Abstract
Background & Aims
Intestinal crypts have a remarkable capacity to regenerate after injury from loss of crypt base columnar (CBC) stem cells. After injury, facultative stem cells (FSCs) are activated to replenish the epithelium and replace lost CBCs. Our aim was to assess the role of insulin-like growth factor-1 (IGF-1) to activate FSCs for crypt repair.
Methods
The intestinal regenerative response was measured after whole body 12-Gy γ-irradiation of adult mice. IGF-1 signaling or its downstream effector mammalian target of rapamycin complex 1 (mTORC1) was inhibited by administering BMS-754807 or rapamycin, respectively. Mice with inducible Rptor gene deletion were studied to test the role of mTORC1 signaling in the intestinal epithelium. FSC activation post-irradiation was measured by lineage tracing.
Results
We observed a coordinate increase in growth factor expression, including IGF-1, at 2 days post-irradiation, followed by a surge in mTORC1 activity during the regenerative phase of crypt repair at day 4. IGF-1 was localized to pericryptal mesenchymal cells, and IGF-1 receptor was broadly expressed in crypt progenitor cells. Inhibition of IGF-1 signaling via BMS-754807 treatment impaired crypt regeneration after 12-Gy irradiation, with no effect on homeostasis. Similarly, rapamycin inhibition of mTORC1 during the growth factor surge blunted the regenerative response. Analysis of Villin-CreERT2;Rptorfl/fl mice showed that epithelial mTORC1 signaling was essential for crypt regeneration. Lineage tracing from Bmi1-marked cells showed that rapamycin blocked FSC activation post-irradiation.
Conclusions
Our study shows that IGF-1 signaling through mTORC1 drives crypt regeneration. We propose that IGF-1 release from pericryptal cells stimulates mTORC1 in FSCs to regenerate lost CBCs.
Keywords: IGF-1, Raptor, Rapamycin, Intestinal Repair, Crypt Regeneration, Intestinal Stem Cells
Abbreviations used in this paper: ANOVA, analysis of variance; CBC, crypt base columnar; DPI, days post-irradiation; EDU, 5-ethynyl-2´-deoxyuridine; FSC, facultative stem cell; HPI, hours post-irradiation; IGF-1, insulin-like growth factor-1; ISC, intestinal stem cell; mTORC1, mammalian target of rapamycin complex 1; PBS, phosphate-buffered saline; qPCR, quantitative reverse transcriptase polymerase chain reaction; SEM, standard error of the mean; UNIRR, unirradiated controls
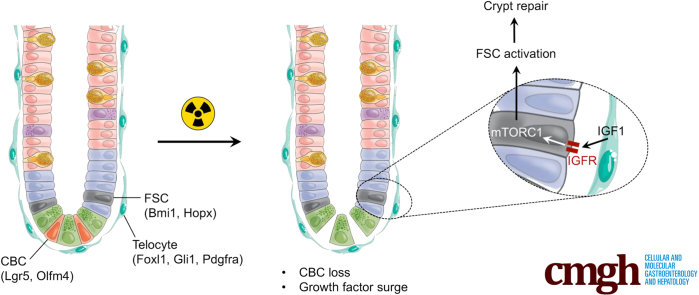
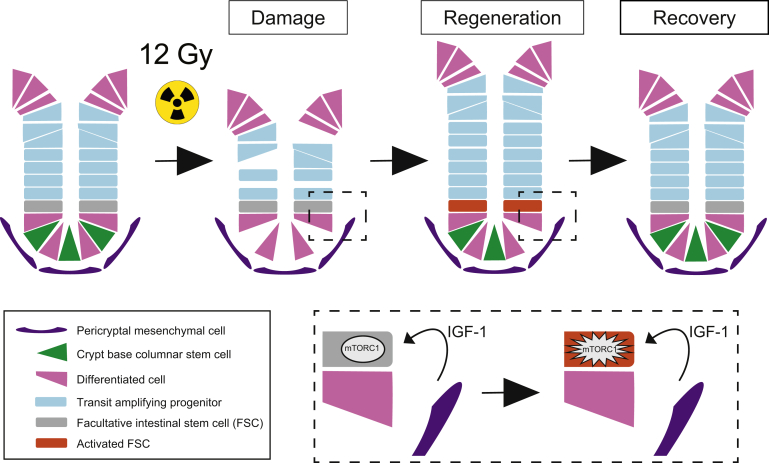
Graphical abstract
Summary.
We show that mesenchymal insulin-like growth factor-1 (IGF-1) promotes intestinal crypt repair following stem cell loss from whole-body irradiation. IGF-1 signals through mTORC1 to activate facultative stem cells to repopulate the damaged intestinal epithelium.
The intestinal epithelium is continually renewed throughout life by adult stem cells. Two intestinal stem cell (ISC) populations have been defined by their distinct roles during homeostasis and after intestinal damage. Active ISCs, also termed crypt base columnar (CBC) stem cells, maintain the intestinal epithelium during homeostasis, fueling cell renewal over one’s lifespan.1 CBC stem cells (marked by Lgr5, Olfm4, and Ascl2) divide approximately once per day to generate highly proliferative transit amplifying progenitors, which differentiate into the various mature epithelial cell types. On the other hand, facultative stem cells (FSCs), also termed +4, quiescent, or reserve stem cells, are crypt epithelial cells that can be activated to repopulate the stem cell niche after injury and loss of CBCs. Whole body exposure to γ-irradiation is commonly used to induce crypt injury, which leads to FSC activation to proliferate and repair the cellular damage and replenish the intestinal epithelium, including replacing lost CBCs.
Many different cells in the intestinal crypt are capable of reprogramming to function as FSCs, including lineage-committed (marked by Alpi and Dll1) or slowly cycling (Bmi1, Hopx, Lrig1, mTert) progenitor cells,2, 3, 4, 5, 6, 7, 8, 9, 10 as well as differentiated Paneth (Defa4, Lyz1) and endocrine (NeuroD1) cells.11, 12, 13 Thus, the crypt shows remarkable cellular plasticity with numerous crypt cell populations capable of remodeling to occupy open stem cell niche space after injury-induced loss of CBC stem cells. The mechanism of FSC activation to regenerate the intestinal epithelium after CBC loss has been a subject of active interest, with mammalian target of rapamycin complex 1 (mTORC1) signaling thought to play a major role.14,15
Intestinal homeostasis and mucosal repair are tightly regulated by the stem cell niche, the crypt/pericryptal microenvironment that consists of signaling factors and cell-to-cell interactions that regulate ISC function. Previously identified niche components include developmental factors such as Wnt/R-spondin and Notch, which are the primary signaling pathways promoting stem cell self-renewal.16 In addition, growth factors such as epidermal growth factor, fibroblast growth factors, and insulin-like growth factor-1 (IGF-1) have been proposed to regulate ISC function.17, 18, 19 To date, niche function has predominantly been defined for CBCs acting during homeostasis, with more limited understanding of the growth factors that orchestrate injury-induced repair.
IGF-1 has been proposed to be important for intestinal regeneration. Exogenous administration or transgenic overexpression of IGF-1 was shown to enhance intestinal epithelial growth and healing under numerous gut injury conditions.17, 18, 19, 20, 21, 22 A potential role for IGF-1 to regulate stem cell function was suggested by a study demonstrating that exogenous IGF-1 increased ISC proliferation and crypt regeneration after injury induced by 14-Gy abdominal irradiation.21 This enhanced regenerative capacity was associated with increased efficiency of progenitor cells to form organoids, suggesting that IGF-1 might activate FSCs to promote crypt repair. IGF-1 signals through mTORC1 via PI3K/Akt signaling. The active mTORC1 complex regulates cellular homeostasis through integration of molecular pathways and environmental cues.23 Although mTORC1 is not essential for intestinal epithelial homeostasis, several studies, some dating back almost 30 years, suggest that mTORC1 functions in crypt repair.14,15,24, 25, 26, 27, 28 Mice with intestinal epithelial deletion of mTOR (which disrupts both mTORC1 and mTORC2) using constitutively expressed Villin-Cre were more sensitive to 10-Gy γ-irradiation, with reduced capacity to regenerate crypts and CBCs.24 Interestingly, this study also showed that crypt epithelial cell deletion of the mTORC2 complex gene Rictor had no effect on crypt regeneration, suggesting that mTORC1 is key for post-irradiation crypt repair.24 The importance of mTORC1 for regeneration after γ-irradiation was also suggested by impaired regeneration in mice treated with the mTORC1 inhibitor rapamycin and after deletion of the mTORC1 complex gene Rptor using inducible Villin-CreERT2.24 Furthermore, manipulation of mTORC1 signaling in FSCs suggested an important role in controlling FSC activity.14,15 Collectively, the findings suggest that epithelial mTORC1 activity plays a key role in crypt regeneration, although the mechanism by which mTORC1 becomes activated after crypt injury is unknown.
In this study we investigated the mechanism of growth factor induction of intestinal crypt regeneration after 12-Gy γ-irradiation. Our findings show that irradiation injury induces the expression of a number of growth factors including IGF-1. Mesenchymal IGF-1 signals to crypt epithelial cells to promote repair via mTORC1 activation. IGF-1/mTORC1 signaling is required for FSC activation to fill open stem cell niche spaces to replace lost CBCs and repair the crypts.
Results
Irradiation Injury Induces a Growth Factor Surge Preceding Crypt Repair
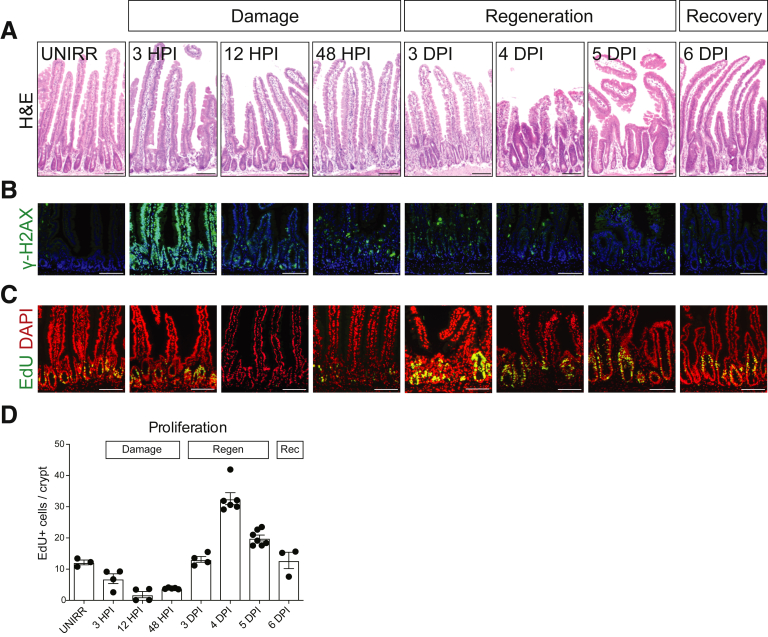
We characterized the intestinal response after 12-Gy whole body γ-irradiation of adult mice. Histologic analysis revealed gross morphologic changes, particularly apparent in the crypt compartment, which allowed categorization of the response into 3 distinct phases: damage, regeneration, and recovery (Figure 1). The damage phase is characterized by rapid cellular injury, followed by loss of proliferating cells and crypt collapse. At 3 hours post-irradiation (HPI), DNA double-strand breaks are demonstrated throughout the epithelium by γ-H2AX staining, which are largely resolved by 12 HPI (Figure 1B). By 12 HPI crypt cell proliferation is markedly reduced (Figure 1C and D), and by 48 HPI crypt architecture is destroyed, with de-cellularization and crypt loss (Figure 1A). The regenerative phase, which is apparent at 3 days post-irradiation (DPI), is characterized by robust crypt recovery. The hallmark of this phase at 3–5 DPI is a hyperproliferative surge and crypt compartment expansion. We denote 6 DPI as the beginning of the recovery phase, where the regenerative response is resolving, with crypt structure and the intestinal epithelium returning to homeostasis.
Figure 1.
Three phases of the intestinal regenerative response post-irradiation. Mice were administered 12-Gy whole body γ-irradiation, and intestinal tissue was collected at various HPI or DPI, as indicated, and compared with intestine from UNIRR mice. (A) Duodenal tissue histology was assessed by H&E staining. (B) DNA damage was detected by immunostaining for γ-H2AX (green) with nuclear counterstain DAPI (blue). (C) Cellular proliferation was assessed by EdU incorporation (green) with DAPI (red). (D) The number of EdU+ cells was counted at the various time points post-irradiation. Proliferating cell number is presented as EdU-positive cells per crypt (mean ± SEM, n = 3–7 mice/group as shown). Scale bars = 100 μm.
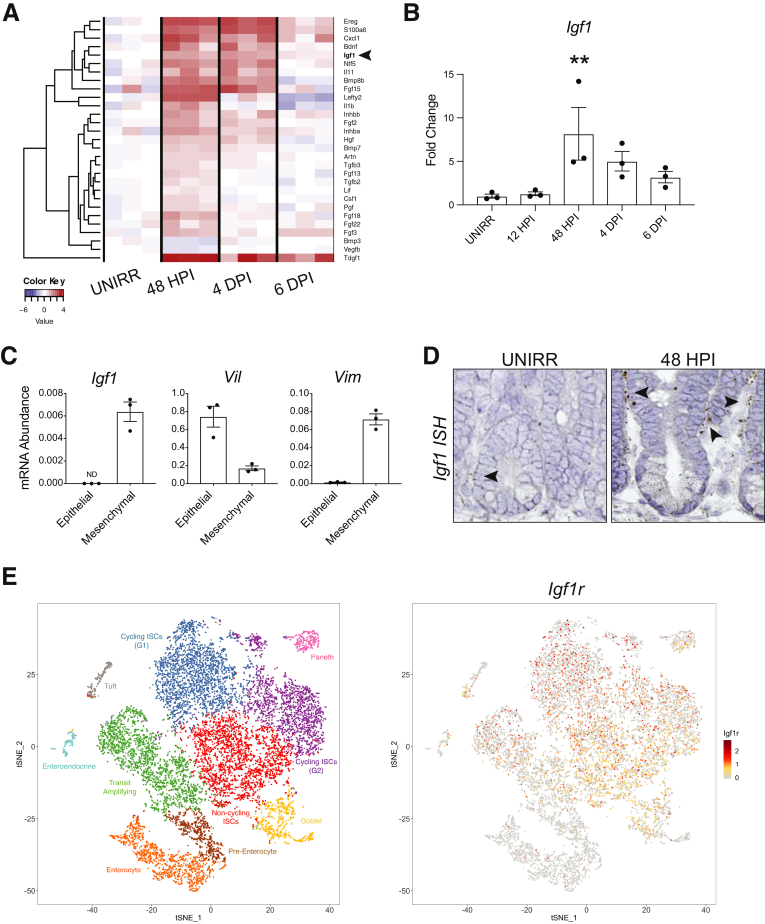
We sought to identify growth factors that might play a role in mediating the response to radiation injury. We assessed growth factor expression signatures using a Qiagen (Hilden, Germany) quantitative reverse transcriptase polymerase chain reaction (qPCR)-based array designed to measure the abundance of 84 mouse growth factor mRNAs. We analyzed time points across the 3 phases of the regenerative response at 48 HPI (damage), 4 DPI (regeneration), and 6 DPI (recovery), compared with mock unirradiated controls (UNIRR). This analysis showed that numerous growth factors were induced at the damage phase, sustained during the regenerative phase, with a return toward baseline during recovery, suggesting that they might play a role in the regenerative response (Figure 2A). Furthermore, several of these growth factors are known to signal through mTORC1, suggesting that they might be involved in FSC activation. Of these, IGF-1 was among the growth factors with the most dramatic change in expression. This growth factor was also of interest because it had been described to enhance crypt regeneration after γ-irradiation,21 yet a role for endogenous IGF-1 had not been defined. Measurement of Igf1 mRNA abundance by qPCR analysis showed an 8-fold increase at 48 HPI during the damage phase, with a gradual return toward baseline with recovery (Figure 2B).
Figure 2.
IGF-1 growth factor expression increases after irradiation-induced intestinal damage. Duodenal tissue from UNIRR and 12-Gy–treated mice at the damage (48 HPI), regeneration (4 DPI), and recovery (6 DPI) phases was isolated and tested for expression of 84 growth factors by RT2 Profiler PCR array analysis. (A) Heat map of growth factor expression after irradiation injury, with red representing higher and blue representing lower expression relative to UNIRR control (n = 3 mice/group). Arrowhead points to growth factor gene of interest Igf1. (B) Quantitative reverse transcriptase polymerase chain reaction (qPCR) analysis of Igf1 mRNA abundance post-irradiation. (C) Analysis of Igf1 mRNA abundance by qPCR in intestinal epithelial or mesenchymal compartments, with validation of tissue fractionation by expression of villin (Vil) or vimentin (Vim), respectively. (D) RNAScope in situ hybridization (ISH) for Igf1 on UNIRR and 48 HPI duodenum. Arrowheads indicate some Igf1-expressing cells at the crypt base. (E) Single-cell RNA-seq analysis of IGF-1 receptor (Igf1r) expression in crypt cells. tSNE plots of cell clusters (left) and Igf1r expression (right). Quantitative data are displayed as mean ± SEM (n = 3 mice/group; ∗∗P < .01 by 1-way ANOVA with Dunnett post-test).
We next defined the intestinal tissue compartments for IGF-1 signaling. Analysis of epithelial versus mesenchymal mRNA by qPCR showed that Igf1 was specifically expressed in the mesenchyme at baseline (Figure 2C). The tissue fractionation was validated by measurement of the epithelial marker villin (Vil) and the mesenchymal marker vimentin (Vim) (Figure 2C). RNAscope in situ hybridization demonstrated that Igf1 transcripts were localized to pericryptal mesenchymal cells, with increased levels at 48 HPI (Figure 2D). This pattern suggested that Igf1 is expressed in telocytes, which have been recently described as crucial niche cells for epithelial homeostasis.29 Furthermore, analysis of our previously published single-cell RNA-seq data30 demonstrated that IGF-1 receptor (Igf1r) was broadly expressed in crypt epithelial cell populations, including stem and transit amplifying progenitor cells (Figure 2E). Together these data suggest a signaling axis from pericryptal stromal cells to crypt epithelial cells. In light of a previous study demonstrating intestinal pro-regenerative properties of IGF-1 in vivo21 and an in vitro study showing that IGF-1 promoted growth of human ISCs,31 we focused the rest of our study on understanding the role of this signaling axis in intestinal repair.
Inhibition of Insulin-like Growth Factor-1/Mammalian Target of Rapamycin Complex 1 Signaling Impairs Intestinal Regeneration
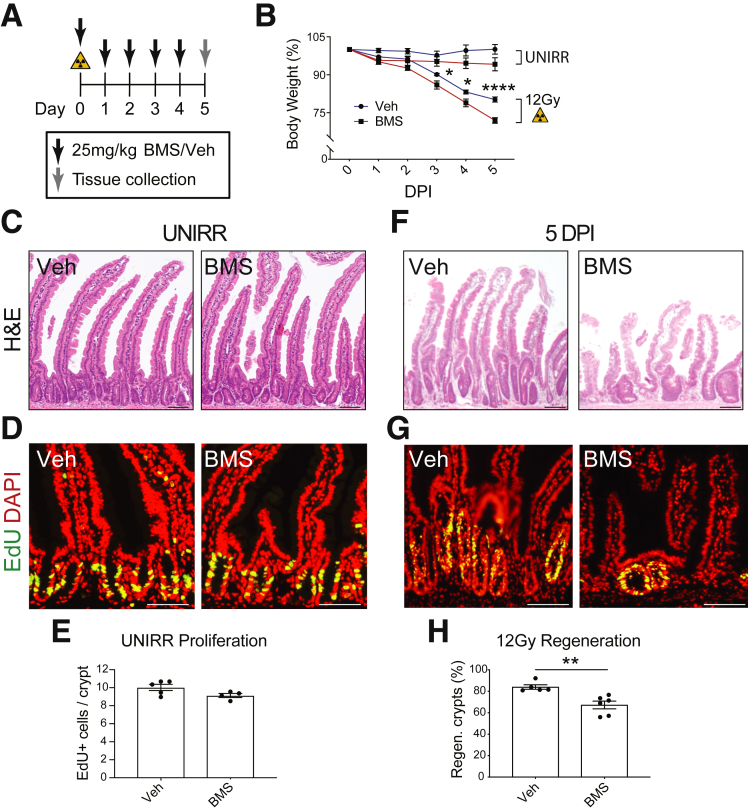
We tested the function of IGF-1 signaling during the regenerative response using the reversible IGF-1 receptor inhibitor BMS-754807 (BMS). Mice were treated daily with either BMS (25 mg/kg) or vehicle after 12-Gy irradiation, and tissue was harvested during the regenerative phase at 5 DPI (Figure 3A). At baseline, inhibition of this pathway for 5 days had no discernible effect on body weight or intestinal homeostasis; tissue architecture and cellular proliferation did not differ between BMS-treated mice and vehicle-treated controls (Figure 3B–E). However, marked differences were observed at 5 DPI between BMS- and vehicle-treated mice. There was a more pronounced post-irradiation weight loss in BMS-treated mice compared with vehicle-treated controls (Figure 3B). Histologic analysis revealed more extensive intestinal damage in BMS-treated mice, with blunted villi and reduced proliferation (Figure 3F and G). Morphometric analysis demonstrated that BMS-treated mice had a significant reduction in crypt regeneration, with 20% fewer regenerating crypts on inhibition of IGF-1 signaling (Figure 3H).
Figure 3.
IGF-1 receptor inhibition impairs intestinal regeneration post-irradiation. (A) Mice were treated with BMS-754807 (BMS; 25 mg/kg) or vehicle (Veh) as shown. BMS was administered to UNIRR or 12-Gy–irradiated mice. (B) Mouse body weight relative to weight at initiation of treatment (n = 5–6 mice/group). (C–H) Duodenal tissue was analyzed at day 5 by H&E staining (C and F) and EdU incorporation (D and G). (E) Proliferation was measured by morphometric analysis of EdU+ cells in UNIRR controls (n = 4–5 mice/group). (H) Crypt regeneration was measured at 5 DPI (n = 5–6 mice/group). Quantitative data are presented as mean ± SEM (∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001 by 2-way ANOVA or by Student t test vs Veh). Scale bars = 100 μm.
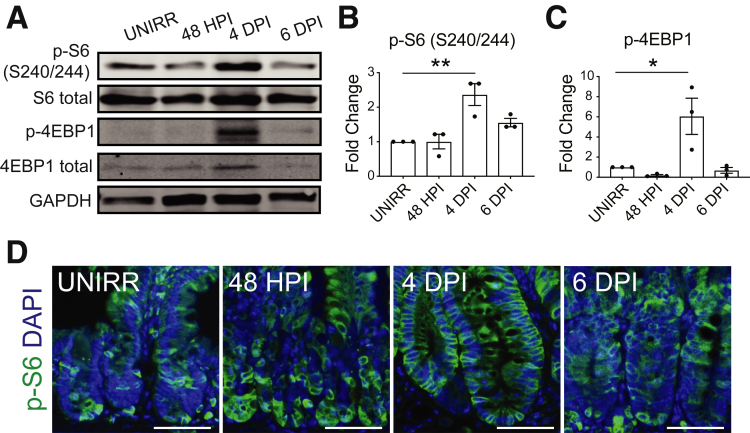
IGF-1 signaling is known to activate mTORC1 complex signaling. Thus, we next examined the role of mTORC1 signaling for crypt regeneration. We first assessed changes to mTORC1 activity post-irradiation by Western blot analysis, observing increased phosphorylation of mTORC1 targets ribosomal protein S6 (p-S6) and 4EBP1 (p-4EBP1) at 4 DPI during the regenerative phase (Figure 4A–C). Immunohistologic analysis of tissue sections confirmed increased S6 phosphorylation (S240/244) in the crypts during regeneration after 12-Gy irradiation (Figure 4D).
Figure 4.
mTORC1 activity increases during the regenerative response. Protein levels of mTORC1 signaling components in duodenum as assessed by Western blotting and immunostaining. (A) Western blot analysis for p-S6 (S240/244), total S6, p-4EBP1, total 4EBP1, and loading control GAPDH. (B) p-S6 and (C) p-4EBP1 band signal was quantified and displayed as mean ± SEM (n = 3 mice/group; ∗P < .05, ∗∗P < .01 by 1-way ANOVA with Dunnett post-test). (D) Immunofluorescent images of p-S6 (green) staining with DAPI counterstain (blue). Scale bars = 100 μm.
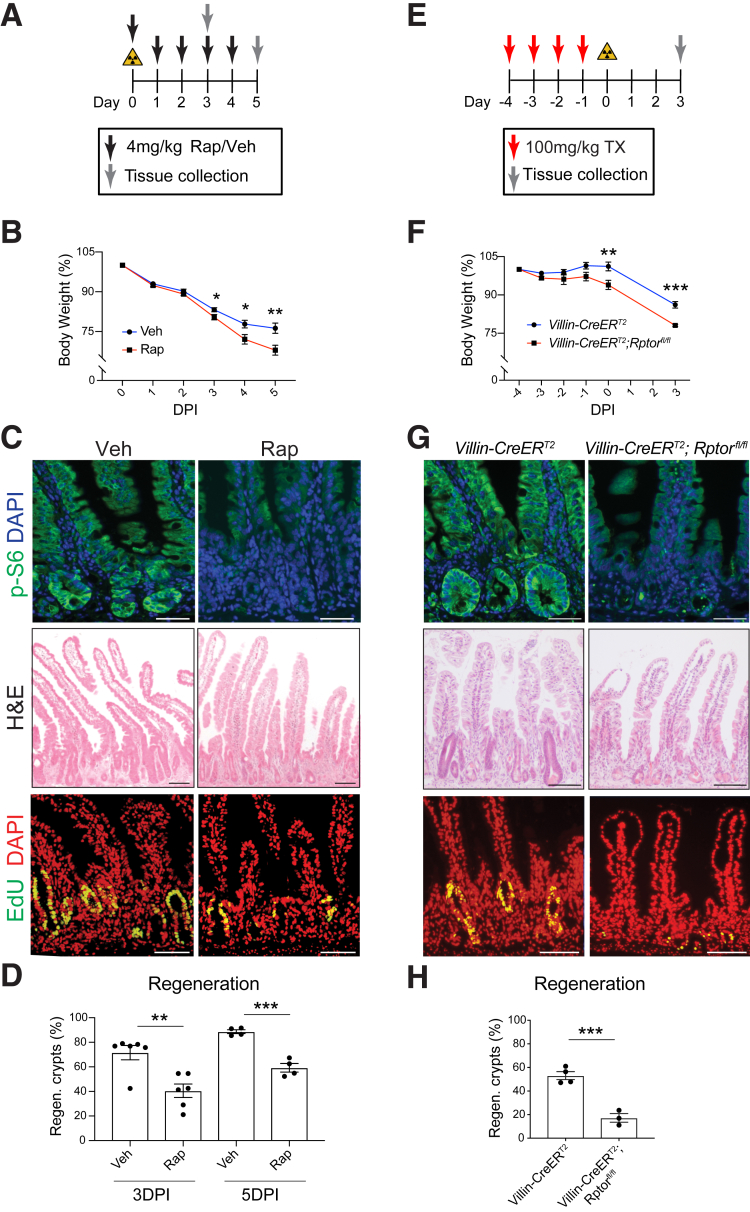
Similar to previously published studies,14,15,26 we showed with our injury model that inhibition of mTORC1 with rapamycin subsequent to 12-Gy irradiation impaired intestinal regeneration. Rapamycin-treated mice exhibited greater loss of body weight compared with irradiation alone (Figure 5A and B). Effective mTORC1 inhibition was confirmed by immunostaining for the downstream mTORC1 target p-S6 (S240/244) (Figure 5C). The effect of the regenerative response was similar to IGF-1 inhibition, with fewer and smaller crypts observed after rapamycin treatment of irradiated mice. Morphometric analysis showed decreased regeneration at both 3 DPI and 5 DPI (Figure 5D). To define the key tissue compartment for mTORC1 signaling, we deleted Rptor from the intestinal epithelium by treating Villin-CreERT2;Rptorfl/fl mice with tamoxifen before 12-Gy irradiation (Figure 5E). Age- and sex-matched Villin-CreERT2;Rptor+/+ mice were used as controls to account for any effects of CreERT2 toxicity to CBC stem cells, which we have previously established.32 We confirmed depleted mTORC1 activity in tamoxifen-treated Villin-CreERT2;Rptorfl/fl mice by immunostaining for p-S6 (S240/244) (Figure 5G). Rptor-deleted mice showed enhanced weight loss post-irradiation (Figure 5F), and histologic analysis revealed impaired crypt regeneration (Figure 5G). At 3 DPI, Villin-CreERT2;RptorF/F crypts appeared few, small, and de-cellularized compared with Villin-CreERT2;Rptor+/+ controls (Figure 5G). Proliferation was reduced, and there was a 3-fold decrease in regenerating crypts compared with controls (Figure 5H). These findings confirm that the impaired regeneration observed with rapamycin administration is attributed to epithelial mTORC1.
Figure 5.
mTORC1 inhibition impairs intestinal regeneration. (A) Mice were irradiated and treated with rapamycin (Rap; 4 mg/kg) or vehicle (Veh), with tissue collection at 3 and 5 DPI. (B) Mouse body weight relative to weight at initiation of Rap treatment (n = 16–25 mice/group). (C) Histologic images of p-S6-, H&E-, and EdU-stained duodenal tissue harvested 5 DPI from Veh- and Rap-treated mice. (D) Crypt regeneration was measured at 3 and 5 DPI (n = 4–6 mice/group). (E) Villin-CreERT2;Rptor+/+ (control) and Villin-CreERT2;Rptorfl/fl mice were tamoxifen (TX)-treated, followed by 12-Gy irradiation, and tissue was collected 3 DPI, as shown. (F) Mouse body weight relative to weight at initiation of treatment (n = 3–4 mice/group). (G) Histologic images of p-S6-, H&E-, and EdU-stained duodenal tissues collected 3 DPI. (H) Crypt regeneration was measured (n = 3–4 mice/group). Quantitative data are presented as mean ± SEM (∗P < .05, ∗∗P < .01, ∗∗∗P < .001 by 2-way ANOVA or Student t test). Scale bars = 100 μm.
Mammalian Target of Rapamycin Complex 1 Signaling Activates Facultative Stem Cells for Intestinal Epithelial Regeneration
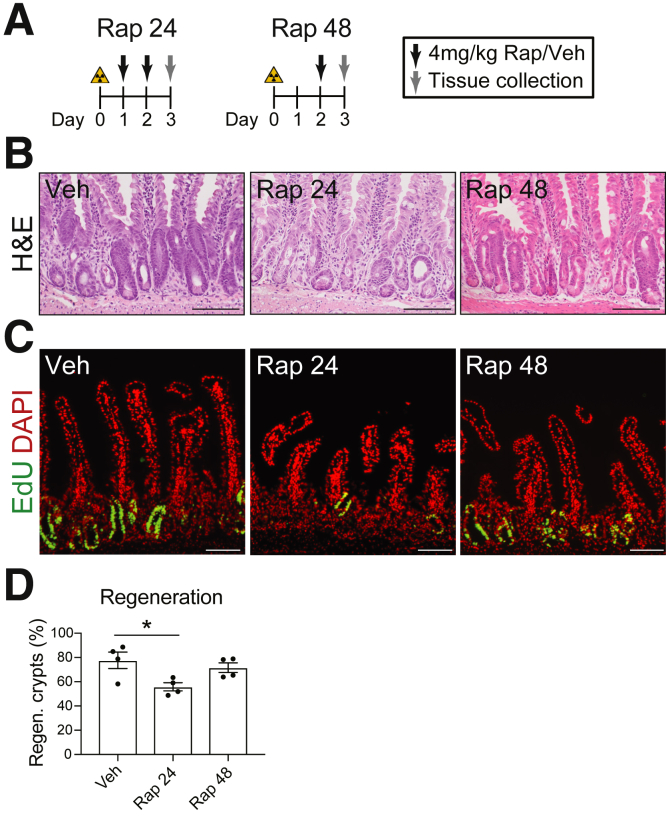
To address the mechanism of activation of mTORC1 signaling, we first investigated the key timing for mTORC1 function in crypt repair. We varied the timing of initiation of rapamycin treatment from 24 to 48 HPI (Figure 6A). Mice with initial rapamycin treatment at 24 HPI exhibited a significant reduction in the number of regenerating crypts (Figure 6B–D). In contrast, mice that received initial rapamycin treatment at 48 HPI exhibited a normal crypt regeneration response (Figure 6B–D). These findings suggest that the key window for mTORC1 action is 24–48 HPI. Importantly, this time window corresponds to the timing for FSC activation, which supports earlier studies suggesting that mTORC1 plays a role in the activation of these cells after irradiation injury.8,10,14
Figure 6.
Key timing for mTORC1 activation post-irradiation. (A) Mice were 12-Gy γ-irradiated and treated with rapamycin (Rap) or vehicle (Veh) starting 24 HPI (Rap 24) or 48 HPI (Rap 48) and analyzed at 3 DPI. (B–D) Duodenal crypt regeneration was assessed by (B) H&E staining and (C) EdU incorporation, with (D) crypt regeneration quantified (n = 4 mice/group). Quantitative data are presented as mean ± SEM (∗P < .05 by 1-way ANOVA with Dunnett post-test). Scale bars = 100 μm.
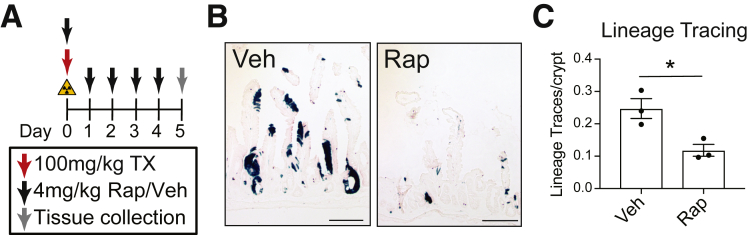
To test whether mTORC1 signaling is important to FSC contribution to the regenerative response, we assessed FSC activity post-irradiation by lineage tracing. Bmi1-CreER;ROSA26-LSL-lacZ mice were irradiated, tamoxifen-treated to induce lineage tracing from Bmi1-positive FSCs, and rapamycin- or vehicle-treated to test the role of mTORC1 for FSC activation (Figure 7A). We observed a significant decrease in lineage traces from rapamycin-treated mice compared with vehicle-treated controls (Figure 7B and C). These findings indicate that mTORC1 is critical for Bmi1-FSC activation after intestinal injury.
Figure 7.
Rapamycin (Rap) impairs facultative stem cell contribution to intestinal regeneration. (A) Bmi1-CreER;ROSA26-LSL-lacZ mice were 12-Gy γ-irradiated, injected with tamoxifen (TX), and treated with Rap or vehicle (Veh), as shown. (B) 5 DPI duodenal sections were stained for β-galactosidase to visualize lineage traces (n = 3 mice/group). (C) Lineage tracing events were counted, and quantitative data are presented as mean ± SEM (∗P < .05 vs Veh by Student t test). Scale bars = 100 μm.
Discussion
Our study shows that the intestinal response to injury induced by 12-Gy whole body irradiation involves a coordinate increase in intestinal growth factor expression, which peaked in the damage phase of the response at 48 HPI. A robust increase in Igf1 expression was observed as a component of the growth factor surge. IGF-1 is known to signal through PI3K/Akt to activate mTORC1, which we observed to be subsequently activated during the regenerative phase of the response at 4 DPI. Inhibition of either IGF-1 or mTORC1 signaling resulted in enhanced loss of body weight and a severe impairment in crypt regeneration post-irradiation. Moreover, targeted genetic deletion studies showed that the intestinal epithelium was the key tissue compartment for mTORC1 signaling during crypt repair after irradiation injury. Our studies also showed that mTORC1 signaling is particularly important 24–48 HPI, which is coincident with the timing of FSC activation after irradiation injury.8,10,14 Accordingly, we showed by using a lineage tracing approach that inhibition of mTORC1 signaling blocked FSC contribution to regeneration. Thus, our studies propose a mechanism by which irradiation injury induces IGF-1 expression, which stimulates mTORC1 signaling in crypt epithelial cells, resulting in FSC activation to re-establish CBC stem cells and repair the crypt (Figure 8). Notably, inhibition of IGF-1 signaling resulted in a more limited effect in comparison with mTORC1 inhibition, suggesting that other growth factors in addition to IGF-1 play a role in the regeneration response post-irradiation (compare Figures 3 and 5).
Figure 8.
Pericryptal IGF-1 secretion stimulates mTORC1-mediated FSC activation. Our model proposes that increased IGF-1 secretion in response to irradiation injury results in mTORC1 signaling in FSCs, leading to their activation to contribute to intestinal regeneration.
We identified the intestinal cellular source of IGF-1 by in situ hybridization. Igf1 transcripts were localized to pericryptal mesenchymal cells in both basal and injury conditions (Figure 2D). This finding agrees with an earlier study that had mapped IGF-1 to a subset of human fibrotic mesenchymal cells in Crohn’s disease patients.33 Interestingly, this expression pattern follows recent studies that identified pericryptal mesenchymal cells that secrete niche factors supporting CBC stem cells to maintain epithelial cell homeostasis.34, 35, 36, 37, 38 Importantly, gene expression profiling studies showed that these cells, termed telocytes, express a number of growth factors including Igf1.37 Telocytes have long cellular processes that lie in close juxtaposition to the basal surface of crypt epithelial cells, which facilitates cross-talk between these mesenchymal cells and epithelial cells to regulate stem/progenitor cell function. Indeed, these cells, marked by Foxl1, Gli1, and Pdgfra, have been identified as the essential Wnt-secreting cells for intestinal crypt maintenance.37, 38, 39 Single cell RNAseq analysis showed that cells marked by Foxl1, but not other mesenchymal cell populations, express Igf1 and other growth factors that would be expected to signal through mTORC1.37 Our findings suggest that these cells may be key for secreting growth factors to orchestrate crypt repair after injury.
In accordance with a signaling axis from mesenchyme to epithelium, we show that Igf1r mRNA is broadly expressed in several crypt stem/progenitor cell populations, including CBCs and transit-amplifying cells (Figure 2E). This agrees with previous reports that demonstrated IGF-1 receptor expression in crypt epithelial cells, including CBCs and FSCs.21,40,41 How specific intestinal stem/progenitor cells might be involved in the growth factor injury response is unresolved. An earlier study addressed the effect of IGF-1 administration on ISCs after 14-Gy abdominal radiation.21 In that study, IGF-1, delivered by implanted minipumps, enhanced crypt regeneration, with increased numbers of actively cycling ISCs observed in the regenerative phase of the response. They further showed increased proliferation of Sox9-EGFPHigh FSCs with IGF-1 treatment. Notably, IGF-1 enhanced the ability of this FSC population to form enteroids,21 supporting a role for IGF-1 signaling to activate FSCs after injury.
Interestingly, IGF-1 does not appear to be required for intestinal homeostasis. We observed normal intestinal morphology and crypt proliferation after 5 days of IGF-1 receptor inhibition, which agrees with previous targeted receptor gene deletion studies showing normal intestinal homeostasis.42,43 However, it has been well-established that exogenous IGF-1 administration can stimulate intestinal mucosal growth under basal conditions as well as after injury in rodent models.44 In addition to irradiation injury,17, 18, 19, 20, 21, 22 exogenous administration of IGF-1 has been shown to enhance intestinal mucosal growth after total parenteral nutrition and small bowel resection.19,45 Our study is the first to show that blocking endogenous IGF-1 impairs intestinal regeneration after irradiation injury and to show evidence for a mechanism by which mesenchymal IGF-1 mediates intestinal regeneration via stimulation of crypt epithelial cell mTORC1. Interestingly, a recent study showed that addition of IGF-1 to human intestinal enteroid culture media enhanced long-term culture, with conservation of normal cellular diversity, suggesting that IGF-1 may also be important for human ISC function.31 Furthermore, up-regulated IGF-1 expression in Crohn’s disease patients also supports a role for this signaling axis in human intestinal disease.33
Our findings demonstrating the importance of IGF-1/mTORC1 signaling during crypt repair agree with previous studies showing that mTORC1 signaling in the intestinal epithelium is required for intestinal crypt regeneration after radiation injury.14,24,26 In addition to our demonstration that activation of Bmi1-CreER–marked FSCs is mTORC1 dependent, a previous study showed that mTORC1 signaling was activated in Hopx-CreERT2–marked FSCs post-irradiation.14 In preliminary studies we also observed that Hopx-CreERT2–marked FSCs require mTORC1 for activation post-irradiation injury. Thus, activation of mTORC1 signaling appears to be a general feature of FSC populations in mounting a regenerative response to crypt injury. We propose that radiation injury induces IGF-1 in pericryptal mesenchymal cells to activate mTORC1, promoting intestinal stem cell potential in various FSC populations (Figure 8).
One study that examined the effect of calorie restriction on FSC function suggested that mTORC1 regulates a change in FSC status from a “dormant” to a “poised” state capable of responding to injury to repopulate the intestinal epithelium.15 This proposed mechanism is also in agreement with the observation of mTORC1-dependent muscle satellite (stem) cell transition from dormancy to functionally poised in response to muscle injury.46 Future studies will be required to understand the mechanism of mTORC1 remodeling of FSC cell state as well as the identification of the injury signal(s) that induce IGF-1 and other growth factor expression in pericryptal mesenchymal cells to orchestrate crypt repair.
Materials and Methods
Mouse Use
Mouse experiments were approved by the Institutional Animal Care & Use Committee at the University of Michigan. Mice were housed in ventilated and automated watering cages with a 12-hour light/dark cycle under specific pathogen-free conditions. The following mouse strains were used: Villin-CreERT2 (gift from Robine lab),47 Rptorfl/fl (JAX 013188),48 Bmi1-CreER (JAX 010531),5 and ROSA26-LSL-lacZ (JAX 003474).49 Mice were maintained on a C57BL/6 strain background. Mice of both sexes aged 1.5–4 months were used.
To induce intestinal injury, mice were exposed to 12-Gy whole body γ-irradiation from a 137Cs source; control mice were mock-irradiated. To activate CreERT2-mediated recombination, mice were injected intraperitoneally with tamoxifen (Sigma-Aldrich, St Louis, MO; 100 mg/kg in 5% ethanol and 95% corn oil). To inhibit mTORC1 activity, mice were injected intraperitoneally with rapamycin (LC Laboratories, Woburn, MA; 4 mg/kg in phosphate-buffered saline [PBS] containing 5% Tween 80 and 5% polyethylene glycol 400). To inhibit IGF-1 signaling, BMS-754807 was administered by oral gavage (MedChemExpress, Monmouth Junction, NJ; 25 mg/kg in 80% polyethylene glycol 400 in water). Intestinal tissue was isolated after ad libitum feeding at various times, as indicated in Figures. For analysis of proliferation, mice were injected intraperitoneally with 5-ethynyl-2´-deoxyuridine (EDU) (Life Technologies, Carlsbad, CA; 25 mg/kg) 2 hours before tissue collection.
Histologic Analysis
Intestinal tissue was fixed in 4% paraformaldehyde in PBS overnight before paraffin processing, as previously described.50 Duodenal paraffin sections (4–5 μm) were stained with H&E to assess intestinal morphology. The EdU Click-it kit (Life Technologies) was used to identify proliferating cells. Regeneration was assessed by using the adapted crypt microcolony survival assay method.51 Regenerating crypts were measured as the number of well-oriented crypts with 5 or more EDU-positive cells divided by the total number of well-oriented crypts. Well-oriented crypts were identified from images of adjacent H&E-stained sections. Immunostaining with rabbit antibodies to γ-H2AX (1:50, Cell Signaling 9718; Cell Signaling Technology, Danvers, MA) and phospho-S6 (S240/244) (1:300, Cell Signaling 5364) was performed as described.52 A goat anti-rabbit immunoglobulin G Alexa Fluor 488 polyclonal secondary antibody was used (1:400, Invitrogen A27034; Carlsbad, CA). Images were captured on a Nikon E800 (Tokyo, Japan) microscope with Olympus DP (Tokyo, Japan) controller software.
Western Blot Analysis
Full-thickness duodenal tissue was homogenized in RIPA buffer (Thermo Fisher Scientific, Waltham, MA; 89900) containing protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, 78440). Cell lysates (40 μg protein) were mixed with NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, NP0007) and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis using NuPAGE MOPS SDS Running Buffer (Thermo Fisher Scientific, NP0001) and NuPAGE 4-12% Bis-Tris gels (Thermo Fisher Scientific, NP0335), following manufacturer’s recommendations. Protein transfer onto 0.45-μm pore size nitrocellulose membrane (GE Healthcare, Chicago, IL) at 100 V for 40 minutes preceded blocking in Odyssey Blocking Buffer (Li-COR Biosciences, Lincoln, NE; 927-40000) for 1 hour at room temperature. Immunoblotting with rabbit antibodies to phospho-S6 (S240/244) (1:500, Cell Signaling 5364) and phospho-4EBP1 (1:200, Cell Signaling 2855) and mouse antibodies to S6 (1:200, Cell Signaling 2317), 4EBP1 (1:200, Cell Signaling 9644), and GAPDH (1:10,000, Thermo Fisher Scientific MA5-15738) was performed on a rocking platform overnight at 4°C. After rinsing the membrane in Tris-buffered saline, 0.1% Tween 20, IRDye 800CW goat α-rabbit (1:10,000, Li-COR 925-32211) and IRDye 680RD goat α-mouse (1:10,000, Li-COR 925-68070) secondary antibodies were used to visualize probed proteins. The membrane was scanned on an Odyssey Imager (Li-COR). Western blot analysis was performed by using the free Image Studio Lite software (Li-COR).
Lineage Tracing
Bmi1-CreER;ROSA26-LSL-lacZ mice were injected with rapamycin or vehicle daily for 5 days, with tamoxifen treatment and 12-Gy whole body irradiation performed on the first day of treatment (Figure 7A). Five DPI intestinal tissue was fixed in 4% paraformaldehyde at 4°C for 1 hour and incubated in 30% sucrose overnight before embedding in OCT. Frozen sections were fixed in 4% paraformaldehyde for 5 minutes, washed 3 times in X-Gal buffer (2 μmol/L MgCl2, 0.02% NP-40 in 0.1 mol/L sodium phosphate, pH 7.3) for 5 minutes, and stained in X-Gal staining solution (Invitrogen 15520-034; 1 mg/mL in 5.2 mmol/L potassium ferrocyanide, 5.2 mmol/L potassium ferricyanide in X-Gal buffer) overnight at 37°C in the dark. The following day, slides were washed 3 times with X-Gal wash buffer and counterstained with neutral red. Stained slides were dehydrated in ethanol/xylene and coverslipped with Permount before imaging. Three serial duodenal sections (10 μm) were imaged from each mouse to count lineage tracing events, which were defined by 4 consecutive lacZ-positive cells across at least 2 serial sections. The number of events was divided by the number of crypt-villus units imaged.
Gene Expression Analysis
RNA from full-thickness duodenal tissue segments was isolated as previously described.53 The mRNA abundance was measured by qPCR as previously described,36 using Igf1 primers CAACTCCCAGCTGTGCAATT (forward) and GCCGAGGTGAACACAAAACT (reverse) to generate a 151 base pair product, Vil primers ATGACTCCAGCTGCCTTCTCT (forward) and GCTCTGGGTTAGAGCTGTAAG (reverse) to generate a 436 base pair product, and Vim primers ACTGCTGCCCTGCGTGATGTG (forward) and GGTACTCGTTTGACTCCTGCTTGG (reverse) to generate a 163 base pair product. Assays were run in triplicate and normalized to Gapdh or Hprt, with Gapdh as described,52 and Hprt primers AGGACCTCTCGAAGTGTTGGATAC (forward) and AACTTGCGCTCATCTTAGGCTTTG (reverse) to generate a 173 base pair product.
For growth factor array analysis, RNAs from UNIRR, 48 HPI, 4 DPI, and 6 DPI were submitted to the University of Michigan DNA Sequencing Core for RT2 Profiler PCR Mouse Growth Factor Array analysis (Qiagen, PAMM-041Z) (Figure 2, Supplementary Tables 1–3). RNA samples from 3 independent mice were analyzed for each time point.
Intestinal Tissue Fractionation
A 6-cm segment of duodenum/jejunum was opened, placed in 15 mL 10 mmol/L EDTA in PBS, and rocked for 10 minutes at 4°C. The tissue was placed villus-side up on a glass plate on ice and lightly scraped 3 times in both directions with a P-100 pipette tip. Scraped tissue was examined by microscopy to ensure that villi and not crypts were removed. Remaining intestine was minced (3–5 mm2) and placed in 15 mL 15 mmol/L EDTA in PBS and rocked for 45 minutes at 4°C. Tissue fragments were moved into a 15-mL conical tube containing 8 mL PBS and shaken by hand for 2 minutes (5 shakes/2 seconds), followed by a short vortex pulse. The solution was filtered through a 70-μm filter into a 50-mL epithelial collection tube. Intact tissue fragments from the filter were placed into 8 mL PBS; shaking/vortexing was repeated, followed by decanting through a new filter into the epithelial collection tube. Tissue fragments (mesenchyme) were picked off the filter, rinsed briefly in PBS, transferred to a 1.5-mL tube, and snap-frozen in liquid N2. The epithelial crypts were collected by centrifugation at 500g at 4°C, transferred to a 1.5-mL tube, and snap-frozen in liquid N2. Tissue fractions were stored at –80°C before RNA isolation.
Single-Cell RNA-seq Analysis
Expression of Igf1r was investigated in the scRNA-seq dataset enriched in ISCs and progenitors.30 The dataset was downloaded from the Gene Expression Omnibus (Accession #GSE92865), and the gene-cell barcode matrices from 14 samples were concatenated. Cells that did not express intestinal epithelial markers or expressed less than 400 genes or more than 4400 genes were filtered out from the matrix. Similarly, genes that were expressed in less than 3 cells were filtered out. The resultant matrix has 13,076 cells and 16,047 genes and was used as input for Seurat R package. The data were log-normalized, and 1477 highly variable genes were identified by using expression cutoff value of 0.0125 and dispersion cutoff value of 0.5. Principal component analysis was performed on the highly variable genes, and the first 15 principal components were used for clustering analysis and tSNE projection. Gene expression heat maps were generated by using the log2 normalized data.
In Situ Hybridization
Tissues were fixed overnight in 10% neutral buffered formalin (Fisher, SF100-4), paraffin embedded, and sectioned to 4 μm. RNAScope Igf1 mouse probe (ACD 443901; ACD Bio, Newark, CA) used the 2.5 HD Brown detection kit (ACD 322310) according to manufacturer’s instructions, with the following specifications. Sections were boiled in target retrieval solution (ACD 322000) for 15 minutes and treated with protease plus (ACD 322340) for 30 minutes at 40°C.
Statistical Analysis
All experiments were performed with at least 3 biological replicates per group. Quantitative data are presented as mean ± standard error of the mean (SEM). Statistical comparisons were conducted with unpaired two-tailed Student t tests, 1-way analysis of variance (ANOVA), or 2-way ANOVA by using the Prism software (Graphpad, San Diego), as indicated in figure legends. Significance is reported as ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗P < .0001.
All authors had access to the study data and had reviewed and approved the final manuscript.
Acknowledgments
The authors thank Yasmine Abushukur for technical help, Erin Collin and Lindsay Griffin for maintaining the mouse colonies, and Dr Douglas Yee (University of Minnesota) for advice on selecting inhibitors of IGF-1 signaling.
CRediT Authorship Contributions
Natacha Bohin (Data curation: Equal; Formal analysis: Equal; Funding acquisition: Supporting; Investigation: Equal; Methodology: Equal; Project administration: Equal; Supervision: Supporting; Writing – original draft: Lead; Writing – review & editing: Supporting)
Kevin McGowan (Conceptualization: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Equal; Writing – review & editing: Supporting)
Theresa Keeley (Formal analysis: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Elizabeth Carlson (Formal analysis: Supporting; Writing – review & editing: Supporting)
Kelley Yan (Formal analysis: Equal; Funding acquisition: Supporting; Methodology: Equal; Writing – review & editing: Supporting)
Linda C. Samuelson, PhD (Conceptualization: Equal; Funding acquisition: Lead; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding NB was supported by the Cellular & Molecular Biology program, a Rackham International Student Fellowship, and the Bernard L. Maas Fellowship; KPM was supported by the Training Program in Organogenesis NIHT32 HD007505. The research was funded by NIHR01-DK118023 to LCS, R03-DK114656 to KSY, BWF CAMS to KSY, and Core support from the Michigan Gastrointestinal Research Center Grant NIHP30-DK34933.
Supplementary Data
References
- 1.de Sousa E.M.F., de Sauvage F.J. Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell. 2019;24:54–64. doi: 10.1016/j.stem.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery R.K., Carlone D.L., Richmond C.A., Farilla L., Kranendonk M.E., Henderson D.E., Baffour-Awuah N.Y., Ambruzs D.M., Fogli L.K., Algra S., Breault D.T. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth S., Franken P., Sacchetti A., Kremer A., Anderson K., Sansom O., Fodde R. Paneth cells in intestinal homeostasis and tissue injury. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H., van Oudenaarden A., Clevers H. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J., Martens A.C.M., Barker N., van Oudenaarden A., Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R., Sangiorgi E., Capecchi M.R., Kuo C.J. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones J.C., Brindley C.D., Elder N.H., Myers M.G., Jr., Rajala M.W., Dekaney C.M., McNamee E.N., Frey M.R., Shroyer N.F., Dempsey P.J. Cellular plasticity of Defa4(Cre)-expressing Paneth cells in response to Notch activation and intestinal injury. Cell Mol Gastroenterol Hepatol. 2019;7:533–554. doi: 10.1016/j.jcmgh.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sei Y., Feng J., Samsel L., White A., Zhao X., Yun S., Citrin D., McCoy J.P., Sundaresan S., Hayes M.M., Merchant J.L., Leiter A., Wank S.A. Mature enteroendocrine cells contribute to basal and pathological stem cell dynamics in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2018;315:G495–G510. doi: 10.1152/ajpgi.00036.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu S., Tong K., Zhao Y., Balasubramanian I., Yap G.S., Ferraris R.P., Bonder E.M., Verzi M.P., Gao N. Paneth cell multipotency induced by Notch activation following injury. Cell Stem Cell. 2018;23:46–59. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yousefi M., Nakauka-Ddamba A., Berry C.T., Li N., Schoenberger J., Simeonov K.P., Cedeno R.J., Yu Z., Lengner C.J. Calorie restriction governs intestinal epithelial regeneration through cell-autonomous regulation of mTORC1 in reserve stem cells. Stem Cell Reports. 2018;10:703–711. doi: 10.1016/j.stemcr.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richmond C.A., Shah M.S., Deary L.T., Trotier D.C., Thomas H., Ambruzs D.M., Jiang L., Whiles B.B., Rickner H.D., Montgomery R.K., Tovaglieri A., Carlone D.L., Breault D.T. Dormant intestinal stem cells are regulated by PTEN and nutritional status. Cell Rep. 2015;13:2403–2411. doi: 10.1016/j.celrep.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos A.J.M., Lo Y.H., Mah A.T., Kuo C.J. The intestinal stem cell niche: homeostasis and adaptations. Trends Cell Biol. 2018;28:1062–1078. doi: 10.1016/j.tcb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howarth G.S., Xian C.J., Read L.C. Insulin-like growth factor-I partially attenuates colonic damage in rats with experimental colitis induced by oral dextran sulphate sodium. Scand J Gastroenterol. 1998;33:180–190. doi: 10.1080/00365529850166923. [DOI] [PubMed] [Google Scholar]
- 18.Lemmey A.B., Ballard F.J., Martin A.A., Tomas F.M., Howarth G.S., Read L.C. Treatment with IGF-I peptides improves function of the remnant gut following small bowel resection in rats. Growth Factors. 1994;10:243–252. doi: 10.3109/08977199409010990. [DOI] [PubMed] [Google Scholar]
- 19.Gillingham M.B., Dahly E.M., Murali S.G., Ney D.M. IGF-I treatment facilitates transition from parenteral to enteral nutrition in rats with short bowel syndrome. Am J Physiol Regul Integr Comp Physiol. 2003;284:R363–R371. doi: 10.1152/ajpregu.00247.2002. [DOI] [PubMed] [Google Scholar]
- 20.Qiu W., Leibowitz B., Zhang L., Yu J. Growth factors protect intestinal stem cells from radiation-induced apoptosis by suppressing PUMA through the PI3K/AKT/p53 axis. Oncogene. 2010;29:1622–1632. doi: 10.1038/onc.2009.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Landeghem L., Santoro M.A., Mah A.T., Krebs A.E., Dehmer J.J., McNaughton K.K., Helmrath M.A., Magness S.T., Lund P.K. IGF1 stimulates crypt expansion via differential activation of 2 intestinal stem cell populations. FASEB J. 2015;29:2828–2842. doi: 10.1096/fj.14-264010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins H.R., Ohneda K., Keku T.O., D'Ercole A.J., Fuller C.R., Williams K.L., Lund P.K. Reduction of spontaneous and irradiation-induced apoptosis in small intestine of IGF-I transgenic mice. Am J Physiol Gastrointest Liver Physiol. 2002;283:G457–G464. doi: 10.1152/ajpgi.00019.2002. [DOI] [PubMed] [Google Scholar]
- 23.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Sampson L.L., Davis A.K., Grogg M.W., Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barron L., Sun R.C., Aladegbami B., Erwin C.R., Warner B.W., Guo J. Intestinal epithelial-specific mTORC1 activation enhances intestinal adaptation after small bowel resection. Cell Mol Gastroenterol Hepatol. 2017;3:231–244. doi: 10.1016/j.jcmgh.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faller W.J., Jackson T.J., Knight J.R., Ridgway R.A., Jamieson T., Karim S.A., Jones C., Radulescu S., Huels D.J., Myant K.B., Dudek K.M., Casey H.A., Scopelliti A., Cordero J.B., Vidal M., Pende M., Ryazanov A.G., Sonenberg N., Meyuhas O., Hall M.N., Bushell M., Willis A.E., Sansom O.J. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francavilla A., Starzl T.E., Scotti C., Carrieri G., Azzarone A., Zeng Q.H., Porter K.A., Schreiber S.L. Inhibition of liver, kidney, and intestine regeneration by rapamycin. Transplantation. 1992;53:496–498. doi: 10.1097/00007890-199202010-00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan Y., Zhang L., Li X., Zhang X., Liu S., Gao N., Li L., Gao G., Wei G., Chen Z., Zheng Y., Ma X., Siwko S., Chen J.L., Liu M., Li D. Repression of mammalian target of rapamycin complex 1 inhibits intestinal regeneration in acute inflammatory bowel disease models. J Immunol. 2015;195:339–346. doi: 10.4049/jimmunol.1303356. [DOI] [PubMed] [Google Scholar]
- 29.Kaestner K.H. The intestinal stem cell niche: a central role for Foxl1-expressing subepithelial telocytes. Cell Mol Gastroenterol Hepatol. 2019;8:111–117. doi: 10.1016/j.jcmgh.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M., Ootani A., Roelf K., Lee M., Yuan J., Li X., Bolen C.R., Wilhelmy J., Davies P.S., Ueno H., von Furstenberg R.J., Belgrader P., Ziraldo S.B., Ordonez H., Henning S.J., Wong M.H., Snyder M.P., Weissman I.L., Hsueh A.J., Mikkelsen T.S., Garcia K.C., Kuo C.J. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujii M., Matano M., Toshimitsu K., Takano A., Mikami Y., Nishikori S., Sugimoto S., Sato T. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 2018;23:787–793. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Bohin N., Carlson E.A., Samuelson L.C. Genome toxicity and impaired stem cell function after conditional activation of CreER(T2) in the intestine. Stem Cell Reports. 2018;11:1337–1346. doi: 10.1016/j.stemcr.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pucilowska J.B., McNaughton K.K., Mohapatra N.K., Hoyt E.C., Zimmermann E.M., Sartor R.B., Lund P.K. IGF-I and procollagen alpha1(I) are coexpressed in a subset of mesenchymal cells in active Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1307–G1322. doi: 10.1152/ajpgi.2000.279.6.G1307. [DOI] [PubMed] [Google Scholar]
- 34.Lahar N., Lei N.Y., Wang J., Jabaji Z., Tung S.C., Joshi V., Lewis M., Stelzner M., Martin M.G., Dunn J.C. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoki R., Shoshkes-Carmel M., Gao N., Shin S., May C.L., Golson M.L., Zahm A.M., Ray M., Wiser C.L., Wright C.V., Kaestner K.H. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol. 2016;2:175–188. doi: 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valenta T., Degirmenci B., Moor A.E., Herr P., Zimmerli D., Moor M.B., Hausmann G., Cantu C., Aguet M., Basler K. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 2016;15:911–918. doi: 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
- 37.Shoshkes-Carmel M., Wang Y.J., Wangensteen K.J., Toth B., Kondo A., Massasa E.E., Itzkovitz S., Kaestner K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degirmenci B., Valenta T., Dimitrieva S., Hausmann G., Basler K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 2018;558:449–453. doi: 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 39.Greicius G., Kabiri Z., Sigmundsson K., Liang C., Bunte R., Singh M.K., Virshup D.M. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A. 2018;115:E3173–E3181. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ney D.M., Huss D.J., Gillingham M.B., Kritsch K.R., Dahly E.M., Talamantez J.L., Adamo M.L. Investigation of insulin-like growth factor (IGF)-I and insulin receptor binding and expression in jejunum of parenterally fed rats treated with IGF-I or growth hormone. Endocrinology. 1999;140:4850–4860. doi: 10.1210/endo.140.10.7029. [DOI] [PubMed] [Google Scholar]
- 41.Munoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., Myant K., Versteeg R., Sansom O.J., van Es J.H., Barker N., van Oudenaarden A., Mohammed S., Heck A.J., Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland K.J., Trivedi S., Lee D., Wan K., Kulkarni R.N., Holzenberger M., Brubaker P.L. Loss of glucagon-like peptide-2-induced proliferation following intestinal epithelial insulin-like growth factor-1-receptor deletion. Gastroenterology. 2011;141:2166–2175. doi: 10.1053/j.gastro.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Santoro M.A., Blue R.E., Andres S.F., Mah A.T., Van Landeghem L., Lund P.K. Obesity and intestinal epithelial deletion of the insulin receptor, but not the IGF 1 receptor, affect radiation-induced apoptosis in colon. Am J Physiol Gastrointest Liver Physiol. 2015;309:G578–G589. doi: 10.1152/ajpgi.00189.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bortvedt S.F., Lund P.K. Insulin-like growth factor 1: common mediator of multiple enterotrophic hormones and growth factors. Curr Opin Gastroenterol. 2012;28:89–98. doi: 10.1097/MOG.0b013e32835004c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knott A.W., Juno R.J., Jarboe M.D., Profitt S.A., Erwin C.R., Smith E.P., Fagin J.A., Warner B.W. Smooth muscle overexpression of IGF-I induces a novel adaptive response to small bowel resection. Am J Physiol Gastrointest Liver Physiol. 2004;287:G562–G570. doi: 10.1152/ajpgi.00438.2003. [DOI] [PubMed] [Google Scholar]
- 46.Rodgers J.T., King K.Y., Brett J.O., Cromie M.J., Charville G.W., Maguire K.K., Brunson C., Mastey N., Liu L., Tsai C.R., Goodell M.A., Rando T.A. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.el Marjou F., Janssen K.P., Chang B.H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 48.Sengupta S., Peterson T.R., Laplante M., Oh S., Sabatini D.M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 49.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 50.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud A., Grosse A.S., Gumucio D.L., Ernst S.A., Tsai Y.H., Dempsey P.J., Samuelson L.C. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohn S.M., Schloemann S., Tessner T., Seibert K., Stenson W.F. Crypt stem cell survival in the mouse intestinal epithelium is regulated by prostaglandins synthesized through cyclooxygenase-1. J Clin Invest. 1997;99:1367–1379. doi: 10.1172/JCI119296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Diaz L., Hinkle K.L., Jain R.N., Zavros Y., Brunkan C.S., Keeley T., Eaton K.A., Merchant J.L., Chew C.S., Samuelson L.C. Parietal cell hyperstimulation and autoimmune gastritis in cholera toxin transgenic mice. Am J Physiol Gastrointest Liver Physiol. 2006;290:G970–G979. doi: 10.1152/ajpgi.00461.2005. [DOI] [PubMed] [Google Scholar]
- 53.Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.