. As the HNN can effectively recover boundary information from multi-scale edge fields, U-Net+HNN+FCN-8s achieves the best overall segmentation performance. To further improve the steatosis region mask prediction, we have modified the standard U-Net module by stacking the dilated convolution layers (depth = 3) at the bottleneck block with the lowest resolution feature maps. This extended U-Net module, named as dil-Unet, presents superior performance to that of the standard U-Net, as demonstrated in Fig. 2(A). The resulting model variation with dil-Unet is named as dil-Unet+HNN+FCN-8s. In our comprehensive comparison study, we have also tested the integration network FCN-4s with an additional layer of skip connection introduced from pool2 for more detailed semantic information. The performance of this model dil-Unet+HNN+FCN-4s is comparable to that of dil-Unet+HNN+FCN-8s, as suggested in Table 1. Finally, we test with dilated FCN module dil-FCN using atrous convolution at the convolutional layer 6 with dilation rate 3 for the integration layer. The resulting model dil-Unet+HNN+dil-FCN, however, does not substantially improve recovery of the weak boundaries of steatosis droplets in clumps. All these models are rigorously evaluated by F-Score, Dice score and Hausdorff-distance, with resulting performance listed in Table 1. By comprehensive evaluations, dil-Unet+HNN+FCN-8s is the best performing model compared to all other variations. This is confirmed with visual segmentation results for overlapping steatosis droplets in Fig. 3. Both quantitative and qualitative results of baseline experiments with DELINEATE model, its network variations, and the state-of-the-art deep learning based segmentation methods demonstrate the superiority of DELINEATE model to others.
Parallel computation and result aggregation
Our study presents a generic analysis work flow to support histology image analysis result assembly and visualization for whole tissue microscopy images. As each liver whole slide histopathology image in our dataset contains several tissue needle biopsy components, we extract each whole tissue biopsy image region by estimating each tissue component location and rotation angle at multiple image resolution levels. The resolution of the resulting image region still remains overwhelmingly large. Therefore, we partition each image region into overlapping patches for analysis. The resulting steatosis contours in neighboring patches are seamlessly assembled with the MaReIA framework that gracefully handles such patch crossing steatosis droplets with efficient spatial indexing based matching and merging [26].
Discovery of a novel indicator for steatosis measurement
With DELINEATE, the resulting quantitative steatosis measures both at the pixel level and steatosis object level from whole slide liver histopathology images of 36 children with the diagnosis of NAFLD are highly correlated with liver tissue pathological grading, radiology readouts, and patient clinical data, respectively. Importantly, the count of recovered steatosis droplets, a promising novel indicator, has enhanced correlation with results from histology review on total and macrovesicular steatosis than pixel level measures. Statistical tests with DELINEATE measures at pixel and object levels between NAFL and NASH diagnostic groups, and groups with and without lobular inflammation produce statistically significant difference. In particular, steatosis object based measure demonstrates its strong discriminative power in these tests. Through statistical tests, DELINEATE steatosis count% (DSC%) is demonstrated as a new promising indicator of histology steatosis profiles predictive of hepatic steatosis grade. These strong correlations and findings of DELINEATE derived steatosis measures with other dimensions of data from the same patient cohort suggest its potential for enhanced clinical decision support.
Method benefits
This steatosis quantitation algorithm can potentially serve as an assistant in the routine steatosis assessment. As our DELINEATE method enables steatosis object level quantitation with recovered contours of individual steatosis droplets in clumps, it enables steatosis measures from the pixel to object level. Thus, DELINEATE allows for individual steatosis size measure and research investigations exploring new steatosis object level morphology features with diagnostic and prognostic value. Our developed DELINEATE method can also alleviate inter- and intra-observer variability in steatosis assessment. Once the DELINEATE model is trained, it can be used as a tool to improve agreement among pathologists and across multiple clinical sites. Furthermore, the DELINEATE method is fully automated. Leveraging parallel computation, DELINEATE model can generate and aggregate steatosis analysis results from whole slide images efficiently. We have made the DELINEATE algorithm open source, which can be modified, upgraded, and customized beyond what can be done with commercial software. Finally, our new algorithm can be readily integrated into existing open-source or commercial software already prevalent in clinical sites.
Limitations and future work
Although our study provides a new framework for quantitative liver steatosis measures, there are some limitations to be addressed in future. As manual annotations on overlapped steatosis droplets are time consuming, only very limited number of tissue regions were included in the training set. Additionally, the training set was composed with randomly selected regions of interest and thus may miss some representative steatosis droplets in clumps. As steatosis droplets have three-dimensional morphology, limiting our current analysis to sampled two-dimensional tissue slides may result in large measure errors and sampling bias. In future study, we will incorporate more annotated tissue samples and explore methods that can identify representative tissue regions for training. For more precise steatosis measure, we plan to extend our analysis from two to three-dimensional tissue space with serial liver slides. In this study, we segment overlapping steatosis droplets in liver tissues. As our proposed deep learning method is generic to histology objects, we will extend our method to segment other overlapping histopathology components that can provide more insights on disease tissues. To better support clinical liver disease diagnosis, we will proceed with measuring morphometric and space organizational features of segmented steatosis droplets. We expect the resulting feature distributions derived from whole slide tissues can be used to indicate liver disease severity quantitatively.
As we use permanent sections of a patient cohort in this study, we do not train the DELINEATE model with frozen section slides enriched with image artifacts. In our future studies, we plan to enhance our method to automatically recognize frozen section artifact with similar appearance to steatosis droplets. Additionally, we plan to extend our analysis to accommodate other stains such as Periodic Acid Schiff (PAS) stains with and without diastase for glycogenation assessment. As permanent sections in this investigation do not present strong microvesicular steatosis components, this study is not designed to detect the microvesicular steatosis. In our follow up study, we will collect microvesicular steatosis cases for model training. We plan to construct steatosis droplet size histograms to determine the optimal cutoff size for microvesicular steatosis, and link this with such outcome parameter as delayed graft function after transplantation.
Supplementary Material
Acknowledgements
This research is supported in part by grants from National Institute of Health 7K25CA181503, 1U01CA242936, 5R01EY028450, and National Science Foundation ACI 1443054 and IIS 1350885.
Footnotes
Author Declaration There is no conflict of interest to report.
Supplementary Information
Supplementary information is available at Laboratory Investigation’s website.
References
- 1.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–71. Epub 2010/01/19. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 2.Brunt EM. Nonalcoholic Fatty Liver Disease: Pros and Cons of Histologic Systems of Evaluation. Int J Mol Sci. 2016;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun HA, Faasse SA, Vos MB. Advances in Pediatric Fatty Liver Disease: Pathogenesis, Diagnosis, and Treatment. Gastroenterology clinics of North America. 2018;47(4):949–68. [DOI] [PubMed] [Google Scholar]
- 4.French MCSG, Bedossa P. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology. 1994;20(1):15–20. [PubMed] [Google Scholar]
- 5.Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Transactions on Biomedical Engineering. 2009;57(4):841–52. [DOI] [PubMed] [Google Scholar]
- 6.Xing F, Xie Y, Yang L. An automatic learning-based framework for robust nucleus segmentation. IEEE transactions on medical imaging. 2015;35(2):550–66. [DOI] [PubMed] [Google Scholar]
- 7.Irshad H, Veillard A, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: a review— current status and future potential. IEEE reviews in biomedical engineering. 2013;7:97–114. [DOI] [PubMed] [Google Scholar]
- 8.Farris AB, Cohen C, Rogers TE, Smith GH. Whole slide imaging for analytical anatomic pathology and telepathology: practical applications today, promises, and perils. Archives of pathology & laboratory medicine. 2017;141(4):542–50. [DOI] [PubMed] [Google Scholar]
- 9.Arnould LCJ, Rondelez L, Courtoy P. Automated morphometric estimation of liver steatosis. Microsc Acta Suppl. 1979;(3):197–203. [PubMed] [Google Scholar]
- 10.Krizhevsky A, Ilya Sutskever, and Geoffrey E. Hinton. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012:1097–105. [Google Scholar]
- 11.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. International Conference on Learning Representations. 2015. https://arxiv.org/abs/1409.1556. [Google Scholar]
- 12.Ronneberger O, Philipp Fischer, and Thomas Brox. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention: Springer, Cham; 2015. p. 234–41. [Google Scholar]
- 13.Dimitriou N, Arandjelovic O, Caie PD. Deep Learning for Whole Slide Image Analysis: An Overview. Frontiers in Medicine. 2019;6. doi: 10.3389/fmed.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan X, Li L, Yang H, Liu Z, Yang J, Zhao L, et al. Accurate segmentation of nuclei in pathological images via sparse reconstruction and deep convolutional networks. Neurocomputing. 2017;229:88–99. [Google Scholar]
- 15.Li W, Manivannan S, Akbar S, Zhang J, Trucco E, McKenna SJ. Gland segmentation in colon histology images using hand-crafted features and convolutional neural networks. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI): IEEE; 2016. p. 1405–8. [Google Scholar]
- 16.Xu Y, Li Y, Wang Y, Liu M, Fan Y, Lai M, et al. Gland instance segmentation using deep multichannel neural networks. IEEE Transactions on Biomedical Engineering. 2017;64(12):2901–12. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Li Y, Liu M, Wang Y, Lai M, Eric I, et al. Gland instance segmentation by deep multichannel side supervision In International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer, Cham; 2016. p. 496–504. [Google Scholar]
- 18.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition. 2015:3431–40. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Qi X, Yu L, Dou Q, Qin J, Heng P-A. DCAN: Deep contour-aware networks for object instance segmentation from histology images. Medical image analysis. 2017;36:135–46. [DOI] [PubMed] [Google Scholar]
- 20.Cui Y, Zhang G, Liu Z, Xiong Z, Hu J. A deep learning algorithm for one-step contour aware nuclei segmentation of histopathology images. Medical & biological engineering & computing. 2019;57(9):2027–43. [DOI] [PubMed] [Google Scholar]
- 21.Roth HR, Lu L, Farag A, Sohn A, Summers RM. Spatial aggregation of holistically-nested networks for automated pancreas segmentation In International conference on medical image computing and computer-assisted intervention: Springer, Cham; 2016. p. 451–9. [Google Scholar]
- 22.Khoshdeli M, Parvin B. Deep Learning Models Delineates Multiple Nuclear Phenotypes in H&E Stained Histology Sections. ArXiv. 2018;abs/1802.04427. https://arxiv.org/abs/1802.04427. [DOI] [PubMed] [Google Scholar]
- 23.Kong J, Lee MJ, Bagci P, Sharma P, Martin D, Adsay NV, et al. Computer-based image analysis of liver steatosis with large-scale microscopy imagery and correlation with magnetic resonance imaging lipid analysis. In 2011 ieee international conference on bioinformatics and biomedicine: IEEE; 2011. p. 333–8. [Google Scholar]
- 24.Otsu N A threshold selection method from gray-level histograms. IEEE transactions on systems, man, and cybernetics. 1979;9(1):62–6. [Google Scholar]
- 25.Altunbay D, Cigir C, Sokmensuer C, Gunduz-Demir C. Color graphs for automated cancer diagnosis and grading. IEEE Transactions on Biomedical Engineering. 2009;57(3):665–74. [DOI] [PubMed] [Google Scholar]
- 26.Vo H, Kong J, Teng D, Liang Y, Aji A, Teodoro G, et al. MaReIA: a cloud MapReduce based high performance whole slide image analysis framework. Distributed and Parallel Databases. 2019;37(2):251–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy M, Wang F, Teodoro G, Vos MB, Farris AB, Kong J. Segmentation of overlapped steatosis in whole-slide liver histopathology microscopy images. In 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC): IEEE; 2018. p. 810–3. [DOI] [PubMed] [Google Scholar]
- 28.Goode A, Gilbert B, Harkes J, Jukic D, Satyanarayanan M. OpenSlide: A vendor- neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Computer graphics and applications. 2001;21(5):34–41. [Google Scholar]
- 30.Yu F, Koltun V. Multi-Scale Context Aggregation by Dilated Convolutions International Conference on Learning Representations (ICLR) 2016. https://arxiv.org/abs/1511.07122. [Google Scholar]
- 31.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the thirteenth international conference on artificial intelligence and statistics. 2010:249–56. [Google Scholar]
- 32.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization 3rd International Conference for Learning Representations 2015. http://arxiv.org/abs/1412.6980. [Google Scholar]
- 33.Xie S, Tu Z. Holistically-nested edge detection. In Proceedings of the IEEE international conference on computer vision. 2015:1395–403. [Google Scholar]
- 34.Wen Q, Chang H, Parvin B. A Delaunay triangulation approach for segmenting clumps of nuclei. In 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro: IEEE; 2009. p. 9–12. [Google Scholar]
- 35.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: A system for large-scale machine learning. In 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16). 2016:265–83.
- 36.Keras. Available from: https://keras.io.
- 37.Roy M, Wang F, Vo H, Teodoro G, Farris AB, Lion EC, et al. Data from “DELINEATE” Github. 2020. Available from: https://github.com/StonyBrookDB/DELINEATE.
- 38.Roy M Steatosis_Dataset_Raw 2020. Available from: 10.6084/m9.figshare.12439886.v1. [DOI]
- 39.Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. Journal of Machine Learning Technologies. 2011;Volume 2(Issue 1):37–63. [Google Scholar]
- 40.Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence. 2017;40(4):834–48. [DOI] [PubMed] [Google Scholar]
- 41.Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Transactions on pattern analysis and machine intelligence. 1993;15(9):850–63. [Google Scholar]
- 42.Leica. Available from: https://www.leicabiosystems.com/digital-pathology/analyze/ihc/aperio-positive-pixel-count-algorithm/. [Google Scholar]
- 43.Lee MJ, Bagci P, Kong J, Vos MB, Sharma P, Kalb B, et al. Liver steatosis assessment: correlations among pathology, radiology, clinical data and automated image analysis software. Pathology-Research and Practice. 2013;209(6):371–9. [DOI] [PubMed] [Google Scholar]
- 44.NASH CRN. Available from: https://jhuccs1.us/nash/default.asp. [Google Scholar]
- 45.Liquori GE, Calamita G, Cascella D, Mastrodonato M, Portincasa P, Ferri D. An innovative methodology for the automated morphometric and quantitative estimation of liver steatosis. Histol Histopathol. 2009;24(1):49–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source codes and annotation data related to this paper are available at GitHub [37]. We share image data in a public repository [38].


