Abstract
Background & Aims
Intestinal epithelial cell (IEC) barrier dysfunction is critical to the development of Crohn’s disease (CD). However, the mechanism is understudied. We recently reported increased microRNA-31-5p (miR-31-5p) expression in colonic IECs of CD patients, but downstream targets and functional consequences are unknown.
Methods
microRNA-31-5p target genes were identified by integrative analysis of RNA- and small RNA-sequencing data from colonic mucosa and confirmed by quantitative polymerase chain reaction in colonic IECs. Functional characterization of activin receptor-like kinase 1 (ACVRL1 or ALK1) in IECs was performed ex vivo using 2-dimensional cultured human primary colonic IECs. The impact of altered colonic ALK1 signaling in CD for the risk of surgery and endoscopic relapse was evaluated by a multivariate regression analysis and a Kaplan–Meier estimator.
Results
ALK1 was identified as a target of miR-31-5p in colonic IECs of CD patients and confirmed using a 3’-untranslated region reporter assay. Activation of ALK1 restricted the proliferation of colonic IECs in a 5-ethynyl-2-deoxyuridine proliferation assay and down-regulated the expression of stemness-related genes. Activated ALK1 signaling increased colonic IEC differentiation toward colonocytes. Down-regulated ALK1 signaling was associated with increased stemness and decreased colonocyte-specific marker expression in colonic IECs of CD patients compared with healthy controls. Activation of ALK1 enhanced epithelial barrier integrity in a transepithelial electrical resistance permeability assay. Lower colonic ALK1 expression was identified as an independent risk factor for surgery and was associated with a higher risk of endoscopic relapse in CD patients.
Conclusions
Decreased colonic ALK1 disrupted colonic IEC barrier integrity and was associated with poor clinical outcomes in CD patients.
Keywords: Inflammatory Bowel Disease, miR-31, ALK1, Intestinal Epithelial Barrier
Abbreviations used in this paper: BMP, bone morphogenetic protein; CD, Crohn’s disease; DM, differentiation medium; EdU, 5-ethynyl-2-deoxyuridine; EM, expansion media; E2F2, E2F Transcription Factor 2; IEC, intestinal epithelial cell; HBSS, Hank’s balanced salt solution; miRNA, microRNA; mRNA, messenger RNA; NIBD, noninflammatory bowel disease; qPCR, quantitative polymerase chain reaction; SES-CD, Simple Endoscopic Score for Crohn's Disease; TEER, transepithelial electrical resistance; TGF-β, transforming growth factor β; 2D, 2-dimensional; UTR, untranslated region
Graphical abstract
Summary.
Activin receptor-like Kinase 1 (ALK1) was identified as a target of microRNA-31-5p in human colonic epithelial cells. Activation of ALK1 enhanced epithelial barrier function ex vivo. Decreased colonic ALK1 was associated with poor clinical outcomes in patients with Crohn’s disease.
Chronic intestinal inflammation in Crohn’s disease (CD) is caused by an aberrant interaction between the mucosal immune system and luminal antigens, often leading to a compromised intestinal epithelial cell (IEC) barrier, in genetically predisposed individuals.1 There is an increasing appreciation for the importance of variable cellular processes in IECs that contribute to the loss of barrier capacity and the development of CD.2,3 We4, 5, 6 and others7 have shown that alterations in IEC gene expression are associated with CD, but the underlying mechanisms driving IEC barrier defects remain unresolved. Current management of CD is largely directed at aberrant immune responses while CD therapy targeting IEC barrier defects remain elusive.8
In contrast to the small intestine where Paneth cells provide additional support for epithelial barrier integrity in part by secreting antimicrobial peptides, the colonic IEC barrier is largely maintained by tight cell-to-cell connections between colonocytes and by the mucus secreted from goblet cells.9 Colonocytes also are enriched with junctional proteins10 that serve as a barrier to luminal contents. Defects in the colonocyte barrier can increase paracellular permeability, which has been shown to lead to intestinal inflammation in several rodent models.11,12 Mechanisms controlling human colonocyte differentiation and survival still are understudied, especially in CD patients.
We recently reported aberrantly increased expression of microRNA-31-5p (miR-31-5p) in colonic IECs of CD patients compared with noninflammatory bowel disease (NIBD) controls.4 MicroRNAs (miRNAs) contribute to the control of various biological processes, including proliferation and differentiation,13 by post-transcriptionally regulating target gene expression.14 We hypothesized that miR-31-5p might contribute to the pathophysiology of CD through regulation of key genes that drive colonic IEC proliferation and differentiation. In this study, we identified and validated a novel miR-31-5p target gene, activin receptor-like kinase 1 (ACVRL1 or ALK1), in human colonic IECs. The functions of ALK1 in the colon are still unknown. We examined the functional impact of ALK1 signaling on IEC biology, notably colonocyte differentiation and barrier integrity, using human primary IECs. We also examined the clinical impact of altered colonic ALK1 levels in patients with CD and showed an association of decreased colonic ALK1 with increased risks of surgery and endoscopic relapse.
Results
ALK1 Is a Putative Target of miR-31-5p in Human Colonic Epithelial Cells
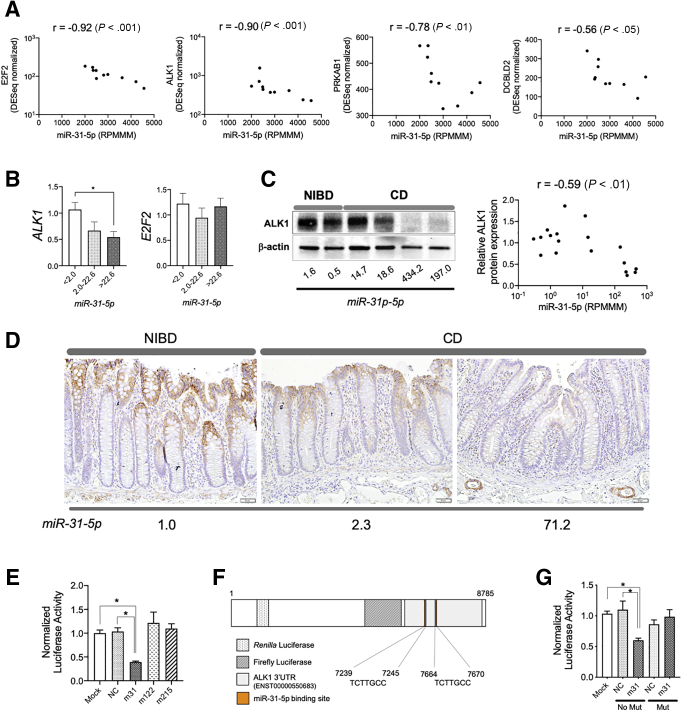
We previously generated and studied expression profiles of messenger RNAs (mRNAs)6 and miRNAs4 in the uninflamed colonic mucosa of 21 adult CD patients and 11 NIBD controls. We found a unique subset of CD patients with especially high miR-31-5p expression (high-miR-31-5p patients), which we determined was driven primarily by miR-31-5p expression in isolated IECs compared with other intestinal immune cell populations.15 Predicted miR-31-5p target genes (n = 27) were enriched significantly among the genes down-regulated in high-miR-31-5p CD patients compared with NIBD controls4 (Table 1), with expression of 4 genes significantly correlated inversely with miR-31 levels (Figure 1A). The strongest 2 were E2F2 and ALK1 (r = -0.92 and -0.90; P < .001 and P < .001, respectively). We next used quantitative reverse-transcription polymerase chain reaction (qPCR) to compare the expression of miR-31-5p with E2F2 and ALK1 specifically in colonic IECs isolated from CD patients and NIBD controls (Figure 1B). Again, we found a significant negative correlation between miR-31-5p and ALK1 (P = .019), but not with E2F2 (P = .982, Jonckheere–Terpstra test). Relative ALK1 protein expression also significantly correlated inversely with miR-31-5p expression (r = -0.59; P < .01), and was decreased remarkably in the colonic mucosa of high-miR-31-5p CD patients (Figure 1C). Low ALK1 levels were confirmed in colonic epithelial cells of CD patients by immunohistochemistry and were shown to be correlated inversely with miR-31-5p levels (Figure 1D). These results provide strong evidence for ALK1 being a miR-31-5p target in human colonic IECs.
Table 1.
Twenty-Seven Predicted miR-31-5p Target Genes That Were Down-Regulated Significantly in Colonic Mucosa of High-miR-31-5p CD Patients Compared With NIBD Controls
| Gene symbol | Average, DESeq normalized count |
Log2 fold change | Adjusted P value | |
|---|---|---|---|---|
| NIBD | High-miR31 CD | |||
| AKAP7 | 290.5 | 141.1 | -1.00 | .00015 |
| ALK1 | 1298.3 | 538.9 | -1.19 | .00027 |
| AK4 | 667.7 | 261.5 | -1.24 | .00075 |
| ARRDC3 | 1211.1 | 771.1 | -0.63 | .00625 |
| ATP10B | 3285.4 | 2057.8 | -0.65 | .01031 |
| COL4A4 | 99.2 | 60.7 | -0.72 | .00628 |
| DCBLD2 | 396.0 | 208.8 | -0.88 | .00266 |
| E2F2 | 297.2 | 115.5 | -1.29 | <.00001 |
| FAM118B | 385.7 | 244.7 | -0.65 | .00007 |
| FMN1 | 1240.0 | 670.1 | -0.86 | .00023 |
| HOMER1 | 98.5 | 35.2 | -1.29 | .00420 |
| KIAA1462 | 509.3 | 152.0 | -1.29 | .04777 |
| KPNA4 | 1808.1 | 1183.4 | -0.59 | .00973 |
| MATR3 | 434.8 | 204.5 | -1.02 | .00136 |
| MBOAT2 | 1019.3 | 390.1 | -1.32 | <.00001 |
| NARS2 | 246.7 | 168.8 | -0.52 | .01588 |
| ORC5 | 165.4 | 128.0 | -0.36 | .03969 |
| PPP1R12B | 3463.1 | 1188.0 | -1.33 | .00589 |
| PPP1R9A | 607.3 | 291.7 | -1.04 | .00002 |
| PRKAB1 | 577.6 | 444.7 | -0.37 | .04906 |
| RAB6B | 71.5 | 33.9 | -0.94 | .03396 |
| SATB2 | 2296.5 | 468.0 | -2.22 | <.00001 |
| SLC26A2 | 22136.2 | 2596.6 | -2.86 | <.00001 |
| TLN2 | 1492.2 | 755.4 | -0.95 | .00004 |
| VAV3 | 1141.7 | 522.4 | -1.07 | .00013 |
| ZC3H12C | 894.9 | 463.3 | -0.91 | .00011 |
| ZNF704 | 1412.6 | 820.8 | -0.77 | .00001 |
DESeq, differential expression analysis for sequence count data.
Figure 1.
ALK1 is a target of miR-31-5p in human colonic epithelial cells. (A) Correlation of expression in the colonic mucosa of CD patients (N = 10) between miR-31-5p (reads per million miRNAs mapped, RPMMM) and predicted targets of miR-31-5p (E2F2, ALK1, PRKAB1, and DCBLD2; DESeq normalized). (B) Association between the expression of miR-31-5p and the expression of ALK1 or E2F2 in isolated colonic epithelial cells (N = 27). Gene expression was quantified by qPCR and samples were split into 3 equally sized groups (N = 9 per group) according to the relative miR-31-5p expression levels. (C) Representative blot of ALK1 expression in the colonic tissue of NIBD and CD patients (left). Correlation between ALK1 protein expression and miR-31-5p in the colonic mucosa (right, N = 18). (D) ALK1 expression by immunohistochemistry in the colonic mucosa of NIBD controls and CD patients. The values shown at the bottom are the matched miR-31-5p expression level normalized to NIBD. (E) 3’UTR reporter assay for ALK1 in the presence or absence of 30 nmol/L miRNA mimics for hsa-miR-31-5p (m31), hsa-miR-122a-5p (m122), or hsa-miR-215-5p (m215) or negative control mimics (NC). N = 6 per group. (F) Schematic representation of the miR-31-5p binding sites in the reporter plasmid. (G) Site-directed mutagenesis assay with 10 nmol/L of m31 or NC mimics (N = 6 per group). All correlation values were calculated by the Spearman correlation coefficient. Each gene expression was normalized to GAPDH (ALK1, E2F2) or RNU48 (miR-31-5p). ∗P < .05. P values were determined by the Kruskal–Wallis test, followed by the Dunn multiple comparison test. Mut, mutation; NC, negative control mimics.
miR-31-5p Directly Suppress ALK1 Expression Through Binding to the 3’ Untranslated Region
To test the direct association between ALK1 mRNA and miR-31-5p, we performed a 3’ untranslated region (UTR) reporter assay in HEK293T cells using synthesized miRNA and negative control mimics (Figure 1E). Mimics for miR-122a-5p and miR-215-5p, which do not have specific binding sites in the 3’UTR of ALK1, were used as negative controls. As expected, miR-31-5p, but not mock miR-122a-5p or miR-215-5p, mimics decreased luciferase activity, suggesting direct regulation of ALK1 by miR-31-5p. To specifically show binding of miR-31-5p to the ALK1 3’UTR, we deleted the predicted miR-31-5p binding sequences in the reporter plasmid by site-directed mutagenesis (Figure 1F) and repeated the reporter assay with the miR-31-5p and negative control mimics (Figure 1G). Exogeneous miR-31-5p mimic no longer reduced luciferase activity of the mutated ALK1 plasmid. These results showed the direct regulation of ALK1 mRNA by miR-31-5p through binding to the 3’UTR.
Decreased ALK1 Expression Is Associated With Reduced NOTCH Activity in Colon
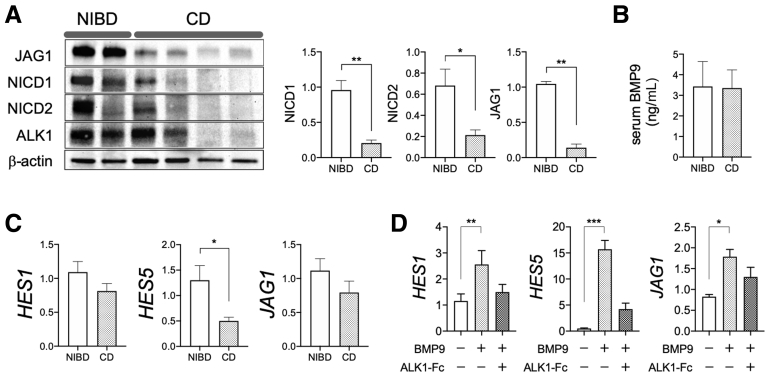
ALK1 is a type 1 receptor for transforming growth factor (TGF)-β signaling molecules16 that specifically binds to the extra-enteric ligand bone morphogenetic protein (BMP)9 and BMP10.17 Although ALK1 is expressed in human colonic IECs (Figure 1D), the role of ALK1-related signaling in IEC biology remains unknown. In the endothelium, ALK1 is known to enhance the expression of NOTCH target genes including JAG1, a ligand for NOTCH.18 Therefore, we examined the expression of JAG1 and NOTCH activity in the colonic mucosa of CD patients. Relative protein expression of JAG1 and the NOTCH intracellular domain was significantly lower in CD patients than in NIBD controls (Figure 2A). The measured serum concentration of ALK1 ligand BMP9 was not different between NIBD and CD patients (Figure 2B). To determine the impact of decreased NOTCH activity on the colonic IECs of CD patients, we next compared NOTCH target gene expression in colonic IECs between NIBD controls and CD patients. The expression of NOTCH targets HES1, HES5, and JAG1 in isolated colonic IECs was lower in CD patients than in NIBD controls (Figure 2C). Furthermore, in primary-cultured colonic IEC monolayers from NIBD patients, BMP9 robustly increased HES1, HES5, and JAG1 (Figure 2D). The increase in NOTCH pathway factors in response to BMP9 was mitigated significantly by addition of the ALK1-Fc chimera protein. These results show an ALK1-low associated decrease in NOTCH activity in the colonic mucosa of CD patients.
Figure 2.
Decreased ALK1 expression is associated with reduced NOTCH activity and NOTCH target gene expression in the colonic epithelial cells of CD patients. (A) Representative blot (left) and the difference of JAG1 and NOTCH intracellular domain (NICD) protein expression between NIBD and CD patients (right). (B) BMP9 concentration in the serum of NIBD controls (N = 17) and CD patients (N = 23). (C) NOTCH target gene expression in colonic epithelial cells from CD patients (N = 15) and NIBD controls (N = 12). (D) NOTCH target gene expression in NIBD patient-derived colonic epithelial cell monolayers. Expanded cells were cultured in expansion media in the presence or absence of BMP9 and ALK1–Fc chimera protein. N = 6 per group. Each gene expression was normalized to (C) GAPDH or (D) RPLP0. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. P values were determined by the (A–C) Mann–Whitney test or the (D) Friedman test followed by the Dunn multiple comparison test.
BMP9-ALK1 Signaling Restricts the Stemness of Human Colonic IECs
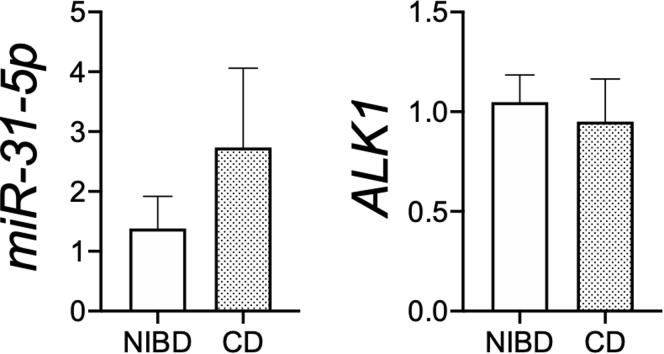
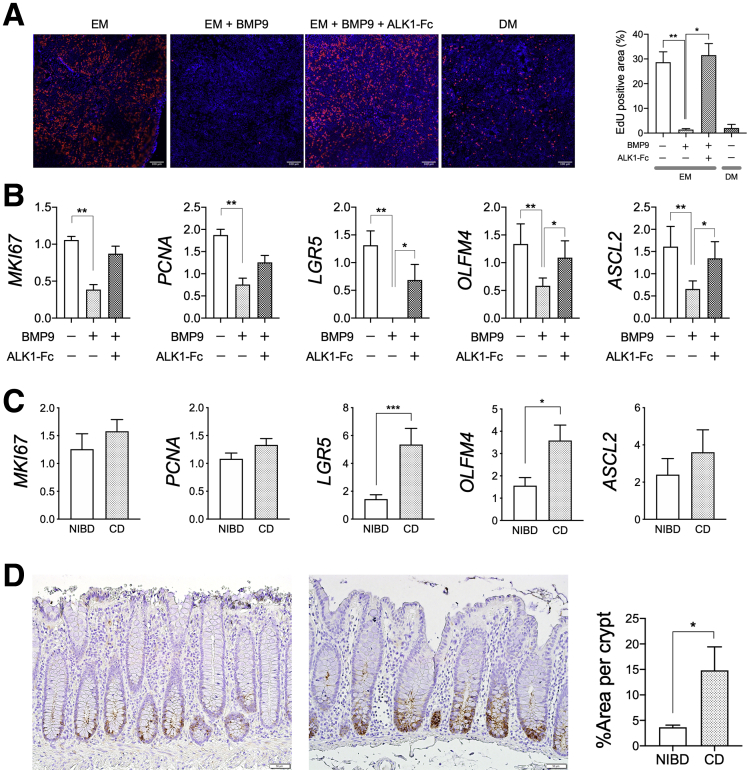
To investigate the impact of decreased ALK1 expression in the colonic IECs of CD patients, we first examined the effect of BMP9 treatment on IEC proliferation ex vivo using patient-derived colonic IEC monolayers. In primary-cultured colonic IEC monolayers derived from CD patients, increased miR-31-5p or decreased ALK1 expression no longer was observed (Figure 3), suggesting that environmental cues are important drivers of the observed miR-31-5p dysregulation in the colonic IECs of CD patients. Therefore, we tested the impact of decreased ALK1 expression on IEC proliferation by stimulating NIBD patient-derived colonic IEC monolayers in the presence or absence of ALK1–Fc chimera protein, which inhibits BMP9-ALK1 interaction and signaling. We found significantly decreased 5-ethynyl-2-deoxyuridine (EdU) incorporation in BMP9-stimulated IECs compared with nonstimulated cells (Figure 4A). BMP9-induced reduction of EdU incorporation was recovered by sequestrating BMP9 using a ALK1–Fc chimera protein, showing the effect of BMP9 in restricting IEC proliferation. We also examined the effect of BMP9 on the expression of IEC proliferation- and stemness-related genes (Figure 4B). Consistent with the results of the EdU assay, the expression of proliferation-related genes, including MKI67 and PCNA, as well as stemness-related genes LGR5, OLFM4, and ASCL2, was decreased significantly in BMP9-stimulated cells. BMP9-induced reduction of expression levels of these genes was recovered by the addition of ALK1–Fc chimera protein.
Figure 3.
Expression of miR-31-5p and ALK1 in primary-cultured colonic epithelial monolayers derived from NIBD controls and CD patients. N = 6 per group. Each gene expression was normalized to RNU48 (miR-31-5p) or GAPDH (ALK1). Statistical significance was determined by the Mann–Whitney test.
Figure 4.
BMP9–ALK1 signaling restricts the stemness of human colonic IECs. (A) EdU assay in NIBD patient-derived colonic epithelial cell monolayers (N = 4–8 per group). Expanded cells were cultured in EM in the presence or absence of BMP9 and ALK1–Fc chimera protein. Red, EdU; blue, Hoechst 33342. (B) Proliferation- and stemness-related gene expression in NIBD patient-derived colonic epithelial cell monolayers (N = 6 per group). (C) Proliferation- and stemness-related gene expression in colonic epithelial cells isolated from CD patients (N = 15) and NIBD controls (N = 12). (D) Representative immunohistochemical images of OLFM4 expression in the colonic crypts of NIBD controls (left) and CD patients (right). The percentage of OLFM4 staining area in colonic crypts was compared between CD patients and NIBD controls (N = 4 per group). Each gene expression was normalized to (B) RPLP0 or (C) GAPDH. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. P values were determined by the (A) Kruskal–Wallis test or the (B) Friedman test followed by the Dunn multiple comparisons test, or the (C and D) Mann–Whitney test.
We hypothesized that colonic IECs of CD patients would maintain higher stemness than IECs of NIBD controls because of decreased ALK1 expression. To test this hypothesis, we compared proliferation- and stemness-related gene expression in isolated colonic IECs between NIBD and CD patients (Figure 4C). As expected, IECs from CD patients showed significantly higher expression of LGR5 and OLFM4, and a trend toward higher levels of MKI67, PCNA, and ASCL2, than IECs from NIBD controls. Increased OLFM4 protein expression in the colonic crypts of CD patients was confirmed by immunohistochemistry (Figure 4D). Taken together, these data suggest a role for BMP9–ALK1 signaling in restricting the stemness of human colonic IECs.
BMP9–ALK1 Signaling Drives Colonic Epithelial Cell Differentiation Toward Colonocytes
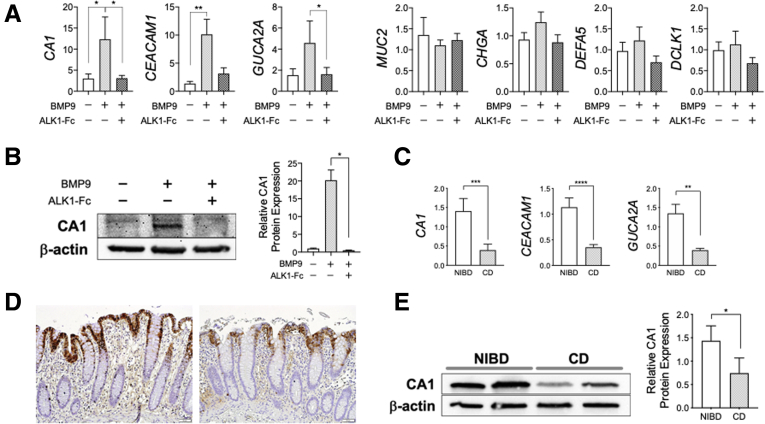
To define the impact of ALK1 signaling on IEC differentiation, we determined the expression of representative IEC lineage-specific genes,10,19 such as CA1 (colonocyte), MUC2 (goblet cell), CHGA (enteroendocrine cell), DEFA5 (Paneth cell), and DCLK1 (tuft cell), in NIBD patient-derived colonic IEC monolayers in the presence or absence of BMP9 stimulation. Treatment with BMP9 significantly up-regulated the expression of CA1 and other colonocyte markers,10 such as CEACAM1 and GUCA2A (Figure 5A), suggesting BMP9–ALK1 signaling impacts colonocyte differentiation. We further confirmed that BMP9-mediated up-regulation of CA1 expression increased CA1 protein levels (Figure 5B). The procolonocytic effect of BMP9 was mitigated significantly by addition of the ALK1–Fc chimera protein (Figure 5A and B). These data show that BMP9–ALK1 signaling preferentially drives colonic IEC differentiation toward colonocytes.
Figure 5.
BMP9–ALK1 signaling is associated with epithelial cell differentiation toward colonocytes. (A) Lineage-specific gene expression in NIBD patient-derived colonic epithelial cell monolayers. Expanded cells were cultured in expansion media in the presence or absence of BMP9 and ALK1–Fc chimera protein. Each gene expression was normalized to RPLP0 (N = 6 per group). (B) CA1 protein expression in NIBD patient-derived colonic epithelial monolayers (N = 4 per group). (C) Colonocyte marker expression in colonic epithelial cells isolated from CD patients (N = 15) and NIBD controls (N = 12). Each gene expression was normalized to GAPDH. (D) CA1 expression by immunohistochemistry in the colonic mucosa of NIBD controls (left) and CD patients (right). (E) CA1 protein expression in the colonic mucosa of NIBD controls and CD patients (N = 5 per group). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. P values were determined by the (A and B) Friedman test followed by the Dunn multiple comparison test, or the (C and E) Mann–Whitney test.
Based on these results, we hypothesized that decreased ALK1 expression in CD patients would result in decreased colonocyte marker expression compared with NIBD controls. To test this hypothesis, we compared expression of colonocyte markers in isolated colonic IECs between NIBD and CD patients (Figure 5C). CD patient-derived IECs showed significantly lower CA1, CEACAM1, and GUCA2A expression than IECs from NIBD controls. Reduced CA1 protein expression also was confirmed in the colonic mucosa of CD patients (Figure 5D and E). Taken together, these data show the role of BMP9–ALK1 signaling in colonocyte differentiation.
BMP9–ALK1 Signaling Does Not Affect Migration of Human Colonic IECs
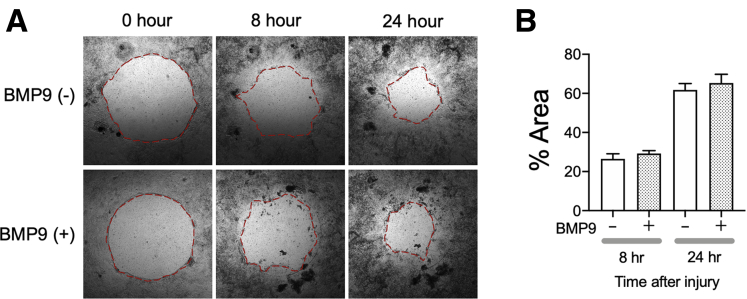
IECs migrate to wound sites and cover the denuded surface, an important step in the wound-healing process.20 We investigated the effect of BMP9–ALK1 signaling on wound healing using NIBD patient-derived colonic IEC monolayers. We measured how much of a wounded area was covered by migrated IECs within 24 hours after BMP9 stimulation compared with nonstimulated controls (Figure 6). The presence of BMP9 did not affect the amount of covered area at either 8 or 24 hours after stimulation. These results suggest that BMP9–ALK1 signaling has little or no impact on cell migration.
Figure 6.
Wound-healing assay in NIBD patient-derived colonic epithelial cell monolayers. (A) Representative images of wound healing in the presence or absence of BMP9. (B) The wounded areas covered with migrated cells were measured at 8 and 24 hours after BMP9 stimulation (N = 7 per group). The red dashed lines show the edge of the cells.
BMP9–ALK1 Signaling Enhances Human Colonic IEC Barrier Integrity
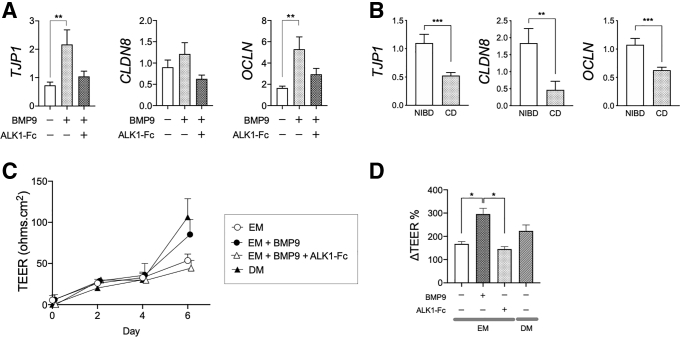
Colonocytes play a key role in barrier function, largely through strong cell–cell interactions mediated by tight junction proteins. Given its effect on colonocyte differentiation, we hypothesized that ALK1 signaling could influence barrier integrity. To test this hypothesis, we first examined gene expression of tight junction proteins, namely TJP1, CLDN8, and OCLN, in NIBD patient-derived colonic IEC monolayers in the presence or absence of BMP9 stimulation (Figure 7A). Tight junction protein gene expression was up-regulated by BMP9 and the effect was muted by the addition of the ALK1–Fc chimera protein. We also compared the expression of these genes in isolated colonic IECs between NIBD and CD patients (Figure 7B). As expected, IECs from CD patients had significantly lower TJP1, CLDN8, and OCLN expression than IECs from NIBD controls. These results suggest that BMP9–ALK1 signaling up-regulates tight junction protein expression, likely through promoting colonocyte differentiation.
Figure 7.
BMP9–ALK1 signaling enhances human colonic IEC barrier integrity. (A) Gene expression of junctional proteins in NIBD patient-derived colonic epithelial cell monolayers. Expanded cells were cultured in expansion media in the presence or absence of BMP9 and ALK1–Fc chimera protein. Each gene expression was normalized to RPLP0 (N = 6 per group). (B) Gene expression of tight junction proteins in colonic epithelial cells isolated from CD patients (N = 15) and NIBD controls (N = 12). Each gene expression was normalized to GAPDH. (C) Epithelial permeability assay in NIBD patient-derived colonic epithelial cells cultured on a collagen scaffold. TEER was measured over time (N = 3 per group). (D) Cells were stimulated with BMP9 in EM in the presence or absence of ALK1–Fc chimera protein or cultured in DM on day 4. The changes in TEER between days 4 and 6 are shown as ΔTEER% (N = 4–8 per group). ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. P values were determined by the (A and D) Kruskal–Wallis test followed by the Dunn multiple comparison test, and the (B) Mann–Whitney test.
We further tested the effect of BMP9–ALK1 signaling on barrier function using transepithelial electrical resistance (TEER) assays. IEC monolayers were generated on a collagen scaffold using NIBD patient-derived colonic crypts and then cultured in the presence or absence of BMP9 (Figure 7C). BMP9 stimulation significantly enhanced IEC barrier integrity as shown by increased ΔTEER% (Figure 7D). This BMP9-induced increase in ΔTEER% was abrogated by addition of the ALK1–Fc chimera protein. Taken together, these results suggest that increased BMP9–ALK1 signaling enhances human colonic IEC barrier integrity at least in part by up-regulating tight junction protein expression.
Decreased Colonic ALK1 Is Associated With Increased Risk of Surgery and Endoscopic Relapse in CD Patients
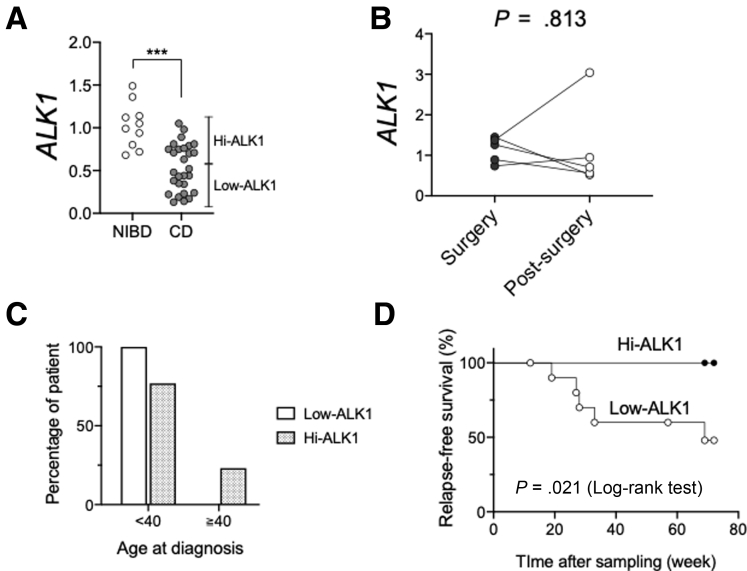
We hypothesized that impaired ALK1 signaling, in part owing to the associated defects in colonocyte maturation and barrier integrity, would be associated with poor clinical outcomes in CD. To study this, we examined ALK1 expression in the colonic mucosa of a new, independent cohort of 28 CD patients and 10 NIBD controls using endoscopically collected biopsy samples (Figure 8A). CD patients showed lower colonic ALK1 expression than NIBD controls as a whole, but a subset of CD patients showed markedly lower ALK1 expression (low-ALK1 CD subset) than the other CD patients (hi-ALK1 CD subset). Therefore, we compared the clinical characteristics between these ALK1-based CD subsets (Table 2). Interestingly, low-ALK1 patients were more likely to experience bowel resections than hi-ALK1 patients (93.3% vs 53.8%; P = .029). We examined the potential effect of surgery on colonic ALK1 expression among 5 matched CD patients by prospectively determining ALK1 expression after bowel resections. Within the median 6-month follow-up period, there was no significant change in the colonic ALK1 expression (Figure 8B). To evaluate the net impact of colonic ALK1 expression on the risk of surgery in CD patients, we conducted a multivariate analysis. In addition to well-accepted risk factors21 that include young age (age, <40 y) at diagnosis, extensive disease (disease location22), use of corticosteroids to control the index flare, and perianal disease at diagnosis, we used membership in the colonic ALK1-based CD subset and use of anti–tumor necrosis factor α agents as additional independent variables that could impact the outcome (risk of surgery). Among these potential risk factors, young age at diagnosis was removed from subsequent analysis because of its significant correlation with the colonic ALK1-based CD subset23 (Table 3 and Figure 8C). A binomial logistic regression analysis showed that having a low-ALK1 CD subtype is an independent risk factor for surgical resection in CD patients (Table 4). Next, we prospectively tracked endoscopic disease activity in 21 patients who were in remission at the time of sample collection. Within a 72-week observational period, 5 of 12 low-ALK1 and 0 of 9 hi-ALK1 CD patients experienced an endoscopic relapse. A significantly higher risk of endoscopic relapse was shown in patients in the low-ALK1 CD subset compared with the hi-ALK1 subset (P < .05, log-rank test) (Figure 8D). All patients showed exacerbated inflammation in the colon except for 1 patient with endoscopic relapse only in ileum. Taken together, these data suggest that decreased colonic ALK1 expression is associated with increased risk of surgery and endoscopic relapse in patients with CD.
Figure 8.
Decreased colonic ALK1 is associated with a poor clinical outcome in CD patients. (A) ALK1 expression was quantified in colonic biopsy samples obtained from NIBD controls (N = 10) and CD patients (N = 28) by qPCR. (B) ALK1 expression in the colonic mucosa of CD patients at the time of surgery and after surgery (N = 5 per group). (C) Percentages of CD patients who were diagnosed as CD before and after age 40 years in low-ALK1 (N = 15) and hi-ALK1 (N = 13) CD subsets. (D) Kaplan–Meier survival analysis to evaluate the impact of colonic ALK1 expression on endoscopic relapse in patients with CD (N = 12 for low-ALK1 and 9 for hi-ALK1 CD subgroups). ALK1 expression was normalized to GAPDH. ∗∗∗P < .001. P values were determined by the (A) Mann–Whitney test, (B) Wilcoxon test, and (D) log-rank test.
Table 2.
Clinical Characteristics at the Time of Sample Collection
| Low-ALK1 | Hi-ALK1 | P value | Total | |
|---|---|---|---|---|
| Total number | 15 | 13 | 28 | |
| Sex, male/female, n (%) | 6 (40)/9 (60) | 11 (84.6)/2 (15.4) | .024a | 17 (60.7)/11 (39.2) |
| Age at sampling, y, mean (SD) | 34.1 (8.8) | 40.4 (14.5) | .310b | 37.0 (12.0) |
| Disease duration, y, mean (SD) | 12.2 (9.1) | 11.6 (8.5) | .856b | 11.9 (8.7) |
| Former/current smoker, n (%) | 7 (46.7) | 4 (30.8) | .460a | 11 (39.3) |
| Disease extent | ||||
| L1/L2/L3, n (%) | 4 (26.7)/1 (6.7)/10 (66.7) | 4 (30.8)/2 (15.4)/7 (53.8) | 8 (28.6)/3 (10.7)/17 (60.7) | |
| L4, n (%) | 3 (20) | 1 (7.7) | .600a | 4 (14.3) |
| Ileal involvement, n (%) | 14 (93.3) | 10 (76.9) | .311a | 24 (85.7) |
| Disease behavior | ||||
| B1/B2/B3, n (%) | 1 (6.7)/11 (73.3)/3 (20.0) | 4 (30.8)/5 (38.5)/4 (30.8) | 5 (17.9)/16 (57.1)/7 (25) | |
| Perianal disease, n (%) | 5 (33.3) | 5 (38.5) | >.999a | 10 (35.7) |
| Stricture, n (%) | 11 (73.3) | 5 (38.5) | .125a | 16 (57.1) |
| Disease activity | ||||
| SES-CD, mean (SD) | 2.9 (3.1) | 4.4 (5.2) | .626b | 3.6 (4.2) |
| Endoscopic remission, n (%) | 12 (80.0) | 9 (69.2) | .670a | 22 (78.6) |
| Abnormal CRP (≥5.0 mg/L), n (%) | 2/11 (18.2) | 0/8 (0.0) | .485a | 2/19 (10.5) |
| Treatment history | ||||
| Aminosalicylates, n (%) | 0 (0.0) | 3 (23.1) | .087a | 3 (10.7) |
| Oral steroids, n (%) | 4 (26.7) | 5 (38.5) | .689a | 9 (32.1) |
| Immunosuppressants, n (%) | 7 (46.7) | 4 (30.8) | .460a | 11 (39.3) |
| Anti-TNFα agents, n (%) | 9 (60.0) | 6 (46.2) | .705a | 15 (53.6) |
| Surgery, n (%) | 14 (93.3) | 7 (53.8) | .029a | 21 (75.0) |
CRP, C-reactive protein; TNF, tumor necrosis factor.
P values were determined by the Fisher exact test.
P values were determined by the Mann–Whitney test.
Table 3.
Correlations Between Explanatory Variables for the Risk of Surgery
| Correlations | ALK1 subset | Age <40 y at diagnosis | Disease location | Corticosteroids | Anti-TNF α | Perianal disease |
|---|---|---|---|---|---|---|
| ALK1 subset | 0.372∗ | 0.161 | 0.045 | 0.138 | 0.053 | |
| Age <40 y at diagnosis | 0.372∗ | 0.270 | 0.292 | 0.141 | 0.017 | |
| Disease location | 0.161 | 0.270 | 0.339 | 0.324 | 0.243 | |
| Corticosteroids | 0.045 | 0.292 | 0.339 | 0.045 | 0.141 | |
| Anti-TNFα | 0.138 | 0.141 | 0.324 | 0.045 | 0.096 | |
| Perianal disease | 0.053 | 0.017 | 0.243 | 0.141 | 0.096 |
NOTE. Correlations between categoric variables were analyzed by the chi-squared test. The strength of correlations measured by Cramer's V test is shown in the table.
TNF, tumor necrosis factor.
P < .05.
Table 4.
Binomial Logistic Regression Analysis for Surgery
| Variables | Regression coefficient | SD | P value | OR | 95% CI of OR |
|---|---|---|---|---|---|
| Low-ALK1 CD subset | 2.91 | 1.37 | .03 | 18.33 | 1.26–266.39 |
| Disease location | 0.85 | 0.68 | .21 | 2.34 | 0.62–8.81 |
| Corticosteroids | -0.38 | 1.46 | .80 | 0.68 | 0.04–11.93 |
| Anti-TNFα agents | -0.86 | 1.17 | .46 | 0.42 | 0.04–4.21 |
| Perianal disease | 0.61 | 1.30 | .64 | 1.85 | 0.14–23.63 |
OR, odds ratio; TNF, tumor necrosis factor.
Discussion
ALK1 is a transmembrane serine/threonine-receptor kinase that belongs to the TGF-β–receptor family. In contrast to the broad ligand-binding specificity of other type 1 receptors for TGF-β signaling molecules (ALK2–7), ALK1 specifically binds to BMP9 and BMP10 secreted from the liver24 and heart,25 respectively. ALK1 is expressed in endothelial cells and plays a crucial role in the development of direct connections between arteries and veins.18 Defects in ALK1 signaling lead to arteriovenous malformations and cause hereditary hemorrhagic telangiectasia, which is characterized by mucocutaneous telangiectasia and gastrointestinal hemorrhage, in genetically predisposed patients.26 Recent tissue proteome analyses showed ALK1 expression in human IECs as well as endothelial cells in small and large intestines.27 However, the role of ALK1 signaling in colonic IEC homeostasis and the clinical impact of attenuated ALK1 signaling in CD have remained unknown. In this study, we identified ALK1 as a target of miR-31-5p.4,5 We showed a dramatic reduction of ALK1 in the colonic IECs of CD patients and showed that ALK1 controls colonocyte differentiation and intestinal epithelial barrier capacity.
BMP4 signaling through ALK3 was reported previously to restrict the stemness of murine leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5)-positive intestinal stem cells.28 Although ALK1 and ALK3 affect downstream Smad signaling pathways similarly to regulate gene expression,16 the role of ALK1-related signaling on IEC stemness was unknown. Therefore, we examined the impact of ALK1 signaling on the stemness of human colonic IECs ex vivo using primary-cultured IECs. BMP9–ALK1 signaling reduced the expression of stemness-related genes including LGR5, restricted proliferation, and directed the fate of human IEC differentiation toward colonocytes. Because differentiation of the LGR5-positive cycling stem cell is accompanied by a loss of proliferative ability, these data clearly show that BMP9–ALK1 signaling restricts human IEC stemness by promoting colonocyte differentiation. Decreased ALK1 expression was associated with increased stemness-related marker expression and decreased colonocyte differentiation in the colonic IECs of CD patients compared with NIBD controls. Although the relative abundance of ALK1 across different colonic epithelial cell types is not yet confidently known, we observed stronger ALK1 expression in the upper half of colonic crypts compared with the lower half. Intriguingly, this ALK1 distribution is mostly matched with that of CA1 in colonic epithelium, implying that BMP9–ALK1 signaling plays an important role not only in the stem cell differentiation toward colonocytes but also the maturation of colonocytes. We also showed that increased BMP9–ALK1 signaling enhanced IEC barrier integrity by up-regulating tight junction protein expression and decreasing paracellular permeability. Although the mechanism of decreased paracellular permeability in BMP9-stimulated IECs needs further investigation, the effect of BMP9–ALK1 signaling in colonocyte differentiation and the role of colonocytes in colonic barrier integrity9 implies that activated ALK1 signaling might enhance barrier integrity by driving colonocyte differentiation and maturation.
Based on this rationale, we hypothesized that attenuated ALK1 signaling might result in a poor clinical outcome in CD patients owing to disrupted colonic barrier function. Association analyses with clinical outcomes showed that decreased colonic ALK1 expression is an independent risk factor for surgery and is associated with an increased risk of endoscopic relapse in the future. Despite the demonstrated role of ALK1 signaling in epithelial barrier function, loss-of-function mutations in ALK1 do not result in spontaneous intestinal inflammation in patients with hereditary hemorrhagic telangiectasia.29 This fact is consistent with previous lines of evidence from rodents30, 31, 32 and asymptomatic first-degree relatives of CD patients,2 suggesting that increased epithelial permeability is necessary but not sufficient for the development of intestinal inflammation and requires another trigger for disease manifestation in CD.33 Nonetheless, our data suggested that disrupted ALK1 signaling might be one of the disease-modifying factors that contribute to the heterogeneity in clinical outcomes among CD patients. Finding markers that predict the natural course of CD is the holy grail of medical researchers in the field.34 Our findings suggest that colonic ALK1 expression levels may help identify these high-risk patients who may require early intensified therapeutic intervention, perhaps staving off a poor clinical outcome. Intriguingly, we found a significant correlation between colonic ALK1-based CD subset and young age at diagnosis as independent variables for the regression analysis. The younger age at diagnosis in the low-ALK1 CD subset compared with the hi-ALK1 subset might partially explain the unknown mechanism why young age at diagnosis can be a risk factor for the disabling course in CD patients. Our findings also show a potential use of ALK1 as a novel target of medical treatment in CD. An increasing number of drugs targeting mucosal immune cells currently are available in the treatment for CD; however, there is no established treatment targeting IEC defects.8 A small biologic potentiator of colonic ALK1 expression could be a treatment targeting IECs in CD.
miR-31-5p expression in primary-cultured colonic IEC monolayers did not recapitulate the whole tissue findings in NIBD and CD patients. This highlights an important role for the environmental factors in regulating miR-31-5p dysregulation in CD patients. Reduced miR-31-5p colonic expression by anti-inflammatory galacto-oligosaccharide exposure35 as well as its enhanced expression in interleukin 10–deficient mice36 may indicate that changes in the intestinal microbiome and/or proinflammatory and anti-inflammatory pathways that are associated with CD may directly affect colonic ALK1 expression via miR-31-5p. In addition, intestinal radiation injury37 promotes small intestinal miR-31-5p expression, which may suggest epithelial injury in CD up-regulates this microRNA and thereby affects ALK1 expression. Nonetheless, the identification of CD-associated external influences on ALK1 awaits further investigation.
Molecular mechanisms downstream of activated ALK1 signaling, especially as they pertain to classic pathways such as WNT/NOTCH/BMP2, 4, 5, along the crypt gradient is an important area of future research. Although these signaling pathways are well studied in the murine model, they remain an active area of research in human beings. We provide evidence for an association between decreased ALK1 expression and NOTCH activity in the colonic mucosa of CD patients. Because the expression of NOTCH target genes including HES1 is crucial for colonocyte differentiation,38 our work implies that ALK1 signaling might promote human colonocyte differentiation at least in part by enhancing NOTCH target gene expression.
We also did not investigate the impact of decreased E2F Transcription Factor 2 (E2F2), another candidate target of miR-31-5p, in CD patients. E2F2 is a transcriptional factor that plays an important role in regulating cell proliferation.39,40 Our data suggested that miR-31-5p regulation of E2F2 expression does not occur in colonic IECs of CD patients. However, increased miR-31-5p also was found in CD14-negative resident macrophages and B lymphocytes in the colon of CD patients.4 Future investigation will be required to understand the potential impact of decreased E2F2 expression in these nonepithelial cells. We did not examine the impact of ALK1 signaling on IECs from the small intestine. We previously reported an up-regulation of miR-31-5p in the ileal tissue of CD patients compared with NIBD controls.4 Given that ALK1 is expressed in the IECs of the small intestine as well as the colon,27 investigation of the role of ALK1 signaling in small intestinal IECs will be informative. We also did not investigate the impact of decreased ALK1 on vascular development in the intestine. ALK1 has a well-accepted role in vascular development, thus increased miR-31-5p might affect vascular formation in the intestine of CD patients. Our immunohistochemistry staining shows ALK1 expression is decreased specifically in colonic IECs, but not in vascular endothelial cells of CD patients (Figure 1D), confirming that IECs are the major target of miR-31-5p in colon. Finally, serum BMP9 levels also were not different in CD vs NIBD patients, suggesting that circulating BMP9 levels may not directly impact local ALK-mediated effects observed in the intestine. This can be the result of a host of reasons stemming from the physiological levels required to mediate effects, individual clearance differences, and so forth. We did not examine the effect of BMP10, another ligand for ALK1, on ALK1 signaling in IECs. Both BMP9 and BMP10 can bind to ALK1 with high affinity in vivo. However, in contrast to BMP9, which circulates in a biologically active form,24 there remains an active debate about the latency of BMP10.41 Therefore, we selected BMP9 for our ex vivo experiments. Nevertheless, distinct roles of these 2 BMPs were reported previously in tumor growth, and thus further investigation of the impact of BMP10 on IEC biology will be necessary in the future.
This study investigated the mechanism of CD using human primary IEC monolayers ex vivo. Our innovative technologies, such as cell proliferation, wound healing, and epithelial permeability assays in 2-dimensional (2D)-cultured primary IECs, will promote further research to understand mechanisms of inflammatory bowel disease and develop novel drugs targeting IECs in the future.
Materials and Methods
Subjects, Samples, and Clinical Information
Colonic mucosa was obtained from surgically resected colon or endoscopically taken biopsy specimens from patients with an established diagnosis of CD and NIBD healthy controls between April 2012 and November 2019. Cross-sectional clinical data were collected at the time of sampling. All samples were collected from disease-unaffected regions without macroscopic inflammation. All biopsy samples were collected from the ascending colon. Additional information for surgical samples is summarized in Table 5. Endoscopic disease activity was evaluated by the Simple Endoscopic Score for Crohn's Disease (SES-CD)42 in CD patients using information extracted retrospectively from endoscopy reports. Endoscopic remission was defined as SES-CD of less than 4.43 In patients with endoscopic remission, endoscopic disease activity was followed up prospectively for up to 72 weeks after sampling to examine endoscopic relapse, defined as SES-CD of 4 or higher.
Table 5.
Clinical Information of Surgical Samples
| Age, y | Sex | Disease | Sample location |
|---|---|---|---|
| 68 | Male | Adenoma/benign tumor | Left side |
| 62 | Male | Adenoma/benign tumor | Right side |
| 74 | Female | Adenoma/benign tumor | Right side |
| 52 | Female | Colon cancer | Right side |
| 51 | Male | Colon cancer | Transverse |
| 80 | Female | Colon cancer | Right side |
| 43 | Male | Colon cancer | Right side |
| 50 | Male | Colon cancer | Left side |
| 83 | Female | Colon cancer | Right side |
| 44 | Male | Colon cancer | Left side |
| 44 | Female | Colon inertia | Left side |
| 41 | Female | Colon inertia | Right side |
| 51 | Female | Constipation | Right side |
| 50 | Female | Crohn's disease | Right side |
| 66 | Female | Crohn's disease | Right side |
| 44 | Male | Crohn's disease | Rectum |
| 21 | Male | Crohn's disease | Left side |
| 13 | Male | Crohn's disease | Right side |
| 49 | Female | Crohn's disease | Right side |
| 28 | Female | Crohn's disease | Right side |
| 20 | Male | Crohn's disease | Right side |
| 45 | Female | Crohn's disease | Right side |
| 18 | Male | Crohn's disease | Right side |
| 49 | Female | Crohn's disease | Right side |
| 47 | Male | Crohn's disease | Right side |
| 29 | Female | Crohn's disease | Right side |
| 26 | Male | Crohn's disease | Right side |
| 75 | Male | Crohn's disease | Right side |
| 26 | Male | Crohn's disease | Right side |
| 45 | Female | Crohn's disease | Transverse |
| 53 | Female | Crohn's disease | Right side |
| 35 | Male | Crohn's disease | Right side |
| 44 | Female | Crohn's disease | Right side |
| 43 | Female | Crohn's disease | Right side |
| 34 | Male | Crohn's disease | Right side |
| 16 | Female | Crohn's disease | Right side |
| 52 | Male | Crohn's disease | Rectum |
| 53 | Female | Crohn's disease | Right side |
| 33 | Male | Crohn's disease | Right side |
| 23 | Male | Crohn's disease | Right side |
| 18 | Male | Crohn's disease | Right side |
| 23 | Male | Crohn's disease | Right side |
| 70 | Male | Crohn's disease | Right side |
| 42 | Male | Crohn's disease | Right side |
| 45 | Female | Crohn's disease | Right side |
| 27 | Female | Crohn's disease | Right side |
| 68 | Male | Diverticulitis | Left side |
| 64 | Female | Diverticulitis | Left side |
| 73 | Female | Diverticulitis | Left side |
| 58 | Female | Diverticulitis | Left side |
| 54 | Male | Diverticulitis | Left side |
| 48 | Female | Diverticulitis | Left side |
| 76 | Male | Diverticulitis | Left side |
| 54 | Female | Diverticulitis | Left side |
| 47 | Male | Diverticulitis | Left side |
| 64 | Female | Diverticulitis | Left side |
| 65 | Female | Diverticulitis | Left side |
| 60 | Male | Ischemic colitis | Right side |
| 76 | Female | Sigmoid volvulus | Left side |
| 49 | Female | Submucosal tumor | Right side |
| 64 | Male | Submucosal tumor | Right side |
NOTE. Right side refers to the cecum to the ascending colon, transverse refers to the transverse colon, and left side refers to the descending colon to the sigmoid colon.
Isolation of Colonic Crypts and Primary Culture of Human IECs
Isolation of colonic crypts and subsequent generation of human primary IEC cultures were performed as we reported previously.44,45 In brief, colonic mucosa was incubated in the isolation buffer44,45 for 75 minutes at room temperature. The tissue was rinsed with phosphate-buffered saline and vigorously shaken by hand to release the crypts. The released crypts were placed into culture immediately or suspended in expansion media (EM)45 containing 20% fetal bovine serum and 10% dimethyl sulfoxide to store at -80°C until use.
For 2D cultures, isolated crypts were spread directly on 48-well plates coated with collagen hydrogel (356236; Corning, Corning, NY).44 Cells were cultured in EM with 10 mmol/L Y-27632 (S1049; Selleckchem, Houston, TX) during the initial 48 hours. The medium was changed every 48 hours. Every 6 days, cells were dissociated in collagenase type IV followed by Accutase (07920; Stemcell Technologies, Vancouver, BC, Canada) and the fragmented cells were passaged to a new collagen-coated plate.
In several experiments, cells were expanded in EM for 4 days and subsequently stimulated with 10 ng/mL BMP9 (553102; BioLegend, San Diego, CA) and 100 ng/mL ALK1–Fc chimera protein (370-AL; R&D Systems, Wayzata, MN), a soluble chimeric protein consisting of the extracellular part of ALK1 fused to a Fc fragment, which inhibits BMP9–ALK1 interaction and signaling.46
Epithelial Permeability Assay
Epithelial permeability was evaluated in 2D-cultured human primary IEC monolayers as we reported previously.47 Primary IECs cultured on 48-well plates were passaged to 12-well cell culture inserts (353180; BD Falcon, San Jose, CA) coated with collagen scaffolds with a gradient of cross-linking48 at a ratio of 1:1. Cells were expanded in EM for 4 days and stimulated with 10 ng/mL BMP9 alone or in combination with 100 ng/mL ALK1–Fc chimera protein in EM or cultured in differentiation medium (DM) as a positive control of colonocyte differentiation44 and TEER increase for an additional 2 days. In contrast to EM, DM lacks exogeneous Wnt family member 3A (WNT3A), R-SPONDIN 3, and NOGGIN produced from L-WRN cells (CRL-3276; ATCC)45 and SB202190 (S1077; Selleckchem), a selective p38 mitogen-activated protein kinase inhibitor. The electrical resistance between the upper and lower surfaces of IEC monolayers was measured before (day 4) and after 2 days of stimulation (day 6) using an EVOM2 epithelial Volt/Ohm meter (World Precision Instruments, FL, USA). IEC monolayer resistance was corrected by subtracting the resistance of scaffolds without cells and normalized by multiplying the effective scaffold surface area to provide a TEER in the unit of Ω cm2. Changes in TEER between day 4 and day 6 were evaluated by calculating ΔTEER% as follows: ΔTEER% =100- ([(TEERday4 – TEERday6)/TEERday4] × 100).47
3’UTR Reporter Assay and Site-Directed Mutagenesis
Plasmid DNA was extracted from a miRNA 3’UTR target clone for ALK1 (HmiT022834-MT06; GeneCopoeia, MD, USA) using the Plasmid Midi Kit (12143; Qiagen, MD, USA). This plasmid DNA then was transfected into HEK293T cells with or without miRNA mimics, double-stranded oligonucleotides designed to mimic the function of endogenous mature miRNAs, for hsa–miR-31-5p, hsa–miR-122a-5p, or hsa–miR-215-5p, or negative control mimics (C-300507-05-0005, C-300591-05, C-300570-05, CN-001000-01; Dharmacon Lafayette, CO). Cells transfected with plasmid DNA in the absence of miRNA mimics were defined as mock. For mutagenesis assays, putative miR-31-5p binding sequences were deleted in the plasmid DNA using PrimeSTAR Mutagenesis Basal Kit (R046A; TaKaRa Bio, CA) according to the manufacturer’s instructions. Successful deletions were checked by Sanger DNA sequencing (Eurofins Genomics Louisvile, KY) and subsequently analyzed with Sequencher 5.4.6 (Gene Codes Ann Arbor, MI). Dual luciferase reporter assays were performed on the Dual Luciferase Reporter Assay System (Promega Madison, WI) using the Luc-Pair Duo-Luciferase HS Assay Kit (LF004; GeneCopoeia) according to the manufacturer’s instructions.
EdU Assay
The EdU-based staining was performed on 2D-cultured primary IECs. Cells were subcultured from 48-well plates to 96-well plates at a ratio of 1:3, expanded in EM for 4 days, followed by either an additional 2 days in EM, with or without 10 ng/mL BMP9 alone or in combination with 100 ng/mL ALK1–Fc chimera protein, or an additional 2 days in DM. Cells were incubated with 10 mmol/K EdU (10540; Lumiprobe, Hunt Valley, MD) for 3 hours at 37°C. DNA-incorporated EdU was detected by sulfo-cyanine5-azide (A3330; Lumiprobe) through a copper-catalyzed covalent reaction between an azide and an alkyne.49 Images were captured using a Nikon Eclipse TE2000-U inverted microscope and processed using NIS-Elements AR version 3.2 software (Nikon, Tokyo, Japan). The EdU fluorescence area was measured by ImageJ software (National Institutes of Health, Bethesda, MD) and normalized by the total cell area occupied by the Hoechst 33342 fluorescence (62249; Thermo Fisher Scientific Waltham, MA), as we described previously.45
Isolation of Colonic Epithelial Cells
Colonic epithelial cells were isolated as previously described.4 In brief, dissected colonic mucosa was cut into small pieces and incubated in magnesium-free Hank’s balanced salt solution (HBSS) containing 2 mmol/L EDTA and 2.5% heat-inactivated fetal bovine serum for 30 minutes with shaking at 37°C. To remove mucus, 1 mmol/L dithiothreitol was added and incubated for an additional 10 minutes with shaking. Collected supernatants were centrifuged, resuspended in HBSS containing 1 mg/mL collagenase type 4 (17104019; Thermo Fisher Scientific), and incubated for 10 minutes at 37°C to further remove the mucus. The fraction was pelleted, resuspended in HBSS, passed through a 40-um filter, and overlayered on 50% Percoll. Cells were centrifuged at 2000 rpm for 20 minutes at room temperature and viable colonic IECs were recovered from the interface layer.
Immunohistochemistry
Immunostaining was performed as described previously.50 Rabbit anti-human ALK1 antibody, rabbit anti-human CA1 antibody, rabbit anti-human OLFM4 antibody, and mouse/rabbit IgG VisUCyte horseradish-peroxidase polymer antibody were purchased from Sigma-Aldrich (St. Louis, MO) (HPA007041), Novus Biologicals (Littleton, CO) (NBP1-88191), Cell Signaling Technology (Beverly, MA) (14369S), and R&D Systems (VC002-025). The images were captured on an inverted microscope (Olympus IX71) with cellSens standard software (Olympus, Tokyo, Japan).
Western Blot Analysis
Western blot analyses were performed on whole-cell extracts.50 Goat anti-human ALK1 antibody (AF370-SP) and goat IgG horseradish-peroxidase–conjugated antibody (HAF109) were purchased from R&D Systems. Rabbit anti-human CA1 antibody was obtained from Novus Biologicals (NBP1-88191). Rabbit anti-human NOTCH1, NOTCH2, JAG1, and β-actin antibodies were obtained from Cell Signaling Technology (3608S, 5732S, 70109S, and 4970S). The images were captured on a chemiluminescent image reader (iBright Imaging Systems; Invitrogen, Waltham, MA) and analyzed by ImageJ software. Each protein expression was normalized to β-actin.
Wound Healing Assay
Colonic IECs were expanded on 12-well cell culture inserts (353180; BD Falcon) and coated with collagen hydrogel (356236; Corning) for 4 days to make confluent IEC monolayers. After culture in DM for 2 days, 1 mm2 of cells were cut off from 8 different portions of monolayers by biopsy punches to generate cell-free wounded areas. To stop cell proliferation and examine the net impact of BMP9 stimulation on cell migration, cells subsequently were cultured in DM lacking fetal bovine serum in the presence or absence of 10 ng/mL BMP9 for an additional 24 hours. The wounded areas were imaged with a confocal microscope at 8 and 24 hours. The average percentages of the initial wounded areas covered by migrating cells were evaluated using ImageJ software.
Enzyme-Linked Immunosorbent Assay
Serum samples were obtained from NIBD and CD patients, and stored at -80°C until use. Before use, frozen serum was thawed on ice and the serum BMP9 concentration was measured on a CLARIOstar microplate reader (BMG Labtech, Cary, NC) by enzyme-linked immunosorbent assay using the human BMP9 enzyme-linked immunosorbent assay kit (HLH-BMP9-1; RayBiotech, Atlanta, GA) according to the manufacturer’s instruction.
Reverse-Transcriptase qPCR Analysis
Total RNA was extracted from dissected colonic mucosa using TRIzol reagent (Thermo Fisher Science) and purified with the Total RNA Purification Kit (17200; Norgen Biotek, Thorold, ON, Canada) according to the manufacturer’s instruction. Total RNA was extracted from isolated and cultured colonic IECs using the Single Cell RNA Purification Kit (51800; Norgen Biotek). Complementary DNA for mRNA was generated from 500 ng of RNA using the High-Capacity Complementary DNA Reverse Transcription Kit (4368814; Thermo Fisher Science). Complementary DNA for miRNAs was generated from 10 ng RNA using the TaqMan MicroRNA Reverse Transcription Kit (4366596; Thermo Fisher Science) with individual miRNA-specific primers (TaqMan miRNA assays, assay ID: 001006 [RNU48], 002279 [miR-31-5p]). qPCR for mRNAs was performed on the QuantStudio 3 RT-PCR system using PowerUp SYBR Green Master Mix (A25776; Thermo Fisher Science) or BrightGreen 2× qPCR MasterMix-Lox ROX (MasterMix-LR; Applied Biological Materials, Richmond, BC, Canada). Each gene expression was normalized to GAPDH or RPLP0. Primer sequences are listed in Table 6.
Table 6.
Primer Sequences for RT-qPCR
| Gene | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| ALK1 | CCTAGCTCAGATGATGCGGG | GGCTTCTCTGGACTGTTGCT |
| ASCL2 | ACGCTTTGGGCCTTACAGAA | GCAGGCGGCTGGTTTTTAAA |
| CA1 | ACAACGATAACCGATCAGTGC | CTTTGCAGAATTCCAGTGAGC |
| CEACAM1 | AACAGCAGAGGTGACAGAGC | GGGTACACGCACTCTGTGAA |
| CHGA | GAGATCCGGAAAGGCGAGAG | GTGCTCCTGTTCTCCCTTCC |
| CLDN8 | GTGGATGAATTGCGTGAGGC | AGCCAAGAAGGACATCACGG |
| DCLK1 | TGAACGTCAAGACCACCTCG | ATGATGGTGACCAGCTTGGG |
| DEFA5 | TCCTTGCTGCCATTCTCCTG | GTCCTGGTTGTCTTCCCCAG |
| E2F2 | TCCACTGGGCTGCCATTTAG | ATTCCCAATGGGTCCTGCAG |
| GAPDH | GACCTGCCGTCTAGAAAAACC | GCTGTAGCCAAATTCGTTGTC |
| GUCA2A | TCTCTGCAAGGAGCCCAATG | CTAGCATCCGGTACAGGCAG |
| LGR5 | TCCAACCTCAGCGTCTTCAC | TTCCCGCAAGACGTAACTCC |
| MKI67 | AAGGAGTGACTCGTGGCTTG | GAGCCAGTTTGAGGTCGTGA |
| MUC2 | ATCTTCATGGGGAGGACGGA | TAGGAGGGAGCCAGCTTGAT |
| OCLN | CATTGCCATCTTTGCCTGTG | AGCCATAACCATAGCCATAGC |
| OLFM4 | AAGGACTGTATTGGGTGGCG | TCCGCAACTCTCGAGCATTT |
| PCNA | TCTGAGGGCTTCGACACCTA | TATCCGCGTTATCTTCGGCC |
| RPLP0 | AATCTCCAGGGGCACCATTG | GAACACCTGCTGGATGACCA |
| TJP1 | TCACGCAGTTACGAGCAAGT | TGAAGGTATCAGCGGAGGGA |
RT, reverse-transcription.
qPCR for miRNAs was performed using the TaqMan Universal PCR Master Mix with individual miRNA-specific probes. Expression of miR-31-5p was normalized to RNU48.
The 2-ΔΔCT method was used to compare relative changes in gene expression across samples. To calculate ΔΔCT values for each sample, the average of ΔCT values in control samples, such as nonstimulated primary-cultured IECs or NIBD-derived isolated IECs, was calculated at first to get the baseline of each gene expression. Then, all the ΔΔCT values were calculated using this averaged ΔCT value to determine the changes of each gene expression in individual samples. Relative mRNA expression was determined by calculating the 2-ΔΔCT value for each sample.
Statistical Analysis
All numeric data in the figures are expressed as means ± SD or SEM. Differences between the 2 groups were analyzed by a Wilcoxon, Mann–Whitney, or the Fisher exact test. Differences among the 3 groups were analyzed by a Kruskal–Wallis or Friedman test, followed by the Dunn multiple comparison test. Trends between variables were analyzed by the Jonckheere–Terpstra test. The Spearman correlation coefficient was used for evaluating correlations between 2 numeric variables. Correlations between 2 categoric variables were evaluated by a chi-square test with Cramer’s V to measure the strength of correlations. P values less than .05 were considered significant. The Kaplan–Meier method was used to generate survival curves and differences between 2 groups were evaluated by a log-rank test. GraphPad Prism (version 8.0 software; GraphPad Software) and R software version 3.5.2 were used for these data analyses. A binomial logistic regression analysis also was performed using R software.
Ethical Statement
This study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice. The study protocol was approved by the Institutional Review Board at the University of North Carolina at Chapel Hill (approval numbers: 19-0819 and 17-0236). All participants provided written informed consent before inclusion in the study. All participants were identified by number and not by name or any protected health information.
All authors had access to the study data and reviewed and approved the final manuscript.
Footnotes
Conflicts of interest These authors disclose the following: Nancy L. Allbritton and Yuli Wang have a financial interest in Altis Biosystems. The remaining authors disclose no conflicts.
CRediT Authorship Contributions Takahiko Toyonaga, MD, PhD (Conceptualization: Lead; Formal analysis: Lead; Investigation: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead); Erin C. Steinbach (Investigation: Supporting); Benjamin P Keith (Data curation: Lead; Formal analysis: Lead; Investigation: Supporting); Jasmine B Barrow (Conceptualization: Supporting; Investigation: Supporting; Resources: Supporting); Matthew R Schaner (Resources: Lead); Elisabeth A Wolber (Resources: Lead); Caroline Beasley (Resources: Lead); Jennifer Huling (Formal analysis: Supporting; Investigation: Supporting; Methodology: Lead; Visualization: Lead); Yuli Wang (Methodology: Lead); Nancy L Allbritton (Methodology: Lead; Supervision: Lead); Nicole Chaumont (Resources: Lead); Timothy S Sadiq (Resources: Lead); Mark J Koruda (Resources: Lead); Animesh Jain (Resources: Supporting); Millie D. Long (Resources: Supporting); Edward L. Barnes (Resources: Supporting); Hans H. Herfarth (Resources: Supporting); Kim L. Isaacs (Resources: Supporting); Jonathan J. Hansen (Resources: Supporting); Michael T. Shanahan (Writing – review & editing: Supporting); Reza Rahbar (Resources: Lead); Terrence S Furey (Data curation: Lead; Formal analysis: Supporting; Funding acquisition: Supporting; Project administration: Supporting; Supervision: Lead; Writing – review & editing: Lead); Praveen Sethupathy (Formal analysis: Supporting; Funding acquisition: Supporting; Project administration: Supporting; Supervision: Lead; Writing – review & editing: Lead); Shehzad Z Sheikh, MD, PhD (Conceptualization: Lead; Formal analysis: Supporting; Funding acquisition: Lead; Methodology: Supporting; Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Supporting; Writing – review & editing: Lead).
Preprint server: This manuscript was posted to bioRxiv (doi: https://doi.org/10.1101/2020.02.21.960070). Transcript profiling was as follows: nucleotide sequence data sets discussed here are available through Gene Expression Omnibus (GEO) under accession number GSE85499 (RNA-seq; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85499) and GSE101819 (smRNA-seq; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101819).
Funding This work was funded in part through Helmsley Charitable Trust (SHARE Project 2); National Institute of Diabetes and Digestive and Kidney Diseases grants P01 DK094779, 1R01DK104828-01A1, R01DK109559, and P30-DK034987; T32 DK007737 (E.C.S); the Thurston Arthritis Research Center and the National Institute of Allergy and Infectious Diseases Loan Repayment Program (E.C.S.); and Career Development Award (567497) and Research Fellow Award (634239) from the Crohn’s and Colitis Foundation. The University of North Carolina Translational Pathology Laboratory is supported in part by grants from the National Cancer Institute (3P30CA016086).
References
- 1.Torres J., Mehandru S., Colombel J.F., Peyrin-Biroulet L. Crohn's disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 2.Hollander D., Vadheim C.M., Brettholz E., Petersen G.M., Delahunty T., Rotter J.I. Increased intestinal permeability in patients with Crohn's disease and their relatives. A possible etiologic factor. Ann Intern Med. 1986;105:883–885. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 3.Martini E., Krug S.M., Siegmund B., Neurath M.F., Becker C. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keith B.P., Barrow J.B., Toyonaga T., Kazgan N., O'Connor M.H., Shah N.D., Schaner M.S., Wolber E.A., Trad O.K., Gipson G.R., Pitman W.A., Kanke M., Saxena S.J., Chaumont N., Sadiq T.S., Koruda M.J., Cotney P.A., Allbritton N., Trembath D.G., Sylvester F., Furey T.S., Sethupathy P., Sheikh S.Z. Colonic epithelial miR-31 associates with the development of Crohn's phenotypes. JCI Insight. 2018;3 doi: 10.1172/jci.insight.122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peck B.C., Weiser M., Lee S.E., Gipson G.R., Iyer V.B., Sartor R.B., Herfarth H.H., Long M.D., Hansen J.J., Isaacs K.L., Trembath D.G., Rahbar R., Sadiq T.S., Furey T.S., Sethupathy P., Sheikh S.Z. MicroRNAs Classify different disease behavior phenotypes of Crohn's disease and may have prognostic utility. Inflamm Bowel Dis. 2015;21:2178–2187. doi: 10.1097/MIB.0000000000000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiser M., Simon J.M., Kochar B., Tovar A., Israel J.W., Robinson A., Gipson G.R., Schaner M.S., Herfarth H.H., Sartor R.B., McGovern D.P.B., Rahbar R., Sadiq T.S., Koruda M.J., Furey T.S., Sheikh S.Z. Molecular classification of Crohn's disease reveals two clinically relevant subtypes. Gut. 2018;67:36–42. doi: 10.1136/gutjnl-2016-312518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCole D.F. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. 2014;20:1829–1849. doi: 10.1097/MIB.0000000000000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neurath M. Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol. 2017;14:688. doi: 10.1038/nrgastro.2017.138. [DOI] [PubMed] [Google Scholar]
- 9.Okumura R., Takeda K. Maintenance of intestinal homeostasis by mucosal barriers. Inflamm Regen. 2018;38:5. doi: 10.1186/s41232-018-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parikh K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H.H., Alham N.K., Ashley N., Johnson E., Hublitz P., Bao L., Lukomska J., Andev R.S., Bjorklund E., Kessler B.M., Fischer R., Goldin R., Koohy H., Simmons A. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 11.Laukoetter M.G., Nava P., Lee W.Y., Severson E.A., Capaldo C.T., Babbin B.A., Williams I.R., Koval M., Peatman E., Campbell J.A., Dermody T.S., Nusrat A., Parkos C.A. JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med. 2007;204:3067–3076. doi: 10.1084/jem.20071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudolph U., Finegold M.J., Rich S.S., Harriman G.R., Srinivasan Y., Brabet P., Boulay G., Bradley A., Birnbaumer L. Ulcerative colitis and adenocarcinoma of the colon in G alpha i2-deficient mice. Nat Genet. 1995;10:143–150. doi: 10.1038/ng0695-143. [DOI] [PubMed] [Google Scholar]
- 13.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 15.Baran-Gale J., Fannin E.E., Kurtz C.L., Sethupathy P. Beta cell 5'-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauklin S., Vallier L. Activin/Nodal signalling in stem cells. Development. 2015;142:607–619. doi: 10.1242/dev.091769. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell D., Pobre E.G., Mulivor A.W., Grinberg A.V., Castonguay R., Monnell T.E., Solban N., Ucran J.A., Pearsall R.S., Underwood K.W., Seehra J., Kumar R. ALK1-Fc inhibits multiple mediators of angiogenesis and suppresses tumor growth. Mol Cancer Ther. 2010;9:379–388. doi: 10.1158/1535-7163.MCT-09-0650. [DOI] [PubMed] [Google Scholar]
- 18.Roman B.L., Hinck A.P. ALK1 signaling in development and disease: new paradigms. Cell Mol Life Sci. 2017;74:4539–4560. doi: 10.1007/s00018-017-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steele S.P., Melchor S.J., Petri W.A., Jr. Tuft cells: new players in colitis. Trends Mol Med. 2016;22:921–924. doi: 10.1016/j.molmed.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leoni G., Neumann P.A., Sumagin R., Denning T.L., Nusrat A. Wound repair: role of immune-epithelial interactions. Mucosal Immunol. 2015;8:959–968. doi: 10.1038/mi.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb C.A., Kennedy N.A., Raine T., Hendy P.A., Smith P.J., Limdi J.K., Hayee B., Lomer M.C.E., Parkes G.C., Selinger C., Barrett K.J., Davies R.J., Bennett C., Gittens S., Dunlop M.G., Faiz O., Fraser A., Garrick V., Johnston P.D., Parkes M., Sanderson J., Terry H., group IBDgec, Gaya D.R., Iqbal T.H., Taylor S.A., Smith M., Brookes M., Hansen R., Hawthorne A.B. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut. 2019;68(Suppl 3):s1–s106. doi: 10.1136/gutjnl-2019-318484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramadas A.V., Gunesh S., Thomas G.A., Williams G.T., Hawthorne A.B. Natural history of Crohn's disease in a population-based cohort from Cardiff (1986-2003): a study of changes in medical treatment and surgical resection rates. Gut. 2010;59:1200–1206. doi: 10.1136/gut.2009.202101. [DOI] [PubMed] [Google Scholar]
- 23.Yoo W., Mayberry R., Bae S., Singh K., Peter He Q., Lillard J.W., Jr. A study of effects of multicollinearity in the multivariable analysis. Int J Appl Sci Technol. 2014;4:9–19. [PMC free article] [PubMed] [Google Scholar]
- 24.Bidart M., Ricard N., Levet S., Samson M., Mallet C., David L., Subileau M., Tillet E., Feige J.J., Bailly S. BMP9 is produced by hepatocytes and circulates mainly in an active mature form complexed to its prodomain. Cell Mol Life Sci. 2012;69:313–324. doi: 10.1007/s00018-011-0751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuhaus H., Rosen V., Thies R.S. Heart specific expression of mouse BMP-10 a novel member of the TGF-beta superfamily. Mech Dev. 1999;80:181–184. doi: 10.1016/s0925-4773(98)00221-4. [DOI] [PubMed] [Google Scholar]
- 26.McDonald J., Wooderchak-Donahue W., VanSant Webb C., Whitehead K., Stevenson D.A., Bayrak-Toydemir P. Hereditary hemorrhagic telangiectasia: genetics and molecular diagnostics in a new era. Front Genet. 2015;6:1. doi: 10.3389/fgene.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 28.Qi Z., Li Y., Zhao B., Xu C., Liu Y., Li H., Zhang B., Wang X., Yang X., Xie W., Li B., Han J.J., Chen Y.G. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat Commun. 2017;8:13824. doi: 10.1038/ncomms13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canzonieri C., Centenara L., Ornati F., Pagella F., Matti E., Alvisi C., Danesino C., Perego M., Olivieri C. Endoscopic evaluation of gastrointestinal tract in patients with hereditary hemorrhagic telangiectasia and correlation with their genotypes. Genet Med. 2014;16:3–10. doi: 10.1038/gim.2013.62. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad R., Chaturvedi R., Olivares-Villagomez D., Habib T., Asim M., Shivesh P., Polk D.B., Wilson K.T., Washington M.K., Van Kaer L., Dhawan P., Singh A.B. Targeted colonic claudin-2 expression renders resistance to epithelial injury, induces immune suppression, and protects from colitis. Mucosal Immunol. 2014;7:1340–1353. doi: 10.1038/mi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khounlotham M., Kim W., Peatman E., Nava P., Medina-Contreras O., Addis C., Koch S., Fournier B., Nusrat A., Denning T.L., Parkos C.A. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity. 2012;37:563–573. doi: 10.1016/j.immuni.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su L., Shen L., Clayburgh D.R., Nalle S.C., Sullivan E.A., Meddings J.B., Abraham C., Turner J.R. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pastorelli L., De Salvo C., Mercado J.R., Vecchi M., Pizarro T.T. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. doi: 10.3389/fimmu.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Assche G., Dignass A., Panes J., Beaugerie L., Karagiannis J., Allez M., Ochsenkuhn T., Orchard T., Rogler G., Louis E., Kupcinskas L., Mantzaris G., Travis S., Stange E., European Crohns and Colitis Organisation The second European evidence-based Consensus on the diagnosis and management of Crohn's disease: Definitions and diagnosis. J Crohn’s Colitis. 2010;4(1):7–27. doi: 10.1016/j.crohns.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Sun J., Liang W., Yang X., Li Q., Zhang G. Cytoprotective effects of galacto-oligosaccharides on colon epithelial cells via up-regulating miR-19b. Life Sci. 2019;231:116589. doi: 10.1016/j.lfs.2019.116589. [DOI] [PubMed] [Google Scholar]
- 36.Schaefer J.S., Montufar-Solis D., Vigneswaran N., Klein J.R. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. J Immunol. 2011;187:5834–5841. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian Y., Ma X., Lv C., Sheng X., Li X., Zhao R., Song Y., Andl T., Plikus M.V., Sun J., Ren F., Shuai J., Lengner C.J., Cui W., Yu Z. Stress responsive miR-31 is a major modulator of mouse intestinal stem cells during regeneration and tumorigenesis. Elife. 2017;6 doi: 10.7554/eLife.29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 39.Iglesias-Ara A., Zenarruzabeitia O., Fernandez-Rueda J., Sanchez-Tillo E., Field S.J., Celada A., Zubiaga A.M. Accelerated DNA replication in E2F1- and E2F2-deficient macrophages leads to induction of the DNA damage response and p21(CIP1)-dependent senescence. Oncogene. 2010;29:5579–5590. doi: 10.1038/onc.2010.296. [DOI] [PubMed] [Google Scholar]
- 40.Infante A., Laresgoiti U., Fernandez-Rueda J., Fullaondo A., Galan J., Diaz-Uriarte R., Malumbres M., Field S.J., Zubiaga A.M. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle. 2008;7:3915–3927. doi: 10.4161/cc.7.24.7379. [DOI] [PubMed] [Google Scholar]
- 41.Sengle G., Ono R.N., Sasaki T., Sakai L.Y. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J Biol Chem. 2011;286:5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daperno M., D'Haens G., Van Assche G., Baert F., Bulois P., Maunoury V., Sostegni R., Rocca R., Pera A., Gevers A., Mary J.Y., Colombel J.F., Rutgeerts P. Development and validation of a new, simplified endoscopic activity score for Crohn's disease: the SES-CD. Gastrointest Endosc. 2004;60:505–512. doi: 10.1016/s0016-5107(04)01878-4. [DOI] [PubMed] [Google Scholar]
- 43.Colombel J.F., Panaccione R., Bossuyt P., Lukas M., Baert F., Vanasek T., Danalioglu A., Novacek G., Armuzzi A., Hebuterne X., Travis S., Danese S., Reinisch W., Sandborn W.J., Rutgeerts P., Hommes D., Schreiber S., Neimark E., Huang B., Zhou Q., Mendez P., Petersson J., Wallace K., Robinson A.M., Thakkar R.B., D'Haens G. Effect of tight control management on Crohn's disease (CALM): a multicentre, randomised, controlled phase 3 trial. Lancet. 2018;390:2779–2789. doi: 10.1016/S0140-6736(17)32641-7. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., DiSalvo M., Gunasekara D.B., Dutton J., Proctor A., Lebhar M.S., Williamson I.A., Speer J., Howard R.L., Smiddy N.M., Bultman S.J., Sims C.E., Magness S.T., Allbritton N.L. Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell Mol Gastroenterol Hepatol. 2017;4:165–182 e7. doi: 10.1016/j.jcmgh.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuli Wang R.K., Gunasekara D.B., Reed M.I., DiSalvo M., Nguyen D.L., Bultman S.J., Sims C.E., Magness S.T., Allbritton N.L. Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell Mol Gastroenterol Hepatol. 2017;5:113–130. doi: 10.1016/j.jcmgh.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Meeteren L.A., Thorikay M., Bergqvist S., Pardali E., Stampino C.G., Hu-Lowe D., Goumans M.J., ten Dijke P. Anti-human activin receptor-like kinase 1 (ALK1) antibody attenuates bone morphogenetic protein 9 (BMP9)-induced ALK1 signaling and interferes with endothelial cell sprouting. J Biol Chem. 2012;287:18551–18561. doi: 10.1074/jbc.M111.338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhatt A.P., Gunasekara D.B., Speer J., Reed M.I., Pena A.N., Midkiff B.R., Magness S.T., Bultman S.J., Allbritton N.L., Redinbo M.R. Nonsteroidal anti-inflammatory drug-induced leaky gut modeled using polarized monolayers of primary human intestinal epithelial cells. ACS Infect Dis. 2018;4:46–52. doi: 10.1021/acsinfecdis.7b00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gunasekara D.B., Speer J., Wang Y., Nguyen D.L., Reed M.I., Smiddy N.M., Parker J.S., Fallon J.K., Smith P.C., Sims C.E., Magness S.T., Allbritton N.L. A monolayer of primary colonic epithelium generated on a scaffold with a gradient of stiffness for drug transport studies. Anal Chem. 2018;90:13331–13340. doi: 10.1021/acs.analchem.8b02845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salic A., Mitchison T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toyonaga T., Matsuura M., Mori K., Honzawa Y., Minami N., Yamada S., Kobayashi T., Hibi T., Nakase H. Lipocalin 2 prevents intestinal inflammation by enhancing phagocytic bacterial clearance in macrophages. Sci Rep. 2016;6:35014. doi: 10.1038/srep35014. [DOI] [PMC free article] [PubMed] [Google Scholar]