ABSTRACT
Pancreatic cancer (PC) is acquired postnatally; to mimic this scenario, we developed an inducible KrasG12D; Ptf1a-CreER™ (iKC) mouse model, in which Kras is activated postnatally at week 16 upon tamoxifen (TAM) administration. Upon TAM treatment, iKC mice develop pancreatic intraepithelial neoplasia (PanIN) lesions and PC with metastasis at the fourth and fortieth weeks, respectively, and exhibited acinar-to-ductal metaplasia (ADM) and transdifferentiation. Kras activation upregulated the transcription factors Ncoa3, p-cJun and FoxM1, which in turn upregulated expression of transmembrane mucins (Muc1, Muc4 and Muc16) and secretory mucin (Muc5Ac). Interestingly, knockdown of KrasG12D in multiple PC cell lines resulted in downregulation of MUC1, MUC4, MUC5AC and MUC16. In addition, iKC mice exhibited ADM and transdifferentiation. Our results show that the iKC mouse more closely mimics human PC development and can be used to investigate pancreatic ductal adenocarcinoma (PDAC) biomarkers, early onset of PDAC, and ADM. The iKC model can also be used for preclinical strategies such as targeting mucin axis alone or in combination with neo-adjuvant, immunotherapeutic approaches and to monitor chemotherapy response.
KEY WORDS: Mucin, Pancreatic ductal adenocarcinoma (PDAC), Inducible KC (iKC) mouse model
Summary: In this study, we developed a tamoxifen-inducible transgenic mouse model that closely mimics the age-related onset of pancreatic cancer (PC). We showed that activation of Kras (iKC) specifically regulates transcription factors that promote mucin expression and acinar to ductal metaplasia, whereas, loss of KrasG12D allele in PC cells resulted in the reduction of mucin proteins such as MUC1, MUC4, MUC5AC and MUC16.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy with an extremely poor prognosis, having a 5-year survival rate of less than 9% (Siegel et al., 2020). The lethality of PDAC is due to late diagnosis, early local invasion, high metastatic potential and resistance to current chemo-radiotherapies (Bailey et al., 2014; Chakraborty et al., 2011). The major risk factors for PDAC initiation and progression include genetic mutations in KRAS, INK4A (p16), Trp53, DPC4 and BRCA2 as well as other risk factors like cigarette smoking, obesity and diabetes mellitus (Guerra et al., 2007; Habbe et al., 2008; Hingorani et al., 2003; Jones et al., 2008). Kirsten rat sarcoma viral oncogene homolog (Kras) is a major driver in PDAC development and the most frequently mutated gene, with constitutive activation in 70–90% of PDAC patients (Bailey et al., 2014; Feldmann et al., 2007; Jones et al., 2008; Prior et al., 2012). Further, accumulation of genetic mutations in other tumor pathway genes ultimately leads to invasive PDAC (Khan et al., 2017).
Early detection of PDAC is difficult because most PDAC patients (>95%) are diagnosed with advanced disease. Since pre-neoplastic human samples are rarely available, genetically engineered mouse models (GEMM) can be used to identify and analyze molecules specifically involved in the initiation, progression and advancement of the disease. Since 2003, the KrasG12D;Pdx1-Cre (KC) and KrasG12D;Trp53R172H;Pdx1-Cre (KPC) mouse models (Hingorani et al., 2003; Hingorani et al., 2005; Rachagani et al., 2012 b) have been used extensively to study the pathogenesis of pancreatic cancer (PC). The tumors and the spectrum of pancreatic intraepithelial neoplasia (PanIN) lesions express a variety of molecules associated with PDAC initiation and progression, including Muc1, Muc4, Muc5Ac, Muc16, Her-1, Her-3, PD-2/hPaf-1, Ncoa3, Mmp-9 and Cox-2 (Dey et al., 2014; Hingorani et al., 2003; Lakshmanan et al., 2015; Rachagani et al., 2012b). However, in these models, the Kras oncogene activation/Trp53 inactivation occurs during prenatal life, throughout all pancreatic cell types; in contrast, PDAC is an age-related disease with Kras activation occurring late in adult life.
In humans, the incidence of PDAC increases sharply after age 50, with most diagnoses at an advanced age (65–85 years), and the earliest reported cases at approximately age 30 (Higuera et al., 2016). The risk of men and women developing PDAC is about 100 and 87 times higher, respectively (Chakraborty et al., 2011; Higuera et al., 2016). This age-related onset is also observed in experimental rodents as the incidence of spontaneous tumors increases after 300 days of age (Anisimov et al., 2005). However, in KC and KPC mouse models, Cre recombinase is induced at embryonic day 8.5, which leads to activation of Kras or inactivation of Trp53 (Dey et al., 2014; Hingorani et al., 2003; Hingorani et al., 2005; Rachagani et al., 2012 b). PanIN lesions develop as early as week 10 in KC and week 5 in KPC models, with tumors and metastasis at 50 and 25 weeks of age, respectively (Hingorani et al., 2003; Hingorani et al., 2005; Rachagani et al., 2012 b; Torres et al., 2013). This early tumor onset may be a possible reason why therapeutics that are successful in preclinical studies fail in clinical trials. In addition, previous studies have shown that PanIN lesions can arise from pancreatic acinar cells through a reprogramming process called acinar-to-ductal metaplasia (ADM) (De La et al., 2008; Feldmann et al., 2007; Habbe et al., 2008), thus serving as a reservoir for PDAC development.
Genes encoding transcription factors critical in pancreatic progenitor cell development have been harnessed to drive Cre recombinase expression in specific progenitor cell populations to conditionally activate gene expression (Magnuson and Osipovich, 2013). These include the well-characterized homeodomain-containing transcription factor, pancreas and duodenum homeobox 1 (Pdx1), and pancreas transcription factor 1a (Ptf1a), which drive formation of endocrine and exocrine pancreatic cell lineages. Lineage tracing with Pdx1-Cre or Ptf1a-Cre (Kawaguchi et al., 2002) Cre-driver lines marks all pancreatic cell types, as both of these transcription factors are expressed in pancreatic multipotent cells (PMC) (Magnuson and Osipovich, 2013). However, since Ptf1a transcription factor expression is retained only by differentiated acinar cells (following embryonic day 8.5–9.5), we acquired a Cre-driver line that allowed us to (1) specifically promote CRE recombination within the acinar cell population and (2) facilitate temporal control of recombination/KRAS expression in postnatal life using CreER (Feil et al., 1996). This would not be possible with Pdx1, which functions both in early pancreas PMC development and later in mature islet endocrine cells.
Kras is the critical driver of several cancers, including PC and it is the most frequently mutated gene (>79%) in PC patients (Haas et al., 2017; Rachagani et al., 2011). Interestingly, mucins were shown to be predominantly expressed in Kras-dependent tumors (Rachagani et al., 2012b). Previously, we showed that genetic ablation of the Kras-specific G12D allele in the MUC4-overexpressing PC cell line (CD18/HPAF) lead to a significant reduction in proliferation, migration, anchorage-dependent growth and ability to form metastasis in mice (Rachagani et al., 2011). Furthermore, a gel-forming mucin, Muc5Ac, was demonstrated to be a critical determinant of poor survival in Kras-mutated lung cancer patients (Bauer et al., 2018). Another membrane-bound mucin, MUC16, is aberrantly overexpressed and is involved in the pathobiological landscapes of pancreatic and lung adenocarcinoma (Haridas et al., 2011; Lakshmanan et al., 2017). Currently, there are no clinically tested Kras inhibitors. Hence, drugs that indirectly affect activated Kras activity through mucins might bring a breakthrough in Kras-dependent tumor treatment. In this context, our inducible model could be used for testing small molecule inhibitors that affect mucins; thereby, indirectly targeting the ‘undruggable’ target, Kras.
In our study design, we generated an inducible KC mouse model, by intercrossing LSL-KrasG12D mice (knockin allele for mutant KrasG12D) (Jackson et al., 2001) with Ptf1a-CreER™ mice (knockin allele for Ptf1a-CreER™) (Kopinke et al., 2012) that have abolished Ptf1a gene function and express the CreERTM-fusion protein from the Ptf1a promoter/enhancer elements. Upon tamoxifen treatment, the PTF1a-CREER-fusion protein moves from the cytoplasm to the nucleus. Cre-mediated recombination results in deletion of the floxed stop sequence Lox-Stop-Lox [(LSL) cassette] and activation of KrasG12D in pancreatic acinar cells. Thus, tamoxifen administration in the compound strain, KrasG12D;Ptf1a-CreER™ (iKC), postnatally, will specifically activate KrasG12D only in the acinar compartment, i.e. after acinar cell terminal differentiation and not in ductal and endocrine cells. We examined the pancreas-specific expression of mucin proteins (Muc1, Muc4, Muc5Ac, and Muc16) and the transcriptions factors (Ncoa3, p-cJun, and FoxM1) involved in their expression. We also investigated if activation of KrasG12D in the acinar compartment could drive reprogramming of acinar cells to express the ductal cell marker cytokeratin-19 (CK19).
RESULTS
PDAC progression in the tamoxifen-induced iKC mouse model
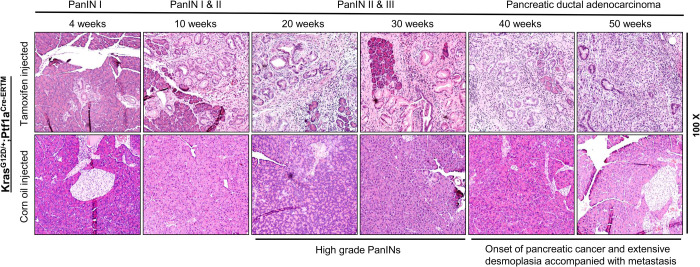
To specifically express KrasG12D in the adult acinar cell lineage, we crossed LSL KrasG12D mice with Ptf1a-CreER™ mice and generated the iKC inducible mouse model. In this model, tamoxifen is used to activate Cre, which induces mutant KrasG12D expression in acinar cells of 16-week-old animals, while littermate controls are injected with corn oil. Mice were euthanized at 4, 10, 20, 30, 40, and 50 weeks (n=4 for each time point) post-treatment. Time to onset of tumor initiation, progression (degree of disease), and the impact of KrasG12D activation on mucin expression were determined. During necropsy, tumors were excised, and distant organs were examined for metastatic lesions. In tamoxifen-treated animals, we observed both micro- and gross metastases to regional lymph nodes, liver, spleen, diaphragm, and mesenteric lymph nodes (data not shown). Pancreata from tamoxifen- and corn-oil-treated mice at each time point were stained with Hematoxylin and Eosin (H&E) to assess differences in histopathology between the groups. Mice treated with tamoxifen displayed pre-neoplastic PanIN I lesions by 4 weeks post-tamoxifen treatment (Fig. 1, upper panel). By 10 weeks, low-grade PanIN lesions (PanIN I and II) were observed, and high-grade (PanIN III) lesions were prevalent at 30 weeks after tamoxifen injection (Fig. 1, upper panel). Pancreatic (PDAC) tumors and extensive desmoplasia were observed at 40–50 weeks post-tamoxifen treatment (Fig. 1, upper panel) and were accompanied with metastasis. Corn-oil-treated mice did not display any histological changes and exhibited normal pancreas histology (Fig. 1, lower panel). Thus, the iKC model closely resembles the clinical scenario of PanIN formation and PDAC development.
Fig. 1.
PC progression in the tamoxifen-induced iKC mouse model. Pancreatic tissues were isolated from corn-oil- and tamoxifen-treated iKC mice at 4, 10, 20, 30 and 50 weeks of age. The tissues were paraffin-embedded and formalin-fixed. Sections (4 µm) were stained with H&E. Pre-cancerous lesions (PanINs) were not detected at any point in corn-oil-treated animals. By 4 weeks after tamoxifen injection, PanINs had formed; they progressed to high-grade lesions by 30 weeks and PC by 50 weeks.
Expression of acinar marker amylase and ductal marker cytokeratin-19 in the iKC mouse model
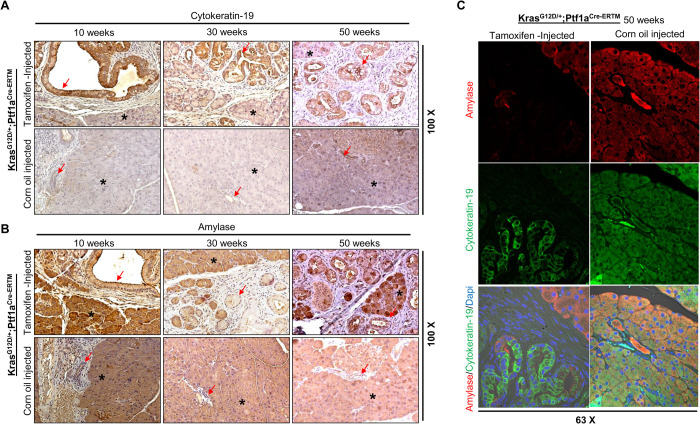
In our previous study (Dey et al., 2014), we reported that PanIN lesions/PDAC can arise from a unique intermediate ADM-like cellular transition; this was shown by localization of epithelial/ductal cell marker, CK-19; acinar cell marker, amylase; and PD2/Paf1 (pancreatic differentiation 2) in KC and cerulin-injected KC mouse models. We showed that knockdown of PD2/Paf1 leads to an increase in the epithelial cell/ductal marker CK-19 and a decrease in acinar cell marker amylase in pancreatic acinar cells (Dey et al., 2014). Thus, we wanted to confirm that the iKC model also exhibits this acinar-to-ductal transdifferentiation process. We performed immunohistochemistry (IHC) and immunolocalization assays in serial sections of pancreas obtained from tamoxifen- and corn-oil-administered (10, 30, and 50 weeks of treatment) mice. As expected, CK-19 expression was observed in ductal epithelium arising from acinar cells undergoing acinar to ductal metaplasia in tamoxifen-treated mice (Fig. 2A upper panel) and was absent in corn-oil-treated mice (Fig. 2A lower panel). Similarly, acinar cells specifically expressed amylase (Fig. 2B upper panel) in the corn-oil-treated and not in tamoxifen-treated mouse pancreas (Fig. 2B lower panel). Next, we examined whether the intermediate acinar-to-ductal-like cells of the newly developing pancreatic ducts co-expressed CK-19 and amylase in tamoxifen-injected mice. CK-19 and amylase frequently co-localized to newly forming ducts that were undergoing the transformation process (Fig. 2C). These results suggest that tamoxifen induces Kras specifically in the acinar compartment of the iKC mouse, and this model will be useful for exploring gene signatures involved in ADM.
Fig. 2.
Expression of acinar marker amylase and ductal marker cytokeratin-19 in iKC mouse model. IHC studies using CK19 and amylase antibodies were performed on pancreatic tissues isolated from both corn-oil- and tamoxifen-treated iKC mice. (A) Expression of CK19 was not detected at any time point in the corn-oil-treated mouse tissues (lower row). In contrast significant upregulation of CK19 was detected in tamoxifen-induced mouse tissues (upper row). (B) The expression of amylase was observed in the acinar compartment of the pancreas (upper and lower panels) and the transdifferentiated ductal compartment (upper panel). (C) Immunofluorescence confocal microscopy revealed that amylase and CK19 were coexpressed in ducts undergoing transdifferentiation in the tamoxifen-induced mice.
Expression of transmembrane mucins Muc1 and Muc4 in the iKC mouse model
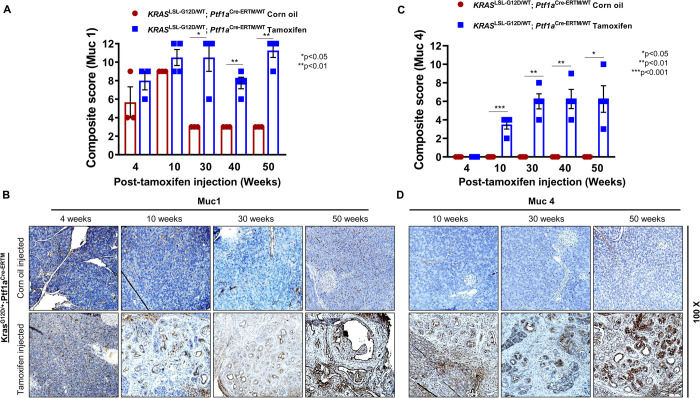
Muc1 is involved in the progression of various cancers, including PC (Andrianifahanana et al., 2001; Bafna et al., 2010; Kaur et al., 2013; Tsutsumida et al., 2006; Westgaard et al., 2009). In our earlier study using the KC mouse model, we showed Muc1 expression is associated with the progression of mouse PDAC (Rachagani et al., 2012b). Therefore, we investigated Muc1 expression in the iKC model. Our results reveal that Muc1 expression increased progressively in the pancreas of tamoxifen-treated iKC mice, starting from 4 weeks to 50 weeks. Average composite IHC scores ranged from 7.5 to 11.25, showing a significant difference from littermate control animals at 30 (P<0.05), 40 (P<0.01) and 50 (P<0.01) weeks post-treatment (Fig. 3A). Muc1 expression was predominantly observed in the epithelial layer of the pancreatic ducts. In contrast, a basal level of Muc1 expression was observed in the pancreas of corn-oil-treated animals and did not increase between 30, 40 and 50 weeks of treatment (Fig. 3B). Muc1 expression in the iKC model reflects what was previously reported in the study of the KC mouse model (Rachagani et al., 2012b).
Fig. 3.
Expression of transmembrane mucins Muc1 and Muc4 in the iKC mouse model. IHC studies were performed to analyze Muc1 and Muc4 protein expression during the progression of PC in the iKC model. (A) Composite IHC scores for Muc1 show a significant increase from 10 to 50 weeks after tamoxifen injection. (B) Muc1 was expressed in iKC mice treated with corn oil or tamoxifen but was higher in tamoxifen-treated mice compared to the control mice at each time point. (C) Composite IHC scores for Muc4 show a significant increase from 10 to 50 weeks after tamoxifen injection, with no expression in corn-oil-treated mice. (D) Muc4 expression was not detected in pancreatic tissues obtained from corn-oil-treated iKC mice (upper row) or iKC mice treated with tamoxifen for only 4 weeks. Muc4 expression was detected in iKC pancreatic tissues at 10, 20, 30 and 50 weeks after tamoxifen treatment (bottom row). Muc4 expression significantly increased with cancer progression.
MUC4 is highly upregulated in pancreatic and ovarian cancers and is associated with poor prognosis (Chaturvedi et al., 2007; Ponnusamy et al., 2010; Rachagani et al., 2012a). Studies from our lab have demonstrated MUC4 is not expressed in the normal pancreas and that MUC4 de novo expression in human PDAC (Andrianifahanana et al., 2001) plays a vital role in the progression and advancement of PC (Bafna et al., 2010; Chaturvedi et al., 2007; Kaur et al., 2013; Lakshmanan et al., 2015; Moniaux et al., 2004; Rachagani et al., 2012a). Study of Muc4 expression in KC mice showed Muc4 RNA and protein are specifically expressed in the ductal epithelial cells of the pancreas (Rachagani et al., 2012b). The expression of Muc4 in the iKC model mirrored this expression pattern. IHC results showed Muc4 is localized to the cell membrane and cytoplasm in the majority of the pancreatic ductal epithelial cells (Fig. 3D lower panel). Muc4 progressively increased with its average IHC composite score increasing from 3.5 at 10 weeks (P<0.001) to 7.33 at 50 weeks (P<0.05) post-treatment (Fig. 3C). This was in sharp contrast to no Muc4 detected in the pancreas of corn-oil-treated littermate control mice of all age groups (10–50 weeks) (Fig. 3D upper panel).
Expression of secreted Muc5Ac in the iKC mouse model
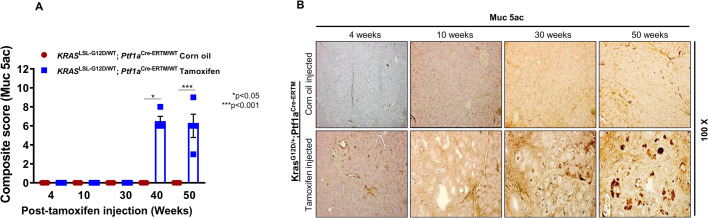
MUC5AC is overexpressed in human PDAC but is undetectable in the normal pancreas (Kaur et al., 2017). These findings are similar to Muc5Ac expression (Bafna et al., 2010; Chaturvedi et al., 2007; Kaur et al., 2013; Rachagani et al., 2012a,b) in the KC model, where Muc5Ac expression is first observed in 10-week-old animals. However, de novo expression of Muc5Ac occurred much later in the iKC model, at 40 weeks of post-tamoxifen treatment (postnatal 56-week-old animals). The Muc5Ac average IHC composite score increased from 6.5 at 40 weeks (P<0.05) to 7.0 at 50 weeks (P<0.001) post-treatment (Fig. 4A and B, lower panel). As observed in KC mice, de novo expression increased progressively in the iKC model. This was in sharp contrast to all corn-oil-treated control groups, where there was no expression of Muc5Ac was observed in the pancreas (Fig. 4B, upper panel).
Fig. 4.
Expression of secreted Muc5Ac in the iKC mouse model. Immunohistochemistry was performed to assess Muc5Ac protein expression during the progression of PC in the iKC model. (A) Composite IHC scores for Muc5Ac expression in tamoxifen- and corn-oil-treated iKC mice from 4 to 50 weeks of age. (B) Muc5Ac was detected in the pancreas of tamoxifen-treated mice only at 40 and 50 weeks after tamoxifen injection and not at 4, 10 and 30 weeks. Pancreata isolated from control animals were negative for Muc5Ac expression.
Expression of Muc16 in the iKC mouse model
Previously, we reported that MUC16 is overexpressed in pancreatic, breast and lung cancer (Haridas et al., 2011; Lakshmanan et al., 2012; Muniyan et al., 2016) and promotes a rapid G2/M checkpoint transition through its interaction with JAK2/STAT3, leading to accelerated proliferation. Further, MUC16 also inhibits apoptosis by suppressing TRAIL-mediated signaling (Lakshmanan et al., 2012). In another study, we showed that the MUC16 C-terminal region (283 amino acids of C-terminus) was responsible for increasing the tumorigenic and metastatic potential of PC cells by promoting the JAK2/STAT3 pathway, that results in upregulation of LMO2 and NANOG genes (Das et al., 2015). Recently, we found that MUC16 can interface between extracellular and intracellular proteins, facilitating PC metastasis (Muniyan et al., 2016). Hence, to build on our previous studies of MUC16 in PC and other cancers, we examined the expression status of Muc16 in the iKC model. Our IHC analysis revealed that Muc16 expression increases progressively from 10 to 50 weeks in tamoxifen-treated mice (Fig. S2, right column). No reactivity was observed in the corn-oil-treated mouse pancreas from 4 to 50 weeks of age (Fig. S2, left column), which may indicate weak immunoreactivity of the antibody in normal pancreas (Wang et al., 2008).
Depletion of mutated Kras decreases mucin expression in human PC cell lines
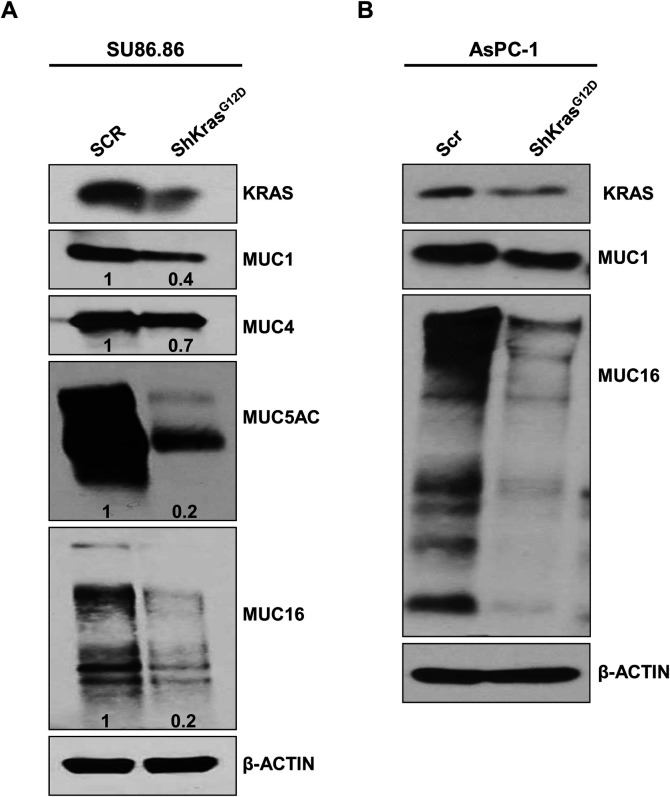
In a previous study, we showed that mosaic Cre activation driven by the Pdx1 promoter results in Kras activation throughout all pancreatic cell types and aberrant mucin expression in the KC mouse model of PDAC (Rachagani et al., 2012b). However, we were uncertain how specific targeting of Kras activation to the pancreatic acinar cell population would impact mucin expression. In addition to observing the resulting increase in mucin expression in iKC mice, we also investigated transient knockdown of mutated KrasG12D in human PC cell lines using shRNA/siRNA, and observed significantly decreased MUC1, MUC4, MUC5AC MUC16 mRNA and protein expression. Our results suggest that mutated Kras either directly or indirectly modulates (increases) mucin expression in human PC cells (Fig. 5A,B; Fig. S3).
Fig. 5.
Depletion of mutated Kras decreases mucin expression in human PC cell lines. (A) Transient knockdown of Kras was performed in Su86.86 cells. Western blot analysis using anti-MUC1, -MUC4, -MUC5AC and -MUC16 antibodies showed downregulation of mucin proteins MUC1, MUC4, MUC5AC and MUC16 upon Kras knockdown. (B) Stable knockdown of Kras in AsPC-1 cells using the pRetro.Puro vector carrying an shRNA against mutated Kras led to significant downregulation of MUC1 and MUC16 in AsPC-1 cells. β-actin served as the internal loading control.
Kras regulates mucins via upregulation of transcription factors
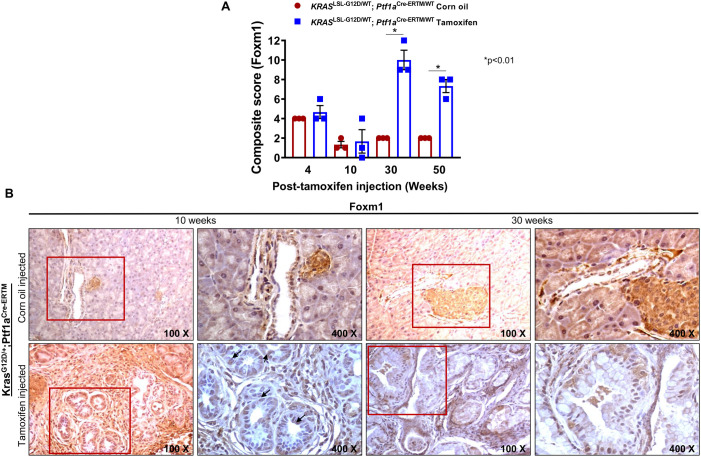
To investigate how mutant Kras regulates mucin expression, we investigated the expression profile of three transcription factors that have been previously shown by us (Kumar et al., 2015) and other groups (Jonckheere and Van Seuningen, 2018) to be associated with mucin regulation in PDAC (Kong et al., 2013; Kumar et al., 2015; Ponnusamy et al., 2010; Rachagani et al., 2012a). Serial sections from the iKC progression model were immunostained for Ncoa3, p-cJun and FoxM1 transcription factors. IHC was performed on pancreatic tissues from iKC mice to confirm that the strong association of NCOA3 with MUC4 and MUC1 expression in human- and mouse- (KC model) PDAC tissues. In iKC PC tissues that were positive for both Muc1 and Muc4, we observed that Ncoa3 was predominantly expressed in the nuclei of pancreatic ductal cells from 10 to 50 weeks and was associated with higher intensity of Muc1- and Muc4-protein expression (increased with Muc1 and Muc4 expression). In contrast, the pancreatic ducts of 10- to 50-week-treated control mice were Ncoa3 negative (Fig. S4A). We also observed upregulation of p-cJun expression in the nucleus of pancreatic ductal cells of iKC mice from 10 to 50 weeks after tamoxifen injection, with no expression of p-cJun detected in the pancreas of control mice (Fig. S4B). A similar upregulation of FoxM1 was observed in mice 30 (P<0.01) to 50 (P<0.01) weeks of age, treated with tamoxifen as compared to control mice (Fig. 6A,B). Overall, our results suggest that the inducible activation of Kras upregulates transcription factors Ncoa3, pcJun and FoxM1, which, in turn, upregulate mucin expression in the pancreas, reflecting our findings that were first reported in the KC model.
Fig. 6.
Expression of FoxM1 in iKC mice after 10 and 30 weeks of tamoxifen treatment. IHC analysis using FoxM1 antibody on pancreatic tissues isolated from corn-oil- and tamoxifen-treated iKC mice. (A) Bar graph showing quantification of immunohistochemical evaluation of FOXM1 expression in pancreas excised from corn-oil- and tamoxifen-treated mice. (B) The expression of FoxM1 in the nucleus was low in the corn-oil group (basal level), but it increased significantly upon tamoxifen injection.
DISCUSSION
For the last two decades, PC researchers have debated the pancreatic cell type from which PDAC originates. Several studies were performed to address this mystery but were inconclusive because human pre-neoplastic tissues were not available (Bailey et al., 2014; Feldmann et al., 2007; Habbe et al., 2008; Higuera et al., 2016) and studies in model systems were not convincing. Our previously published KC mouse model allowed us to capture and study the spectrum of PanIN I-III lesions that progress to PDAC (Hingorani et al., 2003; Rachagani et al., 2012b) with a longer latency than the KPC model. However, certain limitations have been reported in the KC and KPC models such as an ‘extra’-pancreatic phenotype (due to leaky expression of the Pdx1-Cre allele in other tissues such as in the skin, the oral mucosa, the perianal, gastric and adrenal regions) (Hingorani et al., 2003, 2005; Rachagani et al., 2012b). Further, PDAC is an age-related disease that shows a steep increase in incidence after 50 years of age (Chakraborty et al., 2011; Higuera et al., 2016). Similarly, the incidence of spontaneous tumors steadily increases in experimental rodents older than 300 days (Anisimov et al., 2005). Yet, the widely used KC and KPC mouse models activate CRE during embryonic development and animals present with PDAC early in postnatal life (Hingorani et al., 2003, 2005; Rachagani et al., 2012b; Torres et al., 2013). To minimize these concerns, we introduced Cre expression under the control of an alternative promoter (Ptf1a-CreER™), which has allowed us to induce Kras activation postnatally at 16 weeks of life, specifically in acinar cells. This design enabled our study of PanIN progression from the acinar cell lineage and confirmed findings previously made in the KC model under the control of the Pdx1 promoter. The iKC mice developed low-grade PanIN lesions by 4 weeks of treatment, with PDAC and metastasis by 40 weeks post-tamoxifen induction.
A previous study from our lab demonstrated an acinar-to-ductal cell transdifferntiation process in the KC model, using acinar-specific amylase and ductal specific CK-19 as lineage markers (Dey et al., 2014). We assessed these markers using the newly developed iKC model and confirmed this finding. As expected, amylase and CK19 colocalized in metaplastic ducts that were undergoing transdifferentiation following Kras activation. Thus, like the KC model, the iKC model can be appropriately used to explore genes related to the ADM transdifferentiation process (Dey et al., 2014; Rachagani et al., 2012b).
We next evaluated the expression of mucin proteins that are known to have direct roles in PC pathogenesis (Andrianifahanana et al., 2001; Bafna et al., 2010; Kaur et al., 2013). An earlier study showed that Muc1, Muc4 and Muc5Ac were overexpressed during progression of PDAC in the KC model (Rachagani et al., 2012b). This expression pattern mirrored the expression profile of these mucins in human PDAC development (Andrianifahanana et al., 2001; Kaur et al., 2013; Westgaard et al., 2009; Yonezawa et al., 2010). Since the KC model expresses Kras constitutively, we were interested in evaluating mucin expression in the inducible model, with a disease course more similar to human PDAC. Similar to the expression profile in KC mice, Muc1 was elevated in the pancreas of iKC mice (starting at week 4 post-tamoxifen) and was expressed at a basal level in the corn-oil-treated control littermates. The pattern of Muc1 expression in iKC and KC models, and in human PDAC tissues (Andrianifahanana et al., 2001; Kaur et al., 2013; Rachagani et al., 2012b; Tsutsumida et al., 2006; Westgaard et al., 2009; Yonezawa et al., 2010) suggests Muc1 is expressed throughout the pancreas irrespective of the cell type from which PDAC originates.
In contrast to Muc1, Muc4 is not expressed in the normal mouse pancreas or in iKC pancreas until 10 weeks post-tamoxifen treatment; but its expression increases progressively up to 50 weeks of treatment. Our findings in the iKC mouse model is in agreement with what we have observed in the KC mouse model (Rachagani et al., 2012b), where de novo Muc4 expression occurs by 10 weeks of age and progressively increases as disease severity increases to PDAC. De novo expression of Muc5Ac was detected by 40 and 50 weeks post-tamoxifen treatement, with no expression in control animals at any time point. This finding is inconsistent with that observed in the KC model (Rachagani et al., 2012b), where its expression is seen in the pancreas at embryonic day 8.5. This is due to differences between Pdx1 and Ptf1a promoters. Pdx1 Cre-mediated KrasG12D activation occurs during early embryonic development or in early multipotent progenitors; hence Cre activity is widespread, which causes high levels of Muc5Ac. Conversely, the spatial and temporal Ptf1a-Cre activation of KrasG12D occurs only during adult developmental stages or in adult progenitor cells. This difference in promoter function might have direct influence on expression of Muc5Ac or other mucin oncoproteins. Overall, the heavily glycosylated transmembrane mucins, Muc4 and Muc16, are similarly expressed in the KC and iKC models.
Previously, we reported that the transcription factor Ncoa3 regulates mucin expression in the pancreas, and is significantly upregulated in the KC model compared to littermate controls (Kumar et al., 2015). Recapitulating this finding, Ncoa3 expression was elevated by 30 weeks post-tamoxifen treatment in the iKC model. In addition, we analyzed the expression profile of FoxM1 and p-cJun transcription factors in the pancreas of the iKC mouse model. After 10 to 50 weeks of tamoxifen injection, nuclear expression of FoxM1 and p-cJun was observed. This studies demonstrating potential binding sites for FoxM1 and c-Jun in the promotors of mucin genes (Joshi et al., 2016; Kong et al., 2013; Singh et al., 2007). Levels of Ncoa3, FoxM1 and p-cJun were increased in conjunction with Muc1, Muc4, Muc5Ac expression, establishing a potential link between these transcription factors and KrasG12D-regulated mucin expression in PDAC.
Our study characterized Kras-mediated mucin (Muc1, Muc4, Muc5Ac and Muc16) expression during PDAC pathogenesis arising from the acinar cell lineage. The findings in the iKC model support our previous findings in the KC model, and further implicate aberrant mucin expression as a mechanism associated with PDAC initiation, progression and aggressiveness. We plan to develop mouse syngeneic cell lines mimicking the biological features of PDAC progression, such as PanIN and PDAC, from the iKC model, as demonstrated previously (Torres et al., 2013). This mouse model and its syngeneic cell lines will be used to test combinations of approved drugs that can potentially improve PDAC treatment. The developed iKC model will also provide a unique opportunity to identify biomarkers relevant to the early onset of PDAC.
MATERIALS AND METHODS
Mouse model
Animal procedures were performed in accordance with protocols approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee, U.S. Public Health Service guidelines, and the International Council for Laboratory Animal Science (ICLAS). The animals involved in this proposed study were treated humanely and in all situations in which the mice are used, pain and discomfort were minimized. B6.129-Krastm4Tyj (01XJ6) mice were obtained from the National Cancer Institute Mouse Models of Human Cancers Consortium (Frederick, MD, USA). The Ptf1a-CreER™ mice were a kind gift from Dr Christopher V. Wright, Vanderbilt University. To generate iKC mice, we crossed LSLKrasG12D mice with Ptf1aCreERTM mice to generate KrasG12D;Ptf1a-CreER™. The resulting F1 progeny were genotyped for Kras and Ptf1a-CreER by polymerase chain reaction (PCR) using specific primer sets. All positive animals (KrasG12D and Ptf1a-CreER™) carried the mutant KrasG12D allele and Cre recombinase in the pancreas. To activate the Kras (transgenic floxed KrasG12D) alleles in the mouse pancreata postnatally (16 weeks after birth, either sex), we intraperitoneally injected iKC mice (three or four mice/group/time point) with tamoxifen (3 mg/mice for 3 alternative days) or corn oil on three consecutive days, which led to excision of the LSL cassette in the tamoxifen group. Corn-oil-injected mice acted as control with an intact LSL cassette in the pancreas. Animals were housed five mice in a group and were kept under a 12 h light/dark cycle at 22±1°C and 50±10% relative humidity with ad libitum access to food and water. Animals were euthanized by CO2 asphyxiation followed by cervical dislocation at 4, 10, 20, 30, 40 and 50 weeks after tamoxifen/corn-oil injections, and the pancreata were resected and observed for gross metastatic lesions on distant organs. A separate schema demonstrating the design of the study is shown in Fig. S1. A portion of the pancreas tissue was stored in buffered formalin (10%) for IHC analysis. The rest of the pancreas was flash frozen in liquid nitrogen and stored at -80°C for analysis of mucin gene expression (Rachagani et al., 2012b).
DNA isolation and genotyping
Mouse tails were clipped at 15 days of age and DNA was isolated using the Maxwell® 16 mouse tail DNA purification kit (Promega, Madison, USA) according to the manufacturer's protocol. Genotyping PCR was performed using 100 ng of genomic DNA in 2x PCR master mix (Promega, Madison, WI, USA) and 5 pmol of each forward and reverse primers of Kras and Ptf1a-CreER genes: forward (Kras), 5′-CCT TTA CAA GCG CAC GCA GAC TGT AGA-3′; reverse (Kras), 5′-AGC TAG CCA CCA TGG CTT GAG TAA GTC TGC A-3′; forward (Ptf1a-CreER), 5′-GTG ATC ATG CAA GCT GGT GGC T-3′; reverse (Ptf1a-CreER), 5′-CCG ACT TGA CGT AGC AAG CAA C-3′ in a final volume of 20 μl. PCR conditions consisted of 5 min at 95°C, followed by 40 cycles at 94°C for 1 min, annealing at 59°C for 2 min, followed by extension at 72°C for 45 s. A final extension at 72°C was performed for 10 min. PCR products were analyzed using 1.5% agarose gel electrophoresis (Rachagani et al., 2012b).
Cell lines
The human PC cell lines SU86.86, AsPC-1, CD18/HPAF, MIA PaCa-2 and HUPT-3 were obtained from ATCC and authenticated by STR profiling method. These PC cell lines were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics (100 µg/ml of penicillin and streptomycin) at 37°C with 5% CO2 in a humidified atmosphere. Stable KrasG12D allele knockdown PC cell lines were generated by using shRNA specifically targeting the KrasG12D allele as described previously (Rachagani et al., 2011). The human PC cells lines were also routinely validated for mycoplasma contamination.
RNA isolation
Total RNA was extracted from CD18/HPAF-Scr and CD18/HPAF-ShKrasG12D using the mirVana™ miRNA isolation kit (Applied Biosystems, Austin, TX, USA) according to the manufacturer's instructions. RNA purity and integrity was determined by spectrophotometry (NanoDrop™, NanoDrop Technologies Inc., Wilmington, DE, USA) at λ 260/280 and bioanalyzer microfluid chip assays (Agilent Technologies, Waldbronn, Germany), respectively (Rachagani et al., 2012b).
RT-qPCR
Total RNA was reverse transcribed into cDNA. Reactions were prepared with 2 μg total RNA, 10 mM dNTPs, and 1 µl oligo dT. Following incubation at 65°C for 5 min, first-strand RT buffer (5×), DTT (0.1 M) and 1 µl RNase inhibitor were added to the above mixture, and reactions were incubated at 42°C for 2 min. To extend the reaction, each reaction tube received SuperScript II RT (50 units) and was incubated at 42°C for 50 min. RT-PCR was carried out on 2 μl cDNA in 1× SYBRGreen PCR Master Mix (2×, Roche Applied Science, Indianapolis, IN, USA) and primers (MUC5AC/β-actin) in a total volume of 10 μl. PCR conditions consisted of 10 min at 95°C, followed by 40 cycles of annealing and extension at 95°C for 15 s and 60°C for 1 min. Transcript copy numbers were normalized to β-actin, which served as an internal control. PCR products were analyzed using 1.5% agarose gel electrophoresis (Lakshmanan et al., 2012, 2016, 2015; Rachagani et al., 2012b).
Histological analysis
After euthanizing mice at different time intervals (4, 10, 20, 30, 40 and 50 weeks of age), a portion of the pancreas from the F1 progeny of tamoxifen- (floxed KrasG12D) and corn-oil- (unfloxed KrasG12D) administered animals (n=4/group) was fixed in 10% (v/v) buffered formalin (Thermo Fisher Scientific, Fair Lawn, NJ, USA) and embedded in paraffin. Sections of 4 μm thickness were mounted on glass slides, deparaffinized in xylenes, hydrated in a decreasing gradient of ethanol, and stained with H&E as described previously (Rachagani et al., 2012b). The slides were examined under a light microscope (Nikon, Eclipse Ti-S, Japan) (Rachagani et al., 2012b).
IHC
IHC analysis was performed with the following antibodies: anti-mouse Muc1 (1:200, Clone #HMFG1, Abcam, ab70475), Muc5Ac (1:400, clone 45M1, Abcam), Muc16 (1:10,000, RS synthesis, Louisvile, KY, USA), Ncoa3, pcJun and FoxM1, amylase, CK19 (Santa Cruz Biotechnology, CA, USA) and Muc4 (1:4000, 4A rabbit polyclonal), the latter was designed in our lab and developed by GenScript (Piscataway, NJ, USA). IHC analysis was performed on pancreatic tissue from different age groups as mentioned above. Briefly, after formalin fixation of tissues for 48 h, sections of 4 μm thickness were placed on slides. The slides were incubated overnight at 56°C and then were washed with xylene to remove paraffin. Tissue sections were rehydrated with decreasing concentrations of ethanol, and endogenous peroxidase activity was quenched with 5% H202 in methanol for 1 h. Blocking was performed with 2.5% horse serum for 2 h. Primary antibodies were incubated overnight, and tissue sections were washed three times with phosphate buffered saline (PBS) solution containing 0.05% Tween® 20 (PBST). Anti-mouse universal secondary antibody conjugated with horseradish peroxidase was added for 30 min. Following three PBST washes, mucin antibodies were detected using, 3,3′-diaminobenzidine (DAB, Vector Laboratories, Burlingame, CA, USA) prior to counterstaining with Hemotoxylin. The slides were dehydrated with increasing concentrations of ethanol followed by two xylene washes, and mounted with Permount mounting medium (Fisher Scientific Fair Lawn, NJ, USA). Slides were observed and scored by a certified pathologist. The differential expression of mucins was scored from 0–3, where 0 was considered to be negative; 1, weak; 2, moderate and 3, strong. The percentage of positive cells for each respective antibody was given a score on a scale of 1–4 (where 1, 0-25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%). A composite score (0–12) was calculated by multiplying the value of staining intensity by the value representing percentage of positive cells (Rachagani et al., 2012b).
Immunofluorescence
Immunofluoresence was performed as described previously (Lakshmanan et al., 2012, 2015; Ponnusamy et al., 2010; Rachagani et al., 2012a).
Immunoblotting
Immunoblot analysis was performed as described in our previous publications (Lakshmanan et al., 2012, 2015; Ponnusamy et al., 2010; Rachagani et al., 2012a) using the following antibodies: anti-Kras (F234, sc-30) and β-actin (sc-47778) from Santa Cruz Biotechnology MUC5AC (clone CLH2, Novus Biologicals), anti-mouse MUC1 (clone HMFG2), anti-MUC4 (clone 8G7, produced in-house) and anti-MUC16 (clone M11, Dako).
Statistical analysis
Results are expressed as mean±standard error of mean (s.e.m.). The statistical significance of differences was analyzed by Student's t-test to assess staining patterns for each mucin at each interval (different stage) of PC progression.
Supplementary Material
Acknowledgements
We thank Dr Jessica Mercer for her editorial contribution to this paper. We also thank the late Dr Sonny L. Johansson, (Pathologist) University of Nebraska Medical Center, NA, USA, and Dr Christopher V. Wright, Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN, USA for the gift of ptf1a-CreERTM mice.
Footnotes
Competing interests
S.K.B. is one of the co-founders for the Sanguine Diagnostics and Therapeutics, Inc. Other co-authors have no competing interests.
Author contributions
Conceptualization: S.K.B., S.R.; Methodology: K.M., R.P., S.R.K., R.V., S.R.; Validation: S.R.K.; Formal analysis: D.H., P.S., S.M., S.R.; Investigation: K.M., D.H., P.S., S.M., S.R.; Data curation: K.M., P.S.; Writing - original draft: P.S., S.R.; Writing - review & editing: R.P., W.M.J.; Visualization: R.V.; Supervision: S.K.B., S.R.; Project administration: S.K.B., S.R.; Funding acquisition: S.K.B., S.R.
Funding
The authors and work in this manuscript are, in-parts, supported by the National Institutes of Health, National Cancer Institute [P01 CA217798 R21 CA238953, R01, CA183459, R01 CA 195586, and R01 CA206444].
Data availability
All data generated and analyzed during the present study, are included in this article and as supplementary files.
Supplementary information
Supplementary information available online at https://bio.biologists.org/lookup/doi/10.1242/bio.052878.supplemental
References
- Andrianifahanana M., Moniaux N., Schmied B. M., Ringel J., Friess H., Hollingsworth M. A., Buchler M. W., Aubert J. P. and Batra S. K (2001). Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin. Cancer Res. 7, 4033-4040. [PubMed] [Google Scholar]
- Anisimov V. N., Ukraintseva S. V. and Yashin A. I (2005). Cancer in rodents: does it tell us about cancer in humans? Nat. Rev. Cancer 5, 807-819. 10.1038/nrc1715 [DOI] [PubMed] [Google Scholar]
- Bafna S., Kaur S. and Batra S. K (2010). Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 29, 2893-2904. 10.1038/onc.2010.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., DelGiorno K. E. and Crawford H. C (2014). The secret origins and surprising fates of pancreas tumors. Carcinogenesis 35, 1436-1440. 10.1093/carcin/bgu056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. K., Umer M., Richardson V. L., Cumpian A. M., Harder A. Q., Khosravi N., Azzegagh Z., Hara N. M., Ehre C., Mohebnasab M. et al. (2018). Requirement for MUC5AC in KRAS-dependent lung carcinogenesis. JCI Insight 3 10.1172/jci.insight.120941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Baine M. J., Sasson A. R. and Batra S. K (2011). Current status of molecular markers for early detection of sporadic pancreatic cancer. Biochim. Biophys. Acta 1815, 44-64. 10.1016/j.bbcan.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P., Singh A. P., Moniaux N., Senapati S., Chakraborty S., Meza J. L. and Batra S. K (2007). MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol. Cancer Res. 5, 309-320. 10.1158/1541-7786.MCR-06-0353 [DOI] [PubMed] [Google Scholar]
- Das S., Rachagani S., Torres-Gonzalez M. P., Lakshmanan I., Majhi P. D., Smith L. M., Wagner K. U., Batra S. K. (2015). Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells, Oncotarget, 6 5772-5787 10.18632/oncotarget.3308 [DOI] [PMC free article] [PubMed]
- De La O. J.-P., Emerson L. L., Goodman J. L., Froebe S. C., Illum B. E., Curtis A. B. and Murtaugh L. C (2008). Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc. Natl. Acad. Sci. USA 105, 18907-18912. 10.1073/pnas.0810111105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P., Rachagani S., Vaz A. P., Ponnusamy M. P. and Batra S. K (2014). PD2/Paf1 depletion in pancreatic acinar cells promotes acinar-to-ductal metaplasia. Oncotarget 5, 4480-4491. 10.18632/oncotarget.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Brocard J., Mascrez B., LeMeur M., Metzger D. and Chambon P (1996). Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. USA 93, 10887-10890. 10.1073/pnas.93.20.10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann G., Beaty R., Hruban R. H. and Maitra A (2007). Molecular genetics of pancreatic intraepithelial neoplasia. J. Hepatobiliary Pancreat. Surg. 14, 224-232. 10.1007/s00534-006-1166-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Schuhmacher A. J., Cañamero M., Grippo P. J., Verdaguer L., Pérez-Gallego L., Dubus P., Sandgren E. P. and Barbacid M (2007). Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291-302. 10.1016/j.ccr.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Haas M., Ormanns S., Baechmann S., Remold A., Kruger S., Westphalen C. B., Siveke J. T., Wenzel P., Schlitter A. M., Esposito I. et al. (2017). Extended RAS analysis and correlation with overall survival in advanced pancreatic cancer. Br. J. Cancer 116, 1462-1469. 10.1038/bjc.2017.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habbe N., Shi G., Meguid R. A., Fendrich V., Esni F., Chen H., Feldmann G., Stoffers D. A., Konieczny S. F., Leach S. D. et al. (2008). Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl. Acad. Sci. USA 105, 18913-18918. 10.1073/pnas.0810097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haridas D., Chakraborty S., Ponnusamy M. P., Lakshmanan I., Rachagani S., Cruz E., Kumar S., Das S., Lele S. M., Anderson J. M. et al. (2011). Pathobiological implications of MUC16 expression in pancreatic cancer. PLoS ONE 6, e26839 10.1371/journal.pone.0026839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuera O., Ghanem I., Nasimi R., Prieto I., Koren L. and Feliu J (2016). Management of pancreatic cancer in the elderly. World J. Gastroenterol. 22, 764-775. 10.3748/wjg.v22.i2.764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani S. R., Petricoin E. F., Maitra A., Rajapakse V., King C., Jacobetz M. A., Ross S., Conrads T. P., Veenstra T. D., Hitt B. A. et al. (2003). Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437-450. 10.1016/S1535-6108(03)00309-X [DOI] [PubMed] [Google Scholar]
- Hingorani S. R., Wang L., Multani A. S., Combs C., Deramaudt T. B., Hruban R. H., Rustgi A. K., Chang S. and Tuveson D. A (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469-483. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Jackson E. L., Willis N., Mercer K., Bronson R. T., Crowley D., Montoya R., Jacks T. and Tuveson D. A (2001). Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243-3248. 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere N. and Van Seuningen I (2018). Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J. Transl. Med. 16, 259 10.1186/s12967-018-1632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Zhang X., Parsons D. W., Lin J. C.-H., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A. et al. (2008). Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801-1806. 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Cruz E., Rachagani S., Guha S., Brand R. E., Ponnusamy M. P., Kumar S. and Batra S. K (2016). Bile acids-mediated overexpression of MUC4 via FAK-dependent c-Jun activation in pancreatic cancer. Mol. Oncol. 10, 1063-1077. 10.1016/j.molonc.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Kumar S., Momi N., Sasson A. R. and Batra S. K (2013). Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol Hepatol 10, 607-620. 10.1038/nrgastro.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Smith L. M., Patel A., Menning M., Watley D. C., Malik S. S., Krishn S. R., Mallya K., Aithal A., Sasson A. R. et al. (2017). A combination of MUC5AC and CA19-9 improves the diagnosis of pancreatic cancer: a multicenter study. Am. J. Gastroenterol. 112, 172-183. 10.1038/ajg.2016.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R. J. and Wright C. V (2002). The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32, 128-134. 10.1038/ng959 [DOI] [PubMed] [Google Scholar]
- Khan M. A., Azim S., Zubair H., Bhardwaj A., Patel G. K., Khushman M., Singh S. and Singh A. P (2017). Molecular drivers of pancreatic cancer pathogenesis: looking inward to move forward. Int. J. Mol. Sci. 18, 779 10.3390/ijms18040779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Li L., Li Z., Le X., Huang C., Jia Z., Cui J., Huang S., Wang L. and Xie K (2013). Dysregulated expression of FOXM1 isoforms drives progression of pancreatic cancer. Cancer Res. 73, 3987-3996. 10.1158/0008-5472.CAN-12-3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinke D., Brailsford M., Pan F. C., Magnuson M. A., Wright C. V. and Murtaugh L. C (2012). Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Dev. Biol. 362, 57-64. 10.1016/j.ydbio.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Das S., Rachagani S., Kaur S., Joshi S., Johansson S. L., Ponnusamy M. P., Jain M. and Batra S. K (2015). NCOA3-mediated upregulation of mucin expression via transcriptional and post-translational changes during the development of pancreatic cancer. Oncogene 34, 4879-4889. 10.1038/onc.2014.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan I., Ponnusamy M. P., Das S., Chakraborty S., Haridas D., Mukhopadhyay P., Lele S. M. and Batra S. K (2012). MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene 31, 805-817. 10.1038/onc.2011.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan I., Seshacharyulu P., Haridas D., Rachagani S., Gupta S., Joshi S., Guda C., Yan Y., Jain M., Ganti A. K. et al. (2015). Novel HER3/MUC4 oncogenic signaling aggravates the tumorigenic phenotypes of pancreatic cancer cells. Oncotarget 6, 21085-21099. 10.18632/oncotarget.3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan I., Rachagani S., Hauke R., Krishn S. R., Paknikar S., Seshacharyulu P., Karmakar S., Nimmakayala R. K., Kaushik G., Johansson S. L. et al. (2016). MUC5AC interactions with integrin beta4 enhances the migration of lung cancer cells through FAK signaling. Oncogene 35, 4112-4121. 10.1038/onc.2015.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan I., Salfity S., Seshacharyulu P., Rachagani S., Thomas A., Das S., Majhi P. D., Nimmakayala R. K., Vengoji R., Lele S. M. et al. (2017). MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing, p53. Clin. Cancer Res. 23, 3906-3917. 10.1158/1078-0432.CCR-16-2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson M. A. and Osipovich A. B (2013). Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab. 18, 9-20. 10.1016/j.cmet.2013.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniaux N., Andrianifahanana M., Brand R. E. and Batra S. K (2004). Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy. Br. J. Cancer 91, 1633-1638. 10.1038/sj.bjc.6602163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyan S., Haridas D., Chugh S., Rachagani S., Lakshmanan I., Gupta S., Seshacharyulu P., Smith L. M., Ponnusamy M. P. and Batra S. K (2016). MUC16 contributes to the metastasis of pancreatic ductal adenocarcinoma through focal adhesion mediated signaling mechanism. Genes Cancer 7, 110-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy M. P., Lakshmanan I., Jain M., Das S., Chakraborty S., Dey P. and Batra S. K (2010). MUC4 mucin-induced epithelial to mesenchymal transition: a novel mechanism for metastasis of human ovarian cancer cells. Oncogene 29, 5741-5754. 10.1038/onc.2010.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I. A., Lewis P. D. and Mattos C (2012). A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457-2467. 10.1158/0008-5472.CAN-11-2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachagani S., Senapati S., Chakraborty S., Ponnusamy M. P., Kumar S., Smith L. M., Jain M. and Batra S. K (2011). Activated KrasG12D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. Br. J. Cancer 104, 1038-1048. 10.1038/bjc.2011.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachagani S., Macha M. A., Ponnusamy M. P., Haridas D., Kaur S., Jain M. and Batra S. K (2012a). MUC4 potentiates invasion and metastasis of pancreatic cancer cells through stabilization of fibroblast growth factor receptor 1. Carcinogenesis 33, 1953-1964. 10.1093/carcin/bgs225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachagani S., Torres M. P., Kumar S., Haridas D., Baine M., Macha M. A., Kaur S., Ponnusamy M. P., Dey P., Seshacharyulu P. et al. (2012b). Mucin (Muc) expression during pancreatic cancer progression in spontaneous mouse model: potential implications for diagnosis and therapy. J. Hematol. Oncol. 5, 68 10.1186/1756-8722-5-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D. and Jemal A (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70, 7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- Singh A. P., Chauhan S. C., Andrianifahanana M., Moniaux N., Meza J. L., Copin M. C., van Seuningen I., Hollingsworth M. A., Aubert J. P. and Batra S. K (2007). MUC4 expression is regulated by cystic fibrosis transmembrane conductance regulator in pancreatic adenocarcinoma cells via transcriptional and post-translational mechanisms. Oncogene 26, 30-41. 10.1038/sj.onc.1209764 [DOI] [PubMed] [Google Scholar]
- Torres M. P., Rachagani S., Souchek J. J., Mallya K., Johansson S. L. and Batra S. K (2013). Novel pancreatic cancer cell lines derived from genetically engineered mouse models of spontaneous pancreatic adenocarcinoma: applications in diagnosis and therapy. PLoS ONE 8, e80580 10.1371/journal.pone.0080580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumida H., Swanson B. J., Singh P. K., Caffrey T. C., Kitajima S., Goto M., Yonezawa S. and Hollingsworth M. A (2006). RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin. Cancer Res. 12, 2976-2987. 10.1158/1078-0432.CCR-05-1197 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheon D. J., Lu Z., Cunningham S. L., Chen C. M., Luo R. Z., Xing D., Orsulic S., Bast R. C. Jr. and Behringer R. R (2008). MUC16 expression during embryogenesis, in adult tissues, and ovarian cancer in the mouse. Differentiation 76, 1081-1092. 10.1111/j.1432-0436.2008.00295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgaard A., Schjolberg A. R., Cvancarova M., Eide T. J., Clausen O. P. and Gladhaug I. P (2009). Differentiation markers in pancreatic head adenocarcinomas: MUC1 and MUC4 expression indicates poor prognosis in pancreatobiliary differentiated tumours. Histopathology 54, 337-347. 10.1111/j.1365-2559.2009.03227.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa S., Higashi M., Yamada N., Yokoyama S. and Goto M (2010). Significance of mucin expression in pancreatobiliary neoplasms. J. Hepatobiliary Pancreat Sci. 17, 108-124. 10.1007/s00534-009-0174-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.