Abstract
The Bone Morphogenetic Proteins (BMPs) are the largest class signaling molecules within the greater Transforming Growth Factor Beta (TGFβ) family, and are responsible for a wide array of biological functions, including dorsal-ventral patterning, skeletal development and maintenance, as well as cell homeostasis. As such, dysregulation of BMPs results in a number of diseases, including fibrodysplasia ossificans progressiva (FOP) and pulmonary arterial hypertension (PAH). Therefore, understanding BMP signaling and regulation at the molecular level is essential for targeted therapeutic intervention. This review discusses the recent advances in the structural and biochemical characterization of BMPs, from canonical ligand-receptor interactions to co-receptors and antagonists. This work aims to highlight how BMPs differ from other members of the TGFβ family, and how that information can be used to further advance the field. Lastly, this review discusses several gaps in the current understanding of BMP structures, with the aim that discussion of these gaps will lead to advancements in the field.
Keywords: BMP, Structure, Review
Introduction
The Bone Morphogenetic Proteins (BMPs), comprise the largest subgroup of the Transforming Growth Factor β (TGFβ) family of extracellular signaling proteins, consisting of anywhere from 13–20 dimeric proteins [1–5]. Like other TGFβ proteins, BMPs canonically induce signaling by binding to two type I and two type II transmembrane serine-threonine kinase receptors forming heterocomplexes on the cell surface [4–8]. These receptor assemblies, sometimes formed with additional co-receptors, induce the phosphorylation and activation of the type I receptor kinase domain by the type II receptor kinase domain [9–11]. Subsequently, phosphorylation of intracellular SMAD proteins by the type I receptor allows the SMADs to accumulate in the nucleus where they serve as transcription factors [11–15]. Historically, structural studies have been instrumental in understanding of BMP biology by providing the foundation for the mechanistic understanding of growth factor function. While BMP structural biology has been exceedingly well reviewed in the past within the context of the TGFβ family as a whole, this review is meant to focus exclusively on the BMPs, with a special focus on recent structural studies [4–5, 11].
BMP Signaling Ligands
What distinguishes the BMPs from other members of the TGFβ family? The term “BMP class” has at times been used to refer to all named BMPs and Growth and Differentiation Factors (GDFs). However, some of these proteins more closely resemble members of the Activin class of TGFβ ligands (GDF8, and GDF11 for example) in terms of sequence identity, receptor activation and specific SMAD activation [16–17]. As such, a better way to define what a BMP is might be to limit the category to proteins that preferentially signal through a specific set of type I receptors (ACVRL1 {Alk1}, ACVR1 {Alk2}, BMPR1A {Alk3}, and BMPR1B {Alk6}) that ultimately activate transcription through the phosphorylation of SMADs 1, 5 and 9 [4–5, 18–19]. This is distinct from the SMAD2/3 activation that occurs through the type 1 receptors ACVR1B (Alk4), TGFBR1 (Alk5), and ACVR1C (Alk7), which is characteristic of signaling by members of the Activin and TGFβ classes [4–5, 18–19]. However, ligands such as activin A and B through Alk2 and TGFβ through Alk1, possess some situational ability to activate SMAD1/5/9 signaling [20–22]. However, these other proteins generally activate SMAD 1/5/9 in rare situations, leaving us to focus on those TGFβ growth factors that primarily utilize SMAD1/5/9. Of this select group of 13 proteins, crystal structures of six (BMP2, BMP6, BMP7, GDF5, BMP9 and BMP10) have been solved (Fig 1A), either in their unbound state, in complex with binding partners, or both [23–48].
Figure 1.
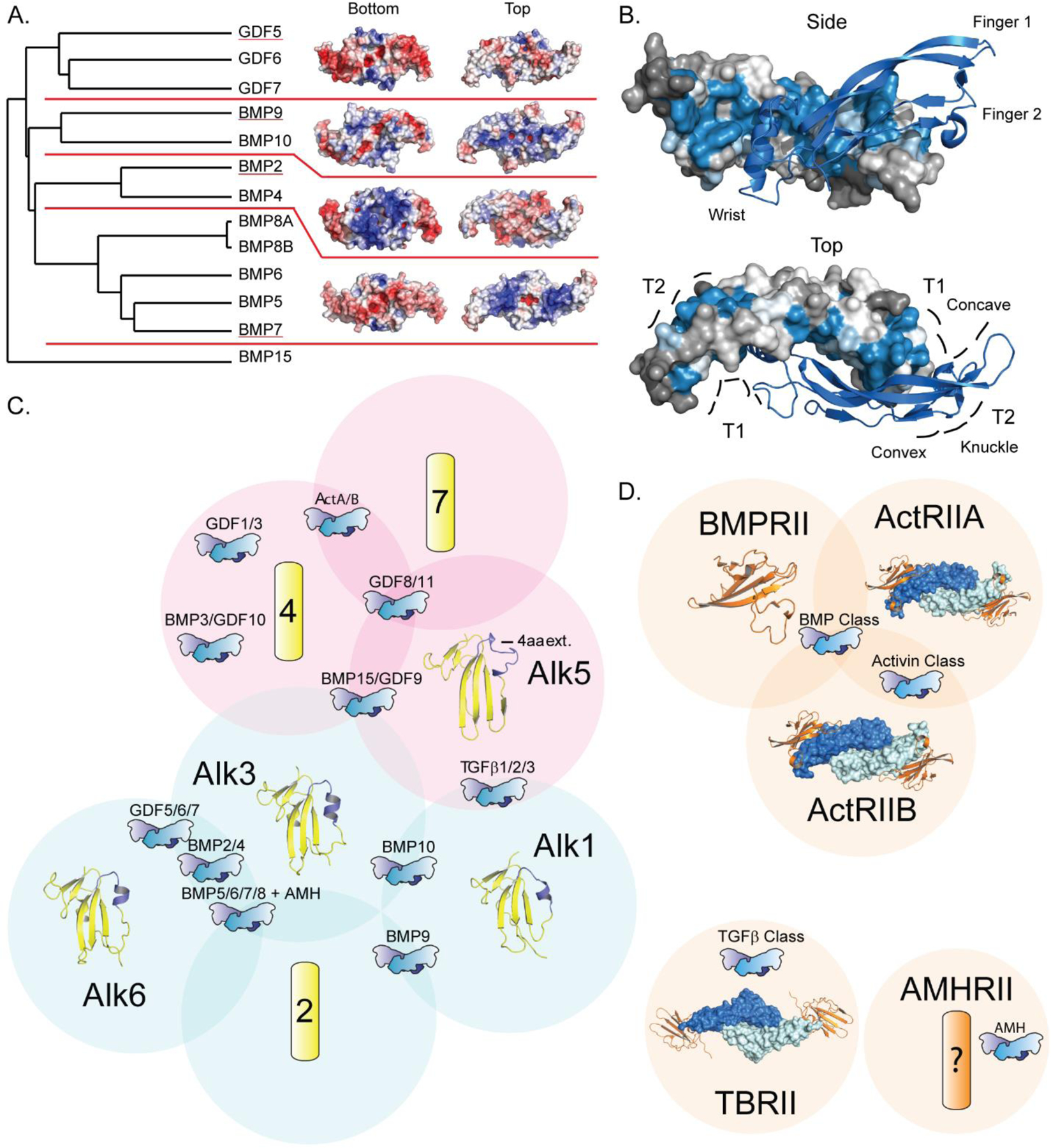
BMP ligand and receptor structures. A) Phylogenetic tree of BMP ligands, separated into subgroups by red lines and electrostatic surfaces of representative structures (underlined) from each available subgroup. Electrostatic surface calculated using APBS Tools (negative potential - colored red, positive potential - colored blue; range from −5 to +5 kbt/ec) [78]. Structure PDB codes: GDF5 (5HK5), BMP9 (4MLP), BMP2 (3BMP), BMP7 (1LXI) [44, 35, 26, 33]. B) BMP ligand residue variance, measured using the ConSurf server, mapped to BMP2 in ribbon (marine) and surface representation (colored based on variance, blue for areas of conservation and gray for areas of high variability) [60–64]. Receptor binding sites for type I (T1) and type II (T2), and structural features are labeled. Structure PDB codes: BMP2 (6OMN) [30]. C) & D) Venn diagram depicting type I (C, yellow) and type II receptor specificity (D, orange) with groups of ligands and structurally unresolved receptors represented schematically. Type I receptor β4-β5 variable loop helix region highlighted in blue. Structure PDB codes: Alk1 (6SF2), Alk3 (2H62), Alk5 (6MAC), Alk6 (3EVS), BMPRII (2HLQ), ActRIIA:BMP2 (2GOO), ActRIIB:BMP9 (4FAO), TBRII:TGFB1 (3KFD) [39, 48, 91, 43, 204, 8, 36, 90].
All BMPs, like other TGFβ family members, are initially produced as a single chain containing a larger N-terminal prodomain and a smaller C-terminal mature signaling domain [4–5]. With a few exceptions, two signaling domains are joined by a disulfide bond with another to form a covalently bound dimer, whereby the prodomains are proteolyzed leaving a mature signaling molecule [49–50]. Classically portrayed as a hand, each chain consists of two sets of anti-parallel β-strands forming finger-like extensions that protrude from a central stabilizing “wrist” α-helix. The structure is stabilized by a conserved cysteine knot formed by three internal disulfide bonds, similar to other growth factors (Fig 1B). All member of the TGFβ family, bind the type I receptors at a composite binding interface formed by the wrist helix of one monomer and the concave face of the finger extensions of the other monomer (Fig 1B) [4–5, 47]. For BMP and Activin class ligands, the type II receptors bind to the convex “knuckle” surface of the finger extensions (Fig 1B) [47]. However, BMPs are distinct from other TGFβ proteins in that they have a higher affinity for the type I receptors than the type II receptors [5, 18, 48, 51]. Additionally, the BMP structures are almost invariant in their adoption of an open or butterfly-like structure, indicating that they are more rigid than other family members [31, 34, 41, 47, 51]. Activin and TGFβ class crystal structures have captured these proteins in many different dimeric orientations [51–58]. The static nature of BMP ligands leads to a rigid binding pocket for the type I receptors, and likely facilitates the high affinity interactions [5, 18, 48, 51].
BMP ligands display a high degree of gross morphological similarity, with a root-mean squared deviation (RMSD) analysis of the Cα atomic positions, performed using the Superpose program in the CCP4 suite, ranging from 1–2 Å [59]. When currently solved structures are analyzed for structural and sequential variations using the ConSurf server, areas of high variability (gray) and high conservation (blue) can be easily visualized [60–64]. The regions of highest variability occur at the fingertip region, which, from receptor-ligand complex structures, aligns with the regions of the type I receptors shown to impart specificity (Fig 1B) [47]. Additionally, there are several small patches of variation in the type 1 receptor binding region, highlighting the minor differences that govern receptor specificity.
The BMP ligands can be subdivided into five different subcategories or clades based on sequence identity (Fig 1A) and receptor specificity (Fig 1C). (1) BMP2/4 signal principally through Alk3 and Alk6 [4–5, 23, 65–67]. (2) BMP5/6/7/8a/8b form a phylogenetically distinct and somewhat more promiscuous subgroup, signaling robustly through Alk2, Alk3, and Alk6 [4–5, 8, 65, 68]. In addition, this subclass is notable for a 21–22 amino acid extension of the N-terminus. (3) The GDF5/6/7 subgroup signals almost exclusively through Alk6, with a lesser affinity for Alk3 [5, 40, 65]. (4) The BMP9/10 subgroup signals principally through Alk1, although BMP9 possesses some affinity for Alk2 and BMP10 for Alk3 [5, 39, 65, 69]. Additionally, BMP9/10 have a truncated N-terminal region, ~10 amino acids less than BMP2/4 and GDF5/6/7, and over 30 less than BMP5/6/7/8. (5) BMP15 is something of an outlier in that while it signals through Alk3 and Alk6, it can form non-disulfide linked heterodimers with GDF9, an Activin class protein that signals through Alk4/5/7 to induce SMAD2/3 [5, 65, 70–71].
One important aspect of BMP biology is their differential interactions with components of the extracellular matrix (ECM), particularly glycosaminoglycan heparan sulfate (HS), which can localize and orient these signaling molecules to the cell surface [72–75]. HS is known to interact preferentially with highly basic, positively charged residues [76–77]. Interestingly, an electrostatic surface representation, calculated using APBS Tools v2.1, shows striking differences between different BMP subgroups (Fig 1A) [78]. BMP2 contains a patch of highly positive amino acid residues on the top of the protein (defined relative to the cell surface when bound to receptors) while BMP7 and BMP9 have (stronger and weaker, respectively) positive patches located on the bottom of the protein, and GDF5 contains no positively charged surface region [78].
Besides helping to localize BMP ligands to the cells surface, HS has been shown to be necessary for robust signaling, as is the case with BMP7 [79]. It is possible that HS helps position BMP ligands for interaction with their target receptors. While the exact HS binding site of BMP7 is unknown, it is thought to be localized on the C-terminal region of the protein [80]. In contrast, HS is known to strongly associate with the N-terminal region of BMP2 [75, 80–81]. HS has also been shown to increase the duration of BMP2 signaling, potentially by regulating turnover of the signaling complex [82–84]. While a HS binding site has been predicted for GDF5, the protein displays little affinity for HS, as consistent with it lack of positive surface potential [75, 85]. However, it has been shown to interact with the related molecule heparin, a highly sulfated variant of HS produced in connective tissue [85]. While it is clear that HS impacts BMP signaling, the complex nature of HS, including chain length, branching and sulfonation patterns makes it difficult to pin down its mechanism(s) of action. More research is needed to understand how HS impacts the affinity of BMP for receptors, prodomain and extracellular antagonists.
Historically, most research on BMP biology has concentrated on homodimeric proteins. However, recent evidence shows that heterodimers plan a major role in BMP biology [69–70, 86–88]. Heterodimeric proteins are novel signaling molecules with altered binding affinities for BMP receptors. BMP7 has been shown to function in development largely as a heterodimer with BMP2 and BMP4 [86]. The BMP2/BMP7, BMP4/BMP7, and BMP2/BMP6 heterodimers have increased signaling activity as compared to homodimer [87–88]. This increase has been ascribed to a relatively higher affinity for type I receptors by BMP2 and BMP4 and a relatively higher affinity for type II receptors by BMP6 and BMP7, generating a hybrid signaling molecule with higher total receptor affinity [87]. Additionally, BMP9/BMP10 heterodimers have been shown to be the primary source of BMP signaling in plasma in vivo, potentially driven by increased binding affinity to different type I receptors, as both proteins signal through Alk1, but BMP9 will also signal through Alk2 and BMP10 through Alk3 [69]. While this area of study is intensely interesting and seemingly of pronounced biological importance, no structure of a BMP heterodimer has been published. Additionally, very few heterodimers have been studied in any detail, leaving open a wide array of questions regarding further unique receptor assemblies, altered antagonist binding, differential heparin interactions, and even which BMPs can form heterodimers.
BMP Receptor Assemblies
Over the last two decades, significant structural insight has been provided, detailing how TGFβ ligands assemble their cognate type I and type II receptors. This work has shown that across the three classes of ligands the mechanisms of receptor assembly differ significantly. Structures within each class have provided insight not only into the different interfaces utilized by each class, but also to the underpinnings of receptor specificity – this is especially of interest since the number of ligands far outweigh the number of receptors. Comparing the structures of the complete signaling complex of each class reveals a consistent type I binding interface, one that is composite and built from interfaces on both ligand monomers (Fig 1B). This is in contrast to type II receptor binding, where positional differences differentiate the TGFβ class from the activins and BMPs [51]. Specifically, TBRII binds at the distal fingertips of the ligand, on its side, while several structures of both activins and BMP complexes show binding of the type II receptor on the convex surface of a single monomer, opposite of the type I interface (Fig 1B, 2A) [8, 89–92]. TBRII binding in this manner contributes an additional interface for type I receptor binding, allowing for a cooperative binding relationship between the receptors not observed in BMP or activin complexes [51, 89, 91].
Figure 2.
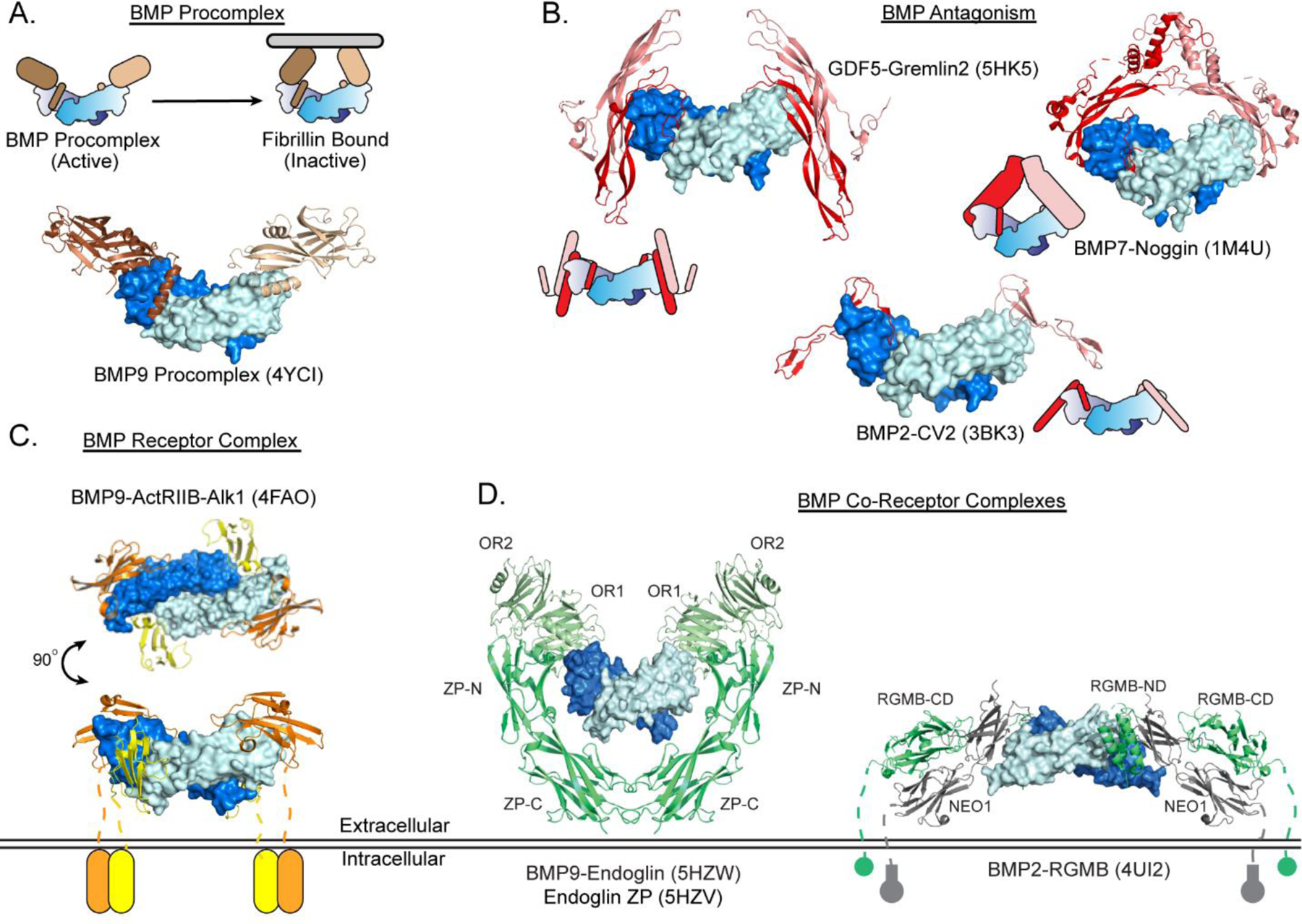
BMP complex structures. A) BMP procomplex containing BMP9 bound to its prodomain. Structure PDB code: proBMP9 (4YCI) [38]. B) BMP antagonist complex structures of GDF5 (marine and cyan) bound to Gremlin2 (red and pink), BMP7 (marine and cyan) bound to Noggin (red and pink) and BMP2 (marine and cyan) bound to CV2 (red and pink). Structure PDB codes: GDF5:Grem2 (5HK5), BMP7:Noggin (1M4U), BMP2:CV2 (3BK3) [44, 32, 25]. C) BMP ternary receptor complex of BMP9 (marine and cyan) bound to ActRIIB (orange) and Alk1 (yellow). Structure PDB code: BMP9:ActRIIB:Alk1 (4FAO) [36]. D) BMP co-receptor complex of BMP9 (marine and cyan) and Endoglin OR domain (pale green) modeled with structure of Endoglin ZP domain (green). BMP co-receptor complex of RGMB (green) bound to BMP2 (marine and cyan) and NEO1 (gray). Structure PDB codes: BMP9:ENDOR (5HZW), ENDZP (5HZV), BMP2:RGMB (4UI2) [37, 45].
Of the five type II receptors available, the BMP class has been shown to interact with three: BMPR2 (BMPRII), ACVR2A (ActRIIA) and ACVR2B (ActRIIB), and generally with lower affinity when compared to the Activin class (Fig 1D) [4–5, 93]. Receptor complex structures of both ActRIIA and ActRIIB with BMP and activin ligands reveal a common mechanism of ligand engagement: a hydrophobic core of type II receptor residues (ie. Tyr42, Trp60, and Phe83 in ActRIIB), which is complemented by a corresponding patch on the knuckle of the ligand (ie. dual Leu residues in BMP2 or an Ile-Ala-Pro motif in activin A) [48, 94]. These similar hydrophobic contacts largely form the basis for the promiscuity of the type II receptors, which rely on these small patches for binding [33, 36, 55, 92, 95]. In fact, specificity for BMP:type II interactions has been shown to arise from the residues surrounding the central interface, where hydrogen bond count is variable and accounts for the affinity differences in ActRIIB between activins (low) and the BMPs (high) [48]. This was shown by Mueller‟s and Nickel‟s groups, where following the introduction of hydrogen bonding amino acids from activin A, BMP2 was converted into a high affinity binder of ActRIIB [48]. This supports the idea that residues outside the hydrophobic core dictate type II receptor binding affinity and specificity. There is no complex structure of BMPRII currently, likely due to its low affinity to BMP ligands.
Unlike the relatively broad specificity of the type II receptors, ActRIIA/B and BMPRII, type I receptors display narrow ranges of promiscuity, interacting with only a subset of the BMP ligands available. This specificity is highlighted here (Fig 1C) and has been reviewed previously [5, 19]. But what separates the seven type I receptors on the molecular level? Structural and biochemical studies of the type I receptors reveal that the loops flanking the three stranded β-sheet core display the largest degree of variation and play large roles in specificity (Fig 1C) [36, 39, 43, 46–47]. The β4-β5 loop of the receptor is positioned toward the finger of one ligand monomer with a short α-helix presented for binding. In fact, in an unbound state, this α-helix of both Alk3 and Alk6 exhibits a large degree of flexibility, becoming ordered upon ligand or antibody binding [42–43, 96]. Furthermore, alanine screening of this region in Alk3 significantly decreased the binding affinity [97].
Structures of both Alk3 and Alk6 show that a single residue in the helix forms a “knob-in-hole”, which anchors the type I ligand interface [43, 46–47, 98]. Here, a Phe from the β4-β5 loop is inserted into a hydrophobic pocket within the ligand dimer interface. Interestingly, the anchoring Phe is conserved in all seven type I receptors, save for Alk1. Recently, the knob-in-hole was shown to be necessary for Alk5 signaling by Activin class member, GDF11, but not for TGFB1, another Alk1 interacting ligand [91, 99]. Interestingly, despite lacking a knob in hole, Alk1 interacts strongly with the BMP9 and BMP10 fingertip 2, where hydrogen bonds and a salt bridge are formed with the bottom of the β4-β5 loop [39].
One major difference between BMP and activin type I receptor binding occurs through contact with the β4-β5 loop. Both Alk4 and Alk5 possess a four amino acid extension in the β4-β5 loop that generates specificity for the Activin class through interaction with ligand fingertip 1, a trend not observed in structures containing Alk1, Alk3 or Alk6 [90]. Biochemical studies performed by Aykul et al. extended this observation to Alk2, and show its importance in binding ActA [100]. However, BMP9 also might use a similar interaction as mutation (Asp366Glu) in fingertip 2 abolished BMP9 binding to Alk2 [39]. Thus, ligand interactions with the β4-β5 loop are important for defining receptor specificity, especially for Alk2 and the activin type I receptors.
Another major difference is that the BMP receptors interact extensively with the wrist region helix and connecting loops (Fig 1B, 2A). The rigid nature of the BMP dimers supports a lock-and-key binding mechanism where the type I binding site is, for the most part, preformed [5, 48, 101]. This contrasts with the activin ligands, which have reduced type I affinity due to a highly flexible wrist region [51, 55]. Domain swapping studies have shown that replacement of the prehelix region can alter type I receptor specificity, consistent with the extensive interactions observed in the crystal structures [102]. The importance of the wrist region in type I receptor binding for the BMP ligands has been demonstrated where a single residue can result in specificity differences. In GDF5, an Arg residue in the prehelix was shown to be important for limiting interaction with Alk6 [40]. Mutation of this residue to an Ala allowed GDF5 to bind and signal much more robustly through Alk3 [102]. Further studies show that Alk2 binding to BMP6 is dependent on the glycosylation state of the wrist helix [29]. Certainly the significance of these interactions is shown where a single point mutation (Leu51Pro) in BMP2 prevents interaction with the prehelix region of Alk3, thereby ablating signaling [23]. Collectively, these studies highlight the prehelix region of the type 1 receptors as a specificity hot zone.
Since the type I receptor binding location is at the composite interface, it is interesting to speculate that heterodimers will display altered type I binding interactions. While the wrist region of one monomer might drive specificity, additional interactions through the fingertips might alter affinity of the receptors. Furthermore, it should be pointed out that within a heterodimer, each type I site is unique (i.e. AtipBwrist; BtipAwrist). For example, the heterodimer cumulin, formed by combining BMP15 with GDF9, forms a receptor complex with two different type I receptors, Alk4 and Alk6 [70]. Future structural studies will be needed to address receptor binding differences to heterodimers.
Co-Receptors of BMP Signaling
BMP ligands share a small subset of receptors, but despite this limitation can produce a wide range of signals. Additional molecules, such as co-receptors increase the diversity of ligand-receptor assembly and provide further cellular regulation. Unlike the type I and type II receptors, co-receptors (sometimes referred to as type III receptors) do not produce a signal directly, but rather promote ligand docking on the cell surface, regulating specificity and likely controlling signaling kinetics [103]. While recent reviews highlighted aspects of co-receptor biology, the following will focus on membrane-bound co-receptors that have been linked to BMP signaling modulation [103].
Endoglin (ENG) and Betaglycan (BG) are membrane glycoproteins that are comprised of N-terminal orphan domains (OR1 and OR2) and C-terminal zona pellucida (ZP-N and ZP-C) domains [104]. Expressed in endothelial cells, ENG functions to modulate BMP9 and BMP10 signaling by interacting with Alk1 [105]. ENG can also modulates the activity of other TGFβ ligands by altering receptor affinity [105–106]. Molecular evidence for how ENG engages BMP9 was provided by the recently solved crystal structure of the complex of BMP9 bound to the OR domains [37]. Here, the OR domains, which are composed of a tandem β-sandwich, bind at the knuckle region of BMP9 through hydrophobic contacts with the OR1 domain while OR2 does not directly interact with the BMP dimer [37]. From this structure, a model of ENG binding to BMP9 was built including the proximal ZP domain, to highlight the symmetrical 2:1 ENG:ligand dimer complex (Fig 2D). This is in contrast to BG which binds ligands in a 1:1 stoichiometry, partially blocking the type I receptor binding site [107–109]. In general, ENG competes for the type II receptor epitope while allowing type I receptor binding [37]. The proposed mechanism suggests that ENG helps to recruit the ligand-type I receptor complex to the cell surface, and is then released, allowing the complex to recruit cognate type II receptors [37]. A complex structure of a ligand:type I receptor:co-receptor ternary complex would greatly enhance our molecular understanding of this mechanism.
Repulsive guidance molecules (RGM) are glycosylphosphatidylinositol (GPI) -anchored receptors that have been found to increase cell sensitivity to low levels of BMP ligands, acting as bona-fide co-receptors [110–111]. RGMC (hemojuvelin) binds BMP6 to induce hepcidin expression in the liver to regulate iron homeostasis, whereas RGMA and RGMB enhance BMP2/4 signaling to enhance bone formation [112–114]. RGMs are divided into two functional components, RGMND and RGMCD. RGMCD binds the cell-surface receptor Neoginin (NEO1) while RGMND binds BMP ligands within the ligand dimer interface (Fig 2D) [115]. RGMND adopts a helical structure that binds in the same location on BMP as the BMP type I receptors, raising the question as to how the co-receptor potentiates BMP signaling [44]. One thought is that low pH, such as that found in the endosome, allows RGM to dissociate, allowing the type I receptor to bind [45]. The binary nature of RGM allows it to bridge BMP2 and NEO1 forming a 2:2:2 stoichiometric complex forming higher order cell surface complexes [45]. However, much remains to be determined as to the functional consequence of this complex interaction and whether an alternative mechanism could also explain RGM‟s ability to potentiate BMP signaling. One possibility is that BMP ligands can simultaneously bind one RGM and one type I receptor, where the resulting complex might have different signaling kinetics due to altered receptor availability. However, further investigation is required to establish the mechanism of RGM-mediated enhancement of BMP signaling.
As BMP ligands have evolved to interact with numerous protein binding partners, it is not surprising that they have been shown to engage other signaling receptors. Besides being involved in their own signaling cascades, certain Receptor Tyrosine Kinases (RTK) have been shown to interact with BMP ligands and receptors. Recently, the RTK Muscle-specific kinase (MuSK) which consists of three immunoglobulin-like domains (Ig1–3) followed by a Frizzled-like CRD domain, was shown to increase BMP signaling [116–119]. This co-receptor activity did not require MuSK to have kinase activity [119]. The study showed that the Ig3 domain of MuSK binds BMP. Interestingly, MuSK was also shown to interact with the type I receptors Alk3 and Alk6 and activin type I receptor Alk4 [120]. The details of how MuSK interacts with BMP ligands and receptors has not been determined.
Receptor tyrosine kinase-like orphan receptors, ROR1 and ROR2, are closely related to MuSK with similar domain architecture apart from an extracellular Kringle domain [121–122]. ROR2 was found to modulate GDF5/Alk6 signaling [123]. ROR2 is specifically trans-phosphorylated by Alk6, independent of BMP ligands, and interacts via the Frizzled-like CRD [123–124]. Further biochemical analysis supports that ROR2 competes with BMPRII for Alk6 binding acting as a negative modulator of the BMP/SMAD pathway [124]. However, beyond these initial studies, limited research has been directed towards uncovering the molecular mechanism for how ROR receptors bind BMP ligands or their cognate receptors.
Prodomain-Ligand Interactions
As mentioned, all TGFβ family ligands are synthesized as precursors composed of an N-terminal prodomain and a C-terminal mature signaling domain or ligand. The prodomain is essential for proper folding and dimeric assembly of the ligand and is cleaved from the mature domain by proprotein convertases (PCs) such as Furin [49–50]. After cleavage, the prodomain often remains non-covalently bound forming a procomplex. Ligand prodomains are more divergent in function and fold than the mature domain and have gained significant attention with respect to their structures and how they regulate ligand signaling [4, 38]. In addition to folding, the prodomains are multi-functional and have roles in ligand latency, localization, ligand stability/bioavailability and homo- and heterodimer formation [50, 86, 125–129]. Given their essential role, it is not surprising that a variety of diseases have been associated with prodomain mutations. For example, cleft palate can be caused by a mutation within the GDF11 prodomain preventing processing by PCs and mutations in the BMP9 prodomain are associated with hereditary hemorrhagic telangiectasia (HHT) [130–131]. While in certain cases, such as for the TGFβ class and Activin class ligands GDF8 and GDF11, the prodomain renders the ligand latent, generally for BMPs the prodomain is thought to be readily displaced by receptor binding and does not interfere with signaling [38, 127–129, 132].
In the last decade, high-resolution structural studies have started to reveal prodomain-ligand interactions. Included are crystallographic structures of TGFβ1 (3RJR), GDF8 (5NTU), activin A (5HLY), and BMP9 (4YCI) shown in Figure 2 [38, 127–128, 132]. These studies have established the general structural features (and terminology) of the prodomain, and have also revealed differences in how they bind ligands. A helix at the N-terminus, termed α1, is highly conserved and is typically positioned at the dimer interface, in a position similar to where the type I receptor binds. For latent complexes, a “latency lasso” wraps around the tip of the ligand fingers to the convex knuckle region, followed by the α2 helix and the rest of the β-structure. The overall architecture of the prodomain can be found in in a “closed” or ring-like state (TGFβ1) or in an “open” conformation where the two prodomains are splayed in a V-conformation (Fig 2A).
BMP9 bound to its prodomain is the only available crystal structure of a BMP procomplex (Fig 2A) [38]. Overall, the BMP9 procomplex was shown to adopt an “open” conformation. Binding data suggested the prodomain of BMP9 and BMP7 interfered with the type II receptor binding, while type I binding was not affected [38, 133]. The structure supported these observations where the type II receptor site was blocked through interactions with the prodomain (α2), but the type I receptors epitope was occupied by the α5 helix, instead of the typical α1 observed in latent procomplexes [127–128]. It was suggested that the α5 helix was displaced readily to allow for type I receptor binding. In fact, recently the BMP9 ligand was solved in complex with both its prodomain and the type I receptor Alk1 [39]. This structure revealed that the α5 helix is indeed displaced, while Alk1 is bound to BMP9. These results suggest that differences in prodomain interactions with the ligand at the type I binding site are important for differentiating the latent and non-latent procomplexes. Interestingly, this might also be a product of the type I receptor affinities, where BMP ligands, due to their higher affinity for the type I receptors, can more readily displace the prodomain, whereas TGFβ1 and GDF8/GDF11 have low affinity for the type I receptor [36, 47–48, 90–91, 101, 134]. Certainly, additional questions arise as to the posttranslational function of the BMP prodomains and whether full dissociation of the prodomain from the ligand occurs during signaling. Interestingly, other studies how shown that BMP7 can transition from an open to closed, latent conformation through interactions with the ECM (Fig 2A) [135].
While the prodomain is essential for proper folding, it also appears to have an important role in facilitating homo- versus heterodimerization of the ligand. Evidence of a cross-over interaction was shown in a recent crystal structure of the TGFβ1 procomplex [136]. Thus, it is logical to think that the prodomain must be “compatible” with its hetero-partner chain. Experimentally, it was shown by the Christensen laboratory that the prodomain of BMP4 was both sufficient and required for the formation of a BMP4/BMP7 heterodimer [126]. Thus, the prodomains appear to be functionally important for the formation of ligand heterodimers, to not only allow for the formation of desired heterodimers (i.e. BMP2/BMP4, BMP2/BMP7, BMP9/BMP10), but are likely to be important for inhibiting the formation of unwanted heterodimers.
Beyond Furin processing at the primary site connecting the prodomain and ligand, other proteolytic events can occur within the prodomain. For instance, the GDF8 and GDF11 prodomains are cleaved by tolloid metalloproteases for activation [129, 137–139]. BMP ligands are also subjected to more non-traditional sites of cleavage by PCs. BMP4 for example has two cleavage sites - one directly N terminal of the mature domain (S1) as expected and another upstream of the S1 site (S2) [140–142]. Sequential cleavage of the BMP4 prodomain is essential for proper development and simultaneous cleavage caused a reduction in mature BMP4 despite no change in precursor products [140]. Likewise, the Drosophila ligand Gbb (a BMP7 orthologue) was shown to have alternate processing sites which impacted activity and receptor preference [142]. This highlights that the prodomain can be cleaved at different locations altering the function of the procomplex.
Extracellular BMP Antagonists
BMP signaling activity is tightly regulated prior to receptor engagement, not only through prodomain interactions, but also by a series of neutralizing antagonists. A variety of structurally distinct molecules regulate the activities of BMP ligands by effectively blocking the receptor-ligand interactions [143]. Extracellular BMP antagonists are diverse, ranging from single-domain to larger multidomain proteins and bind distinct members of the BMP class with different specificities and affinities. While this aspect of BMP biology has been reviewed previously, this section will provide a general summary and highlight recent research on both low and high-resolution structures [4, 144–149].
Secreted by the Spemann organizer, Noggin is covalently linked homodimer that potently antagonizes BMP2 and BMP4, but can also target other ligands with lower affinity (e.g. BMP6 and BMP7) [150–151]. In 2002, Groppe et al. resolved the high resolution crystal structure of Noggin bound to BMP7 revealing that a Noggin dimer forms a symmetrical caliper-like structure, making extensive contacts with the knuckle of each ligand [32]. The N-terminus of Noggin threads into the convex surface of the dimer interface (Fig 2B), which together effectively occludes both type I and type II receptors from binding BMP. The N-terminal segment interacts with hydrophobic residues in the composite interface of the BMP ligand reminiscent of the type I receptor “knob-in-hole” binding mechanism [27]. The binding at the type II receptor site is blocked by the C-terminal half of the clip domain and by two fingers via predominantly hydrophobic interactions. Thus, a single Noggin dimer blocks all four receptor binding epitopes of a BMP ligand.
Chordin, also secreted by the Spemann organizer, is a multi-domain protein that antagonizes BMP signaling. Chordin contains four cysteine-rich von willebrand factor C (VWC) domains of ~80 amino acids each, important for BMP binding. The VWC domains are located at the ends of Chordin with VWC1 located in the N-terminus and VWC2–4 located in series at the C-terminus. The specificity of the individual VWC domains of Chordin with different BMP ligands was characterized previously [152]. Multiple domains are important for antagonism as Chordin can be cleaved at two sites (N-terminal and C-terminal) by the BMP1/tolloid metalloprotease reducing the affinity of individual domain interactions and liberating BMP. Currently, there is no crystal structure of Chordin alone or in complex with BMP. However, Troilo et al. used transmission electron microscopy and small angle X-ray scattering to generate a low-resolution structure of Chordin [153]. The structure shows a horseshoe-like shape that supports a model where VWC1 binds one end of the BMP dimer with VWC2–4 binding at the other end. This model is significantly different from Noggin, as Chordin binds to the dimeric ligand in an asymmetric manner.
Phylogenetically similar to Chordin, Crossveinless-2 (CV2) is another extracellular BMP antagonist that contains VWC domains [154–155]. Depending on cell context, CV2 can either enhance or inhibit BMP activity [154]. At the N-terminus CV2 contains five VWC domains followed by a von Willebrand factor type D (VWD) and a trypsin inhibitor-like cysteine-rich domain. Domain analysis revealed that the first VWC domain (VWC1) is responsible for the majority of BMP binding [152]. Leveraging this information, Zhang et al., solved the structure of the VWC1 bound to BMP2 (Fig 2B) [25]. Two VWC1 domains bound to each side of the ligand dimer. The VWC1 domain was shown to consist of three parts, an ~8 amino acid N-terminus region referred as the „Clip”, a ~31 amino acid N-terminal subdomain SD1, and a ~22 amino acid C-terminal subdomain SD2. The structure showed that the clip domain securely occupies the wrist region of BMP2 and, similar to Noggin, blocks the type I receptor binding site [25]. The subdomain SD1 binds the knuckle epitope obstructing the type II receptor binding epitope. Thus, while structurally unrelated to Noggin, the VWC1 domain of CV2 adopts a similar strategy where the bulk of interactions occur on the knuckle region with a coil segment that binds the dimer interface.
Twisted gastrulation (TSG) is another BMP modulator but is much smaller than the above inhibitors. The N-terminal cysteine-rich domain, analogous to a VWC domain, binds BMP ligands with high affinity [156–157]. Only a low-resolution structure of TSG alone is available [158]. Interestingly, TSG not only binds BMP ligands, but also interacts with the C-terminal region of Chordin through largely uncharacterized protein motifs in the TSG C-terminal domain. This causes the formation of higher-order complexes which, similar to CV2, can either enhance or inhibit BMP signaling [158]. However, the details of the interactions of TSG, Chordin and BMP ligands remain unresolved at the atomic level.
Lastly, the DAN family is the largest family of BMP antagonists, containing seven members: Gremlin1, Gremlin2, DAND5, NBL1, Cerberus, Sclerostin (SOST) and SOSTDC1 [148]. These single domain antagonists exhibit varying specificities for different BMPs and can bind other ligands of the TGFβ family (e.g Cerberus neutralizes Nodal and activin B) [159–161]. Besides BMP inhibition, both SOST and SOSTDC1 can inhibit Wnt signaling by binding with LRP5/6 receptors [162–165]. NMR structures of SOST revealed that DAN proteins contain a core growth-factor like domain (central cystine-knot paired with extended β-strands) with flanking N- and C- termini [166–167]. Subsequent crystal structures of Gremlin1, Gremlin2, and NBL1 reveal that most members form highly stable non-disulfide linked homodimers. Distinct from Noggin which forms a head-to-head dimer, the DAN dimer is arranged in a head-to-tail fashion with extensive β-strand contacts holding the two monomer together [168–170]. In 2016, our laboratory published the crystal structure of the Gremlin2-GDF5 complex (Fig 2B) [44]. The complex revealed that Gremlin2 binds perpendicular to the GDF5 ligand with a dimer of Gremlin2 on each side, forming an H-like structure. The core growth-factor like domain binds the ligand knuckle and the N-terminus threads into the type I epitope site, similar to Noggin and CV2. Thus, Gremlin2 effectively blocks both type I and type II receptor binding sites on GDF5. Having both the ligand-bound and unbound structures illuminates the molecular transition of the N-terminus from a helix that shields the hydrophobic core domain to a β-strand and coil that bind the top ligand finger and dimer interface, respectively. Interestingly, while other antagonists such as Noggin and Follistatin form terminal inhibitory complexes, the nature of Gremlin2 binding lends itself to a repeating structure, or a form of “daisy-chain” repeat [44]. While the structure of Gremlin2 revealed the type I site is blocked, NBL1 which has a shorter N-terminal extension does not inhibit type I binding [168]. The functional consequences of these structural findings are difficult to ascertain, however, it brings to light the structural diversity within the DAN family.
Here we have focused on BMP modulators that have, to some extent, been characterized structurally. However, there are numerous other modulators that lack significant structural characterization with respect to BMP inhibition. For example, Thrombospondin-1 (TSP-1), SMOCs, and CCN family members have all been functionally characterized as BMP-antagonists [171–175]. Furthermore, similar to prodomain interactions, the mechanism by which these modulators interact with the ECM or other BMP binding proteins still needs significant investigation.
Intracellular Receptor Structures
Following the binding of a BMP ligand to the extracellular domain of both type I and type II receptors, the intracellular kinase domain of the type II receptor will phosphorylate the type I receptor. Thereafter, the activated type I receptor will bind and phosphorylate SMAD proteins, which then accumulate in the nucleus to regulate transcription. In this section structures of the intracellular kinase domains will be discusses as they have allowed us to better understand this mechanism on a molecular level. While kinase domain structures have been previously reviewed, a special focus will be placed on the receptors specific to BMPs and how they differ from the other receptors in the family [5, 11, 51]. Also, several naturally occurring mutations within BMP receptors which result in a wide array of diseases will be reviewed, and the structures of these receptors are able to reveal about these mutations.
As mentioned, the receptors of the TGFβ family, both type I and type II, are serine-threonine protein kinases. The first type I and type II structures of a TGFβ receptor kinases, the structures of Alk5 and ActRIIB, revealed that these receptors adopt a canonical protein kinase fold, consisting of a five-stranded β-sheet N-lobe, a hinge region linker, and an α-helical C-lobe (Fig 3A) [176–177]. However, some differences between the type I and type II structures exist. Type I receptors are regulated by a small 12kDa immunophilin protein, FK-506 binding protein 1A (FKBP12), which sits atop the receptor, holding it in an inactive state [176, 178–180]. Upon ligand mediated receptor assembly, the constitutively active type II receptor will phosphorylate the type I receptor within the GS domain, a ~30 residue glycine-serine (GS) rich domain positioned between the cell membrane and N-lobe of type I receptors [181]. Structures of Alk5 bound and unbound by FKBP12, have helped to explain this mechanism of type I receptor activation [176, 178]. These structures show that displacement of FKBP12 allows the GS domain and a conserved α-helix, termed αC, within the N-lobe to reposition to an open active confirmation. It is thought that this conformational switch allows for direct binding between the type I receptor and SMAD proteins [178]. To date, the intracellular kinase domain has been structurally resolved for the BMP type I receptors Alk1, Alk2, and Alk6 and the type II receptors ActRIIA, ActRIIB, and BMPRII [177, 182–192]. All receptors share the mentioned conserved kinase fold, however, as discussed in the sections below, slight changes in structural conformation and sequence allow for specificity between receptors.
Figure 3.
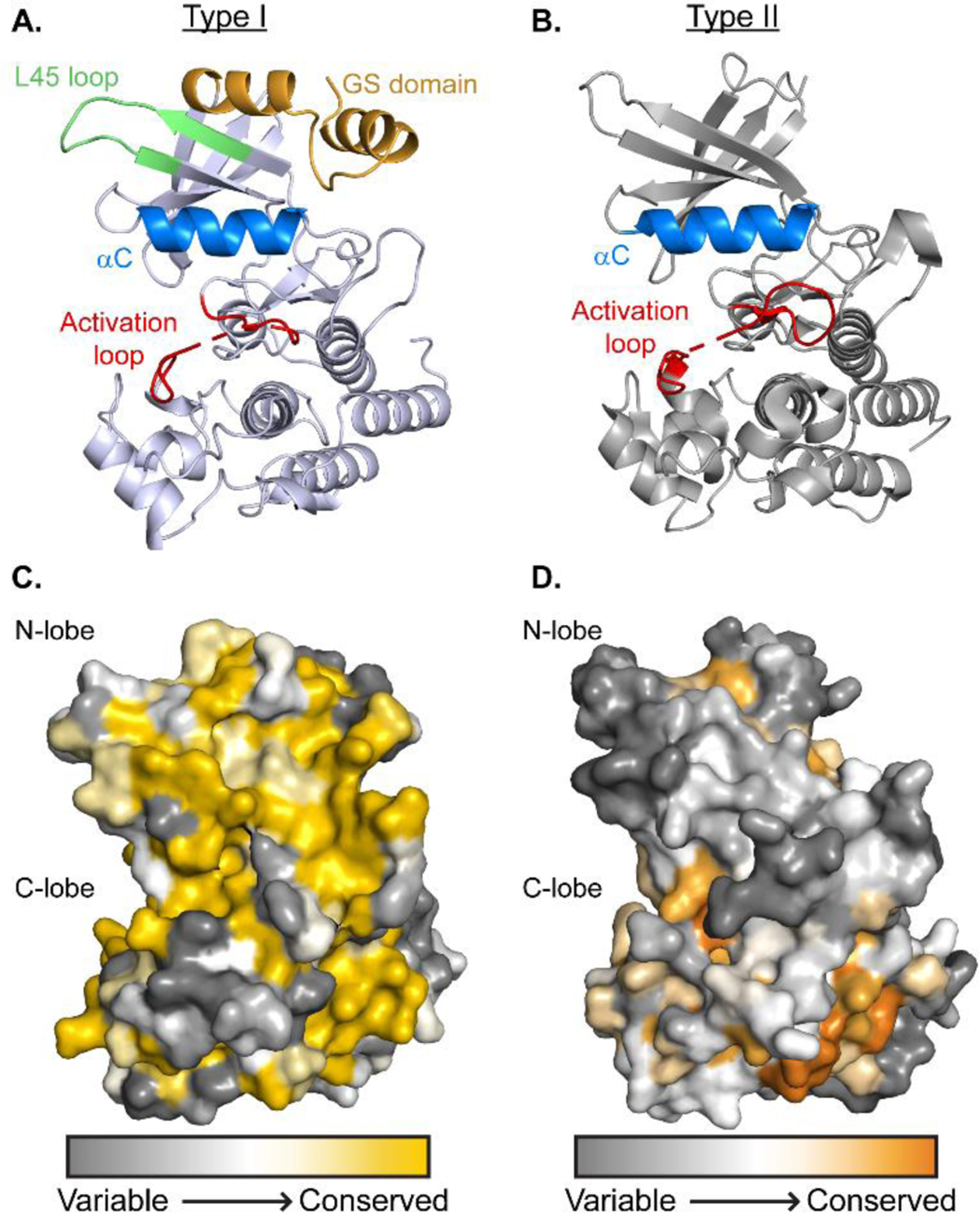
Structural features of BMP intracellular kinase domains. A) Structure of type I receptor Alk6 kinase domain (3MDY) with GS domain (gold), L45 loop (green), αC (blue), and Activation loop (red) highlighted and labelled [189]. B) Structure of type II receptor BMPRII kinase domain (3G2F) with αC (blue) and Activation loop (red) highlighted and labelled [192]. C) & D) Sequence conservation of intracellular kinase domains. Using ConSurf software and Clustal sequence alignment sequence conservation of type I (C) and type II (D) receptors is shown [60–64]. Conservation is colored from dark gray (variable) to yellow (conserved) for type I receptors and orange (conserved) for type II receptors.
Much like their extracellular domains, the kinase domains all share a high level of structural conservation. Despite this, these receptors are able to activate different SMAD proteins leading to various signaling outcomes. SMAD specificity was first investigated by both Feng and Derynck and Chen et al. who showed through generation of mutant or chimera receptors, SMAD activation was specific to a loop in the N-lobe of the receptor, termed the L45 loop (Fig 3A) [193–194]. For example, swapping the L45 loop of Alk5 for that of Alk3 resulted in an exchange of function, in which Alk5 was able to phosphorylate SMAD1 and ALk3 was able to phosphorylate SMAD2 [194]. Using a combination of structures and sequences we are able to visualize these regions of specificity using the ConSurf server to color amino acids based on sequence similarity, grouping type I receptors (Fig 3C) and type II receptors (Fig 3D) [60–64]. Regions with high sequence conservation are colored in yellow (type I) or orange (type II) whereas low conservation is colored in gray. In general, the type I receptor sequences are more conserved than the type II receptor. Regions with high conservation in the type I receptor include the GS domain and much of the C-lobe. However, as expected, the L45 loop as well as other regions predicted to interact with SMAD proteins within the N-lobe differ between type I receptors (Fig 3C) [178]. The overall high level of conservation of the type I receptors is reasonable, as the phosphorylation sites of these receptors is conserved within the GS domain. Alternatively, the type II receptors contain several phosphorylation sites throughout their kinase domains as well as other sites for posttranslational modifications and binding partners [195–196]. For example, Xu et al. showed that the arginine methyltransferase PRMT1, which associates with BMPRII, is required to initiate SMAD mediated BMP signaling and may justify the slow kinetics of SMAD activation [196].
Within the context of BMP signaling, several pathogenic mutations are observed in the kinase domains. Fortunately, structural studies of these receptors and their mutations have aided in the careful design of small molecule inhibitors. A single point mutation in the kinase domain of Alk2, R206H, which is located just after the GS domain, results in aberrant signaling of Alk2, and the development of fibrodysplasia ossificans progressiva (FOP). From the structure, it is believed that this mutation does not allow the binding of FKBP12, thus resulting in an active receptor. Furthermore, the crystal structure of the Alk2 kinase domain bound to small molecule inhibitors has allowed for the design of a highly specific inhibitor, LDN-212854, with enhanced selectivity for Alk2 [185]. Other studies have used this structure-based drug design method to develop more potential therapies for FOP and improve upon the pharmacological properties of these small molecules [190, 197]. Additional structural studies comparing the conformation of the ATP-binding pockets of Alk2 and Alk3 have also been used to generate a small molecule therapy for treatment of gliomas, E6201, and potentially explain the observed selectivity of this molecule for Alk2 [189].
Mutations in BMPRII are the predominant risk factor for 53–96% of heritable cases and responsible for nearly 25% of idiopathic cases of pulmonary arterial hypertension (PAH) [178]. A recent study by Chaikuad et al. solved the crystal structure of the conserved BMPRII kinase domain and mapped the mutations found in PAH patients onto the structure. Using this information, it is clear why these cause a non-functional BMPRII, as many of these mutations are located in the hydrophobic protein core and would likely result in severe steric clashes, disrupting the fold of BMPRII. However, some mutations were present on the protein surface, suggesting that patients with these variants may be more responsive to BMPRII targeted therapies [192].
Uniquely, BMPRII has a C-terminal tail extension of its intracellular domain, which is not observed in any other type II receptor. Studies conducted within the last 20 years have shown that this region interacts with LIM kinase 1 (LIMK1) and Tctex-1, but for unknown purposes [198–201]. Interestingly, PAH causing mutations are also found in the cytoplasmic tail, suggesting that this unique feature of BMPRII plays an important functional role [202]. Furthermore, a more recent study investigating both a “short” BMPRII, consisting only of the conserved kinase domain, and a “long” BMPRII, containing the full receptor, has shown that the long form quickly undergoes clathrin-mediated endocytosis, while the short form has higher SMAD activity. Thus, it has been proposed that this cytoplasmic tail may be important for regulation of BMPRII levels [203]. However, this region is still structurally unresolved, and the mechanism for how it may be regulating BMP signaling remains elusive.
Large structural gaps still exist within the field, including the lack both the intracellular component and the full length receptor complex structures. Exactly how the type II receptor engages the type I receptor for activation remains unknown, as well as how SMAD proteins specifically bind to type I receptors. Surely, complex structures of the intracellular components would help to elucidate major questions remaining in the field. While structural studies have formed the foundation for our mechanistic understanding of BMP signaling and regulation, numerous gaps in our knowledge exists where future studies will be aimed at understanding larger complex assemblies. For example, a structure of the full-length ligand: receptor complex.
Highlights:
Structural biology studies have been foundational in the understanding of BMP biology.
BMP growth factors are structurally and biochemically distinct from other members of the TGFβ family.
These structural features inform the distinct functional mechanisms of signaling, processing, and inhibition of these fundamental signaling pathways.
Acknowledgments
The following work was funded by an R35 grant (GM134923) awarded to T.B.T.
Abbreviations:
- TGFβ
transforming growth factor beta
- BMP
bone morphogenetic protein
- ECM
extracellular matrix
- HS
heparan sulfate
- ENG
endoglin
- BG
betaglycan
- PC
proprotein convertase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Urist MR, Strates BS. Bone morphogenetic protein. J Dent Res 1971;50(6):1392–1406. doi: 10.1177/00220345710500060601 [DOI] [PubMed] [Google Scholar]
- 2.Reddi AH. Bone morphogenetic proteins: from basic science to clinical applications. J Bone Joint Surg Am 2001;83-A Suppl 1(Pt 1):S1–S6. doi: 10.2106/00004623-200100001-00001 [DOI] [PubMed] [Google Scholar]
- 3.Wozney JM. The bone morphogenetic protein family and osteogenesis. Mol Reprod Dev 1992;32(2):160–167. doi: 10.1002/mrd.1080320212 [DOI] [PubMed] [Google Scholar]
- 4.Hinck AP, Mueller TD, Springer TA. Structural Biology and Evolution of the TGF-β Family. Cold Spring Harb Perspect Biol 2016;8(12):a022103 Published 2016 Dec 1. doi: 10.1101/cshperspect.a022103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yadin D, Knaus P, Mueller TD. Structural insights into BMP receptors: Specificity, activation and inhibition. Cytokine Growth Factor Rev 2016;27:13–34. doi: 10.1016/j.cytogfr.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 6.Yamashita H, ten Dijke P, Huylebroeck D, et al. Osteogenic protein-1 binds to activin type II receptors and induces certain activin-like effects. J Cell Biol 1995;130(1):217–226. doi: 10.1083/jcb.130.1.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev 1998;9(1):49–61. doi: 10.1016/s1359-6101(97)00036-1 [DOI] [PubMed] [Google Scholar]
- 8.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci U S A 2006;103(20):7643–7648. doi: 10.1073/pnas.0602558103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massagué J TGFβ signalling in context. Nat Rev Mol Cell Biol 2012;13(10):616–630. doi: 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity 2019;50(4):924–940. doi: 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaikuad A, Bullock AN. Structural Basis of Intracellular TGF-β Signaling: Receptors and Smads. Cold Spring Harb Perspect Biol 2016;8(11):a022111 Published 2016 Nov 1. doi: 10.1101/cshperspect.a022111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdollah S, Macías-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem 1997;272(44):27678–27685. doi: 10.1074/jbc.272.44.27678 [DOI] [PubMed] [Google Scholar]
- 13.Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci 2001;114(Pt 24):4359–4369. [DOI] [PubMed] [Google Scholar]
- 14.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 1996;383(6603):832–836. doi: 10.1038/383832a0 [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 1998;94(5):585–594. doi: 10.1016/s0092-8674(00)81600-1 [DOI] [PubMed] [Google Scholar]
- 16.Walker RG, Poggioli T, Katsimpardi L, et al. Biochemistry and Biology of GDF11 and Myostatin: Similarities, Differences, and Questions for Future Investigation. Circ Res 2016;118(7):1125–1142. doi: 10.1161/CIRCRESAHA.116.308391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh SP, Yeo CY, Lee Y, Schrewe H, Whitman M, Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev 2002;16(21):2749–2754. doi: 10.1101/gad.1021802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickel J, Mueller TD. Specification of BMP Signaling. Cells 2019;8(12):1579 Published 2019 Dec 5. doi: 10.3390/cells8121579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heldin CH, Moustakas A. Signaling Receptors for TGF-β Family Members. Cold Spring Harb Perspect Biol 2016;8(8):a022053 Published 2016 Aug 1. doi: 10.1101/cshperspect.a022053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canali S, Core AB, Zumbrennen-Bullough KB, et al. Activin B Induces Noncanonical SMAD1/5/8 Signaling via BMP Type I Receptors in Hepatocytes: Evidence for a Role in Hepcidin Induction by Inflammation in Male Mice. Endocrinology 2016;157(3):1146–1162. doi: 10.1210/en.2015-1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen OE, Hella H, Elsaadi S, Jacobi C, Martinez-Hackert E, Holien T. Activins as Dual Specificity TGF-β Family Molecules: SMAD-Activation via Activin- and BMP-Type 1 Receptors. Biomolecules 2020;10(4):519 Published 2020 Mar 29. doi: 10.3390/biom10040519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramachandran A, Vizán P, Das D, et al. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. Elife 2018;7:e31756 Published 2018 Jan 29. doi: 10.7554/eLife.31756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD. Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol 2004;11(5):481–488. doi: 10.1038/nsmb756 [DOI] [PubMed] [Google Scholar]
- 24.Esquivies L, Blackler A, Peran M, et al. Designer nodal/BMP2 chimeras mimic nodal signaling, promote chondrogenesis, and reveal a BMP2-like structure. J Biol Chem 2014;289(3):1788–1797. doi: 10.1074/jbc.M113.529180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang JL, Qiu LY, Kotzsch A, et al. Crystal structure analysis reveals how the Chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev Cell 2008;14(5):739–750. doi: 10.1016/j.devcel.2008.02.017 [DOI] [PubMed] [Google Scholar]
- 26.Scheufler C, Sebald W, Hülsmeyer M. Crystal structure of human bone morphogenetic protein-2 at 2.7 A resolution. J Mol Biol 1999;287(1):103–115. doi: 10.1006/jmbi.1999.2590 [DOI] [PubMed] [Google Scholar]
- 27.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol 2000;7(6):492–496. doi: 10.1038/75903 [DOI] [PubMed] [Google Scholar]
- 28.Allendorph GP, Isaacs MJ, Kawakami Y, Izpisua Belmonte JC, Choe S. BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors [published correction appears in Biochemistry. 2007 Oct 30;46(43):12246]. Biochemistry 2007;46(43):12238–12247. doi: 10.1021/bi700907k [DOI] [PubMed] [Google Scholar]
- 29.Saremba S, Nickel J, Seher A, Kotzsch A, Sebald W, Mueller TD. Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J 2008;275(1):172–183. doi: 10.1111/j.1742-4658.2007.06187.x [DOI] [PubMed] [Google Scholar]
- 30.Seeherman HJ, Berasi SP, Brown CT, et al. A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix. Sci Transl Med 2019;11(489):eaar4953. doi: 10.1126/scitranslmed.aar4953 [DOI] [PubMed] [Google Scholar]
- 31.Griffith DL, Keck PC, Sampath TK, Rueger DC, Carlson WD. Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily. Proc Natl Acad Sci U S A 1996;93(2):878–883. doi: 10.1073/pnas.93.2.878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groppe J, Greenwald J, Wiater E, et al. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature 2002;420(6916):636–642. doi: 10.1038/nature01245 [DOI] [PubMed] [Google Scholar]
- 33.Greenwald J, Groppe J, Gray P, et al. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 2003;11(3):605–617. doi: 10.1016/s1097-2765(03)00094-7 [DOI] [PubMed] [Google Scholar]
- 34.Brown MA, Zhao Q, Baker KA, et al. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem 2005;280(26):25111–25118. doi: 10.1074/jbc.M503328200 [DOI] [PubMed] [Google Scholar]
- 35.Wei Z, Salmon RM, Upton PD, Morrell NW, Li W. Regulation of bone morphogenetic protein 9 (BMP9) by redox-dependent proteolysis. J Biol Chem 2014;289(45):31150–31159. doi: 10.1074/jbc.M114.579771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Townson SA, Martinez-Hackert E, Greppi C, et al. Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J Biol Chem 2012;287(33):27313–27325. doi: 10.1074/jbc.M112.377960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito T, Bokhove M, Croci R, et al. Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1. Cell Rep 2017;19(9):1917–1928. doi: 10.1016/j.celrep.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi LZ, Brown CT, Gao Y, et al. Structure of bone morphogenetic protein 9 procomplex. Proc Natl Acad Sci U S A 2015;112(12):3710–3715. doi: 10.1073/pnas.1501303112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmon RM, Guo J, Wood JH, et al. Molecular basis of ALK1-mediated signalling by BMP9/BMP10 and their prodomain-bound forms. Nat Commun 2020;11(1):1621 Published 2020 Apr 1. doi: 10.1038/s41467-020-15425-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickel J, Kotzsch A, Sebald W, Mueller TD. A single residue of GDF-5 defines binding specificity to BMP receptor IB. J Mol Biol 2005;349(5):933–947. doi: 10.1016/j.jmb.2005.04.015 [DOI] [PubMed] [Google Scholar]
- 41.Schreuder H, Liesum A, Pohl J, Kruse M, Koyama M. Crystal structure of recombinant human growth and differentiation factor 5: evidence for interaction of the type I and type II receptor-binding sites. Biochem Biophys Res Commun 2005;329(3):1076–1086. doi: 10.1016/j.bbrc.2005.02.078 [DOI] [PubMed] [Google Scholar]
- 42.Klammert U, Mueller TD, Hellmann TV, et al. GDF-5 can act as a context-dependent BMP-2 antagonist. BMC Biol 2015;13:77 Published 2015 Sep 18. doi: 10.1186/s12915-015-0183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotzsch A, Nickel J, Seher A, Sebald W, Müller TD. Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity. EMBO J 2009;28(7):937–947. doi: 10.1038/emboj.2009.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nolan K, Kattamuri C, Rankin SA, Read RJ, Zorn AM, Thompson TB. Structure of Gremlin-2 in Complex with GDF5 Gives Insight into DAN-Family-Mediated BMP Antagonism. Cell Rep 2016;16(8):2077–2086. doi: 10.1016/j.celrep.2016.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Healey EG, Bishop B, Elegheert J, Bell CH, Padilla-Parra S, Siebold C. Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat Struct Mol Biol 2015;22(6):458–465. doi: 10.1038/nsmb.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotzsch A, Nickel J, Seher A, et al. Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome. J Biol Chem 2008;283(9):5876–5887. doi: 10.1074/jbc.M706029200 [DOI] [PubMed] [Google Scholar]
- 47.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci U S A 2006;103(20):7643–7648. doi: 10.1073/pnas.0602558103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber D, Kotzsch A, Nickel J, et al. A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol 2007;7:6 Published 2007 Feb 12. doi: 10.1186/1472-6807-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J Biol Chem 2011;286(7):5087–5099. doi: 10.1074/jbc.M110.188615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison CA, Al-Musawi SL, Walton KL. Prodomains regulate the synthesis, extracellular localisation and activity of TGF-β superfamily ligands. Growth Factors 2011;29(5):174–186. doi: 10.3109/08977194.2011.608666 [DOI] [PubMed] [Google Scholar]
- 51.Goebel EJ, Hart KN, McCoy JC, Thompson TB. Structural biology of the TGFβ family. Exp Biol Med (Maywood) 2019;244(17):1530–1546. doi: 10.1177/1535370219880894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinck AP, Archer SJ, Qian SW, et al. Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2. Biochemistry 1996;35(26):8517–8534. doi: 10.1021/bi9604946 [DOI] [PubMed] [Google Scholar]
- 53.Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex. Nat Struct Biol 2002;9(3):203–208. doi: 10.1038/nsb766 [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Fischer G, Hyvönen M. Structure and activation of pro-activin A. Nat Commun 2016;7:12052 Published 2016 Jul 4. doi: 10.1038/ncomms12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenwald J, Vega ME, Allendorph GP, Fischer WH, Vale W, Choe S. A flexible activin explains the membrane-dependent cooperative assembly of TGF-beta family receptors. Mol Cell 2004;15(3):485–489. doi: 10.1016/j.molcel.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 56.Stamler R, Keutmann HT, Sidis Y, Kattamuri C, Schneyer A, Thompson TB. The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity. J Biol Chem 2008;283(47):32831–32838. doi: 10.1074/jbc.M801266200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell 2005;9(4):535–543. doi: 10.1016/j.devcel.2005.09.008 [DOI] [PubMed] [Google Scholar]
- 58.Lerch TF, Shimasaki S, Woodruff TK, Jardetzky TS. Structural and biophysical coupling of heparin and activin binding to follistatin isoform functions. J Biol Chem 2007;282(21):15930–15939. doi: 10.1074/jbc.M700737200 [DOI] [PubMed] [Google Scholar]
- 59.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 60.Ashkenazy H, Abadi S, Martz E, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 2016;44(W1):W344–W350. doi: 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 2010;38(Web Server issue):W529–W533. doi: 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Celniker G, Nimrod G, Ashkenazy H, et al. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Isr. J. Chem 53 (2013) 199–206. 10.1002/ijch.201200096 [DOI] [Google Scholar]
- 63.Glaser F, Pupko T, Paz I, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 2003;19(1):163–164. doi: 10.1093/bioinformatics/19.1.163 [DOI] [PubMed] [Google Scholar]
- 64.Landau M, Mayrose I, Rosenberg Y, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 2005;33(Web Server issue):W299–W302. doi: 10.1093/nar/gki370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller TD, Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett 2012;586(14):1846–1859. doi: 10.1016/j.febslet.2012.02.043 [DOI] [PubMed] [Google Scholar]
- 66.Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. Receptor oligomerization and beyond: a case study in bone morphogenetic proteins. BMC Biol 2009;7:59 Published 2009 Sep 7. doi: 10.1186/1741-7007-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang HM, Cheng JC, Fang L, et al. Recombinant BMP4 and BMP7 downregulate pentraxin 3 in human granulosa cells. J Clin Endocrinol Metab 2015;100(3):E365–E374. doi: 10.1210/jc.2014-2496 [DOI] [PubMed] [Google Scholar]
- 68.Ebisawa T, Tada K, Kitajima I, et al. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J Cell Sci 1999;112 ( Pt 20):3519–3527 [DOI] [PubMed] [Google Scholar]
- 69.Tillet E, Ouarné M, Desroches-Castan A, et al. A heterodimer formed by bone morphogenetic protein 9 (BMP9) and BMP10 provides most BMP biological activity in plasma. J Biol Chem 2018;293(28):10963–10974. doi: 10.1074/jbc.RA118.002968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng J, Li Q, Wigglesworth K, et al. Growth differentiation factor 9:bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc Natl Acad Sci U S A 2013;110(8):E776–E785. doi: 10.1073/pnas.1218020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang HM, Cheng JC, Klausen C, Leung PC. BMP15 suppresses progesterone production by down-regulating StAR via ALK3 in human granulosa cells. Mol Endocrinol 2013;27(12):2093–2104. doi: 10.1210/me.2013-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuo WJ, Digman MA, Lander AD. Heparan sulfate acts as a bone morphogenetic protein coreceptor by facilitating ligand-induced receptor hetero-oligomerization. Mol Biol Cell 2010;21(22):4028–4041. doi: 10.1091/mbc.E10-04-0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fisher MC, Li Y, Seghatoleslami MR, Dealy CN, Kosher RA. Heparan sulfate proteoglycans including syndecan-3 modulate BMP activity during limb cartilage differentiation. Matrix Biol 2006;25(1):27–39. doi: 10.1016/j.matbio.2005.07.008 [DOI] [PubMed] [Google Scholar]
- 74.Hu Z, Wang C, Xiao Y, et al. NDST1-dependent heparan sulfate regulates BMP signaling and internalization in lung development. J Cell Sci 2009;122(Pt 8):1145–1154. doi: 10.1242/jcs.034736 [DOI] [PubMed] [Google Scholar]
- 75.Gandhi NS, Mancera RL. Prediction of heparin binding sites in bone morphogenetic proteins (BMPs). Biochim Biophys Acta 2012;1824(12):1374–1381. doi: 10.1016/j.bbapap.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 76.Caldwell EE, Nadkarni VD, Fromm JR, Linhardt RJ, Weiler JM. Importance of specific amino acids in protein binding sites for heparin and heparan sulfate. Int J Biochem Cell Biol 1996;28(2):203–216. doi: 10.1016/1357-2725(95)00123-9 [DOI] [PubMed] [Google Scholar]
- 77.Fromm JR, Hileman RE, Caldwell EE, Weiler JM, Linhardt RJ. Pattern and spacing of basic amino acids in heparin binding sites. Arch Biochem Biophys 1997;343(1):92–100. doi: 10.1006/abbi.1997.0147 [DOI] [PubMed] [Google Scholar]
- 78.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Irie A, Habuchi H, Kimata K, Sanai Y. Heparan sulfate is required for bone morphogenetic protein-7 signaling. Biochem Biophys Res Commun 2003;308(4):858–865. doi: 10.1016/s0006-291x(03)01500-6 [DOI] [PubMed] [Google Scholar]
- 80.Billings PC, Yang E, Mundy C, Pacifici M. Domains with highest heparan sulfate-binding affinity reside at opposite ends in BMP2/4 versus BMP5/6/7: Implications for function. J Biol Chem 2018;293(37):14371–14383. doi: 10.1074/jbc.RA118.003191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem 1996;237(1):295–302. doi: 10.1111/j.1432-1033.1996.0295n.x [DOI] [PubMed] [Google Scholar]
- 82.Kanzaki S, Takahashi T, Kanno T, et al. Heparin inhibits BMP-2 osteogenic bioactivity by binding to both BMP-2 and BMP receptor. J Cell Physiol 2008;216(3):844–850. doi: 10.1002/jcp.21468 [DOI] [PubMed] [Google Scholar]
- 83.Kanzaki S, Ariyoshi W, Takahashi T, et al. Dual effects of heparin on BMP-2-induced osteogenic activity in MC3T3-E1 cells. Pharmacol Rep 2011;63(5):1222–1230. doi: 10.1016/s1734-1140(11)70642-9 [DOI] [PubMed] [Google Scholar]
- 84.Hu Z, Wang C, Xiao Y, et al. NDST1-dependent heparan sulfate regulates BMP signaling and internalization in lung development. J Cell Sci 2009;122(Pt 8):1145–1154. doi: 10.1242/jcs.034736 [DOI] [PubMed] [Google Scholar]
- 85.Ayerst BI, Smith RA, Nurcombe V, Day AJ, Merry CL, Cool SM. Growth Differentiation Factor 5-Mediated Enhancement of Chondrocyte Phenotype Is Inhibited by Heparin: Implications for the Use of Heparin in the Clinic and in Tissue Engineering Applications. Tissue Eng Part A 2017;23(7–8):275–292. doi: 10.1089/ten.TEA.2016.0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim HS, Neugebauer J, McKnite A, Tilak A, Christian JL. BMP7 functions predominantly as a heterodimer with BMP2 or BMP4 during mammalian embryogenesis. Elife 2019;8:e48872 Published 2019 Sep 30. doi: 10.7554/eLife.4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morimoto T, Kaito T, et al. The bone morphogenetic protein-2/7 heterodimer is a stronger inducer of bone regeneration than the individual homodimers in a rat spinal fusion model. Spine J 2015. June 1;15(6):1379–90. doi: 10.1016/j.spinee.2015.02.034. Epub 2015 Feb 28. [DOI] [PubMed] [Google Scholar]
- 88.Valera E, Isaacs MJ, Kawakami Y, Izpisúa Belmonte JC, Choe S. BMP-2/6 heterodimer is more effective than BMP-2 or BMP-6 homodimers as inductor of differentiation of human embryonic stem cells. PLoS One 2010;5(6):e11167 Published 2010 Jun 17. doi: 10.1371/journal.pone.0011167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zúñiga JE, Groppe JC, Cui Y, et al. Assembly of TbetaRI:TbetaRII:TGFbeta ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent. J Mol Biol 2005;354(5):1052–1068. doi: 10.1016/j.jmb.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 90.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem 2010;285(19):14806–14814. doi: 10.1074/jbc.M109.079921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goebel EJ, Corpina RA, Hinck CS, et al. Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity. Proc Natl Acad Sci U S A 2019;116(31):15505–15513. doi: 10.1073/pnas.1906253116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand:receptor interactions. EMBO J 2003;22(7):1555–1566. doi: 10.1093/emboj/cdg156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olsen OE, Wader KF, Hella H, et al. Activin A inhibits BMP-signaling by binding ACVR2A and ACVR2B. Cell Commun Signal 2015;13:27 Published 2015 Jun 6. doi: 10.1186/s12964-015-0104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirsch T, Nickel J, Sebald W. BMP-2 antagonists emerge from alterations in the low-affinity binding epitope for receptor BMPR-II. EMBO J 2000;19(13):3314–3324. doi: 10.1093/emboj/19.13.3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science 1995;267(5196):383–386. doi: 10.1126/science.7529940 [DOI] [PubMed] [Google Scholar]
- 96.Klages J, Kotzsch A, Coles M, et al. The solution structure of BMPR-IA reveals a local disorder-to-order transition upon BMP-2 binding. Biochemistry 2008;47(46):11930–11939. doi: 10.1021/bi801059j [DOI] [PubMed] [Google Scholar]
- 97.Hatta T, Konishi H, Katoh E, et al. Identification of the ligand-binding site of the BMP type IA receptor for BMP-4. Biopolymers 2000;55(5):399–406. doi: [DOI] [PubMed] [Google Scholar]
- 98.Kirsch T, Nickel J, Sebald W. Isolation of recombinant BMP receptor IA ectodomain and its 2:1 complex with BMP-2. FEBS Lett 2000;468(2–3):215–219. doi: 10.1016/s0014-5793(00)01214-x [DOI] [PubMed] [Google Scholar]
- 99.Oh SP, Seki T, Goss KA, et al. Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci U S A 2000;97(6):2626–2631. doi: 10.1073/pnas.97.6.2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aykul S, Corpina RA, Goebel EJ, et al. Activin A forms a non-signaling complex with ACVR1 and type II Activin/BMP receptors via its finger 2 tip loop [published online ahead of print, 2020 Jun 9]. Elife 2020;9:e54582. doi: 10.7554/eLife.5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Groppe J, Hinck CS, Samavarchi-Tehrani P, et al. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 2008;29(2):157–168. doi: 10.1016/j.molcel.2007.11.039 [DOI] [PubMed] [Google Scholar]
- 102.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: insights into receptor utilization and heparin binding. EMBO J 2009;28(17):2662–2676. doi: 10.1038/emboj.2009.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nickel J, Ten Dijke P, Mueller TD. TGF-β family co-receptor function and signaling. Acta Biochim Biophys Sin (Shanghai) 2018;50(1):12–36. doi: 10.1093/abbs/gmx126 [DOI] [PubMed] [Google Scholar]
- 104.Kim SK, Henen MA, Hinck AP. Structural biology of betaglycan and endoglin, membrane-bound co-receptors of the TGF-beta family. Exp Biol Med 2019;244(17):1547–1558. doi: 10.1177/1535370219881160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alt A, Miguel-Romero L, Donderis J, et al. Structural and functional insights into endoglin ligand recognition and binding. PLoS One 2012;7(2). doi: 10.1371/journal.pone.0029948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barbara NP, Wrana JL, Letarte M. Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor-β superfamily. J Biol Chem 1999;274(2):584–594. doi: 10.1074/jbc.274.2.584 [DOI] [PubMed] [Google Scholar]
- 107.Scherner O, Meurer SK, Tihaa L, Gressner AM, Weiskirchen R. Endoglin differentially modulates antagonistic transforming growth factor-β1 and BMP-7 signaling. J Biol Chem 2007;282(19):13934–13943. doi: 10.1074/jbc.M611062200 [DOI] [PubMed] [Google Scholar]
- 108.Kim SK, Whitley MJ, Krzysiak TC, et al. Structural Adaptation in Its Orphan Domain Engenders Betaglycan with an Alternate Mode of Growth Factor Binding Relative to Endoglin. Structure 2019;27(9):1427–1442.e4. doi: 10.1016/j.str.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee NY, Kirkbride KC, Sheu RD, Blobe GC. The Transforming Growth Factor-β Type III Receptor Mediates Distinct Subcellular Trafficking and Downstream Signaling of Activin-like Kinase (ALK)3 and ALK6 Receptors. Luo K, ed. Mol Biol Cell 2009;20(20):4362–4370. doi: 10.1091/mbc.e09-07-0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Corradini E, Babitt JL, Lin HY. The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev 2009;20(5–6):389–398. doi: 10.1016/j.cytogfr.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siebold C, Yamashita T, Monnier PP, Mueller BK, Pasterkamp RJ. RGMs: Structural Insights, Molecular Regulation, and Downstream Signaling. Trends Cell Biol 2017;27(5):365–378. doi: 10.1016/j.tcb.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Babitt JL, Huang FW, Wrighting DM, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 2006;38(5):531–539. doi: 10.1038/ng1777 [DOI] [PubMed] [Google Scholar]
- 113.Xia Y, Yu PB, Sidis Y, et al. Repulsive guidance molecule RGMa alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J Biol Chem 2007;282(25):18129–18140. doi: 10.1074/jbc.M701679200 [DOI] [PubMed] [Google Scholar]
- 114.Samad TA, Rebbapragada A, Bell E, et al. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem 2005;280(14):14122–14129. doi: 10.1074/jbc.M410034200 [DOI] [PubMed] [Google Scholar]
- 115.Bell CH, Healey E, Van Erp S, et al. Structure of the Repulsive Guidance Molecule (RGM)-neogenin signaling hub. Science (80-) 2013;341(6141):77–80. doi: 10.1126/science.123232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Healey EG, Bishop B, Elegheert J, Bell CH, Padilla-Parra S, Siebold C. Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat Struct Mol Biol 2015;22(6):458–465. doi: 10.1038/nsmb.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fish LA, Fallon JR. Multiple MuSK signaling pathways and the aging neuromuscular junction. Neurosci Lett 2020;102:135014. doi: 10.1016/j.neulet.2020.135014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Till JH, Becerra M, Watty A, et al. Crystal structure of the MuSK tyrosine kinase: Insights into receptor autoregulation. Structure 2002;10(9):1187–1196. doi: 10.1016/S0969-2126(02)00814-6 [DOI] [PubMed] [Google Scholar]
- 119.Yilmaz A, Kattamuri C, Ozdeslik RN, et al. MuSK is a BMP co-receptor that shapes BMP responses and calcium signaling in muscle cells. Sci Signal 2016;9(444):1–13. doi: 10.1126/scisignal.aaf0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuehn R, Eckler SA, Gautam M. Multiple alternatively spliced transcripts of the receptor tyrosine kinase MuSK are expressed in muscle. Gene 2005;360(2):83–91. doi: 10.1016/j.gene.2005.07.009 [DOI] [PubMed] [Google Scholar]
- 121.Stricker S, Rauschenberger V, Schambony A. ROR-Family Receptor Tyrosine Kinases Vol 123 1st ed. Elsevier Inc.; 2017. doi: 10.1016/bs.ctdb.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 122.Debebe Z, Rathmell WK. Ror2 as a therapeutic target in cancer. Pharmacol Ther 2015;150:143–148. doi: 10.1016/j.pharmthera.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sammar M, Stricker S, Schwabe GC, et al. Modulation of GDF5/BRI-b signalling through interaction with the tyrosine kinase receptor Ror2. Genes to Cells 2004;9(12):1227–1238. doi: 10.1111/j.1365-2443.2004.00799.x [DOI] [PubMed] [Google Scholar]
- 124.Sammar M, Sieber C, Knaus P. Biochemical and functional characterization of the Ror2/BRIb receptor complex. Biochem Biophys Res Commun 2009;381(1):1–6. doi: 10.1016/j.bbrc.2008.12.162 [DOI] [PubMed] [Google Scholar]
- 125.Gregory KE, Ono RN, Charbonneau NL, et al. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J Biol Chem 2005;280(30):27970–27980. doi: 10.1074/jbc.M504270200 [DOI] [PubMed] [Google Scholar]
- 126.Neugebauer JM, Kwon S, Kim HS, et al. The prodomain of BMP4 is necessary and sufficient to generate stable BMP4/7 heterodimers with enhanced bioactivity in vivo. Proc Natl Acad Sci U S A 2015;112(18):E2307–E2316. doi: 10.1073/pnas.1501449112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shi M, Zhu J, Wang R, et al. Latent TGF-β structure and activation. Nature 2011;474(7351):343–349. Published 2011 Jun 15. doi: 10.1038/nature10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cotton TR, Fischer G, Wang X, et al. Structure of the human myostatin precursor and determinants of growth factor latency. EMBO J 2018;37(3):367–383. doi: 10.15252/embj.201797883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Walker RG, McCoy JC, Czepnik M, et al. Molecular characterization of latent GDF8 reveals mechanisms of activation. Proc Natl Acad Sci U S A 2018;115(5):E866–E875. doi: 10.1073/pnas.1714622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wooderchak-Donahue WL, McDonald J, O’Fallon B, et al. BMP9 mutations cause a vascular-anomaly syndrome with phenotypic overlap with hereditary hemorrhagic telangiectasia. Am J Hum Genet 2013;93(3):530–537. doi: 10.1016/j.ajhg.2013.07.004\ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cox TC, Lidral AC, McCoy JC, et al. Mutations in GDF11 and the extracellular antagonist, Follistatin, as a likely cause of Mendelian forms of orofacial clefting in humans. Hum Mutat 2019;40(10):1813–1825. doi: 10.1002/humu.23793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Fischer G, Hyvönen M. Structure and activation of pro-activin A. Nat Commun 2016;7:12052 Published 2016 Jul 4. doi: 10.1038/ncomms12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sengle G, Ono RN, Lyons KM, Bächinger HP, Sakai LY. A new model for growth factor activation: type II receptors compete with the prodomain for BMP-7. J Mol Biol 2008;381(4):1025–1039. doi: 10.1016/j.jmb.2008.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Walker RG, Czepnik M, Goebel EJ, et al. Structural basis for potency differences between GDF8 and GDF11. BMC Biol 2017;15(1):19 Published 2017 Mar 3. doi: 10.1186/s12915-017-0350-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wohl AP, Troilo H, Collins RF, Baldock C, Sengle G. Extracellular Regulation of Bone Morphogenetic Protein Activity by the Microfibril Component Fibrillin-1. J Biol Chem 2016;291(24):12732–12746. doi: 10.1074/jbc.M115.704734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao B, Xu S, Dong X, Lu C, Springer TA. Prodomain-growth factor swapping in the structure of pro-TGF-β1. J Biol Chem 2018;293(5):1579–1589. doi: 10.1074/jbc.M117.809657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ge G, Hopkins DR, Ho WB, Greenspan DS. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol 2005;25(14):5846–5858. doi: 10.1128/MCB.25.14.5846-5858.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Le VQ, Iacob RE, Tian Y, et al. Tolloid cleavage activates latent GDF8 by priming the pro-complex for dissociation. EMBO J 2018;37(3):384–397. doi: 10.15252/embj.201797931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ge G, Greenspan DS. Developmental roles of the BMP1/TLD metalloproteinases. Birth Defects Res C Embryo Today 2006;78(1):47–68. doi: 10.1002/bdrc.20060 [DOI] [PubMed] [Google Scholar]
- 140.Tilak A, Nelsen SM, Kim HS, et al. Simultaneous rather than ordered cleavage of two sites within the BMP4 prodomain leads to loss of ligand in mice. Development 2014;141(15):3062–3071. doi: 10.1242/dev.110130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Künnapuu J, Shimmi O. Evolutional imprints on the sequences of BMP2/4/DPP type proteins. Fly (Austin) 2010;4(1):21–23. doi: 10.4161/fly.4.1.10652 [DOI] [PubMed] [Google Scholar]
- 142.Anderson EN, Wharton KA. Alternative cleavage of the bone morphogenetic protein (BMP), Gbb, produces ligands with distinct developmental functions and receptor preferences. J Biol Chem 2017;292(47):19160–19178. doi: 10.1074/jbc.M117.793513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Walsh DW, Godson C, Brazil DP, Martin F. Extracellular BMP-antagonist regulation in development and disease: tied up in knots. Trends Cell Biol 2010;20(5):244–256. doi: 10.1016/j.tcb.2010.01.008 [DOI] [PubMed] [Google Scholar]
- 144.Rider CC, Mulloy B. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J 2010;429(1):1–12. doi: 10.1042/BJ20100305 [DOI] [PubMed] [Google Scholar]
- 145.Todd GM, Gao Z, Hyvönen M, Brazil DP, Ten Dijke P. Secreted BMP antagonists and their role in cancer and bone metastases [published online ahead of print, 2020 May 28]. Bone 2020;137:115455. doi: 10.1016/j.bone.2020.115455 [DOI] [PubMed] [Google Scholar]
- 146.Mulloy B, Rider CC. The Bone Morphogenetic Proteins and Their Antagonists. Vitam Horm 2015;99:63–90. doi: 10.1016/bs.vh.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 147.Brazil DP, Church RH, Surae S, Godson C, Martin F. BMP signalling: agony and antagony in the family. Trends Cell Biol 2015;25(5):249–264. doi: 10.1016/j.tcb.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 148.Nolan K, Thompson TB. The DAN family: modulators of TGF-β signaling and beyond. Protein Sci 2014;23(8):999–1012. doi: 10.1002/pro.2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zinski J, Tajer B, Mullins MC. TGF-β Family Signaling in Early Vertebrate Development. Cold Spring Harb Perspect Biol 2018;10(6):a033274 Published 2018 Jun 1. doi: 10.1101/cshperspect.a033274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 1992;70(5):829–840. doi: 10.1016/0092-8674(92)90316-5 [DOI] [PubMed] [Google Scholar]
- 151.Zimmerman LB, De Jesús-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 1996;86(4):599–606. doi: 10.1016/s0092-8674(00)80133-6 [DOI] [PubMed] [Google Scholar]
- 152.Zhang JL, Huang Y, Qiu LY, Nickel J, Sebald W. von Willebrand factor type C domain-containing proteins regulate bone morphogenetic protein signaling through different recognition mechanisms. J Biol Chem 2007;282(27):20002–20014. doi: 10.1074/jbc.M700456200 [DOI] [PubMed] [Google Scholar]
- 153.Troilo H, Zuk AV, Tunnicliffe RB, et al. Nanoscale structure of the BMP antagonist chordin supports cooperative BMP binding. Proc Natl Acad Sci U S A 2014;111(36):13063–13068. doi: 10.1073/pnas.1404166111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang JL, Patterson LJ, Qiu LY, Graziussi D, Sebald W, Hammerschmidt M. Binding between Crossveinless-2 and Chordin von Willebrand factor type C domains promotes BMP signaling by blocking Chordin activity. PLoS One 2010;5(9):e12846 Published 2010 Sep 23. doi: 10.1371/journal.pone.0012846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Binnerts ME, Wen X, Canté-Barrett K, et al. Human Crossveinless-2 is a novel inhibitor of bone morphogenetic proteins. Biochem Biophys Res Commun 2004;315(2):272–280. doi: 10.1016/j.bbrc.2004.01.048 [DOI] [PubMed] [Google Scholar]
- 156.Oelgeschläger M, Larraín J, Geissert D, De Robertis EM. The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature 2000;405(6788):757–763. doi: 10.1038/35015500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gazzerro E, Canalis E. Bone morphogenetic proteins and their antagonists. Rev Endocr Metab Disord 2006;7(1–2):51–65. doi: 10.1007/s11154-006-9000-6 [DOI] [PubMed] [Google Scholar]
- 158.Troilo H, Barrett AL, Zuk AV, et al. Structural characterization of twisted gastrulation provides insights into opposing functions on the BMP signalling pathway. Matrix Biol 2016;55:49–62. doi: 10.1016/j.matbio.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Piccolo S, Agius E, Leyns L, et al. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 1999;397(6721):707–710. doi: 10.1038/17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Aykul S, Martinez-Hackert E. New Ligand Binding Function of Human Cerberus and Role of Proteolytic Processing in Regulating Ligand-Receptor Interactions and Antagonist Activity. J Mol Biol 2016;428(3):590–602. doi: 10.1016/j.jmb.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Aykul S, Ni W, Mutatu W, Martinez-Hackert E. Human Cerberus prevents nodal-receptor binding, inhibits nodal signaling, and suppresses nodal-mediated phenotypes. PLoS One 2015;10(1):e0114954 Published 2015 Jan 20. doi: 10.1371/journal.pone.0114954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Semënov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem 2005;280(29):26770–26775. doi: 10.1074/jbc.M504308200 [DOI] [PubMed] [Google Scholar]
- 163.Li X, Zhang Y, Kang H, et al. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem 2005;280(20):19883–19887. doi: 10.1074/jbc.M413274200 [DOI] [PubMed] [Google Scholar]
- 164.Faraahi Z, Baud’huin M, Croucher PI, Eaton C, Lawson MA. Sostdc1: A soluble BMP and Wnt antagonist that is induced by the interaction between myeloma cells and osteoblast lineage cells [published correction appears in Bone. 2019 Jul;124:166]. Bone 2019;122:82–92. doi: 10.1016/j.bone.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lintern KB, Guidato S, Rowe A, Saldanha JW, Itasaki N. Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J Biol Chem 2009;284(34):23159–23168. doi: 10.1074/jbc.M109.025478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Veverka V, Henry AJ, Slocombe PM, et al. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem 2009;284(16):10890–10900. doi: 10.1074/jbc.M807994200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weidauer SE, Schmieder P, Beerbaum M, Schmitz W, Oschkinat H, Mueller TD. NMR structure of the Wnt modulator protein Sclerostin. Biochem Biophys Res Commun 2009;380(1):160–165. doi: 10.1016/j.bbrc.2009.01.062 [DOI] [PubMed] [Google Scholar]
- 168.Nolan K, Kattamuri C, Luedeke DM, et al. Structure of protein related to Dan and Cerberus: insights into the mechanism of bone morphogenetic protein antagonism. Structure 2013;21(8):1417–1429. doi: 10.1016/j.str.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nolan K, Kattamuri C, Luedeke DM, et al. Structure of neuroblastoma suppressor of tumorigenicity 1 (NBL1): insights for the functional variability across bone morphogenetic protein (BMP) antagonists. J Biol Chem 2015;290(8):4759–4771. doi: 10.1074/jbc.M114.628412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kišonaitė M, Wang X, Hyvönen M. Structure of Gremlin-1 and analysis of its interaction with BMP-2. Biochem J 2016;473(11):1593–1604. doi: 10.1042/BCJ20160254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Amthor H, Christ B, Rashid-Doubell F, Kemp CF, Lang E, Patel K. Follistatin regulates bone morphogenetic protein-7 (BMP-7) activity to stimulate embryonic muscle growth. Dev Biol 2002;243(1):115–127. doi: 10.1006/dbio.2001.0555 [DOI] [PubMed] [Google Scholar]
- 172.Glister C, Kemp CF, Knight PG. Bone morphogenetic protein (BMP) ligands and receptors in bovine ovarian follicle cells: actions of BMP-4, −6 and −7 on granulosa cells and differential modulation of Smad-1 phosphorylation by follistatin. Reproduction 2004;127(2):239–254. doi: 10.1530/rep.1.00090 [DOI] [PubMed] [Google Scholar]
- 173.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol 2012;44(3):480–488. doi: 10.1016/j.biocel.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Canalis E Nephroblastoma overexpressed (Nov) is a novel bone morphogenetic protein antagonist. Ann N Y Acad Sci 2007;1116:50–58. doi: 10.1196/annals.1402.055 [DOI] [PubMed] [Google Scholar]
- 175.Sallon C, Callebaut I, Boulay I, et al. Thrombospondin-1 (TSP-1), a new bone morphogenetic protein-2 and −4 (BMP-2/4) antagonist identified in pituitary cells. J Biol Chem 2017;292(37):15352–15368. doi: 10.1074/jbc.M116.736207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Huse M, Chen YG, Massagué J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell 1999;96(3):425–436. doi: 10.1016/s0092-8674(00)80555-3 [DOI] [PubMed] [Google Scholar]
- 177.Han S, Loulakis P, Griffor M, Xie Z. Crystal structure of activin receptor type IIB kinase domain from human at 2.0 Angstrom resolution. Protein Sci 2007;16(10):2272–2277. doi: 10.1110/ps.073068407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massagué J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell 2001;8(3):671–682. doi: 10.1016/s1097-2765(01)00332-x [DOI] [PubMed] [Google Scholar]
- 179.Wang T, Li BY, Danielson PD, et al. The immunophilin FKBP12 functions as a common inhibitor of the TGF beta family type I receptors. Cell 1996;86(3):435–444. doi: 10.1016/s0092-8674(00)80116-6 [DOI] [PubMed] [Google Scholar]
- 180.Chen YG, Liu F, Massague J. Mechanism of TGFbeta receptor inhibition by FKBP12. EMBO J 1997;16(13):3866–3876. doi: 10.1093/emboj/16.13.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature 1994;370(6488):341–347. doi: 10.1038/370341a0 [DOI] [PubMed] [Google Scholar]
- 182.Kerr G, Sheldon H, Chaikuad A, et al. A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis. Angiogenesis 2015;18(2):209–217. doi: 10.1007/s10456-014-9457-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hudson L, Mui J, Vázquez S, et al. Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept. J Med Chem 2018;61(16):7261–7272. doi: 10.1021/acs.jmedchem.8b00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chaikuad A, Alfano I, Kerr G, et al. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J Biol Chem 2012;287(44):36990–36998. doi: 10.1074/jbc.M112.365932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Williams E, Bullock AN. Structural basis for the potent and selective binding of LDN-212854 to the BMP receptor kinase ALK2. Bone 2018;109:251–258. doi: 10.1016/j.bone.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mohedas AH, Wang Y, Sanvitale CE, et al. Structure-activity relationship of 3,5-diaryl-2-aminopyridine ALK2 inhibitors reveals unaltered binding affinity for fibrodysplasia ossificans progressiva causing mutants. J Med Chem 2014;57(19):7900–7915. doi: 10.1021/jm501177w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sanvitale CE, Kerr G, Chaikuad A, et al. A new class of small molecule inhibitor of BMP signaling. PLoS One 2013;8(4):e62721 Published 2013 Apr 30. doi: 10.1371/journal.pone.0062721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ensan D, Smil D, Zepeda-Velázquez CA, et al. Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma. J Med Chem 2020;63(9):4978–4996. doi: 10.1021/acs.jmedchem.0c00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fortin J, Tian R, Zarrabi I, et al. Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas. Cancer Cell 2020;37(3):308–323.e12. doi: 10.1016/j.ccell.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sekimata K, Sato T, Sakai N, et al. Bis-Heteroaryl Pyrazoles: Identification of Orally Bioavailable Inhibitors of Activin Receptor-Like Kinase-2 (R206H). Chem Pharm Bull (Tokyo) 2019;67(3):224–235. doi: 10.1248/cpb.c18-00598 [DOI] [PubMed] [Google Scholar]
- 191.Horbelt D, Boergermann JH, Chaikuad A, et al. Small molecules dorsomorphin and LDN-193189 inhibit myostatin/GDF8 signaling and promote functional myoblast differentiation. J Biol Chem 2015;290(6):3390–3404. doi: 10.1074/jbc.M114.604397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chaikuad A, Thangaratnarajah C, von Delft F, Bullock AN. Structural consequences of BMPR2 kinase domain mutations causing pulmonary arterial hypertension. Sci Rep 2019;9(1):18351 Published 2019 Dec 4. doi: 10.1038/s41598-019-54830-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Feng XH, Derynck R. A kinase subdomain of transforming growth factor-beta (TGF-beta) type I receptor determines the TGF-beta intracellular signaling specificity. EMBO J 1997;16(13):3912–3923. doi: 10.1093/emboj/16.13.3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen YG, Hata A, Lo RS, et al. Determinants of specificity in TGF-beta signal transduction. Genes Dev 1998;12(14):2144–2152. doi: 10.1101/gad.12.14.2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Luo K, Lodish HF. Signaling by chimeric erythropoietin-TGF-beta receptors: homodimerization of the cytoplasmic domain of the type I TGF-beta receptor and heterodimerization with the type II receptor are both required for intracellular signal transduction. EMBO J 1996;15(17):4485–4496. [PMC free article] [PubMed] [Google Scholar]
- 196.Xu J, Wang AH, Oses-Prieto J, et al. Arginine Methylation Initiates BMP-Induced Smad Signaling. Mol Cell 2013;51(1):5–19. doi: 10.1016/j.molcel.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sato T, Sekimata K, Sakai N, et al. Structural Basis of Activin Receptor-Like Kinase 2 (R206H) Inhibition by Bis-heteroaryl Pyrazole-Based Inhibitors for the Treatment of Fibrodysplasia Ossificans Progressiva Identified by the Integration of Ligand-Based and Structure-Based Drug Design Approaches. ACS Omega 2020;5(20):11411–11423. Published 2020 May 12. doi: 10.1021/acsomega.9b04245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Machado RD, Southgate L, Eichstaedt CA, et al. Pulmonary Arterial Hypertension: A Current Perspective on Established and Emerging Molecular Genetic Defects. Hum Mutat 2015;36(12):1113–1127. doi: 10.1002/humu.22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Machado RD, Rudarakanchana N, Atkinson C, et al. Functional interaction between BMPR-II and Tctex-1, a light chain of Dynein, is isoform-specific and disrupted by mutations underlying primary pulmonary hypertension. Hum Mol Genet 2003;12(24):3277–3286. doi: 10.1093/hmg/ddg365 [DOI] [PubMed] [Google Scholar]
- 200.Foletta VC, Lim MA, Soosairajah J, et al. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1 [published correction appears in J Cell Biol. 2003 Oct 27;163(2):421. Soosairaiah Juliana [corrected to Soosairajah Juliana]]. J Cell Biol 2003;162(6):1089–1098. doi: 10.1083/jcb.200212060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J 2004;23(24):4792–4801. doi: 10.1038/sj.emboj.7600418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cogan J, Austin E, Hedges L, et al. Role of BMPR2 alternative splicing in heritable pulmonary arterial hypertension penetrance. Circulation 2012;126(15):1907–1916. doi: 10.1161/CIRCULATIONAHA.112.106245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Amsalem AR, Marom B, Shapira KE, et al. Differential regulation of translation and endocytosis of alternatively spliced forms of the type II bone morphogenetic protein (BMP) receptor. Mol Biol Cell 2016;27(4):716–730. doi: 10.1091/mbc.E15-08-0547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mace PD, Cutfield JF, Cutfield SM. High resolution structures of the bone morphogenetic protein type II receptor in two crystal forms: implications for ligand binding. Biochem Biophys Res Commun 2006;351(4):831–838. doi: 10.1016/j.bbrc.2006.10.109 [DOI] [PubMed] [Google Scholar]


