ABSTRACT
Among the three embryonic germ layers, the mesoderm plays a central role in the establishment of the vertebrate body plan. The mesoderm is specified by secreted signaling proteins from the FGF, Nodal, BMP and Wnt families. No new classes of extracellular mesoderm-inducing factors have been identified in more than two decades. Here, we show that the pinhead (pnhd) gene encodes a secreted protein that is essential for the activation of a subset of mesodermal markers in the Xenopus embryo. RNA sequencing revealed that many transcriptional targets of Pnhd are shared with those of the FGF pathway. Pnhd activity was accompanied by Erk phosphorylation and required FGF and Nodal but not Wnt signaling. We propose that during gastrulation Pnhd acts in the marginal zone to contribute to mesoderm heterogeneity via an FGF receptor-dependent positive feedback mechanism.
KEY WORDS: Wnt, FGF, Erk1, Akt, Brachyury, Nodal, Antero-posterior axis, Xenopus
Summary: Xenopus Pinhead is a secreted cystine-knot protein that functions in a positive feedback loop with FGF to activate Erk signaling and induce mesoderm.
INTRODUCTION
The vertebrate embryonic body plan forms via the specification of three germ layers: the ectoderm, mesoderm and endoderm. The mesoderm plays a central role in this process, being responsible for tissue patterning and cell movements during gastrulation. At late blastula stages, the mesoderm [or more precisely, the mesendoderm (Rodaway and Patient, 2001)] is characterized by the dorsoanterior and ventroposterior domains of gene expression. The dorsoanterior domain marks the signaling center known as the Spemann organizer, which gives rise to the dorsal mesoderm and modulates all three germ layers. Factors secreted from the organizer induce neural tissue in the ectoderm and subdivide the mesoderm into dorsal (notochord), paraxial (somites), intermediate (kidney and gonads) and lateral/ventral (e.g. blood) types. Signals from the ventrolateral marginal zone and vegetal endoderm also contribute to mesoderm patterning (De Robertis and Kuroda, 2004; Harland and Gerhart, 1997; Kiecker et al., 2016; Langdon and Mullins, 2011; Niehrs, 2004; Zorn and Wells, 2009).
Studies from the past three decades extensively characterized the signaling pathways that contribute to the formation of the three germ layers and specify the primary embryonic axis (Harland and Gerhart, 1997). Only a handful of secreted signaling molecules from the FGF, Wnt, Nodal and BMP families have been shown to be involved in mesoderm induction and patterning (Christen and Slack, 1999; Kiecker et al., 2016; Kimelman, 2006; Schohl and Fagotto, 2002). Pinhead (pnhd) was originally described in Xenopus tropicalis as a gene controlling head development; however, its roles in specific developmental processes and the underlying signaling mechanisms remain uncharacterized (Kenwrick et al., 2004). In Xenopus gastrulae, pnhd is expressed in a broad ventrolateral domain in the marginal zone (Kenwrick et al., 2004; Kjolby and Harland, 2017), suggesting a role in mesoderm specification and patterning.
In this study, we evaluate a function of pnhd in mesoderm development. We show that pnhd is dynamically expressed in many tissues during the early development of Xenopus. Pnhd protein is readily secreted from frog gastrula cells and mammalian tissue culture cells. We also show that Pnhd is both necessary and sufficient for the activation of many mesoderm-specific genes. This functional activity of Pnhd required both FGF and Nodal signaling, and has been manifested by the phosphorylation of Erk1. These observations lead us to propose that Pnhd is a secreted factor that controls mesoderm formation in an FGF-receptor-dependent manner.
RESULTS
Pnhd is a secreted signaling protein that modulates axial development
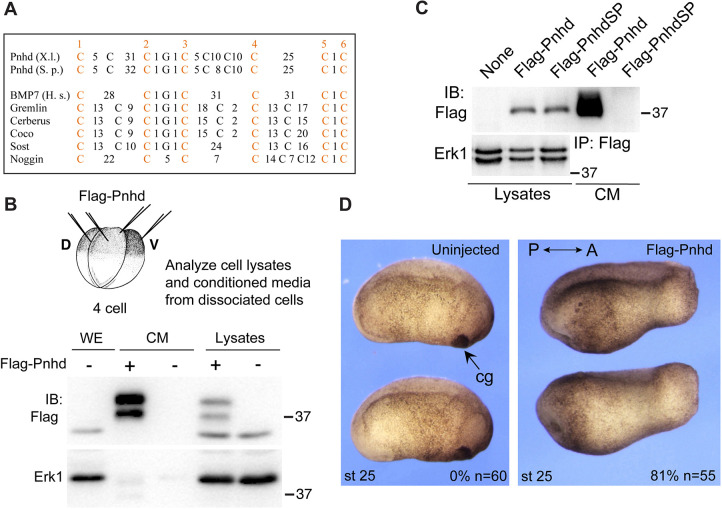
The pnhd gene encodes a conserved protein containing three cystine knot (CK) motifs (Avsian-Kretchmer and Hsueh, 2004; Imai et al., 2012; Isaacs, 1995). There are no other identifiable protein domains. The CK is a common feature of many extracellular proteins and has been proposed to stabilize protein tertiary structure via disulfide bonds (Roch and Sherwood, 2014). Pnhd is present in many animals, from insects to amniotes. Sequence alignment shows the conservation of the protein throughout all three CK domains (Fig. S1). The CKs of Pnhd significantly deviate from those of other molecules (Fig. 1A), indicating that it belongs to a new class of proteins with yet uncharacterized signaling properties.
Fig. 1.
Pnhd is a secreted protein that promotes posterior development. (A) Alignment of CK domains from Pnhd and other secreted proteins. Spacing is indicated by numbers of non-conserved amino acids between conserved cysteine residues. X.l., Xenopus laevis; S.p., Stegastes partitus; H.s., Homo sapiens. (B) Secretion of Pnhd by Xenopus gastrula cells. Four-cell embryos were injected with 0.5 ng of Flag-Pnhd RNA for each blastomere, cultured to the onset of gastrulation and dissociated to individual cells. Pnhd levels were compared in the media conditioned for 3 h and the corresponding cell lysates. (C) Pnhd is secreted by transfected HEK293Tcells. Deletion of the putative signal peptide in PnhdSP prevents secretion. (D) Head defects in embryos injected dorsally with 2 ng of pnhd RNA at the four-cell stage. Frequencies of embryos with head defects and their total number are indicated. The results are representative of more than five independent experiments. A, anterior; cg, cement gland; CM, conditioned medium; IB, immunoblot; IP, immunoprecipitation; P, posterior.
The presence of the highly hydrophobic N-terminal 22-amino acid stretch in the deduced Pnhd protein sequence suggested a candidate signal peptide. We therefore examined whether the product of pnhd is secreted from the embryonic cells. To accomplish this, RNA encoding Flag-tagged Pnhd was microinjected into Xenopus embryos, which were then dissociated into single cells at the beginning of gastrulation, and the medium, conditioned by the dissociated cells for 3 h, was examined by immunoblotting (Fig. 1B). Pnhd protein was predominantly found in the conditioned medium, supporting the hypothesis that it is secreted into the extracellular space. Immunoblotting revealed two bands of ∼39 kDa and ∼42 kDa. The ratio of the upper and lower band intensities was variable, suggesting that the protein undergoes post-translational modifications, such as glycosylation. These observations have been confirmed in transfected HEK293T cells (Fig. S2, Fig. 1C). Full-length Pnhd protein was found largely in the medium conditioned by the transfected HEK293T cells, whereas the Pnhd construct lacking the signal peptide remained in the cell lysates (Fig. 1C). We estimate that at least 70-90% of Pnhd is secreted, whereas 10-30% is associated with the cell pellet fraction.
We next evaluated the embryonic phenotype caused by pnhd RNA injected into Xenopus early blastomeres. At tailbud stages, embryos expressing pnhd RNA consistently developed an enlarged trunk and tail, but contained reduced or no head structures compared with uninjected siblings (Fig. 1D). We conclude that Pnhd is a secreted protein that can modulate head and axis formation in Xenopus embryos.
Pnhd is dynamically expressed in the early embryo
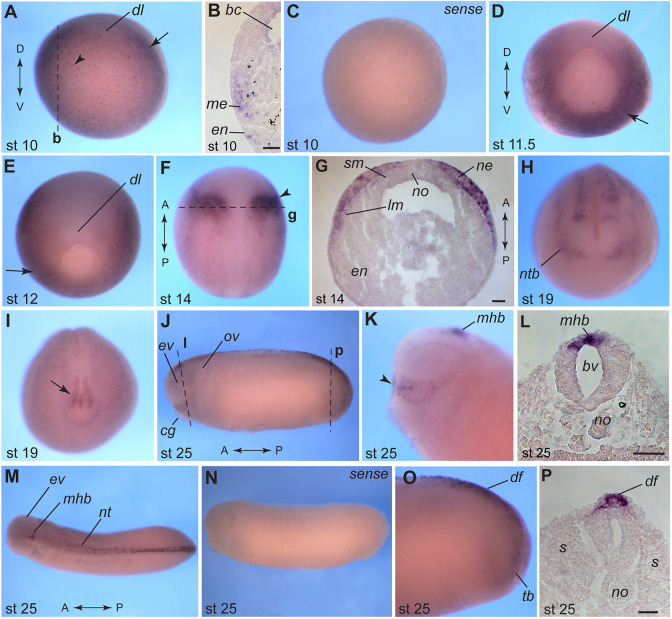
Previous reports indicate that pnhd RNA is enriched in the marginal zone at the onset of gastrulation (Kenwrick et al., 2004; Kjolby and Harland, 2017). To gain further insights into Pnhd function, we carried out whole-mount in situ hybridization (WISH) with embryos taken at different developmental stages. At the onset of gastrulation, Pnhd transcripts were detected both in the mesoderm and vegetal endoderm (Fig. 2A-D). Notably, pnhd RNA was excluded from the dorsal midline (Fig. 2A,D,E) as reported for many Wnt and FGF target genes (Kjolby et al., 2019). Additionally, we observed strong bilateral expression domains in the anterior neuroectoderm and weaker staining in the lateral plate mesoderm at stages 13 and 14 (Fig. 2F,G), consistent with previous studies (Bae et al., 2014; Plouhinec et al., 2014). The neural plate domain appeared to correspond to the future midbrain-hindbrain boundary (MHB). At later stages, pnhd RNA was also enriched in the anterior preplacodal ectoderm, the dorsal neural tube and its boundary (Fig. 2H), and the presumptive tailbud area demarcated by the chordoneural hinge (Fig. 2I). At stage 25, pnhd transcripts were evident at the MHB (Fig. 2J-M), along the dorsal midline and in the dorsal fin (Fig. 2J,M-P). The identified predominant pnhd expression domains correspond to regions with high levels of FGF and Wnt signaling, and suggest functions in the mesoderm, neural tissue, neural crest and placodes, and the dorsal fin.
Fig. 2.
Pnhd transcript localization at different developmental stages. WISH was carried out with albino embryos using pnhd antisense and sense RNA probes. (A) Vegetal view of a stage 10 embryo (dorsal is up). Arrowhead points to vegetal endoderm. (B) Cross-section of a stage 10 embryo. (C) Control embryo, stage 10 (sense probe). (D) Vegetal view of a stage 11.5 embryo (dorsal is up). (E) Vegetal view of a stage 12 embryo (dorsal side is up). Arrows in A,D,E indicate mesodermal expression. (F) Dorsal view of a stage 14 embryo (anterior is up). Arrowhead marks the neuroectoderm. (G) Cross-section of the embryo shown in F. (H) Anterior view of a stage 19 embryo (dorsal is up). (I) Posterior view of a stage 19 embryo (dorsal is up). Arrow indicates staining in the tailbud. (J) Side view of a stage 25 embryo. (K) Head of a stage 25 embryo (anterior is left). Arrowhead points to the signal in the superficial ectoderm cells that are dorsal to the cement gland. (L) Cross-section corresponding to the midbrain level of embryo in J. (M) Dorsal view of a stage 25 embryo. (N) Stage 25, control sense probe. (O) Lateral view of a stage 25 embryo tailbud. Anterior is to the left in J,K,M-O. (P) Transverse section of a stage 25 embryo corresponding to J and O. Dashed lines mark the approximate level of corresponding sections (indicated by bold letters). Dorsoventral (D/V) and anteroposterior (A/P) embryonic axes are indicated. Abbreviations: bc, blastocoel; bv, brain ventricle; cg, cement gland; df, dorsal fin; dl, dorsal blastopore lip; en, endoderm; ev, eye vesicle; lm, lateral mesoderm; me, mesoderm; mhb, midbrain-hindbrain boundary; ne, neuroectoderm; nt, neural tube; ntb, neural tube border; no, notochord; ov, otic vesicle; s, somites; sm, somitic mesoderm; tb, tailbud. Scale bars: 50 µm (B,G,L); 25 µm (P).
Pnhd induces mesodermal markers in animal pole explants
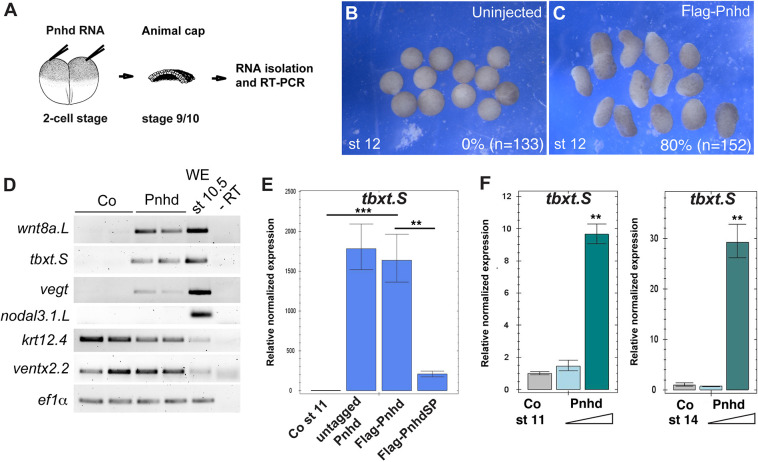
The morphological phenotype of embryos injected with pnhd RNA is consistent with enhanced ventroposterior development. Pnhd expression in the ventrolateral marginal zone suggests that it might participate in mesoderm formation. We therefore assessed whether pnhd can induce the mesoderm in the animal pole ectoderm, a tissue lacking mesodermal gene expression. Although uninjected animal cap explants retained their spherical shape at stage 12, the explants isolated from embryos injected with pnhd RNA have visibly elongated (Fig. 3A-C). Notably, explant elongation frequently accompanies mesoderm induction, mimicking convergent extension movements during gastrulation (Howard and Smith, 1993).
Fig. 3.
Pnhd induces mesoderm in ectodermal explants. (A-E) Early embryos were injected with 1-2 ng of Pnhd RNA. Ectoderm explants were dissected at late blastula stages and cultured until stage 12 to examine morphology (B,C) and gene expression by RT-PCR (D,E). (B,C) Pnhd RNA induced animal cap elongation by stage 12. Frequencies of elongated explants and their total number are indicated. The results represent more than five independent experiments. (D) Induction of selected mesodermal markers by Flag-Pnhd RNA (1 ng). (E) Flag-Pnhd RNA has the same ability to induce tbxt as untagged Pnhd RNA in RT-qPCR, but this activity is lost in Flag-PnhdSP lacking the signal peptide (2 ng of each RNA). (F) Pnhd protein was purified from the supernatants of transfected HEK293T cells. To assess its mesoderm-inducing activity, ectoderm explants were dissected from stage 10 embryos and cultured in 0.6×MMR solution containing 1.5 µg/ml or 6.5 µg/ml of Pnhd. RT-qPCR was carried out for tbxt at stage 11 or stage 14. Data are mean±s.d. Significance was determined by an unpaired two-tailed Student's t-test. **P<0.01, ***P<0.001.
Indeed, RT-PCR demonstrated the upregulation of several mesodermal markers, including tbxt/brachyury (Smith et al., 1991), wnt8a (Christian et al., 1991) and vegt (Gentsch et al., 2013; Zhang et al., 1998), indicating that Pnhd can induce mesodermal progenitor fates (Fig. 3D). However, other markers, including the dorsal mesoderm markers nodal3 (Smith et al., 1995) and gsc (Cho et al., 1991), were not induced by Pnhd, demonstrating target selectivity. To ensure that the tag does not affect Pnhd biological activity, we used RT-qPCR to confirm that Flag-Pnhd induced tbxt to the same degree as the original untagged construct (Fig. 3E). The mutant lacking the signal peptide was significantly less active than Pnhd (Fig. 3E), indicating that Pnhd secretion is crucial for its function.
To unequivocally establish that Pnhd functions in the extracellular space, we tested Pnhd-Flag protein that was affinity-purified from the medium conditioned by transfected HEK293T cells. When added to ectoderm explants, the purified protein induced tbxt in a dose-dependent manner (Fig. 3F). These observations indicate that Pnhd is a new mesoderm-inducing factor that exerts its biological effects in the extracellular space.
RNA sequencing (RNA-seq) defines candidate transcriptional targets of Pnhd
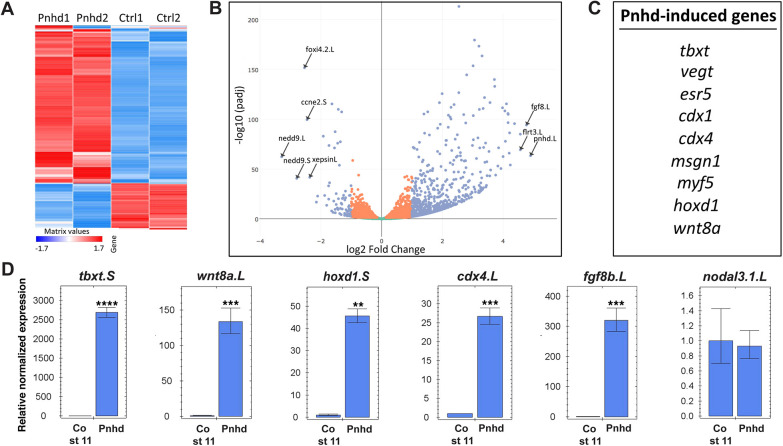
An unbiased transcriptome-wide approach was taken to identify the genes differentially regulated by Pnhd RNA in ectoderm cells (Fig. 4A, Table S1). Gene set enrichment analysis (GSEA) revealed strong enrichment of mesoderm-specific markers among the top 100 upregulated genes. No specific trend was observed for the downregulated genes. Highly ranked among the upregulated genes were known FGF and Wnt targets, including tbxt, cdx4, hoxd1, wnt8a and msgn1 (Fig. 4B,C) (Branney et al., 2009; Chung et al., 2004; Ding et al., 2018; Kjolby and Harland, 2017; Nakamura et al., 2016), which were validated by RT-qPCR (Fig. 4D). These genes are known to be expressed in the marginal zone, and most of them are excluded from the organizer region at the onset of gastrulation (Kjolby et al., 2019; Nakamura et al., 2016). By contrast, other mesodermal genes, such as the dorsal markers nodal, gsc or noggin, and the ventral mesoderm (blood) markers szl, bambi and ventx1.2, have not been significantly changed, indicating that only a subset of ventroposterior mesodermal genes is sensitive to pnhd. These findings suggest that Pnhd is involved in paraxial mesoderm formation.
Fig. 4.
RNA sequencing defines Pnhd target genes. (A) Heatmap of gene expression in the Pnhd-expressing and control uninjected animal pole cells that were cultured until stages 11 and 12. The duplicate samples are highly similar. (B) Volcano plot shows top genes upregulated by Pnhd. (C) Differentially expressed genes that are induced by Pnhd RNA (1.5 ng) in animal caps. The list was derived from the top 100 genes induced by Pnhd in four independent RNA-seq experiments. (D) qRT-PCR validation of Pnhd targets in animal caps. Data are mean±s.d. Significance was determined by an unpaired two-tailed Student's t-test. **P<0.01, ***P<0.001, ****P<0.0001.
For loss-of-function analysis, two morpholino oligonucleotides (MOs) have been designed and validated in separate experiments, verifying their efficiency and specificity. Pnhd MOatg efficiently blocked Pnhd RNA translation, whereas the splicing-blocking MO (Pnhd MOsp) with an unrelated sequence interfered with endogenous Pnhd transcript splicing (Fig. S3A,B). Both MOs caused the ‘pinhead’ phenotype, i.e. deficiency in head structures (Fig. S3C-E). Importantly, the defect caused by Pnhd MOsp has been rescued by Pnhd RNA (Fig. S3F,G).
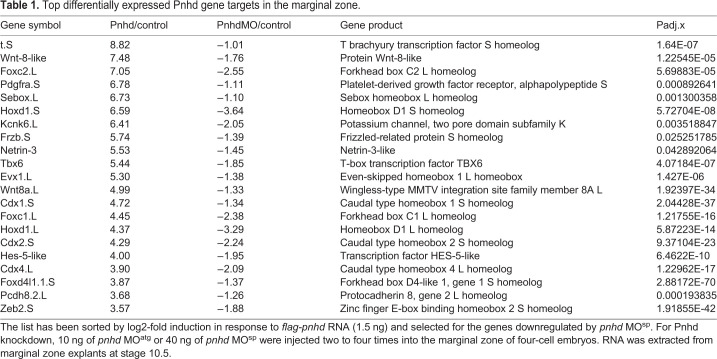
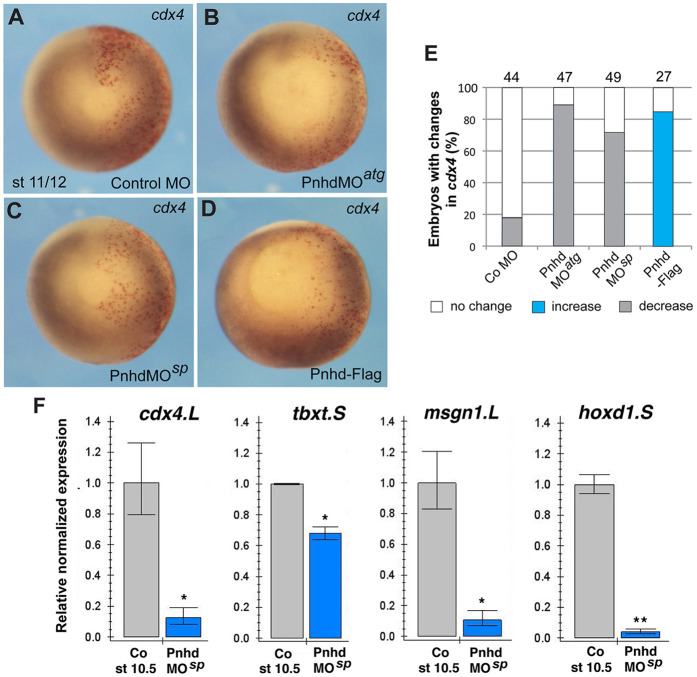
RNA-seq revealed putative Pnhd targets that were downregulated in marginal zone cells depleted of Pnhd. The results from the gain-of-function and the loss-of-function studies were combined to determine consensus gene targets. We found a total of 71 pnhd ‘signature’ target genes, defined as the genes upregulated in ectoderm explants by Pnhd RNA and downregulated in the marginal zone by Pnhd MOsp (Table 1, Table S2). To confirm the requirement of Pnhd in mesoderm formation, selected candidate gene targets from RNA-seq data were validated by WISH. Both cdx4 and wnt8 transcripts were upregulated by Pnhd RNA overexpression and downregulated in the cells depleted of Pnhd (Fig. 5A-E, Fig. S4A-E). RT-qPCR further confirmed the downregulation of cdx4, hoxd1, tbxt, msgn1 and wnt8 transcripts in the marginal zone after pnhd depletion, although tbxt decreased only mildly, possibly because this gene is controlled by multiple signaling pathways, and the ventral marker admp2 was unaffected (Fig. 5F, Fig. S4F).
Table 1.
Top differentially expressed Pnhd gene targets in the marginal zone.
Fig. 5.
Pnhd is required for mesoderm formation. (A-D) WISH validates changes in cdx4 expression in embryos with manipulated Pnhd levels in one half of each embryo. Red-gal was used as a β-galactosidase substrate (red) for lineage tracing of the injected area. Compare gene expression (dark staining) between the injected (red) and uninjected sides. (E) Quantification of changes in cdx4 RNA in Pnhd-depleted or overexpressing embryos. (F) RT-qPCR confirmation of the downregulation of cdx4, hoxd1, msgn1 and tbxt in stage 10.5 marginal zone explants depleted of pnhd. Data are mean±s.d. Significance was determined by an unpaired two-tailed Student's t-test. *P<0.05, **P<0.01.
At later stages, WISH analysis demonstrated the disrupted and reduced expression of myod in pnhd-depleted embryos indicating abnormal somite segmentation (Fig. S5A,B). The chordin domain appeared narrower compared with uninjected embryos (Fig. S5E,F). In contrast, the blood marker α-globin was not significantly changed (Fig. S5C,D). Taken together, our gain- and loss-of-function experiments show that Pnhd signaling is involved in mesodermal fate specification during gastrulation.
Pnhd signaling depends on the FGF pathway
As Pnhd target genes are similar to the ones activated by the FGF and Wnt pathways (Kjolby et al., 2019), we assessed whether these pathways play a role in Pnhd signaling. Importantly, we observed that pnhd transcription is induced in ectodermal cells by FGF and Wnt proteins (Fig. S6A-C), as reported by earlier studies (Branney et al., 2009; Chung et al., 2004; Ding et al., 2018; Kjolby and Harland, 2017; Nakamura et al., 2016).
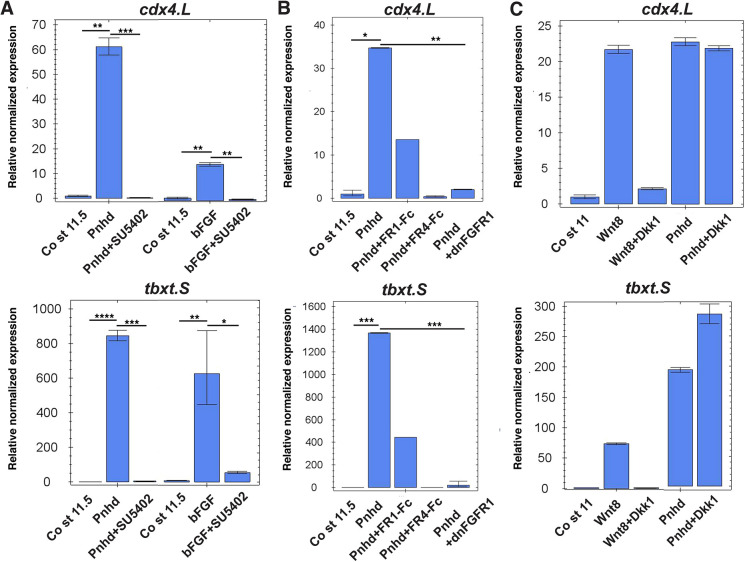
FGF proteins are known to play key roles in mesoderm development by activating tyrosine kinase receptors (FGFRs), the Akt protein kinase and extracellular signal-regulated kinases (Erk, also known as mitogen-activated protein kinase, MAPK) (Christen and Slack, 1999; Dubrulle and Pourquié, 2004; Manning and Toker, 2017; Ornitz and Itoh, 2015). To interfere with the FGF pathway, we used SU5402, a pharmacological inhibitor of FGF receptor activity (Mohammadi et al., 1997), a dominant negative form of FGFR1 that forms nonfunctional dimers with wild-type receptors (Amaya et al., 1991), and secreted inhibitory forms of FGF receptors (Marics et al., 2002). Pnhd failed to activate tbxt and cdx4 in the presence of SU5402 (Fig. 6A). Similarly, Pnhd activity was compromised by dominant interfering FGFR constructs (Fig. 6B).
Fig. 6.
Pnhd response requires FGF but not Wnt signaling. (A-C) Embryos were injected in the animal pole region at the two-cell stage with 1-2 ng of Pnhd RNA, FGFR1-Fc, FGFR4-Fc or dnFGFR1 RNA (2 ng each), 1 ng of Wnt8 or 300 pg of Dkk1 RNA, as indicated. Ectoderm explants were dissected at stages 9-10 and cultured until stages 11-11.5 for gene expression analysis by RT-qPCR. (A) The induction of tbxt and cdx4 by Pnhd is blocked by the FGF inhibitor SU5402 (100 µm). Stimulation with bFGF was used as a positive control. (B) Gene target activation by Pnhd was prevented by DN-FGFR1 and secreted forms of FGFR1-Fc and FGFR4-Fc. (C) The Wnt inhibitor Dkk1 did not affect Pnhd signaling but effectively blocked Wnt8 responses. Data are mean±s.d. Significance was determined by an unpaired two-tailed Student's t-test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To inhibit Wnt signaling, we used Dkk1, which physically associates with and inhibits the signaling through the Wnt co-receptor LRP5/6 (Bafico et al., 2001; Mao et al., 2001; Semënov et al., 2001). As expected, Dkk1 inhibited Wnt-dependent activation of gene targets (Fig. 6C). Notably, Dkk1 did not suppress the response to Pnhd (Fig. 6C), indicating that Pnhd signaling does not require Wnt proteins. Consistent with this interpretation, the headless phenotype of Pnhd RNA-injected embryos was rescued by inhibiting FGF signaling with DN-FGFR4-Fc (Fig. S7). These studies suggest that the stimulation of target genes by Pnhd requires FGF but not Wnt activity.
Erk1 but not Akt might mediate Pnhd effects on transcription
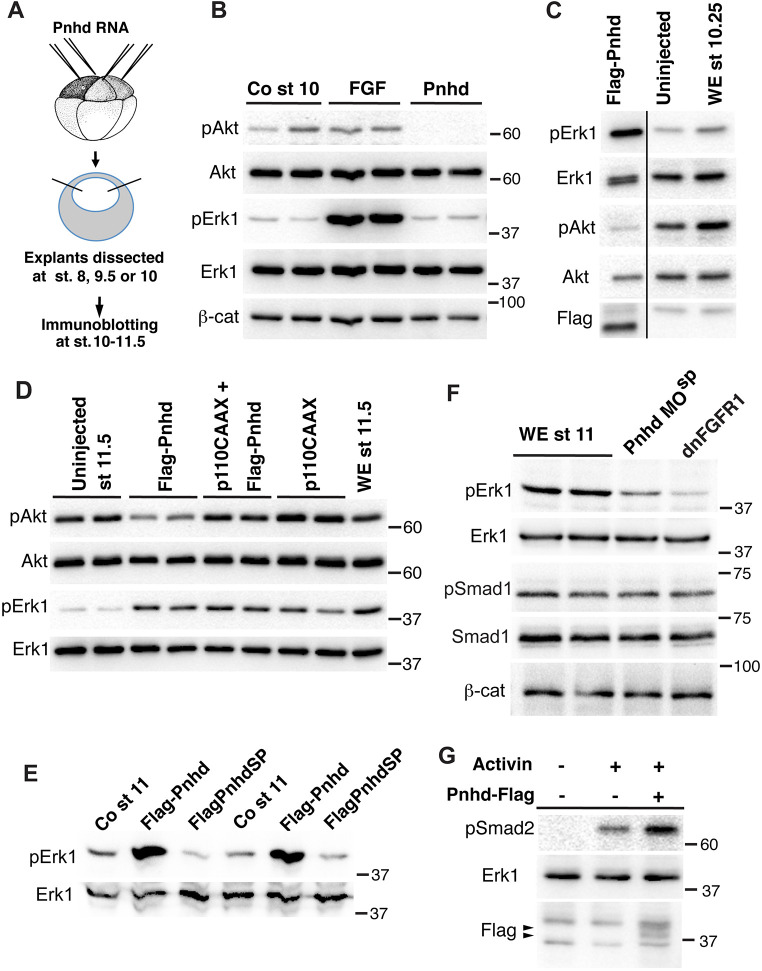
To further investigate the pathways that are modulated by Pnhd, we examined the abundance and the phosphorylation status of the common cytoplasmic signaling mediators in pnhd-expressing cells. The direct comparison of FGF and Pnhd effects on the early ectoderm confirmed that, unlike FGF, Pnhd does not activate Erk1 in the early explants that are isolated at stage 8 and cultured until stage 10 (Fig. 7A,B). Interestingly, in the same explants, we found that Pnhd, but not FGF, led to the pronounced inhibition of Akt, a kinase implicated in many pathways, including receptor tyrosine kinase signaling (Manning and Toker, 2017). By contrast, β-catenin levels did not change (Fig. 7B). The negative effect of Pnhd on Akt was reproducible and stage-dependent, and it was less pronounced at the end of gastrulation and correlated with Erk phosphorylation (Fig. 7C,D).
Fig. 7.
Pnhd inhibits Akt, but activates Erk1. (A) Schematic of the experiments shown in B-D. Embryos were injected with RNAs encoding Flag-Pnhd (1.5 ng), p110CAAX (0.5 ng), or treated with FGF protein, as indicated. Ectoderm explants were dissected at stages 8, 9.5 or 10, and cultured until the desired stage for immunoblotting with indicated antibodies. (B) Comparison of Pnhd and FGF effects on blastula ectoderm. Pnhd inhibits Akt phosphorylation in ectoderm explants isolated at stage 8 and analyzed at stage 10. FGF has no effect on Akt, but activates Erk. (C) Pnhd inhibits Akt but induces Erk phosphorylation in ectoderm isolated at stage 9.5 and cultured until stage 10.25. This result has been obtained in at least ten experiments. The separator line serves to indicate that several irrelevant gel lanes have been omitted. (D) Pnhd-dependent stimulation of Erk is not affected by the Akt activator p110CAAX. (E) Erk1 phosphorylation in Pnhd-expressing embryos at stage 11. Embryos were injected with RNAs encoding Flag-Pnhd or Flag-PnhdSP (1.5 ng each). (F) Downregulation of Erk1 phosphorylation in lysates of stage 11 embryos injected with Pnhd MOsp. Embryos were injected with RNAs encoding dnFGFR1 (1.5 ng each) or 40 ng of Pnhd MOsp, as indicated. There are no detectable changes in β-catenin or phospho-Smad1. (G) Pnhd promotes Smad2 phosphorylation by Activin. Ectoderm explants were dissected from the injected embryos at stage 8 and cultured for 1 h with or without Activin. Immunoblot analysis with anti-pSmad2 antibodies is shown. Pnhd is detected by anti-Flag antibodies (arrowhead). Erk1 is a control for loading. WE, whole embryo controls in C,D,F.
To assess the importance of the observed phospho-Akt downregulation for Pnhd-dependent Erk activation, we modulated the function of PI3 kinase, an upstream activator of Akt (Manning and Toker, 2017). Neither stimulation of Akt by the constitutively active PI3K (p110CAAX) (Carballada et al., 2001) nor its inhibition by the phosphatase PTEN, influenced the ability of Pnhd to stimulate Erk phosphorylation (Fig. 7D, Fig. S8). Although Akt does not appear to mediate Pnhd signaling to Erk in these experiments, it might function in a parallel pathway that affects mesoderm development independently of Erk (Carballada et al., 2001; Dubrulle and Pourquié, 2004).
By contrast, overexpressed Pnhd caused robust phospho-Erk1 accumulation at gastrula stages (Fig. 7C-E). Of note, the Pnhd construct lacking the signal peptide failed to activate Erk1 (Fig. 7E). Conversely, the level of phospho-Erk1 decreased in Pnhd-depleted embryos, similar to the effect of DN-FGFR1 (Fig. 7F). No significant changes in β-catenin levels or Smad1 phosphorylation were detected in Pnhd-depleted embryos, supporting specificity (Fig. 7F). These observations are consistent with the idea that Pnhd promotes mesoderm formation via the FGF- and Erk-dependent pathway.
We next evaluated whether Pnhd signaling is affected by the Nodal/Activin pathway. Notably, the stimulation of animal cap explants with Pnhd enhanced Activin-dependent Smad2 phosphorylation, indicating crosstalk (Fig. 7G). On its own, Pnhd did not change phospho-Smad2 levels (data not shown). Notably, SB505124, an inhibitor of Nodal/Activin receptor signaling (DaCosta Byfield et al., 2004), interfered with Pnhd-dependent target gene activation and explant elongation (Fig. S9). These observations indicate that both FGF and Nodal pathways contribute to the ability of Pnhd to activate mesodermal gene targets.
Pnhd expression in the marginal zone is essential for mesoderm formation during gastrulation
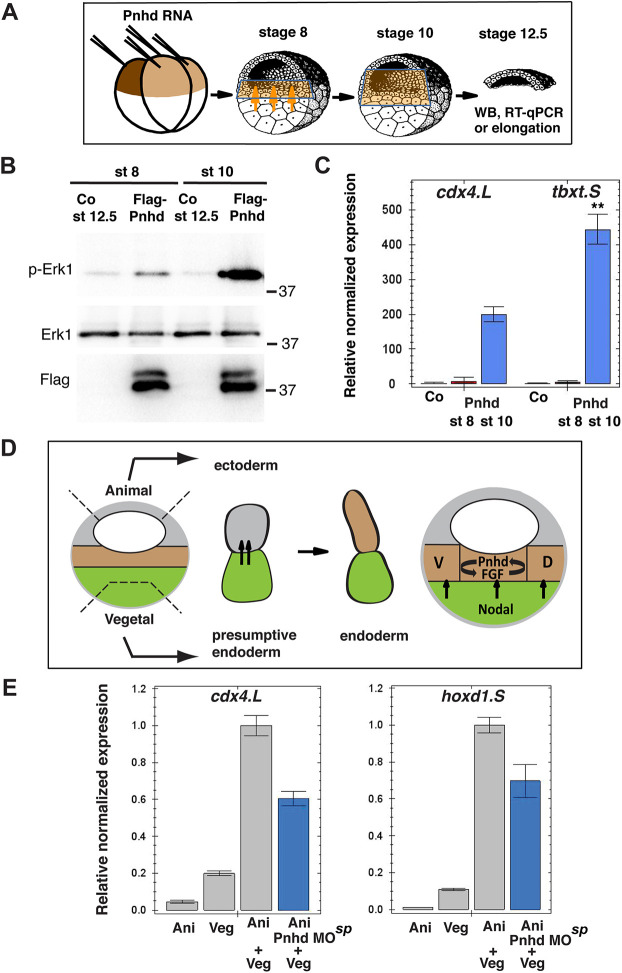
To get insights into the developmental stage at which Pnhd operates, we examined whether the Pnhd effect depends on the time of animal cap isolation (Fig. 8A). When the explants were isolated at stage 10 and cultured to stage 12, they robustly elongated in response to pnhd RNA (Fig. S10). However, no response was detected in the explants prepared at stage 8 (Fig. S10). Moreover, the explants isolated at stage 10, unlike the ones isolated at stage 8, revealed preferential phosphorylation of Erk1 (Fig. 8B) and selective activation of Pnhd gene targets (Fig. 8C). The only difference between the two groups of explants containing Pnhd is the time of their contact with the inducing tissue, i.e. the adjacent mesendoderm. Therefore, between stage 8 and stage 10, the explanted ectoderm must have received additional signals from the marginal zone (Sokol, 1993). Thus, Pnhd induces cdx4 and tbxt synergistically with these additional signals and this might involve the Nodal and/or the FGF pathway, as predicted by our inhibitor studies.
Fig. 8.
Pnhd cooperates with endogenous-inducing signals to promote mesoderm formation during gastrulation. (A) Schematic of the experiments presented in B,C. Mesoderm-inducing signals are indicated by orange arrows, and the cells receiving them are in light brown. Each animal blastomere of two-cell embryos received 1 ng of Pnhd RNA. Pnhd-expressing or control ectoderm explants were isolated at stage 8 or stage 10, as indicated. When the control embryos reached stage 12.5, the explants were lysed for immunoblotting with antibodies specific for pErk1 and Erk (B) and for RT-qPCR analysis of cdx4 and tbxt transcripts (C). (B) Erk is synergistically activated by Pnhd and endogenous signals in ectoderm explants isolated at stage 10. (C) Cooperative activation of mesodermal gene targets by Pnhd and endogenous signals in ectoderm dissected at stage 10. (D,E) Pnhd is required for mesoderm formation in response to endogenous-inducing signals. (D) Schematic of the experiment shown in E. Model for Pnhd function (right). Pnhd is activated in the marginal zone by Nodal and FGF signaling and functions within the newly induced mesodermal layer. D, dorsal mesoderm; V, ventral mesoderm. The green color indicates presumptive endoderm in the vegetal pole, the gray color indicates presumptive ectoderm at the animal pole, and the future mesoderm is represented by brown shading. (E) Pnhd is required for mesoderm formation in animal-vegetal conjugates. Pnhd MOsp-injected or control animal pole explants were combined with vegetal explants at stage 8. After culture until stage 11, levels of cdx4 and hoxd1 transcripts were determined in the conjugates by RT-qPCR. Data are mean±s.d. Significance was determined by an unpaired two-tailed Student's t-test. **P<0.01.
We next investigated whether Pnhd is required for mesoderm formation in response to the endogenous-inducing signals in animal-vegetal conjugates. Mesoderm-specific gene activation was suppressed in pnhd-depleted conjugates compared with wild-type controls (Fig. 8D,E). This result supports our model that Pnhd functions in mesoderm specification in response to initial mesoderm-inducing signals. We also assessed whether Pnhd is required for mesoderm induction by FGF. FGF-dependent induction of cdx4 and tbxt was strongly inhibited in Pnhd morphants (Fig. S11), consistent with the positive feedback between Pnhd and FGF. Based on these findings, we propose that Pnhd is activated in the marginal zone by early vegetal-inducing signals and, in turn, functions in the marginal zone by triggering multiple mesodermal markers (Fig. 8D).
DISCUSSION
This study identifies pnhd as a secreted regulator of mesoderm formation during Xenopus gastrulation. Many genes induced by Pnhd are FGF-dependent markers and modulators of posterior mesoderm, such as tbxt, msgn1 or cdx4. By contrast, many dorsal markers, including nodal, gsc and noggin, and ventral genes, such as szl, bambi and ventx1.2, were not affected in Pnhd-depleted marginal zone explants based on our RNA-seq analysis. Being produced in the marginal zone during gastrulation, pnhd appears to predominantly affect presumptive somitic mesoderm. Nevertheless, the analysis of single cell transcriptome data using the SPRING tool (Briggs et al., 2018) indicates that pnhd itself is not transcribed in the cdx4-, hoxd1- or msgn1-expressing cells, suggesting that it modulates target genes in the paracrine rather than autocrine manner. The activation of caudal-related (cdx) genes, tbx genes (tbxt and vegt) and posteriorly expressed hox genes is characteristic of the tail organizer (De Robertis and Kuroda, 2004; Harland and Gerhart, 1997; Niehrs, 2004), and provides an explanation for the headless phenotype of Pnhd-expressing embryos. We note that this phenotype is consistent with the increased posteriorizing activity of FGF (Cox and Hemmati-Brivanlou, 1995; Lamb and Harland, 1995) and can be rescued by inhibiting FGF signaling (Fig. S7). Our marker analysis at later stages support the view that Pnhd primarily affects paraxial mesoderm, as evident by the disrupted segmentation of myod+ somites at later stages and the lack of effect on α-globin (Fig. S5). The narrow chordin domain in pnhd-depleted embryos leaves open the possibility that pnhd might have a role in notochord development. Nevertheless, the late developmental defects of pnhd morphants are modest, suggesting that other pathways maintain the mesoderm when pnhd is no longer expressed. This conclusion reiterates the existence of multiple signaling pathways operating during mesoderm specification (Gentsch et al., 2013; Kimelman, 2006; Loose and Patient, 2004; Morley et al., 2009).
Many putative Pnhd target genes are expressed in the ventrolateral marginal zone during gastrulation and largely overlap with FGF and Wnt targets (Branney et al., 2009; Chung et al., 2004; Ding et al., 2018; Kjolby and Harland, 2017; Nakamura et al., 2016). The DNA regulatory sequences of these target genes include T-cell factor (TCF) and Ets DNA-binding sites that are engaged, and contribute to the transcriptional regulation (Kjolby et al., 2019). Consistent with the crosstalk with the FGF pathway, Pnhd-mediated target gene transcription strongly correlates with Erk1 phosphorylation. Although PI3K-Akt signaling does not modulate Erk activation by Pnhd, it might be involved in a parallel pathway leading to mesoderm development (Carballada et al., 2001; Dubrulle and Pourquié, 2004). We note that Pnhd functions differently from canonical Wnt ligands because the cell response to Pnhd cannot be blocked by the Wnt antagonist Dkk1. Pnhd RNA does not trigger secondary axis formation or activate the direct Wnt target nodal3. Also, there is no decrease in the level of β-catenin, a common Wnt component.
Our observations contrast those of Kenwrick et al. (2004) who observed enlarged anterior structures in X. tropicalis embryos injected with pnhd RNA (Kenwrick et al., 2004). The study hypothesized that the ‘pinhead’ phenotype of the morphants is due to the inhibitory effect of Pnhd on the Wnt pathway. Although pnhd-overexpressing embryos sometimes appear anteriorized at tailbud stages due to the developmental delay, we find that Wnt target genes are commonly stimulated rather than repressed by Pnhd. Two other studies proposed that Pnhd inhibits (Imai et al., 2012) or promotes (Yan et al., 2019) the activity of Admp, a BMP-related protein (Dosch and Niehrs, 2000; Joubin and Stern, 1999; Moos et al., 1995). Although both Xenopus Admp proteins stimulate ventral mesoderm formation characterized by active Smad1 (Kumano et al., 2006), pnhd morphants had no changes in Smad1 phosphorylation. Thus, crosstalk of Pnhd with distinct signaling pathways remains to be investigated in more detail in future studies.
We have demonstrated that Pnhd is essential for the response of embryonic cells to exogenous FGF and endogenous vegetal-inducing signals. Conversely, blocking FGF and Nodal signaling interferes with Pnhd-mediated activation of mesodermal genes. Notably, Nodal/Activin signaling has been shown to require the FGF pathway in Xenopus embryos (Cornell et al., 1995; LaBonne and Whitman, 1994). The interdependence of pnhd, FGF and Nodal signaling highlights the positive regulatory feedback during posterior mesoderm development and, possibly, later at the MHB and other sites of pnhd expression.
Although the pnhd gene has been conserved in many animals from insects to reptiles and birds, it is absent in mammals, which suggests there are differences in mesoderm specification. It is currently unknown how the Pnhd signal is transmitted inside the cell. Presumably, the CK motif of Pnhd interacts with a receptor at the cell surface. Although our experiments did not detect the association with FGF receptors, it is possible that Pnhd forms a complex with a heparan sulfate proteoglycan that functions as an FGF co-receptor (Lin, 2004; Ornitz, 2000). Alternatively, Pnhd might act by binding to molecules in the extracellular space, as shown for other pleiotropic modulators containing the CK motif, such as Cerberus or Wise (Imai et al., 2012; Lintern et al., 2009; Piccolo et al., 1999). Future analysis of Pnhd interactions with various signaling proteins is warranted to elucidate the mechanism underlying its effects on embryonic mesoderm.
MATERIALS AND METHODS
Plasmids, in vitro RNA synthesis and MOs
pCS2-Pnhd, pCS2-Flag-Pnhd, pCS2-Flag-PnhdSP, pCS2-Pnhd-Flag and pCS2-HA-Pnhd plasmids were generated by PCR from the X. laevis DNA clone for pnhd.L (accession number NM_001127751) obtained from Dharmacon. A pnhd.L fragment was subcloned into the Bgl2/BamH1 sites of pXT7 to produce pXT7-Pnhd. pCS2-mFGF8-HA was generated by subcloning BamH1-Cla1 DNA fragment from pBSSK-mFGF8 variant I (Crossley and Martin, 1995) into pCS2-HA. pCS2-PTEN-HA was obtained from Jeff Wrana (University of Toronto, Canada) (Shnitsar et al., 2015), constitutively active pI3K (p110CAAX) in pCS2 (Carballada et al., 2001) was obtained from Chenbei Chang (University of Alabama, USA), and pBSSK-chordin was obtained from Eddy De Robertis (University of California, Los Angeles, USA). Plasmids containing α-globin, myod, wnt8a and cdx4 anti-sense probes were generated by PCR. Details of cloning are available upon request.
Capped mRNAs were synthesized using an mMessage mMachine kit (Ambion). In addition to Pnhd and FGF8 plasmids, the following plasmids were used: pCS2-hDkk1 (Krupnik et al., 1999); pSP64T-Wnt3a (Wolda et al., 1993); pXT7-GFP-C1; pCS2-nucβGal pSP64T-Wnt8 (Sokol et al., 1991); DN-FGFR1 (Amaya et al., 1991); and the secreted inhibitory forms of FGFR1 and 4 (FGFR1-Fc and FGFR4-Fc) (Marics et al., 2002). The pCS2-HA-XVangl2 construct has been described previously (Ossipova et al., 2015). The following MOs were purchased from Gene Tools: Pnhd MOatg (translation-blocking), 5′-ACAAGAAAAGATGTTCCATGTCTG-3′; Pnhd MOsp (splicing-blocking), 5′-CCTGTTCATCACGCTACCATCTAAA-3′; and control MO (CoMO), 5′-GCTTCAGCTAGTGACACATGCAT-3′.
Xenopus embryo culture and microinjections, explants, secreted protein production and treatment
In vitro fertilization and culture of X. laevis embryos were carried out as previously described (Dollar et al., 2005). Staging was determined according to Nieuwkoop and Faber (1967). For microinjections, two- to four-cell embryos were transferred into 2-3% Ficoll in 0.5× Marc's modified Ringer's (MMR) solution [(50 mM NaCl, 1 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2 and 2.5 mM HEPES (pH 7.4)] (Peng, 1991) and 5-10 nl of mRNA or MO solution was injected into one or more blastomeres. The amounts of injected mRNA per embryo have been optimized in preliminary dose-response experiments and are indicated in the figure legends. For animal cap experiments, both blastomeres at the two-cell stage were injected into the animal pole region.
Ectoderm (animal caps), vegetal or marginal zone explants were prepared at stages 8 to 11, and cultured in 0.6× MMR solution until the indicated time for morphological observations, RNA extraction or immunoblot analysis. Stimulation of ectoderm explants with 50 ng/ml of Xenopus recombinant bFGF or 1 ng/ml of human Activin βA was performed as described previously (Itoh and Sokol, 1994). For stimulation with Pnhd, stage 10 ectoderm explants were cultured with 1.5 µg/ml or 6.5 µg/ml of Flag-Pnhd in 0.6× MMR solution until stage 11 or stage 14. Animal-vegetal conjugates were prepared immediately after dissection and cultured until stage 11 for RNA extraction.
Secreted proteins were produced after dissociating animal pole cells in Ca/Mg-free medium, culturing them for 2-3 h and collecting the supernatant for analysis. SU5402 (Calbiochem), a pharmacological inhibitor of FGF signaling, and type I TGFβ receptor inhibitor SB505124 (Sigma-Aldrich) were prepared as 10 mM stock solutions in DMSO and used at a final concentration of 100 µM.
Cell culture and transfection
Human embryonic kidney 293T cells were maintained in Dulbecco's modified eagle media (Corning) with 10% fetal bovine serum (Gemini) and penicillin/streptomycin (Sigma-Aldrich). Cells growing at 70% confluence were transiently transfected using linear polyethylenimine (MW 25,000, Polysciences) as described previously (Ossipova et al., 2009). Briefly, each 35-mm dish with cells received 1.5 µg of pCS2 plasmids encoding Flag-Pnhd or Flag-GFP as a control. Vector DNA (pCS2) was added to the plasmid DNA mixture to bring the total DNA amount to 3 µg. Cell supernatants and lysates were collected 24 h to 48 h after transfection. The Flag-Pnhd protein was produced from a 50 ml culture of transiently transfected HEK293T cells (Bon Opus) and was stored at −80°C. Flag-Pnhd protein levels were estimated by comparison with known amounts of bovine serum albumin on a Coomassie blue-stained gel.
RNA-seq
Pnhd-expressing (1.5 ng) and control uninjected animal pole cells were cultured until stage 11. For pnhd knockdown, 10 ng of pnhd MOatg or 40 ng of pnhd MOsp were injected two to four times into the marginal zone of four-cell embryos. RNA was extracted from marginal zone explants at stage 10.5 or ectoderm explants at stages 11-11.5 using an RNeasy kit (Qiagen). cDNA library preparation and paired-end 150 bp sequencing were performed by Novogene using Illumina HiSeq2000 analyzers. The raw reads (FASTQ files) were filtered to remove reads containing adapters or reads of low quality. The sequences were mapped to the Xenopus genome version XL-9.1_v1.8.3.2 (Xenbase) using HISAT2 (Kim et al., 2015). The total mapped reads were larger than 73% for all samples and the multiple mapped reads were lower than 9%, which is within the generally accepted limits of higher than 70% and lower than 10%, respectively. The files were sorted using the Samtools package (www.htslib.org/doc/samtools-1.2.html). The sequences were counted using the HTSeq package (Anders et al., 2015). The differentially expressed genes (DEGs) were detected using DESeq (Anders and Huber, 2010) with a twofold change cutoff. The P-value estimation was based on the negative binomial distribution, using the Benjamini-Hochberg estimation model with an adjusted P-value of <0.05. The heatmap and volcano plots were generated using the publicly available BioJupies software (Torre et al., 2018). DEGs were evaluated using the GSEA software (Subramanian et al., 2005). Pnhd-induced DEGs were assessed from four separate RNA-seq experiments using independent samples.
Ectoderm explants, RT-PCR and qRT-PCR
RNA was extracted from a group of 3-5 mixed embryos, 6-10 marginal zone or 10-30 animal pole explants, using an RNeasy kit (Qiagen). For RT-PCR, cDNA was made from 1-2 µg of total RNA using the first strand cDNA kit (Invitrogen) or iScript (Bio-Rad) according to the manufacturer's instructions. PCR using Taq polymerase was carried out based on standard protocols. For RT-qPCR, the reactions were amplified using a CFX96 light cycler (Bio-Rad) with Universal SYBR Green Supermix (Bio-Rad). Primer sequences used for RT-PCR and RT-qPCR have been described previously (Ossipova and Sokol, 2011) and are listed in Table S3. The reaction mixture consisted of 1× Power SYBR Green PCR Master Mix, 0.3 μM primers and 1 μl of cDNA in a total volume of 10 μl. The cycling conditions used for RT-qPCR were optimized in separate experiments. The ΔΔCT method was used to quantify the results. All samples were normalized to control uninjected embryos or explants. Transcripts for elongation factor 1α1 [also known as eukaryotic translation elongation factor 1α1 (eef1a1)] were used for normalization. Data are representative of two to three independent experiments and shown as mean±s.d. Statistical significance was assessed by an unpaired two-tailed Student's t-test (*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001).
WISH
WISH and X-gal staining were carried out as described previously (Harland, 1991), except when Red-gal substrate was used instead of X-gal. Diogoxigenin-rUTP-labeled RNA probes were prepared by in vitro transcription of linearized pnhd.L, chordin, α-globin, myod, cdx4 and wnt8a DNA templates with T7 or SP6 RNA polymerases and the RNA labeling mix containing digoxigenin-rUTP (Roche). Nuclear β-galactosidase (β-gal) RNA (50 pg) was a lineage tracer. For the sense probe, the same pnhd construct was linearized with Bgl2 and transcribed with Sp6 polymerase. A second anti-sense pnhd probe was prepared by linearizing the same plasmid with EcoRV. For pnhd.L, the same expression pattern after in situ hybridization was obtained using two different anti-sense RNA probes. In situ stained embryos were embedded in cold water fish gelatin-sucrose mixture and sectioned (25 μm) using a Leica CM3050 cryostat (Ossipova et al., 2009). Images were digitally acquired on a Zeiss Axio Imager microscope. All data are representative of two to three independent experiments.
Immunoprecipitation and immunoblot analysis
For immunoprecipitation, cells transfected for 24 h were lysed in IP buffer [10 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1% Triton X-100, 1 mM Na3VO4, 10 mM NaF and 25 mM β-glycerol phosphate], containing protease inhibitor cocktail (cOmplete Mini, EDTA-free, Roche). After centrifugation for 5 min at 16,000 g, the supernatant was incubated with anti-Flag agarose beads (Sigma-Aldrich) at 4°C for 2-3 h. The beads were washed three times with IP buffer and boiled in the SDS-PAGE sample buffer. Immunoblot analysis was carried out essentially as described previously (Itoh et al., 2005). Briefly, five embryos at stage 10.5 were homogenized in 75 µl of lysis buffer [50 mM Tris-HCl (pH 7.6), 50 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerol phosphate and 1 mM phenylmethylsulfonyl fluoride]. After centrifugation for 3 min at 16,000 g, the supernatant was subjected to SDS-PAGE and western blot analysis following standard protocols. The following primary antibodies were used: mouse anti-Flag M2 (1:2000; Sigma-Aldrich, F3165), rabbit anti-HA (1:2000; Bethyl Laboratories, A190-108A), rabbit anti-pErk1 (1:1000; Phospho-p44/42, T202/Y204, Cell Signaling Technology, 4370S), rabbit anti-Erk1 (1:1000; Santa Cruz Biotechnology, K23, SC-94), anti-pSmad1/5 S463/465 (1:2000; 41D10, Cell Signaling Technology, 9516S) and anti-Smad1 (1:500; Invitrogen Life Technologies, 38-5400), rabbit anti-pSmad2 S465/467 (1:1000; 138D4, Cell Signaling Technology, 3108S), rabbit anti-pAkt S473 (1:2000; Cell Signaling Technology, 4060T), anti-Akt (pan) (1:2000; Cell Signaling Technology, 4691T) and mouse anti-β-catenin (E-5) (1:2000; Santa Cruz Biotechnology, SC-7963). The detection was carried out by enhanced chemiluminescence as described previously (Itoh et al., 2005), using the ChemiDoc MP imager (Bio-Rad). Every immunoblotting result was repeated three to 12 times.
Supplementary Material
Acknowledgements
We thank Rohan Bareja, Olivier Elemento and the Mount Sinai Genomics Core Facility for help with RNAseq analysis at the early stages of this project; Phil Soriano and David Kimelman for the FGF plasmids; Eddy De Robertis for the chordin plasmid; Jeff Wrana for the PTEN plasmid; and Chenbei Chang for the pI3 kinase plasmids. We are grateful to Kety Giannetti for confirming PnhdMOsp efficiency by RT-PCR. We also thank Miho Matsuda for the critical reading of the manuscript and members of the Sokol laboratory for discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: O.O., S.Y.S.; Methodology: O.O., K.I., A.R., J.E., S.Y.S.; Software: S.Y.S.; Validation: O.O., K.I., J.E.; Formal analysis: O.O., A.R., S.Y.S.; Investigation: O.O., K.I., J.E., S.Y.S.; Data curation: O.O., K.I., A.R., S.Y.S.; Writing - original draft: O.O., S.Y.S.; Writing - review & editing: O.O., K.I., A.R., J.E.; Visualization: O.O.; Supervision: S.Y.S.; Project administration: S.Y.S.; Funding acquisition: S.Y.S.
Funding
This study was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (National Institutes of Health) (HD092990 to S.Y.S.). Deposited in PMC for release after 12 months.
Data availability
The RNA-seq data reported in this paper have been deposited in GEO under accession number GSE143795.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.188094.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.188094.reviewer-comments.pdf
References
- Amaya E., Musci T. J. and Kirschner M. W. (1991). Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 66, 257-270. 10.1016/0092-8674(91)90616-7 [DOI] [PubMed] [Google Scholar]
- Anders S. and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avsian-Kretchmer O. and Hsueh A. J. W. (2004). Comparative genomic analysis of the eight-membered ring cystine knot-containing bone morphogenetic protein antagonists. Mol. Endocrinol. 18, 1-12. 10.1210/me.2003-0227 [DOI] [PubMed] [Google Scholar]
- Bae C.-J., Park B.-Y., Lee Y.-H., Tobias J. W., Hong C.-S. and Saint-Jeannet J.-P. (2014). Identification of Pax3 and Zic1 targets in the developing neural crest. Dev. Biol. 386, 473-483. 10.1016/j.ydbio.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A., Liu G., Yaniv A., Gazit A. and Aaronson S. A. (2001). Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 3, 683-686. 10.1038/35083081 [DOI] [PubMed] [Google Scholar]
- Branney P. A., Faas L., Steane S. E., Pownall M. E. and Isaacs H. V. (2009). Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS ONE 4, e4951 10.1371/journal.pone.0004951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs J. A., Weinreb C., Wagner D. E., Megason S., Peshkin L., Kirschner M. W. and Klein A. M. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballada R., Yasuo H. and Lemaire P. (2001). Phosphatidylinositol-3 kinase acts in parallel to the ERK MAP kinase in the FGF pathway during Xenopus mesoderm induction. Development 128, 35-44. [DOI] [PubMed] [Google Scholar]
- Cho K. W. Y., Blumberg B., Steinbeisser H. and De Robertis E. M. (1991). Molecular nature of Spemann's organizer: the role of the Xenopus homeobox gene goosecoid. Cell 67, 1111-1120. 10.1016/0092-8674(91)90288-A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B. and Slack J. M. (1999). Spatial response to fibroblast growth factor signalling in Xenopus embryos. Development 126, 119-125. [DOI] [PubMed] [Google Scholar]
- Christian J. L., McMahon J. A., McMahon A. P. and Moon R. T. (1991). Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development 111, 1045-1055. [DOI] [PubMed] [Google Scholar]
- Chung H. A., Hyodo-Miura J., Kitayama A., Terasaka C., Nagamune T. and Ueno N. (2004). Screening of FGF target genes in Xenopus by microarray: temporal dissection of the signalling pathway using a chemical inhibitor. Genes Cells 9, 749-761. 10.1111/j.1356-9597.2004.00761.x [DOI] [PubMed] [Google Scholar]
- Cornell R. A., Musci T. J. and Kimelman D. (1995). FGF is a prospective competence factor for early activin-type signals in Xenopus mesoderm induction. Development 121, 2429-2437. [DOI] [PubMed] [Google Scholar]
- Cox W. G. and Hemmati-Brivanlou A. (1995). Caudalization of neural fate by tissue recombination and bFGF. Development 121, 4349-4358. [DOI] [PubMed] [Google Scholar]
- Crossley P. H. and Martin G. R. (1995). The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439-451. [DOI] [PubMed] [Google Scholar]
- DaCosta Byfield S., Major C., Laping N. J. and Roberts A. B. (2004). SB-505124 is a selective inhibitor of transforming growth factor-β type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 65, 744-752. 10.1124/mol.65.3.744 [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. and Kuroda H. (2004). Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 20, 285-308. 10.1146/annurev.cellbio.20.011403.154124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Colozza G., Sosa E. A., Moriyama Y., Rundle S., Salwinski L. and De Robertis E. M. (2018). Bighead is a Wnt antagonist secreted by the Xenopus Spemann organizer that promotes Lrp6 endocytosis. Proc. Natl. Acad. Sci. USA 115, E9135-E9144. 10.1073/pnas.1812117115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollar G. L., Weber U., Mlodzik M. and Sokol S. Y. (2005). Regulation of Lethal giant larvae by Dishevelled. Nature 437, 1376-1380. 10.1038/nature04116 [DOI] [PubMed] [Google Scholar]
- Dosch R. and Niehrs C. (2000). Requirement for anti-dorsalizing morphogenetic protein in organizer patterning. Mech. Dev. 90, 195-203. 10.1016/S0925-4773(99)00245-2 [DOI] [PubMed] [Google Scholar]
- Dubrulle J. and Pourquié O. (2004). fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 427, 419-422. 10.1038/nature02216 [DOI] [PubMed] [Google Scholar]
- Gentsch G. E., Owens N. D. L., Martin S. R., Piccinelli P., Faial T., Trotter M. W. B., Gilchrist M. J. and Smith J. C. (2013). In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep 4, 1185-1196. 10.1016/j.celrep.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. M. (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. In Methods Cell Biol. (ed. Kay B. K. and Peng H. B.), pp. 685-695. San Diego: Academic Press Inc. [DOI] [PubMed] [Google Scholar]
- Harland R. and Gerhart J. (1997). Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13, 611-667. 10.1146/annurev.cellbio.13.1.611 [DOI] [PubMed] [Google Scholar]
- Howard J. E. and Smith J. C. (1993). Analysis of gastrulation: different types of gastrulation movement are induced by different mesoderm-inducing factors in Xenopus laevis. Mech. Dev. 43, 37-48. 10.1016/0925-4773(93)90021-O [DOI] [PubMed] [Google Scholar]
- Imai K. S., Daido Y., Kusakabe T. G. and Satou Y. (2012). Cis-acting transcriptional repression establishes a sharp boundary in chordate embryos. Science 337, 964-967. 10.1126/science.1222488 [DOI] [PubMed] [Google Scholar]
- Isaacs N. W. (1995). Cystine knots. Curr. Opin. Struct. Biol. 5, 391-395. 10.1016/0959-440X(95)80102-2 [DOI] [PubMed] [Google Scholar]
- Itoh K. and Sokol S. Y. (1994). Heparan sulfate proteoglycans are required for mesoderm formation in Xenopus embryos. Development 120, 2703-2711. [DOI] [PubMed] [Google Scholar]
- Itoh K., Brott B. K., Bae G.-U., Ratcliffe M. J. and Sokol S. Y. (2005). Nuclear localization is required for Dishevelled function in Wnt/β-catenin signaling. J. Biol. 4, 3 10.1186/jbiol20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubin K. and Stern C. D. (1999). Molecular interactions continuously define the organizer during the cell movements of gastrulation. Cell 98, 559-571. 10.1016/S0092-8674(00)80044-6 [DOI] [PubMed] [Google Scholar]
- Kenwrick S., Amaya E. and Papalopulu N. (2004). Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev. Dyn. 229, 289-299. 10.1002/dvdy.10440 [DOI] [PubMed] [Google Scholar]
- Kiecker C., Bates T. and Bell E. (2016). Molecular specification of germ layers in vertebrate embryos. Cell. Mol. Life Sci. 73, 923-947. 10.1007/s00018-015-2092-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B. and Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D. (2006). Mesoderm induction: from caps to chips. Nat. Rev. Genet. 7, 360-372. 10.1038/nrg1837 [DOI] [PubMed] [Google Scholar]
- Kjolby R. A. S. and Harland R. M. (2017). Genome-wide identification of Wnt/β-catenin transcriptional targets during Xenopus gastrulation. Dev. Biol. 426, 165-175. 10.1016/j.ydbio.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjolby R. A. S., Truchado-Garcia M., Iruvanti S. and Harland R. M. (2019). Integration of Wnt and FGF signaling in the Xenopus gastrula at TCF and Ets binding sites shows the importance of short-range repression by TCF in patterning the marginal zone. Development 146, dev179580 10.1242/dev.179580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnik V. E., Sharp J. D., Jiang C., Robison K., Chickering T. W., Amaravadi L., Brown D. E., Guyot D., Mays G., Leiby K. et al. (1999). Functional and structural diversity of the human Dickkopf gene family. Gene 238, 301-313. 10.1016/S0378-1119(99)00365-0 [DOI] [PubMed] [Google Scholar]
- Kumano G., Ezal C. and Smith W. C. (2006). ADMP2 is essential for primitive blood and heart development in Xenopus. Dev. Biol. 299, 411-423. 10.1016/j.ydbio.2006.08.010 [DOI] [PubMed] [Google Scholar]
- LaBonne C. and Whitman M. (1994). Mesoderm induction by activin requires FGF-mediated intracellular signals. Development 10, 463-472. 10.1016/0168-9525(94)90085-X [DOI] [PubMed] [Google Scholar]
- Lamb T. M. and Harland R. M. (1995). Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development 121, 3627-3636. [DOI] [PubMed] [Google Scholar]
- Langdon Y. G. and Mullins M. C. (2011). Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu. Rev. Genet. 45, 357-377. 10.1146/annurev-genet-110410-132517 [DOI] [PubMed] [Google Scholar]
- Lin X. (2004). Functions of heparan sulfate proteoglycans in cell signaling during development. Development 131, 6009-6021. 10.1242/dev.01522 [DOI] [PubMed] [Google Scholar]
- Lintern K. B., Guidato S., Rowe A., Saldanha J. W. and Itasaki N. (2009). Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J. Biol. Chem. 284, 23159-23168. 10.1074/jbc.M109.025478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M. and Patient R. (2004). A genetic regulatory network for Xenopus mesendoderm formation. Dev. Biol. 271, 467-478. 10.1016/j.ydbio.2004.04.014 [DOI] [PubMed] [Google Scholar]
- Manning B. D. and Toker A. (2017). AKT/PKB signaling: navigating the network. Cell 169, 381-405. 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B., Wu W., Li Y., Hoppe D., Stannek P., Glinka A. and Niehrs C. (2001). LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411, 321-325. 10.1038/35077108 [DOI] [PubMed] [Google Scholar]
- Marics I., Padilla F., Guillemot J. F., Scaal M. and Marcelle C. (2002). FGFR4 signaling is a necessary step in limb muscle differentiation. Development 129, 4559-4569. [DOI] [PubMed] [Google Scholar]
- Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R. and Schlessinger J. (1997). Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955-960. 10.1126/science.276.5314.955 [DOI] [PubMed] [Google Scholar]
- Moos M. Jr, Wang S. and Krinks M. (1995). Anti-dorsalizing morphogenetic protein is a novel TGF-beta homolog expressed in the Spemann organizer. Development 121, 4293-4301. [DOI] [PubMed] [Google Scholar]
- Morley R. H., Lachani K., Keefe D., Gilchrist M. J., Flicek P., Smith J. C. and Wardle F. C. (2009). A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA 106, 3829-3834. 10.1073/pnas.0808382106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., de Paiva Alves E., Veenstra G. J. C. and Hoppler S. (2016). Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules. Development 143, 1914-1925. 10.1242/dev.131664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. (2004). Regionally specific induction by the Spemann-Mangold organizer. Nat. Rev. Genet. 5, 425-434. 10.1038/nrg1347 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D. and Faber J. (1967). Normal Table of Xenopus laevis (Daudin). Amsterdam: North Holland. [Google Scholar]
- Ornitz D. M. (2000). FGFs, heparan sulfate and FGFRs: complex interactions essential for development. BioEssays 22, 108-112. [DOI] [PubMed] [Google Scholar]
- Ornitz D. M. and Itoh N. (2015). The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215-266. 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O. and Sokol S. Y. (2011). Neural crest specification by noncanonical Wnt signaling and PAR-1. Development 138, 5441-5450. 10.1242/dev.067280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O., Ezan J. and Sokol S. Y. (2009). PAR-1 phosphorylates Mind bomb to promote vertebrate neurogenesis. Dev. Cell 17, 222-233. 10.1016/j.devcel.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O., Chu C.-W., Fillatre J., Brott B. K., Itoh K. and Sokol S. Y. (2015). The involvement of PCP proteins in radial cell intercalations during Xenopus embryonic development. Dev. Biol. 408, 316-327. 10.1016/j.ydbio.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. B. (1991). Xenopus laevis: practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol. 36, 657-662. 10.1016/S0091-679X(08)60301-5 [DOI] [PubMed] [Google Scholar]
- Piccolo S., Agius E., Leyns L., Bhattacharyya S., Grunz H., Bouwmeester T. and De Robertis E. M. (1999). The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397, 707-710. 10.1038/17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouhinec J.-L., Roche D. D., Pegoraro C., Figueiredo A. L., Maczkowiak F., Brunet L. J., Milet C., Vert J.-P., Pollet N., Harland R. M. et al. (2014). Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev. Biol. 386, 461-472. 10.1016/j.ydbio.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch G. J. and Sherwood N. M. (2014). Glycoprotein hormones and their receptors emerged at the origin of metazoans. Genome Biol. Evol. 6, 1466-1479. 10.1093/gbe/evu118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaway A. and Patient R. (2001). Mesendoderm: an ancient germ layer? Cell 105, 169-172. 10.1016/S0092-8674(01)00307-5 [DOI] [PubMed] [Google Scholar]
- Schohl A. and Fagotto F. (2002). Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development 129, 37-52. [DOI] [PubMed] [Google Scholar]
- Semënov M. V., Tamai K., Brott B. K., Kuhl M., Sokol S. and He X. (2001). Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr. Biol. 11, 951-961. 10.1016/S0960-9822(01)00290-1 [DOI] [PubMed] [Google Scholar]
- Shnitsar I., Bashkurov M., Masson G. R., Ogunjimi A. A., Mosessian S., Cabeza E. A., Hirsch C. L., Trcka D., Gish G., Jiao J. et al. (2015). PTEN regulates cilia through Dishevelled. Nat. Commun. 6, 8388 10.1038/ncomms9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. C., Price B. M. J., Green J. B. A., Weigel D. and Herrmann B. G. (1991). Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67, 79-87. 10.1016/0092-8674(91)90573-H [DOI] [PubMed] [Google Scholar]
- Smith W. C., McKendry R., Ribisi S. Jr and Harland R. M. (1995). A nodal-related gene defines a physical and functional domain within the Spemann organizer. Cell 82, 37-46. 10.1016/0092-8674(95)90050-0 [DOI] [PubMed] [Google Scholar]
- Sokol S. Y. (1993). Mesoderm formation in Xenopus ectodermal explants overexpressing Xwnt8: evidence for a cooperating signal reaching the animal pole by gastrulation. Development 118, 1335-1342. [DOI] [PubMed] [Google Scholar]
- Sokol S., Christian J. L., Moon R. T. and Melton D. A. (1991). Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell 67, 741-752. 10.1016/0092-8674(91)90069-B [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre D., Lachmann A. and Ma'ayan A. (2018). BioJupies: automated generation of interactive notebooks for RNA-Seq data analysis in the cloud. Cell Syst. 7, 556-561.e553. 10.1016/j.cels.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolda S. L., Moody C. J. and Moon R. T. (1993). Overlapping expression of Xwnt-3A and Xwnt-1 in neural tissue of Xenopus laevis embryos. Dev. Biol. 155, 46-57. 10.1006/dbio.1993.1005 [DOI] [PubMed] [Google Scholar]
- Yan Y., Ning G., Li L., Liu J., Yang S., Cao Y. and Wang Q. (2019). The BMP ligand Pinhead together with Admp supports the robustness of embryonic patterning. Sci. Adv. 5, eaau6455 10.1126/sciadv.aau6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Houston D. W., King M. L., Payne C., Wylie C. and Heasman J. (1998). The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell 94, 515-524. 10.1016/S0092-8674(00)81592-5 [DOI] [PubMed] [Google Scholar]
- Zorn A. M. and Wells J. M. (2009). Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221-251. 10.1146/annurev.cellbio.042308.113344 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.