ABSTRACT
Despite the known importance of the transcription factors ATOH1, POU4F3 and GFI1 in hair cell development and regeneration, their downstream transcriptional cascades in the inner ear remain largely unknown. Here, we have used Gfi1cre;RiboTag mice to evaluate changes to the hair cell translatome in the absence of GFI1. We identify a systematic downregulation of hair cell differentiation genes, concomitant with robust upregulation of neuronal genes in the GFI1-deficient hair cells. This includes increased expression of neuronal-associated transcription factors (e.g. Pou4f1) as well as transcription factors that serve dual roles in hair cell and neuronal development (e.g. Neurod1, Atoh1 and Insm1). We further show that the upregulated genes are consistent with the NEUROD1 regulon and are normally expressed in hair cells prior to GFI1 onset. Additionally, minimal overlap of differentially expressed genes in auditory and vestibular hair cells suggests that GFI1 serves different roles in these systems. From these data, we propose a dual mechanism for GFI1 in promoting hair cell development, consisting of repression of neuronal-associated genes as well as activation of hair cell-specific genes required for normal functional maturation.
KEY WORDS: GFI1, Hair cells, Inner ear
Summary: GFI1 serves a dual function in hair cell maturation, by both promoting hair cell gene expression and repressing a neuronal-associated transcriptional profile that is expressed during early development.
INTRODUCTION
Hair cells (HCs) are the sensory cells of the inner ear that are necessary for hearing and balance. Studies of HC development have demonstrated that progression of precursor cells to HCs relies on the transcription factors ATOH1, POU4F3 and GFI1 (Bermingham et al., 1999; Costa et al., 2015; Hertzano et al., 2004; Wallis et al., 2003). During cochlear development, ATOH1 protein is expressed in a basal to apical strip of cells between embryonic days (E) 13.5 and 14.5, committing these cells to a HC fate and also activating expression of POU4F3 between E14.5 and E16 (Hertzano et al., 2004; Mulvaney and Dabdoub, 2012). POU4F3 then activates the expression of GFI1 at E16.5 (Hertzano et al., 2004; Wallis et al., 2003). Mice deficient in these transcription factors exhibit severe defects in HC development, ranging from no HC formation in the inner ears of Atoh1 mutants, to delayed degeneration of HCs in Pou4f3 and Gfi1 mutants (Bermingham et al., 1999; Hertzano et al., 2004; Wallis et al., 2003). In the Gfi1−/− mouse inner ears, first the cochlear outer HCs (OHCs) and then the inner HCs (IHCs) degenerate in a basal-to-apical gradient, while dysfunctional vestibular HCs survive up to 5 months of age, resulting in both auditory and vestibular deficits (Fiolka et al., 2006; Wallis et al., 2003). Additionally, studies that identified Gfi1 as a downstream target of POU4F3 found that Gfi1 expression was nearly undetectable in POU4F3-deficient HCs, and similarities between the OHC degradation pattern of Pou4f3 and Gfi1 mutant mice suggest that the Pou4f3−/− OHC phenotype is mainly a result of Gfi1 deficiency (Hertzano et al., 2004). However, despite the known importance of these transcription factors, little is known of their downstream transcriptional cascades in developing HCs.
Here, we define the role of GFI1 in the developing mouse inner ear by analyzing the translatome of Gfi1 mutant HCs. Our studies reveal that Gfi1 mutant HCs exhibit significantly decreased expression of genes associated with normal HC development and function, as well as significantly increased expression of genes involved in neuronal differentiation. Further analysis of the upregulated genes during HC development indicates that this neuronal-associated gene expression pattern is normally expressed within the cochlear HCs early in development, and their downregulation corresponds to the onset to Gfi1 expression.
RESULTS AND DISCUSSION
HC degeneration and TUBB3 expression in newborn outer but not inner or vestibular Gfi1cre/cre HCs
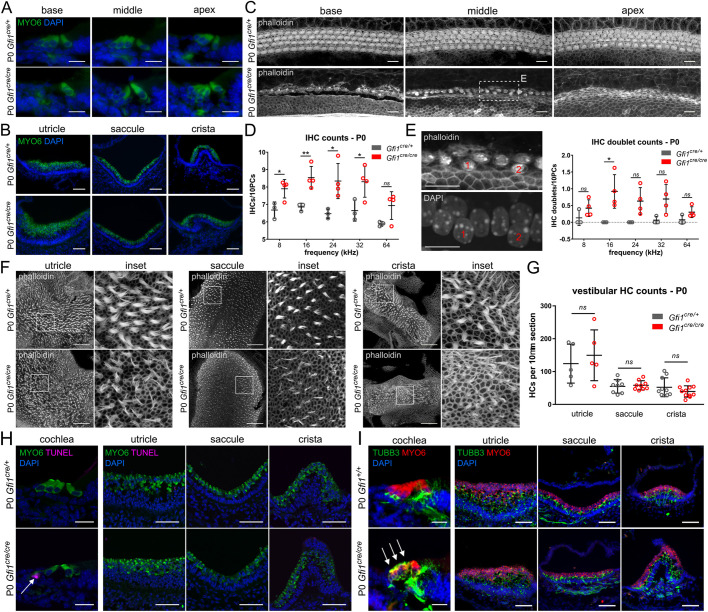
To analyze GFI1-deficient HCs, we used the Gfi1cre knock-in mouse in which the coding regions of Gfi1 exons 1-5 have been replaced with Cre recombinase (Yang et al., 2011). Homozygotes display both loss of GFI1 and efficient recombination in auditory and vestibular HCs, as well as inner ear macrophages (Matern et al., 2017; Yang et al., 2011). Our earlier work showed that Gfi1cre/cre mice are deaf and exhibit severe vestibular dysfunction, comparable with the phenotype reported for Gfi1−/− mice (Matern et al., 2017). Staining for the HC marker MYO6 further demonstrates that early postnatal (P0) Gfi1cre/cre cochlear OHCs but not IHCs degenerate in a basal to apical gradient, whereas mutant vestibular HCs are not significantly different in density compared with controls (Fig. 1A-B,G). P0-P5 whole-mounted cochleae also show a gradient pattern of OHC degeneration, with progressive loss of OHCs from base to apex, while the IHCs are present but appear immature (more like apical IHCs) and ultimately degenerate by P32 (Fig. 1C, Figs S1A and S2A). We also observed an increase in the number of IHCs in mutants at P0 and P5 (Fig. 1D, Fig. S1B), which we attribute to an increased incidence of IHC doublets (Fig. 1E). In the P0, P5 and P32 utricle, saccule and crista, stereociliary bundles appear thinner compared with controls (Fig. 1F, Figs S1C and S2B). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) revealed that mutant OHCs degenerate by apoptosis, whereas no TUNEL-positive HCs were detected in the mutant vestibular organs (Fig. 1H). Finally, it has previously been reported that Gfi1−/− OHCs aberrantly express the neuron-specific marker TUBB3 at E17.5 (Wallis et al., 2003). Staining of P0 Gfi1cre/cre inner ears showed that the OHCs of this model also aberrantly express TUBB3, while TUBB3 staining was not observed in the IHCs or vestibular HCs (Fig. 1I). When combined, these results indicate that the Gfi1cre/cre mouse can be used as a reliable model to study Gfi1 deficiency in the inner ear and suggest different functional roles for GFI1 in the cochlear and vestibular HCs.
Fig. 1.
HC degeneration and TUBB3 expression in outer but not inner or vestibular HCs of newborn GFI1 mutant ears. (A-C) Gfi1cre/cre cochlear OHCs degenerate in a basal to apical gradient at P0 (A,C), whereas vestibular HCs persist (B) (n=6). (D,E) Inner hair cell (IHC) counts at P0 revealed an increase in IHCs between 8 and 32 kHz (D), as well as increased IHC doublets at 16 kHz (E) in the Gfi1cre/cre cochlea (Gfi1cre/+ n=3, Gfi1cre/cre n=4). (F) Gfi1cre/cre vestibular HCs possess thinner stereocilia bundles (n=3). (G) P0 vestibular HC counts revealed no significant difference in HC number between genotypes (n=3). (H) Positive TUNEL staining is present in the P0 Gfi1cre/cre cochlea, which is indicative of OHC death by apoptosis, while no TUNEL staining is observed in the vestibular system (n=3). (I) Gfi1cre/cre OHCs abnormally expressed the neuronal marker TUBB3 (n=3). Scale bars: 20 µm (A,C,E,H,I; cochlea); 50 µm (B,F,H,I; vestibule). Data are mean±s.d. *P<0.05, **P<0.01; ns, not significant. Statistical significance assessed using a two-tailed Welch's t-test.
Enrichment of the inner ear HC translatomes using RiboTag
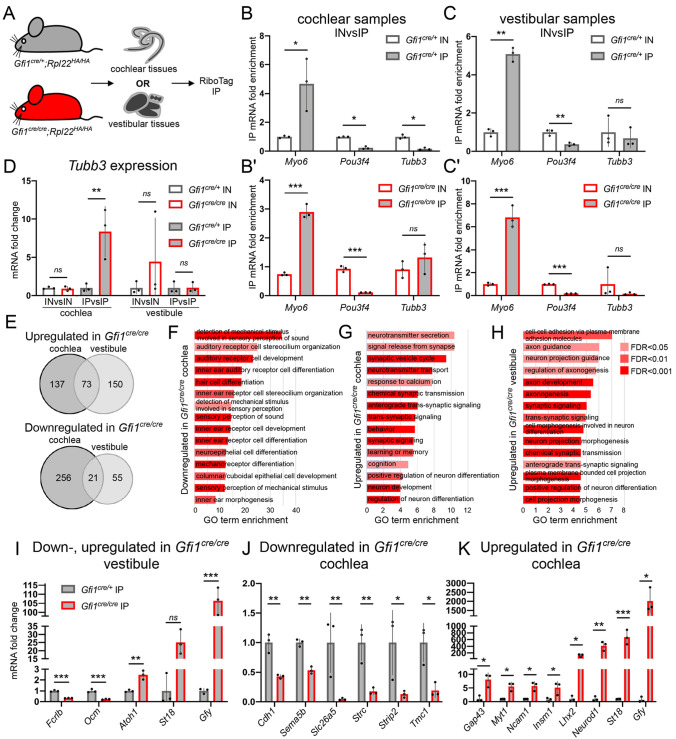
As GFI1 is known to function as a transcriptional repressor (Suzuki et al., 2016), we hypothesized that GFI1 targets would be upregulated in Gfi1cre/cre HCs. We therefore took advantage of Cre recombinase expression in the Gfi1cre model to perform a comprehensive analysis of GFI1-deficient HC gene expression using the RiboTag mouse (see Materials and Methods). This model allows for Cre-dependent expression of hemagglutinin-tagged (HA) ribosomes that can then be used to immunoprecipitate actively translated mRNA from cell types of interest (i.e. the translatome) (Sanz et al., 2009). Cochlear and vestibular tissues (utricles, saccules and cristae) were collected from newborn Gfi1cre/cre;Rpl22HA/HA mutants and Gfi1cre/+;Rpl22HA/HA controls for immunoprecipitation of HC mRNA (Fig. 2A). qPCR analysis indicated that immunoprecipitation from both mutants and controls increased mRNA levels of the HC marker Myo6 while depleting mRNA levels of the mesenchymal marker Pou3f4 (Fig. 2B,C). Tubb3 mRNA was not enriched by RiboTag immunoprecipitation compared with whole-tissue inputs across all samples (Fig. 2B,C). However, comparing transcript levels of Tubb3 between Gfi1cre/cre and Gfi1cre/+ immunoprecipitated mRNA identified a significant increase in Tubb3 in the mutant cochlear HCs (∼7-fold, P=0.0046), while the same comparison using Gfi1cre/cre and Gfi1cre/+ whole-cochlea input samples was not sufficiently sensitive to detect this difference (Fig. 2D). This highlights the utility of RiboTag for studying HC-specific changes in gene expression, as differences in HC transcripts that are also expressed in other cell types (e.g. Tubb3 in the cochleovestibular ganglion) may not be detected when comparing mRNA from whole tissues.
Fig. 2.
Translatome analysis reveals significant upregulation of neuronal mRNA in the HCs of GFI1 mutant mice. (A) P0 cochlear and vestibular tissues from mice expressing Rpl22HA in HCs were collected separately for RiboTag immunoprecipitation. (B,C) In both cochlear (B, Gfi1cre/+; B', Gfi1cre/cre) and vestibular (C, Gfi1cre/+; C', Gfi1cre/cre) samples, immunoprecipitates (IPs) had higher levels of transcripts for the HC-expressed gene Myo6 compared with input (IN), but had lower levels of the transcripts for the mesenchymal-expressed gene Pou3f4. Immunoprecipitates were not enriched with Tubb3 (n=3). Dots represent individual replicates. Data are mean±s.d. (D) Tubb3 is upregulated in the mutant cochlear IPs compared with control IPs (fold change=7.87, P=0.0046), but not in mutant cochlear IN compared with control IN (n=3). (E) Number of genes upregulated and downregulated in the Gfi1cre/cre cochlear and vestibular HCs. (F-H) Top 15 enriched gene ontology (GO) terms from genes downregulated (F) or upregulated (G) in Gfi1cre/cre cochlear HCs, or upregulated in Gfi1cre/cre vestibular HCs (H). (I-K) qPCR validation of dysregulated genes in vestibular (I) and cochlear (J,K) Gfi1cre RiboTag immunoprecipitation samples (n=3). *P<0.05, **P<0.01, ***P<0.001, ns, not significant. Statistical significance assessed by a two-tailed Welch's t-test. Data are mean±s.d.
GFI1-deficient HCs downregulate HC genes and upregulate genes associated with neuronal differentiation
To further assess global differences in Gfi1cre/cre HC gene expression, we performed RNA sequencing (RNA-seq) of the HC translatomes (Tables S1 and S2). Overall, we detected 210 upregulated and 277 downregulated genes in the Gfi1cre/cre cochlear HCs, and 223 upregulated and 76 downregulated genes in the Gfi1cre/cre vestibular HCs [log2 fold change (LFC) >1 or <−1, false discover rate (FDR) <0.001, full separation – see Materials and Methods) (Fig. 2E). The HC-expressed genes Chrna1, Atp2a3 and Fcrlb were found to be downregulated in both systems, whereas the HC-expressed gene Myo6, which normally precedes GFI1 in expression, was not significantly changed (Chessum et al., 2018; Liu et al., 2014; Scheffer et al., 2007). This suggests that the differences in HC gene expression observed between Gfi1cre/cre and Gfi1cre/+ are not a result of differences in HC number. Consistent with our qPCR and immunostaining results, Tubb3 was upregulated in the Gfi1cre/cre cochlear HC translatome only (LFC=3.49, FDR=6.59E-40). Interestingly, among the genes detected as upregulated or downregulated in either the cochlear or vestibular HCs, only 73 (25.17%) upregulated and 21 (6.33%) downregulated genes were shared between the two systems (Fig. 2E). Finally, as Gfi1cre also drives recombination in inner ear macrophages, we interrogated our dataset for changes to 360 previously defined macrophage-expressed genes (Matern et al., 2017). Of these, only four genes (Nceh1, Rnf128, Fgd3 and Spp1) were dysregulated in either the Gfi1cre/cre cochlear or vestibular samples, suggesting that the observed differences in gene expression are likely a result of global changes to HCs rather than macrophages.
To identify major dysregulated biological processes in the Gfi1-deficient HCs, we performed Gene Ontology (GO) term enrichment analyses. These revealed that genes downregulated in the mutant cochlear HCs are significantly enriched for genes involved in sensory perception of sound and inner ear development, such as Slc26a5 (LFC=−4.24, FDR=3.28E-33), Strc (LFC=−3.07, FDR=6.07E-37) and Tmc1 (LFC=−1.28, FDR=1.74E-06), suggesting Gfi1-deficient cochlear HCs have undergone a maturation arrest (Fig. 2F). A similar analysis of genes downregulated in the Gfi1cre/cre vestibular HCs revealed only one significant GO term (‘myeloid leukocyte migration’, enrichment=14.95, adjusted P=0.0347), possibly highlighting the limited published knowledge of vestibular HC gene function. Supporting this, one gene within this GO term (Spp1, encoding osteopontin: LFC=−2.03, FDR=2.70E-13) was recently identified as a marker for type I HCs in the mouse utricle (McInturff et al., 2018; Wang et al., 2019).
Analysis of genes upregulated in the mutant cochlear and vestibular HCs revealed a striking enrichment for genes involved in neuronal processes, including GO terms such as ‘neuron development’ and ‘positive regulation of neuron differentiation’ (Fig. 2G,H). Interestingly, three transcriptional regulators associated with neuronal development fall within the top five upregulated genes in the Gfi1cre/cre cochlear HCs: Lhx2 (LFC=7.1, FDR=1.22E-125), Neurod1 (LFC=6.73, FDR=2.62E-132) and St18 (LFC=6.72, FDR=4.75E-133) (Kameyama et al., 2011; Matsushita et al., 2014; Pataskar et al., 2016; Subramanian et al., 2011). Of particular interest is Neurod1, a known driver of neuronal fate that is also important for early HC development (Jahan et al., 2010, 2013). NEUROD1 has been shown to target and upregulate other transcriptional regulators of neuronal development, such as St18, which was also significantly upregulated in Gfi1cre/cre vestibular HCs (LFC=4.86, FDR=8.66E-42), and Myt1, which was significantly upregulated in Gfi1cre/cre cochlear HCs (LFC=1.1, FDR=8.43E-9) (Lizio et al., 2015; Pataskar et al., 2016). Additionally, the transcription factor gene Insm1, another NEUROD1 target important for spiral ganglion and OHC development, was upregulated in the Gfi1cre/cre cochlear HCs (LFC=1.16, FDR=1.36E-7), and Atoh1 was upregulated in the Gfi1cre/cre vestibular HCs (LFC=2.04, FDR=1.45E-17) (Jahan et al., 2010; Lizio et al., 2015; Lorenzen et al., 2015; Wiwatpanit et al., 2018).
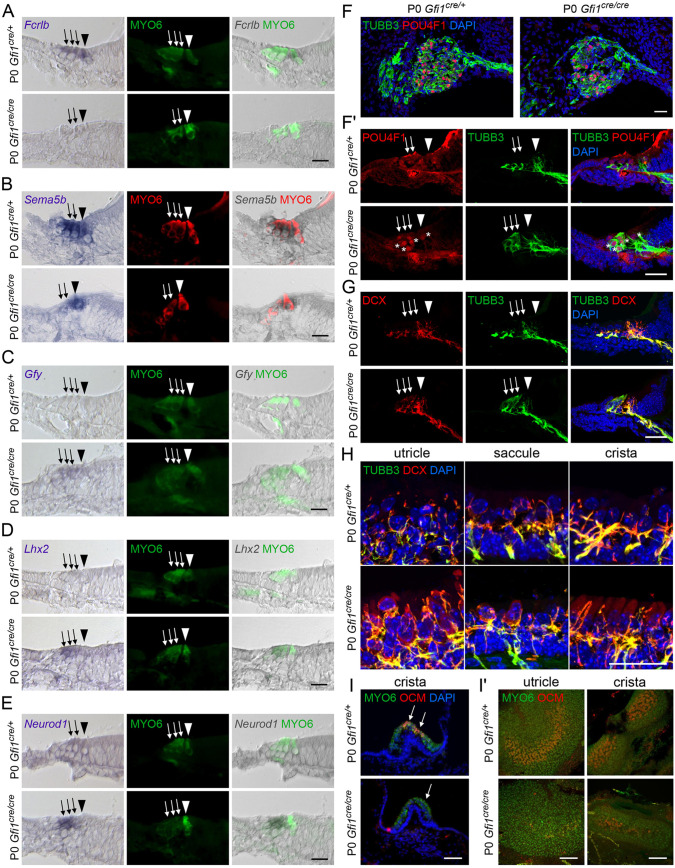
To validate the RNA-seq results, we chose 17 genes for analysis by qPCR in independent RiboTag samples. These showed that the HC-expressed genes Cdh1, Sema5b, Slc26a5, Strc, Strip2 and Tmc1 were downregulated in the Gfi1cre/cre cochleae, and Fcrlb and Ocm were downregulated in the Gfi1cre/cre vestibular system (Fig. 2I,J). Additionally, the neuronal-associated genes Gap43, Gfy, Myt1, Ncam1, Insm1, Neurod1, St18 and Lhx2 were upregulated in the mutant cochlear HCs, and Atoh1 and Gfy were upregulated in the mutant vestibular HCs (Fig. 2I,K). Of note, upregulation of St18 in the mutant vestibular HCs did not reach statistical significance, most likely due to variability in fold change between the mutant samples (St18 fold change ranged from 18-33× upregulated in Gfi1cre/cre, P=0.051) (Fig. 2I). In situ hybridization and immunostaining further revealed HC-specific differences in expression of the HC genes Fcrlb and Sema5b, as well as the neuronal-associated genes Gfy, Neurod1, Lhx2, doublecortin (DCX) and POU4F1 (normally a specific marker of type IC neurons) in the mutant cochlear HCs compared with controls (Fig. 3A-G, Figs S3 and S4) (Petitpré et al., 2018; Shrestha et al., 2018; Sun et al., 2018). Interestingly, changes in mRNA and protein expression varied between mutant IHCs and OHCs, confirming that GFI1 plays distinct roles in the development of these two cell types. In the vestibular system, immunostaining revealed upregulation of the neuronal marker DCX (Fig. 3H), as well as downregulation of the striolar HC-expressed protein OCM (oncomodulin) in Gfi1cre/cre ears (Fig. 3I). These results further validate that, in the absence of GFI1, HC-specific gene expression is significantly disrupted, and HCs express multiple markers of neuronal cells.
Fig. 3.
Downregulation of HC differentiation genes and upregulation of neuronal-associated genes in the GFI1 mutant HCs. (A-F) HC-specific downregulation of Fcrlb (A) and Sema5b (B), and upregulation of the neuronal markers Gfy (C), Lhx2 (D), Neurod1 (E), POU4F1 (normally Type IC neuron-specific, F,F′) and DCX (doublecortin, normally neuron-specific, G) in the Gfi1cre/cre cochlea (n=3). Arrowheads indicate IHCs; arrows indicate OHCs; asterisks indicate nuclear POU4F1 staining. Scale bars: 20 µm. (H) Upregulation of DCX in Gfi1cre/cre vestibular HCs (n=3). Scale bar: 20 µm. (I,I′) Downregulation of OCM in the Gfi1cre/cre striolar vestibular HCs (n=3, arrows indicate loss of OCM expression). Scale bars: 50 µm. See also Figs S3 and S4.
GFI1 plays a role in repressing early neuronal gene expression in developing HCs
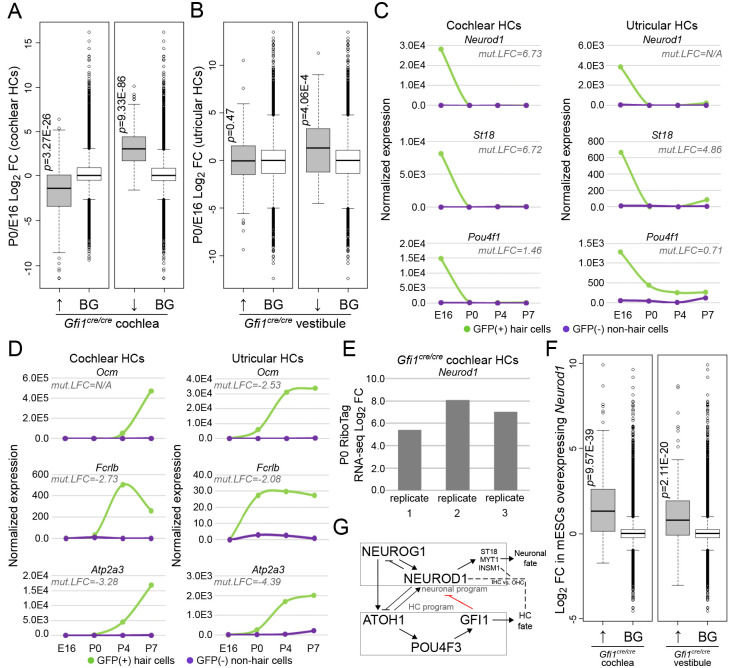
Given the upregulation of neuronal-associated genes in Gfi1cre/cre HCs, we assessed whether these genes are aberrantly activated in Gfi1cre/cre HCs or whether they are normally present in early HCs but fail to be downregulated without GFI1. For this, we examined a dataset that recorded gene expression profiles in sorted cochlear and utricular HCs at four developmental time points (E16, P0, P4 and P7) (data available via the gEAR portal; Scheffer et al., 2015). Importantly, this dataset shows that the neuronal-associated genes upregulated in Gfi1cre/cre cochlear HCs are normally expressed at E16, preceding the onset of Gfi1 expression at E16.5, and are subsequently downregulated by P0 (Fig. 4A,C). This suggests that a neuronal transcriptional profile is indeed expressed in early differentiating cochlear HCs but fails to be repressed in the Gfi1cre/cre mice. Importantly, the expression of genes upregulated in Gfi1cre/cre vestibular HCs did not change between E16 and P0 in utricular HCs (Fig. 4B), reflecting either a different mechanism of vestibular HC development or the earlier onset of Gfi1 expression in the vestibular system (E14.5, a time point not captured in the interrogated dataset) (Wallis et al., 2003). Additionally, we found that genes downregulated in Gfi1cre/cre cochlear and vestibular HCs are normally upregulated during development from E16 to P0 (Fig. 4A,B,D).
Fig. 4.
GFI1 represses a neuronal-associated transcriptional profile in developing HCs. (A,B) As a group, genes upregulated (↑) in the Gfi1cre/cre cochlear HCs (n=198) are normally repressed during HC development between E16 and P0 (A). Genes upregulated in Gfi1cre/cre vestibular HCs (n=207) did not show a difference in expression between E16 and P0 (B). Genes downregulated (↓) in Gfi1cre/cre cochlear (n=248) and vestibular (n=73) HCs are normally upregulated during HC development between E16 and P0 (A,B). (C) Examples of neuronal genes downregulated during HC development but upregulated in Gfi1cre/cre HCs. (D) Examples of HC genes upregulated during HC development but downregulated in Gfi1cre/cre HCs. (E) Neurod1 is the second most strongly upregulated gene in Gfi1cre/cre cochlear HCs. (F) Genes upregulated in Gfi1cre/cre cochlear (n=194) and vestibular (n=189) HCs are upregulated by mESCs overexpressing Neurod1. Statistical significance for A-C,F was assessed by a two-tailed Wilcoxon's test, comparing the Log2 fold change (FC) values of each gene group to the Log2 FC values of background (BG), which is all other genes expressed: B and C, cochlea↑BG, n=20,009, cochlea↓BG, n=19,959, vestibule↑BG, n=20,000, vestibule↓BG, n=20,134; and D, cochlea↑BG, n=21,743, vestibule↑BG, n=21,748. Center line represents median Log2 FC, gray box demarcates first and third quartiles, whiskers demarcate first and third quartiles±1.5× interquartile range values, dots represent single outliers. (G) The proposed mechanism by which GFI1 promotes HC fate is by repressing a neuronal-associated transcriptional profile during development.
NEUROD1 is a known transcriptional regulator of neuronal programs (Pataskar et al., 2016). As Neurod1 is detected in our dataset as the third highest upregulated gene in Gfi1cre/cre cochlear HCs (Fig. 4E), and is also the most repressed gene in normal cochlear HCs between E16.5 and P0 (Fig. 4C; ∼1000-fold repression), we investigated whether NEUROD1 or its targets could be responsible for driving neuronal-associated gene expression in Gfi1 mutant HCs. For this, we compared the genes upregulated in Gfi1cre/cre HCs with a dataset measuring changes in gene expression in mouse embryonic stem cells (mESCs) induced to neurodifferentiate through NEUROD1 overexpression (Pataskar et al., 2016). We found that the neuronal-associated genes upregulated in the Gfi1cre/cre HCs are also significantly induced by Neurod1 expression in mESCs (Fig. 4F). Overall, these analyses suggest that GFI1-deficient cochlear and vestibular HCs undergo a maturation arrest and maintain expression of a neuronal transcriptional profile that is likely normally present immediately after HC specification.
Conclusions
We have used the Gfi1cre/cre mouse model to perform a comprehensive analysis of GFI1 loss on neonatal inner ear HC translatomes. Our gene expression results show that in the absence of GFI1, HCs within the cochlear and the vestibular organs undergo a maturation arrest, failing to upregulate known markers of mature HCs, such as Strc, Tmc1 and Ocm. Additionally, our results suggest that an important role of GFI1 in early HC differentiation is to repress neuronal-associated gene expression (Fig. 4G), adding to the accumulating evidence of GFI1s ability to serve this function (see Lee et al., 2019). The abundant upregulation of neuronal-associated genes in the Gfi1cre/cre HCs is likely secondary to the maintained expression of key transcriptional drivers of neuronal fate, such as Neurod1, which appear to be normally expressed in early developing HCs. Finally, this identified role for GFI1 in repressing neuronal-associated gene expression has an important translational impact, explaining part of its functional significance in directing stem cells towards the HC fate.
MATERIALS AND METHODS
Animals
The RiboTag (C57BL/6N background) and Gfi1cre mice (C57BL/6J background) have been described previously (Sanz et al., 2009; Yang et al., 2011). To generate animals for the Gfi1cre;RiboTag RNA-seq dataset, RiboTag mice (Rpl22HA/HA) were crossed to Gfi1cre/+ mice to produce Gfi1cre/+;Rpl22HA/+ mice. These mice were further crossed to obtain Gfi1cre/+;Rpl22HA/HA breeding pairs, which were used to generate Gfi1cre/+;Rpl22HA/HA and Gfi1cre/cre;Rpl22HA/HA neonates for RiboTag immunoprecipitation. Both male and female mice were used for all experiments. All procedures involving animals were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and have been approved by the Institutional Animal Care and Use Committee at the University of Maryland School of Medicine (protocol numbers 1015003 and 0918005).
Immunostaining and image acquisition
Inner ears from Gfi1cre mice were dissected between P0 and P32 and fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4°C. Ears were then either stored in PBS for further dissection or processed and embedded in OCT compound (Tissue-Tek). Immunostaining on sectioned or whole-mounted tissues was performed using the following primary antibodies: mouse anti-TUBB3 (1:500, BioLegend, 801213); goat anti-oncomodulin N-19 (1:100, Santa Cruz Biotechnology, sc-7446, Lot L0814); mouse anti-POU4F1 (1:50, Millipore, MAB1585); guinea pig anti-DCX (1:5000, Millipore, AB2253); rabbit anti-myosin-VI (1:1000, Proteus BioSciences, 25-6791). Complementary Alexa Fluor 488 and 546 (1:800, Invitrogen) antibodies were used for secondary detection, Alexa Fluor 488 Phalloidin (1:1000, Invitrogen) was used to stain F-actin, and DAPI (25 µg/ml, Invitrogen) was used to stain cell nuclei. TUNEL staining was performed using the Click-iT Plus TUNEL Assay (Invitrogen) following the manufacturer's instructions. An average of between zero and one TUNEL-positive HC nuclei were observed per Gfi1cre/cre cochlear cross-section (∼12 cross-sections analyzed per animal, three animals per genotype), always within the middle and basal turns where OHCs are actively degenerating. Images were acquired using either a Nikon Eclipse E600 microscope equipped with a Lumenera Infinity 3 camera or a Nikon CSU-W1 spinning disk confocal microscope located at the University of Maryland School of Medicine Center for Innovative Biomedical Resources, Confocal Microscopy Core, Baltimore, Maryland.
RiboTag immunoprecipitations
RiboTag immunoprecipitations from inner ear tissues were performed as previously described (Chessum et al., 2018). Of note, Gfi1cre/+ animals were used as controls in this experiment out of necessity to drive Cre recombination, despite their age-related hearing loss phenotype (Matern et al., 2017). Only minimal differences in gene expression have been previously noted between Gfi1+/+ and Gfi1cre/+ at P8 (all attributed to potential sex bias in samples analyzed), suggesting that Gfi1cre/+ can serve as a proxy for Gfi1+/+ (Matern et al., 2017). For each biological sample, cochlear ducts or vestibular tissues (utricles, saccules and cristae) from five P0 Gfi1cre/+;Rpl22HA/HA or Gfi1cre/cre;Rpl22HA/HA mice were pooled and homogenized in 1 ml of supplemented homogenization buffer [50 mM Tris-HCl (pH 7.4), 100 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM 1,4-Dithiothreitol, 1× protease inhibitor cocktail, 200 U/ml RNAseOUT, 100 μg/ml cycloheximide and 1 mg/ml heparin]. Careful dissection was performed to minimize collection of neuronal tissues. Homogenates were then centrifuged to remove cell debris (9400 g for 10 min at 4°C) and 40 μl reserved as input control. HA antibody (5 μg; BioLegend, 901502, Lot# B220767) was added to the remaining homogenate and incubated under gentle rotation for 4-6 h at 4°C before adding 300 μl of rinsed Invitrogen Dynabeads Protein G magnetic beads (Thermo Fisher). Samples were then incubated under gentle rotation overnight at 4°C. Bound beads were rinsed 3× with 800 μl high salt buffer [50 mM Tris-HCl (pH 7.4), 300 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM 1,4-Dithiothreitol, 100 μg/ml cycloheximide] under gentle rotation at 4°C. Buffer RLT from the RNeasy Plus Micro kit (Qiagen) (350 μl) was used to release bound ribosomes and associated RNAs from the beads. RNA was extracted using the RNeasy Plus Micro kit (Qiagen), using 16 µl of nuclease free water for elution. RNA quality was assessed using the Agilent RNA Pico kit (Agilent Technologies), and only samples with RIN >8 were used for RNA-sequencing.
RNA-sequencing and data analysis
Gfi1cre;RiboTag IP RNA-seq libraries were prepared in biological triplicates from cochlear tissues using the Ovation Ultralow Library Preparation Kit (NuGEN), and vestibular tissues using the NEBNext Ultra Directional RNA Library Prep Kit (New England BioLabs) as per the manufacturers’ instructions. Samples were then sequenced on a HiSeq 2500 system (Illumina) using a 100 bp paired end read configuration at the University of Maryland School of Medicine Institute for Genome Sciences. Reads were aligned to the mouse reference genome (mm10) using TopHat (Trapnell et al., 2009), and the number of reads aligning to predicted coding regions was quantified using HTSeq (Anders et al., 2015). See Table S3 for alignment statistics. Significant differential gene expression between Gfi1cre/+ and Gfi1cre/cre RiboTag IP samples was assessed using DEseq (Anders and Huber, 2010). In addition to a cutoff of LFC >1 or <−1 and FDR<0.001, we required a full separation of normalized expression values (counts per million, CPM) between replicates to refer to a gene as differentially expressed. For example, for a gene to be referred to as downregulated in the Gfi1cre/cre samples compared with Gfi1cre/+, the normalized expression levels measured in each replicate of Gfi1cre/cre must be lower than the lowest expression level measured in a Gfi1cre/+ replicate. Gene ontology analyses were performed using the Gene Ontology database (www.geneontology.org) (Harris et al., 2004).
Quantitative PCR
RNA from independent Gfi1cre;RiboTag IP and input samples was reverse-transcribed using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher), and preamplified using TaqMan PreAmp Master Mix (Applied Biosystems). qPCR was performed using TaqMan Fast Advanced Master Mix (Applied Biosystems). In cases where no amplification was detected, threshold values were set to the maximum cycle (40). Cycle threshold levels were normalized to averaged Actb and Tbp expression, with the exception of Fig. 2J,K, where cochlear samples were normalized to Myo6 to control for possible differences in HC number. qPCR was performed in three biological replicates, and statistical significance between samples was assessed by Welch's t-test. See Table S4 for TaqMan probes.
In situ hybridization
In situ hybridization was performed as previously described (Chessum et al., 2018). Briefly, 10 µm inner ear sections mounted on positively charged glass slides were refixed with 4% PFA before a 10 min treatment with 2 µg/ml Proteinase-K (New England Biolabs). The Proteinase-K reaction was stopped using 4% PFA, and tissues were soaked in acetylation and permeabilization buffers. Digoxigenin-labeled probes for Fcrlb, Sema5b, Gfy, Lhx2 and Neurod1 were hybridized to sections overnight at 65°C. See Table S4 for primer sequences. After washing, slides were incubated with sheep anti-digoxigenin antibody conjugated to alkaline phosphatase (Sigma-Aldrich, 11093274910, Lot# 14608125) diluted to 1:100 overnight at 4°C. Colorimetric visualization of hybridized probes was performed using BM purple AP substrate precipitating solution (Roche). The colorimetric reaction was halted by soaking slides in 1× TBS, after which slides were reblocked and immunostained with the rabbit anti-myosin-VI antibody (1:1000, Proteus BioSciences, Cat# 25-6791) and a corresponding Alexa Fluor 488 or 546 secondary (1:800, Invitrogen).
Supplementary Material
Acknowledgements
The authors thank Drs M. K. Lobo and J. Zuo for providing the RiboTag and Gfi1cre mouse models for this study, and Dr A. Cheng for critically reviewing the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.S.M., B.M., R.E., R.H.; Methodology: B.M., R.H.; Software: Y.S., R.E.; Validation: M.S.M.; Formal analysis: M.S.M., Y.S., R.E.; Investigation: M.S.M., B.M., E.L.L., M.M., Y.O., A.T., R.E.; Writing - original draft: M.S.M., R.H.; Writing - review & editing: M.S.M., B.M., R.E., R.H.; Visualization: M.S.M., B.M.; Supervision: B.M., R.E., R.H.; Project administration: B.M., R.H.; Funding acquisition: R.E., R.H.
Funding
This work was supported by the National Institute on Deafness and Other Communication Disorders (NIDCD)/National Institutes of Health (R01DC013817 and R01DC03544 to R.H.; T32DC00046 and F31DC016218 to M.S.M.); and by the United States - Israel Binational Science Foundation (2017218 to R.H. and R.E.). Deposited in PMC for release after 12 months.
Data availability
RNA-seq data have been deposited in the GEO under accession number GSE135760 and are also available for viewing and analysis on the gEAR portal (https://umgear.org/p?s=fd78e02b).
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.186015.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.186015.reviewer-comments.pdf.
References
- Anders S. and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. and Huber W. (2015). Genome analysis HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham N. A., Hassan B. A., Price S. D., Vollrath M. A., Ben-Arie N., Eatock R. A., Bellen H. J., Lysakowski A. and Zoghbi H. Y. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837-1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Chessum L., Matern M. S., Kelly M. C., Johnson S. L., Ogawa Y., Milon B., McMurray M., Driver E. C., Parker A., Song Y. et al. (2018). Helios is a key transcriptional regulator of outer hair cell maturation. Nature 563, 696-700. 10.1038/s41586-018-0728-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A., Sanchez-Guardado L., Juniat S., Gale J. E., Daudet N. and Henrique D. (2015). Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142, 1948-1959. 10.1242/dev.119149 [DOI] [PubMed] [Google Scholar]
- Fiolka K., Hertzano R., Vassen L., Zeng H., Hermesh O., Avraham K. B., Dührsen U. and Möröy T. (2006). Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 7, 326-333. 10.1038/sj.embor.7400618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshal L. B., Mungall C. et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, 258-261. 10.1093/nar/gkh066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R., Montcouquiol M., Rashi-elkeles S., Elkon R., Yücel R., Frankel W. N., Rechavi G., Möröy T., Friedman T. B., Kelley M. W. et al. (2004). Transcription profiling of inner ears from Pou4f3 ddl/ddl identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum. Mol. Genet. 13, 2143-2153. 10.1093/hmg/ddh218 [DOI] [PubMed] [Google Scholar]
- Jahan I., Pan N., Kersigo J. and Fritzsch B. (2010). Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS ONE 5, e11661 10.1371/journal.pone.0011661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I., Pan N., Kersigo J. and Fritzsch B. (2013). Beyond generalized hair cells: molecular cues for hair cell types. Hear. Res. 297, 30-41. 10.1016/j.heares.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janky R., Verfaillie A., Imrichová H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Sanchez M. N., Potier D. et al. (2014). iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama T., Matsushita F., Kadokawa Y. and Marunouchi T. (2011). Myt/NZF family transcription factors regulate neuronal differentiation of P19 cells. Neurosci. Lett. 497, 74-79. 10.1016/j.neulet.2011.04.033 [DOI] [PubMed] [Google Scholar]
- Lee C., Rudneva V. A., Erkek S., Zapatka M., Chau L. Q., Tacheva-Grigorova S. K., Garancher A., Rusert J. M., Aksoy O., Lea R. et al. (2019). Lsd1 as a therapeutic target in Gfi1-activated medulloblastoma. Nat. Commun. 10, 332 10.1038/s41467-018-08269-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Pecka J. L., Zhang Q., Soukup G. A., Beisel K. W., David X. and He Z. Z. (2014). Characterization of transcriptomes of cochlear inner and outer hair cells. J. Neurosci. 34, 11085-11095. 10.1523/JNEUROSCI.1690-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M., Ishizu Y., Itoh M., Lassmann T., Hasegawa A., Kubosaki A., Severin J., Kawaji H., Nakamura Y., Suzuki H. et al. (2015). Mapping mammalian cell-type-specific transcriptional regulatory networks using KD-CAGE and ChIP-seq data in the TC-YIK cell line. Front. Genet. 6, 331 10.3389/fgene.2015.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen S. M., Duggan A., Osipovich A. B., Magnuson M. A. and García-Añoveros J. (2015). Insm1 promotes neurogenic proliferation in delaminated otic progenitors. Mech. Dev. 138, 233-245. 10.1016/j.mod.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern M. S., Vijayakumar S., Margulies Z., Milon B., Song Y., Elkon R., Zhang X., Jones S. M. and Hertzano R. (2017). Gfi1Cre mice have early onset progressive hearing loss and induce recombination in numerous inner ear non-hair cells. Sci. Rep. 7, 42079 10.1038/srep42079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita F., Kameyama T., Kadokawa Y. and Marunouchi T. (2014). Spatiotemporal expression pattern of Myt/NZF family zinc finger transcription factors during mouse nervous system development. Dev. Dyn. 243, 588-600. 10.1002/dvdy.24091 [DOI] [PubMed] [Google Scholar]
- McInturff S., Burns J. C. and Kelley M. W. (2018). Characterization of spatial and temporal development of Type I and Type II hair cells in the mouse utricle using new cell-type-specific markers. Biol. Open 7, bio038083 10.1242/bio.038083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney J. and Dabdoub A. (2012). Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J. Assoc. Res. Otolaryngol. 13, 281-293. 10.1007/s10162-012-0317-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataskar A., Jung J., Smialowski P., Noack F., Calegari F., Straub T. and Tiwari V. K. (2016). NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 35, 24-45. 10.15252/embj.201591206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitpré C., Wu H., Sharma A., Tokarska A., Fontanet P., Wang Y., Helmbacher F., Yackle K., Silberberg G., Hadjab S. et al. (2018). Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 9, 3691 10.1038/s41467-018-06033-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E., Yang L., Su T., Morris D. R., Mcknight G. S. and Amieux P. S. (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 106, 13939-13944. 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer D., Sage C., Plazas P. V., Huang M., Wedemeyer C., Zhang D.-S., Chen Z.-Y., Elgoyhen A. B., Corey D. P. and Pingault V. (2007). The α1 subunit of nicotinic acetylcholine receptors in the inner ear: transcriptional regulation by ATOH1 and co-expression with the γ subunit in hair cells. J. Neurochem. 103, 2651-2664. 10.1111/j.1471-4159.2007.04980.x [DOI] [PubMed] [Google Scholar]
- Scheffer I., Shen X. J., Corey X. D. P. and Chen X. Z. (2015). Gene expression by mouse inner ear hair cells during development. J. Neurosci. 35, 6366-6380. 10.1523/JNEUROSCI.5126-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B. R., Chia C., Wu L., Kujawa S. G., Liberman M. C. and Goodrich L. V. (2018). Sensory neuron diversity in the inner ear is shaped by activity. Cell 174, 1229-1246.e17. 10.1016/j.cell.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Sarkar A., Shetty A. S., Muralidharan B., Padmanabhan H., Piper M., Monuki E. S., Bach I., Gronostajski R. M., Richards L. J. et al. (2011). Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc. Natl. Acad. Sci. USA 108, E265-E274. 10.1073/pnas.1101109108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Babola T., Pregernig G., So K. S., Nguyen M., Su S. S.-M., Palermo A. T., Bergles D. E., Burns J. C. and Müller U. (2018). Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174, 1247-1263.e15. 10.1016/j.cell.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Maruyama S., Tamauchi H., Kuwahara M., Horiuchi M., Mizuki M., Ochi M., Sawasaki T., Zhu J., Yasukawa M. et al. (2016). Gfi1, a transcriptional repressor, inhibits the induction of the T helper type 1 programme in activated CD4 T cells. Immunology 147, 476-487. 10.1111/imm.12580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis D., Hamblen M., Zhou Y., Venken K. J. T., Schumacher A., Grimes H. L., Zoghbi H. Y., Orkin S. H. and Bellen H. J. (2003). The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221-232. 10.1242/dev.00190 [DOI] [PubMed] [Google Scholar]
- Wang T., Niwa M., Sayyid Z. N., Hosseini D. K., Pham N., Jones S. M., Ricci A. J. and Cheng A. G. (2019). Uncoordinated maturation of developing and regenerating postnatal mammalian vestibular hair cells. PLoS Biol. 17, e3000326 10.1371/journal.pbio.3000326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwatpanit T., Lorenzen S. M., Cantú J. A., Foo C. Z., Hogan A. K., Márquez F., Clancy J. C., Schipma M. J., Cheatham M. A., Duggan A. et al. (2018). Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563, 691-695. 10.1038/s41586-018-0570-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Gan J., Xie X., Deng M., Feng L., Chen X., Gao Z. and Gan L. (2011). Gfi1-Cre knock-in mouse line: a tool for inner ear hair cell-specific gene deletion. Genesis 48, 400-406. 10.1002/dvg.20632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.